Abstract
The immunoglobulin heavy-chain (IgH) locus undergoes large-scale contraction in B cells poised to undergo IgH V(D)J recombination. We considered the possibility that looping of distinct IgH V regions plays a role in promoting long-range interactions. Here, we simultaneously visualize three subregions of the IgH locus, using three-dimensional fluorescence in situ hybridization. Looping within the IgH locus was observed in both B- and T-lineage cells. However, monoallelic looping of IgH V regions into close proximity of the IgH DJ cluster was detected in developing B cells with significantly higher frequency when compared with hematopoietic progenitor or CD8+ T-lineage cells. Looping of a subset of IgH V regions, albeit at lower frequency, was also observed in RAG-deficient pro-B cells. Based on these observations, we propose that Ig loci are repositioned by a looping mechanism prior to IgH V(D)J rearrangement to facilitate the joining of Ig variable, diversity, and joining segments.
Keywords: Immunoglobulin heavy-chain, V(D)J rearrangement, locus contraction, long-range interactions, lymphocytes, E2A-deficient hematopoietic progenitors
Eukaryotic chromosomes are assembled into higher-order structures that are tightly packaged inside the nucleus. Recent evidence strongly suggests that these complex chromatin structures are both dynamic and critical for appropriate regulation of gene expression. The organization of chromatin into a repressive or permissive state is regulated by noncoding elements such as promoters, enhancers, insulators, locus control regions, and matrix attachment regions (Baxter et al. 2002; Felsenfeld and Groudine 2003). These cis-acting elements are selectively occupied by tissue- and stage-specific transcription factors that often serve as docking sites for chromatin remodeling factors. The precise mechanism regulating long-range interactions of cis-acting elements remains to be elucidated. Recently, a looping model, in which regulatory elements separated by relative long distances in the β-globin locus are brought into physical proximity, with the intervening DNA looping out, has been suggested to establish an open chromatin domain and to activate β-globin expression (Carter et al. 2002; Tolhuis et al. 2002; Ostermeier et al. 2003).
In B cells, the gene encoding the immunoglobulin heavy chain (IgH) is assembled by combinatorial joining of variable (V), diversity (D), and joining (J) DNA segments (Jones and Gellert 2004; Jung and Alt 2004). This process, known as V(D)J recombination, is critically dependent on the expression of the recombination activating genes 1 and 2 (RAG-1 and RAG-2) (Mombaerts et al. 1992; Shinkai et al. 1992). In-frame IgH V(D)J rearrangements leads to the formation of the pre-B-cell receptor. Pre-BCR-mediated signaling inhibits RAG gene expression, preventing further IgH gene rearrangement. B cells at this stage undergo rapid cellular expansion, ultimately followed by growth arrest, induction of RAG gene expression, and Ig light-chain (IgL) gene rearrangement. Once a productive IgL chain has been formed, RAG gene expression and IgL rearrangements are inhibited in the absence of auto-reactivity.
Both the IgH and IgL loci span a large region, which, in principle, forms a mechanistic barrier for efficient Ig rearrangement. Elegant studies, using three-dimensional (3D) fluorescent in situ hybridization analysis have shown that Ig gene rearrangement correlates with alterations in the nuclear location of the IgH locus. Both IgH alleles are localized in the proximity of the nuclear membrane in non-B-lineage cells. In contrast, in B cells, the IgH locus is relocated to the center of the nucleus, where it also undergoes large-scale condensation (Kosak et al. 2002).
Rearrangement of the IgH gene is tightly regulated by the combined activities of various transcription factors, including E2A, Pax5, and EBF (Romanow et al. 2000; Fuxa et al. 2004; Seet et al. 2004). E2A and EBF have been shown to act in concert with RAG proteins to activate IgH DJ gene rearrangement when overexpressed in nonlymphoid cells (Romanow et al. 2000; Goebel et al. 2001). Furthermore, EBF has been shown to directly or indirectly promote IgH V(D)J recombination upon enforced expression in E47-deficient hematopoietic progenitor cells (Seet et al. 2004). B-cell development in Pax-5-deficient mice is blocked at an early stage and show impaired distal V-region usage (Urbanek et al. 1994; Schebesta et al. 2002; Hesslein et al. 2003; Fuxa et al. 2004). Additionally, recent studies have indicated that Pax-5 acts to promote long-range contraction of the IgH locus in B cells poised to undergo IgH V(D)J gene rearrangement (Fuxa et al. 2004). Here, we visualize three domains present in the IgH locus, using 3D fluorescence in situ hybridization. We detect looping of the IgH locus in developing B cells involving the juxtaposition of IgH V regions into proximity of the IgH D/J regions. Interestingly, looping of V regions was also observed in RAG-deficient pro-B cells. These data indicate that in developing B cells, IgH V regions loop in the proximity of the IgH D/J cluster prior to the onset of Ig gene recombination. We propose that prior to Ig gene rearrangement, Ig loci are repositioned by a looping mechanism to facilitate joining of Ig gene segments.
Results and Discussion
Long-range 3D complexity in lymphocytes
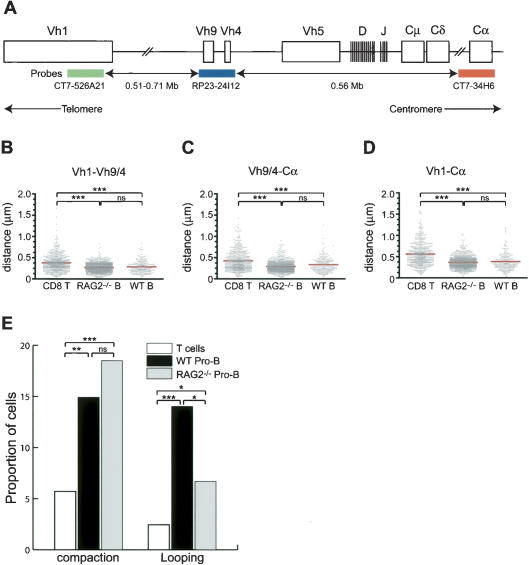
The murine IgH locus spans over 3 Mb of DNA, containing an estimated 200 V segments, 16 D segments, and four J segments. The V segments have been grouped in 15 families based on sequence identity (Mainville et al. 1996). The largest family, Vh1 (J558), contains at least 100 V members, occupies ∼1 Mb of DNA, and is located in the D-distal half of the V region. The Vh4 and Vh9 (gam3-8 and X-24) region is located ∼560 kb upstream of Ig Cα (Chevillard et al. 2002; Fig. 1A). To explore the mechanisms contributing to the large-scale contraction of the IgH locus observed in developing B cells (Baxter et al. 2002; Kosak et al. 2002), four-color 3D immunofluorescence in situ hybridization (3D FISH) was used to directly visualize three regions of the IgH locus. Three specific BAC probes were simultaneously used to label the 3′ region of Vh1 (green), the Vh 4 and Vh 9 (Vh9/4) region (blue), and the Ig Cα exons (red) (Fig. 1A). Lamin-specific antibodies were used to label the nuclear membrane (gray).
Figure 1.
Compaction of distinct subregions of the IgH locus. (A) The murine IgH locus and positions of the three BAC probes are indicated (not drawn to scale). The distances separating each of the three BAC probes—CT7-526A21, CT7-34H6, and RP23-24I12—and their positions within the IgH locus were determined using the Ensembl mouse genome database. Note that the exact location of the CT7-526A21 BAC remains to be determined. The colors of the three probes are shown as follows: CT7-526A21 (green), RP23-24I12 (blue), and CT7-34H6 (red). (B-D) Scatter-plots of the distances separating the V and Cα regions in T cells, in vitro-cultured wild-type, and RAG2-deficient pro-B cells. Y-axis indicates distances separating the loci in microns. Red line indicates the average distance in each group. (★★★) Significant difference (p < 0.0001) of the averages compared with that obtained from T-lineage cells. (E) Proportion of cells (Y-axis) in which the IgH locus is compacted or in a looping configuration in T cells (white), wild-type pro-B cells (black), or RAG2-/- pro-B cells (gray). (nd) Not determined; (★) p < 0.05; (★★) p < 0.01.
Four cell types were used in the following experiments: in vitro expanded pro-B cells derived from either wild-type mice or RAG2-deficient mice, in vitro-activated CD8+ splenic T cells, and E2A-deficient hematopoietic progenitor cells. Nuclei were hybridized with the Ig specific probes and analyzed by deconvolution microspcopy. A fraction of nuclei derived from pro-B cells did not show hybridization to either the Vh 9/4 or to both Vh1 and Vh9/4 probes. These were most likely derived from pro-B cells that had completed IgH V(D)J rearrangement, since such nuclei were not observed in RAG2-/- pro-B cells and T cells (data not shown). Only cells in which all three signals could be detected from both alleles were included in the analysis. To precisely calculate the distances separating each probe, the 3D coordinates of the center of mass of each probe were obtained by fitting each signal in each z-section into a polygon (Fig. 2D,G,J,M). Note that often only one out of two IgH allele are shown because of the optical sectioning. Polygons obtained for each probe were then interpolated into a 3D object for which the coordinates of the center of mass were calculated. This analysis revealed striking features of the interphase organization of the IgH locus in the various cell types.
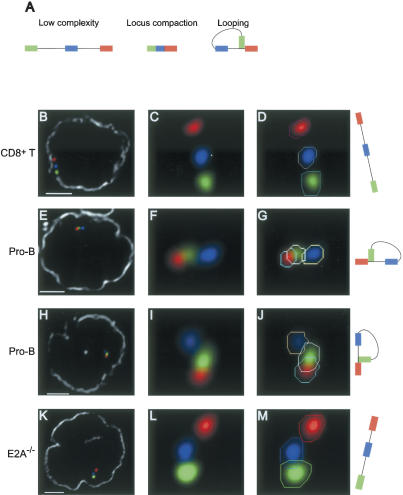
Figure 2.
Looping involving the distal IgH Vh1 and the Cα regions. (A) Schematic diagram of three possible distinct IgH configurations. (B-M) Three-dimensional configuration of the IgH locus resolved using four-color FISH. Digitally magnified pictures of IgH alleles are shown. Polygons (in one selected z-section) that were used to identify the coordinates of the center of mass for each signal are indicated (D,G,J,M). The nuclear membrane stained using lamin antibodies is shown in gray (B,E,H,K). Bar, 2 μm. (B-D) IgH locus in CD8+ T cells. (E-J) Looping involving the Vh1 and the Cα region in wild-type pro-B cells. (K-M) Configuration of the IgH locus in E2A-deficient hematopoietic progenitor cells.
The distances separating the Vh1 and Vh9/4 regions were significantly reduced in wild-type and RAG2-deficient pro-B cells (p < 0.001), representing a 41% and 31% contraction, respectively, when compared with the distance separating these probes in T cells (Fig. 1B; Table 1). Additionally, a significant long-range contraction (p < 0.001) of the average distance separating the Vh9/4 and Cα regions was observed in wild-type (27%) and RAG2-deficient pro-B cells (46%) when compared with that of T-lineage cells (Fig. 1C; Table 1). Consistently, we observed a smaller proportion of IgH alleles in pro-B cells showing distances >0.4 μm and an increase in the frequency of alleles showing a distance between 0 and 0.2 μm when compared with CD8+ T cells (Supplementary Fig. S1). Additionally, the relative distribution of distances separating each of the three regions appeared equally scattered in pro-B cells and T cells, suggesting that the IgH locus undergoes contraction in most pro-B cells (Supplementary Fig. S2).
Table 1.
Distances separating the IgH subregions, Vh1, Vh9/4, and Cα in pro-B cells, RAG-deficient pro-B cells, CD8+ T cells, and in E2A-deficient hematopoietic progenitor cells
| Pro-B | RAG2−/− | CD8+ T | E2A−/− | |
|---|---|---|---|---|
| n | 114+ | 372 | 245 | 200 |
| Vh1-Vh9/4 | 0.29 ± 0.14*** | 0.27 ± 0.11*** | 0.38 ± 0.20 | 0.33 ± 0.15 |
| Vh9/4-Cα | 0.33 ± 0.19*** | 0.29 ± 0.12*** | 0.42 ± 0.23 | 0.42 ± 0.23 |
| Vh1-Cα | 0.38 ± 0.21*** | 0.37 ± 0.14*** | 0.56 ± 0.28 | 0.51 ± 0.24 |
| Vh1-Vh9/4 + Vh9/4-Cα | 0.62 ± 0.25 | 0.56 ± 0.17 | 0.81 ± 0.31 | 0.74 ± 0.28 |
Shown are averages ± SD expressed in microns. (n) The number of cells analyzed for each cell type. (+) Some cells may have undergone Ig V(D)J recombination. (***) p < 0.001 when compared with CD8+ T cells.
It could be argued that IgH locus contraction in pro-B cells was caused by nuclear size differences. To address this question, the nuclear diameter of the cells was examined. The diameter of CD8+ T cells and pro-B cells derived from RAG-deficient mice is comparable, 7.47 ± 1.00 and 7.74 ± 0.83 μm respectively. In contrast, the diameter of wild-type pro-B cells was significantly reduced to 5.72 ± 0.62 μm when compared with pro-B cells derived from RAG-deficient mice or CD8+ T cells (Supplementary Table S1). To determine whether contraction of the IgH locus in pro-B cells derived from wild-type mice is caused by a reduction in nuclear size, we measured the distances separating two control probes, RP23-132L4 and RP23-478G17, located opposite of the Igh locus, at 12A1.3 on the arm of chromosome 12. We note that the region delineated by the two control probes comprises at least one housekeeping gene (Supplementary Fig. S3). Interestingly, the distances separating these loci, on average 1.76 Mb per μm, were equivalent in all three cell types (Supplementary Table S1). Taken together, these data indicate that locus contraction in developing B cells is not a general property of pro-B-cell nuclei.
If the spatial organization of the IgH locus were to be linear, then it would be expected that the sum of the distances separating the Vh1 and Vh9/4 probes and Vh9/4 and Cα probes would be equal to the distances separating the Vh1 and Cα regions. However, in all cell types analyzed, the distance separating the Vh1-Cα probes was substantially less than predicted from a linear configuration (Fig. 1D; Table 1). Taken together, these observations indicate that the IgH chain gene locus displays long-range 3D complexity in both T and B lymphocytes.
Condensation of the IgH locus in lymphoid cells
Two possible configurations that can account for long-range 3D complexity, locus compaction, and looping were distinguished from the low-complexity configuration and examined in detail (Fig. 2A). Both of the observations presented here and in previous studies showed large-scale contraction of the IgH locus in developing B cells (Kosak et al. 2002). To examine whether the reduction in distances reflects chromatin condensation, we used the following numerical criteria. Locus compaction was defined when the distance separating the Vh1 from the Vh9/4 regions was <0.2 μm and the distance separating Vh1 and Cα was <0.25 μm, representing at least a 2.5-fold contraction. Using these criteria, our analysis revealed that 5.7% of the CD8+ T cells contained alleles in a compacted conformation (Fig. 1E). In contrast, pro-B cells derived from RAG-deficient or wild-type mice contained significantly more alleles in a compacted state, the frequency being 18.5% and 14.9%, respectively (Figs. 1E, 3D-F). We note that we cannot rule out the possibility that wild-type pro-B cells contained alleles in which recombination occurred downstream of the Vh9/4 region.
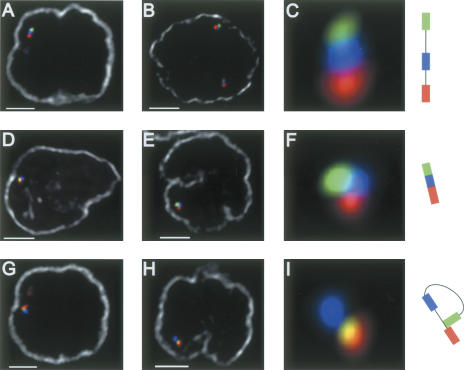
Figure 3.
Looping involving the IgH locus in RAG-deficient pro-B cells. Three distinct configurations of the IgH locus were detected Three-dimensional FISH in nuclei derived RAG deficient pro-B cells. (A-C) Low-complexity. (D-F) Locus compaction. (G-I) Looping. The nuclear membrane is shown in gray. Bar, 2 μm.
Looping involving the IgH locus in developing B cells
Upon visualizing the IgH locus in nuclei derived from pro-B cells and CD8+ T, we observed a substantial fraction of alleles, 24.6% and 19.6%, respectively, showing the Vh1 (labeled in green) region in closer proximity to the Cα (labeled in red) region relative to the Vh9/4 (labeled in blue) region. This observation suggested the presence of looped IgH domains in a large fraction of both B- and T-lineage cells (Supplementary Fig. S4). Interestingly, closer examination revealed that, in contrast to CD8+ T cells, a substantial proportion of looped IgH alleles derived from wild-type pro-B cells showed the Vh1 and Cα regions (labeled with green and red, respectively) localized in close proximity, with the two signals often overlapping, whereas the Vh9/4 (labeled in blue) region was distinctly separated (Fig. 2A,E-J).
In order to quantitatively determine the degree of looped structures in which the Vh1 regions was localized in close proximity to the Cα region and the IgH DJ cluster, we used the following numerical criteria: (1) The distance separating Vh1 and Cα is <0.2 μm, corresponding to ∼250 kb. The latter is determined from the distance separating the Vh9/4 from the Cα region, estimated at 1.28 Mb/μm, measured in CD8+ T cells, which represent a cell type in which the IgH locus is present in relatively “low-complexity uncompacted” 3D configuration. We note that the distance separating the IgH D cluster from the IgH Cα exons is ∼200 kb. Thus, 0.2 μm is consistent with localization of the Vh1 region in proximity to the IgH D cluster. (2) The distance separating Vh9/4 and Cα is greater than the distance separating Vh1 and Cα by at least 0.15 μm. Using these criteria, looping was observed in ∼14% of interphase nuclei of pro-B cells (Figs. 1E, 2E-J). We note that such loops occurred monoallelically (Supplementary Table S2). Using a BAC probe located at the telomeric border of the Vh1 region, we observed that this region also underwent looping in developing B cells, indicating that different Vh1 regions have the ability to form loops with DNA segments located in the proximity of the IgH Cα domain (data not shown).
Using the numerical criteria described above, such loops were also detected in interphase nuclei of CD8 T-lineage cells (Fig. 1E). However, the frequency of loops in T cells was significantly lower, 2.4% versus 14.0%, respectively, when compared with the one observed in pro-B cells (p < 0.0001) (Fig. 1E). Similarly, loops in E2A-deficient hematopoetic progenitor cells were detected at low frequency (1.5%, C. Sayegh, unpubl.). Taken together, these observations demonstrate that loops involving IgH V regions and DNA segments localized in close proximity to the IgH DJ cluster can be detected with relatively high frequency in developing B cells.
IgH locus undergoes looping in RAG-deficient B-lineage cells
In vitro studies have demonstrated that the RAG proteins have the ability to recruit recombination substrates into a synaptic complex. In this complex, RAG-mediated DNA double-stranded breaks are introduced and the broken ends subsequently fused by general DNA repair mechanisms (Eastman et al. 1996; Steen et al. 1996). Synapse formation has not been directly observed in B-lineage cells. Nevertheless, if such complexes occurred, then recruitment of V segments belonging to the Vh1 family into synaptic complexes could give rise to loop structures as detected by our assay. To determine whether looping of the Vh1 region occurs as a consequence of RAG-mediated synapse formation during IgH V-DJ rearrangement, we analyzed the 3D organization of the IgH locus in interphase nuclei derived from RAG2-deficient pro-B cells.
Interestingly, the fraction of pro-B nuclei derived from RAG2-deficient mice exhibiting looped alleles (6.7%), while significantly lower than that found in wild-type pro-B cells (p = 0.0197) (14.0%), was significantly greater than the fraction observed in CD8+ T cells (p = 0.0224) (2.4%) (Figs. 1E, 3G-I). It is conceivable that the difference in the proportion of looped alleles observed in wild-type pro-B cells and RAG2-/- pro-B cells is caused by the absence of D-J joints, which normally precede IgH rearrangement and/or by the deficiency in RAG activity. Nevertheless, these observations indicate that looping in the IgH locus occurs in B cells, poised to undergo rearrangement, even in the absence of RAG activity.
Looping of Ig variable regions in developing B cells
Recent studies have begun to elucidate the mechanisms by which long-range interactions of cis-regulatory elements promote the regulation of gene expression. A model has been proposed for the developmental regulation of expression of β-globin genes, where the repressive chromatin state is maintained by attaching regulatory regions to the nuclear matrix, thus preventing long-range promoter-enhancer interactions (Ostermeier et al. 2003). Activation of gene expression would require the expression of specific transcription factors, the acetylation of histones at hypersensitive sites, and finally, the clustering of distally located regulatory regions through a looping mechanism. The extended structure of the IgH V region, encompassing ∼2 Mb of DNA, is a potential barrier to the efficient synapsis and rearrangement of distally located elements. In support of this, Alt and colleagues have proposed a chromosomal proximity hypothesis, stating that the rearrangement mechanism preferentially favors genes that are in close proximity (Yancopoulos et al. 1984; Yancopoulos and Alt 1986). This is best demonstrated in pro-B cells derived from IL7R-deficient mice where a gradient of usage of V elements is observed; D-proximal V elements rearranging normally, whereas usage of D-distal V-segments is impaired (Chowdhury and Sen 2001). Such a mechanism has also been proposed to regulate rearrangement of the T-cell receptor β gene (Spicuglia et al. 2002). We propose that, similar to the regulation of β-globin gene expression, looping presents a mechanism by which elements that are physically separated by large distances are brought into proximity during Ig gene rearrangement.
Our observations indicate that in both B- and T-lineage cells, loops within the IgH locus can be detected with relative high frequency. However, the frequency of loops involving IgH V regions that are located in close proximity to the IgH DJ cluster is significantly higher in B cells as compared with T-lineage cells. Based on these observations, we propose that the IgH locus is organized in both B and T cells in looped-like structures. Prior to the onset of IgH gene rearrangements, IgH V-region segments are moved into close proximity of the DJ cluster to promote IgH V(D)J gene rearrangement.
Our observations also demonstrate that looping into close proximity of the IgH DJ cluster is monoallelic. It is conceivable that monoallelic looping contributes to allelic exclusion (Bergman and Cedar 2004). However, we note that our observations do not exclude the possibility that IgH V regions, distinct from the IgH Vh1, undergo looping on the second allele.
How are such loops organized and formed? It is plausible that looping of distinct Ig Vh regions may require the interactions of specific matrix attachement region (MAR)-binding proteins with MARs that are interspersed in the IgH locus (Goebel et al. 2002). Alternatively, reorganization of a matrix to which the IgH locus is tethered may regulate looping. It should now be possible using the strategy described here to examine how Ig loops are formed and how they are regulated.
Materials and Methods
Mice and cell culture
All mice used were maintained onto a C57BL/6 background. Pro B cells were generated from femoral bone marrow suspensions by positive enrichment of B220+ cells using magnetic separation (Miltenyi biotec) and cultured for 3-4 d in Optimem medium containing 10% fetal calf serum, 200 U/mL penicillin, 200 μg/mL streptomycin, 4 mM L-glutamine, and 50 μM β-mercaptoethanol supplemented with IL-7 and SCF at 2 ng/mL. CD8+ T cells were isolated form splenic cell suspensions by positive enrichment of CD8+ cells using magnetic separation. Cells were plated at 0.1 × 106 cells/mL in 24-well plates previously coated with anti-CD3ε antibody (2C11) and cultured for 3 d in RPMI 1640 containing 10% fetal calf serum, 200 U/mL penicillin, 200 μg/mL streptomycin, and 4 mM L-glutamine to which anti-CD28 antibodies were added. E2A-/- hematopoietic progenitors were grown as described previously (Ikawa et al. 2004).
3D immuno FISH
Approximately 40 μL of a 1 × 106 cells/mL suspension of cells grown in vitro was directly attached to coverslips. Cells were fixed in 4% paraformaldehyde for 10 min at room temperature (RT). Coverslips were treated with 0.1 M Tris-Cl (pH 7.2) for 10 min at RT and the cells were subsequently washed in 1× PBS. Cells were permeabilized for 10 min at RT with 1× PBS + 0.1% Triton X-100 + 0.1% saponin (WB1), and then incubated in 20% glycerol 1× PBS solution for 20 min. Coverslips were immersed in liquid nitrogen and allowed to thaw at RT. Two additional freeze-thaw cycles were performed. Coverslips were then washed once in 1× PBS and incubated at 37°C in 1× PBS, 0.1% Triton X-100, 5% bovine serum albumin (BB1). The nuclear membrane was stained using antiLamin A and B-specific antibodies (Santa Cruz) diluted to 2 μg/mL in BB1 for 30 min at 37°C. Coverslips were washed twice in WB1 for 10 min at RT. Cells were then incubated for 30 min at 37°C with biotinylated goat-specific donkey antibodies (Jackson Immunoresearch) diluted to 30 μg/mL in BB1 supplemented with normal donkey serum to a 5% v/v concentration. Coverslips were washed twice in WB1 for 10 min at RT and once in 1× PBS. Cells were subsequently fixed in a 2% PFA in 1× PBS solution for 10 min at RT. Coverslips were treated for 5 min in 0.1 M Tris-Cl (pH 7.2), and then washed once in 1× PBS. Cells were incubated in a 0.1 N HCl solution for 30 min at RT, washed once in 1× PBS, and treated with 0.1?g/L DNase-free RNase in BB1 for 1 h at 37°C. Coverslips were washed once in 1× PBS and in a 1× PBS, 0.5% Triton X-100, 0.5% saponin for 30 min at RT. Coverslips were then washed once in 1× PBS. The genomic DNA was denatured by incubating coverslips at 73°C in 2× SSC, 70% formamide solution for 2-1/2 min, followed by a 1-min incubation in 2× SSC, 50% formamide. Excess liquid was then rapidly dabbed and 10 μL of hybridization cocktail was added to each coverslip. Coverslips were mounted on slides, sealed with rubber cement, and incubated overnight at 37°C. In all experiments, the hybridization cocktail contained 400 ng of each labeled probe, 4 μg of mouse C0t DNA, 1 μg of sheared salmon-sperm DNA in 50% formamide, 2× SSC, 10% dextran sulfate. The probes were denatured for 5 min at 70°C and chilled on ice prior to their addition to the coverslips. BAC CT7-526A21 and CT7-34H6 were labeled using a nick-translation kit (Roche) with alexa-fluor 488-5 dUTP and alexa-fluor 568-5 dUTP, respectively (molecular probes). BAC RP23-24I12 was labeled with a digoxigenin (DIG) nick-translation kit (Roche). On the following day, coverslips were delicately removed from the slides, and then washed once in 2× SSC, 50% formamide for 15 min, and three times in 2× SSC 5 min each at 37°C. Coverslips were then incubated in 2× SSC, 3% BSA, 0.1% Tween-20 (BB2) for 30 min at RT. Dig-labeled probes and lamin antibodies were detected by incubating the cells with Cy5-conjugated mouse anti-DIG antibodies (Jackson Immunoresearch) at 20 μg/mL and Marina blue-conjugated neutravidin (Molecular probes) at 10 μg/mL in BB2 supplemented with normal mouse serum to 3% v/v for 30 min at 37°C. Following this, coverslips were washed twice in 2× SCC + 0.1% Tween-20 for 10 min each and once in 1× PBS at RT. Excess PBS was gently wiped out and coverslips mounted on slides.
Image acquisitions, distance calculations, and statistics
Images were captured with a DeltaVision deconvolution microscope system (Applied Precision, Inc.) located at the UCSD cancer center microscope facility. Using a 100× (NA 1.4) lens, images of ∼40 serial optical sections, spaced by 0.2 μm, were acquired. The data sets were deconvolved and optical sections merged to produce 3D pictures using SoftWorx software (Applied Precision, Inc) on a Silicon Graphics Octane workstation. The 3D coordinates of the center of mass of each probe were input into Microsoft Excel, and the distances separating each probe were calculated using the equation
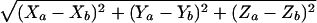 |
where X, Y, and Z are the coordinates of object a or b. In order to assess the different 3D structures, specific logical commands were written in Excel to specify each structure: (1) Locus compaction = AND(distance Vh1 to Vh9/4 < 0.2 μm, distance Vh1 to Cα < 0.25 μm). (2) Looping = AND(distance Vh1 to Cα < 0.2 μm, distance Vh9/4 to Cα - Vh1 to Cα > 0.15 μm).
In order to assess the statistical significance of the difference in distances between proB cells and CD8+ T cells, we performed an ordinary ANOVA test using nonparametric methods, as most data did not fit a Gaussian distribution. Dunn's tests were performed for multiple comparison post-tests. In order to assess the statistical significance of the difference in looping observed between RAG2-/- pro B cells and CD8+ T cells, contingency tables were established (Supplementary Table S3). All statistical tests were performed using Graphpad's Prism version 4.0.
Acknowledgments
We thank Karine Monier, James Feramisco, and Steve McMullen for valuable help in 3D FISH techniques, image acquisition, and data analysis. We are grateful to Isaac Engel, Albert Westerman, and Maho Niwa for comments on the manuscript. C.E.S. was supported by a postdoctoral fellowship from the Canadian Institute of Health Research (CIHR). This work was supported by grants from the NIH to C.M.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1254305.
References
- Baxter J., Merkenschlager, M., and Fisher, A.G. 2002. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 14: 372-376. [DOI] [PubMed] [Google Scholar]
- Bergman Y. and Cedar, H. 2004. A stepwise epigenetic process controls immunoglobulin allelic exclusion. Nat. Rev. Immunol. 4: 753-761. [DOI] [PubMed] [Google Scholar]
- Carter D., Chakalova, L., Osborne, C.S., Dai, Y.F., and Fraser, P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32: 623-626. [DOI] [PubMed] [Google Scholar]
- Chevillard C., Ozaki, J., Herring, C.D., and Riblet, R. 2002. A three-megabase yeast artificial chromosome contig spanning the C57BL mouse Igh locus. J. Immunol. 168: 5659-5666. [DOI] [PubMed] [Google Scholar]
- Chowdhury D. and Sen, R. 2001. Stepwise activation of the immunoglobulin μ heavy chain gene locus. EMBO J. 20: 6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M., Leu, T.M., and Schatz, D.G. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature 380: 85-88. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. and Groudine, M. 2003. Controlling the double helix. Nature 421: 448-453. [DOI] [PubMed] [Google Scholar]
- Fuxa M., Skok, J., Souabni, A., Salvagiotto, G., Roldan, E., and Busslinger, M. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes & Dev. 18: 411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P., Janney, N., Valenzuela, J.R., Romanow, W.J., Murre, C., and Feeney, A.J. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 194: 645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P., Montalbano, A., Ayers, N., Kompfner, E., Dickinson, L., Webb, C.F., and Feeney, A.J. 2002. High frequency of matrix attachment regions and cut-like protein x/CCAAT-displacement protein and B cell regulator of IgH transcription binding sites flanking Ig V region genes. J. Immunol. 169: 2477-2487. [DOI] [PubMed] [Google Scholar]
- Hesslein D.G., Pflugh, D.L., Chowdhury, D., Bothwell, A.L., Sen, R., and Schatz, D.G. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes & Dev. 17: 37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T., Kawamoto, H., Wright, L.Y., and Murre, C. 2004. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity 20: 349-360. [DOI] [PubMed] [Google Scholar]
- Jones J.M. and Gellert, M. 2004. The taming of a transposon: V(D)J recombination and the immune system. Immunol. Rev. 200: 233-248. [DOI] [PubMed] [Google Scholar]
- Jung D. and Alt, F.W. 2004. Unraveling V(D)J recombination; insights into gene regulation. Cell 116: 299-311. [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok, J.A., Medina, K.L., Riblet, R., Le Beau, M.M., Fisher, A.G., and Singh, H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158-162. [DOI] [PubMed] [Google Scholar]
- Mainville C.A., Sheehan, K.M., Klaman, L.D., Giorgetti, C.A., Press, J.L., and Brodeur, P.H. 1996. Deletional mapping of fifteen mouse VH gene families reveals a common organization for three Igh haplotypes. J. Immunol. 156: 1038-1046. [PubMed] [Google Scholar]
- Mombaerts P., Iacomini, J., Johnson, R.S., Herrup, K., Tonegawa, S., and Papaioannou, V.E. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68: 869-877. [DOI] [PubMed] [Google Scholar]
- Ostermeier G.C., Liu, Z., Martins, R.P., Bharadwaj, R.R., Ellis, J., Draghici, S., and Krawetz, S.A. 2003. Nuclear matrix association of the human β-globin locus utilizing a novel approach to quantitative real-time PCR. Nucleic Acids Res. 31: 3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanow W.J., Langerak, A.W., Goebel, P., Wolvers-Tettero, I.L., van Dongen, J.J., Feeney, A.J., and Murre, C. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell 5: 343-353. [DOI] [PubMed] [Google Scholar]
- Schebesta M., Heavey, B., and Busslinger, M. 2002. Transcriptional control of B-cell development. Curr. Opin. Immunol. 14: 216-223. [DOI] [PubMed] [Google Scholar]
- Seet C.S., Brumbaugh, R.L., and Kee, B.L. 2004. Early B cell factor promotes B lymphopoiesis with reduced Interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 199: 1689-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun, G., Lam, K.P., Oltz, E.M., Stewart, V., Mendelsohn, M., Charron, J., Datta, M., Young, F., Stall, A.M., et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68: 855-867. [DOI] [PubMed] [Google Scholar]
- Spicuglia S., Kumar, S., Yeh, J.H., Vachez, E., Chasson, L., Gorbatch, S., Cautres, J., and Ferrier, P. 2002. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol. Cell 10: 1479-1487. [DOI] [PubMed] [Google Scholar]
- Steen S.B., Gomelsky, L., and Roth, D.B. 1996. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells 1: 543-553. [DOI] [PubMed] [Google Scholar]
- Tolhuis B., Palstra, R.J., Splinter, E., Grosveld, F., and de Laat, W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10: 1453-1465. [DOI] [PubMed] [Google Scholar]
- Urbanek P., Wang, Z.Q., Fetka, I., Wagner, E.F., and Busslinger, M. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79: 901-912. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D. and Alt, F.W. 1986. Regulation of the assembly and expression of variable-region genes. Annu. Rev. Immunol. 4: 339-368. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D., Desiderio, S.V., Paskind, M., Kearney, J.F., Baltimore, D., and Alt, F.W. 1984. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature 311: 727-733. [DOI] [PubMed] [Google Scholar]