Abstract
The small Hsp DmHsp27 from Drosophila melanogaster is one of the few small heat shock proteins (sHsps) found within the nucleus. We report that its dimerization is independent of disulfide bond formation and seems to rely on salt bridges. Unlike metazoan sHsps, DmHsp27 forms two populations of oligomers not in equilibrium. Mutations at highly conserved arginine residues in mammalian sHsps have been reported to be associated with protein conformational defects and intracellular aggregation. Independent mutation of three highly conserved arginines (R122, R131, and R135) to glycine in DmHsp27 results in only one population of higher molecular weight form. In vitro, the chaperone-like activity of wild-type DmHsp27 was comparable with that of its two isolated populations and to the single population of the R122G, R131G, and R135G using luciferase as substrate. However, using insulin, the chaperone-like activity of wild-type DmHsp27 was lower than that of R122G and R131G mutants. Altogether, the results characterize wild-type DmHsp27 and its alpha-crystallin domain (ACD) arginine mutants and may give insight into protection mechanism of sHsps.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0748-7) contains supplementary material, which is available to authorized users.
Keywords: Small heat shock protein (sHsp), DmHsp27, Chaperone, Alpha-crystallin domain (ACD), Drosophila melanogaster
Introduction
The small heat shock proteins (sHsps) are a ubiquitous family of ATP-independent stress proteins found in all domains of life (Caspers et al. 1995; de Jong et al. 1998; Fu et al. 2006; Maaroufi and Tanguay 2013). These proteins are upregulated in response to a variety of stresses that negatively impact protein homeostasis.
Small Hsps are characterized by the presence of a conserved α-crystallin domain (ACD), which is flanked by a variable N-terminal region (NTR) and a C-terminal extension (CTE). The ACD represents the conserved signature motif of sHsps. Structurally, it forms a compact β-sandwich, consisting of two antiparallel sheets made of three and four β-strands, respectively, connected by a short inter-domain loop (Delbecq and Klevit 2013; Van Montfort et al. 2001). 3D structures of isolated ACDs demonstrate that they usually form stable dimers, but the ACD alone is not sufficient for oligomer formation (Bagneris et al. 2009; Clark et al. 2011; Laganowsky et al. 2010). Mutation of conserved arginines within the ACD may cause neurological disease and cataract (Benndorf et al. 2014; Boncoraglio et al. 2012; Datskevich et al. 2012; Evgrafov et al. 2004; Gentil and Cooper 2012; Inagaki et al. 2006; Treweek et al. 2005; Vicart et al. 1998).
The genome of Drosophila melanogaster encodes for 12 proteins containing an ACD (Morrow and Tanguay 2015). Here, we investigate D. melanogaster Hsp27 (DmHsp27), a nuclear-localized sHsp (Beaulieu et al. 1989; Michaud et al. 2008). In addition to its upregulation in response to stress, DmHsp27 also shows tissue- and stage-specific expression patterns during development (Michaud et al. 2002).
The structure of DmHsp27 has not yet been reported. Here, we show that DmHsp27 forms dimer independently of disulfide bond formation and rather seems dependent on salt bridges. Surprisingly, DmHsp27 forms two populations of oligomers. We have studied the structure and chaperone-like activity of wild-type DmHsp27 (DmHsp27WT) and three mutants of arginine within the ACD domain. Although the three different arginine mutants form high molecular weight oligomers distinct from the ones of the wild-type protein, they display either a similar chaperone-like activity as the wild-type protein or even a better one.
Materials and methods
Cloning and expression of recombinant DmHsp27
The cDNA of DmHsp27WT (Morrow et al. 2006) was cloned into the small ubiquitin-related modifier (SUMO) fusion vector pETHSUK (a gift from Dr. S. Weeks (Heirbaut et al. 2014; Weeks et al. 2007)) using Gibson Assembly (NEB). The expression of the recombinant proteins as fusions with SUMO protein increased protein expression significantly. This is enhanced by the availability of SUMO-hydrolase, which removes specifically SUMO from the fusion protein (Amerik and Hochstrasser 2004; Weeks et al. 2007). Point mutations were introduced by PCR, using Gibson Assembly (NEB).
The Escherichia coli BL21 (DE3) pLysS strain (NEB) was used for expression of DmHsp27WT and mutants. Briefly, transformed bacteria were grown in 10 ml of Luria-Bertani (LB) broth miller (EMD Millipore) containing 100 μg/ml ampicillin (Biobasic Canada) at 37 °C overnight. After a 1:50 dilution, the bacteria were grown 3 h at 37 °C until an OD600 around 0.6. Protein expression was then induced by 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Roche) 5 h at 30 °C. Bacterial cells were collected by centrifugation, washed twice with binding buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris–HCl pH 8), and stored at −80 °C until purification.
Purification of recombinant DmHsp27
The cell pellet was suspended in 10 ml of binding buffer supplemented with 0.01 mg/ml of DNase (Sigma, Aldrich) and 4 mM of phenylmethanesulfonyl fluoride (PMSF) (Sigma, Aldrich). The cells were lysed in a cold room (+7 °C) using French press (SLM AMINCO). Cell debris were removed by centrifugation at 10,000×g at 4 °C for 30 min. The soluble fraction was incubated with 1 ml bed volume of Ni-NTA agarose (Qiagen) to separate recombinant protein from contaminating proteins by chelate affinity chromatography. The column was washed with 30 volumes of washing buffer (60 mM imidazole, 500 mM NaCl, 20 mM Tris–HCl pH 8). Bound protein was eluted with elution buffer (500 mM imidazole, 500 mM NaCl, 20 mM Tris–HCl pH 8). The eluate was collected in 1-ml fractions. The fractions containing the pure protein were dialyzed against storage buffer (20 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM dithiothreitol (DTT)), concentrated, and pooled. Pooled fractions were digested with SUMO-hydrolase and additionally purified by size exclusion chromatography (SEC) using a Superose 6 10/300 column (GE Life Sciences) equilibrated with 20 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM DTT to remove SUMO-hydrolase, HIS-SUMO-tag, and undigested protein. Protein fractions were verified by SDS-PAGE, and pure samples were pooled together and concentrated using centrifugal filters (Amicon, EMD Millipore).
Fluorescence spectroscopy
Intrinsic tryptophan fluorescence was measured in fluorescence buffer (150 mM NaCl, 20 mM HEPES-KOH pH 8) at 20 °C. Fluorescence of protein samples (0.1 mg/ml) was excited at 295 nm and recorded in the range 300–400 nm (both slits width 5 nm).
Temperature-induced conformational changes of DmHsp27WT were followed by recording intrinsic tryptophan fluorescence. The pure protein samples (0.1 mg/ml) in fluorescence buffer were heated with the constant rate 1 °C/min from 20 up to 90 °C and cooled back at the same rate using Varian Cary Eclipse spectrofluorometer. Tryptophan was excited at 297 nm and fluorescence recorded at 332 nm (slits 5 nm).
Analytical SEC
The quaternary structure of DmHsp27WT was analyzed by SEC. Different concentrations of samples were loaded on Superose 6 10/300 column (GE Life Sciences) equilibrated with 20 mM Tris–HCl pH 8 and 150 mM NaCl and eluted at 0.5 ml/min. Fractions of 500 μl were collected and their protein composition was analyzed by SDS–gel electrophoresis. For estimating the molecular weight, the column was calibrated with protein markers immunoglobulin M (IGM) from bovine serum (900 kDa) (Sigma), thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), and Blue Dextran 2000 to determine the void volume (GE Life Sciences).
Native gel electrophoresis
The quaternary structure was also resolved by non-denaturing native gel electrophoresis. DmHsp27 was run on pre-cast 4–12% gradient native Tris–glycine gels (Thermo Fisher Scientific) in Mini-Cell electrophoresis system (XCell SureLock, Life Technology). Gels were run at 150 V for 90 min. After electrophoresis, the protein complexes were stained with Coomassie blue.
Luciferase heat-induced aggregation assay
The heat-induced aggregation assay was conducted essentially as described in Morrow et al. (2006). Luciferase (0.1 μM, Promega) was heat-denatured at 42 °C in the absence or presence of DmHsp27WT or its arginine mutants for 30 min (0.4 μM). Aggregation was determined by light scattering at 320 nm on a spectrophotometer with thermostated cells (Varian Cary 100, Montreal, Quebec, Canada). Data are representative of three different assays and expressed as the mean ± standard deviation.
Insulin DTT-induced aggregation assay
Chaperone-like activity was also determined by aggregation of insulin as a result of the reduction of disulfide bonds (Holmgren 1979). Insulin (52 μM, Sigma) in 10 mM potassium phosphate and 100 mM NaCl (pH 8) was pre-incubated alone or in the presence of (13 μM) DmHsp27WT or mutants for 30 min. The reaction was started by addition of DTT up to the final concentration of 20 mM, and aggregation of insulin was followed by an increase in the optical density at 320 nm on a spectrophotometer with thermostated cells. Data are representative of three different assays and expressed as the mean ± standard deviation.
Molecular modeling
The 3D models of ACD of DmHsp27WT and its mutants were built with SWISS-MODEL (Biasini et al. 2014) using as template the ACD of HspB1 (PDB: 4MJH). The obtained models were energy minimized by KoBaMIN (Rodrigues et al. 2012), and their quality was assessed by Ramachandran plot analysis through PROCHECK (Laskowski et al. 1993). Ramachandran plot revealed that 96 and 4 % of residues are in favored and allowed region, respectively. Visualization of 3D models was realized with PyMOL software (http://pymol.org/).
Dynamic light scattering
For complex characterization, dynamic light scattering (DLS) experiments were performed with 800 nm radiation on a DynaPro instrument (Wyatt Technology Corp., Santa Barbara, CA) as described previously (Michiel et al. 2009; Skouri-Panet et al. 2012). All samples were filtered through 0.22 μm filters. The protein concentration was adjusted to 0.2 to 2 mg/ml. The sample volume was 80 μl, and the acquisition time was 10 s for a total measurement of up to 1 h. The sample temperature was controlled with a Peltier cell with a precision of 0.1 °C. The hydrodynamic radius (Rh) and the polydispersity (% Pd) were obtained using regularization methods (Dynamics software, version 6).
Cell culture and transfection conditions
HeLa cells were maintained in MEM Alpha (Gibco) supplemented with 5 % FBS penicillin and streptomycin. For transfections, cells were plated 24 h in advance at a confluence of 175,000 cells/well (six-well plate) containing glass coverslip. Then, cells were washed three times in OptiMEM (Gibco) and incubated for 4 h in OptiMEM containing the plasmid (pcDNA-DmHsp27)/Lipofectamine (Invitrogen) complex (1.5 μl Lipofectamine/1.5 μg DNA) prepared according to the manufacturer’s protocol. After the incubation, cells were washed three times with complete medium and without antibiotics and left to express for 24 to 72 h before being processed for immunofluorescence.
Immunofluorescence analyses
Cells were washed three times in PBS and fixed by incubation in pre-chilled methanol at −20 °C for 20 min. They were washed three times with PBS containing 0.1 % Tween 20-X (PBST) and blocked for 1 h in PBST containing 5 % BSA (PBST-BSA), for 1 h at room temperature. After blocking, cells were incubated in primary antibody (monoclonal anti-DmHsp27 (2C8E11) (Michaud et al. 2008)) diluted (1/20) in PBST-BSA for 1 h at room temperature. After washing with PBST, cells were incubated with secondary antibody (goat anti-mouse Alexa 488 (Invitrogen)) for 45 min. Coverslips were mounted in Vectashield mounting medium (Vector Laboratories). Samples were visualized using an Axio Observer Z1 microscope equipped with an Axiocam camera (Carl Zeiss, Canada).
Results
Dimerization of DmHsp27
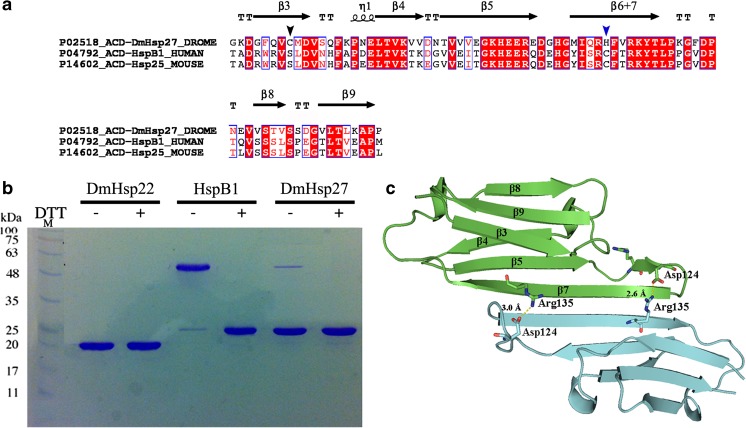
sHsps show a significant underrepresentation of cysteines. The reduction of the number of cysteines in these sequences may be important to prevent unwanted cross-linking under oxidative conditions, which would prevent subunit exchange and thus hinder the dynamics of sHsps (Kriehuber et al. 2010). The sequence of DmHsp27 has a single cysteine residue at position 93, located in the β3 strand (Fig. 1a). Human HspB1 and murine Hsp25 can form dimers by an intermolecular disulfide bond between cysteine residues at positions 137 and 141, respectively, located in the β6+7 strand (Baranova et al. 2011; Zavialov et al. 1998) (Fig. 1a).
Fig. 1.
Dimeric interface of DmHsp27. a Amino acid sequence alignment of ACD of DmHsp27, human HspB1, and mouse Hsp25. Alignment was generated using multiple sequence comparison by log-expectation (MUSCLE). Secondary structures indicated in the alignment were assigned according to the determined crystal structure of ACD HspB1 (PDB: 4MJH). Black and blue arrowheads indicate cysteine positions. b Non-reducing SDS-PAGE, showing disulfide cross-linking of full-length DmHsp22, DmHsp27, and human HspB1. The proteins were incubated at 20 °C for 21 h with 5 mM DTT (+) or without any reducing agent (−) loaded on non-reducing SDS-PAGE and detected by Coomassie blue staining. c Predicted 3D structure of ACD of DmHsp27 generated with SWISS-MODEL (Biasini et al. 2014) based on defined structure of HspB1 ACD (PDB 4MJH). Visualization of 3D models was realized with PyMOL software. Monomers represented as blue and green. The salt bridge between conserved arginine 135 and aspartic acid 124 is shown
To assess if the disulfide bond is involved in dimeric interface between cysteines residues in the β3 strand of DmHsp27, we examined its migration on a native gel under reductive and non-reductive conditions compared to human HspB1 and DmHsp22. Human HspB1 was used as a positive control (Baranova et al. 2011) and DmHsp22 that does not have cysteine was used as a negative control. As seen in Fig. 1b, DmHsp27 behaves like DmHsp22 suggesting that the dimeric interface does not involve disulfide bond.
A 3D structure model of ACD-DmHsp27WT was generated based on the defined structure of ACD-HspB1 (PDB 4MJH; Hochberg et al. 2014), sharing 70 % similarity and 53 % identity (Fig. 1a). According to the resulting model of DmHsp27, ACD monomer is folded into a compact, six-stranded β-sandwich composed of two β-sheets. The first contains β3, β8, and β9 strands and, therefore, both termini of ACD, and the second β-sheet contains β4, β5, and β6+7 strands (Fig. 1c). The β-strands are interconnected by three- to ten-residue-long loops. The largest loop, L4 (residues 119–128), connects the β5 and β7 strands. DmHsp27 has the metazoan dimer structure, in which the β6- and β7-strands are fused into an elongated strand that forms the dimer interface with its counterpart from the neighboring monomer in an antiparallel orientation.
Thermal transition of DmHsp27
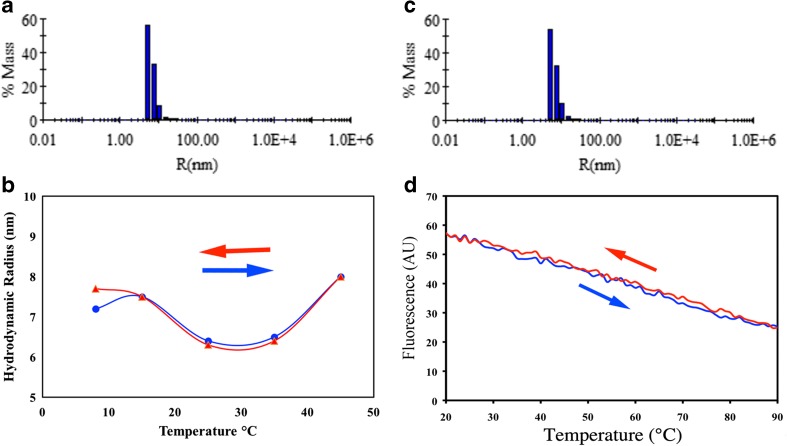
The rate of exchange of sHsps subunits is strongly dependent on temperature (Bova et al. 1997; Bova et al. 2002). We used DLS to analyze the thermal stability of DmHsp27WT by separating soluble populations of different hydrodynamic radius (Rh). At 8 °C, DmHsp27WT has a Rh about 7.2 nm and percentage of polydispersity of 51.5 % (Fig. 2a). The heat-induced changes in DmHsp27WT oligomeric size were followed from 8 up to 45 °C. At each temperature, the scattered intensity was recorded up to 1 h, and the corresponding Rh is shown in Fig. 2b. At 8 °C, the Rh is equal to 7.2 nm, with a high polydispersity, more than 50 %. At 25 and 35 °C, the Rh slightly decreases to 6.4 and 6.5 nm but increases to 8.0 nm at 45 °C. Cooling back the temperature from 45 to 8 °C resulted in a decrease of the oligomeric size to values similar to the one observed prior to heating (Fig. 2b, c). This behavior is compatible with sHsp assemblies composed by an average number of subunits that could correspond to different populations present in solution.
Fig. 2.
Thermal transition of DmHsp27 wild type. Histogram of hydrodynamic radius (Rh) for DmHsp27 WT at 8 °C (a) and return from 45 °C to 8 °C (c), given an average Rh (in nm) and the corresponding percentage of polydispersity (%Pd). b Hydrodynamic radius for DmHsp27 WT as a function of temperature. The arrows indicate directions of heating or cooling. d Changes of tryptophan fluorescence of DmHsp27 induced by temperature. The arrows indicate directions of heating or cooling
DmHsp27WT contains one tryptophan residue located in the NTR at position 21. We took advantage of the fluorescence of this amino acid to follow heat-induced changes in the oligomeric size. Heating sample from 20 up to 90 °C and cooling it back to 20 °C was accompanied by reversible changes of intrinsic fluorescence of tryptophan (Fig. 2d), suggesting that thermal transitions of the DmHsp27 structure are reversible even at extreme temperature.
Dissection of DmHsp27 wild type
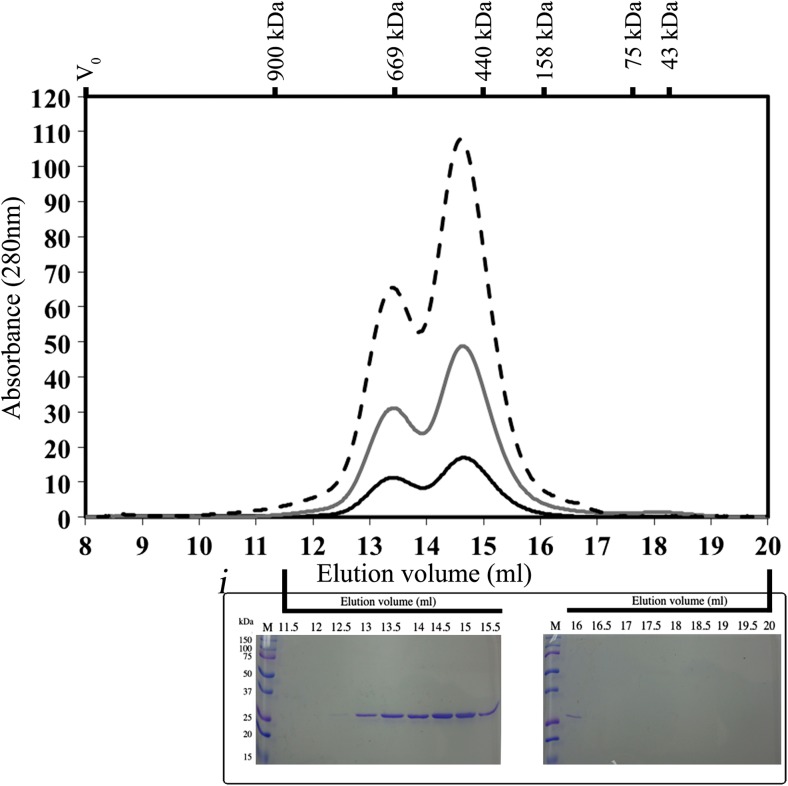
While DLS gives a general idea of the oligomeric size, SEC is recognized as a more sensitive method to determine the oligomeric structure. The profile of DmHsp27WT elution on a Superose 6 10/300 GL gives two peaks. The first one elutes at 13.4 ml (peak no. 1) and corresponds to a molecular masses of 725 kDa, while the second one (peak no. 2) was eluted at 14.6 ml and has a molecular masses of 540 kDa (Fig. 3). The position of the two peaks was independent of the quantity of protein loaded on the column (Fig. 3). As shown on the Coomassie blue-stained gels of the collected fractions, peak no. 2 is 1.8 times more abundant than peak no. 1.
Fig. 3.
SEC analysis of DmHsp27 WT. Size exclusion chromatography (SEC) analysis using a Superose 6 10/300 GL (GE Life Sciences) column, which was calibrated by using IGM (900 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (43 kDa) to generate a calibration curve. Blue Dextran (2000 kDa) was used to determine the void volume of the column V 0. a Profile on column of 600 μg (dashed line), 300 μg (gray line), and 100 μg (black line) of pure DmHsp27W. Insert (i) present SDS electrophoresis of samples collected (500 μl for each fraction) after size exclusion chromatography. The proteins were detected by Coomassie blue staining. The elution volume is indicated above the gel
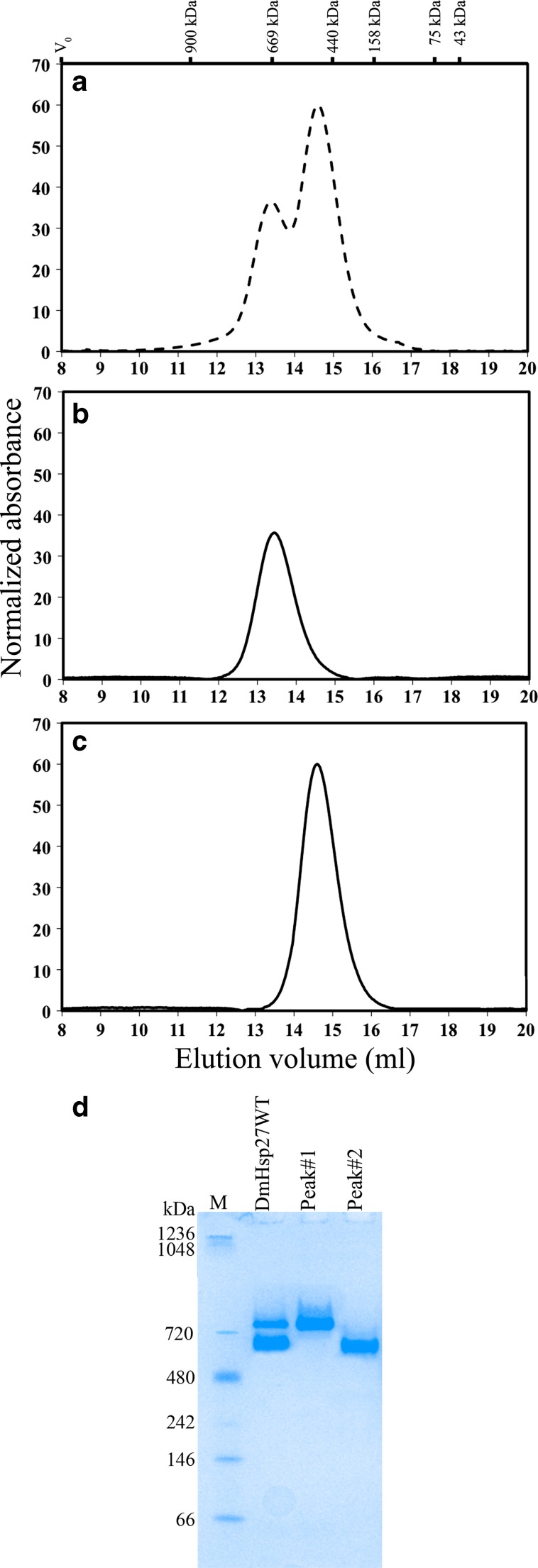
To determine if the populations of the two DmHSP27 peaks were in equilibrium, each was isolated and loaded separately on the Superose 6 column. When peak no. 1 was reloaded on the column, a single peak with the same volume of elution was obtained (Fig. 4b). Similarly, peak no. 2 also gave a single peak eluting at the same position of 14.6 ml (Fig. 4c). This result was confirmed by the profile of migration of DmHsp27WT on a native gel. Indeed, as seen in Fig. 4d, the migration of DmHSP27WT results in two bands and each of the isolated population migrates to its original position.
Fig. 4.
Complexes of DmHsp27 WT. Size exclusion chromatography of DmHsp27WT (a), peak no. 1 (b), and peak no. 2 (c). Normalized elution profiles obtained after loading 600 μg of pure protein on the Superose 6 column and re-injection of peak no. 1 and peak no. 2 obtained after this injection. Elution volumes of markers are shown above and include Blue Dextran to indicate the void volume V 0. d Native gradient (4–12 %) polyacrylamide gel electrophoresis of 25 μg of different species of DmHsp27WT and isolated peak no. 1 and peak no. 2. Positions of standard protein markers with known molecular weights are shown on the left
Study of three arginines in ACD
As mentioned in the “Introduction,” the ACD is the hallmark of the sHsp family. Arginine mutants from the ACD of HspB1, HspB4, HspB5, and HspB8 have been reported to be involved in different diseases such as distal hereditary motor neuropathy, Charcot–Marie–Tooth disease, desmin-related myopathy, and cataracts (Benndorf et al. 2014; Boncoraglio et al. 2012; Datskevich et al. 2012; Evgrafov et al. 2004; Gentil and Cooper 2012; Inagaki et al. 2006; Irobi et al. 2004; Litt et al. 1998; Treweek et al. 2005; Vicart et al. 1998).
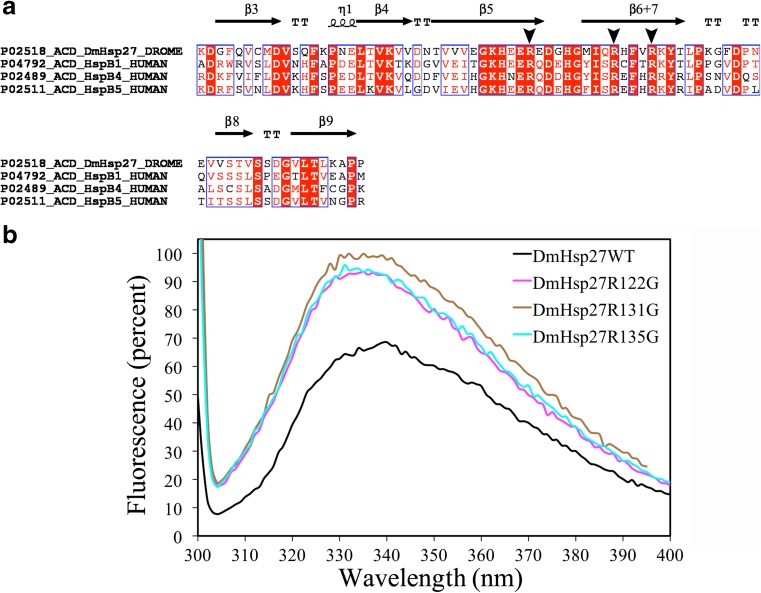
Based on the importance and conservation of arginine residues for other sHsps (Fig. 5a), arginine at positions 122, 131, and 135 of DmHSP27 was substituted with glycine and purified. Their amplitude of intrinsic tryptophan fluorescence was recorded by a spectrofluorometer. As mentioned earlier, DmHsp27 contains one tryptophan residue located in the NTR at position 21. The fluorescence spectrum of DmHsp27WT is characterized by a maximum in the range of 330–350 nm (Fig. 5b). The intensity of fluorescence of DmHsp27WT was 1.5 times smaller than that of ACD mutants without significant change in position of maximum of fluorescence (Fig. 5b). This might indicate that the tryptophan residue of DmHsp27 wild type is more accessible to quenching than the corresponding residue in ACD mutants.
Fig. 5.
Analysis of three conserved arginine in ACD. a Amino acid sequence alignment of ACD of DmHsp27 and humans HspB1, HspB4, and HspB5. Alignment was generated using multiple sequence comparison by log-expectation (MUSCLE). Secondary structures indicated in the alignment were assigned according to the determined crystal structure of ACD HspB1 (PDB: 4MJH). Black arrowheads indicate the conserved arginines. b Fluorescence spectra of the DmHsp27WT and its ACD mutants. Intrinsic tryptophan fluorescence was excited at 295 nm and recorded in the range of 300–400 nm (both slits width 5 nm)
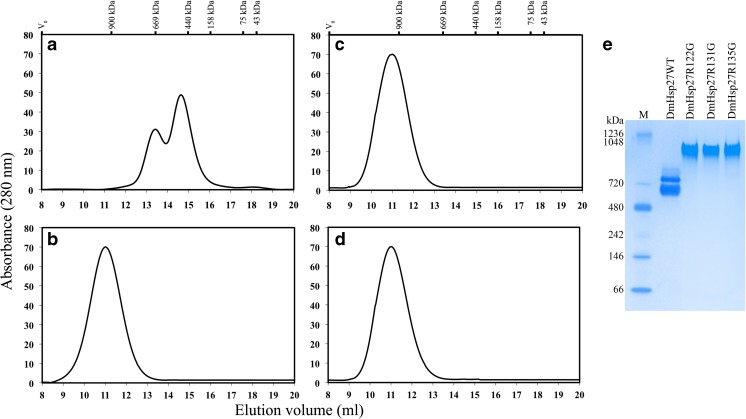
The oligomeric structure was analyzed by SEC and polyacrylamide native gel electrophoresis. In contrast to DmHsp27WT, the profile of elution of each arginine mutant resulted in one single peak eluting at 11 ml and corresponding to a molecular mass of 1100 kDa (Fig. 6a–d). Similar to the SEC analysis, the mutants also migrate as single band on a native gel. This band migrated at a higher molecular weight than the two bands observed for DmHsp27WT (Fig. 6e). Since ACD mutants had increased oligomeric size, we tested the ability to aggregate in vivo in cultured cells as shown for mutant R148G in Hsp27 of Chinese hamster and R120G, R120C, and R120D in HspB5 by (Chavez Zobel et al. 2005 and Simon et al. 2007). Localization of DmHSP27WT and ACD mutants was determined 24, 48, and 72 h after transfection HeLa cells. The DmHsp27 mutants presented the same type of nuclear localization in the transfected cell line, as expected from previous work (Beaulieu et al. 1989b; Michaud et al. 2008) (data not shown).
Fig. 6.
SEC analysis of DmHsp27 ACD mutants. Size exclusion chromatography of wild-type DmHsp27 and ACD mutants. Three micrograms of different mutants DmHsp27WT (a), R122G (b), R131G (c), and R135G (d) were loaded on a Superose 6 column. Elution volumes of markers are shown above and include Blue Dextran to indicate the void volume V 0. e Native gradient (4–12 %) polyacrylamide gel electrophoresis of 25 μg of different species of DmHsp27WT, R122G, R131G, and R135G. Positions of standard protein markers with known molecular weights are shown on the left
Chaperone-like activity
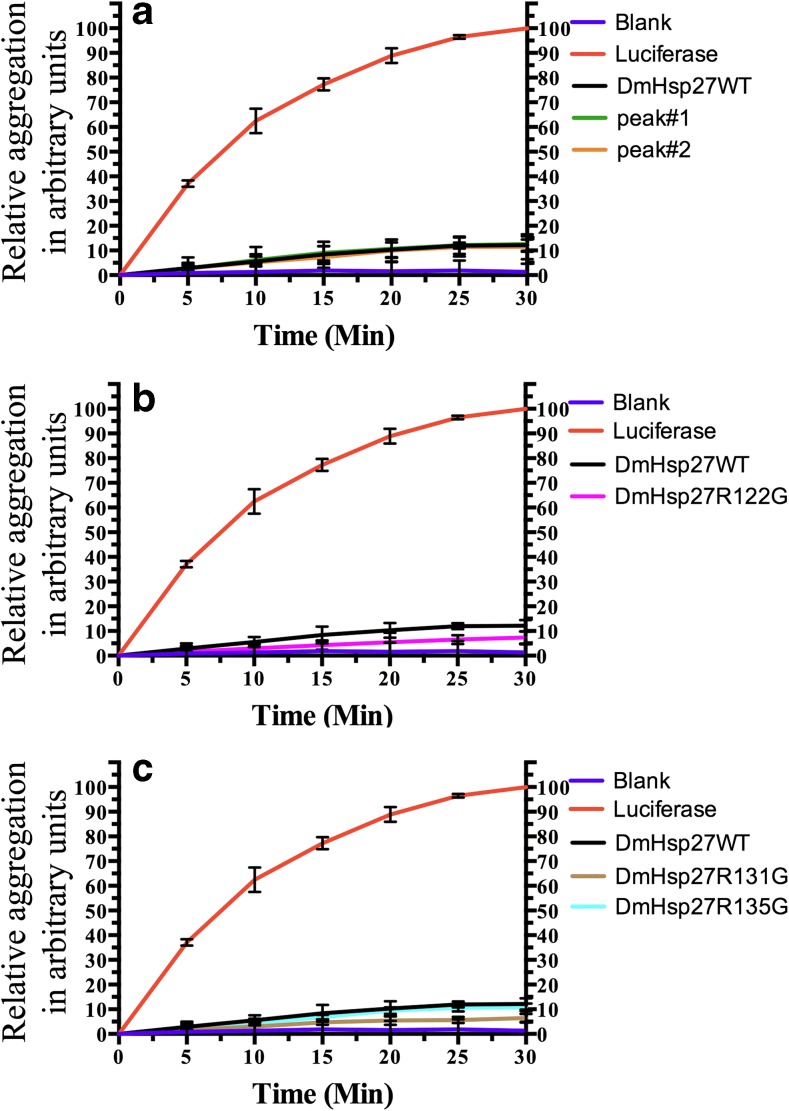
Several sHsps can maintain substrates in a folding-competent state (Mogk et al. 2003; Morrow et al. 2006). To determine if the different forms of DmHsp27WT and the arginine residue mutants within the ACD influence chaperone function, their ability to prevent aggregation on two distinct substrates, luciferase and insulin, was investigated. As shown in Fig. 7, heating of luciferase at 42 °C is accompanied by its aggregation which can be measured by an increase in the optical density at 320 nm. The presence of DmHsp27WT or its two isolated populations (peak no. 1 and no. 2) resulted in a similar prevention of aggregation (Fig. 7a). The R122G, R131G, and R135G mutants were as efficient as the wild-type protein to prevent luciferase heat-induced aggregation (Fig. 7b, c).
Fig. 7.
Effect of DmHsp27, its two peaks, and ACD mutants on the heat-induced aggregation of luciferase. The heat-induced aggregation of luciferase was observed by recording the optical density at 320 nm. The curve labeled luciferase correspond to aggregation (0.1 μM) of isolated protein substrates in the absence of DmHsp27, whereas the curves labeled DmHsp27WT or by peak no. 1 or no. 2 or DmHsp27R122G, DmHsp27R131G, and DmHsp27R135G correspond to aggregation in the presence of (0.4 μM) the DmHsp27WT or its peak fractions (a) or different mutants (b, c). Data are representative of three independent experiments with error bars corresponding to the standard error of the mean
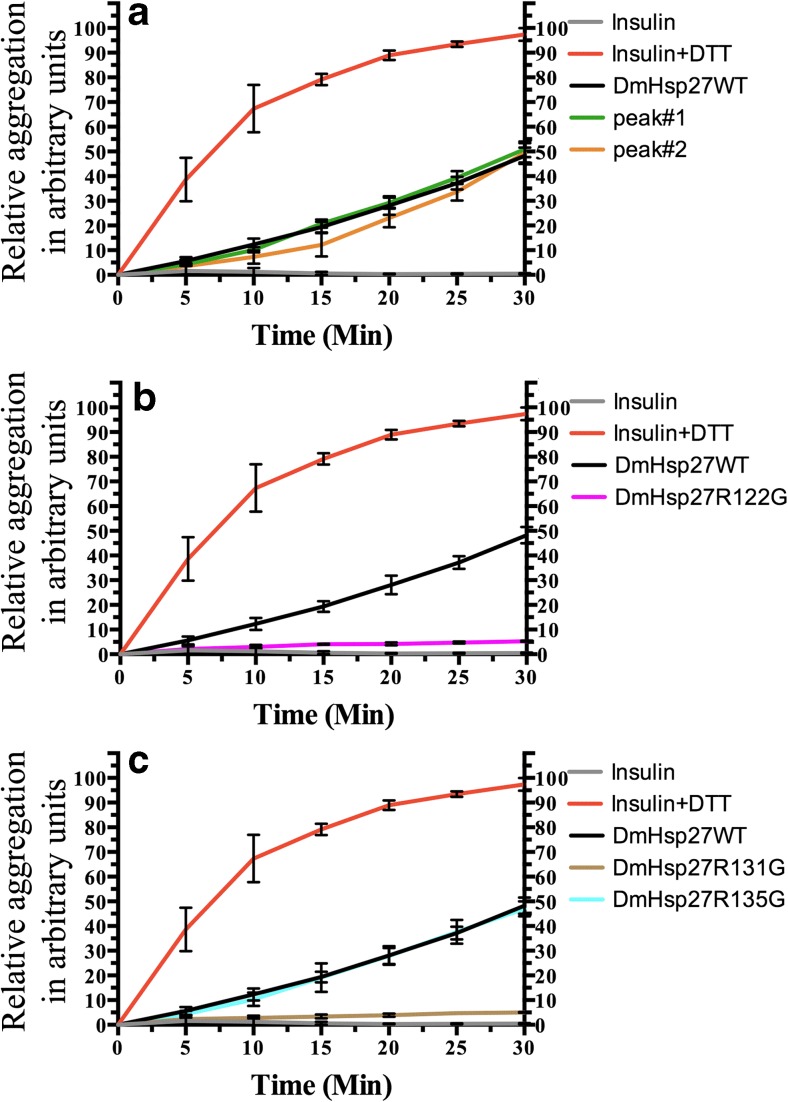
The ability of DmHSP27WT and its arginine mutants to prevent aggregation of insulin under reductive conditions was also measured (Fig. 8). Similar to the heat-induced aggregation assay, DmHsp27WT, from either of the two isolated populations (peak nos. 1 and 2), and DmHsp27R135G gave similar results. All these preparations were able to prevent the aggregation of half on the insulin present (Fig. 8a, c). Interestingly, DmHsp27R122G and DmHsp27R131G mutants were more efficient and completely prevented aggregation of insulin (Fig. 8b, c).
Fig. 8.
Effect of DmHsp27, its two peaks, and ACD mutants on reduction-induced aggregation of insulin. The reduction-induced aggregation of insulin was observed by recording the optical density at 320 nm. The reaction was started by addition of DTT up to the final concentration of 20 mM and aggregation of insulin was followed by an increase in the optical density at 320 nm (curve insulin + DTT). The curves labeled DmHsp27WT or by peak no. 1 or no. 2 or DmHsp27R122G, DmHsp27R131G, and DmHsp27R135G correspond to aggregation of 52 μM of insulin in the presence of DTT and 13 μM of the DmHsp27WT or its peak fractions (a) or different mutants (b, c). The concentration of insulin and DmHsp27 variants was equal to 0.3 mg/ml. Data are representative of three independent experiments with error bars corresponding to the standard error of the mean
Discussion
Small Hsps constitute a diverse and widespread family of molecular chaperones. They are virtually ubiquitous proteins, being present in all kingdoms of life. The D. melanogaster genome encodes 12 sHsps, which have distinctive developmental expression patterns, intracellular localizations, and substrate specificities. Here, the structure and chaperone-like function of DmHsp27 nuclear sHsp and arginine mutants within the ACD was investigated.
DmHsp27 contains a single cysteine residue at position 93. While a cysteine residue is involved in the dimerization of HspB1 and murine Hsp25 (Baranova et al. 2011; Zavialov et al. 1998), the results presented here demonstrate that DmHSP27WT does not form dimeric interface through the disulfide bond. We suggest that DmHsp27 has the metazoan dimer structure dependent on salt bridges in the β6+7 strand. This dimer structure was referred as “β7-interface dimer” in which the β6- and β7-strands are fused into an elongated strand that forms the dimer interface with its counterpart from the neighboring monomer in an antiparallel orientation (Haslbeck and Vierling 2015).
DmHsp27WT forms large soluble oligomers in physiological conditions. These oligomers tend to be larger and more polydisperse with increasing temperature. Interestingly, the change in oligomeric size observed with increasing temperature is completely reversible when the protein is cooled down. Similar results have been observed for HspB1 (Skouri-Panet et al. 2012). These results illustrate the possibility of an important reversible structural reorganization of DmHsp27 during thermal stress. The data of intrinsic tryptophan fluorescence showed that heating was accompanied by a monotonous decrease. Moreover, the amplitude of fluorescence of DmHsp27WT returns to its initial value upon cooling, supporting an important reversible structural reorganization during stress around tryptophan residue.
The usual profile of sHsps on SEC is one symmetric peak corresponding to the oligomeric form. It has been reported that HspB1 and HspB5 form large oligomers with apparent molecular weight of approximately 560 and 660 kDa, respectively (Mymrikov et al. 2012; Nefedova et al. 2013). Our data suggest that unlike mammalian sHsps, DmHsp27WT forms two populations of oligomers with apparent molecular weight of 725 and 540 kDa. While sHsps are recognized to be dynamic and in constant equilibrium (Basha et al. 2012; Stengel et al. 2010), this does not appear to be the case for the oligomers of DmHsp27. Indeed, each isolated population gave a single peak with the same elution volume when separated and reanalyzed by SEC or native gel. Moreover, the chaperone activity of these populations was comparable to that of DmHsp27WT under reductive and heat stress conditions. The question arises why DmHsp27WT has two functional populations of oligomers. At present, there is no a clear answer to this question. However, the recently published data obtained on AgsA and sHsp from the pathogenic bacterium Salmonella enterica showed that it exists in equilibrium between two oligomeric states (18-mer and 24-mer). This equilibrium seems essential for chaperone activity in vivo (Mani et al. 2016). The results presented here suggest that the equilibrium between DmHsp27 populations is not important for its in vitro chaperone activity. Further experimentations will be needed to assess the in vivo function of the two populations of DmHsp27.
In the hope of getting inactive mutants, three different arginine residues located in the ACD were mutated and characterized. DmHsp27 arginine 122 is equivalent to HspB1 arginine 127, whose mutation to tryptophan causes autosomal dominant distal hereditary motor neuropathy. Moreover, DmHsp27 arginine 131 is equivalent to HspB1 arginine 136, whose mutation to tryptophan or leucine is associated with autosomal dominant Charcot–Marie–Tooth disease of the second type (Evgrafov et al. 2004). Finally, DmHsp27 arginine 135 is equivalent to HspB1 arginine 140, HspB5 arginine 120, and HspB4 arginine 116, for which mutation to glycine and cysteine (HspB1-R140G, HspB4-R116C and HspB5-R120G) causes autosomal dominant distal hereditary motor neuropathy, Charcot–Marie–Tooth disease of the second type, dominant desmin-related myopathy, and cataracts (Bera and Abraham 2002; Gentil and Cooper 2012; Inagaki et al. 2006; Litt et al. 1998; Vicart et al. 1998).
Our data show that mutation of arginine residues within the ACD to glycine increased intrinsic fluorescence of tryptophan. This might signify that the tryptophan residue of DmHsp27 wild type is more accessible than the corresponding residue in ACD mutants, suggesting that the tryptophan environment within the (NTR) is more flexible.
Native electrophoresis and SEC analyses show that ACD mutants induce important changes in the quaternary structure of DmHsp27. All mutants form large oligomers of approximately 1100 kDa. This property of DmHsp27 arginine mutants to form large oligomers is similar to HspB1-R140G, HspB4-R116C, and HspB5-R120G (Bera et al. 2002; Bova et al. 1999; Cobb and Petrash 2000; Kumar et al. 1999; Michiel et al. 2009; Nefedova et al. 2013).
On the other hand, the mutant HspB1-R136W equally displayed an elution profile similar to wild-type protein with two major peaks, corresponding to 700 and 60 kDa (Almeida-Souza et al. 2010), although the mutant HspB1-R127W eluted predominantly in two peaks corresponding to 700 and 30 kDa (Almeida-Souza et al. 2010).
In vivo, DmHSP27WT, R122G, R131G, and R135G mutants presented the same intranuclear localization when expressed in HeLa cells and seemed associated with nuclear speckles.
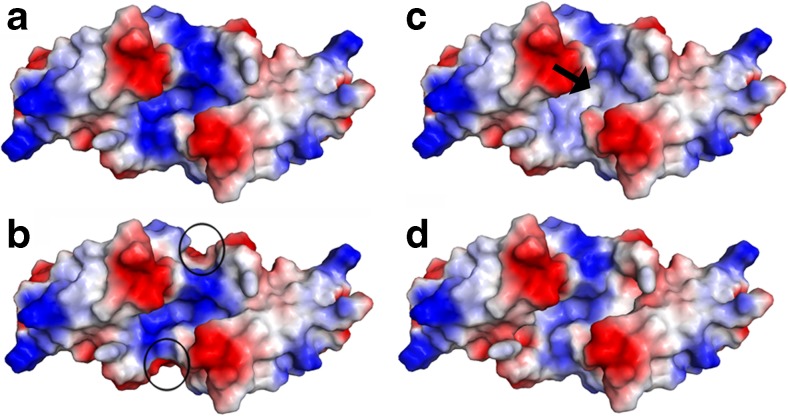
The chaperone activity of R135 was comparable to that of DmHsp27WT under reductive stress and heat stress conditions, while R122G and R131G mutants were more efficient than DmHsp27WT to prevent aggregation of insulin. Some studies have shown that HspB4-R116C, HspB5-R120G, and HspB1-R140G decrease the chaperone-like activity using a multitude of substrates (Bova et al. 1999; Kumar et al. 1999; Nefedova et al. 2013; Simon et al. 2007). Others have revealed that HspB5-R120G was comparable to HspB5WT in its ability to prevent aggregation of insulin (Treweek et al. 2005). General chaperone activity models suggest that sHsps dissociated to small oligomeric forms, presumably dimers, which re-associate to a new oligomeric form containing the bound substrate (Haslbeck and Vierling 2015). In an attempt to understand how ACD mutations in DmHsp27 induce changes in the structure and chaperone activity, we used the 3D electrostatic surface representations of DmHsp27WT, R122G, R131G, and R135G. In DmHsp27WT, the groove antiparallel dimer interface is positively charged (Fig. 9a). The mutated DmHspR122G presents a change of charge from positive to negative in the top of the central region from the face view (Fig. 9b, arrow). This change is clearer in the central part from side view (Supplementary Fig. 1), while for DmHspR131G, positively charged groove becomes slightly positive to hydrophobic (Fig. 9c). For these two mutants, the mutation of arginine (a bulky and positively charged amino acid) to glycine (a small and non-charged amino acid) uncovered negative and hydrophobic regions and could explain the increase of chaperone activity. The electrostatic potential of DmHspR135G revealed a slight change in the surface charge comparatively to DmHsp27WT (Fig. 9d). This could explain why its chaperone activity is similar to that of DmHsp27WT. A study by Clark et al. (2011) shows that mutant HspB5-R120G can rearrange to form dimer by pairing between histidine 83 and aspartic acid 80. Such rearrangement could dissociate DmHsp27R135G in the same way to form dimer by pairing between glutamine 98 and aspartic acid 95 based on sequence alignment. It is possible that ACD dimer in DmHsp27 is able to have more than one conformation, and mutation changes would be compensated. Further crystallographic structures could give us such answers. Altogether, these results characterize DmHsp27WT and its ACD arginine mutants. However, more investigations are required for the understanding of the molecular mechanisms responsible of this difference.
Fig. 9.
Electrostatic surfaces and charge distribution of DmHsp27WT and its ACD mutants. Face view comparison of the electrostatic surfaces and charge of DmHsp27WT (a), R122G (b), R131G (c), and R131G (d). Black circles indicate differences between DmHsp27WT and R122G, and black arrow indicates difference between DmHsp27WT and R131G
Electronic supplementary material
(GIF 4427 kb).
Acknowledgements
We would like to thank Jérémie Hamel from the Institute for Integrative Systems Biology (IBIS) for help with SEC, Fériel Skouri-Panet from the Institute of Mineralogy, Materials Physics and Cosmochemistry, and Ahmed Haouz from the crystallogenesis platform at the Pasteur Institute for technical help. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to RMT. MTM held a studentship from PROTEO, Québec.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-016-0748-7) contains supplementary material, which is available to authorized users.
References
- Almeida-Souza L, et al. Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charcot-Marie-Tooth neuropathy. J Biol Chem. 2010;285:12778–12786. doi: 10.1074/jbc.M109.082644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Bagneris C, Bateman OA, Naylor CE, Cronin N, Boelens WC, Keep NH, Slingsby C. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392:1242–1252. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV. Three-dimensional structure of alpha-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J Mol Biol. 2011;411:110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JF, Arrigo AP, Tanguay RM. Interaction of Drosophila 27,000 Mr heat-shock protein with the nucleus of heat-shocked and ecdysone-stimulated culture cells. J Cell Sci. 1989;92(Pt 1):29–36. doi: 10.1242/jcs.92.1.29. [DOI] [PubMed] [Google Scholar]
- Benndorf R, Martin JL, Kosakovsky Pond SL, Wertheim JO. Neuropathy- and myopathy-associated mutations in human small heat shock proteins: characteristics and evolutionary history of the mutation sites. Mutat Res Rev Mutat Res. 2014 doi: 10.1016/j.mrrev.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera S, Abraham EC. The alphaA-crystallin R116C mutant has a higher affinity for forming heteroaggregates with alphaB-crystallin. Biochemistry. 2002;41:297–305. doi: 10.1021/bi011010v. [DOI] [PubMed] [Google Scholar]
- Bera S, Thampi P, Cho WJ, Abraham EC. A positive charge preservation at position 116 of alpha A-crystallin is critical for its structural and functional integrity. Biochemistry. 2002;41:12421–12426. doi: 10.1021/bi0204140. [DOI] [PubMed] [Google Scholar]
- Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncoraglio A, Minoia M, Carra S. The family of mammalian small heat shock proteins (HSPBs): implications in protein deposit diseases and motor neuropathies. Int J Biochem Cell Biol. 2012;44:1657–1669. doi: 10.1016/j.biocel.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Bova MP, Ding LL, Horwitz J, Fung BK. Subunit exchange of alphaA-crystallin. J Biol Chem. 1997;272:29511–29517. doi: 10.1074/jbc.272.47.29511. [DOI] [PubMed] [Google Scholar]
- Bova MP, Yaron O, Huang Q, Ding L, Haley DA, Stewart PL, Horwitz J. Mutation R120G in alphaB-crystallin, which is linked to a desmin-related myopathy, results in an irregular structure and defective chaperone-like function. Proc Natl Acad Sci U S A. 1999;96:6137–6142. doi: 10.1073/pnas.96.11.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bova MP, Huang Q, Ding L, Horwitz J. Subunit exchange, conformational stability, and chaperone-like function of the small heat shock protein 16.5 from Methanococcus jannaschii. J Biol Chem. 2002;277:38468–38475. doi: 10.1074/jbc.M205594200. [DOI] [PubMed] [Google Scholar]
- Caspers GJ, Leunissen JA, de Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain”. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- Clark AR, Naylor CE, Bagneris C, Keep NH, Slingsby C. Crystal structure of R120G disease mutant of human alphaB-crystallin domain dimer shows closure of a groove. J Mol Biol. 2011;408:118–134. doi: 10.1016/j.jmb.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BA, Petrash JM. Structural and functional changes in the alpha A-crystallin R116C mutant in hereditary cataracts. Biochemistry. 2000;39:15791–15798. doi: 10.1021/bi001453j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha-crystallin—small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/S0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Datskevich PN, Nefedova VV, Sudnitsyna MV, Gusev NB. Mutations of small heat shock proteins and human congenital diseases. Biochemistry (Mosc) 2012;77:1500–1514. doi: 10.1134/S0006297912130081. [DOI] [PubMed] [Google Scholar]
- Delbecq SP, Klevit RE. One size does not fit all: the oligomeric states of alphaB crystallin. FEBS Lett. 2013;587:1073–1080. doi: 10.1016/j.febslet.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgrafov OV, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class. A J Mol Evol. 2006;62:257–266. doi: 10.1007/s00239-005-0076-5. [DOI] [PubMed] [Google Scholar]
- Gentil BJ, Cooper L. Molecular basis of axonal dysfunction and traffic impairments in CMT. Brain Res Bull. 2012;88:444–453. doi: 10.1016/j.brainresbull.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Vierling E. A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol. 2015;427:1537–1548. doi: 10.1016/j.jmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirbaut M, Beelen S, Strelkov SV, Weeks SD. Dissecting the functional role of the N-terminal domain of the human small heat shock protein HSPB6. PLoS One. 2014;9:e105892. doi: 10.1371/journal.pone.0105892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg GK, et al. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci U S A. 2014;111:E1562–E1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- Inagaki N, et al. Alpha B-crystallin mutation in dilated cardiomyopathy. Biochem Biophys Res Commun. 2006;342:379–386. doi: 10.1016/j.bbrc.2006.01.154. [DOI] [PubMed] [Google Scholar]
- Irobi J, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- Kumar LV, Ramakrishna T, Rao CM. Structural and functional consequences of the mutation of a conserved arginine residue in alphaA and alphaB crystallins. J Biol Chem. 1999;274:24137–24141. doi: 10.1074/jbc.274.34.24137. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, et al. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19:1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- Maaroufi H, Tanguay RM (2013) Analysis and phylogeny of small heat shock proteins from marine viruses and their cyanobacteria host. PLoS One 8:e81207. doi:10.1371/journal.pone.0081207 [DOI] [PMC free article] [PubMed]
- Mani N, Bhandari S, Moreno R, Hu L, Prasad BV, Suguna K. Multiple oligomeric structures of a bacterial small heat shock protein. Sci Rep. 2016;6:24019. doi: 10.1038/srep24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- Michaud S, Lavoie S, Guimond MO, Tanguay RM. The nuclear localization of Drosophila Hsp27 is dependent on a monopartite arginine-rich NLS and is uncoupled from its association to nuclear speckles. Biochim Biophys Acta. 2008;1783:1200–1210. doi: 10.1016/j.bbamcr.2008.01.031. [DOI] [PubMed] [Google Scholar]
- Michiel M, Skouri-Panet F, Duprat E, Simon S, Ferard C, Tardieu A, Finet S. Abnormal assemblies and subunit exchange of alphaB-crystallin R120 mutants could be associated with destabilization of the dimeric substructure. Biochemistry. 2009;48:442–453. doi: 10.1021/bi8014967. [DOI] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Friedrich KL, Schonfeld HJ, Vierling E, Bukau B. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem. 2003;278:31033–31042. doi: 10.1074/jbc.M303587200. [DOI] [PubMed] [Google Scholar]
- Morrow G, Tanguay RM (2015) Drosophila small heat shock proteins: an update on their features and functions. In: Tanguay RM, Hightower LE (eds) The big book on small heat shock proteins, vol 8. Heat shock proteins. Springer International Publishing, pp 579–606
- Morrow G, Heikkila JJ, Tanguay RM. Differences in the chaperone-like activities of the four main small heat shock proteins of Drosophila melanogaster. Cell Stress Chaperones. 2006;11:51–60. doi: 10.1379/CSC-166.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymrikov EV, Seit-Nebi AS, Gusev NB. Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones. 2012;17:157–169. doi: 10.1007/s12192-011-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedova VV, Datskevich PN, Sudnitsyna MV, Strelkov SV, Gusev NB. Physico-chemical properties of R140G and K141Q mutants of human small heat shock protein HspB1 associated with hereditary peripheral neuropathies. Biochimie. 2013;95:1582–1592. doi: 10.1016/j.biochi.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Rodrigues JP, Levitt M, Chopra G. KoBaMIN: a knowledge-based minimization web server for protein structure refinement. Nucleic Acids Res. 2012;40:W323–W328. doi: 10.1093/nar/gks376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Michiel M, Skouri-Panet F, Lechaire JP, Vicart P, Tardieu A. Residue R120 is essential for the quaternary structure and functional integrity of human alphaB-crystallin. Biochemistry. 2007;46:9605–9614. doi: 10.1021/bi7003125. [DOI] [PubMed] [Google Scholar]
- Skouri-Panet F, Michiel M, Ferard C, Duprat E, Finet S. Structural and functional specificity of small heat shock protein HspB1 and HspB4, two cellular partners of HspB5: role of the in vitro hetero-complex formation in chaperone activity. Biochimie. 2012;94:975–984. doi: 10.1016/j.biochi.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Stengel F, et al. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci U S A. 2010;107:2007–2012. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek TM, et al. R120G alphaB-crystallin promotes the unfolding of reduced alpha-lactalbumin and is inherently unstable. FEBS J. 2005;272:711–724. doi: 10.1111/j.1742-4658.2004.04507.x. [DOI] [PubMed] [Google Scholar]
- Van Montfort R, Slingsby C, Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv Protein Chem. 2001;59:105–156. doi: 10.1016/S0065-3233(01)59004-X. [DOI] [PubMed] [Google Scholar]
- Vicart P, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007;53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov A, Benndorf R, Ehrnsperger M, Zav’yalov V, Dudich I, Buchner J, Gaestel M. The effect of the intersubunit disulfide bond on the structural and functional properties of the small heat shock protein Hsp25. Int J Biol Macromol. 1998;22:163–173. doi: 10.1016/S0141-8130(98)00014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(GIF 4427 kb).