Abstract
In human cells, a critical pathway in gene regulation subjects mRNAs with AU-rich elements (AREs) to rapid decay by a poorly understood process. AREs have been shown to directly activate deadenylation, decapping, or 3′-to-5′ exonucleolytic decay. We demonstrate that enzymes involved in all three of these mRNA decay processes, as well as 5′-to-3′ exonucleolytic decay, associate with the protein tristetraprolin (TTP) and its homolog BRF-1, which bind AREs and activate mRNA decay. TTP and BRF-1 each contain two activation domains that can activate mRNA decay after fusion to a heterologous RNA-binding protein, and inhibit ARE-mediated mRNA decay when overexpressed. Both activation domains employ trans-acting factors to trigger mRNA decay, and the N-terminal activation domain functions as a binding platform for mRNA decay enzymes. Our data suggest that the TTP protein family functions as a molecular link between ARE-containing mRNAs and the mRNA decay machinery by recruitment of mRNA decay enzymes, and help explain how deadenylation, decapping, and exonucleolytic decay can all be independently activated on ARE-containing mRNAs. This describes a potentially regulated step in activation of mRNA decay.
Keywords: mRNA turnover, AU-rich elements, TTP, decapping, deadenylation, exonuclease
Gene expression can be regulated at many levels, including mRNA turnover. An important signal for rapid mRNA turnover in mammalian cells is the AU-rich element (ARE), which is found in the 3′UTR of many highly regulated mRNAs (Chen and Shyu 1995; Wilusz et al. 2001; Shim and Karin 2002). AREs are characterized by sequence regions rich in adenosines and uridines and often include one or more copies of the pentamer sequence AUUUA (Chen and Shyu 1995; Wilusz et al. 2001; Shim and Karin 2002). AREs are prevalent in mRNAs that encode proto-oncogenes, interleukins, and cytokines (Chen and Shyu 1995; Shim and Karin 2002). In addition, an extensive compilation of ARE-containing mRNAs has shown that AREs are found in mRNAs encoding many different classes of proteins (Bakheet et al. 2001). Importantly, while ARE-containing mRNAs are normally rapidly degraded, specific cell signals can trigger mRNA stabilization and result in protein production. For example, activation of T cells results in stabilization of the mRNA encoding the interleukin IL-2, which leads to IL-2 production and secretion (Shim and Karin 2002). The importance of AREs is underscored by the observation that the proto-oncogene c-fos can be transformed into an oncogene by the removal of the AREs from the c-fos mRNA 3′UTR (Miller et al. 1984).
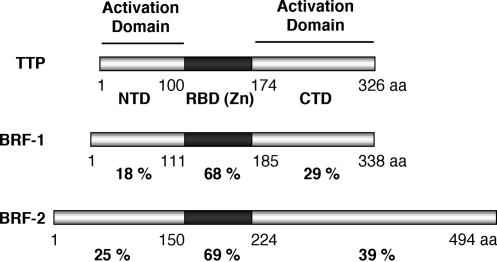
Several proteins that interact with AREs have been isolated. The most intensely studied of these is tristetraprolin (TTP) (Fig. 1; Blackshear 2002). TTP contains two CCCH-type zinc-finger domains that are necessary and sufficient for ARE binding (Lai et al. 2000). Several lines of evidence suggest that TTP is involved in ARE-mediated decay. For example, overexpression of TTP results in reduced accumulation of many, but not all, ARE-containing mRNAs (Lai and Blackshear 2001). In addition, a mouse, which is homozygous for a deletion of the TTP gene, accumulates in the blood stream high levels of TNF-α, a protein encoded by an ARE-containing mRNA (Taylor et al. 1996). Two paralogs of TTP involved in ARE-mediated decay are BRF-1 and BRF-2 (also named hsERF1/ZFP36L1/TIS11b and hsERF2/ZFP36L2/TIS11d, respectively) (Fig. 1; Lai et al. 2000; Stoecklin et al. 2002). Human HT1080 cells that contain a mutation in the BRF-1 gene show low levels of ARE-mediated decay that can be rescued by expression of exogenous BRF-1 or TTP (Stoecklin et al. 2002). In addition, similar to TTP, overexpression of BRF-1 and BRF-2 results in reduced accumulation of ARE-containing mRNAs (Lai et al. 2000; Stoecklin et al. 2002). In contrast to TTP, both BRF-1 and BRF-2 are essential for mouse early development (Taylor et al. 1996; Ramos et al. 2004; Stumpo et al. 2004).
Figure 1.
Schematic representation of TTP, BRF-1, and BRF-2. The RNA-binding zinc-finger domain is shown in black (RBD-Zn). The N-terminal (NTD) and C-terminal (CTD) mRNA decay activation domains defined in this study are indicated. The percent amino acid identity to TTP, of the NTD, RNA-binding domain, and CTD of BRF-1 and BRF-2 is given. (aa) Amino acids.
Other ARE-binding proteins have also been implicated in ARE-mediated decay. For example, overexpression of the ARE-binding protein hnRNP D/AUF1 can modulate the decay rate of ARE-containing mRNAs but shows destabilizing or stabilizing effects depending on the cell type (DeMaria and Brewer 1996; Xu et al. 2001; Raineri et al. 2004). Another ARE-binding protein, HuR, stabilizes ARE-containing mRNAs when overexpressed (Fan and Steitz 1998; Peng et al. 1998; Raineri et al. 2004). Finally, the ARE-binding protein KSRP is associated with mRNA decay enzymes, and its depletion leads to a decrease in ARE-mediated decay in human cells and cell extracts (Chen et al. 2001; Gherzi et al. 2004).
In Saccharomyces cerevisiae, most mRNAs targeted for mRNA decay are believed to undergo rapid deadenylation, followed by deadenylation-dependent decapping and exonucleolytic decay from the 5′ and 3′ ends (Tharun and Parker 2001; Parker and Song 2004). In contrast, several mRNA decay processes have been observed to be activated independently of each other by AREs in human mRNAs, including rapid deadenylation and 3′- to-5′ exonucleolytic decay in cells, as well as deadenylation, decapping, and 3′-to-5′ exonucleolytic decay in cell extracts (Chen et al. 1995, 2001; Voeltz and Steitz 1998; Ford et al. 1999; Gao et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002; Lai et al. 2003). These observations suggest that mRNA decay enzymes are separately activated to degrade ARE-containing mRNAs. However, it is currently unknown how this is accomplished in the human cell. Several human mRNA decay enzymes have been characterized. A decapping complex that includes the proteins hDcp2 and hDcp1a/1b (two paralogs exist) (Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002), a 5′-to-3′ exonuclease called hXrn1 (Bashkirov et al. 1997), and a complex of ≥10 3′-to-5′ exonucleases termed the exosome (Mitchell et al. 1997; Chen et al. 2001) were all identified on the basis of their similarity to S. cerevisiae mRNA decay enzymes. In addition, human deadenylases have been identified, including PARN (Korner and Wahle 1997) and hCcr4 (Chen et al. 2002; Chang et al. 2004; Cougot et al. 2004).
Although proteins and mRNA decay processes responsible for recognition and degradation of ARE-containing mRNAs have been studied in some detail, very little is known about the mechanism by which ARE-binding proteins, such as TTP, BRF-1, and BRF-2, activate the mRNA decay enzymes on target mRNAs in the human cell. This is a key step in mRNA decay, which is a potential target for regulation. Here we show evidence that activation of ARE-mediated decay by the TTP protein family involves direct recruitment of mRNA decay enzymes to ARE-containing mRNAs. We show that TTP and BRF-1 employ two activation domains that both trigger ARE-mediated mRNA decay using trans-acting factors, which in the case of the N-terminal activation domain includes mRNA decay enzymes responsible for decapping, deadenylation, and exonucleolytic decay from the 5′ and 3′ ends. This helps explain how several mRNA decay processes can be independently activated by AREs in the human cell. We propose that regulation of ARE-mediated mRNA decay may involve the modulation of these mRNA decay activation domains of TTP and BRF-1.
Results
Exogenously expressed TTP and BRF-1 exist in complex with mRNA decay enzymes
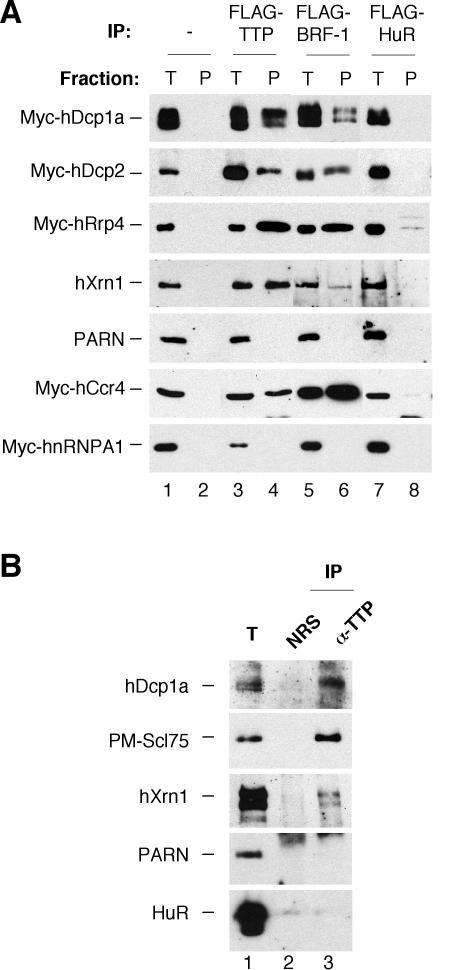
How can AREs activate several different mRNA degradation processes, including rapid deadenylation, decapping, and 3′-to-5′ exonucleolytic decay? We considered the possibility that ARE-binding proteins, such as TTP and BRF-1 (Fig. 1), function to recruit mRNA decay enzymes to target mRNAs. To address this question, we first tested whether TTP and BRF-1 exist in complex with mRNA decay enzymes responsible for these processes. The coimmunopurification assays in Figure 2A demonstrate that transiently expressed, myc-tagged decapping complex subunits, hDcp1a and hDcp2, as well as the 5′-to-3′ exonuclease hXrn1 and the deadenylase hCcr4, all coimmunopurify with Flag-tagged TTP and BRF-1 proteins from RNase-treated human embryonic kidney (HEK) 293T cell extracts (Fig. 2A, lanes 4,6). Moreover, a component of the human exosome, hRrp4, also copurifies with TTP and BRF-1, whereas the deadenylase PARN does not (Fig. 2A, lanes 4,6), which confirms earlier studies (Chen et al. 2001; Lai et al. 2003). None of the mRNA decay enzymes copurify at significant levels with the ARE-binding protein HuR (Fig. 2A, lane 8), which is not an activator of mRNA decay (Fan and Steitz 1998; Peng et al. 1998). In addition, no nonspecific binding to the anti-Flag resin was observed (Fig. 2A, lane 2). The RNA-binding hnRNP A1 protein, which associates with TTP and BRF-1 in a strictly RNA-dependent manner (data not shown), served as a control for efficient RNase treatment and does not copurify with TTP and BRF-1 (Fig. 2A, lower panel, lanes 4,6). We conclude that exogenously expressed TTP and BRF-1 are associated with mRNA decay enzymes in an RNA-independent manner in human cell extracts.
Figure 2.
TTP and BRF-1 exist in complex with mRNA decay enzymes. (A) Western blots showing coimmunoprecipitation assays. Transiently expressed Flag-tagged TTP, BRF-1, and HuR were immunoprecipitated from HEK 293T cell extracts, and precipitates were probed for the presence of coexpressed Myc-tagged or endogenous mRNA decay proteins as indicated. (Lanes 1,3,5,7) Five percent total extract (T). (Lanes 4,6,8) Immunoprecipitate pellets (P). (Lane 2) Immunoprecipitate from cells that do not express a Flag-tagged protein. (B) Western blots showing endogenous coimmunoprecipitation assays. (Lane 3) Endogenous TTP was immunoprecipitated from marmoset T-cell extracts, and the precipitate was probed for the presence of endogenous mRNA decay proteins as indicated. (Lane 1) Five percent total cell extract (T). (Lane 2) Immunoprecipitation reaction using normal rabbit serum (NRS) in place of anti-TTP antiserum.
Endogenous TTP exist in complex with mRNA decay enzymes
To test whether endogenous TTP exists in complex with endogenous mRNA decay enzymes in cell extracts, we used polyclonal antibodies in coimmunopurification assays from a marmoset T-cell line, which was previously shown to express TTP (Cook et al. 2004). The results in Figure 2B demonstrate that the mRNA decay enzymes hDcp1a, PM-Scl75 (a component of the exosome), and hXrn1 all copurify with endogenous TTP from RNase-treated marmoset T cell extracts. Again, no association was observed with PARN. The RNA-binding protein HuR served as a negative control and was not associated with TTP (Fig. 2B, bottom panel). These results confirm the observations with tagged exogenous proteins in Figure 2A. We conclude that the ARE-binding proteins TTP and BRF-1 associate, either directly or via unknown proteins, with mRNA decay enzymes responsible for decapping, deadenylation, and exonucleolytic decay from the 5′ and 3′ ends.
Overexpression of the decapping enzyme hDcp2 results in enhanced ARE-mediated decay
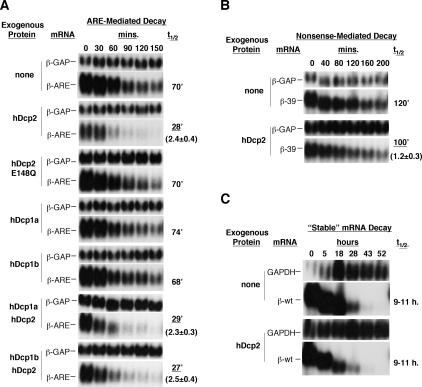
It was previously shown that deadenylation and 3′-to-5′ exonucleolytic decay are both activated by AREs in vitro and in human tissue culture cells (Chen et al. 1995, 2001; Voeltz and Steitz 1998; Ford et al. 1999; Gao et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002; Lai et al. 2003), and that decapping is activated in vitro (Gao et al. 2001). However, it remained to be established whether decapping is activated by AREs in the human cell. To address this question, we established an ARE-mediated mRNA decay assay in human HeLa Tet-off cells, using a β-globin reporter mRNA (β-ARE) that contains the potent ARE from GM-CSF mRNA in the 3′UTR. Transcription of the β-ARE mRNA is controlled by a tetracycline regulatory promoter, which allows for measurement of the mRNA decay rate in pulse-chase experiments in human HeLa Tet-off cells. A β-globin mRNA with an extended 3′UTR (β-GAP) (Lykke-Andersen et al. 2000), which is expressed from a constitutive CMV promoter, served as an internal control for the calculation of mRNA half-lives. The results in Figure 3 show that the ARE-containing β-globin mRNA (β-ARE; t½ = 70 min) (Fig. 3A) displays an eight- to ninefold enhanced mRNA decay rate compared with that of β-globin mRNA with no ARE (β-wt, t½ = 9-11 h) (Fig. 3C).
Figure 3.
Overexpression of the decapping enzyme, hDcp2, activates ARE-mediated mRNA decay. (A) Northern blots showing decay rates in HeLa Tet-off cells of a β-globin reporter mRNA (β-ARE) that contains an AU-rich element from GM-CSF mRNA in the 3′UTR, in the absence of coexpressed protein (none) or in the presence of coexpressed decapping complex subunits, as indicated on the left. Time points above the panels refer to minutes after transcriptional repression. β-ARE levels were normalized to the internal control β-GAP mRNA, and β-ARE half-lives were calculated and are shown on the right. The average fold of change in half-lives (three experiments) that were significantly different from the assay with no exogenous protein is given with standard deviation on the right. (B) Northern blots showing decay rates of an NMD β-globin reporter mRNA, in the absence or presence of coexpressed hDcp2. (C) Northern blots showing decay rates of wild-type β-globin mRNA, in the absence or presence of coexpressed hDcp2. The internal control was endogenous GAPDH mRNA.
We next wished to test the role of decapping enzymes in the decay of the β-ARE reporter mRNA. We were unsuccessful in establishing RNA interference conditions in HeLa Tet-off cells that reduce the level of the catalytic hDcp2 subunit of the decapping complex, even using an siRNA that has previously been successfully used in another human cell line (Lejeune et al. 2003). We therefore tested instead the effect of decapping enzyme overexpression. The results in Figure 3A show that overexpression of hDcp2 causes a two- to threefold reduction in the half-life of the β-ARE reporter mRNA (t½ = 28 min; 2.4 ± 0.4-fold reduction in three experiments). hDcp2 was overexpressed approximately two- to threefold as estimated by Western blotting (data not shown). Importantly, the effect of hDcp2 requires decapping activity, because a catalytically inactive hDcp2 mutant protein (E148Q) (Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002) was not capable of activating mRNA decay (t½ = 70 min). In contrast to hDcp2, overexpression of another decapping complex subunit hDcp1a, as well as its paralog hDcp1b, had no effect on decay of the β-ARE mRNA (t½ = 74 min and 68 min, respectively), even though they were expressed at three- to fivefold higher levels than endogenous proteins (data not shown). Moreover, the effect of hDcp2 was not enhanced by coexpressed hDcp1a or hDcp1b (Fig. 3A, two lower panels). Overexpression of hDcp2 had little or no effect on the decay rate of a β-globin mRNA that undergoes nonsensemediated decay (NMD) (Fig. 3B, β-39) or on wild-type β-globin mRNA (Fig. 3C, β-wt). We conclude that overexpression of hDcp2 enhances decay of the ARE-mediated decay reporter β-ARE mRNA. This suggests that decapping can be a limiting step in ARE-mediated decay in human cells. In contrast to hDcp2, the hDcp1a/hDcp1b subunit of the decapping complex appears to not be limiting in the cell.
The NTD of TTP is responsible for the association with mRNA decay enzymes
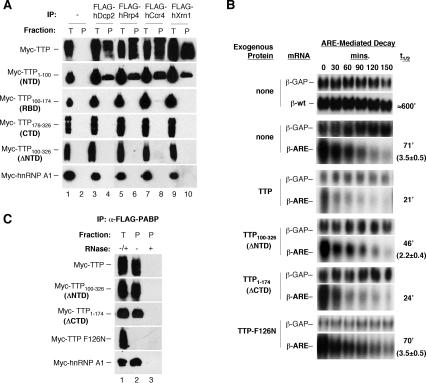
We next wished to test which regions of TTP are responsible for the observed association with individual mRNA decay enzymes. TTP, BRF-1, and BRF-2 each consist of an RNA-binding zinc-finger domain flanked by N-terminal (NTD) and C-terminal (CTD) domains of unknown function (Fig. 1A). We tested each of these domains for their ability to copurify with mRNA decay enzymes from RNase-treated human cell extracts. Remarkably, the coimmunoprecipitation assays in Figure 4A show that all of the decay enzymes tested, hDcp2, hRrp4, hCcr4, and hXrn1, copurify exclusively with the NTD of TTP (Myc-TTP1-100). In contrast, the zinc-finger domain (Myc-TTP100-174) and CTD (Myc-TTP176-326) are not found in complex with any of the mRNA decay enzymes tested. Importantly, deletion of the TTP NTD (Myc-TTP100-326) prevents association with the mRNA decay enzymes. Full-length TTP served as a positive control and was found associated with all decay enzymes, whereas the negative control, hnRNP A1, showed no association. TTP, and all C-terminal fragments thereof, appear as streaks in the immunoblots, probably due to phosphorylation (Chrestensen et al. 2004). We conclude that the NTD is both necessary and sufficient for the association of TTP with mRNA decay enzymes, whereas the TTP zinc-finger domain and CTD do not stably associate with any tested mRNA decay enzyme.
Figure 4.
The TTP NTD is necessary and sufficient for the association with mRNA decay enzymes and is important for optimal mRNA decay stimulation. (A) Western blots showing the association of TTP deletion mutant proteins with mRNA decay enzymes. (Lanes 4,6,8,10) Transiently expressed Flag-tagged hDcp2, hRrp4, hCcr4, or hXrn1 was immunoprecipitated from HEK 293T cell extracts and probed for the presence of coexpressed Myc-tagged TTP, TTP fragments, or hnRNP A1 as indicated. (Lanes 1,3,5,7,9) Five percent total cell extract (T). (Lane 2) Immunoprecipitates from HEK 293T cells with no Flag-tagged protein. (B) Northern blots showing decay rates of the β-ARE reporter mRNA, in the absence of coexpressed protein (none), or in the presence of TTP, TTP100-326, TTP1-174, or TTP-F126N as indicated. Time points above the panels refer to minutes after transcriptional repression. β-ARE half-lives were calculated and are shown on the right, together with the average fold of change in half-lives, with standard deviation (three experiments) for those that were significantly different from those in the presence of TTP. (C) Western blots showing the association of TTP and TTP mutant proteins with poly-A mRNA, isolated through association with Flag-PABP. (Lane 1) Five percent total extract (T). (Lanes 2,3) Anti-Flag-PABP immunoprecipitates after treatment without (-, lane 2) or with (+, lane 3) RNase A.
The NTD of TTP is important for full activation of mRNA decay
We next wished to test whether the NTD is important for TTP activity. TTP and TTP from which the NTD or CTD has been deleted were previously shown to efficiently bind ARE-RNA in vitro, and to promote reduced accumulation of ARE-containing mRNAs when expressed in human cells (Lai et al. 2000; Lai and Blackshear 2001). To directly test the effect of exogenously expressed TTP and TTP lacking the NTD on ARE-mediated mRNA decay, we used a titration experiment to establish levels of TTP that enhance decay of the β-globin ARE-mediated decay reporter mRNA (β-ARE), but allow accumulation of sufficient β-ARE mRNA to accurately measure the rate of decay (data not shown). In the mRNA decay assays in Figure 4B, expression of TTP causes a 3.5-fold decrease in the half-life of the β-ARE mRNA (from 71 to 21 min). However, the TTP100-326 mutant that lacks the NTD responsible for mRNA decay enzyme association shows impaired activity (t½ = 46 min; 2.2 ± 0.4-fold higher than TTP in three experiments). This protein is expressed at similar levels as full-length TTP as determined by Western blotting (data not shown). The TTP1-174 mutant protein that lacks the CTD expresses at two- to threefold higher levels than full-length TTP (data not shown) and shows activity similar to that of full-length TTP (t½ =24 min). Both of the TTP deletion mutant proteins have intact RNA-binding domains (Fig. 1), and they both bind efficiently to poly-A mRNA as shown by their RNA-dependent copurification with poly-A-binding protein (PABP) in Figure 4C (second and third panels). An RNA-binding-deficient TTP-F126N mutant protein (Lai et al. 2002), that served as a negative control is inactive in mRNA decay (Fig. 4B; t½ = 70 min), and does not associate with mRNA (Fig. 4C). The exogenously expressed TTP and TTP mutant proteins all localize uniformly in the cytoplasm as determined by indirect immunofluorescence assays (data not shown). Taken together, these results demonstrate that the NTD of TTP is important for full activation of ARE-mediated decay, and that a correlation exists between the ability of TTP to associate with mRNA decay enzymes and to efficiently activate mRNA decay (Fig. 4A,B). However, it is important to note that deletion of the NTD does not render TTP completely inactive (Fig. 4B; Lai et al. 2000). Therefore, regions of TTP other than the NTD appear to play an important role in activation of mRNA decay (see below).
The NTD of TTP and BRF-1 is an mRNA decay activation domain
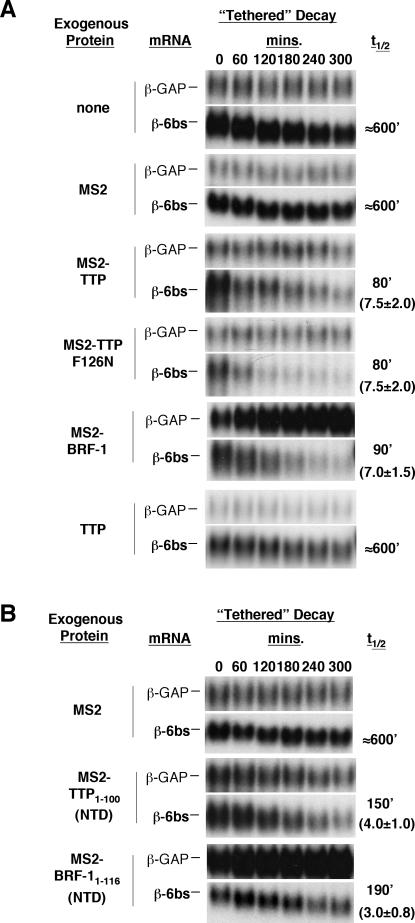
We next wished to test whether the NTD of TTP constitutes an mRNA decay activation domain. Many proteins involved in gene expression exhibit modular structures. For example, many transcription factors contain DNA-binding domains with separate transcription activation domains that are functional after fusion with heterologous DNA-binding proteins. In a similar manner, we wished to test if the NTD of TTP and BRF-1 can activate mRNA decay after fusion with a heterologous RNA-binding protein. To test this, we took advantage of a tethering assay where TTP, BRF-1, and fragments thereof were fused to the MS2 coat protein and tested for their ability to activate mRNA decay of a β-globin reporter mRNA that contains six MS2 coat protein-binding sites in the 3′UTR (β-6bs) (Lykke-Andersen et al. 2000).
The mRNA decay assays (one of four independent experiments is shown in Fig. 5A) show that the decay rate of the β-6bs reporter mRNA is enhanced six- to ninefold upon expression of MS2-TTP (t½ = 80 min, 7.5 ± 2.0-fold reduction), compared with the absence of an exogenous protein (t½ ≈ 600 min, half-life measured in a longer time-course). The MS2 coat protein alone has no effect on the decay rate (t½ ≈ 600 min). Importantly, the enhanced decay rate is a consequence of tethering, because expression of TTP that lacks the MS2 coat protein fusion has no effect on the half-life of β-6bs mRNA (t½ ≈ 600 min). Interestingly, the RNA-binding-deficient mutant TTP protein, TTP-F126N, which is unable to activate ARE-mediated decay (Fig. 4B; Lai et al. 2002), enhances mRNA decay to a similar extent as wild-type TTP after tethering (t½ = 80 min) (Fig. 5A). This demonstrates that RNA binding and mRNA decay activation are two separate activities in the TTP protein. We also tested the effect of tethered BRF-1, and it resulted in a six- to eightfold enhanced decay rate (t½ = 90 min, 7.0 ± 1.5-fold reduction compared with MS2 alone). We note that the rate of β-globin mRNA decay induced by tethered TTP and BRF-1 is comparable to that triggered by the potent ARE from GM-CSF mRNA (cf. Figs. 5A and 3A, 4B).
Figure 5.
The NTD of TTP and BRF-1 can activate mRNA decay when tethered via the MS2 coat protein. (A) Northern blots showing decay rates of a β-globin mRNA with six MS2 coat protein-binding sites in the 3′UTR (β-6bs), in the presence of exogenous MS2, TTP, MS2-TTP, and MS2-BRF-1 proteins indicated on the left. Calculated β-6bs mRNA half-lives and the average fold change from the assay with the MS2 protein alone are given on the right (four experiments). (B) Northern blots showing decay rates of β-6bs mRNA, in the presence of tethered NTDs of TTP and BRF-1 proteins indicated on the left. Calculated β-6bs mRNA half-lives and average fold change are given on the right (three and four experiments).
We next tested the activity of the NTD of TTP and BRF-1. The mRNA decay assays in Figure 5B show that the decay rate of the β-6bs reporter is enhanced three- to fivefold and two- to fourfold upon expression of MS2-TTP1-100 (t½ = 150 min, 4.0 ± 1.0-fold reduction in four independent experiments) and MS2-BRF-11-116 (t½ = 190 min; 3.0 ± 0.8-fold reduction in three experiments), respectively, compared with the MS2 coat protein alone (t½ ≈ 600 min). We conclude that the NTD of TTP and BRF-1 constitutes an activation domain that is capable of triggering mRNA decay when associated with a heterologous RNA-binding protein, although to a lesser extent than full-length TTP and BRF-1.
The CTD of TTP and BRF-1 constitutes a second mRNA decay activation domain
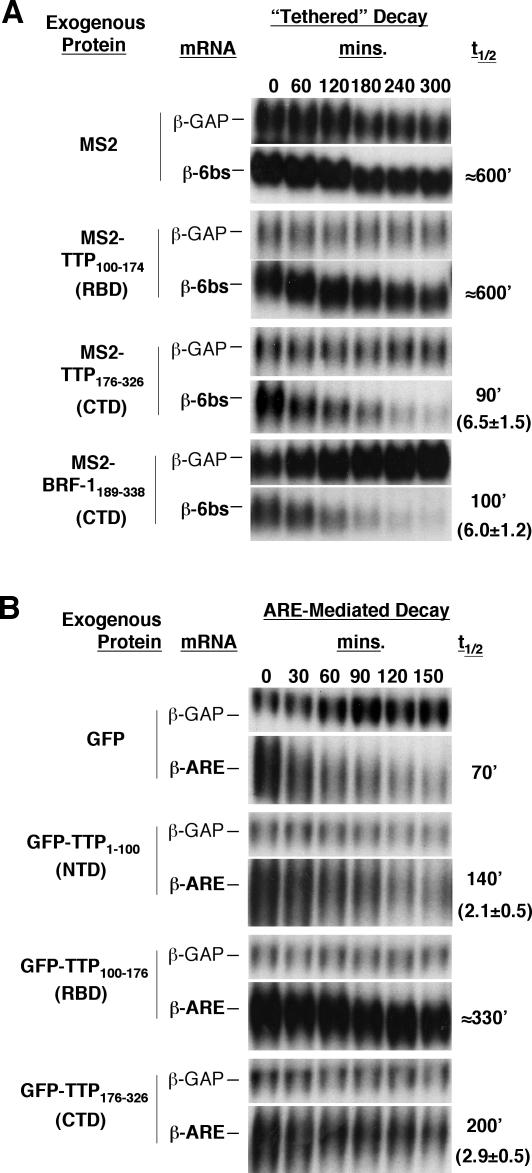
The data shown above demonstrate the importance of the NTD of TTP and BRF-1 in mRNA decay. However, we noticed that the NTD was not as active as the fulllength proteins in the tethering assays (Fig. 5), and that TTP ΔNTD shows partial activity (Fig. 4B). We therefore suspected that other regions of TTP may play a role in activation of mRNA decay. To test this, we used the tethering assays to ask if the RNA-binding domain and CTD of TTP also function to activate mRNA decay. The results in Figure 6A show that the decay rate of the β-6bs reporter is enhanced five- to eightfold upon tethering of the TTP CTD (MS2-TTP176-326; t½ = 90 min; 6.5 ± 1.5- fold reduction in four experiments), compared with the MS2 coat protein alone (t½ ≈ 600 min). In contrast, the RNA-binding domain of TTP, which expresses at similar levels as the other TTP fragments (data not shown), was not capable of activating mRNA decay in the tethering assay (MS2-TTP100-174;t½ ≈ 600 min). We also tested the CTD of BRF-1, and it shows mRNA decay activation activity similar to that of the TTP CTD (t½ = 100 min, 6.0 ± 1.2-fold activation in three experiments). We conclude that the CTD of TTP and BRF-1 can activate mRNA decay (Fig. 6A), despite its inability to stably associate with mRNA decay enzymes (Fig. 4A).
Figure 6.
The CTD of TTP and BRF-1 are mRNA decay activation domains, and overexpression of TTP NTD and CTD inhibits ARE-mediated mRNA decay. (A) Northern blots showing decay rates of β-6bs mRNA in the presence of tethered TTP and BRF-1 domains as indicated on the left. Calculated half-lives and average fold change are given on the right (four experiments). (B) Northern blots showing the decay rate of the ARE-mediated decay reporter mRNA, β-ARE, in the presence of coexpressed GFP, GFP-TTP1-100, GFP-TTP100-174, or GFP-TTP176-326. Half-lives of the reporter mRNAs and average fold change are given on the right (three experiments).
Overexpression of the TTP NTD, RNA-binding domain, and CTD inhibits ARE-mediated decay
If the NTD and CTD of TTP associate with trans-acting factors (such as mRNA decay enzymes) that are important for ARE-mediated decay in the human cell, their overexpression in the absence of an RNA-binding domain would be predicted to inhibit ARE-mediated decay. To test this, we assayed the ability of exogenously expressed TTP NTD, RNA-binding domain, and CTD to impair ARE-mediated mRNA decay in HeLa Tet-off cells. In these experiments, TTP fragments were fused with green fluorescent protein (GFP) for detection. The mRNA decay assays in Figure 6B demonstrate that overexpression of the TTP NTD (GFP-TTP1-100) and TTP CTD (GFP-TTP176-326) leads to an increase in the half-life of the β-ARE reporter mRNA (t½ = 140 and 200 min, 2.1 ± 0.5-fold and 2.9 ± 0.5-fold increase, respectively, in three experiments) compared with the expression of GFP alone (t½ = 70 min). As a positive control, expression of the RNA-binding domain of TTP, which can bind ARE sequences (Lai et al. 2000), but does not activate mRNA decay (Fig. 6A), strongly inhibits decay of the β-ARE mRNA (four- to fivefold; t½ ≈ 330 min), most likely due to competitive RNA binding. We conclude that the NTD and CTD of TTP both associate with titratable trans-acting factors that are limiting for ARE-mediated decay in the human cell.
Discussion
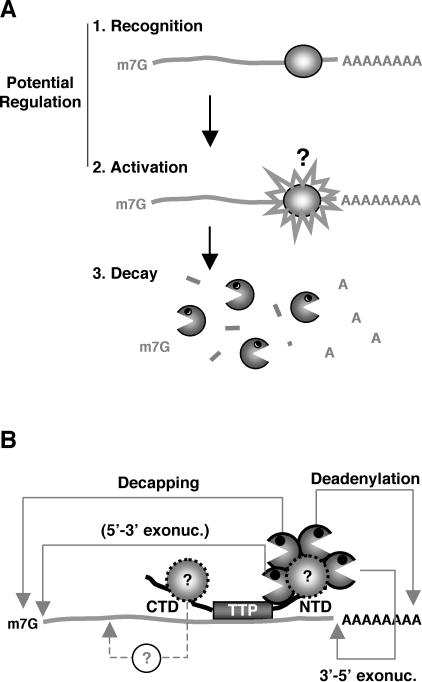
The process of mRNA decay can be divided into three steps: (1) target mRNA recognition, (2) activation of the mRNA decay machinery, and (3) degradation (Fig. 7A). The first two steps are potential targets for regulation, and it is therefore essential to elucidate these to understand how mRNA turnover can be regulated to modulate gene expression. In the case of ARE-mediated mRNA decay, several proteins that function in the target mRNA recognition step have been characterized, including the TTP-family of zinc-finger proteins (Fig. 1). However, the mechanism by which these ARE-binding proteins activate mRNA decay is poorly understood. In this study we have presented experiments aimed at elucidating the activation step in ARE-mediated decay.
Figure 7.
mRNA decay activation by the TTP family of ARE-binding proteins. (A) Three steps of mRNA decay. Activation of mRNA decay is poorly understood, indicated by a question mark. Regulation of mRNA decay is hypothesized to take place on the recognition or activation steps. mRNA decay enzymes are shown as “Pac-men.” (B) How TTP may activate ARE-mediated decay by recruitment of trans-acting factors, which includes mRNA decay enzymes via the NTD. The putative activation of 5′-to-3′ exonucleolytic decay is shown in brackets because it is based solely on the interaction between hXrn1 and TTP/BRF-1 shown in this study. All other indicated decay processes are based on both interaction and decay data. For details, see Discussion. The NTD is shown facing the poly-A tail based on the solution structure of the BRF-2 zinc-finger complexed with RNA (Hudson et al. 2004).
Decapping can be a limiting step in ARE-mediated mRNA decay
Several mRNA decay processes have previously been shown to be implicated in the degradation phase of ARE-mediated decay (Fig. 7A, step 3), including deadenylation and 3′-to-5′ exonucleolytic decay in tissue culture cells and cell extracts, as well as decapping in cell extracts (Chen et al. 1995, 2001; Voeltz and Steitz 1998; Ford et al. 1999; Gao et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002; Lai et al. 2003). However, it was not previously tested if decapping plays a role in ARE-mediated decay in the human cell. Here we have shown evidence that decapping enzymes are implicated in ARE-mediated decay in human cells: First, overexpression of a catalytic subunit, hDcp2, of the decapping complex leads to enhanced ARE-mediated mRNA decay, only if hDcp2 is catalytically active (Fig. 3A). Importantly, the effect of overexpressed hDcp2 is not a result of general activation of mRNA decay in the cell, because a stable mRNA and an mRNA that undergoes NMD were not significantly influenced by hDcp2 overexpression (Fig. 3B,C). Second, decapping complex subunits exist in complex with the ARE-binding proteins TTP and BRF-1 (Fig. 2), and associate with a domain of TTP and BRF-1 that is capable of activating mRNA decay (Figs. 4A, 5B). Unfortunately we were unable to test if reduction of cellular hDcp2 levels correlates with reduced ARE-mediated decay, because tested siRNAs did not deplete hDcp2 from the HeLa Tet-off cells used for the mRNA decay assays. It is perhaps surprising that overexpressed hDcp2 does not significantly activate NMD, because decapping has been shown to play a major role in NMD in S. cerevisiae (Muhlrad and Parker 1994), and the decapping complex is known to associate with Upf1, a central component in the NMD pathway (He and Jacobson 1995; Lykke-Andersen 2002). It is possible that decapping plays a less prominent role in human than in yeast NMD, or that hDcp2 levels are not limiting for NMD in the human cell. In contrast to hDcp2, overexpression of hDcp1a/hDcp1b did not enhance ARE-mediated decay. Perhaps hDcp1a/hDcp1b levels are not limiting for decapping activity in the cell. Our results suggest that decapping is another mRNA decay process activated by AREs in human cells, in addition to deadenylation and 3′-to-5′ exonucleolytic decay. It remains to be established whether AREs can also activate 5′-to-3′ exonucleolytic decay. These results set the stage to test how ARE-binding proteins activate mRNA decay.
The NTD and CTD of TTP and BRF-1 are mRNA decay activation domains
The following observations suggest that TTP and BRF-1 contain two domains that can individually activate mRNA decay when fused with an RNA-binding domain: First, the NTD and CTD of TTP and BRF-1 each activate mRNA decay when tethered to a target mRNA via an MS2 coat protein fusion (Figs. 5B, 6A). In contrast, the TTP zinc-finger domain showed no activity (Fig. 6A). Second, TTP lacking either the NTD or CTD retains the ability to activate ARE-mediated decay, although TTP ΔNTD functions at a reduced level compared with fulllength TTP (Fig. 4B). In contrast, TTP lacking both of the NTD and CTD inhibits decay of a coexpressed ARE-mediated decay reporter mRNA (Fig. 6B). Based on these observations, we term the NTD and CTD of TTP and BRF-1 “mRNA decay activation domains” (Fig. 1). The NTD and CTD of BRF-2 are 41% and 58% identical to BRF-1, respectively, and therefore most likely function in a similar manner. We conclude that the TTP family of ARE-binding zinc-finger proteins consist of an RNA-binding domain in fusion with two mRNA decay activation domains (Figs. 1, 7B). This is reminiscent of transcriptional activator proteins that in many cases consist of DNA-binding and transcription activation domains.
The NTD of TTP is a binding platform for mRNA decay enzymes
How does the NTD activation domain of TTP and BRF-1 trigger mRNA decay? Several observations suggest that the NTD functions as a binding platform for enzymes involved in mRNA decay. First, both TTP and BRF-1 coimmunopurify with mRNA decay enzymes involved in decapping, deadenylation and exonucleolytic decay from the 5′ and 3′ ends (Fig. 2). Second, the NTD of TTP is necessary and sufficient for the association of TTP with mRNA decay enzymes (Fig. 4A). Third, deletion of the NTD of BRF-1 prevents the association with mRNA decay enzymes (J. Lykke-Andersen and E. Wagner, unpubl.). A simple model based on these data is that the NTD of the TTP protein family serves to recruit mRNA decay enzymes responsible for decapping, deadenylation, and exonucleolytic decay to ARE-containing mRNAs (Fig. 7B). Importantly, this helps explain how AREs can independently activate mRNA deadenylation, decapping, and 3′-to-5′ exonucleolytic decay, and suggests that the TTP protein family can perform all of these functions (Chen et al. 1995, 2001; Voeltz and Steitz 1998; Ford et al. 1999; Gao et al. 2001; Wang and Kiledjian 2001; Mukherjee et al. 2002; Lai et al. 2003). A prediction from our data is that AREs can also activate 5′-to-3′ exonucleolytic decay independently of other mRNA decay processes. It is important to note that our data do not discriminate whether mRNA decay enzymes interact directly or indirectly, via other proteins, with the TTP NTD. For example, it is possible that an NTD-associated protein is responsible for the association with mRNA decay enzymes (Fig. 7B). We did not observe a direct interaction between bacterially expressed hDcp1a or hDcp2 fused to glutathione-S-transferase and TTP translated in a reticulocyte lysate (data not shown). An important goal for future studies should be to determine the relative importance of each of the mRNA decay enzymes in mRNA decay triggered by the TTP protein family.
How does the CTD of TTP and BRF-1 activate mRNA decay?
What is the function of the CTD of the TTP family of ARE-binding proteins? The following observations suggest that the CTD of TTP activates mRNA decay by the association with trans-acting factors that are distinct from known mRNA decay enzymes: First, overexpression of the TTP CTD impairs ARE-mediated decay, which suggests that a limiting factor in ARE-mediated decay exists in complex with the TTP CTD (Fig. 6B). Second, in contrast to the NTD of TTP, the CTD shows no stable association with mRNA decay enzymes (Fig. 4A). These observations suggest that the CTD of TTP activates mRNA decay without directly recruiting known mRNA decay enzymes but by instead employing different trans-acting factors. However, it cannot be completely ruled out that the CTD exhibits unstable association with mRNA decay enzymes, which was not detectable in coimmunoprecipitation assays. An important goal for future experiments should be to identify the trans-acting factors employed by the CTD. We speculate that the CTD-interacting factors may be involved in mRNP remodeling or in localization of mRNA to the cytoplasmic processing bodies that are believed to form sites of mRNA decay (Sheth and Parker 2003; Cougot et al. 2004), or they may constitute yet uncharacterized mRNA decay enzymes such as, for example, endonucleases. However, more experiments are needed to discriminate between these possibilities. We found no RNA-independent interaction between TTP and the RNA helicase RHAU (J. Lykke-Andersen and E. Wagner, unpubl.), a potential mRNP remodeling enzyme recently implicated in ARE-mediated decay (Tran et al. 2004). We considered the possibility that the TTP CTD functions as a dimerization domain and thereby recruits endogenous TTP, BRF-1, or BRF-2 in the tethering assays. However, while we observed that myc-TTP could be coimmunoprecipitated with Flag-TTP, this association was dependent on the TTP NTD and zinc-finger domain but not on the CTD (data not shown).
Regulation of ARE-mediated decay through the NTD and CTD mRNA decay activation domains?
A simple model based on our data is that the TTP family of ARE-binding proteins use two activation domains to trigger ARE-mediated mRNA decay, one that recruits mRNA decay enzymes to target mRNAs and another that employs distinct trans-acting factors, perhaps to help render the target mRNP accessible for degradation by recruited decay enzymes (Fig. 7B). It is tempting to speculate that regulation of ARE-mediated decay involves the repression of one or both of these mRNA decay activation domains on specific mRNAs. For example, the activity of the NTD and CTD of the TTP protein family could be modulated by associated proteins or by protein modifications, which each in turn could be regulated by specific cell signals. Alternatively, the activity of the TTP protein family could be regulated by the modulation of their RNA-binding affinity. Intriguingly, two phosphorylation events that result in 14- 3-3 protein recruitment and repression of TTP activity have been mapped to the NTD and immediately after the RNA-binding domain, respectively (Johnson et al. 2002; Chrestensen et al. 2004; Stoecklin et al. 2004), and these modifications do not appear to modulate RNA binding by TTP (Johnson et al. 2002; Stoecklin et al. 2004; J. Lykke-Andersen and E. Wagner, unpubl.). It is an exciting possibility that these modifications interfere with the activity of the NTD and CTD activation domains. An important future goal will be to determine how the activity of the TTP family of ARE-binding proteins is controlled to regulate ARE-mediated decay and gene expression.
Materials and methods
Plasmid constructs
mRNA decay reporter plasmids are based on the human β-globin gene inserted between HindIII and XbaI sites of a derivative of pcDNA3 (Invitrogen) that have the CMV promoter replaced by a CMV minimal promoter with six upstream tetracycline-responsive elements (pPC, sequence available upon request). pPC-β6bs contains six MS2 coat protein-binding sites (Lykke-Andersen et al. 2000) inserted into the β-globin reporter 3′UTR between XbaI and ApaI sites. pPC-βARE contains a 62-bp ARE from the GM-CSF mRNA 3′UTR (Voeltz and Steitz 1998) inserted in the same place. pcβ-GAP was described earlier (Lykke-Andersen et al. 2000).
Plasmids named pcDNA3-Myc and pcDNA3-Flag, used for expression of N-terminally myc- or Flag-tagged exogenous mRNA decay enzymes and RNA-binding proteins, are based on pcDNA3, which has a sequence encoding a Myc- or a Flag-tag inserted immediately upstream of the HindIII site. The open reading frames of hDcp2, hRrp4, hXrn1, hCcr4, hnRNP A1, HuR, BRF-1, and BRF-1 fragments were inserted between BamHI and NotI sites, whereas open reading frames of hDcp1a, TTP, and TTP fragments were inserted between EcoRI and NotI sites. The open reading frames of TTP, BRF-1, and TTP/BRF-1 fragments were also inserted into plasmids based on pcDNA3 (called pcNMS2-Flag and pcNEGFP), which contain an upstream in-frame Flag-tagged MS2 coat protein or enhanced GFP open reading frame, respectively. The MS2 coat protein contains two point mutations to avoid protein multimerization (Lykke-Andersen et al. 2000).
Antibodies
Rabbit polyclonal antisera were raised (Cocalico Biologicals Inc.) against hDcp1a (amino acids 1-251), hXrn1 (amino acids 1232-1354), and PARN (amino acids 1-196) fused to N-terminal glutathione S-transferase tags. Rabbit anti-PM-Scl75 (Mukherjee et al. 2002) and anti-TTP (Brooks et al. 2002) polyclonal antisera were generous gifts from Drs. Jeff Wilusz (Colorado State University, Fort Collins, CO) and William Rigby (Dartmouth College, Hanover, NH), respectively.
mRNA decay assays
Human HeLa Tet-off cells (Clontech) in DMEM/10% fetal bovine serum (FBS) at ≈30%-50% confluency in 3.5-cm wells were transfected in the presence of 50 ng/mL tetracycline, using TransIT HeLaMonster reagent according to manufacturers protocols (Mirus), with a total of 2 μg of plasmid, including 25 ng internal control expression plasmid, pcβG, and 0.5 μg reporter mRNA expression plasmid, pPC-βARE (Figs. 3A, 4B, 6B), pPC- β39 (Fig. 3B), pPC-βwt (Fig. 3C), or pPC-β6bs (Figs. 5, 6A). In addition, exogenous protein expression plasmids were included as follows: 1.0 μg pcDNA3-Flag-hDcp2, pcDNA3-Flag-hDcp2 E148Q, and/or 0.5 μg pcDNA3-Flag-hDcp1a or pcDNA3-Flag-hDcp1b (Fig. 3); 5 ng pcDNA3-myc-TTP or pcDNA3-myc-TTP derivative (Fig. 4B); 0.25 μg pcNMS2-Flag-TTP, pcNMS2-Flag-BRF-1, or pcNMS2-Flag-TTP/BRF-1 derivatives (Figs. 5, 6A); or 1.5 μg pcNEGFP-TTP derivative (Fig. 6B). In each transfection, pcDNA3 was added to a total of 2 μg plasmid. Thirty to forty hours after transfection, a transcriptional pulse from the reporter mRNA expression plasmid was initiated by washing cell with phosphate buffered saline (PBS) and feeding with 2 mL DMEM/10% FBS, containing no tetracycline. Six hours later, tetracycline was added to 1.0 μg/mL to stop transcription. Cells were washed with PBS and taken up in 1 mL Trizol (Invitrogen) starting (0-min time point) 30 min after addition of tetracycline, and then every 30, 40, or 60 min as indicated in Figures 3, 4, 5, 6, 7. Total RNA was prepared according to manufacturer's protocols, and Northern blots were performed as described earlier (Lykke-Andersen et al. 2000).
Immunoprecipitation assays
Anti-Flag immunopurification assays were performed from HEK 293T cells transfected in 3.5-cm wells, as described earlier (Lykke-Andersen 2002). Immunoprecipitation of endogenous TTP was performed from herpesvirus saimiri-transformed marmoset T cells (a generous gift from Dr. Ron Desrosiers, Harvard Medical School, Boston, MA), in which TTP expression was induced as described by Cook et al. (2004). TTP was immunopurified from RNase-treated marmoset T-cell extracts from ≈2 × 107 cells, using 23 μg of affinity purified anti-TTP antibody (Brooks et al. 2002), and immunoprecipitates were analyzed for coprecipitating mRNA decay enzymes by Western blotting.
Acknowledgments
We thank Drs. Jeff Wilusz and William Rigby for antibodies; Dr. Ron DesRosiers for a marmoset T-cell line; and Drs. Tom Blumenthal, Ravinder Singh, and Heidi Cook for careful reading of the manuscript. This research was supported by National Institutes of Health grant GM 066811 and the Pew Scholars Program in the Biological Sciences, sponsored by the Pew Charitable Trusts.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1282305.
References
- Bakheet T., Frevel, M., Williams, B.R., Greer, W., and Khabar, K.S. 2001. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29: 246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V.I., Scherthan, H., Solinger, J.A., Buerstedde, J.M., and Heyer, W.D. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136: 761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P.J. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30: 945-952. [DOI] [PubMed] [Google Scholar]
- Brooks S.A., Connolly, J.E., Diegel, R.J., Fava, R.A., and Rigby, W.F. 2002. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 46: 1362-1370. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Yamashita, A., Chen, C.Y., Yamashita, Y., Zhu, W., Durdan, S., Kahvejian, A., Sonenberg, N., and Shyu, A.B. 2004. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes & Dev. 18: 2010-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y. and Shyu, A.B. 1995. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 20: 465-470. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Xu, N., and Shyu, A.B. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15: 5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451-464. [DOI] [PubMed] [Google Scholar]
- Chen J., Chiang, Y.C., and Denis, C.L. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21: 1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrestensen C.A., Schroeder, M.J., Shabanowitz, J., Hunt, D.F., Pelo, J.W., Worthington, M.T., and Sturgill, T.W. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 279: 10176-10184. [DOI] [PubMed] [Google Scholar]
- Cook H.L., Mischo, H.E., and Steitz, J.A. 2004. The herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol. Cell. Biol. 24: 4522-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko, S., and Seraphin, B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria C.T. and Brewer, G. 1996. AUF1 binding affinity to A + U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271: 12179-12184. [DOI] [PubMed] [Google Scholar]
- Fan X.C. and Steitz, J.A. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17: 3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Watson, J., Keene, J.D., and Wilusz, J. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes & Dev. 13: 188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Wilusz, C.J., Peltz, P.W., and Wilusz, J. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20: 1134-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherzi R., Lee, K.Y., Briata, P., Wegmuller, D., Moroni, C., Karin, M., and Chen, C.Y. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14: 571-583. [DOI] [PubMed] [Google Scholar]
- He F. and Jacobson, A. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes & Dev. 9: 437-454. [DOI] [PubMed] [Google Scholar]
- Hudson B.P., Martinez-Yamout, M.A., Dyson, H.J., and Wright, P.E. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11: 257-264. [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Stehn, J.R., Yaffe, M.B., and Blackwell, T.K. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 277: 18029-18036. [DOI] [PubMed] [Google Scholar]
- Korner C.G. and Wahle, E. 1997. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 272: 10448-10456. [DOI] [PubMed] [Google Scholar]
- Lai W.S. and Blackshear, P.J. 2001. Interactions of CCCH zinc finger proteins with mRNA: Tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 276: 23144-23154. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo, E., Thorn, J.M., Kennington, E.A., and Blackshear, P.J. 2000. Interactions of CCCH zinc finger proteins with mRNA: Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 275: 17827-17837. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Kennington, E.A., and Blackshear, P.J. 2002. Interactions of CCCH zinc finger proteins with mRNA: Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 277: 9606-9613. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23: 3798-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Li, X., and Maquat, L.E. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12: 675-687. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu, M.D., and Steitz, J.A. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121-1131. [DOI] [PubMed] [Google Scholar]
- Miller A.D., Curran, T., and Verma, I.M. 1984. c-fos protein can induce cellular transformation: A novel mechanism of activation of a cellular oncogene. Cell 36: 51-60. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457-466. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker, R. 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578-581. [DOI] [PubMed] [Google Scholar]
- Mukherjee D., Gao, M., O'Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21: 165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. and Song, H. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121-127. [DOI] [PubMed] [Google Scholar]
- Peng S.S., Chen, C.Y., Xu, N., and Shyu, A.B. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17: 3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri I., Wegmueller, D., Gross, B., Certa, U., and Moroni, C. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32: 1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos S.B., Stumpo, D.J., Kennington, E.A., Phillips, R.S., Bock, C.B., Ribeiro-Neto, F., and Blackshear, P.J. 2004. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131: 4883-4893. [DOI] [PubMed] [Google Scholar]
- Sheth U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. and Karin, M. 2002. The control of mRNA stability in response to extracellular stimuli. Mol. Cell 14: 323-331. [PubMed] [Google Scholar]
- Stoecklin G., Colombi, M., Raineri, I., Leuenberger, S., Mallaun, M., Schmidlin, M., Gross, B., Lu, M., Kitamura, T., and Moroni, C. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21: 4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W.F., Blackwell, T.K., and Anderson, P. 2004. MK2-induced tristetraprolin: 14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23: 1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo D.J., Byrd, N.A., Phillips, R.S., Ghosh, S., Maronpot, R.R., Castranio, T., Meyers, E.N., Mishina, Y., and Blackshear, P.J. 2004. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Mol. Cell. Biol. 24: 6445-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G.A., Carballo, E., Lee, D.M., Lai, W.S., Thompson, M.J., Patel, D.D., Schenkman, D.I., Gilkeson, G.S., Broxmeyer, H.E., Haynes, B.F., et al. 1996. A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4: 445-454. [DOI] [PubMed] [Google Scholar]
- Tharun S. and Parker, R. 2001. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell 8: 1075-1083. [DOI] [PubMed] [Google Scholar]
- Tran H., Schilling, M., Wirbelauer, C., Hess, D., and Nagamine, Y. 2004. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell 13: 101-111. [DOI] [PubMed] [Google Scholar]
- van Dijk E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, B. 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K. and Steitz, J.A. 1998. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol. Cell. Biol. 18: 7537-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Kiledjian, M. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751-762. [DOI] [PubMed] [Google Scholar]
- Wang Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 99: 12663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz C.J., Wormington, M., and Peltz, S.W. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2: 237-246. [DOI] [PubMed] [Google Scholar]
- Xu N., Chen, C.Y., and Shyu, A.B. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21: 6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]