Abstract
Protein degradation is an essential and highly regulated process. The proteasomal degradation of the tumor suppressors p53 and p73 is regulated by both polyubiquitination and by an ubiquitin-independent process. Here, we show that this ubiquitin-independent process is mediated by the 20S proteasomes and is regulated by NQO1. NQO1 physically interacts with p53 and p73 in an NADH-dependent manner and protects them from 20S proteasomal degradation. Remarkably, the vast majority of NQO1 in cells is found in physical association with the 20S proteasomes, suggesting that NQO1 functions as a gatekeeper of the 20S proteasomes. We further show that this pathway plays a role in p53 accumulation in response to ionizing radiation. Our findings provide the first evidence for in vivo degradation of p53 and p73 by the 20S proteasomes and its regulation by NQO1 and NADH level.
Keywords: NQO1, p73, p53, 20S proteasome, protein degradation, NADH
Protein degradation determines the outcome of many cellular physiological processes (Coux et al. 1996). Degradation of proteins by the proteasomes occurs via various pathways (Verma and Deshaies 2000; Pickart and Cohen 2004). The most intensely studied one is the ubiquitin-26S proteasome pathway (Hershko 1996; Hershko and Ciechanover 1998; Goldberg 2003). The tumor suppressor p53 is a very labile protein that undergoes Mdm2 and ubiquitin-dependent 26S proteasomal degradation (Haupt et al. 1997; Kubbutat et al. 1997). Recently, we reported that degradation of p53 also occurs in an Mdm2 and ubiquitin-independent manner (Asher et al. 2002b). This pathway of p53 degradation is regulated by NAD(P)H quinone oxidoreductase 1 (NQO1) (Asher et al. 2001, 2002a,b, 2003, 2004), yet the underlying molecular mechanisms that control p53 degradation remained elusive.
Results and Discussion
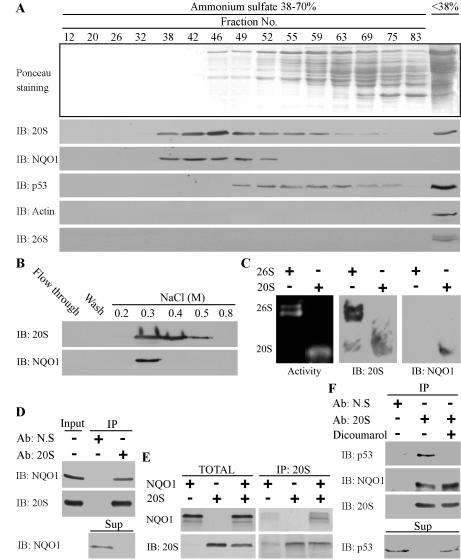
To investigate the role of NQO1 in proteasomal degradation, we followed NQO1 distribution in fractionated mouse liver extracts. Ammonium sulfate precipitation and gel-filtration chromatography of liver extracts revealed that the majority of NQO1 cofractionated with the 20S proteasomes (Fig. 1A). These fractions are devoid of the 26S proteasomes that were excluded by the differential ammonium sulfate precipitation (Fig. 1A, IB: 26S, anti TBP1 a subunit of the 19S). These results suggest that the vast majority of cellular NQO1 is found in a large protein complex that possibly includes the 20S proteasomes.
Figure 1.
NQO1 is physically associated with 20S but not 26S proteasomes. (A) Mouse liver extract was precipitated at 38%-70% ammonium sulfate and subjected to Sepharose 6B gel-filtration chromatography. Fractions were collected and analyzed by SDS-PAGE and immunoblotting (IB). (B) Fractions from Sepharose 6B gel-filtration column containing 20S proteasomes were loaded on Resource Q anion-exchange column and eluted with different concentrations of NaCl. Fractions were analyzed by SDS-PAGE and immunoblotting (IB). (C) Purified 26S and 20S proteasomes (0.3 M NaCl fraction) were subjected to nondenaturing PAGE and analyzed for peptidase activity or by immunoblotting (IB). (D) Fractions from Sepharose 6B gel-filtration column containing 20S proteasomes were pooled (Input), and immunoprecipitation experiments were performed with mouse anti-Flag antibody as a control for nonspecific binding (Ab: N.S) or with rabbit anti-C9 antibody (Ab: 20S). The immunoprecipitants (IP) and the supernatant (Sup) were analyzed by SDS-PAGE and immunoblotting (IB) (E) 35S-labeled NQO1 was incubated alone or mixed together with 20S proteasomes (TOTAL). The 20S proteasomes were immunoprecipitated with rabbit anti-C9 antibody (IP: 20S). (F) Fractions from Sepharose 6B gel-filtration column containing 20S proteasomes were pooled and immunoprecipitation experiments were performed with mouse anti-Flag antibody as a control for nonspecific binding (Ab: N.S) or with rabbit anti-C9 antibody (Ab: 20S) in the absence (-) or presence (+) of 200 μM dicoumarol. The immunoprecipitants (IP) and the supernatant (Sup) were analyzed by SDS-PAGE and immunoblotting (IB). Immunoblot analysis (IB) was performed with rabbit anti-C9 antibody to identify the 20S proteasomes, rabbit anti-TBP1 antibody to identify the 26S proteasomes, goat anti-NQO1 antibody, mouse anti-mouse p53 antibody, and with mouse anti-Actin. 35S-labeled NQO1 was detected by autoradiography. Proteasome peptidase activity was determined by their ability to hydrolyze the flurogenic peptide suc-LLVY-AMC, as described in Materials and Methods.
To further study this possibility, the 20S-containing fractions were pooled and fractionated by anion exchange chromatography according to a standard 20S purification protocol (Friguet et al. 2002). Remarkably, NQO1 was detected in the 0.3 M NaCl fraction containing the 20S proteasomes (Fig. 1B). Electrophoresis of the 0.3 M NaCl fraction on a nondenaturing PAGE, followed by peptidase activity assay, showed that the purified 20S is functional (Fig. 1C, Activity panel). Immunoblot analysis with anti NQO1 antibody revealed that NQO1 comigrated with the 20S proteasomes, but not with the 26S proteasomes (Fig. 1C). Finally, a coimmunopercipitation experiment showed that NQO1 coimmunopreciptated with 20S proteasomes (Fig. 1D). Altogether, these results suggest that NQO1 and 20S proteasomes are in physical association. Immunoprecipitation of the 20S proteasomes depleted most of the NQO1 from the supernatant (Fig. 1D), further indicating that the vast majority of cellular NQO1 is bound to the 20S proteasomes. Consistently, reticulocyte lysate translated 35S-labeled NQO1-bound purified 20S proteasomes in vitro (Fig. 1E).
Analysis of p53 distribution in the gel-filtration fractions showed that a portion of p53 cofractionated with 20S and NQO1 (Fig. 1A). Furthermore, both NQO1 and p53 were brought down following immunoprecipitation of the 20S proteasomes, indicating the existence of a triplet p53-NQO1-20S complex (Fig. 1F).
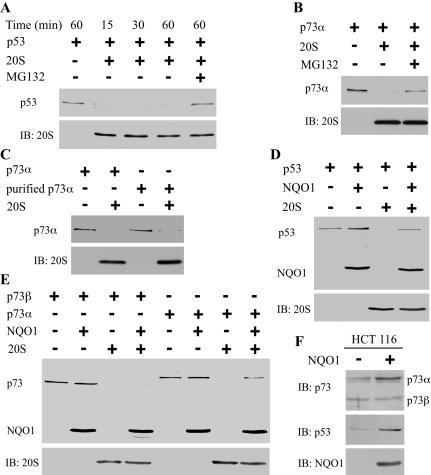
The observed association of NQO1 with 20S proteasomes and our previous findings that NQO1 regulates p53 and p73 degradation prompted us to investigate the role of 20S proteasomes in degradation of these proteins in vitro. To this end, reticulocyte lysate-translated 35S-labeled p53 was incubated with purified 20S proteasomes and the level of 35S-labeled p53 was determined by SDS-PAGE and autoradiography. A substantial amount of p53 was degraded already after 15 min, and this was inhibited in the presence of the proteasome inhibitor MG132 (Fig. 2A). Similar results were obtained with 35S-labeled p73α (Fig. 2B). p53 and p73 degradation was not observed in the presence of purified 26S proteasomes (data not shown). To rule out the involvement of reticulocyte lysate components in degradation by the 20S proteasomes, the 35S-labeled p73 was first immunoaffinity purified (Supplementary Fig. 1) and then subjected to degradation by the 20S proteasomes. Under these conditions, a similar degree of p73 degradation was observed (Fig. 2C).
Figure 2.
NQO1 selectively protects p73α, but not p73β, from 20S proteasomal degradation. (A) 35S-labeled p53 was incubated without (-) or with (+) 20S proteasomes at 37°C for 15, 30, and 60 min, without (-) or with (+) 50 μM MG132. (B) 35S-labeled p73α was incubated without (-) or with (+) 20S proteasomes without (-) or with (+) 50 μM MG132 at 37°C for 1 h. (C) 35S-labeled Flag-p73α or immunoaffinity-purified 35S-labeled Flag-p73α were incubated without (-) or with (+) 20S proteasomes at 37°C for 1 h. (D) 35S-labeled p53 was incubated in the presence of 1 mM NADH without (-) or with (+) 20S proteasomes, without (-) or with (+) 35S-labeled NQO1 (E) 35S-labeled p73α or p73β were incubated in the presence of 1 mM NADH without (-) or with (+) 20S proteasomes, without (-) or with (+) 35S-labeled NQO1. 35S-labeled p73 and NQO1 were analyzed by SDS-PAGE and detected by autoradiography. The 20S proteasomes were detected by immunoblot analysis (IB) with rabbit anti C9 antibody. (F) Cell extracts of HCT116 cells and HCT116 cells that stably express pEFIRES NQO1 were fractionated by SDS-PAGE; immunoblot analysis (IB) was carried out with mouse anti-p53, with rabbit anti-p73 antibody, and with goat anti-NQO1 antibody.
Next, we addressed the involvement of NQO1 in 20S proteasomal degradation. The in vitro degradation assays were repeated in the absence or presence of in vitro-translated NQO1. Remarkably, p53 and p73α degradation by the 20S proteasomes was inhibited in the presence of excess NQO1 and NADH, a cofactor of NQO1 (see below), suggesting that NQO1 directly regulates the degradation of these proteins by the 20S proteasomes (Fig. 2D,E). In contrast, 20S degradation of p73β, a p73 isoform that lacks the C-terminal SAM domain region, was not affected by NQO1 (Fig. 2E). This distinctive behavior was also observed in vivo in cells overexpressing NQO1 by accumulation of endogenous p73α and p53, but not of p73β (Fig. 2F). These observations are in agreement with the finding that NQO1 knockout mice display reduced levels of p73α and p53 (Long et al. 2002).
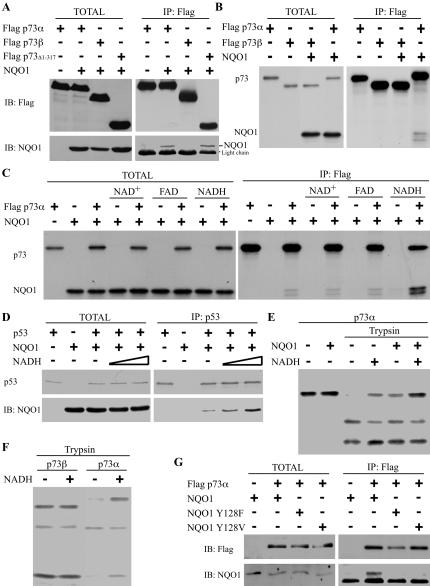
The observation that p73β degradation is NQO1 refractory led us to investigate the requirement of the C-terminal SAM domain region in p73α stabilization by NQO1. We and others have previously reported that NQO1 binds p53 (Anwar et al. 2003; Asher et al. 2003). To examine whether NQO1 binds p73α through its unique C-terminal region, Flag-p73α, Flag-p73β, and Flag-p73Δ1-317 were expressed either alone or together with NQO1 in 293 human kidney (HEK) cells; cell extracts were subjected to immunoprecipitation with an anti-Flag antibody. NQO1 coimmunoprecipitated with wild-type p73α and with the N-terminal truncated p73Δ1-317 mutant, but not with p73β (Fig. 3A). Similar results were obtained using in vitro binding assays with 35S-labeled NQO1, p73α, and p73β (Fig. 3B). Thus, the C-terminal SAM domain region of p73α is required for direct binding to NQO1 both in vivo and in vitro. These results suggest that the interaction between NQO1 and p73α protect p73α from degradation by the 20S proteasomes.
Figure 3.
NQO1 binds the SAM domain of p73α in an NADH-dependent manner. (A) 293 HEK cells were transiently transfected with pEFIRES Flag-p73α, pEFIRES Flag-p73β, or pEFIRES Flag-p73Δ1-317 without (-) or with pEFIRES-NQO1 (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). (B) In vitro 35S-labeled Flag-p73α or Flag-p73β were incubated alone or mixed together with 35S-labeled NQO1 (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). (C) 35S-labeled Flag-p73α and NQO1 were incubated alone or mixed together without or with NAD+, FAD, or NADH (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). (D) In vitro 35S-labeled p53 was incubated alone (-) or together with (+) recombinant NQO1 in the absence (-) or presence of 100 μM or 1 mM NADH. p53 was immunoprecipitated with mouse anti-human p53 antibody (IP: p53). (E)In vitro 35S-labeled Flag-p73α was incubated without (-) or with (+) 1 mM NADH, in vitro-translated NQO1 or 1 mM NADH together with in vitro-translated NQO1, and partially digested with trypsin. (F) In vitro 35S-labeled Flag-p73α or Flag-p73β was incubated without (-) or with (+) 1 mM NADH and partially digested with trypsin. (G) 293 HEK cells were transiently transfected with pEFIRES Flag-p73α alone or together with pEFIRES NQO1, pEFIRES NQO1-Y128F, or pEFIRES NQO1-Y128V (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). Immunoblot analysis (IB) was carried out with mouse monoclonal anti-Flag antibody and with goat anti-NQO1 antibody. 35S-labeled NQO1, p53, and p73 were detected by autoradiography.
Next, we performed in vitro binding assays in the presence of the NQO1 cofactors NAD, FAD, or NADH. A remarkable increase in the binding of NQO1 to p73α was observed only in the presence of NADH (Fig. 3C). Similarly, the binding of NQO1 to p53 was increased in the presence of NADH in a dose-dependent manner (Fig. 3D). To test the possible effect of NQO1 and NADH on p73 conformation, we performed partial trypsin digestion experiments. Partial trypsin digestion of 35S-labeled p73α was inhibited in the presence of NADH or NQO1, and to a greater extent, in the presence of both NADH and NQO1 (Fig. 3E). These results suggest that NQO1 introduces a conformational change in p73α that is augmented in the presence of NADH. The effect of NADH alone may be due to NQO1 found in the lysate (data not shown), however, a direct effect of NADH on p73α conformation cannot be ruled out. Digestion of p73α, but not p73β, was inhibited in the presence of NADH, indicating that this effect is mediated via the p73α C-terminal SAM domain region (Fig. 3F). To examine the requirement of NADH for NQO1 binding to p73α in vivo, we utilized both genetic and pharmacological approaches. Tyr 128 of NQO1 is peripherally involved in the binding of NADH (Ma et al. 1992). Therefore, we examined the ability of p73α to bind two different NQO1 mutants with reduced NADH-binding capacity, NQO1-Y128F and NQO1-Y128V (Ma et al. 1992). In contrast to wild-type NQO1, neither NQO1-Y128F nor NQO1-Y128V coimmunoprecipitated with p73α in 293 HEK cells (Fig. 3G), confirming the requirement of NQO1-associated NADH for binding to p73α in vivo.
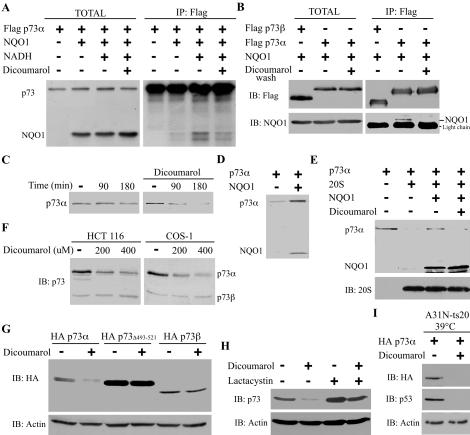
Dicoumarol, a drug that competes specifically with NADH for binding to NQO1 (Hosoda et al. 1974), was used to further confirm the requirement of NADH for the p73-NQO1 interaction. The in vitro binding assay with 35S-labeled NQO1 and p73α was performed in the absence or presence of NADH, and without or with dicoumarol. The binding of p73α to NQO1 was increased in the presence of NADH and decreased upon addition of dicoumarol (Fig. 4A). To recapitulate these experiments in cells, coimmunoprecipitated p73α-NQO1 complex was washed with dicoumarol before its elution and separation on SDS-PAGE. The level of NQO1 associated with p73α was strongly reduced by dicoumarol (Fig. 4B), suggesting that dicoumarol dissociates the preformed p73α-NQO1 complex. Similar results were obtained with p53 (see Fig. 5A).
Figure 4.
Dicoumarol disrupts the binding of NQO1 to p73α and induces ubiquitin-independent proteasomal degradation of p73α.(A) 35S-labeled Flag-p73α was incubated alone or with NQO1 in the absence (-) or presence of (+) NADH or NADH together with 300 μM dicoumarol (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). (B) 293 HEK cells were transiently transfected with pEFIRES Flag-p73β or pEFIRES Flag-p73α together with pEFIRES NQO1 (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). The beads were washed without (-) or with (+) 300 μM dicoumarol (dicoumarol wash). (C) 35S-labeled p73α was incubated in reticulocyte lysate degradation mixture at 37°C for 90 and 180 min in the absence or presence of 300 μM dicoumarol. (D) 35S-labeled Flag-p73α was incubated alone or together with 35S-labeled NQO1 in reticulocyte lysate degradation mixture at 37°C for 90 min. (E) 35S-labeled p73α was incubated in the presence of NADH without (-) or with (+) 20S proteasomes without (-) or with (+) 35S-labeled NQO1 and without (-) or with (+) 200 μM dicoumarol. (F) HCT116 or COS 1 cells were cultured without (-) or with 200 or 400 μM dicoumarol for 5 h. (G) HCT116 cells were transiently transfected with pSG5 HA-p73α, pSG5 HA-p73Δ493-521, or pSG5 HA-p73β, and 24-h post transfection, cells were cultured without (-) or with (+) 300 μM dicoumarol for 5 h. (H) HCT116 cells were cultured without (-) or with (+) 200 μM dicoumarol and without (-) or with (+) 50 μM lactacystin for 5 h. (I) A31N-ts20 cells were transiently transfected with pSG5 HA-p73α, incubated for 24 h at the restrictive temperature (39°C), and then cultured for 5 h without (-) or with (+) 300 μM dicoumarol. Immunoblot analysis (IB) was carried out with the following antibodies: mouse anti-Flag, mouse anti-HA, goat anti-NQO1, rabbit anti-p73, mouse anti-mouse p53, mouse anti-Actin, and rabbit anti-C9 to identify the 20S proteasomes. 35S-labeled p73 and NQO1 were detected by autoradiography.
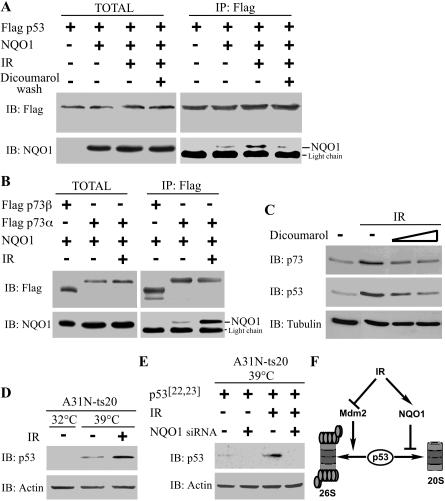
Figure 5.
NQO1 stabilizes p73α and p53 following γ-irradiation. (A) 293 HEK cells were transiently transfected with pRc/CMV Flag-p53 without (-) or with (+) pEFIRES NQO1. Twenty-four hours following transfection, cells were γ-irradiated at 4 Gy (IR) and cultured for an additional 4 h (TOTAL). Flag-p53 was immunoprecipitated with anti-Flag beads (IP: Flag). The beads were washed without (-) or with (+) 300 μM dicoumarol (dicoumarol wash). (B) 293 HEK cells were transiently transfected with pEFIRES Flag-p73β or pEFIRES Flag-p73α, together with pEFIRES NQO1. Twenty-four hours following transfection, cells were γ-irradiated at 4 Gy (IR) and cultured for an additional 4 h (TOTAL). Flag-p73 was immunoprecipitated with anti-Flag beads (IP: Flag). (C) HCT116 cells were γ-irradiated at 6 Gy (IR) and cultured without (-) or with 200 or 400 μM dicoumarol for 4 h. (D) A31N-ts20 cells were incubated for 24 h at the permissive (32°C) or restrictive (39°C) temperature. Cells cultured at 39°C were then γ-irradiated at 4 Gy (IR) and cultured for additional 4 h at 39°C. (E) A31N-ts20 cells were transiently transfected with pRc/CMV human p53[22,23] without or with pSUPER NQO1 encoding NQO1-specific siRNA. Cells were then incubated for 24 h at 39°C, γ-irradiated at 4 Gy (IR), and cultured for an additional 4 h at 39°C. (F) A schematic model of p53 accumulation following IR. Protein extraction and immunoblot analysis (IB) were carried out as described in the Materials and Methods with mouse anti-mouse p53, mouse anti-human p53, mouse anti-Flag antibody, goat anti-NQO1 antibody, rabbit anti-p73, and mouse anti-Actin or anti-Tubulin antibodies.
Our findings suggest that the NADH-dependent binding of NQO1 to p73α protect p73α from 20S proteasomal degradation. Since dicoumarol dissociates the NQO1-p73α complex, it is expected to promote p73α degradation. Indeed, in vitro degradation of 35S-labeled p73α was enhanced in the presence of dicoumarol (Fig. 4C). Conversely, p73α was stabilized in the presence of NQO1 (Fig. 4D), but not by NQO1-Y128F or NQO1-Y128V mutants that did not bind p73α (Supplementary Fig. 2). Furthermore, in the presence of dicoumarol, NQO1 no longer protected p73α from degradation by the 20S proteasomes (Fig. 4E). The effect of dicoumarol was also tested in vivo. Treatment of either HCT116 human colon carcinoma cells or COS-1 cells with dicoumarol resulted in a dose-dependent decrease in the level of endogenous p73α, but not of p73β, which does not bind NQO1 (Fig. 4F). Consistently, dicoumarol induced degradation of HA-p73α, but not of HA-p73β or HA-p73Δ493-521, a deletion mutant that lacks the C-terminal SAM domain expressed in 293 HEK cells (Fig. 4G). The decrease in the level of p73α following dicoumarol treatment was prevented by the presence of the proteasome inhibitor lactacystin (Fig. 4H). Finally, by using the A31N-ts20 BALB/c cell line that harbors a temperature-sensitive E1 variant and, therefore, defective in polyubiquitination at the restrictive temperature (Chowdary et al. 1994), we show that dicoumarol-induced p73α and p53 degradation is ubiquitin independent (Fig. 4I; Asher et al. 2002b), supporting the possibility that this process is executed in vivo by the 20S proteasomes.
Dicoumarol inhibits the accumulation of p53 following γ-irradiation (IR) and blocks p53-dependent apoptosis in mouse thymocites (Asher et al. 2001). To examine whether the binding of p53 and p73α to NQO1 is increased following IR, Flag-p53 or Flag-p73α and NQO1 were expressed in 293 HEK cells and cells were exposed to IR. Coimmunoprecipitation experiments revealed that under these conditions, the binding of p53 and p73α to NQO1 is increased (Fig. 5A,B). The increase in NQO1-p53 binding is NADH dependent, as the complex dissociated in the presence of dicoumarol (Fig. 5A). Dicoumarol treatment also reduced p53 and p73α accumulation following IR (Fig. 5C), suggesting that NQO1 binding is important for p53 and p73α stabilization under these conditions. The role of Mdm2 in ubiquitin and 26S proteasomal p53 degradation is well documented. Upon γ-irradiation, p53 undergoes post-translational modifications and escapes Mdm2-mediated degradation (Vogelstein et al. 2000). Interestingly, when A31N-ts20 cells were γ-irradiated under conditions defective in polyubiquitination, whereby Mdm2 can no longer promote p53 degradation (Asher et al. 2002b), the level of p53 following IR was further increased (Fig. 5D), suggesting that p53 accumulates independently of ubiquitin-mediated degradation. Furthermore, similar results were obtained with human p53[22,23], a p53 mutant that does not bind Mdm2 (Fig. 5E). Coexpression of NQO1-specific siRNA (Asher et al. 2002b) with human p53[22,23] prevented p53 accumulation following IR (Fig. 5E). Our results indicate that NQO1 plays a role in p53 and p73α accumulation following γ-irradiation. Escaping Mdm2-mediated degradation is probably not sufficient for efficient p53 stabilization following IR, because p53 is still susceptible to 20S proteasomal degradation. We propose that in order to achieve efficient p53 accumulation following γ-irradiation, NQO1-p53 interaction is increased to eliminate p53 degradation by the 20S proteasomes (Fig. 5F).
This study provides evidence for in vivo 20S proteasomal degradation of specific short-lived proteins. First, unlike the 26S proteasomal degradation, the pathway described here does not require polyubiquitination. Second, this pathway is responsive in vivo to inhibitors of the core 20S proteasomes, such as MG132 and lactacystin, but is not inhibited by glucoseamine (data not shown), an inhibitor of the 26S proteasomes (Zhang et al. 2003). Third, the observed in vivo process was fully and reliably recapitulated in vitro using purified 20S proteasomes. Under both conditions, p73α and p53 degradation was blocked by NQO1 in the presence of NADH and stimulated by dicoumarol. Recent experiments in our lab suggest that NQO1 also binds and stabilizes ornithine decarboxilase (ODC) in vivo (data not shown). Remarkably, all of these NQO1-binding proteins undergo 20S proteasomal degradation in vitro and are protected by NQO1. Unlike p53, p73α, and ODC, the stability of other short-lived proteins, such as the cyclin D1 and p27kip, was not affected by NQO1 (Supplementary Fig. 3).
Our findings suggest that NQO1 functions as a gatekeeper of the 20S proteasomes. NQO1 is associated with the 20S proteasomes and interacts with p73α and p53 in an NADH-dependent manner to protect them from 20S proteasomal degradation. Dicoumarol, by competing with NADH, abolishes the NQO1-p73α and p53 interaction and, therefore, sensitizes these proteins to degradation by the 20S proteasomes. Dicoumarol does not alter the binding of NQO1 to the 20S proteasomes (Supplementary Fig. 4). Indeed, treatment of the 20S-NQO1-p53 ternary complexes with dicoumarol resulted in dissociation only of p53, but not of NQO1 and 20S (Fig. 1F). The interaction of NQO1 with p53 and p73, potential 20S substrates, is modulated under various physiological conditions, such as IR (Fig. 5F).
Materials and methods
Cells and cell culture
The cell lines used were as follows: 293 human kidney cells (HEK), COS-1 monkey kidney, HCT116 human colon carcinoma, p53 null HCT116 cells, HCT116 stably transfected with NQO1 (Asher et al. 2001), M1-t-p53 mouse myeloid leukemic cells (Asher et al. 2001), and A31N-ts20, a BALB/c mouse cell line that harbors a temperature-sensitive E1 ubiquitin-activating enzyme (Chowdary et al. 1994). Cells were grown as previously described (Asher et al. 2002b).
Compounds
Dicoumarol (Sigma) was dissolved in 0.13N NaOH, lactacystin (Sigma) in DMSO. β-NADH, β-NAD+, and FAD (Sigma) were dissolved in water.
Plasmids and transfection
The plasmids used were as follows: pEFIRES and pSG5 NQO1 encoding wild-type human NQO1, pEFIRES and pSG5 NQO1-Y128F, pEFIRES and pSG5 NQO1-Y128V, pSUPER NQO1 encoding NQO1-specific siRNA, pSG5 HA-p73α encoding wild-type monkey p73, pSG5 HA-p73β, pSG5 HA-p73Δ493-521, pEFIRES Flag-p73α, pEFIRES Flag-p73β, pEFIRES Flag-p73Δ1-317, pRc/CMV p53 encoding wild-type human p53, and pRc/CMV p53[22,23]. Transient transfections of 293 human kidney cells were carried out by the calcium phosphate method. Transfection of A31N-ts20 cells was carried out with jetPEI, (Poly Transfection).
Immunoblot analysis
Cell extracts and immunoblot analysis were carried out as previously described (Asher et al. 2001). The antibodies used were as follows: goat anti-NQO1 C19 and R20 (Santa Cruz), mouse anti-human p53 (Pab 1801), mouse anti-mouse and human p53 (Pab 240), mouse anti-cyclin D1 (Santa Cruz), rabbit anti-p27kip antibody (Santa Cruz), rabbit anti-p73 (Asher et al. 2002b), rabbit anti-TBP1 (a subunit of the 19S), rabbit anti-C9 subunit of the 20S proteasomes (Mamroud-Kidron et al. 1994), mouse monoclonal anti-HA (Sigma), mouse monoclonal anti-Flag (Sigma), and mouse monoclonal anti-Actin (Sigma).
In vitro protein degradation assays
The in vitro degradation assay of in vitro reticulocyte lysate-translated [35S]methionine p73α was performed as previously described (Asher et al. 2002b). Degradation of in vitro-translated [35S]methionine p73 or p53 with 1 μg of purified 20S proteasome (from Sigma, or prepared by us, or kindly provided by Dr. Y. Reiss, Proteologies, Ltd., Rehovot, Israel) was carried out in degradation buffer (100 mM Tris-HCL at pH 7.5, 150 mM NaCl, 5 mM MgCl, 2 mM DTT), at 37°C for 1 h. Samples were mixed with Laemmli sample buffer, heated at 95°C for 5 min, and electrophoresed on SDS-PAGE. Following electrophoresis, proteins were transferred to cellulose nitrate membranes and detected by autoradiography. Purified p73α was generated by immunoprecipitation of in vitro-translated [35S]methionine Flag-p73α with Flag beads (Sigma), followed by elution with Flag peptide (Sigma).
Coimmunoprecipitation studies
Coimmunoprecipitation experiments from cell extracts and from in vitro35S-labeled proteins were carried out as previously described (Asher et al. 2003).
Trypsin digestion
Trypsin digestion of [35S]methionine Flag-p73 was carried out in digestion buffer (100 mM Tris-HCL at pH 7.5, 150 mM KCl, 0.1% NP40, 10% glycerol, 0.2 ng/μL trypsin [Sigma]), at 37°C for 10 min. Samples were then mixed with Laemmli sample buffer, heated at 95°C for 5 min, electrophoresed on SDS-PAGE, and detected by autoradiography.
Proteasome purification and analysis
Mouse livers were homogenized in buffer containing 20 mM Tris-HCL (pH 7.5), 1 mM EDTA, 1 mM DTT, and 250 mM sucrose. Following 38%-70% ammonium sulfate precipitation and centrifugations, the precipitate was dissolved in buffer containing 20 mM Tris-HCL (pH 7.5), 1 mM DTT, 20% glycerol, and loaded on a Sepharose 6B column. Fractions of 2 mL were collected and analyzed for the presence of NQO1 and 20S proteasomes by Western blot analysis. The proteasome-containing fractions were combined and loaded onto a Resource-Q column. Elution was performed with 0.35-0.4 M NaCl, and samples were concentrated and loaded on a 10%-40% glycerol gradient. The purified proteasomes were analyzed by SDS-polyacrylamide gels or by nondenaturing polyacrylamide gel (Glickman et al. 1998). Proteasome peptidase activity was determined by their ability to hydrolyze the flurogenic peptide suc-LLVY-AMC as previously described (Glickman et al. 1998).
Purification of recombinant NQO1
pET28-His-TEV-NQO1 was expressed in bacteria, and the bacteria were then lysed by sonication in 50 mM Tris-HCL (pH 7.5), 150 mM NaCl, and 1 mM PMSF. Soluble His-TEV-NQO1 was purified using a Ni-NTA column (HiTrap chelating HP) followed by gel-filtration chromatography (HiLoad 16/60 superdex 200). Purified His-TEV-NQO1 was further cleaved by TEV protease, and the His-TEV was removed upon binding to a Ni-NTA column.
Acknowledgments
We thank Dr. Y. Reiss for the purified 20S, S. Budilovsky and V. Reiss for their assistance, and Dr. J. Lotem and A. Cooper for their critical review and advice. This work was supported by a research grant from the Israel Academy of Sciences and Humanities to C.K. and to Y.S., and a grant to Y.S. from the Laub Fund for Oncogene Research.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.319905.
References
- Anwar A., Dehn, D., Siegel, D., Kepa, J.K., Tang, L.J., Pietenpol, J.A., and Ross, D. 2003. Interaction of human NAD(P)H:quinone oxidoreductase 1 (NQO1) with the tumor suppressor protein p53 in cells and cell-free systems. J. Biol. Chem. 278: 10368-10373. [DOI] [PubMed] [Google Scholar]
- Asher G., Lotem, J., Cohen, B., Sachs, L., and Shaul, Y. 2001. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc. Natl. Acad. Sci. 98: 1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Lotem, J., Kama, R., Sachs, L., and Shaul, Y. 2002a. NQO1 stabilizes p53 through a distinct pathway. Proc. Natl. Acad. Sci. 99: 3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Lotem, J., Sachs, L., Kahana, C., and Shaul, Y. 2002b. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. 99: 13125-13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Lotem, J., Tsvetkov, P., Reiss, V., Sachs, L., and Shaul, Y. 2003. P53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc. Natl. Acad. Sci. 100: 15065-15070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G., Lotem, J., Sachs, L., and Shaul, Y. 2004. p53-dependent apoptosis and NAD(P)H:quinone oxidoreductase 1. Methods Enzymol. 382: 278-293. [DOI] [PubMed] [Google Scholar]
- Chowdary D.R., Dermody, J.J., Jha, K.K., and Ozer, H.L. 1994. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14: 1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O., Tanaka, K., and Goldberg, A.L. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65: 801-847. [DOI] [PubMed] [Google Scholar]
- Friguet B., Bulteau, A.L., Conconi, M., and Petropoulos, I. 2002. Redox control of 20S proteasome. Methods Enzymol. 353: 253-262. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615-623. [DOI] [PubMed] [Google Scholar]
- Goldberg A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895-899. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya, R., Kazaz, A., and Oren, M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296-299. [DOI] [PubMed] [Google Scholar]
- Hershko A. 1996. Lessons from the discovery of the ubiquitin system. Trends Biochem. Sci. 21: 445-449. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover, A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67: 425-479. [DOI] [PubMed] [Google Scholar]
- Hosoda S., Nakamura, W., and Hayashi, K. 1974. Properties and reaction mechanism of DT diaphorase from rat liver. J. Biol. Chem. 249: 6416-6423. [PubMed] [Google Scholar]
- Kubbutat M.H., Jones, S.N., and Vousden, K.H. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299-303. [DOI] [PubMed] [Google Scholar]
- Long D.J., Gaikwad, A., Multani, A., Pathak, S., Montgomery, C.A., Gonzalez, F.J., and Jaiswal, A.K. 2002. Disruption of the NAD(P)H-:quinone oxidoreductase 1 (NQO1) gene in mice causes myelogenous hyperplasia. Cancer Res. 62: 3030-3036. [PubMed] [Google Scholar]
- Ma Q., Cui, K., Xiao, F., Lu, A.Y., and Yang, C.S. 1992. Identification of a glycine-rich sequence as an NAD(P)H-binding site and tyrosine 128 as a dicumarol-binding site in rat liver NAD(P)H:quinone oxidoreductase by site-directed mutagenesis. J. Biol. Chem. 267: 22298-22304. [PubMed] [Google Scholar]
- Mamroud-Kidron E., Omer-Itsicovich, M., Bercovich, Z., Tobias, K.E., Rom, E., and Kahana, C. 1994. A unified pathway for the degradation of ornithine decarboxylase in reticulocyte lysate requires interaction with the polyamine-induced protein, ornithine decarboxylase antizyme. Eur. J. Biochem. 226: 547-554. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. and Cohen, R.E. 2004. Proteasomes and their kin: Proteases in the machine age. Nat. Rev. Mol. Cell. Biol. 5: 177-187. [DOI] [PubMed] [Google Scholar]
- Verma R. and Deshaies, R.J. 2000. A proteasome howdunit: The case of the missing signal. Cell 101: 341-344. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane, D., and Levine, A.J. 2000. Surfing the p53 network. Nature 408: 307-310. [DOI] [PubMed] [Google Scholar]
- Zhang F., Su, K., Yang, X., Bowe, D.B., Paterson, A.J., and Kudlow, J.E. 2003. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115: 715-725. [DOI] [PubMed] [Google Scholar]