Abstract
The holdase activity and oligomeric propensity of human small heat shock proteins (sHSPs) are regulated by environmental factors. However, atomic-level details are lacking for the mechanisms by which stressors alter sHSP responses. We previously demonstrated that regulation of HSPB5 is mediated by a single conserved histidine over a physiologically relevant pH range of 6.5–7.5. Here, we demonstrate that HSPB1 responds to pH via a similar mechanism through pH-dependent structural changes that are induced via protonation of the structurally analogous histidine. Results presented here show that acquisition of a positive charge, either by protonation of His124 or its substitution by lysine, reduces the stability of the dimer interface of the α-crystallin domain, increases oligomeric size, and modestly increases chaperone activity. Our results suggest a conserved mechanism of pH-dependent structural regulation among the human sHSPs that possess the conserved histidine, although the functional consequences of the structural modulations vary for different sHSPs.
Keywords: Heat shock protein, Acidosis, Nuclear magnetic resonance (NMR), α-crystallin domain (ACD), Oligomeric structure, Structural regulation, HSPB1, Hsp27
Introduction
Several human small heat shock proteins (sHSPs) form polydisperse oligomers, ranging from dimers to ∼50mers that interchange subunits dynamically. Their oligomeric propensity and distribution is strongly regulated by environmental factors, but atomic-level mechanistic details remain enigmatic due to their size and plasticity. HSPB1 (Hsp27) is one of the ubiquitously expressed human sHSPs and is implicated in numerous diseases (Datskevich et al. 2012). HSPB1 expression is upregulated in response to certain stressors but little is known about how oligomeric HSPB1 structure and dynamics are affected by changing conditions. Recent structures of the truncated α-crystallin domain (ACD) of HSPB1 (Baranova et al. 2011; Hochberg et al. 2014; Rajagopal et al. 2015a) provide a platform on which to probe effects of environmental changes and disease-associated mutations at residue-level resolution in the ACD.
As the defining domain of sHSPs, the ACD sequences of human sHSPs (HSPB1–9) are modestly conserved. NMR and crystal structures of truncated and full-length mammalian sHSPs, including HSPB1, 4, 5, and 6, (Bagneris et al. 2009; Baranova et al. 2011; Hochberg et al. 2014; Jehle et al. 2010, 2011; Laganowsky et al. 2010; Rajagopal et al. 2015a, b; Weeks et al. 2013) all have a conserved β-sandwich IgG-like fold. In almost all cases, two ACDs form a homodimer via antiparallel alignment of their β6+7 strands. The observed structural conservation suggests that sHSPs may share behavior that arises from conserved residues. We have reported pH-dependent properties of HSPB5 that include destabilization of its ACD dimer and, paradoxically, expansion of the number of subunits in its oligomeric species as a function of decreasing pH over a range of pH 7.5–6.5 (Rajagopal et al. 2015b). A decrease in pH within this range is consistent with pH measurements in ischemic tissue in mouse brains (McVicar et al. 2014) and may destabilize some cellular proteins, particularly those whose pI values fall within this pH range. Additionally, HSPB1 was found to be phosphorylated in response to low pH incubation of cultured AGS gastric cancer cells (Singh et al. 2011). The source of the pH-dependent behavior was found to be a histidine residue within the HSPB5 ACD, His104, that is conserved in all human sHSPs except HSPB7 and HSPB9. Mutation of His104 to lysine (H104K) to mimic a perpetually protonated state yields an HSPB5 that closely simulates both the dramatic structural and functional effects observed in the wild-type (WT) protein at low pH (i.e., pH 6.5). In the presence of a destabilized model client protein, α-lactalbumin, large H104K-HSPB5 oligomers reorganize into smaller oligomers, form long-lived co-complexes with the client, and exhibit enhanced chaperone activity by delaying the appearance of light-scattering aggregates for longer than WT-HSPB5. The high conservation of histidine at the structurally analogous position in six other human sHSPs suggests that similar pH-dependent properties may also play a role in the function and regulation of these sHSPs. Here, we investigate the analogous histidine in HSPB1, His124, and find that it modulates the protein’s structure in a similar manner to His104 in HSPB5 but has a much more modest effect on function.
Results
The dimer interface of HSPB1-ACD is destabilized by decreased pH
HSPB1-ACD (residues 80–176) forms a homodimer and is predominantly dimeric (>90%) at 100 μM protein under reducing conditions at pH 7.5 (Rajagopal et al. 2015a). The antiparallel β-strand alignment that occurs at the dimer interface places the sole Cys residue (Cys137) in HSPB1 directly across from the corresponding Cys137 residue in the other subunit and a disulfide bond readily forms in the homodimer. Reduction of the disulfide bond is accompanied by minor perturbations and broadening of some peaks in the ACD NMR spectra that arise primarily from residues along the dimer interface, attributed to an exchange process that is inhibited in the oxidized (disulfide-bonded) dimer (Rajagopal et al. 2015a). Under reducing conditions, a decrease in pH from 7.5–6.5 is accompanied by additional broadening of peaks that arise from residues along the dimer interface (β6+7 strand), in the neighboring β5 strand, and in the loop connecting these strands (Fig. 1a, e, and f). As in the case of HSPB5, the disappearance of these peaks is most likely due to intermediate exchange (on the NMR timescale) between at least one monomeric and one dimeric state (multiple registers are possible).
Fig. 1.
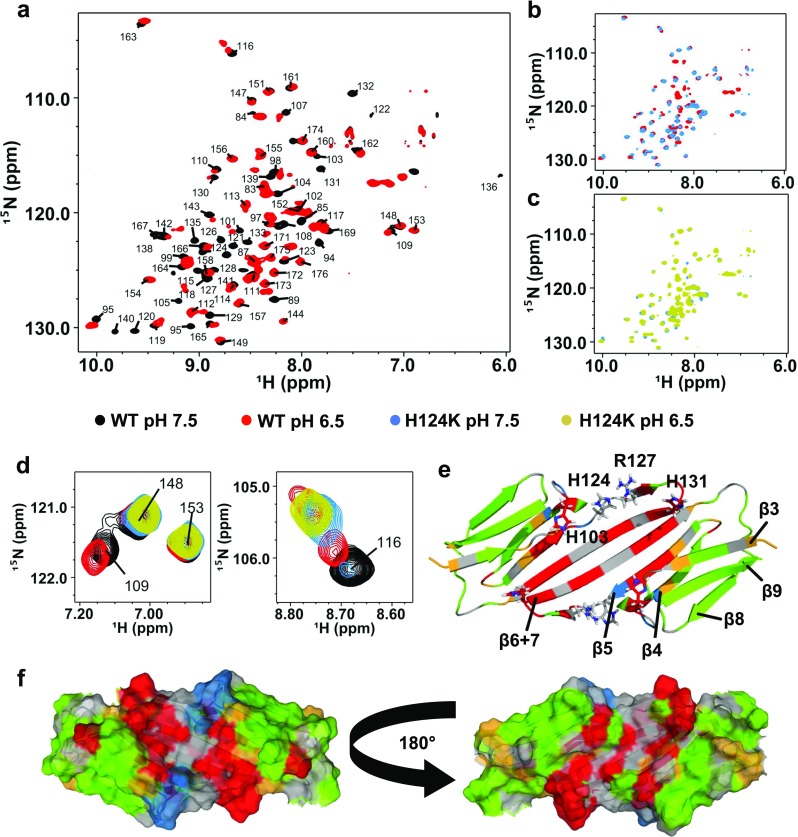
Low pH or the H124K mutation destabilizes the dimer interface of HSPB1-ACD. a–c (1H-15N)-HSQC-TROSY NMR spectra of HSPB1-ACD WT or H124K at pH 7.5 or 6.5. Spectra were collected on 100-μM samples at 25 °C. a WT protein at pH 7.5 (black) and pH 6.5 (red). Peaks corresponding to residues along the dimer interface disappear (broaden) at pH 6.5, indicating exchange between monomeric and dimeric states or multiple states. b WT at pH 6.5 (red) and H124K at pH 7.5 (blue). Coincidence of peaks in the WT and mutant spectra at different pH indicates that the H124K mutant mimics the low pH state of the WT. c H124K protein at pH 7.5 (blue) and pH 6.5 (yellow). Only minimal spectral changes are observed as a function of pH. d Examples of peaks undergoing different spectral behaviors as a function of pH, maintaining the same color scheme as a–c. Peaks arising from residues 148 and 153 are unperturbed; the residue 109 peak is not observed in either H124K spectra, as it is close spatially to the site of mutation and has likely changed chemical environment substantially; the peak from residue 116 shows doubling and substantial chemical shift perturbations in the WT spectrum at pH 6.5 and in the mutant. e Major perturbations in the NMR spectra are mapped onto a cartoon representation of the HSPB1-ACD dimer solution NMR structure (2N3J). Two ACD monomers form a dimer via interaction of their β6+7 strands, arranged in an antiparallel fashion. Three conserved histidines are located in different loops in the ACD of HSPB1. His124 is within hydrogen-bonding distance of Arg127 of the same protomer, both within the loop connecting the β5 and β6+7 strands. Residues mapped in red have peaks that disappear when the pH is lowered or the H124K mutation is introduced at pH 7.5. Residues in yellow shift more than 0.2 ppm and/or double when the pH is lowered or the mutation is introduced. Residues mapped in blue are perturbed specifically in the spectrum of H124K-ACD and are all in the immediate vicinity of the site of mutation. Unperturbed peaks are shown in green and residues without assigned peaks are in gray. f Surface rendering of the HSPB1-ACD structure, maintaining the same color scheme as e
In addition to the peaks that broaden, some peaks exhibit chemical shift perturbations (CSPs) without substantial broadening at pH 6.5 (Fig. 1e and f, yellow). Moderate CSPs are observed in strands β3, 4, and 5, flanking residues whose peaks have disappeared. These CSPs indicate a change in the local chemical environment, most likely due to the protonation of His124 and/or other histidines (see next section for discussion of minor CSPs and other histidines). A few of the shifting peaks also exhibit peak doubling at pH 6.5 (Fig. 1d). Peak doubling is indicative of the presence of two states that interchange slowly. Peak doubling was also observed as a function of pH in the HSPB5-ACD where it was attributed to exchange between the dimeric and monomeric species (Rajagopal et al. 2015b). HSPB1-ACD peaks that clearly undergo doubling arise from residues within the β-strands 4 and 5 and the loop connecting them. They are surface-exposed and away from the dimer interface, suggesting small adjustments within the protomer upon dissociation. Nevertheless, the general lack of perturbations observed for resonances arising from the β3, β4, β8, and β9 strands indicates that the overall structure of the protomer is retained at the lower pH.
H124K mutation mimics pH-dependent changes in HSPB1-ACD
The pH-dependent spectral changes at and near the HSPB1 dimer interface confirm that HSPB1 shares properties with HSPB5. To ask if the behavior observed is due to the analogous histidine, we made the analogous mutation in the HSPB1-ACD, namely H124K. As shown in Fig. 1b, the spectrum of H124K-ACD at pH 7.5 is remarkably similar to the spectrum of WT-ACD at pH 6.5. In particular, peaks that disappear or are shifted in the spectrum of WT-ACD at pH 6.5 are already perturbed at pH 7.5 in the spectrum of the mutant. Mapping the residues that are perturbed in the mutant onto the dimer structure shows that they arise from residues in the dimer interface and neighboring strand (Fig. 1e and f, red and yellow). The same residues that undergo moderate CSPs and peak doubling at pH 6.5 for the WT protein show the same behavior in the H124K mutant at pH 7.5. Thus, the substitution of lysine for His124 appears to recapitulate the effects of protonating His124 by lowering the pH.
A few differences are observed among the mutant H124K spectra and the pH 6.5 WT spectrum. A few peaks disappear or have shifted beyond easy recognition in the mutant spectrum compared to that of the WT at pH 6.5, which all correspond to residues in the immediate vicinity of the mutation. Mutation of histidine to lysine presents a different chemical environment to residues that are spatially adjacent (Fig. 1e and f, blue). Importantly, only very minor CSPs are observed in the spectrum of the mutant at pH 7.5 versus pH 6.5 (Fig. 1c) and these are consistent with the very small CSPs of the same residues between pH states of the WT. These slight shifts are attributable to partial protonation of other histidines (His103 or His131) over the pH range of 7.5–6.5. The pK R values of the analogous histidines in HSPB5 are 6.6 and <6, respectively, suggesting that His103 will be partly protonated at pH 6.5 (Rajagopal et al. 2015b). Similar to the case for HSPB5, substitution of His124 with a constitutive positive charge (lysine) closely mimics the WT state at pH 6.5. Altogether, the NMR data confirm that replacement of His124 with a perpetually charged lysine mimics the “low pH” structural changes and stabilizes that form so that it is predominant even at pH 7.5. From the standpoint of ACD structure and dimer-to-monomer equilibria, the role of His124 (His104 in HSPB5) appears to be conserved.
HSPB1-FL oligomers are pH-sensitive
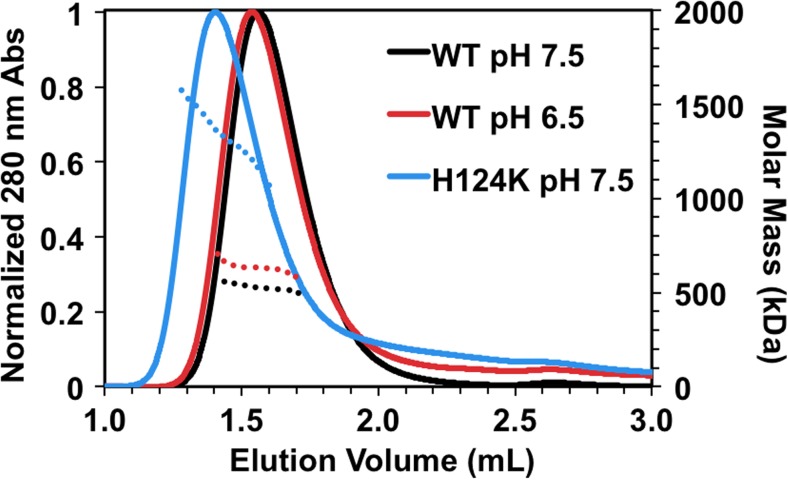
WT-HSPB1 forms large (∼450 kDa), polydisperse oligomers that are highly sensitive to factors such as temperature (Hayes et al. 2009; Lelj-Garolla and Mauk 2006), phosphorylation (Shashidharamurthy et al. 2005), and disease-associated missense mutations (Almeida-Souza et al. 2010; Muranova et al. 2015; Nefedova et al. 2013a, b). Earlier elution of HSPB1 oligomers at low pH in size-exclusion chromatography (SEC) has been previously reported (Chernik et al. 2004). To assess whether the earlier elution of HSPB1 oligomers is due to a change in shape or an increase in the average number of subunits at low pH, WT-HSPB1 was analyzed by SEC coupled with multi-angle light scattering (MALS). The protein elutes earlier at pH 6.5 than at pH 7.5 as expected, indicating that the oligomers that exist at the lower pH have, on average, a larger hydrodynamic radius (Fig. 2). MALS reveals that the average molar mass of the oligomer increases from 520 to 630 kDa at pH 6.5, corresponding to an increase in the average number of subunits from 23 to 28 at pH 6.5 (Table 1). This is a smaller expansion than was observed over the same pH range for HSPB5, where the average number of subunits in its oligomers increases from 24 to 36 (Rajagopal et al. 2015b). In the HSPB1 case, there is also a small but reproducible increase in species that elute after the main peak, as evidenced by the absorbance not returning to baseline. This “bleeding” behavior in which protein intensity is observed through the SEC run indicates that smaller species dissociate on the timescale of the experiment. Increased bleeding is observed for WT-HSPB1 at pH 6.5, consistent with previous results (Chernik et al. 2004), and far more dramatic effects are seen for H124K-HSPB1 at pH 7.5. The behavior is consistent with a fraction of WT-HSPB1 existing as smaller oligomers, as observed by mass spectrometry and SEC (Jovcevski et al. 2015). Additionally, the oligomers of disease-associated HSPB1 mutants were reported to elute earlier than WT by SEC but to dissociate at relatively high concentrations, implying weakened interactions within the oligomers (Nefedova et al. 2013b). A majority of the mutant protein elutes as very large oligomers, with an average molar mass and number of subunits of 1330 kDa and 58, respectively, and there is even more bleeding of smaller species throughout the elution profile for the mutant. Together, the data suggest that destabilization of the dimer interface of HSPB1 by decreasing pH or mutation has two effects: (1) it increases the average number of subunits in the oligomers and (2) it alters the exchange rate between large oligomers and smaller species.
Fig. 2.
The H124K mutation induces formation of expanded oligomers but also increases the proportion of smaller oligomers. One-hundred micromolar samples were incubated at 37 °C prior to injection onto a Superose 6 SEC column and analyzed by multi-angle light scattering (MALS). Calculated molar mass is shown (dotted lines) for the top 50% of the UV chromatogram for each sample. WT-HSPB1 elutes at a slightly earlier volume at pH 6.5 relative to pH 7.5 and has a larger average molar mass, corresponding to an increase in the number of subunits. At pH 7.5, H124K-HSPB1 elutes earlier than WT with a substantial increase in molar mass (see Table 1). The elution profiles of both WT-HSPB1 at pH 6.5 and H124K-HSPB1 at pH 7.5 have long tails at longer elution times, indicting an increase in the proportion of small oligomers. These data together suggest that destabilization of the dimer interface destabilizes the oligomers while paradoxically expanding their size
Table 1.
Average molar masses across the top 50% of UV chromatograms and corresponding number of subunits (monomer =22.8 kDa)
| Average molar mass (kDa) | Average number of subunits | |
|---|---|---|
| WT pH 7.5 | 520 | 23 |
| WT pH 6.5 | 630 | 28 |
| H124K pH 7.5 | 1330 | 58 |
The low pH-mimicking mutant of HSPB1-FL has modestly increased chaperone activity
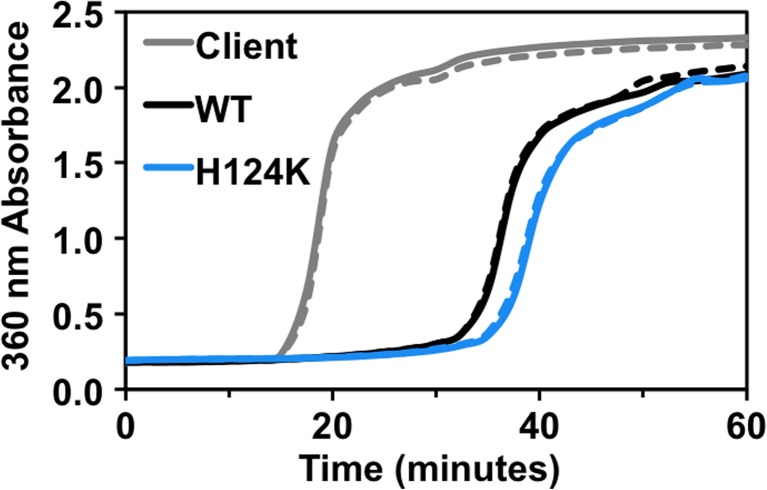
The low-pH mimic of HSPB5, H104K-HSPB5, exhibits highly enhanced chaperone activity against the client protein, α-lactalbumin (Rajagopal et al. 2015b), so we asked whether the same was true for the analogous mimic in HSPB1. WT-HSPB1 delays aggregation of several destabilized, aggregation-prone model client proteins (Hayes et al. 2009; Muranova et al. 2015; Shashidharamurthy et al. 2005), and as with oligomeric propensity, this behavior is dependent upon several factors. Under the conditions of our assays, WT-HSPB1 substantially delays the onset of large light scattering species of α-lactalbumin that has been destabilized by a reducing agent (Fig. 3). Although the time to onset of observable aggregation is reproducibly delayed by H124K-HSPB1 longer than by WT-HSPB1, the difference in this activity is quite modest. This result is in distinct contrast to the “hyperactive” activity exhibited by H104K-HSPB5.
Fig. 3.
Low pH-mimicking mutant of HSPB1 is not highly activated relative to WT-HSPB1. WT and H124K HSPB1 delay aggregation of a model client protein, α-lactalbumin. The final sHSP and client concentrations were 60 and 600 μM, respectively, and aggregation of α-lactalbumin was induced by addition of 20 mM DTT. Duplicate samples are shown. Absorbance at 360 nm was measured over time at 37 °C. The H124K mutant delays aggregation of α-lactalbumin slightly longer than WT protein. This contrasts with the dramatic increase in chaperone activity reported for the analogous H104 mutation in HSPB5 (Rajagopal et al. 2015b)
Discussion
We have previously shown that a relatively small, physiologically relevant drop in pH has a dramatic impact on the structure and function of the human sHSP HSPB5 (Rajagopal et al. 2015b). Here, we confirm similar pH-dependent properties for another ubiquitously expressed human sHSP, HSPB1. The ACDs of these two sHSPs (and other human sHSPs) are conserved both in sequence and structure, notably in the abundance of charged residues that form a tenuous and dynamic network across the dimer interface of two ACDs. A decrease in pH to 6.5, in the range of a cell undergoing acidosis, destabilizes the interaction between ACDs along the long antiparallel β6+7 strands that compose the dimer interface. The monomeric structure of the ACD appears to be maintained in its low-pH state as evidenced by the observation that residues far from the dimer interface are relatively unperturbed in the NMR spectra. A conserved histidine in the loop preceding the β6+7 strand was shown to be the pH-conformational switch in HSPB5 (His104), changing protonation states within the pH range of interest. Mutation of this histidine to lysine recapitulated local and global structural changes seen in the WT protein at pH 6.5, providing a mimic of the low pH state that is stable at higher pH. We show here that HSPB1 also undergoes a pH-dependent change in structure and that the analogous substitution in HSPB1, H124K, can mimic the low-pH state. At the level of ACD structure, the HSPB1-ACD dimer is destabilized at pH 6.5 or by the H124K mutation. Similar to HSPB5, HSPB1 oligomers expand at low pH from an average of 23 subunits to 28 subunits, but this is a less dramatic expansion than that for HSPB5, which increases from 24 to 36 subunits. Substitution of His124 with a perpetually protonated lysine dramatically increases the size of HSPB1 oligomers that exist at pH 7.5 from an average of 23 subunits to an average of 58 subunits, which is a larger expansion than that elicited by the H104K mutation in HSPB5, where oligomers expand to 40 subunits on average. Another notable difference between the consequences of the analogous His-to-Lys mutation is that it leads to only a modest increase in the chaperone activity of HSPB1 in contrast to the strongly enhanced activity in HSPB5. Altogether, the results suggest that pH modulation of structure is conserved between HSPB5 and HSPB1 and likely other sHSPs that possess the conserved histidine, but the extent of the modulation varies among sHSPs. It is tempting to propose that certain sHSPs, such as HSPB5, may be “key players” for maintaining proteostasis during acidosis, while other sHSPs, such as HSPB1, are secondary in their importance to acidosis and are reserved for regulation by other stress factors.
Mutations in charged ACD residues have been reported for human sHSPs, both as disease-associated (inherited mutations) and for structural manipulation and study. There are currently four known disease-associated mutations at charged sites in the ACD of HSPB1-R127W, R136W/L, R140G, and K141Q. Although, to our knowledge, none of these mutations has been structurally characterized in the context of a truncated ACD domain, these mutations produce notably altered oligomeric properties and chaperone function in full-length HSPB1 (Almeida-Souza et al. 2010; Nefedova et al. 2013a). Whether the mutations destabilize the dimer interface via a mechanism similar to H124K remains to be seen, but given the close proximity of His124 to Arg127, a mutation of this arginine might have a similar effect. Intriguingly, a crystal structure of HSPB1-ACD that contained mutations in two charged residues proximal to His124, namely E125A/E126A, is monomeric, with monomers arranged in a hexameric ring formed by contacts between β4 and β6+7 strands of neighboring ACDs (Baranova et al. 2011). The two mutated glutamate residues are not directly involved in the new interface, suggesting that their mutation destabilized the canonical dimer interface, allowing alternative ACD interactions to come into play.
Change in pH is one of the many environmental stressors experienced in a cell and is implicated in sHSP regulation and function. Both HSPB1 and HSPB5 have been reported to coordinate metal ions (Asthana et al. 2014; Mainz et al. 2012), the concentration of which may depend on cellular stress conditions. Several charged residues in the HSPB5-ACD, including His104, are implicated in copper and zinc ion binding, and this binding affects dimer stability, oligomeric propensity, and chaperone function (Mainz et al. 2012). Although residue-specific interactions with metal ions have not been determined for HSPB1, the residues proposed as ligands in HSPB5 are conserved in HSPB1. HSPB1 is unique among human sHSPs in that it readily forms a disulfide bond between ACDs at the dimer interface (Cys137) under non-reducing conditions, implying that this is a likely state for HSPB1 in a cell undergoing oxidative stress. Thus, it appears that the stability of the dimer interface can be tuned by a growing number of environmental conditions, including pH, metal ion concentration, and oxidative stress. These effects at the dimer interface lead to global changes in oligomeric properties, be they changes in size, polydispersity, overall stability, or subunit exchange kinetics. Presumably, the net effect on function is an alteration of exposed surfaces or availability of the surfaces by which sHSPs interact with client proteins under normal versus stress conditions. The effect is clearly evident in the low pH-mimicking form of HSPB5 (H104K) that reorganizes in the presence of client to form smaller, longer-lived complexes compared to the WT counterpart (Rajagopal et al. 2015b). A full understanding of the structure and function of HSPB1 must ultimately include the effects of each of these stress-related factors. There is likely to be strong interplay among these modes of sHSP regulation, resulting in carefully tuned sHSP function under the myriad conditions that cells must endure.
Methods
Protein expression and purification
Human HSPB1-ACD (Gln80 through Ser176), full-length HSPB1 (P04792), and the H124K mutant forms of both were expressed from pET23a plasmid in BL21(DE3) Escherichia coli cells. Non-isotopically labeled protein was grown in standard LB with 100 μg/mL ampicillin at 37 °C to an OD600 of ∼0.6 and expressed using 0.5–1.0 mM IPTG (ACD vs. full-length) at 22 °C for ∼20 h. 15N–isotopically labeled protein was grown in MOPS minimal media with 1 g/L 15NH4Cl and 100 μg/mL ampicillin, and expression was carried out identically to the non-labeled protein. Cells were lysed by freeze-thaw and incubation with lysozyme. Impurities were precipitated from the ACD protein supernatant by adding ammonium sulfate to 35% saturation, and the ACD protein was then precipitated from the resulting supernatant by adding ammonium sulfate to 70%. Full-length protein was simply precipitated by adding ammonium sulfate to 40% saturation. HSPB1-ACD was then desalted using a G25 column in 20 mM Tris at pH 8.0 and purified by diethylaminoethyl (DEAE) and monoQ columns by eluting with NaCl. HSPB1-FL was desalted using a G25 column in 20 mM Tris, 10 mM MgCl2, and 30 mM NH4Cl at pH 7.6 and purified by a DEAE column by eluting with NaCl. The ACD and full-length proteins were concentrated and further purified by SEC on Superdex 75 and 200 columns, respectively, in 50 mM NaPi, 100 mM NaCl, and 0.5 mM EDTA at pH 7.5. Purity of all samples was >95%, as confirmed by SDS-PAGE.
NMR spectroscopy
1H-15N HSQC-TROSY experiments were carried out on a 600 MHz Bruker Avance spectrometer equipped with a cryoprobe. Spectra were collected at 25 °C on 100-μM protein samples in 50 mM sodium phosphate, 100 mM NaCl, 0.5 mM EDTA, and 5 mM DTT at pH 7.5 or 6.5. Spectra were analyzed using NMRView (Johnson 2004). Assignments at pH 6.5 were confirmed by titration (intermediate pH points) and triple resonance experiments (HNCO and HNCACB).
SEC-MALS
Chromatograms were collected on a GE AKTA Pure coupled to a Wyatt miniDAWN TREOS and Optilab T-rEX differential refractive index detector. Molar mass was calculated from the Raleigh ratio based on multi-angle (static) light scattering and protein concentration from the change in refractive index (dn/dc = 0.185). Analysis was performed using Wyatt ASTRA VI software, and curves were calibrated with apoferritin. 100-μM samples of HSPB1-FL were prepared in 50 mM sodium phosphate, 100 mM NaCl, 0.5 mM EDTA, and 2 mM DTT at pH 7.5 or 6.5. Samples were incubated at 37 °C for 1 hour prior to placement in a 4 °C autosampler and were injected onto a GE Superose 6 Increase 3.2/300 mm column equilibrated with the matched buffer at room temperature (27–28 °C).
Chaperone assay
Samples were prepared in a 96-well plate with final protein concentrations of 600 uM bovine α-lactalbumin and 60 μM HSPB1-FL in 50 mM sodium phosphate, 100 mM NaCl, and 0.5 mM EDTA at pH 7.5. Samples were incubated at 37 °C for 30 min prior to triggering aggregation with final concentrations of 20 mM DTT and 5 mM EDTA. Absorbance at 360 nm was measured in a BioTek plate reader at 37 °C every 2.5 min, with shaking prior to each measurement. Absorbance of each sHSP alone was measured to confirm that sHSPs were not aggregating under these conditions.
Acknowledgements
This work was supported by NIH grant 1R01 EY017370 to REK. AFC is supported in part by NIH MBTG T32 GM008268 and the Hurd Fellowship in Biophysics from the UW School of Medicine.
References
- Almeida-Souza L, Goethals S, de Winter V, Dierick I, Gallardo R, Van Durme J, Irobi J, Gettemans J, Rousseau F, Schymkowitz J, Timmerman V, Janssens S. Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charot-Marie-Tooth neuropathy. J Biol Chem. 2010;285:12778–12786. doi: 10.1074/jbc.M109.082644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana A, Bollapalli M, Tangirala R, Bakthisaran R, Rao CM. Hsp27 suppressed the Cu2+-induced amyloidogenicity, redox activity, and cytotoxicity of α-synuclein by metal ion stripping. Free Radical Bio Med. 2014;72:176–190. doi: 10.1016/j.freeradbiomed.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Bagneris C, Bateman OA, Naylor CE, Cronin N, Boelens WC, Keep NH, Slingsby C. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392:1242–1252. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV. Three-dimensional structure of α-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J Mol Biol. 2011;411:110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Chernik IS, Panasenko OO, Li Y, Marston SB, Gusev NB. pH-induced changes of the structure of small heat shock proteins with molecular mass 24/27 kDa (HspB1) Biochem Bioph Res Co. 2004;324:1199–1203. doi: 10.1016/j.bbrc.2004.09.176. [DOI] [PubMed] [Google Scholar]
- Datskevich PN, Nefedova VV, Sudnitsyna MV, Gusev NB. Mutations of small heat shock proteins and human congenital diseases. Biochemistry-Moscow. 2012;77:1500–1514. doi: 10.1134/S0006297912130081. [DOI] [PubMed] [Google Scholar]
- Hayes D, Napoli V, Mazurkie A, Stafford WF, Graceffa P. Phosphorylation dependence of Hsp27 multimeric size and chaperone function. J Biol Chem. 2009;284:18801–18807. doi: 10.1074/jbc.M109.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg GKA, Ecroyd H, Liu C, Cox D, Cascio D, Sawaya M, Collier MP, Stroud J, Carver JA, Baldwin AJ, Robinson CV, Eisenberg DS, Benesch JL, Laganowsky A. The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity. P Natl Acad Sci USA. 2014;111:E1562–E1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle S, Rajagopal P, Bardiaux B, Markovic S, Kuhne R, Stout JR, Higman VA, Klevit RE, van Rossum BJ, Oschkinat H. Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers. Nat Struct Mol Biol. 2010;17:1037–1042. doi: 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle S, Vollmar BS, Bardiaux B, Dove KK, Rajagopal P, Gonen T, Oschkinat H, Klevit RE. N-terminal domain of αB-crystallin provides a conformational switch for multimerization and structural heterogeneity. P Natl Acad Sci USA. 2011;108:6409–6414. doi: 10.1073/pnas.1014656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Jovcevski B, Kelly MA, Rote AP, Berg T, Gastall HY, Benesch JLP, Aquilina JA, Ecroyd H. Phosphomimics destabilize Hsp27 oligomeric assemblies and enhance chaperone activity. Cell Chem Biol. 2015;22:186–195. doi: 10.1016/j.chembiol.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Benesch JL, Landau M, Ding L, Sawaya MR, Cascio D, Huang Q, Robinson CV, Horwitz J, Eisenberg D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19:1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelj-Garolla B, Mauk AG. Self-association and chaperone activity of Hsp27 are thermally activated. J Biol Chem. 2006;281:8169–8174. doi: 10.1074/jbc.M512553200. [DOI] [PubMed] [Google Scholar]
- Mainz A, Bardiaux B, Kuppler F, Multhaup G, Felli IC, Pierattelli R, Reif B. Structural and mechanistic implications of metal binding in the small heat-shock protein αB-crystallin. J Biol Chem. 2012;287:1128–1138. doi: 10.1074/jbc.M111.309047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicar N, Li AX, Goncalves DF, Bellyou M, Meakin SO, Prado MA, Bartha R. Quantitative tissue pH measurement during cerebral ischemia using amine and amide concentration-independent detection (AACID) with MRI. J Cerebr Blood F Met. 2014;34:690–698. doi: 10.1038/jcbfm.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranova LK, Weeks SD, Strelkov SV, Gusev NB. Characterization of mutants of human small heat shock protein HspB1 carrying replacements in the N-terminal domain and associated with hereditary motor neuron diseases. PLoS One. 2015 doi: 10.1371/journal.pone.0126248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedova VV, Datskevich PN, Sudnitsyna MV, Strelkov SV, Gusev NB. Physio-chemical properties of R140G and K141Q mutants of human small heat shock protein HspB1 associated with hereditary peripheral neuropathies. Biochimie. 2013;95:1582–1592. doi: 10.1016/j.biochi.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Nefedova VV, Sudnitsyna MV, Strelkov SV, Gusev NB. Structure and properties of G84R and L99M mutants of human small heat shock protein HspB1 correlating with motor neuropathy. Arch Biochem Biophys. 2013;538:16–24. doi: 10.1016/j.abb.2013.07.028. [DOI] [PubMed] [Google Scholar]
- Rajagopal P, Liu Y, Shi L, Clouser AF, Klevit RE. Structure of the α-crystallin domain from the redox-sensitive chaperone, HSPB1. J Biomol NMR. 2015;63:223–228. doi: 10.1007/s10858-015-9973-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal P, Tse E, Borst AJ, Delbecq SP, Shi L, Southworth DR, Klevit RE. A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. elife. 2015 doi: 10.7554/eLife.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidharamurthy R, Koteiche HA, Dong J, Mchaourab HS. Mechanism of chaperone function in small heat shock proteins. J Biol Chem. 2005;280:5281–5289. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- Singh AK, Manns MP, Seidler U. Cytoprotective effects of acidosis via heat shock protein HSP27 against the anticancer drug doxorubicin. Cell Mol Life Sci. 2011;68:1041–1051. doi: 10.1007/s00018-010-0503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SD, Baranova EV, Heirbaut M, Beelen S, Shkumatov AV, Gusev NB, Strelkov SV. Molecular structure and dynamics of the dimeric human small heat shock protein HSPB6. J Struct Biol. 2013;185:342–354. doi: 10.1016/j.jsb.2013.12.009. [DOI] [PubMed] [Google Scholar]