Abstract
IMPORTANCE
Lung cancer is the leading cause of cancer death in the United States in all ethnic and racial groups. The overall death rate from lung cancer is higher in black patients than in white patients.
OBJECTIVE
To compare the prevalence and types of somatic alterations between lung cancers from black patients and white patients. Differences in mutational frequencies could illuminate differences in prognosis and lead to the reduction of outcome disparities by more precisely targeting patients’ treatment.
DESIGN, SETTING, AND PARTICIPANTS
Tumor specimens were collected from Baptist Cancer Center (Memphis, Tennessee) over the course of 9 years (January 2004-December 2012). Genomic analysis by massively parallel sequencing of 504 cancer genes was performed at Dana-Farber Cancer Institute (Boston, Massachusetts). Overall, 509 lung cancer tumors specimens (319 adenocarcinomas; 142 squamous cell carcinomas) were profiled from 245 black patients and 264 white patients.
MAIN OUTCOMES AND MEASURES
The frequencies of genomic alterations were compared between tumors from black and white populations.
RESULTS
Overall, 509 lung cancers were collected and analyzed (273 women [129 black patients; 144 white patients] and 236 men [116 black patients; 120 white patients]). Using 313 adenocarcinomas and 138 squamous cell carcinomas with genetically supported ancestry, overall mutational frequencies and copy number changes were not significantly different between black and white populations in either tumor type after correcting for multiple hypothesis testing. Furthermore, specific activating alterations in members of the receptor tyrosine kinase/Ras/Raf pathway including EGFR and KRAS were not significantly different between populations in lung adenocarcinoma.
CONCLUSIONS AND RELEVANCE
These results demonstrate that lung cancers from black patients are similar to cancers from white patients with respect to clinically actionable genomic alterations and suggest that clinical trials of targeted therapies could significantly benefit patients in both groups.
Lung cancer remains the leading cause of death from cancer in the United States.1 Black populations have the highest lung cancer mortality rate of any racial or ethnic group in the United States.2 The average annual age-adjusted incidence is 65.0 and 58.8 per 100000 in black patients and white patients, respectively.2 Many factors may contribute to the disparities in both incidence and outcome including differences in access to health care, smoking behavior, and other socioeconomic variables.3–5 However, these known risk factors do not fully explain the differences in the incidence and outcome between these 2 populations.6–8
Acquired alterations to the cancer genome—including somatic single-nucleotide variants, insertions and deletions (indels), copy number variations (CNVs), and structural rearrangements—contribute to tumorigenesis by activating pathways involved in cell growth and proliferation, resistance to apoptosis, and immune cell invasion.9 Alterations in receptor tyrosine kinases (RTKs) such as EGFR, ALK, RET, and ROS1 are clinically important as they confer sensitivity to kinase inhibitors in lung adenocarcinomas.10,11 Recent studies12–14 led by our group and The Cancer Genome Atlas (TCGA) have performed comprehensive molecular characterization of lung tumors to identify other potentially targetable alterations. We have shown that the frequency of acquired alterations in lung cancer driver genes are largely distinct between the 2 largest subclasses of non–small-cell lung cancer (NSCLC), lung adenocarcinoma, and lung squamous cell carcinoma.15
In addition to histology, the frequency of acquired alterations in lung tumors can vary across sex, smoking status, and ancestral populations.10 For example,EGFR kinase domain mutations occur at a significantly higher frequency in lung adenocarcinomas from women compared with men, never-smokers compared with smokers, and East Asian populations compared with non-Asian populations.10,16,17 The higher rates of EGFR mutations in these populations have resulted in a corresponding higher rate of patients with clinical response to EGFR inhibitors.18–20 Studies examining mutational profiles of EGFR in tumors from patients with black ancestry have been less conclusive. Some studies have reported significantly lower frequencies of EGFR mutations in black patients compared with whites,21–24 while other studies did not observe any association of EGFR mutation status with ancestry or self-reported race.25–28The interpretation of these findings have been limited by the presence of confounding factors such as sex and smoking status or by the fact that cohorts were pooled together from different clinical settings across different institutions. Moving beyond EGFR, amore recent study25 examining a panel of known driver mutations across several hundred NSCLC samples found a lower overall frequency of mutations in any gene in tumors from black patients compared with white patients.25 However, no studies have been performed that comprehensively characterize genomic alterations across hundreds of well-known cancer genes in a large number of tumors from both black and white populations.
To understand the relationship between tumoral genomic alterations and black ancestry, we profiled the exome sequences of 504 cancer genes in 509 lung cancers from black patients and white patients. These tumors were collected from patients in the same clinical setting and were well matched for clinical covariates. We did not identify enrichment of mutations or copy number changes in either population. These results suggest that targeted therapies and biomarker-driven clinical trials should significantly benefit patients in both groups.
Methods
Sample Collection
Formalin-fixed paraffin-embedded (FFPE) lung tumor specimens were collected from black and white patients from Baptist Cancer Center in Memphis, Tennessee. All specimens were obtained from patients with approval from the relevant institutional review boards. Lung cancer cases from January 2004 to December 2012 were identified from a search of pathology department records across 3 health care systems (Baptist Memorial Health Care, Methodist-LeBonheur Healthcare, and St Francis Healthcare) in metropolitan Memphis. Race was categorized by patient self-identification. From each institution, specimens from black patients were paired with specimens from white patients by matching for histology, sex, and age.
Sequencing and Analysis
In total, 607 tumors underwent DNA isolation and sequencing of 504 cancer-related genes using the OncoPanel platform29 (eMethods in Supplement 1 and eTable 1 in Supplement 2). Overall, 550 libraries with sufficient material were submitted for sequencing. Thirty tumors were excluded for failing sequencing quality control (eMethods in Supplement 1), and an additional 11 tumors were excluded due to insufficient clinical annotation, leaving a total of 509 tumors for analysis. Data were aligned and processed as previously described (eMethods in Supplement 1).30–33 Principal component analysis (PCA)34,35 and other statistical analyses are described in eMethods in Supplement 1. Because we did not have paired normal tissue for these patients, we excluded variants found in exomes of noncancer tissue using the Exome Aggregation Consortium database (eFigure 1 in Supplement 1).
Results
Study Cohort
In total, 509 tumor specimens from245 black patients and 264 white patients were analyzed (Table) (eTable 2 in Supplement 2). This included 319 adenocarcinomas, 142 squamous cell carcinomas, 47 NSCLC of other histology, and 1 small-cell carcinoma. Twenty-nine samples were obtained from a biopsy of a nonlung site for a putative lung cancer metastasis. The majority of tumors were stage I (n = 305) or stage II (n = 98). We performed secondary pathological review for 380 lung cancers (eMethods in Supplement 1) and confirmed histological classification in 223 of 247 lung adenocarcinomas (90%) and 80 of 97 squamous cell carcinomas (82%).
Table.
Cohort Demographics by Self-reported Racea
| Characteristic | Black (n = 245) | White (n = 264) | All | P Value |
|---|---|---|---|---|
| Sex | ||||
| Male | 116 | 120 | 236 | .72 |
| Female | 129 | 144 | 273 | |
| Pack-years | ||||
| Median (range) | 30 (0–120) | 40 (0–120) | 38 (0–120) | <.001 |
| NA | 35 | 35 | 70 | |
| Smoking status | ||||
| Never | 26 | 23 | 49 | .33 |
| Former | ||||
| Heavy | 92 | 116 | 208 | |
| Light | 18 | 12 | 30 | |
| Active | ||||
| Heavy | 86 | 88 | 174 | |
| Light | 19 | 14 | 33 | |
| NA | 4 | 11 | 15 | |
| Histology | ||||
| NSCLC | ||||
| Adenocarcinoma | 150 | 169 | 319 | .94 |
| Squamous | 69 | 73 | 142 | |
| Large-cell undifferentiated | 8 | 6 | 14 | |
| Adenosquamous | 6 | 7 | 13 | |
| Large-cell neuroendocrine | 5 | 5 | 10 | |
| Undefined | 6 | 4 | 10 | |
| Small-cell carcinoma | 1 | 0 | 1 | |
| Stage | ||||
| I | 142 | 163 | 305 | .60 |
| II | 53 | 45 | 98 | |
| III | 28 | 32 | 60 | |
| IV | 10 | 9 | 19 | |
| NA | 12 | 15 | 27 |
Abbreviations: NA, not applicable or no data available; NSCLC, non–small-cell lung cancer.
Unless otherwise indicated, data are reported as number of patients.
Association of Self-reported Race With Germline Variation
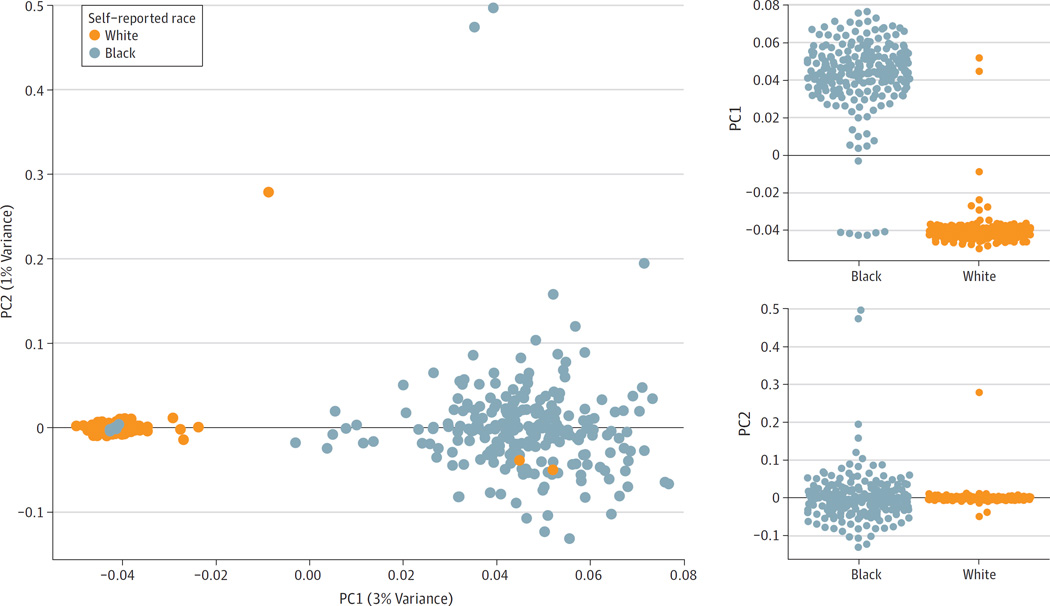
Previous studies have noted that self-reported clinical variables such as race and sex are not always congruent with underlying biological ancestry or sex as defined by genetic variation and structure.36To examine the relationship between self-reported race and biological ancestry, we performed PCA on single-nucleotide variants (SNVs) likely to be inherited and not somatically acquired (eMethods and eFigure 1 in Supplement 1). Principal component 1 (PC1) was strongly associated with self-reported race (Wilcoxon rank-sum test P = 7.6 × 10−79) (Figure 1). Two SNV profiles from reported white patients clustered with the samples from black patients, and 6 SNV profiles from reported black patients clustered with the white patient samples. Additionally, 3 samples were outliers along PC2. These 11 samples were excluded from all further analyses examining the association between ancestry and genomic alterations.
Figure 1. Association of Germline Variation With Self-reported Race Using PCA.
We identified single-nucleotide variations (SNVs) that were likely to be germline (ie, inherited and not acquired) due to their presence in African American or non-Finnish European (NFE) populations at a frequency of at least 1%. We performed principal component analysis (PCA) on these SNVs (n = 3517) and found that principal component 1 (PC1) was strongly correlated with self-reported race (P = 7.6 × 10−79). Two reported white patients strongly clustered with the black population, and 6 reported black patients strongly clustered with the white population. The misclustering of these patients could be due to errors in clinical data collection or differences in the patient’s self-perceived race from their biological ancestry. The samples from these patients were excluded from further analyses examining the relationship between ancestry and genomic alterations.
Mutational Signatures
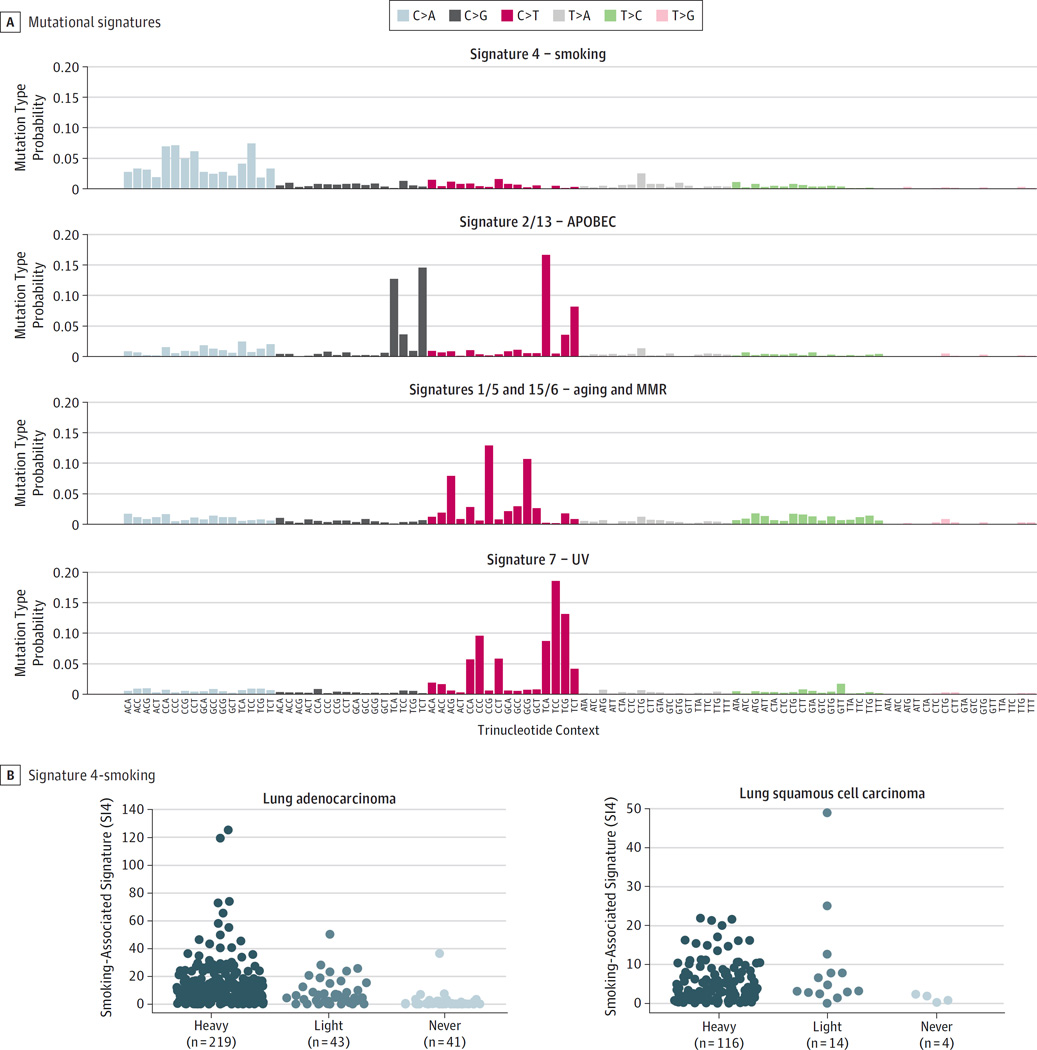
Various exogenous exposures or endogenous biological processes can contribute to the overall mutational load observed in lung tumors. We identified 4 mutational signatures present in the cohort37 (eMethods in Supplement 1). These signatures were strongly correlated with previously characterized signatures of smoking (designated as SI4), aberrant APOBEC (apolipoprotein B messenger RNA editing enzyme, catalytic polypeptide-like) activity (SI2 and SI13), mismatch repair (MMR) and aging (SI6/SI15 and SI1/SI5), and ultraviolet (UV) radiation exposure (SI7) (Figure 2) (eFigure 2 in Supplement 1).38 The smoking-related signature SI4 was enriched for cytosine (C)>adenine (A) transversions and strongly associated with smoking status in lung adenocarcinoma (P = 9.1 × 10−15) (Figure 2) and to a lesser extent in lung squamous cell carcinoma (P = .08). No signatures were associated with ancestry, suggesting that the mutagenic processes are similar in lung tumors from black and white populations in lung adenocarcinoma or lung squamous cell carcinoma (P > .05 for all) (eFigure 3 in Supplement 1).
Figure 2. Mutational Signatures in Tumors From Black and White Populations.
A, Four mutational signatures were identified using nonnegative matrix factorization on 96 trinucleotide mutation types across 509 lung tumors including those related to smoke exposure (SI4), aberrant APOBEC activation (SI13 and SI2), aging (and SI1/5) and mismatch repair (SI15/6), and UV exposure (SI7). Each bar corresponds to the probability of observing a particular mutation in a trinucleotide context within each signature. B, The smoking signature was associated with smoking status in lung adenocarcinoma (P = 9.1 × 10−15) and to a lesser extent lung squamous cell carcinomas (P = .08). MMR indicates mismatch repair.
Lack of Association Between Genomic Alterations and Ancestry
To assess the relationship between ancestry and mutation status, we examined the mutational frequencies in 313 lung adenocarcinomas and 138 lung squamous cell carcinomas. This set excluded 6 adenocarcinomas and 4 squamous cell carcinomas from patients whose ancestry was not confirmed on PC1 or were outliers on PC2 in the PCA analysis (Figure 1). Clinical variables were not significantly associated with ancestry in either set of tumors with the exception of smoking pack-years (P < .05 for all) (eTables 3 and 4 in Supplement 2). The total number of putative somatic mutations, the percentage of genes amplified, or the percentage of genes deleted per tumor were not associated with ancestry (eFigure 4 in Supplement 1). Similarly, other quality control metrics were not associated with ancestry (eFigure 4 in Supplement 1).
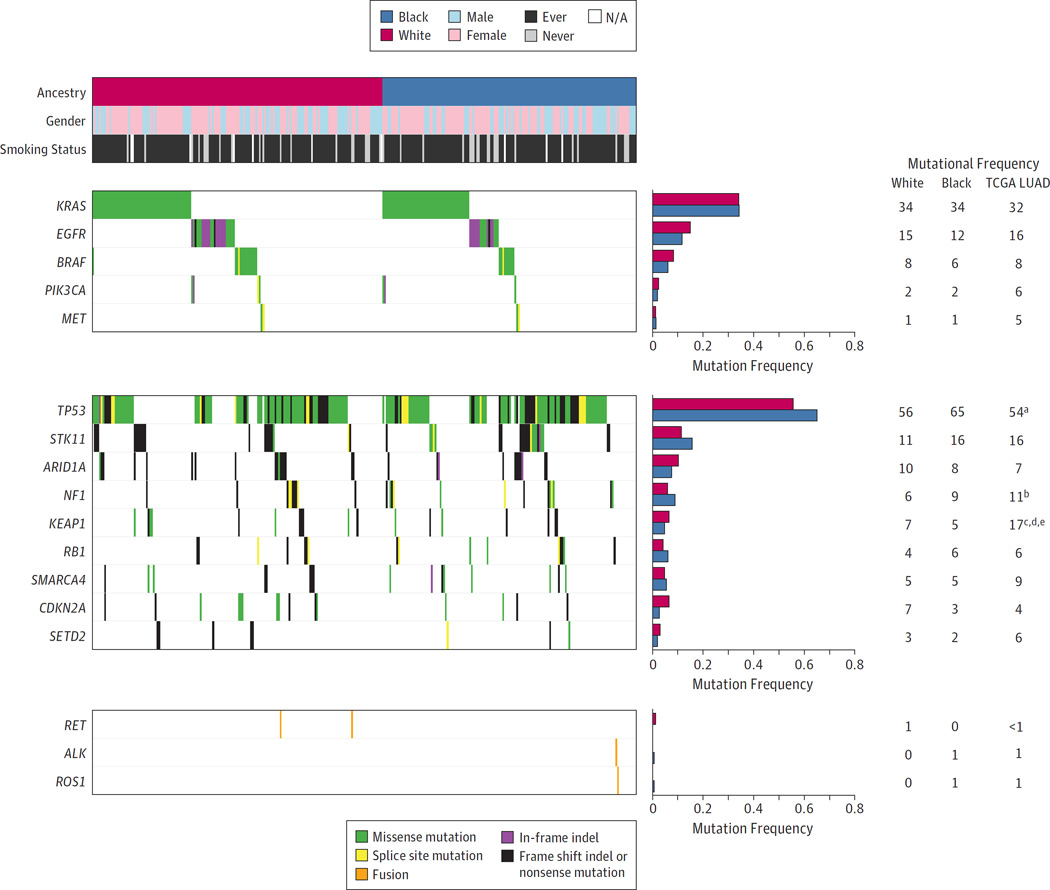
When examining previously characterized cancer genes in lung adenocarcinoma, the most commonly mutated oncogenes included KRAS (34%), EGFR (14%), and BRAF (7%), while the most commonly mutated tumor suppressors included TP53 (60%), STK11 (14%), and ARID1A (9%) (Figure 3). The most commonly mutated lung squamous cell carcinoma oncogenes included NFE2L2 (12%), and PIK3CA (9%) while tumor suppressors included TP53 (88%), CDKN2A (17%), and PTEN (14%) (eFigure 5 in Supplement 1). Using the Fisher exact test, we did not observe any statistical association between ancestry and mutation frequencies for any gene after correction for multiple hypothesis testing in either tumor type (FDR Q > .05) (eTables 5 and 6 in Supplement 2). We also compared the mutational frequencies in this cohort with the mutational frequencies found in tumors from TCGA and related studies.15 In lung adenocarcinoma, TP53 had a higher mutation frequency in tumors from black patients (P = .02), NF1 had a lower mutation frequency in tumors from white patients (P = .045), and KEAP1 had lower mutational frequencies in tumors from both populations (P < .001) in this cohort compared with tumors from TCGA (n = 660) (Figure3). No significant differences were observed between the lung squamous cell carcinomas from this cohort and those from TCGA (n = 484) using the Fisher exact test (P > .05for all) (eFigure 5 in Supplement 1).
Figure 3. Frequencies of Mutations and Fusions for Known Lung Adenocarcinoma Driver Genes.
The frequencies of putative acquired alterations in each gene were compared between tumors from black and white ancestry using the Fisher exact test. No lung cancer genes reached statistical significance after correction for multiple hypothesis testing (FDR Q > .05). Frequencies of alterations are shown for previously characterized oncogenes and tumor suppressors in lung adenocarcinoma.13,15 These frequencies are also compared with those found in a cohort of 660 lung adenocarcinomas (LUAD) from TCGA and related studies.13,15 Footnotes indicate a significant difference between the mutational frequencies in tumors from black patients in this cohort from those from TCGA using the Fisher exact test (aP < .05; bP < .01; cP < .001) or a significant difference between tumors from white patients in this cohort and those from TCGA (dP < .05; eP < .01). Columns correspond to tumors and rows correspond to genes or clinical annotation. The individual boxes are colored according to the type of alteration for that gene in that tumor. NA indicates not applicable.
We also determined if the number of copy number gains or losses for individual genes was significantly different between populations in either tumor type. Similar to the mutational profiles, we did not observe significant differences after correction for multiple hypothesis testing for the majority of genes (eFigures 6 and 7 in Supplement 1; eTables 7–10 in Supplement 2). The only exception is SULT1A1, which displayed a modestly significantly higher number of gains in tumors from black patients (FDR Q = .06). Finally, while the numbers were too small for statistical evaluation in lung adenocarcinoma, we observed one ALK rearrangement and one ROS1 rearrangement in tumors from black patients (Figure 3) (eTable 11 in Supplement 2). Two RET rearrangements were observed in tumors from white patients.
Ras/Raf/RTK Pathway Alterations in Lung Adenocarcinomas From Black Patients
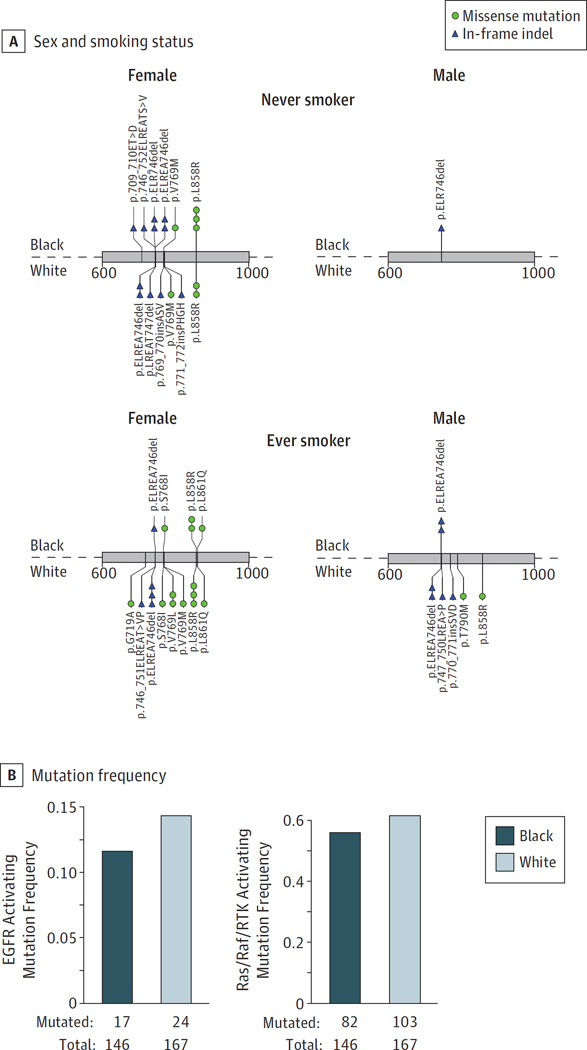
In lung adenocarcinoma, alterations in members of the Ras/Raf/RTK signaling pathways are of particular interest because they confer sensitivity to clinically approved targeted therapeutics.39 We first examined the mutational profile of EGFR and observed 46 mutations across 41 tumors (Figure4A). Five tumors had more than one EGFR mutation including p.V769M/p.ELREA746del, p.S768I/p.V769L, p.V769M/p.L858R, p.T790M/p.L858R, and p.G719A/p.L861Q. In-frame indels in exon 19 were observed in both men and women and did not show enrichment in black vs white women as has been previously reported23 (Fisher exact P = .80). The overall frequency of activating EGFR mutations was not associated with ancestry (P = .51) (Figure 4B). Using lung adenocarcinomas from patients with sufficient clinical annotation (n = 303), we performed logistic regression examining EGFR mutation status as a function of ancestry, sex, and smoking status. A significant increase was observed in never-smokers compared with ever-smokers (P = 2.85 × 10−6), and a modest increase in females compared with males (P = .08). However, no significant association was observed between EGFR mutation status and ancestry (P = .35) (eFigure 8 in Supplement 1 and eTable 12 in Supplement 2).
Figure 4. Known Activating Ras/Raf/RTK Pathway Alterations in Tumors From Black and White Populations.
We examined previously characterized activating mutations in genes that are members of the Ras/Raf/RTK pathway.13,15 A, A total of 23 activating single-nucleotide variations (SNVs) and 23 in-frame indels were found in EGFR across 313 lung adenocarcinomas. These mutations are grouped by smoking status, sex, and ancestry. One EGFR p.L858R mutation in tumor from a white female is not shown in this figure due to lack of smoking annotation. B, We did not observe a significant enrichment or depletion of known activating mutations in EGFR (left) or all activating mutations in the pathway (right) between tumors from black and white populations (P > .05 for all).
To determine if the overall frequency of mutations from any gene in the pathway was associated with race, we designated lung adenocarcinomas that had a previously characterized activating somatic single-nucleotide variants, indel, amplification, or gene fusion in a known RTK/Ras/Raf driver13,40,41 as “oncogene positive” (n = 128) while the remaining lung adenocarcinomas were considered “oncogene negative” (n = 185). Only fusions with canonical breakpoints were considered. MET and ERBB2 were considered amplified if the tumor had a log2 copy number ratio greater than 1.15 Ancestry was not significantly associated with oncogene negative status using a Fisher exact test (P = .36) (Figure 4B). This lack of association was also observed in a logistic regression model controlling for smoking status and sex (P = .36) (eFigure 8 in Supplement 1 and eTable 13 in Supplement 2). These results indicate that the proportions of Ras/Raf/RTK pathway alterations are similar between lung cancer patients from black and white populations in this study population.
Discussion
The primary goal of this study was to determine if the frequencies of putative acquired alterations in lung tumors are different between black and white ancestries. We examined variants in 504 genes from 313 lung adenocarcinomas and 138 lung squamous cell carcinomas and did not observe associations between ancestry and somatic genomic alterations within each histological subtype. As the targeted sequencing panel used in this study was limited to cancer genes, we did not have the ability to perform comprehensive ancestral inference and classification. However, we were able to capture SNVs in the cancer genes likely to be inherited due to their relatively high frequency in nondisease tissue from the Exome Aggregation Consortium database. We observed that the variation among these markers was strongly associated with self-reported race and largely confirmed the status of the self-reported race annotation in the majority of cases. The misclustering of tumors from 8 patients could be owing to errors in clinical data collection or differences in the patient’s self-identified race from their biological ancestry.
We observed 4 mutational signatures that have been previously characterized in cancer38 including those related to smoke exposure, aberrant APOBEC activity,42 MMR deficiency or aging, and UV exposure. Similar to 3 tumors from TCGA,15 we observed a single lung squamous cell carcinoma from a white male patient that had a pattern of UV-associated mutations often observed in skin cancers and may represent a metastasis to the lung.
Several previous studies have compared mutational frequencies between lung tumors from black and white patients and have reported conflicting results. Most of these earlier studies have focused on EGFR owing to its importance as a clinically actionable target and its higher prevalence in East Asian populations. Studies by Leidner et al24 and Yang et al21 reported statistically significant lower frequencies of EGFR mutations in black patients compared with white patients; Bauml et al22 reported lower frequencies in black female smokers compared with white female smokers; and Bollig-Fischer et al23 found higher rates of EGFR exon19 indels in black females compared with white females. In contrast, Cote et al,28 Reinersman et al,27 Yamaguchi et al,26 and Araujo et al25 did not observe any association of EGFR mutations with race. Of the studies that did not reach statistical significance, Reinersman et al27 found a higher frequency of EGFR mutations in blacks, while the others found lower frequencies.25,26,28 Overall, the frequency of EGFR mutations in black patients across all of these studies range from2%to 19%.25 Several confounding factors may contribute to these discrepancies. First, EGFR mutation frequency varies by both smoking history and by sex.36 While some studies were balanced for clinical covariates between populations or used logistic regression to control for unbalanced variables,22–25 other studies did not take this into consideration.21,26,27 Furthermore, some studies used lung tumors from black patients that were collected in different clinical settings21,24,25,27 or were profiled with a different genomic technology25 than the tumors collected from white patients. Comparisons of populations collected in different clinical settings may be influenced by factors such as sample quality, inclusion criteria, regional smoking behavior, and accessibility to health care, which are difficult to quantify and control in statistical models.
More recent studies have moved beyond EGFR and KRAS to profiling mutations across panels of genes. Bollig-Fischer et al23 examined 214 coding mutations in 26 cancer genes and observed that EGFR exon 19 indels were enriched in black women (P = .048). Araujo et al25 examined 38 SNVs across 8 genes as well as EGFR and ERBB2 indels and ALK translocations. They did not find any significant differences in individual genes, but did observe an overall reduction in the frequency of tumors from black patients that had any driver alteration (P = .04). Due to the exploratory nature of these studies, no correction for multiple hypothesis testing was performed. However, foregoing this correction increases the risk of observing false positives due to chance and reduces the likelihood of replication in future studies. This may be a contributing factor in our inability to replicate the findings from these studies.
Our cohort of lung cancers was collected from institutions in the same region and clinical setting, profiled with the same DNA sequencing assay and was well matched for clinical variables with the exception of pack-years of smoking. Although the value of pack-years was associated with ancestry, we did not observe significant differences in the smoking-associated signature (SI4) suggesting that differences between the populations in smoke exposure are minimal in our cohort. While this study profiled the largest number of genes compared with any of the previous studies, we cannot rule out the possibility that another gene outside of our assayed regions will have substantially different mutational frequencies between black and white populations. However, since the previously characterized recurrently altered genes in both tumor types12,13,15 did not display significant differences, we believe that the possibility of another gene displaying strong differences between populations is unlikely. We did observe a modest increase in the proportion of tumors with copy number gains in SULT1A1 from black patients. However, the variability in this region is likely germline as previously reported.43
Limitations
The mutational frequencies in this cohort were not significantly different from those observed in TCGA cohorts for most genes suggesting that the tumors in our cohort are not substantially different from those in TCGA. The only exceptions were in lung adenocarcinoma and included the tumor suppressors TP53, NF1, and KEAP1. Because we did not have access to paired normal tissue, we limited the missense and splice site mutations in our analysis to those previously observed in human cancers as annotated in the COSMIC (Catalogue Of Somatic Mutations In Cancer) database. This may have led to the exclusion of some true novel somatic mutations and contributed to the lower mutational frequencies observed in these genes.
Conclusions
In addition to the mutational frequencies of individual genes, we examined the differences in the frequency of previously curated Ras/Raf/RTK pathway alterations known to drive cellular transformation in lung adenocarcinoma. We again did not observe significant associations between ancestry and the mutational frequencies of the pathway members included in this panel. Therefore, we believe that clinical sequencing coupled with targeted therapies against these alterations will benefit patients in both populations. Overall, these results suggest that differences in exomic mutations do not contribute to the observed racial disparities in incidence and mortality between black and white populations. Further investigation into other genomic factors such as mutations in noncoding regions, epigenetic alterations, and gene expression changes or other socioeconomic variables beyond smoking behavior and access to health care may be required to fully explain these disparities.
Supplementary Material
Key Points.
Question Are the mutational profiles of lung cancers different between black and white patients?
Findings In this genomic analysis of a cohort of patients with lung cancer, mutational frequencies were not significantly different between black and white populations in both lung adenocarcinoma and lung squamous cell carcinoma.
Meaning Differences in mutational profiles do not likely contribute to the increased incidence and mortality of lung cancer in black populations.
Acknowledgments
Funding/Support: This work was conducted with support from the National Cancer Institute Lung Cancer Disparities Center (grants 1P50CA148596 and R01CA172253 to Dr Osarogiagbon).
Role of the Funder/Sponsor: The funders/sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: We thank Phani Davineni, MSc; Bruce Wollison, MSc; Sam Hunter, PhD; and Ryan Abo, PhD; for bioinformatics assistance. They were not compensated for their contributions beyond their established salaries.
Footnotes
Author Contributions: Drs MacConaill and Meyerson had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Concept and design: Lathan, Sholl, Williams, Meyerson, MacConaill.
Acquisition, analysis, or interpretation of data: Campbell, Sholl, Ducar, Vega, Sunkavalli, Lin, Hanna, Schubert, Thorner, Faris, Osarogiagbon, van Hummelen, MacConaill.
Drafting of the manuscript: Campbell, Sunkavalli, Thorner, Osarogiagbon, Meyerson, MacConaill.
Critical revision of the manuscript for important intellectual content: Campbell, Lathan, Sholl, Ducar, Vega, Lin, Hanna, Schubert, Faris, Williams, Osarogiagbon, van Hummelen, Meyerson, MacConaill.
Statistical analysis: Campbell, Hanna.
Obtained funding: Williams, Meyerson, MacConaill.
Administrative, technical, or material support: Lathan, Ducar, Vega, Sunkavalli, Lin, Hanna, Thorner, Faris, Williams, Osarogiagbon.
Supervision: van Hummelen.
Tissue acquisition and data management: Faris.
Conflict of Interest Disclosures: Dr Meyerson is a founder of Foundation Medicine; was a consultant and equity holder in Foundation Medicine within the last 3 years; is an inventor on a patent for the analysis of EGFR mutations in lung cancer diagnostics; and receives research support from Bayer. Dr Osarogiagbon is a stockholder of Eli Lilly, Foundation Medicine and Pfizer; a consultant and speaker for Eli Lilly and Genentech; and has a patent application for a lymph node specimen collection kit. No other conflicts are reported.
REFERENCES
- 1.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al., editors. National Cancer Institute; [Accessed December 12, 2016]. SEER Cancer Statistics Review, 1975–2013. (based on November 2015 SEER data submission, posted to the SEER web site, April 2016). http://seer.cancer.gov/csr/1975_2013/ [Google Scholar]
- 3.Geyer S. Social inequalities in the incidence and case fatality of cancers of the lung, the stomach, the bowels, and the breast. Cancer Causes Control. 2008;19(9):965–974. doi: 10.1007/s10552-008-9162-5. [DOI] [PubMed] [Google Scholar]
- 4.Albano JD, Ward E, Jemal A, et al. Cancer mortality in the United States by education level and race. J Natl Cancer Inst. 2007;99(18):1384–1394. doi: 10.1093/jnci/djm127. [DOI] [PubMed] [Google Scholar]
- 5.Coughlin SS, Matthews-Juarez P, Juarez PD, Melton CE, King M. Opportunities to address lung cancer disparities among African Americans. Cancer Med. 2014;3(6):1467–1476. doi: 10.1002/cam4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bach PB, Schrag D, Brawley OW, Galaznik A, Yakren S, Begg CB. Survival of blacks and whites after a cancer diagnosis. JAMA. 2002;287(16):2106–2113. doi: 10.1001/jama.287.16.2106. [DOI] [PubMed] [Google Scholar]
- 7.Blackstock AW, Herndon JE, II, Paskett ED, et al. Cancer and Leukemia Group B. Similar outcomes between African American and non-African American patients with extensive-stage small-cell lung carcinoma: report from the Cancer and Leukemia Group B. J Clin Oncol. 2006;24(3):407–412. doi: 10.1200/JCO.2005.02.1436. [DOI] [PubMed] [Google Scholar]
- 8.Haiman CA, Stram DO, Wilkens LR, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354(4):333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 11.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell JD, Alexandrov A, Kim J, et al. Cancer Genome Atlas Research Network. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 17.Arrieta O, Cardona AF, Martin C, et al. Updated Frequency of EGFR and KRAS Mutations in NonSmall-Cell Lung Cancer in Latin America: The Latin-American Consortium for the Investigation of Lung Cancer (CLICaP) J Thorac Oncol. 2015;10(5):838–843. doi: 10.1097/JTO.0000000000000481. [DOI] [PubMed] [Google Scholar]
- 18.Fong T, Morgensztern D, Govindan R. EGFR inhibitors as first-line therapy in advanced non-small cell lung cancer. J Thorac Oncol. 2008;3(3):303–310. doi: 10.1097/JTO.0b013e3181645477. [DOI] [PubMed] [Google Scholar]
- 19.Blackhall F, Ranson M, Thatcher N. Where next for gefitinib in patients with lung cancer? Lancet Oncol. 2006;7(6):499–507. doi: 10.1016/S1470-2045(06)70725-2. [DOI] [PubMed] [Google Scholar]
- 20.Yang CH. EGFR tyrosine kinase inhibitors for the treatment of NSCLC in East Asia: present and future. Lung Cancer. 2008;60(suppl 2):S23–S30. doi: 10.1016/S0169-5002(08)70102-8. [DOI] [PubMed] [Google Scholar]
- 21.Yang SH, Mechanic LE, Yang P, et al. Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res. 2005;11(6):2106–2110. doi: 10.1158/1078-0432.CCR-04-1853. [DOI] [PubMed] [Google Scholar]
- 22.Bauml J, Mick R, Zhang Y, et al. Frequency of EGFR and KRAS mutations in patients with non small cell lung cancer by racial background: do disparities exist? Lung Cancer. 2013;81(3):347–353. doi: 10.1016/j.lungcan.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bollig-Fischer A, Chen W, Gadgeel SM, et al. Racial diversity of actionable mutations in non-small cell lung cancer. J Thorac Oncol. 2015;10(2):250–255. doi: 10.1097/JTO.0000000000000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leidner RS, Fu P, Clifford B, et al. Genetic abnormalities of the EGFR pathway in African American Patients with non-small-cell lung cancer. J Clin Oncol. 2009;27(33):5620–5626. doi: 10.1200/JCO.2009.23.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araujo LH, Lammers PE, Matthews-Smith V, et al. Somatic mutation spectrum of non-small-cell lung cancer in african americans: a pooled analysis. J Thorac Oncol. 2015;10(10):1430–1436. doi: 10.1097/JTO.0000000000000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi N, Vanderlaan PA, Folch E, et al. Smoking status and self-reported race affect the frequency of clinically relevant oncogenic alterations in non-small-cell lung cancers at a United States-based academic medical practice. Lung Cancer. 2013;82(1):31–37. doi: 10.1016/j.lungcan.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinersman JM, Johnson ML, Riely GJ, et al. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in African Americans. J Thorac Oncol. 2011;6(1):28–31. doi: 10.1097/JTO.0b013e3181fb4fe2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cote ML, Haddad R, Edwards DJ, et al. Frequency and type of epidermal growth factor receptor mutations in African Americans with non-small cell lung cancer. J Thorac Oncol. 2011;6(3):627–630. doi: 10.1097/JTO.0b013e31820a0ec0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36(4):E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abo RP, Ducar M, Garcia EP, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2014;43(3):e19. doi: 10.1093/nar/gku1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904– 909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9(4):e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toyooka S, Matsuo K, Shigematsu H, et al. The impact of sex and smoking status on the mutational spectrum of epidermal growth factor receptor gene in non small cell lung cancer. Clin Cancer Res. 2007;13(19):5763–5768. doi: 10.1158/1078-0432.CCR-07-0216. [DOI] [PubMed] [Google Scholar]
- 37.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oxnard GR, Binder A, Jänne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;31(8):1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 41.PaoW, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;18(3):349–351. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- 42.Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45(9):970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16(5):463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.