Abstract
The ten mammalian small heat shock proteins (sHSPs/HSPBs) show a different expression profile, although the majority of them are abundant in skeletal and cardiac muscles. HSPBs form hetero-oligomers and homo-oligomers by interacting together and complexes containing, e.g., HSPB2/HSPB3 or HSPB1/HSPB5 have been documented in mammalian cells and muscles. Moreover, HSPB8 associates with the Hsc70/Hsp70 co-chaperone BAG3, in mammalian, skeletal, and cardiac muscle cells. Interaction of HSPB8 with BAG3 regulates its stability and function. Weak association of HSPB5 and HSPB6 with BAG3 has been also reported upon overexpression in cells, supporting the idea that BAG3 might indirectly modulate the function of several HSPBs. However, it is yet unknown whether other HSPBs highly expressed in muscles such as HSPB2 and HSPB3 also bind to BAG3. Here, we report that in mammalian cells, upon overexpression, HSPB2 binds to BAG3 with an affinity weaker than HSPB8. HSPB2 competes with HSPB8 for binding to BAG3. In contrast, HSPB3 negatively regulates HSPB2 association with BAG3. In human myoblasts that express HSPB2, HSPB3, HSPB8, and BAG3, the latter interacts selectively with HSPB8. Combining these data, it supports the interpretation that HSPB8-BAG3 is the preferred interaction.
Keywords: Small heat shock proteins/HSPBs, BAG3, Interaction, Competition
Introduction
The family of mammalian small heat shock proteins (HSPs) comprises ten members that according to the new nomenclature have been named HSPB1-HSPB10. Although these 10 HSPBs share a high sequence homology, they can widely differ in term of expression profile and biochemical properties. Concerning the expression profile, while some members like, e.g. HSPB1, HSPB5, and HSPB8 are widely expressed, other members such as, e.g. HSPB4, HSPB9, and HSPB10 are selectively expressed in the eye lens (HSPB4) or male germ cells (HSPB9 and HSPB10). Moreover, HSPB2 and HSPB3 are selectively expressed in differentiated skeletal and cardiac muscles, where they form a complex (Sugiyama et al. 2000). Curiously, the muscles express the largest variety of HSPBs, namely HSPB1, HSPB2, HSPB3, HSPB5, HSPB6, HSPB7, and HSPB8 (Fontaine, Sun et al. 2005). This suggests that HSPB functions are important for the development and/or maintenance of muscles. This interpretation is supported by the findings that mutations in several HSPBs have been found associated with neuromuscular and muscular diseases, including HSPB1, HSPB3, HSPB5, and HSPB8 (Vicart, Caron et al. 1998; Evgrafov, Mersiyanova et al. 2004; Irobi, Van Impe et al. 2004; Kolb, Snyder et al. 2010; Ghaoui, Palmio et al. 2015). Moreover, double knockout in mice of HSPB2 and HSPB5 leads to severe muscle degeneration with aging, further supporting the notion that these chaperones are required for muscle health and maintenance (Brady, Garland et al. 2001).
Concerning the biochemical properties, some HSPBs form large oligomers by interacting with themselves or other members of the HSPB family. For example, HSPB1 and HSPB5 form large homo-oligomers; however, mixed HSPB1 and HSPB5 assemblies have also been reported both in mammalian cells, tissue, or in vitro (Zantema, Verlaan-De Vries et al. 1992; Liu and Welsh 1999; Sugiyama et al. 2000). By interacting together, HSPB2 and HSPB3 form hetero-oligomeric complexes with a specific subunit ratio of 3:1; in particular, 4-, 8-, 12-, 16- and 24-mer with the fixed 3:1 ratio have been documented (den Engelsman, Boros et al. 2009). HSPB2-HSPB3 complex formation has been shown not only in test tube, but also in cells and muscles (Sugiyama et al. 2000; den Engelsman, Boros et al. 2009). Besides being able to homo- and hetero-oligomerize, HSPB8 stably interacts with BAG3, a co-chaperone of Hsc70/Hsp70 (Carra, Seguin et al. 2008). In particular, in mammalian cells and muscle, HSPB8 forms a stable complex with BAG3 with a stoichiometry of 2:1 (Carra, Seguin et al. 2008; Fuchs, Poirier et al. 2010). Since the discovery of the HSPB8-BAG3 complex, binding to BAG3 has been documented in mammalian cells also for HSPB5 (Hishiya, Salman et al. 2011) and HSPB6 (Fuchs, Poirier et al. 2010); however, their binding affinity to BAG3 is much weaker compared to the one of HSPB8, and competition for binding to BAG3 between HSPB8 and these HSPBs occurs (Fuchs, Poirier et al. 2010).
Here, we investigated in mammalian cells whether other HSPBs can bind to BAG3 with affinities similar to the one of HSPB8. We focused on the HSPBs that are expressed in muscles, and we excluded from our study HSPB4, HSPB9, and HSPB10 (due to their restricted expression profile). We confirm that HSPB5 weakly binds to BAG3 in mammalian cells (Hishiya, Salman et al. 2011). Interestingly, we found that HSPB2, but not HSPB3, can weakly bind to BAG3 upon overexpression in mammalian cells. HSPB2, and not HSPB2-HSPB8 hetero-dimers, directly binds to BAG3; moreover, HSPB2 competes with HSPB8, since increasing the expression levels of HSPB8 displaces HSPB2 from its association with BAG3. Similarly, co-expression of HSPB3 with HSPB2 negatively regulates its association with BAG3. The weak and competitive nature of HSPB2 binding to BAG3 was further confirmed in human differentiated myoblasts, where HSPB8-BAG3 is the preferred interaction, and no stable association of HSPB2 with BAG3 was observed.
Methods
Cell culture and transfection
HEK293T cells (human embryonic kidney-293 cells expressing the large T-antigen of SV40/simian virus 40) were grown in Dulbecco’s modified Eagle’s medium with a high glucose concentration (Euroclone) supplemented with 10% (v/v) fetal bovine serum (Sigma-Aldrich). Cells were transfected by calcium phosphate precipitation as described previously (Carra, Seguin et al. 2008). Human immortalized myoblasts (LHCNM2 cells) were a kind gift from Prof. E. Pegoraro (Italy). Cycling LHCNM2 cells were cultured in HAM’s F12 (EuroClone) supplemented with 2-mM L-glutamine, 100-U/mL penicillin/streptomycin, 20% FBS (Gibco) and 25 ng/mL of rh FGF-b/FGF-2 (ImmunoTools). Differentiated LHCNM2 cells were cultured in DMEM supplemented with 2-mM L-glutamine, 100-U/mL penicillin/streptomycin, 2% horse serum (Gibco), and 30 μg/mL of insulin. siRNA (small interfering RNA) for HSPB8 (target sequence: AGAGCAGUUUCAACAACGA) and control sequence (siCONTROL non-targeting siRNA) were from Dharmacon. siRNAs were transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. The cDNAs used in this study are the following: pci-His-BAG3 and pci-His-dB8-BAG3 (Fuchs et al. 2010); FRT-TO-V5-HSPB1-HSPB8, FRT-TO-HSPB2 and FRT-TO-HSPB3 (Vos, Zijlstra et al. 2010). FRT-TO-myc-HSPB3 was generated by PCR using FRT-TO-HSPB3 as template. Transfections of the above-mentioned cDNAs were performed using the calcium phosphate method (Carra, Seguin et al. 2008).
Purification of His-tagged BAG3 with Ni-NTA beads
HEK293T cells were transfected with cDNA encoding for His-tagged BAG3 (His-BAG3) and either an empty vector (control) or vectors encoding for V5-tagged HSPBs (Vos, Zijlstra et al. 2010). Twenty-four-hours post-transfection, cells were scraped and homogenized in lysis buffer containing 20-mM Tris/HCl, pH 7.4, 2.5-mM MgCl2, 3% (v/v) glycerol, 0.5% NP40 (Nonidet P40), 150-mM NaCl, 10-mM imidazole, 1-mM DTT and 1× complete protease inhibitor cocktail (Roche). The cell lysates were centrifuged at 14.000g at 4 °C to pellet the NP40 insoluble proteins. His-BAG3 was purified from NP40-soluble lysates using Ni-NTA agarose beads (Qiagen). After 1 h of incubation at 4 °C, the Ni-NTA beads were washed three times with lysis buffer, followed by two other washes using a washing buffer enriched in imidazol (20-mM Tris/HCl, pH 7.4, 2.5-mM MgCl2, 3% (v/v) glycerol, 0.5% NP40, 300-mM NaCl, 20-mM imidazol, and 1× complete protease inhibitor cocktail). The proteins bound to the beads were recovered by boiling in 2% SDS sample buffer. The input and the bead fractions were then separated by SDS/PAGE (12.5% gel) and analyzed by Western blotting. BAG3 was used as a loading control.
Co-immunoprecipitation with anti-V5 and anti-myc antibodies
HEK293T cells were transfected with cDNA encoding for His-BAG3, V5-tagged HSPBs (Carra, Seguin et al. 2008; Vos, Zijlstra et al. 2010) or myc-tagged HSPB3. Twenty-four hours post-transfection, cells were scraped and homogenized in lysis buffer containing 20-mMTris/HCl, pH 7.4, 1.25-mM MgCl2, 3.3% (v/v) glycerol, 0.5% NP40, 150-mM NaCl, 1-mM DTT and 1× complete protease inhibitor cocktail). The cell lysates were centrifuged at 14.000g at 4 °C to pellet the NP40 insoluble proteins; the NP40 soluble fraction was subjected to co-immunoprecipitation. V5-tagged HSPBs or myc-tagged HSPB3 were immunoprecipitated using protein A/G sepharose beads coated with anti-V5 or anti-myc antibodies, respectively. After 1 h of incubation at 4 °C, the beads were extensively washed in lysis buffer, and the immunocomplexes were recovered by boiling in 2% SDS sample buffer. The input and the bead fractions were separated by SDS/PAGE (12.5% gel) and analyzed by Western blotting. Unless otherwise indicated, BAG3 was used as a loading control.
Preparation of samples for western blotting
HEK293T or LHCNM2 cells were lysed in Laemmli sample buffer containing 2% SDS and homogenized by sonication. Protein samples were boiled for 3 min at 100 °C, reduced with β-mercaptoetanol and separated by SDS-PAGE.
Antibodies
The antibodies used in this study are the following: mouse monoclonal anti-HSPB2 (sc-136,339, Santa Cruz Biotechnology), rabbit polyclonal anti-HSPB3 (SAB1100972, Sigma-Aldrich), rabbit polyclonal anti-Desmin (sc-14,026, Santa Cruz Biotechnology), mouse monoclonal anti-α-tubulin (T6074, Sigma-Aldrich), rabbit polyclonal anti-Myogenin (sc-576, Santa Cruz Biotechnology), mouse monoclonal anti-V5 (R960–25; Invitrogen), mouse monoclonal anti-myc (9E10; sc-40, Santa Cruz Biotechnology), and mouse monoclonal anti-myc (9E10; kindly provided by Prof. R.M. Tanguay). Rabbit polyclonal anti-HSPB8 and rabbit polyclonal anti-BAG3 were homemade antibodies kindly provided by Prof. J. Landry (Carra, Seguin et al. 2008).
Mouse and rabbit HRP-conjugated secondary antibodies for western blot were from GE Healthcare Europe GmbH.
Immunofluorescence microscopy
Cycling and differentiated LHCNM2 cells were grown on glass coverslip or plastic chamber slides, respectively. Cells were washed with cold PBS prior to fixation with 3.7% formaldehyde in PBS for 9 min at room temperature, followed by permeabilization with cold acetone for 5 min at −20 °C. Cells were blocked in PBS containing 3% BSA and 0.1% Triton X-100. This blocking solution was also used for incubation with primary and secondary antibodies, which were performed overnight at 4 °C and for 1 h at room temperature, respectively. Analysis of the cells was done by confocal imaging using a Leica SP2 AOBS system (Leica Microsystems) equipped with a ×63 oil-immersion lens.
Results
Overexpressed HSPB5 weakly binds to BAG3 in HEK293T cells
As previously mentioned, binding of HSPB5, HSPB6, and HSPB8 to BAG3 has been demonstrated under overexpression conditions in HEK293 and HEK293T cells (Carra, Seguin et al. 2008; Fuchs, Poirier et al. 2010; Hishiya, Salman et al. 2011). To compare the binding affinity to BAG3 of different HSPBs, we overexpressed in HEK293T cells HSPB1, HSPB2, HSPB3, HSPB5, HSPB6, HSPB7, and HSPB8 together with BAG3. We mainly used V5-tagged versions of these HSPBs in order to compare their expression levels. V5-tagged HSPBs have been previously generated, and their anti-aggregation and pro-degradative properties towards mutant Huntingtin exon 1 (Htt) or a fragment of Ataxin-3 (SCA3) containing an extended polyglutamine (polyQ) stretch was tested in HEK293 cells (Vos, Zijlstra et al. 2010).
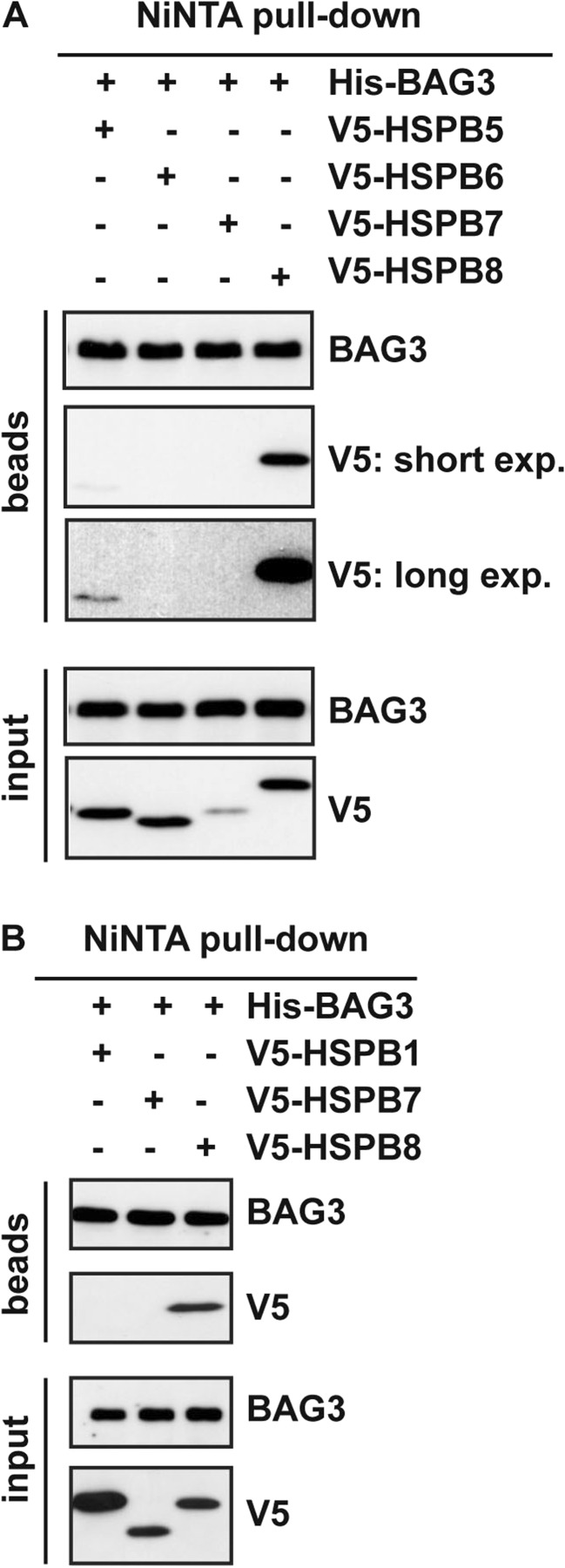
First, we co-transfected HEK293T cells with His-BAG3 and V5-tagged HSPB1, HSPB5, HSPB6, HSPB7, and HSPB8 (Fig. 1a, b). Twenty-four hours post-transfection, the cell lysates were subjected to Ni-NTA pull-down. We confirmed that V5-tagged HSPB8 binds to BAG3 (Fig. 1a, b ). Although expressed at similar levels, HSPB6 (Fig. 1a) and HSPB7 (Fig. 1b) were not pulled-down by His-BAG3. Instead, a weak binding was observed for V5-tagged HSPB5 (Fig. 1a). In contrast, although expressed at higher levels than HSPB8, V5-tagged HSPB1 did not interact with His-BAG3 under these conditions (Fig. 1b). HSPB1, HSPB5, and HSPB6 have been previously shown to weakly interact with BAG3 (Fuchs, Poirier et al. 2010; Hishiya, Salman et al. 2011). We were able under these conditions to see some association of HSPB5 to BAG3, while we had no signal for HSPB1 and HSPB6. Interestingly, in vitro studies confirmed by size-exclusion chromatography and chemical crosslinking that HSPB6 weakly interacts with BAG3; however, the complex resulting from this interaction is much less stable than the HSPB8-BAG3 complex (Shemetov and Gusev 2011). Considering the weak and unstable nature of HSPB6 binding to BAG3, our results suggest that the expression levels of HSPB6 reached in our experiment were not sufficient to yield stable and detectable interaction with BAG3. Similarly, HSPB1 binding to BAG3 was detected in cells using mass spectrometry followed by quantitative high-throughput LUMIER assays (Taipale, Tucker et al. 2014) or in vitro, using recombinant proteins (Rauch, Tse et al. 2016). Under these conditions, the affinity of HSPB1 for BAG3 was much weaker compared to the one of HSPB8 (Rauch, Tse et al. 2016). Collectively, these results support that, in HEK293T cells, HSPB8 has a greater binding affinity to BAG3 compared to HSPB5 (Fig. 1a) and, especially, HSPB6 and HSPB1.
Fig. 1.
Overexpressed HSPB5 weakly binds to BAG3. a HEK293T cells were transfected with cDNAs encoding for V5-tagged HSPB5–8 and His-tagged BAG3. Twenty-four hours post-transfection, the NP-40 soluble fractions were subjected to Ni-NTA purification. b HEK293T cells were transfected with cDNAs encoding for V5-tagged HSPB1, HSPB7, HSPB8, and His-tagged BAG3. Twenty-four hours post-transfection, the NP-40 soluble fractions were subjected to Ni-NTA purification. Beads and input fractions from A and B were processed for western blotting using anti-V5 and anti-BAG3 specific antibodies. BAG3 protein levels in the input fraction were used as internal loading control
Overexpressed HSPB2, but not HSPB3, weakly binds to BAG3
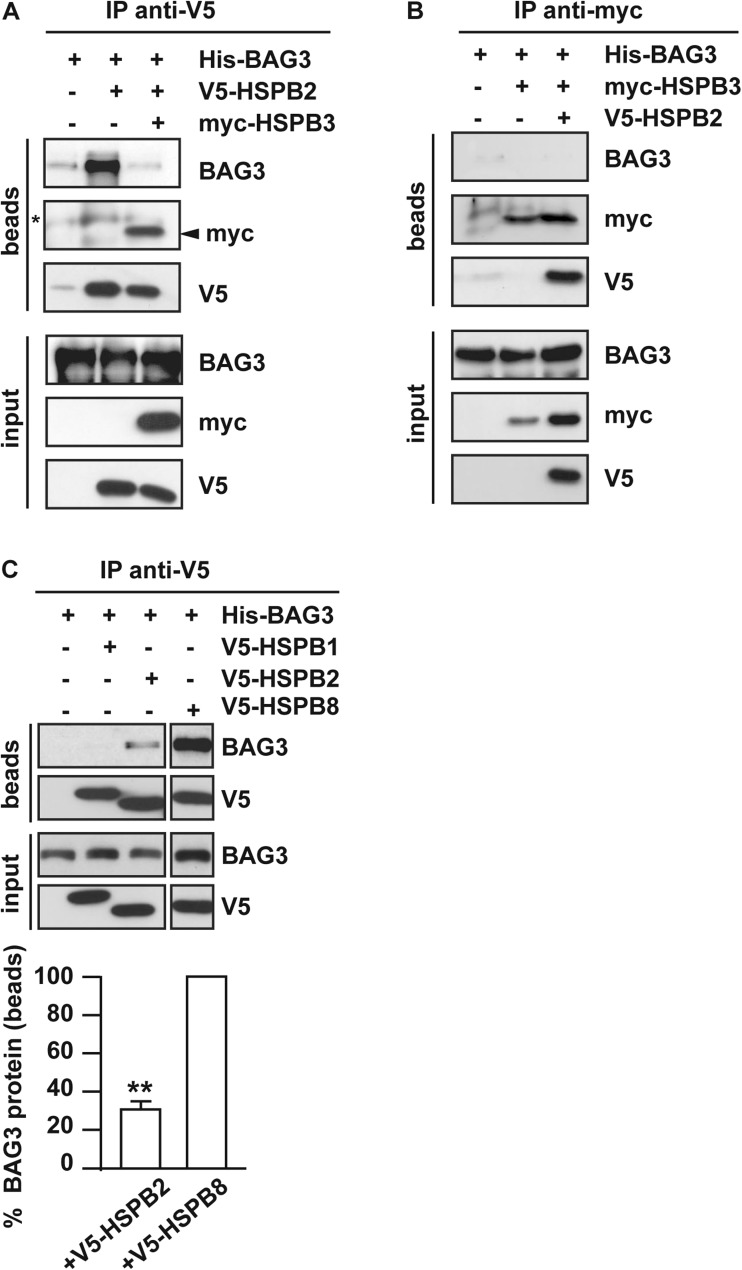
We next asked whether HSPB2 and HSPB3, which are expressed in differentiated skeletal and cardiac muscle cells (Sugiyama et al. 2000), can bind to BAG3. To test if HSPB2 binds to BAG3, we transfected HEK293T cells with His-BAG3 alone, used as control, or with V5-tagged HSPB2, and 24-h post-transfection, we performed an immunoprecipitation using a V5 specific antibody. Considering that HSPB2 and HSPB3 form a stoichiometric complex (den Engelsman, Boros et al. 2009), we also co-transfected V5-HSPB2 and myc-HSPB3 together with BAG3 to test whether HSPB2 and HSPB3 in complex might interact with BAG3. Fig. 2a shows that HSPB2 interacts with BAG3. Interestingly, when HSPB2 and HSPB3 were co-expressed with BAG3, HSPB3 co-immunoprecipitated with HSPB2, is abrogating the HSPB2-BAG3 binding (Fig. 2a). We also tested whether HSPB3 alone might associate with BAG3. We found that myc-tagged HSPB3 either expressed alone or with HSPB2 did not interact with BAG3 (Fig. 2b). Combining these experiments demonstrates that HSPB3 competes with BAG3 and abrogates its weak association with HSPB2 (Fig. 2a, b).
Fig. 2.
Overexpressed HSPB2, but not HSPB3, binds to BAG3 with an affinity lower than HSPB8. a, b HEK293T cells were transfected with cDNAs encoding for V5-tagged HSPB2, myc-tagged HSPB3 and His-tagged BAG3, alone or combined. Twenty-four hours post-transfection, cell lysates were subjected to co-immunoprecipitation using an anti-V5 antibody (a) and or anti-myc antibody (b). Beads and input fractions were processed for western blotting using anti-myc, anti-V5, and anti-BAG3 specific antibodies. BAG3 protein levels in the input fraction were used as internal loading control. A: asterisk corresponds to IgG non-specific signal; arrowhead indicates myc-HSPB3. c HEK293T cells were transfected with cDNAs encoding for V5-tagged HSPB1, HSPB2, HSPB8, and his-tagged BAG3. Twenty-four hours post-transfection, cells were processed as described in A. Quantitation of the total amount of BAG3 co-immunoprecipitated by HSPB8 and HSPB2 (**p = 0.00191; average values ± s.e.m. of n = 3 independent experiments)
We next compared the binding affinity to His-BAG3 of V5-tagged HSPB2 and HSPB8. We overexpressed in HEK293T cells His-BAG3 with V5-tagged HSPB1, HSPB2, or HSPB8. Twenty-four hours post-transfection, the cells were subjected to immunoprecipitation using the V5 antibody. The expression levels of these three HSPBs were similar. We confirmed that V5-tagged HSPB2 binds to BAG3, although with a weaker affinity compared to HSPB8 (Fig. 2c). No interaction was detected between HSPB1 and BAG3 under these conditions (Fig. 2c).
Overexpressed HSPB2 competes with HSPB8 for binding to BAG3
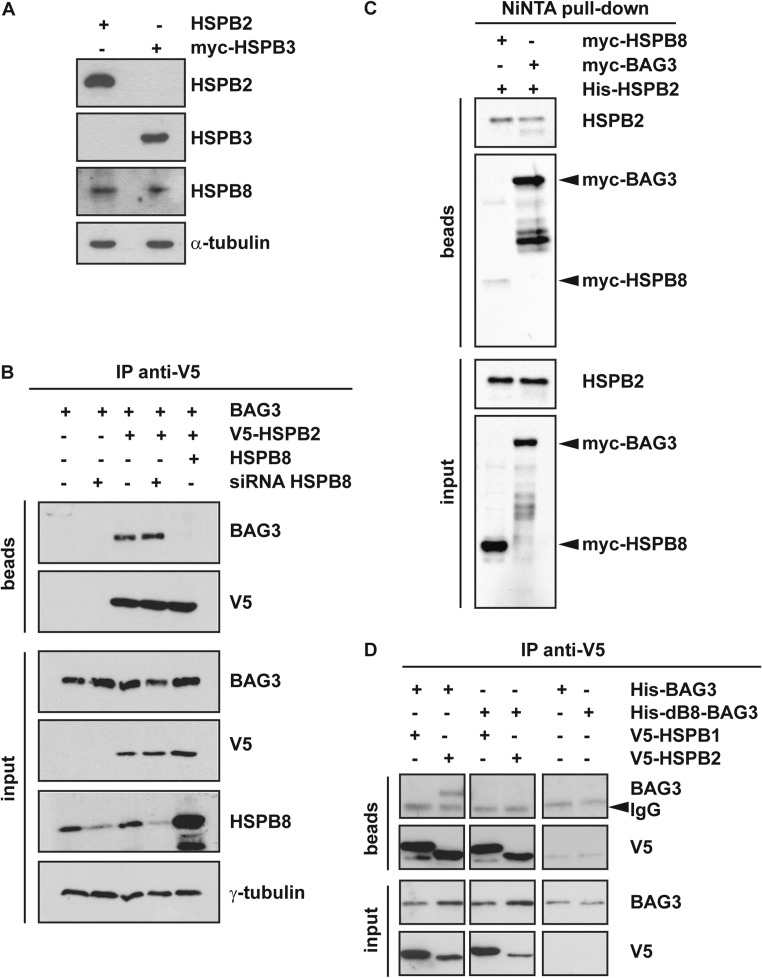
In contrast to HSPB3 (Fontaine, Sun et al. 2005), HSPB2 has been shown to interact with HSPB8 (Sun, Fontaine et al. 2004). We verified that the HSPB8 protein is endogenously expressed in HEK293T cells at detectable levels (Fig. 3a); instead, HSPB2 and HSPB3 were detected by western blot only upon transient transfection, further confirming their highly restricted expression profile (Fig. 3a). We then asked whether the association of overexpressed HSPB2 with BAG3 was direct or could be mediated by the formation of HSPB2-HSPB8 heterodimers (due to interaction of overexpressed HSPB2 with endogenous HSPB8). To address this question, we overexpressed His-BAG3 with V5-tagged HSPB2 in control HEK293T cells expressing endogenous HSPB8 or in HSPB8-depleted HEK293T cells (which were previously transfected with a specific RNAi targeting HSPB8). In parallel, we co-expressed in control HEK293T, His-BAG3 with V5-tagged HSPB2 and untagged HSPB8. Seventy-two hours after the transfection of the HSPB8 RNAi (or a nontargeting sequence used as control) and 24 h after the overexpression of His-BAG3, V5-HSPB2, and/or HSPB8, we performed an immunoprecipitation using the V5 antibody. V5-tagged HSPB2 co-immunoprecipitated equal amounts of BAG3 in both control, and HSPB8-depleted HEK293T cells, suggesting that HSPB2 directly binds to BAG3, and that the interaction is not mediated by the formation of HSPB2-HSPB8 heterodimers (Fig. 3b). Moreover, HSPB2 binding to BAG3 was abrogated when HSPB8 was overexpressed. This result opens the possibility that (1) HSPB2 and HSPB8 compete for binding to BAG3, with HSPB8 showing a stronger binding affinity to BAG3 than HSPB2 or (2) overexpressed HSPB8 could bind to HSPB2, thereby impeding on its association with BAG3, similarly to HSPB3 (Fig. 2a). To test this hypothesis, we compared the binding affinity of HSPB8 or BAG3 to HSPB2. HEK293T cells were transfected with his-HSPB2 and myc-tagged HSPB8 or BAG3, and 24-h post-transfection, cell lysates were subjected to Ni-NTA pull-down. Fig. 3c shows, using an anti-myc antibody, that myc-BAG3 binds to his-HSPB2 with a higher affinity compared to myc-HSPB8. This result strongly supports our interpretation that, overexpressed HSPB8 competes with HSPB2 for binding to BAG3.
Fig. 3.
HSPB8 strongly competes with HSPB2 for binding to BAG3. a HEK293T cells express endogenous HSPB8, but not HSPB2 nor HSPB3. HEK293T cells were transfected for 24 h with cDNAs encoding for HSPB2 or myc-tagged HSPB3. Protein extracts were subjected to western blotting using antibodies specific for HSPB2, HSPB3, and HSPB8. α-Tubulin was used as internal loading control. b HEK293T cells were left untreated or transfected with a specific siRNA against HSPB8. 48 h later, cells were transfected with cDNAs encoding for his-tagged BAG3, V5-HSPB2, and/or HSPB8. Twenty-four hours post-transfection, the cells were subjected to co-immunoprecipitation using an anti-V5 antibody. Beads and input fractions were processed for western blotting using anti-V5, anti-HSPB8, and anti-BAG3 specific antibodies. γ-Tubulin was used as internal loading control. c HEK293T cells were transfected with cDNAs encoding for his-HSPB2 and myc-HSPB8 or myc-BAG3. Twenty-four hours post-transfection, cells were subjected to Ni-NTA purification. Beads and input fractions were processed for western blotting using anti-HSPB2 and anti-myc specific antibodies. d HEK293T cells were transfected with cDNAs encoding for V5-tagged HSPB1, HSPB2, and His-tagged BAG3, or the deletion mutant dB8-BAG3 (unable to bind to HSPB8). Cells expressing his-tagged BAG3 or dB8-BAG3 alone were used as negative controls (right panel). Twenty-four hours post-transfection, the NP-40 soluble fractions were subjected to co-immunoprecipitation using an anti-V5 antibody. Beads and input fractions were processed for western blotting using anti-V5 and anti-BAG3 specific antibodies. BAG3 protein levels were used as internal loading control. Arrowheads indicates IgG non-specific signal
Based on these results, one would expect that HSPB2 and HSPB8 bind to the same binding region to BAG3. We previously identified two stretches of amino acids containing two conserved IPV (Ile-Pro-Val) motifs in BAG3 that mediate its binding to HSPB8 (Fuchs, Poirier et al. 2010). Deletion of these specific sequences (amino-acid residues 87–101 and 200–213; dB8-BAG3) abrogates the association between HSPB8 and BAG3 (Fuchs, Poirier et al. 2010). To test whether HSPB2 binds to the same residues as HSPB8, we co-transfected V5-tagged HSPB2 with either His-BAG3 or its deletion mutant unable to bind to HSPB8 (His-dB8-BAG3), and 24-h post-transfection, we performed an immunoprecipitation using the V5 antibody. We also co-transfected V5-tagged HSPB1 with His-BAG3, which, according to our findings, should not stably and detectably interact in HEK293T cells. HSPB2, but not HSPB1, weakly co-immunoprecipitated BAG3. Deletion of the amino-acid residues 87–101 and 200–213 of BAG3 abrogated its association with HSPB2 (Fig. 3d). Combining these results demonstrates that HSPB2 can interact with BAG3 with a weaker affinity than HSPB8 and that HSPB2 and HSPB8 compete for binding to the same region.
HSPB8-BAG3 is the preferred interaction also in differentiated human myoblasts that express HSPB2 and HSPB3
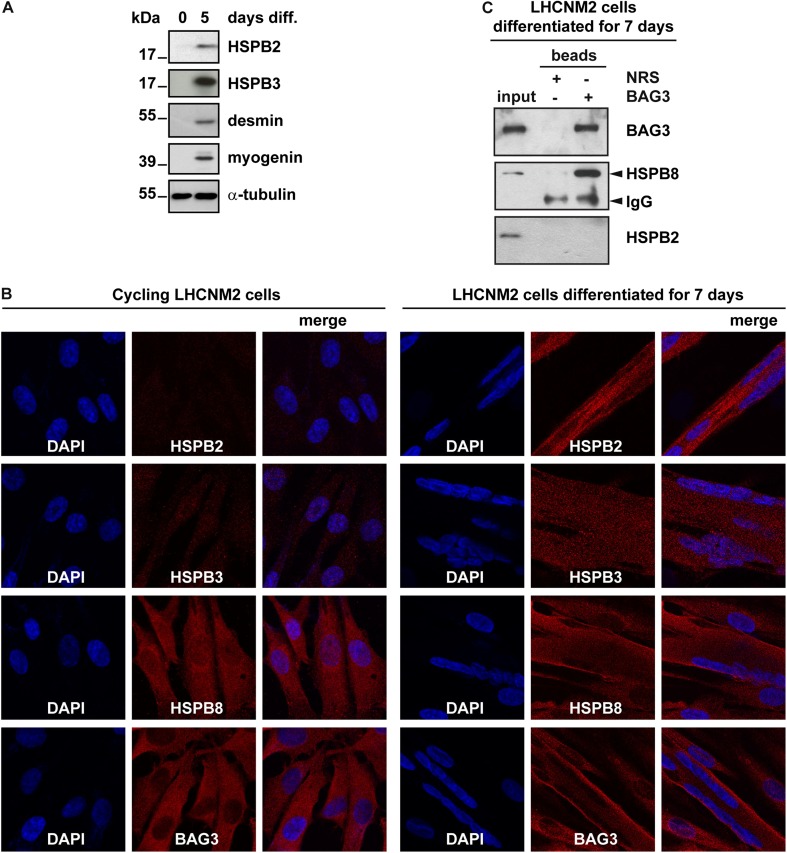
Besides skeletal and cardiac muscle cells, HSPB2 is also expressed in glioma and breast cancer cells, where it exerts anti-apoptotic functions (Oshita, Chen et al. 2010). Instead, HSPB3 is expressed in muscles and in several fetal tissues, where it has been suggested to participate to embryonic stem cell differentiation, although experimental evidence supporting such a function is still lacking (Lam, Wing Tsui et al. 1996; Pennings, van Dartel et al. 2011). First, here we verified whether human immortalized myoblasts that are grown under cycling conditions express endogenous HSPB2 and HSPB3 proteins. In line with previous reports using mouse immortalized cycling muscle cells (C2C12 cells) (Sugiyama, Suzuki et al. 2000), no HSPB2 and HSPB3 were detected in cycling LHCNM2 cells by western blotting (Fig. 4a). Instead, both HSPB2 and HSPB3 proteins were induced in LHCNM2 cells differentiated for 5 days (Fig. 4a; similarly to what previously found in C2C12 cells or muscle tissue extracts) (Sugiyama et al. 2000). Desmin and myogenin, which are induced during muscle differentiation, were used as positive controls to monitor LHCNM2 cell differentiation (Fig. 4a). We next performed immunofluorescence studies, in human cycling and differentiated myoblasts to analyze the expression and distribution of endogenous HSPB2, HSPB3, HSPB8, and BAG3. We confirmed that HSPB2 and HSPB3 are absent in cycling LHCNM2 cells, while, upon differentiation, they are distributed throughout the cells (Fig. 4b, compare cycling and differentiated LHCNM2 cells). In contrast, HSPB8 and BAG3 are expressed in both cycling and differentiated LHCNM2 cells, where they are mainly distributed in the cytosol (Fig. 4b).
Fig. 4.
HSPB8-BAG3 is the preferred complex also in differentiated human myoblasts. a Human myoblasts (LHCNM2 cells) were grown under cycling conditions (0 days diff.) or let to differentiate for 5 days (5 days diff.), prior to extraction of total proteins. Expression levels of endogenous HSPB2, HSPB3, desmin, and myogenin (used as differentiation markers) were evaluated by western blotting. α-Tubulin was used as internal loading control. b Fluorescent microscopy on cycling and differentiated LHCNM2 cells showing expression and subcellular distribution of endogenous HSPB2, HSPB3, HSPB8, and BAG3, using specific antibodies. DAPI was used to stain nucleic acid. Note that HSPB2 and HSPB3 are detected only in differentiated LHCNM2 cells, in line with the results shown in panel A. c LHCNM2 cells were differentiated for 7 days and were then subjected to co-immunoprecipitation using anti-BAG3 specific antibody. Beads coated with normal rabbit serum (NRS) were used as negative control. Beads and input fractions were processed for western blotting using anti-BAG3, anti-HSPB2, or anti-HSPB8 antibodies. Arrowheads indicates IgG non-specific signal
Next, we asked whether endogenous HSPB2 can interact with endogenous BAG3 in differentiated LHCNM2 cells. LHCNM2 cells were grown in differentiating medium for 7 days, and the cell extracts were subjected to immunoprecipitation using the BAG3 antibody. We found that BAG3 co-immunoprecipitated HSPB8, but not HSPB2 in differentiated human myoblasts (Fig. 4c). This result is in line with our data obtained under overexpression conditions showing that HSPB8 has a higher binding affinity for BAG3 than HSPB2. Based on these findings, we conclude that also in human differentiated myoblasts HSPB8-BAG3 is the preferred interaction.
Discussion
In the present study, we compared the binding affinity to BAG3 of several HSPB members upon overexpression in HEK293T cells. Since BAG3 is highly expressed in cardiac and skeletal muscle cells and its mutations leads to cardiomyopathy and muscular dystrophy (Selcen, Muntoni et al. 2009; Arimura, Ishikawa et al. 2011; Norton, Li et al. 2011; Villard, Perret et al. 2011; Chami, Tadros et al. 2014; Franaszczyk, Bilinska et al. 2014; Toro, Perez-Serra et al. 2016), we selected for our study the HSPB proteins that are expressed in skeletal and/or cardiac muscle cells, namely HSPB1, HSPB2, HSPB3, HSPB6, HSPB7, and HSPB8 (Fontaine, Sun et al. 2005). We mainly used V5-tagged forms of these HSPBs in order to directly compare their expression levels and their binding affinities to BAG3, using an antibody specific for the V5 tag. In agreement with published data (Carra, Seguin et al. 2008; Hishiya, Salman et al. 2011), we confirmed that HSPB8 binds to BAG3, while HSPB5 only weakly interacted with it. We found that upon overexpression in HEK293T cells, also HSPB2, but not HSPB3, showed weak interaction with BAG3, compared to HSPB8. Interestingly, such interaction was regulated by both HSPB8 and HSPB3. In particular, HSPB8 directly competed with HSPB2 for binding to the same IPV domains on BAG3, showing a higher affinity and displacing HSPB2 from such interaction. Instead, HSPB3 interacted with HSPB2, displacing HSPB2 from its weak association with BAG3.
Under our experimental conditions, we were unable to detect interaction between HSPB1, HSPB3, HSPB6, or HSPB7, and BAG3; however, binding of HSPB1 to BAG3 was documented, in human cells, by mass spectrometry and quantitative high-throughput LUMIER assays (Taipale, Tucker et al. 2014). Moreover, weak binding of HSPB6 to BAG3 was previously described both in cells and in vitro, using recombinant proteins (Fuchs, Poirier et al. 2010; Shemetov and Gusev 2011). This apparent discrepancy suggests that HSPB1 and HSPB6 would bind to BAG3 with an affinity that is weaker than the ones of HSPB5 and HSPB2, at least in HEK293T cells using V5-tagged proteins. In particular, binding of HSPB1 or HSPB6 to BAG3 could be below detection level using conventional western blotting, rather than more sensitive techniques like mass spectrometry and quantitative high-throughput LUMIER assays (Taipale, Tucker et al. 2014). Combined with the published data, our results in HEK293T cells suggest the following hierarchy of affinities for BAG3: HSPB8 > > HSPB2, HSPB5 and HSPB6, which all show weaker association (data presented here and previous reports; (Fuchs, Poirier et al. 2010; Shemetov and Gusev 2011)). This interpretation is in line with recent findings from Rauch et al., who, using recombinant proteins, also found that HSPB8 binds to BAG3 with an affinity that is much higher compared to the ones of HSPB6 and, especially, HSPB1 (Rauch, Tse et al. 2016). However, our results also support that, based only on co-immunoprecipitation assays performed in mammalian cells under overexpression conditions, it is not possible to conclude whether one specific HSPB can or cannot bind to BAG3, and caution must be taken when interpreting these data. To firmly establish if a given HSPB displays some binding capacity to BAG3, studies in vitro with pure recombinant proteins are required (similarly to what has been done with HSPB6 and, recently with HSPB1; (Shemetov and Gusev 2011; Rauch, Tse et al. 2016)). Thus, although by co-immunoprecipitation in HEK293T cells, we had no evidence of binding of HSPB3 and HSPB7, one cannot conclude that such interaction is not occurring; in other words, these data do not exclude the possibility that using pure recombinant proteins also HSPB3 and HSPB7 could weakly interact with BAG3. However, although we cannot exclude that some transient binding of several HSPBs to BAG3 might occur, our data strongly demonstrate that HSPB8 has a high binding affinity for BAG3, and that HSPB8-BAG3 is the preferred interaction, both in cells and in vitro (Shemetov and Gusev 2011; Rauch, Tse et al. 2016). This conclusion is further supported by our experimental findings in human myoblasts (LHCNM2 cells). In fact, we found that in differentiated LHCNM2 cells that express HSPB2, HSPB3, HSPB8, and BAG3, only HSPB8, and not HSPB2, was co-immunoprecipitated by BAG3. Similarly, when investigating endogenous proteins, the HSPB8-BAG3, but not the HSPB6-BAG3 complex, was documented in muscle extracts (Fuchs, Poirier et al. 2010). Taken together, these data demonstrate that independent on the system used, being it an in vitro system with pure recombinant proteins, non-muscular cells (Carra, Seguin et al. 2008) and muscular cells (Fig. 4 and (Fuchs, Poirier et al. 2010)) expressing endogenous or transiently transfected proteins, the HSPB8-BAG3 complex is the preferred one, and the other HSPBs need to compete with endogenous HSPB8 for weak and transient binding to BAG3.
Finally, our results open the possibility that under specific conditions where HSPB8 levels would be decreased or in presence of HSPB8 mutants that have decreased binding affinity for BAG3, such as for K141E and K141N-HSPB8 associated with motor neuropathy (Carra et al. 2010; Shemetov and Gusev 2011), other HSPBs might compete with HSPB8 for interaction with BAG3, and both HSPB8-BAG3 and, e.g., HSPB2-BAG3, HSPB5-BAG3, or HSPB6-BAG3 complexes might transiently co-exist, with potential consequences on their functions.
Acknowledgements
We thank Prof. E. Pegoraro and Dr. E. Galletta for providing the LHCNM2 cells and for useful technical support for their use. SC is grateful to Prof. Harm H. Kampinga for useful discussions and tools. SC is grateful to Telethon (GEP12008 and GGP15001) and Association Francaise contres les Myopathies (grant number 15999) for financial support. SC also thanks the Centro Interdipartimentale Grandi Strumenti (CIGS) of the University of Modena and Reggio Emilia for support with confocal microscopy.
Footnotes
Laura Mediani and Federica F. Morelli contributed equally to this work.
References
- Arimura T, Ishikawa T, et al. Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat. 2011;32(12):1481–1491. doi: 10.1002/humu.21603. [DOI] [PubMed] [Google Scholar]
- Brady JP, Garland DL, et al. AlphaB-crystallin in lens development and muscle integrity: a gene knockout approach. Invest Ophthalmol Vis Sci. 2001;42(12):2924–2934. [PubMed] [Google Scholar]
- Carra S, Boncoraglio A, et al. Identification of the drosophila ortholog of HSPB8: implication of HSPB8 loss of function in protein folding diseases. J Biol Chem. 2010;285(48):37811–37822. doi: 10.1074/jbc.M110.127498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, et al. HspB8 chaperone activity toward poly (Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283(3):1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- Chami N, Tadros R, et al. Nonsense mutations in BAG3 are associated with early-onset dilated cardiomyopathy in French Canadians. Can J Cardiol. 2014;30(12):1655–1661. doi: 10.1016/j.cjca.2014.09.030. [DOI] [PubMed] [Google Scholar]
- den Engelsman J, Boros S, et al. The small heat-shock proteins HSPB2 and HSPB3 form well-defined heterooligomers in a unique 3 to 1 subunit ratio. J Mol Biol. 2009;393(5):1022–1032. doi: 10.1016/j.jmb.2009.08.052. [DOI] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36(6):602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Fontaine JM, Sun X, et al. Interactions of HSP22 (HSPB8) with HSP20, alphaB-crystallin, and HSPB3. Biochem Biophys Res Commun. 2005;337(3):1006–1011. doi: 10.1016/j.bbrc.2005.09.148. [DOI] [PubMed] [Google Scholar]
- Franaszczyk M, Bilinska ZT, et al. The BAG3 gene variants in polish patients with dilated cardiomyopathy: four novel mutations and a genotype-phenotype correlation. J Transl Med. 2014;12:192. doi: 10.1186/1479-5876-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Poirier DJ, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2010;425(1):245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- Ghaoui R, Palmio J, et al. Mutations in HSPB8 causing a new phenotype of distal myopathy and motor neuropathy. Neurology. 2015;86(4):391–398. doi: 10.1212/WNL.0000000000002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiya A, Salman MN, et al. BAG3 directly interacts with mutated alphaB-crystallin to suppress its aggregation and toxicity. PLoS One. 2011;6(3):e16828. doi: 10.1371/journal.pone.0016828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irobi J, Van Impe K, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet. 2004;36(6):597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- Kolb SJ, Snyder PJ, et al. Mutant small heat shock protein B3 causes motor neuropathy: utility of a candidate gene approach. Neurology. 2010;74(6):502–506. doi: 10.1212/WNL.0b013e3181cef84a. [DOI] [PubMed] [Google Scholar]
- Lam WY, Wing Tsui SK, et al. Isolation and characterization of a human heart cDNA encoding a new member of the small heat shock protein family—HSPL27. Biochim Biophys Acta. 1996;1314(1–2):120–124. doi: 10.1016/S0167-4889(96)00121-8. [DOI] [PubMed] [Google Scholar]
- Liu C, Welsh MJ. Identification of a site of Hsp27 binding with Hsp27 and alpha B-crystallin as indicated by the yeast two-hybrid system. Biochem Biophys Res Commun. 1999;255(2):256–261. doi: 10.1006/bbrc.1999.0174. [DOI] [PubMed] [Google Scholar]
- Norton N, Li D, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshita SE, Chen F, et al. The small heat shock protein HspB2 is a novel anti-apoptotic protein that inhibits apical caspase activation in the extrinsic apoptotic pathway. Breast Cancer Res Treat. 2010;124(2):307–315. doi: 10.1007/s10549-010-0735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings JL, van Dartel DA, et al. Identification by gene coregulation mapping of novel genes involved in embryonic stem cell differentiation. Stem Cells Dev. 2011;20(1):115–126. doi: 10.1089/scd.2010.0181. [DOI] [PubMed] [Google Scholar]
- Rauch, J. N., E. Tse, et al. (2016). BAG3 Is a Modular, Scaffolding Protein that physically Links Heat Shock Protein 70 (Hsp70) to the Small Heat Shock Proteins. J Mol Biol. [DOI] [PMC free article] [PubMed]
- Selcen D, Muntoni F, et al. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009;65(1):83–89. doi: 10.1002/ana.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemetov AA, Gusev NB. Biochemical characterization of small heat shock protein HspB8 (Hsp22)-Bag3 interaction. Arch Biochem Biophys. 2011;513(1):1–9. doi: 10.1016/j.abb.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Suzuki A, et al. Muscle develops a specific form of small heat shock protein complex composed of MKBP/HSPB2 and HSPB3 during myogenic differentiation. J Biol Chem. 2000;275(2):1095–1104. doi: 10.1074/jbc.275.2.1095. [DOI] [PubMed] [Google Scholar]
- Sun X, Fontaine JM, et al. Interaction of human HSP22 (HSPB8) with other small heat shock proteins. J Biol Chem. 2004;279(4):2394–2402. doi: 10.1074/jbc.M311324200. [DOI] [PubMed] [Google Scholar]
- Taipale M, Tucker G, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158(2):434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Perez-Serra A, et al. Familial dilated cardiomyopathy caused by a novel Frameshift in the BAG3 Gene. PLoS One. 2016;11(7):e0158730. doi: 10.1371/journal.pone.0158730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicart P, Caron A, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20(1):92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Villard E, Perret C, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MJ, Zijlstra MP, et al. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum Mol Genet. 2010;19(23):4677–4693. doi: 10.1093/hmg/ddq398. [DOI] [PubMed] [Google Scholar]
- Zantema A, Verlaan-De Vries M, et al. Heat shock protein 27 and alpha B-crystallin can form a complex, which dissociates by heat shock. J Biol Chem. 1992;267(18):12936–12941. [PubMed] [Google Scholar]