Abstract
DNA barcoding of United Arab Emirates (UAE) native plants is of high practical and scientific value as the plants adapt to very harsh environmental conditions that challenge their identification. Fifty-one plant species belonged to 22 families, 2 monocots, and 20 eudicots; a maximum number of species being legumes and grasses were collected. To authenticate the morphological identification of the wild plant taxa, rbcL and matK regions were used in the study. The primer universality and discriminatory power of rbcL is 100%, while it is 35% for matK locus for these plant species. The sequences were submitted to GenBank; accession numbers were obtained for all the rbcL sequences and for 6 of matK sequences. We suggest rbcL as a promising barcode locus for the tested group of 51 plants. In the present study, an inexpensive, simple method of identification of rare desert plant taxa through rbcL barcode is being reported.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0746-1) contains supplementary material, which is available to authorized users.
Keywords: United Arab Emirates, Monocots, Eudicots, Native plants, rbcL, matK, Barcode
Introduction
The UAE is a dry land, covered with wadis, waterless riverbeds, sand dunes, plains, and mountains. Diverse flora and fauna exist in the desert, the diversity of which is due to phyto-geographical variations of the desert. Identification of UAE native plant and animal species is vital for preservation as well as developing biodiverse resources. Relying solely on morphological characteristics of plants for taxonomic classification is difficult due to geographical variations and it mandates specialized taxonomists (Chase and Fay 2009). Under such situations, identification of plant taxa even from a small part of tissue from any geographical location is possible with the aid of barcoding regions, suggested by Consortium of Barcode of Life (CBOL) in 2009. Besides, DNA barcoding and genomics share an emphasis on large-scale genetic data acquisition that offer new answers to genotypic and phenotypic questions beyond the reach of the traditional taxonomic disciplines.
Ideally, a DNA barcode consists of a standardized short sequence of DNA (400–800 bp) that, in principle, can be easily generated and would be unique for every species on the planet. The most commonly used barcode for animals is a small region in cytochrome oxidase gene (COI) in the mitochondria. However, for plants, many plastid sequences have been studied and validated, and were recommended as possible barcode loci (Kress et al. 2005; Chase et al. 2007; Ford et al. 2009; Pennisi 2007). A pair of such recommended sequences are segments of two plastid genes, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL), and maturase K (matK). These two sequences have been adopted as standard plant DNA barcodes by the international body of the Plant Working Group (PWG) of the Consortium for the Barcoding of Life (CBOL 2009). Universal primer sequences designed by CBOL were used successfully for identification and authentication of some international as well as regional plant species; which are tried as precedence for the present plant taxa.
DNA barcoding, a two decade old science reviewed extensively (Pecnikar and Buzan 2013; Ali et al. 2014), has applications in many branches of science other than identification of organisms plants/animals. Some of the applied areas of barcoding are recognizing insect–host relationship (Jurado-Rivera et al. 2009), analyzing the diet of herbivores (Valentini et al. 2009; Staudacher et al. 2011; Stech et al. 2011), scrutinizing the components of herbal medicines (Srirama et al. 2010), food products (Jaakola et al. 2010) and in ecological forensics to identify the plant from a small tissue of root, or seedling or cryptic life stages (e.g., of fern gametophytes) and endangered species.
The plant taxa of UAE include salt, heat, and drought tolerant, and some of which are medicinally important (Sakkir et al. 2012); there are no barcoding studies reported on such plant taxa so far. In an attempt to develop barcode for some of the natural flora of UAE, plants were collected from three Emirates which have different bioclimatic zones. Dubai and Umm Al-Quwain are hotter and with iron rich red/blue colored soils, while Fujairah is cooler, mountainous with black colored soils (Basha et al. 2015). In Dubai Emirate, plants were collected from three regions, viz., Mushrif park where the natural flora of Dubai are retained and also from urbanized regions Al twar and Al Quasais where the Experimental school University of Modern sciences (UMS) is located. We made an endeavor to identify and authenticate plant species collected from these five regions in UAE and evaluated the potentiality of plastid rbcL and matK regions for possible barcoding. This is the first report on DNA barcoding, which facilitates species-level taxonomy of known and unknown species of UAE.
Materials and methods
Plant sample collection
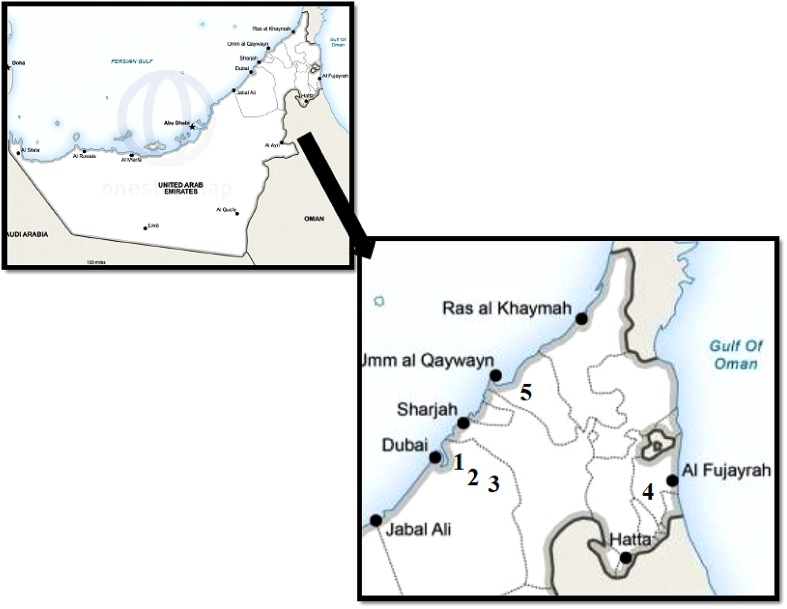
A total of 51 plants were randomly collected from five geographical locations and also on the way to Fujairah from the University; the sample collection included 20 medicinally important plant species (Fig. 1, Supplementary Table S1). The collected plant samples were placed in zip lock plastic bags containing silica gel with sample ID until they are transported to the laboratory and kept in freezer (−80 °C) for further analysis. A part of the twig is prepared for herbarium, which is referred for plant identification and not for DNA extraction. The plants were identified based on the morphology referring to different books (Karim et al. 2013; Jongbloed 2003; Bolus 2000) and were assigned to the respective families.
Fig. 1.
Geographical location of plant sample collection: 1 Al Qusais, 2 Al twar, 3 Mushrif park, 4 Wadis (Fujairah), and 5 Umm al-Quwain
DNA extraction, PCR amplification, and sequencing success
Total genomic DNA of plant samples were extracted using Plant/Fungal DNA extraction kit (Norgen Bioteck, Canada) and DNA was preserved at −20 °C for further analysis. The target DNA regions, namely rbcL and matK, were amplified with respective universal DNA barcoding primers (CBOL 2009). For rbcL gene, rbcLa-F: 5′-ATGTCACCACAAACAGAGACTAAAGC-3′ and rbcLa-R: 5′-GTAAAATCAAGTCCACCRCG-3′; for matK gene, and matK-KIM1R: 5′-ACCCAGTCCATCTGGAAATCTTGGTTC-3′ and matK-KIM3F: 5′-CGTACAGTACTTTTGTGTTTACGAG-3′ were used. PCR was performed using a reaction mixture of a total volume of 50:25 µl of Taq PCR Master Mix (Norgen Biotek, Canada), 22 µl distilled water, 1 µl forward primer (10 µM), 1 µl reverse primer (10 µM), and 1 µl of the DNA template (50–80 ng/µl). The PCR conditions were as follows: 1 cycle (94 °C for 3 min), 35 cycles (94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min), and 1 cycle 72 °C for 7 min. Amplified PCR products of rbcL and matK were checked on 1.5% agarose gel electrophoresis for the respective bands (600 and 900 bps) which were sent to Macrogen (Seoul, South Korea) for DNA sequencing.
Sequence alignment and data analyses
The obtained forward and reverse sequences were aligned using online pairwise alignment tool and were submitted to NCBI BLAST, (www.ncbi.com) and/or BOLD. The query sequences were identified considering E value a <1 × 10−5 and maximum hits (99 or 100%) with a species in the reference database. In addition to BLAST, MEGA 6 (Tamura et al. 2013) was used for phylogenetic tree analysis employing Neighbor-Joining (NJ) method (Saitou and Nei 1987).
Results
Taxonomical identification
The collected 51 plants from the five regions (Fig. 1) were identified based on morphology and floral structures referring to the text books (Jongbloed 2003; Bolus 2000), and research papers (Karim et al. 2013) were assigned to the respective families (Fig. 2). The collected flora belonged to 22 families; maximum number of species belonged to Poaceae and Fabaceae (Supplementary Table S1). Although Poaceae has maximum number of plants in the collection (≈18%), it does not reflect the actual distribution of the flora of the areas, as the samples were mostly collected after a shower of rain enabling the seeds to germinate and grow; while among legumes, the collected plant samples were mostly trees except Hippocrepis comosa. Thirteen families are represented each by a single species. Euphorbia larica is being reported for the first time from UAE (Supplementary Fig. S1 D).
Fig. 2.
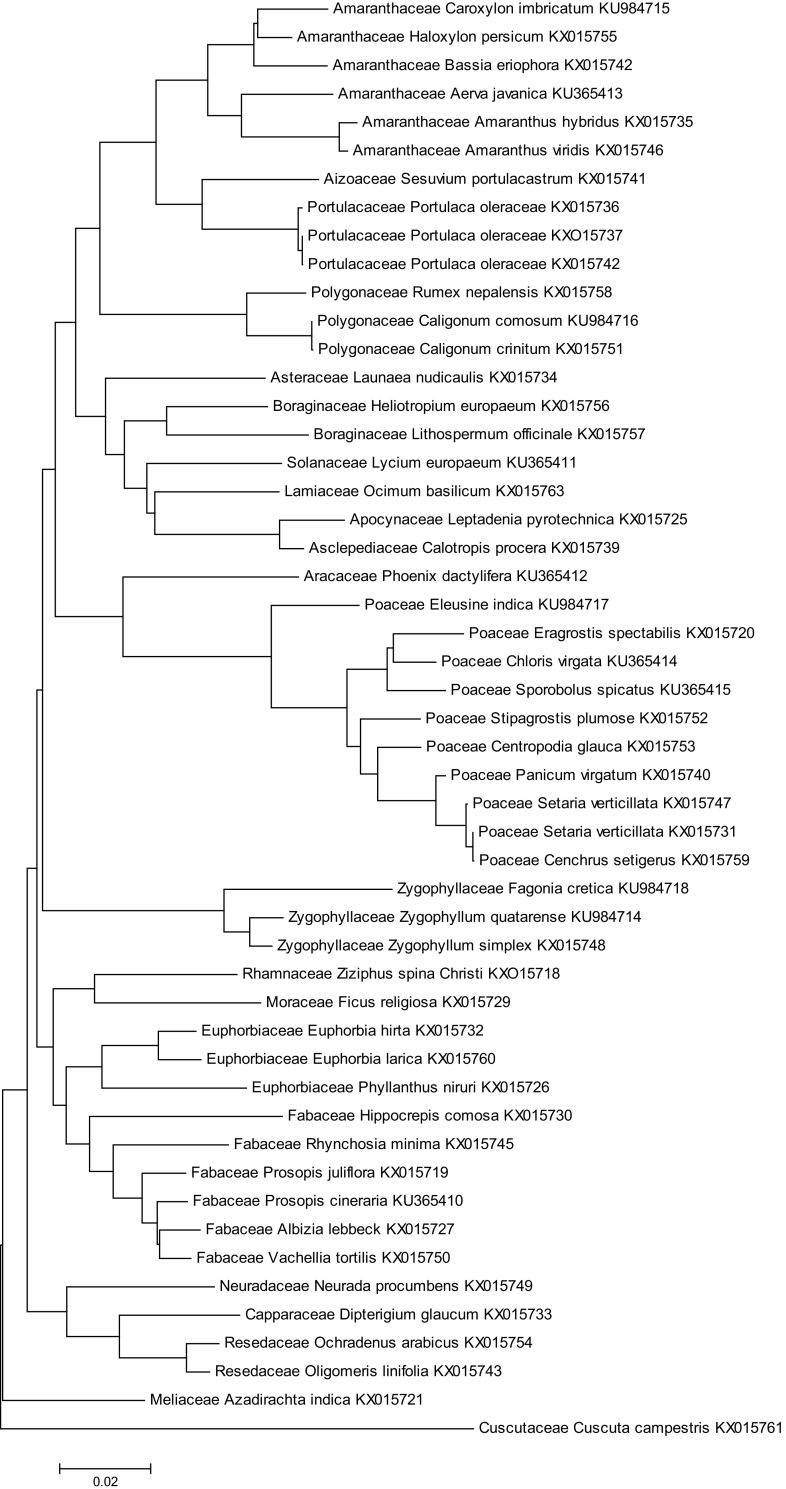
Molecular phylogenetic analysis using rbcL for 51 taxa by maximum-likelihood method: the evolutionary history was inferred using the neighbor-joining method based on Tamura et al. (2013). The optimal tree with the sum of branch length = 1.31,846,941 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The differences in the composition bias among sequences were considered in evolutionary comparisons. The analysis involved 51 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All ambiguous positions were removed for each sequence pair. There were a total of 620 positions in the final data set. Evolutionary analyses were conducted in MEGA6
Plastid rbcL: DNA extraction, PCR amplification, and sequencing
The standard extraction protocol suggested by Norgen, Bioteck worked for 100% (50 of autotrophs +1 parasite) plants. Amplification of rbcL yielded PCR products for all the taxa. The generated query sequences of 51 plants were matched with the reference sequences in BLAST (Altschul et al. 1990) and BOLD (Ratnasingham and Hebert 2007). The query sequences of 51 plants were identified up to species level with 99 or 100% in either of the algorithms. The identification success was equally good for 11 monocots and 40 eudicots using rbcL locus. The present study included five genera from five families, viz., Prosopis (Fabaceae), Euphorbia (Euphorbiaceae), Amaranthus (Amaranthaceae), Calligonum (Polygonaceae), and Zygophyllum (Zygophyllaceae), each represented by two species; could be resolved to individual species (Fig. 2). The plant taxa collected from different areas with different morphologies assumed to be different species were identified as varieties of the same species through sequence alignment, Portulaca oleracea and Setaria verticillata (Supplementary Fig S1A-C). Euphorbia larica, a new plant from UAE, is being reported for the first time; a few more plant sequences of UAE natives Zygophyllum qatarense, Caroxylon imbricatum, Caligonum comosum, and Stipagrostis plumosa are deposited in GenBank; accession numbers were obtained for the respective plant species (Fig. 2).
Plastid matK: DNA extraction, PCR, and sequencing
The set of matK primers amplified only 35.29% (18/51) of the tested plant taxa. The failure of amplification in more than half of the plants is not due to the poor quality of the extracted DNA, which were amplified 100% with rbcL primer set. When the matK sequences were aligned with the reference sequences in NCBI and/BOLD, only 27.45% (14/51) resulted in correct species identification; four of the query sequences mis-matched: 1. Zygophyllum qatarense with Z. rosowii or Z. fabago, 2. Sporobolus spicatus with S. cryptandrus and Salsola inermis with Salsola kali and Lycium europaeum with Lycium barbarum. For these four plants, matK algorithm is not able to identify the species due to the absence of species specific unique regions. The idea that the plant is a hybrid of the two species can be negated as the same plant was correctly identified with rbcL marker that corresponded with the morphological description. The matched sequences were submitted to Bankit and accession numbers were obtained only for six plant taxa—Phoenix dactilifera (KU204814), Aerva javanica (KU 204823), Ziziphus spina Christi (KX015722), Prosopis juliflora (KX015723), Azadirachta indica (KX015724), and Albizia lebbeck (KX015728).
Phylogenetic tree: Families and clustering
The phylogenetic tree was constructed using Neighbor-Joining method and the evolutionary distances were computed employing maximum composite likelihood method. The collected plant taxa were grouped into monocots and eudicots. Monocots represented a monophyletic clade, included sub-division commelinids that contained two families, Poaceae and Arecaceae. The eudicots included plant taxa that are grouped into three subdivisions, viz., rosids, asterids, and superasterids that contained the remaining 20 families. Those families that are represented by two species/family such as Amaranthaceae, Polygonaceae, Zygophyllaceae, Euphorbiaceae, and Fabaceae were grouped into clades, while the remaining families were each contained only one representative species which were as monophyletic group (Fig. 2). Cuscuta campestris is a parasitic plant and its family is arranged as an out group (Fig. 2).
Discussion
Plastid rbcL
In this study, we tested the potentiality of plastid rbcL region as a barcode for 51 UAE native plant species. Identification using rbcL region matched with morphological identification 100% to species level; it appears to be an accurate estimate of species identification with this single locus for the group of tested plants. Universal bar code should possess three characteristics, short sequence, universality, and unique identifiers (Stoeckle 2003), which was more applicable to algae than to land plants due to the absence of variable identifiers (Presting 2006). So far, single barcode region suitable to all plant taxa is not reported (Ballardini et al. 2013); different barcodes are successful to different taxa, for example, rbcL locus for palmae (Naeem et al. 2014; Bafeel et al. 2012); ITS for Asteraceae (Gong et al. 2015); multiple loci, ITS + rpoB and rpoC1 for Resedaceae (Khan et al. 2012); rbcL + matK for flora of South central Ontario, Canada (Burgess et al. 2011), and rbcL + matK + trnH-psbA for Arecaceae (Yang et al. 2012). Morphological identification of grasses is difficult because of their reduced reproductive structures and rbcL + matK were used to resolve ambiguous species (Awad et al. 2015). Nevertheless, in the present study, 10 Poaceae genera could be resolved with rbcL region alone. Our study although is of small sample size, the members span across 22 families, including diverse orders Asterales, Caryophyllales, Zygophyllales, Malphigiales, and commelinids in which rbcL is well conserved locus enabling authentication of species identification.
Plastid matK
In the current investigation, matK amplified 35% of the taxa and, among those, identified only 27% species correctly. This indicates nucleotide substitutions in the annealing sites and also in the remaining amplified region. Low amplification of matK locus is not unique to the present desert plants, but was also reported to other arid plants (Bafeel et al. 2011), angiosperms (Kool et al. 2012) and gymnosperms (Saas et al. 2007). The failure of matK amplification in plant taxa could be due to the large size of the amplification product (≈900 bps) that is liable to degradation (Fazekas et al. 2012); or due to nucleotide variations as single-nucleotide substitutions that prevent PCR amplification (Bru et al. 2008). Our study with 27% species resolution is in accordance with nucleotide substitutions. An ecotype of Caroxylon imbricatum collected from Saudi Arabia yielded an amplicon >900 bps with the primer set matK-A-matK-2.1F and matK 5R (Bafeel et al. 2011), while the UAE cultivars yielded appropriate band (900 bps) with a different set of primers (MatK-KIM1R and KIM3F). Although other sets of primers have not been tried, we recommend this primer set a suitable primer for amplification of matK region for this species. MatK region has high rate of nucleotide substitutions (Hilu et al. 2003) or restructuring of the locus (de Groot et al. 2011) probably alternate primer sequences may improve the success rate of matK amplification for some of the present taxa, hence as barcoding locus. However, the species in which the matK region is amplified had a broad taxonomic coverage including families Rhamnaceae to Poaceae, indicating that the conserved sequence of the locus is noteworthy.
It can be deduced from the obtained data that between the two loci, rbcL region is more stable and conserved than matK locus for the tested taxa. Multiple barcode loci were suggested for proper discrimination of species that would minimize interspecific sharing of sequences (Hollingsworth et al. 2009), supplementary barcode loci trnH-psb-A and nuclear ITS were tried for a few plant taxa, and the amplification of former yielded multiple bands and of the latter did not discriminate species, (data not shown). The results are encouraging, which provide a backbone of information in the data set; further studies for species resolution of a genus can be carried as and when more species are available.
Phylogenetic tree: families and clustering
In the present study, partial amplification sequences of rbcL were further used to understand the evolutionary linkage of the collected UAE plants. Using neighbor-joining method, the evolutionary distances for the 51 plant species were distinguished into individual clades, which are consistent with molecular classification of Angiosperms (APG IV 2016). Neighbor-joining method is reported to discriminate most of the species better compared to other methods (de Vere et al., 2012). Our study included many genera from different families, and only two species/genus in five taxa. Three varieties of Portulaca oleraceae, morphologically different (Fig. S1), were identified as the same species; in the phylogenetic tree, they were closely arranged but distinguished (Fig. 2). Also the species of the genera Caligonum, Zygophyllum, and Euphorbia are well resolved in the phylogenetic tree without overlaps; inferring that the rbcL locus could be used to assess the evolutionary linkage of the collected samples.
Conclusions
Plastid rbcL stands as a promising barcode locus for the group of 51 UAE native plants, based on the assessments of recoverability, good sequence quality, universality, and high levels of species discrimination. Plastid matK region has more nucleotide substitutions, which evolves faster than rbcL region among the tested UAE local plants. Between the two bar coding regions, rbcL is better conserved hence the preferred code, which resolved diverse taxa that belonged to 22 families including monocots and eudicots, which is an easy-to-use inexpensive identification system that would enable non-experts to identify these rare desert species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work is funded by the National Research Foundation of UAE under Grant (No. UIRCA 2013-24951). The authors express their gratitude to NRF and to the University of Modern sciences for providing them with the equipment and infrastructure to carry the research work.
Footnotes
Lina Maloukh and Alagappan Kumarappan are co-first authors.
Highlight
Universal barcode locus rbcL is the recommended barcode for diverse desert (UAE) plants for species resolution.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0746-1) contains supplementary material, which is available to authorized users.
Contributor Information
Lina Maloukh, Email: l.maloukh@ums.ae.
Alagappan Kumarappan, Email: alagumicro@yahoo.co.in.
Mohammad Jarrar, Email: M.jarrar@ums.ae.
Jawad Salehi, Email: j.salehi@ums.ae.
Houssam El-wakil, Email: Hosamelwakil@gmail.com.
T. V. Rajya Lakshmi, Phone: 971 4 2646506, Email: v.lakshmi@ums.ae
References
- Ali AM, Gyulai G, Hidvegi N, Kerti B, Al Hemaid FMA, Pandey AK, Lee J. The changing epitome of species identification-DNA barcoding. Saudi J Biol sci. 2014;21:204–231. doi: 10.1016/j.sjbs.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- APG An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot J Linean Soc. 2016;181(1):1–20. doi: 10.1111/boj.12385. [DOI] [Google Scholar]
- Awad M, Fahmy RM, Mosa KA, Helmy M, El-Feky FA. Assessment of candidate DNA barcoding loci for the wheat and grass family Poaceae in Egypt. Genome. 2015;58:190–191. [Google Scholar]
- Bafeel SO, Arif IA, Bakir MA, Khan HA, Al Farhan AH, Al Homaidan AA, Ahamed A, Thomas J. Comparative evaluation of PCR success with universal primers of maturase K (matK) and ribulose-1,5-bisphosphate carboxylase oxygenase large subunit (rbcL) for barcoding of some arid plants. Plant OMICS. 2011;4:195–198. [Google Scholar]
- Bafeel SO, Arif IA, Bakir MA, Al Homaidan AA, Al Farhan AH, Khan HA. DNA barcoding of arid wild plants using rbcL gene sequences. Genet Mol Res. 2012;11:1934–1941. doi: 10.4238/2012.July.19.12. [DOI] [PubMed] [Google Scholar]
- Ballardini M, Mercuri A, Littardi C, Abbas S, Couderc M, Ludeña B, Pintaud JC. The chloroplast DNA locus psbZ-trnfM as a potential barcode marker in Phoenix L. (Arecaceae) Zookeys. 2013;365:71–82. doi: 10.3897/zookeys.365.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha G, Ouarda TBMJ, Marpu PR. Long term projections of temperature, precipitation and soil moisture using non-stationary oscillation processes over the UAE region. Int J Climatol. 2015;15:4606–4618. doi: 10.1002/joc.4310. [DOI] [Google Scholar]
- Bolus L. Flora of Egypt. Cairo: Al Hadara Publishing; 2000. [Google Scholar]
- Bru D, Martin-Laurent F, Philippot L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl Environ Microbiol. 2008;74:1660–1663. doi: 10.1128/AEM.02403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess KS, Fazekas AJ, Kesanakurti PR, Graham SW, Husband BC, Newmaster SG, Percy DM, Hajibabaei M, Barrett SCH. Discriminating plant species in a local temperate flora using the rbcL + matK DNA barcode. Methods Ecol Evol. 2011;2:333–340. doi: 10.1111/j.2041-210X.2011.00092.x. [DOI] [Google Scholar]
- CBOL Plant Working Group A DNA barcode for land plants. PNAS. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Fay MF. Barcoding of plants and fungi. Science. 2009;325:682–683. doi: 10.1126/science.1176906. [DOI] [PubMed] [Google Scholar]
- Chase MW, Cowan RS, Hollingsworth PM, Van den Berg C, Madriñán S, Petersen G, Seberg O, Jørgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson T, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M. A proposal for a standardized protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- de Groot GA, During HJ, Maas JW, Schneider H, Vogel JC, Erkens R. Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PLoS One. 2011;6:e16371. doi: 10.1371/journal.pone.0016371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vere N, Rich TCG, Ford CR, Trinder SA, Long C, et al. DNA barcoding the native flowering plants and conifers of wales. PLoS One. 2012;7(6):37945. doi: 10.1371/journal.pone.0037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas AJ, Kuzmina ML, Newmaster SG, Hollingsworth PM. DNA barcoding methods for land plants. Methods Mol Biol. 2012;858:223–252. doi: 10.1007/978-1-61779-591-6_11. [DOI] [PubMed] [Google Scholar]
- Ford CS, Ayres KL, Toomey N, Haider N, Alphen Stahl JV, Kelly LJ, Wikström N, Hollingsworth PM, Duff RJ, Hoot SB, Cowan RS, Chase MW, Wilkinson MJ. Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc. 2009;159:1–11. doi: 10.1111/j.1095-8339.2008.00938.x. [DOI] [Google Scholar]
- Gong W, Liu Y, Chen J, Hong Y, Kong HH. DNA barcodes identify Chinese medicinal plants and detect geographical patterns of Sinosenecio (Asteraceae) J Syst Evol. 2015;54:83–91. doi: 10.1111/jse.12166. [DOI] [Google Scholar]
- Hilu KW, Borsch T, Müller K, Soltis DE, Soltis PS, Savolainen V, Chase MW, Powell MP, Alice LA, Evans R, Sauquet H, Neinhuis C, Slotta T, Rohwer JG, Campbell CS, Chatrou LW. Angiosperm phylogeny based on matK sequence information. Am J Bot. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, Cowan R, Chase MW, Gaudeul M, Hollingsworth PM. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Res. 2009;9:439–457. doi: 10.1111/j.1755-0998.2008.02439.x. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Suokas M, Häggman N. Novel approaches based on DNA barcoding and high-resolution melting of amplicons for authenticity analyses of berry species. Food Chem. 2010;123:494–500. doi: 10.1016/j.foodchem.2010.04.069. [DOI] [Google Scholar]
- Jongbloed M. The comprehensive guide to the wild flowers of the United Arab Emirates. Abu Dhabi: Environmental Research and Wild Life Development Agency; 2003. [Google Scholar]
- Jurado-Rivera JA, Vogler AP, Reid C, Petitpierre E, Gómez-Zurita J. DNA barcoding insect–host plant associations. Proc Biol Sci R Soc. 2009;276:639–648. doi: 10.1098/rspb.2008.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FM, Dakheel AJ, Kameswara Rao N. Salt-tolerant plants of the United Arab Emirates. Abu Dhabi: Abu Dhabi Food Control Authority and International Center for Biosaline Agriculture; 2013. [Google Scholar]
- Khan S, Al-Qurainy F, Nadeem M, Tarroum M. Development of genetic markers for Ochradenus arabicus (Resedaceae), an endemic medicinal plant of Saudi Arabia. Genet Mol Res. 2012;11:1300–1308. doi: 10.4238/2012.May.14.4. [DOI] [PubMed] [Google Scholar]
- Kool A, de Boer HJ, Kruger A, Rydberg A, Abbad A, Bjork L, Martin G. Molecular identification of commercialized medicinal plants in Southern Morocco. PLoS One. 2012 doi: 10.1371/journal.pone.0039459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem A, Khan AA, Cheema HMN, Khan IA, Buerkert A. DNA barcoding for species identification in the palmae family. Genet Mol Res. 2014;13:10341–10348. doi: 10.4238/2014.December.4.29. [DOI] [PubMed] [Google Scholar]
- Pecnikar ZF, Buzan EV. 20 Years since the introduction of DNA barcoding from theory to application. J Appl Genet. 2013 doi: 10.1007/s13353-013-0180-y. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Taxonomy: wanted: a barcode for plants. Science. 2007;318:190–191. doi: 10.1126/science.318.5848.190. [DOI] [PubMed] [Google Scholar]
- Presting GG. Identification of conserved regions in the plastid genome: implications for DNA barcoding and biological function. Can J Bot. 2006;84:1434–1443. doi: 10.1139/b06-117. [DOI] [Google Scholar]
- Ratnasingham S, Hebert PDN (2007) BOLD: The bar code of life data system (http://www.barcodinglife.org). Mol Ecol Notes 7:355–364 [DOI] [PMC free article] [PubMed]
- Saas C, Little DP, Stevenson DW, Specht CD. DNA bar coding in the Cycadales: testing the potential of proposed barcoding markers for species identification cycads. PLoS One. 2007;11:e1154. doi: 10.1371/journal.pone.0001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakkir S, Kabshawi M, Mehairbi M. Medicinal plants diversity and their conservation status in the United Arab Emirates (UAE) J Med Plants Res. 2012;6(7):1304–1322. [Google Scholar]
- Srirama R, Senthilkumar U, Sreejayan N, Ravikanth G, Gurumurthy BR, Shivanna MB, Sanjappa M, Ganeshaiah KN, Shaanker RU. Assessing species admixtures in raw drug trade of Phyllanthus, a hepato-protective plant using molecular tools. J Ethnopharmacol. 2010;130:208–215. doi: 10.1016/j.jep.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Staudacher K, Wallinger C, Schallhart N, Traugott M. Detecting ingested plant DNA in soil-living insect larvae. Soil Biol Biochem. 2011;43:346–350. doi: 10.1016/j.soilbio.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stech M, Kolvoort E, Loone MJ, Vrieling K, Kruijer JD. Bryophyte DNA sequences from faeces of an arctic herbivore, barnacle goose (Branta leucopsis) Mol Ecol Res. 2011;11:404–408. doi: 10.1111/j.1755-0998.2010.02938.x. [DOI] [PubMed] [Google Scholar]
- Stoeckle M. Taxonomy, DNA, and the bar code of Life. Bioscience. 2003;53:796. doi: 10.1641/0006-3568(2003)053[0796:TDATBC]2.0.CO;2. [DOI] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F, Gielly L, Cruaud C, Nascetti G, Wincker P, Swenson JE, Taberlet P. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol Res. 2009;9:51–60. doi: 10.1111/j.1755-0998.2008.02352.x. [DOI] [PubMed] [Google Scholar]
- Yang HQ, Dong YR, Gu ZJ, Liang N, Yang JB. A Preliminary Assessment of matK, rbcL and trnH—psbA as DNA Barcodes for Calamus (Arecaceae) Species in China with a note on ITS. Ann Bot Fenn. 2012;49:319–330. doi: 10.5735/085.049.0603. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.