Abstract
Basic medical research critically depends on the finished human genome sequence. Two types of gaps are known to exist in the human genome: those associated with heterochromatic sequences and those embedded within euchromatin. We identified and analyzed a euchromatic island within the pericentromeric repeats of the human Y chromosome. This 450-kb island, although not recalcitrant to subcloning and present in 100 tested males from different ethnic origins, was not detected and is not contained within the published Y chromosomal sequence. The entire 450-kb interval is almost completely duplicated and consists predominantly of interchromosomal rather than intrachromosomal duplication events that are usually prevalent on the Y chromosome. We defined the modular structure of this interval and detected a total of 128 underlying pairwise alignments (≥90% and ≥1 kb in length) to various autosomal pericentromeric and ancestral pericentromeric regions. We also analyzed the putative gene content of this region by a combination of in silico gene prediction and paralogy analysis. We can show that even in this exceptionally duplicated region of the Y chromosome, eight putative genes with open reading frames reside, including fusion transcripts formed by the splicing of exons from two different duplication modules as well as members of the homeobox gene family DUX.
Last year the male-specific sequence of the human Y chromosome was announced (Skaletsky et al. 2003). Determining the sequence of the human Y was an enormous task due to the rather highly repetitive genomic landscape of this chromosome shaped by an extraordinary evolutionary history. The majority (41 Mb) of the entire chromosome (63 Mb) is comprised of three blocks of highly reiterated satellites as well as other repeat sequences. Even the 23-Mb male-specific euchromatic region appears to have an unusual genetic structure with very large gene-rich palindromes. Near-perfect sequence duplications appear to preserve their structural integrity due to gene conversion events. Within this complex genomic environment, the chromosome generated its present gene repertoire by gene acquisition from the autosomes and the X chromosome, followed by selective gene amplification.
A surprising finding from the analysis of the human genome sequence was that 5% of our genome consists of segmental duplications. The size, fraction, and degree of sequence identity of these segmental duplications are unique to the human genome. Analysis of these regions has also revealed that they are composed of DNA containing partial copies or complete genic sequences composed of exons and introns. Indeed, these highly active regions have also been demonstrated to be the birthplace of novel genes (Samonte and Eichler 2002). We have isolated and sequenced a 554-kb genomic segment from the human Y chromosome that contains a 450-kb euchromatic island hidden between satellite 3 sequence, adjacent to the centromere on the long arm of the chromosome. Analysis of the Y-specific pericentromeric sequence revealed that it is almost entirely composed of blocks of duplicated genomic segments sharing 95%-99% homology to multiple human autosomes. Segmental duplications with nearly identical sequence of this range have been detected throughout the genome within exceptional, mostly pericentromeric regions (Horvath et al. 2000b; Bailey et al. 2002; Eichler et al. 2004; Rudd and Willard 2004; She et al. 2004). Here, we provide a detailed analysis of the structural composition of this euchromatic island in pericentromeric Yq11. The extent and degree of homology between this region and paralogous segments elsewhere in the genome were evaluated and the mosaic modular structure defined. Eight putative genes with open reading frames have been identified.
Results
The pericentromeric region in Yq11
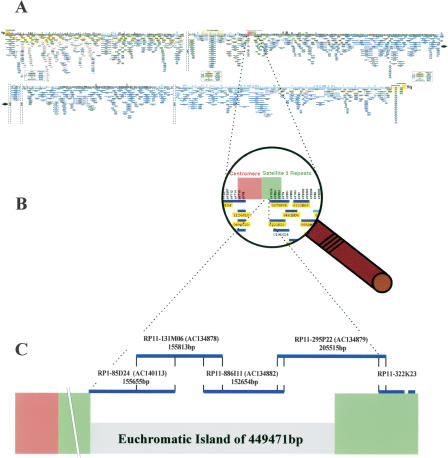
The Y chromosomal centromere is essentially composed of alphoid DNA surrounded by a range of other satellite and nonsatellite repeated sequences in a complex region spanning several hundred kilobases. To analyze the region next to the centromere unambiguously, we generated several Y-specific sequence-tagged sites (STSs) (Kirsch et al. 2002). STS SKY1 (AJ487121) is contained within the P1-derived artificial chromosome clone (PAC) RP1-85D24 (AC140113). The Y-specific STS SKY2 (AJ487122) was identified within bacterial artificial chromosome clone (BAC) RP11-295P22 (AC134879). Additional Y-specific STSs, SKY5-7, were generated by sequencing either BAC- or PAC-end fragments as well as internal sequences from YAC 17C12C (Kirsch et al. 2002). Using RP1-85D24 and RP11-295P22 as seed clones, we set out to close a gap of unknown size by a combination of PCR-based screening and hybridization to a single BAC library generated from one male individual, to avoid allelic variation (Fig. 1C). BAC clones RP11-131M06 (AC134878) and RP11-886I11 (AC134882) overlap with RP1-85D24 and RP11-295P22 and completely cover the genomic region between RP1-85D24 and RP11-295P22. The entire region of overlap between RP11-295P22 and RP11-322K23 (the most proximal clone in the Y-chromosomal clone-based map; Tilford et al. 2001) is restricted to satellite sequences, and proof of overlap was confirmed by sequence-family variant (SFV) typing (for details see Kirsch et al. 2002). This method relies on subtle variations as characteristic features of closely related but nonallelic sequences. Satellite probes from a previous pulsed-field-derived map (Cooper et al. 1992) were subsequently used to verify the integrity of the contig. Attempts to extend the contig towards the centromere resulted in the identification of heavily unstable genomic clones which consisted almost exclusively of satellite sequences. A minimum tiling path for sequencing (Fig. 1C) consisting of three BAC clones and one PAC clone was determined. Sequencing was carried out at the Washington University Genome Sequencing Center (WUGSC, St. Louis, Missouri). The four genomic clones include Y-specific STSs (Kirsch et al. 2002). Finished BAC clones share a 100% identity over their entire regions of overlap, whereas PAC clone RP1-85D24, which was derived from a different library (individual), shares 99.9% sequence identity (RP1-85D24 ↔ RP11-131M06 71279 bp; RP11-131M06 ↔ RP11-886I11 33248 bp; RP11-886I11 ↔ RP11-295P22 10705 bp). The entire overlap of 10417 bp between RP11-295P22 and RP11-322K23 consists of satellite 3 repeats. Sequences were assembled to form a contiguous sequence of 554,625 bp. Comparison of the complete sequence with the current human genome assembly of the National Center for Biotechnology Information (NCBI) indicated that this pericentromeric Yq11 contig is not part of the publicly available Y chromosome sequence. To determine whether this region reflects a low-frequency polymorphism or a constant part of the human Y chromosome, we studied 100 male individuals from different ethnic backgrounds for the presence of the Y-derived STSs SKY1, SKY2, and DUXY. All tested male individuals were scored positive for all three markers, whereas none of the markers was found in female controls.
Figure 1.
Location of a euchromatic island flanked by centromeric satellite 3 repeats on the long arm of the Y chromosome. (A) Minimum tiling path for sequencing the Y chromosome as published by Tilford et al. (2001). (B) Enlarged view of the genomic region encompassing the centromere and satellite 3 repeats (Tilford et al. 2001). (C) Illustration of the P1-derived artifical chromosome (PAC) and bacterial artificial chromosome (BAC) clones assembled into the pericentromeric Yq11 contig. Blue lines indicate name and sequence length of respective clones. Clone names include library origin; accession nos. are in parentheses. PAC RP1-85D24 extends 2 kb into the satellite 3 sequence block forming a constant part of the human Y chromosome centromere (Tyler-Smith 1987). The overlap sizes of the clones are as follows: RP1-85D24 ↔ RP11-131M06 71,279 bp; RP11-131M06 ↔ RP11-886I11 33,248 bp; RP11-886I11 ↔ RP11-295P22 10,705 bp. RP11-295P22 overlaps by 10,417 bp with RP11-322K23, the most centromeric clone presented in Tilford et al. (2001). The distal half of RP11-295P22 consists exclusively of satellite 3 repeat sequence. Subtracting satellite 3 segments from the entire 554-kb sequence discloses previously unknown 450 kb of euchromatic DNA sequence.
Segmental duplications in the pericentromeric Yq11 region
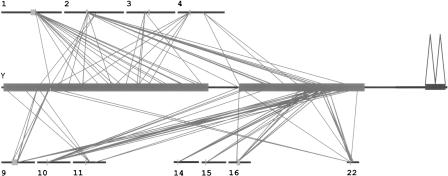
To investigate the molecular and chromosomal structure of this region, we examined the duplication content of the 554-kb segment by comparing it to the human genome (Build 34). To detect extensive internal and external pairwise chromosomal similarities, whole genome assembly comparison (WGAC) was used whereby simultaneously large gaps or insertions/deletions within the DNA were bridged (Bailey et al. 2001; see Methods). This analysis revealed that 80.2% (444,601/554,625 bp) of the pericentromeric sequence was composed of segmental duplications. In addition, 73.8% (409,187 bp) and 5.3% (29,289 bp) of the DNA are duplicated interchromosomally and intrachromosomally, respectively. An exceptional type of duplicated sequence was detected in the center of the analyzed sequence. We found that 30,323 bp (5.5%) of the euchromatic island are homologous to the 3.3-kb repeat family associated with hetero-chromatin (Lyle et al. 1995). After subtracting satellite sequences, 394,666 bp (71.2%) of duplicated sequence not including simple repeat structures remained. The majority (64%) of duplicated sequences is located within alignments ≥10 kb. Most of the interchromosomal duplications map to pericentromeric autosomal regions, e.g., chromosomes 1, 2, 3, 4, 9, 10, 11, 14, 15, 16, and 22 (Fig. 2). Others map to ancestral pericentromeric regions, e.g., 2q14.3/q21 (Avarello et al. 1992), 4q22-24 (Horvath et al. 2000a), and 9q12/q13 (Baldini et al. 1993). Sequence divergence estimates ranged from 93% to 97%, implying a recent origin within the last 30 million years of primate evolution.
Figure 2.
Human Y chromosome pericentromeric segmental duplications. A simplified version of the 554-kb sequenced contig is shown in the middle. The two large boxes represent the genomic segments composed of interchromosomal duplications, the small box that of intrachromosomal duplications. Other chromosomes are represented as horizontal black lines, above and below. Centromeres and acrocentric p arms are indicated as tiny boxes. All diagonal lines represent pairwise sequence comparisons ≥10 kb of DNA. The majority of Y pericentromeric duplications localize to the pericentromeric regions of autosomes. On chromosomes 2 and 4, the ancestral pericentromeric regions also show significant pairwise alignments. The coordinates are based on the published NCBI human genome assembly (January 2004, Build 34, Vers. 2).
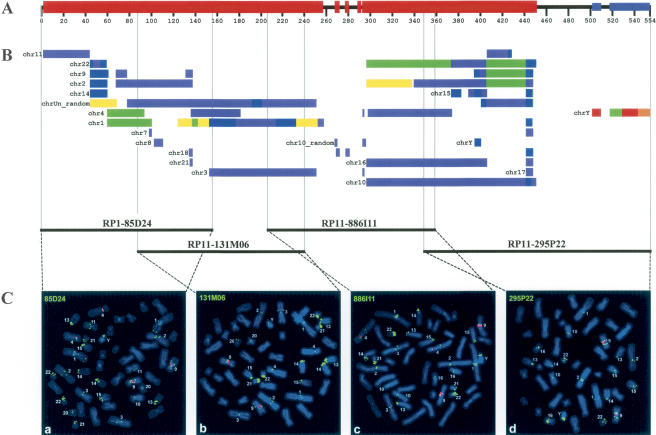
To analyze the segmental duplications by an independent experimental approach, we performed fluorescence in situ hybridization (FISH) analysis with all four clones forming the minimal pericentromeric contig. FISH mapping confirmed that the entire 554-kb segment is highly duplicated. PAC clone RP1-85D24 hybridizes to 27, BAC RP11-131M06 to 24, BAC RP11-886I11 to 25, and BAC RP11-295P22 to 18 different chromosomal segments besides the Y chromosome. The majority of the signals is confined to centromeric locations. Figure 3 shows the direct comparison of the in silico predicted duplication pattern (colored bars) and the FISH pattern for each individual clone. The results are summarized in Table 1. The pericentromeric Yq11 region shares remarkably long stretches of sequence (≥ 100 kb) with chromosomes 1, 2, 3, 10, 16, and 22. In addition, six completely sequenced clones whose chromosomal origins are not identified yet share similarities of similar length to pericentromeric Yq11. Compared to WGAC, FISH analysis detected more extensive interchromosomal duplications, suggesting either a poor representation or a misassignment of these sequences in the current human genome assembly. These may map to gaps or pericentromeric regions. For example, all four clones show cross-hybridization to the 13cen/13p11-13 region, yet paralogous sequences were not detected there by WGAC (Table 1). Strikingly, all four clones label the short arms of the five acrocentric chromosomes.
Figure 3.
Summary scheme depicting homologies between the pericentromeric region of Yq11 and other chromosomes. (A) The black horizontal bar shows the 554-kb sequenced region. Segments with interchromosomal (red boxes) and intrachromosomal (blue boxes) duplications are indicated. (B) All colored bars represent sequence homologies between the Y-chromosomal pericentromeric region and autosomes as determined by standard whole-genome analysis comparison (WGAC). Each color indicates a specific degree of homology: red, 100%-99%; orange, 99%-98%; yellow, 98%-97%; green, 97%-96%; blue, 96%-95%; indigo, 95%-94%; and violet, 93%. Each bar is preceded by the corresponding chromosome number. Bars that correspond to different chromosomes are indicated separately. Paralogies to sequenced genomic regions not assigned to a specific chromosome are summarized as chrUn_random. (C) Two-color FISH of human Y-chromosomal PAC (85D24) and BAC (131M06, 886I11, 295P22) clones (labeled in green) to human male metaphase spreads is shown below. Centromeres of chromosomes 4 and the constitutive heterochromatic region of the long arm of chromosomes 9 are labeled in red. Metaphases shown in a-d reflect the most proximal (a) to distal (d) order in the contig. Chromosomes with specific hybridization signals are tagged, respectively. The in silico identified paralogous segments and the chromosomal band localizations of the specific signals are listed in Table 1.
Table 1.
Sequence and comparative FISH results for interchromosomal duplications
| Boundary
|
Paralogous regions detected by
|
||||
|---|---|---|---|---|---|
| Clone | Library | Beginning (bp) | Ending (bp) | WGAC | FISH |
| 85D24 | RPCI-1 | 1 | 155655 | 1q21, 2p11, 2q11, 2q21, 3p13, 4p12, 9p11, 9q13, 9q22, 11q13, 14cen, 15q11, 18p11, 21q11, 22q11 | 1p11/p12, 1q12/q21, 1q44, 2cen, 2q14.3/q21, 3cen, 4cen, 9p11/p12, 9q12/q13, 10cen, 11q13, 13cen, 13p11.2, 13p13, 14cen, 14p11.2, 14p13, 15cen, 15p11.2, 15p13, 20cen, 21cen, 21p11.2, 21p13, 22cen, 22p11.2, 22p13 |
| 131M06 | RPCI-11 | 84376 | 240189 | 1q21, 2q11, 2q21, 3p13, 4p12, 9q13, 14cen, 15q11 | 1q12/q21, 2cen, 2q14.3/q21, 3cen, 3p12, 4cen, 9p11/p12, 9q12/q13, 13cen, 13p11.2, 13p13, 14cen, 14p11.2, 14p13, 15cen, 15p11.2, 15p13, 20q11.2, 21cen, 21p11.2, 21p13, 22cen, 22p11.2, 22p13 |
| 886I11 | RPCI-11 | 206941 | 359595 | 1q21, 2p11, 3p13, 4q23, 10p11, 16p11, 22q11 | 1q12/q21, 2cen, 3cen, 3p12, 4cen, 4q22-24, 9cen, 9q12/q13, 10cen, 13cen, 13p11.2, 13p13, 14cen, 14p11.2, 14p13, 15cen, 15p11.2, 15p13, 16cen, 21cen, 21p11.2, 21p13, 22cen, 22p11.2, 22p13 |
| 295P22 | RPCI-22 | 348890 | 554625 | 1q21, 2p11, 2q21, 3p26, 4q23, 7p11, 7q11, 9p11, 9q13, 10p11, 11cen, 15q11, 16p11, 17cen, 18p11, 21q11, 22q11, 22q12, Yq11 | 1q12/q21, 2cen, 9p11/p12, 9q12/q13, 10cen, 13cen, 13p11.2, 13p13, 14cen, 14p11.2, 14p13, 15cen, 15p11.2, 15p13, 16cen, 22cen, 22p11.2, 22p13 |
Modular structure and gene content of the pericentromeric region in Yq11
Successive rearrangements account for the complex structure of genomic segments consisting of segmental duplications. To define the modular structure of pericentromeric Yq11, we set out to trace back each distinct sequence block of the 554-kb sequence and its paralogous counterparts on other chromosomes to a common ancestral progenitor. Due to the mosaic structure of this region of the human Y chromosome, the definition of modules based on the identification of junctional boundaries shared with other chromosomes alone is not sufficient to decode its patchwork structure completely. Because gene sequences were used successfully in previous analyses to delineate modules in more complex duplications (Eichler et al. 1996; Shaikh et al. 2000), we used the same strategy for the pericentromeric Yq11 region. In total, we defined 37 modules that were distributed inter- (36 modules) and intrachromosomally (one). All modules were present only once in the Yq11 region. Twenty-nine modules were identified solely on the basis of well demarcated junctional boundaries. Ten of them presented one genic sequence, whereas four showed two genic features derived from different ancestral loci. Two further genic features were shown to spread across junctional boundaries. No transcriptional activity was documented among these 20 duplication modules. Several degenerative processed pseudogenes have been detected in Yq11 with multiple copies in other pericentromeric regions.
Of the 20 identified genic sequences sharing significant homology to the 554-kb sequence, all are present in at least one other location in the human genome. The principle gene-related features of each genic sequence are summarized in Table 2. Direct comparison with the NCBI dbEST database by best expressed sequence tag (EST) placement showed significant nucleotide identity to the Y chromosome.
Table 2.
Genic paralogies in pericentromeric Yq11
| Region of homology | Gene | Name | GenBank acc. no. | Ancestral locus | Reference |
|---|---|---|---|---|---|
| Degenerated processed pseudogenes | |||||
| 55219-58321 | FLJ42128 | Testis cDNA clone | AK124122 | Unknown | Unpublished |
| 68740-70354 | LOC339742 | Image cDNA clone | BC045732 | Chr 2 | Unpublished |
| 100202-106214 | ASNS | Asparagine synthetase | NM_133436 | Chr 7q21 | Arfin et al. 1983 |
| 142459-148130 | FLJ35140 | Kazusa cDNA clone | AK092459 | Unknown | Unpublished |
| 164406-174880 | FLJ00310 | Kazusa cDNA clone | AK090412 | Chr 1 | Unpublished |
| 296713-299847 | PABPC1 | Poly(A) binding protein, cytoplasmic 1 | NM_002568 | Chr 8q23 | Grange et al. 1987 |
| 357596-359239 | ARP3ß | Actin-related protein 3-beta | NM_020445 | Chr 7q32-36 | Machesky and Gould 1999 |
| 407551-409679 | TRIM43 | Tripartite motif-containing 43 | NM_138800 | Chr 2 | Unpublished |
| ESTs | |||||
| 32480-33082 | Hs.252460 | UNIGENE EST cluster | / | Chr 11 | Unpublished |
| 236680-242264 | THC1666755 | TIGR EST cluster | / | Unknown | Unpublished |
| Genes with partial exon-intron structure | |||||
| 46985-47778 | AF038169 | IMAGE cDNA clone | BC043584 | Chr 1 | Unpublished |
| 134919-135544 | C21orf81 | Chromosome 21 unknown ORF 81 | AF426257 | Chr 21 | Reymond et al. 2002 |
| 302288-347418 | LOC150159 | CG10806-like IMAGE cDNA clone | NM_139173 | Chr 4 | Unpublished |
| 374270-380489 | CHEK2 | CHK2 checkpoint homolog | NM_007194 | Chr 22q12 | Matsuoka et al. 1998 |
| 435493-435986 | MGC32713 | IMAGE cDNA clone | BC034141 | Unknown | Unpublished |
| Potential coding gene | |||||
| 115523-120823 | FLJ00219 | Kazusa cDNA clone | AK074146 | Chr 13 | Unpublished |
| 115523-176727 | FLJ35473 | Kazusa cDNA clone | AK092792 | Unknown | Unpublished |
| 115523-176733 | FLJ39663 | Kazusa cDNA clone | AK096952 | Unknown | Unpublished |
| 115520-176692 | pp5644 | cDNA clone | AF289559 | Unknown | Unpublished |
| 268641-269264 | DUX1 | Double homeobox, 1 | NM_012146 | 4q35 | Ding et al. 1998 |
| 276644-276976 | DUX1 | Double homeobox, 1 | NM_012146 | 4q35 | Ding et al. 1998 |
| 283281-283613 | DUX1 | Double homeobox, 1 | NM_012146 | 4q35 | Ding et al. 1998 |
| 294054-294476 | DUX1 | Double homeobox, 1 | NM_012146 | 4q35 | Ding et al. 1998 |
| 357403-359228 | FKSG74 | FKSG74 | AY026352 | Chr 16 | Unpublished |
The position of each feature within the 554kb contiguous sequence is given, together with the accession number and EST cluster information. Unknown ancestral locus indicates either that BAC clones corresponding to that sequence are not chromosomally assigned or a corresponding genomic sequence is not available. References and ancestral locus of each genic paralogy is shown.
Degenerated processed pseudogenes and genes with partial exon-intron structure
Of the 20 gene segments found on Yq11, 13 are unlikely to be functional genes. Eight have features of degenerated processed pseudogenes, and five genes show only partial exon-intron structure (Fig. 4B,C). All of them were derived from more complete genes elsewhere in the genome and propagated as part of segmental duplications to the human Y chromosome. They present an overall nucleotide sequence identity of 82%-97%.
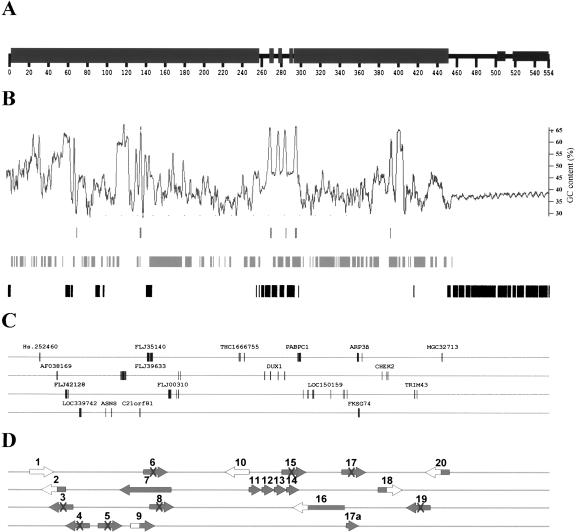
Figure 4.
Putative gene content of the euchromatic island in the pericentromeric region of the human Y. (A) The structure of the pericentromeric region of Yq11 is presented as a horizontal line with boxes representing segmental duplications. (B) Genomic properties of the Yq11 region: from top to bottom—(G+C) content, CpG islands, interspersed repeats including Alu, LINE, and HERV, satellite sequences including 5-bp and 68-bp repeats. (C) Only Y-specific sequences corresponding to exons of known autosomal genes or EST clusters with exon/intron boundaries are shown. Exons of the identified genes or pseudogenes identified are drawn to scale. For the ease of illustration, genic sequences were spread over four horizontal lines. (D) Large arrows indicate the predicted transcriptional direction. The GenBank accession nos. for sequences shown are Hs.252460, AF038169 (BC043584), FLJ42128 (AK124122), LOC339742 (BC045732), ASNS (NM_133436), FLJ39633 (AK096952), C21orf81 (AF426257), FLJ35140 (AK092459), FLJ00310 (AK090412), THC 1666755, DUX1 (NM_012146), PABPC1 (NM_002568), LOC150159 (NM_139173), ARP3β (NM_020445), FKSG74 (AY026352), CHEK2 (BC004207), TRIM43 (BC015353), and MGC32713 (BC034141). The state of each genic sequence is characterized as follows:  Gene with intact ORF (7, 11-14, 17a);
Gene with intact ORF (7, 11-14, 17a);  EST cluster with unidentified ORF (1,10);
EST cluster with unidentified ORF (1,10);  Partial gene (2, 9, 16, 18, 20);
Partial gene (2, 9, 16, 18, 20);  Degenerated processed pseudogene (3, 4, 5, 6, 8, 15, 17, 19).
Degenerated processed pseudogene (3, 4, 5, 6, 8, 15, 17, 19).
A striking example is the processed pseudogene of asparagine synthetase (ASNS). This processed pseudogene consists of a proximal part encompassing exons 1 and 2, and a distal part comprising exons 5 and 6 of the functional ASNS gene. Whereas the distal part has a nucleotide identity of 96%, the proximal part has merely 85%. Thus, the fragments of the ASNS processed pseudogene result from temporally different retrotranspositions subsequently juxta-posed through the process of paralogous recombination.
A second interesting case involves a Y-chromosomal member of the recently identified ARP3β pseudogenes-derived gene family locating to different chromosomes (FKSG72-74) (Fig. 4C). Genic sequence 17a, comprising only a portion of the degenerated processed pseudogene 17, may therefore also represent a functional gene. It has experienced a one-base pair deletion resulting in an altered carboxy-terminal portion after amino acid 88. This frameshift leads to a premature termination of the open reading frame (ORF), resulting in a protein of only 107 amino acids. All paralogs are predicted to encode single-exon ORFs of 193-199 amino acids with 98%-100% of identity to the well defined ARP3β ORF (transcripts 17 and 17a; Fig. 4C).
ESTs and candidate genes
The other genic segments are likely to be functional. Besides two EST clusters (UniGene EST AI678041 and TIGR THC1666755) with unknown function, several interesting candidate genes were analyzed in more detail.
The four paralogous mRNAs FLJ39633, FLJ00219, FLJ35473, and pp5644 with a nucleotide identity between 94% and 100% are weakly homologous to tektin A1 (A46170), a cytoskeletal protein from Strongylocentrotus purpuratus (Norrander et al. 1992). SIM4 analyses revealed that FLJ39633 (AK096952) is composed of eight exons (Fig. 4B,C), whereas the remaining three mRNAs only contain parts of paralogous copies on other chromosomes. The genomic structure discloses that exons 1 and 2 and exons 3 to 8 reside within a few kilobases, whereas the distance between exon 2 and 3 is more than 50 kb. Remarkably, this intron extends over five different defined modules, of which three were identified by the presence of genic sequences. All three internal genic sequence paralogs show an inverted transcriptional orientation relative to the tektin A1-homologous mRNAs, thereby excluding the incidental incorporation into a growing transcript.
We also found four copies of the DUX (double homeobox) gene family (Fig. 4). DUX genes encode single-exon ORFs (Beckers et al. 2001) with highest homologies to the paired-type homeobox genes PAX3 and PAX7. The Y-derived copies (DUXY1-DUXY4) are organized as a tandem repeat (Fig. 5). The DUXY gene family members are predicted to encode ORFs of ≥110 amino acids with conserved amino termini, including the first homeodomain (Fig. 6). None of the Y-derived copies encodes a complete second homeodomain. A nuclear localization signal was defined in the amino terminal part of the paired-type homeodomain in several proteins. A similar stretch of basic amino acids is still present in all DUXY genes. A TATA box and a transcription start site were found upstream of the predicted translation start site of DUXY1-4. There is no poly(A) signal in DUXY genes, which might explain the absence of DUX mRNA sequences in EST databases.
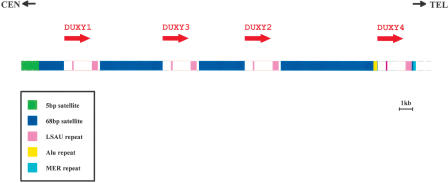
Figure 5.
Sequential organization of the DUXY locus in the pericentromeric region of Yq11. The orientation of centromere and telomere is shown at the top. Four copies of the DUX gene family (DUXY1-Y4) are clustered as an imperfect tandem repeat within a genomic segment of 30,323 bp. The transcriptional orientation of each copy is indicated by an arrow. Each DUXY copy is enclosed by repeated elements of tandemly repeated simple sequences (68-bp satellite and LSAU repeat; see legend in figure). Whereas the two types of LSAU repeats are constant in size (120-122 bp and 494-497 bp), the 68-bp satellite sequence is highly variable (2004, 1921, 3707, and 7538 bp). At the distal end of the most telomeric 68-bp satellite block, an Alu repeat has been integrated. The centromeric boundary of the DUXY locus is defined by a block of 5-bp satellite sequence, whereas the telomeric boundary is defined by a MER7 repeat.
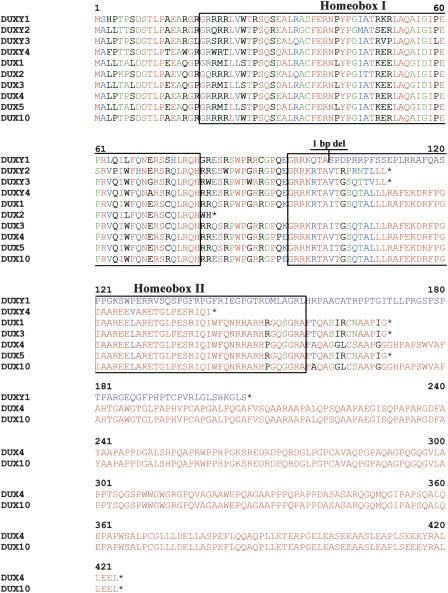
Figure 6.
Comparison of predicted amino-acid sequence of cDNAs and paralogous genomic copies of double homeobox DUX-like genes. DUX1, DUX2, DUX3, DUX4, DUX5, and DUX10 represent human double homeobox-containing genes from 4q35. All genes consist only of a single exon. The color code corresponds to the CLUSTALW default for amino-acid sequence comparison. The boxes indicate the 60-amino-acid conserved homeodomain. Analogous to the DUX gene family member DUX2, none of the Y-derived DUX family members contain a complete copy of the second homeodomain (Homeobox II). The location of a 1-bp deletion in DUXY1 relative to all other family members is indicated, resulting in a frameshift and a C-terminally altered amino acid sequence (purple). We resequenced the DUXY1 copy from a PCR product amplified from BAC RP11-886I11 (AC134882) and a normal male individual and confirmed the accuracy of the 1-bp deletion. Black stars indicate stop codons.
Discussion
Fundamentally, the pericentromeric region in Yq11 can be sub-divided into satellite 3 sequences and a euchromatic island flanked by these repeats. The presence of the satellite 3 sequence was interpreted by Skaletsky and colleagues (2003) as the end of the euchromatin, but new additional Y-specific markers revealed this euchromatic island. Investigating this euchromatic island encompassed by satellite sequences has illuminated its complex structure and the dynamic history of sequences located in this region. We have characterized all the segmental duplications and provide a genome-wide view of the results. The comparison of FISH and sequence homology analysis of this region strongly suggests an underrepresentation of pericentromeric regions of the acrocentric chromosomes in the current human genome sequence.
Ninety-three percent of the sequenced euchromatic island was shown to be involved in segmental duplications (≥90% identity and ≥1 kb in length). Its highly duplicative nature was used to organize the genomic segment into minimal evolutionary shared segments (modules) and to assess the transcriptional potential of the duplications. Our analysis shows quite a striking correspondence to the whole-chromosome study by Bailey et al. (2002). The balanced distribution of pairwise genetic-distance estimates (K values) supports the observation of frequently occurring duplications over the past 30 million years of evolution. The euchromatic island in pericentromeric Yq11 shares duplications ≥100 kb with autosomes 1, 10, and 16, and three chromosomally unassigned clones. These larger segments consist of numerous smaller duplication modules of diverse evolutionary origin. Taken together, these findings fit with the proposed two-step model of pericentromeric duplication wherein an initial process of transposition seeding (accumulation of smaller duplications within pericentromeric regions) is followed by pericentromeric exchange (spreading of larger patchwork blocks to pericentromeric regions of nonhomologous chromosomes) (Eichler et al. 1997; Horvath et al. 2000b; Luijten et al. 2000; Samonte and Eichler 2002). In contrast, the most pericentromeric position in the euchromatic island is exclusively present on chromosome 11. This paralogous segment shows a sequence identity of 94.2% to pericentromeric Yq11 (Fig. 2). Our analysis cannot preclude that this region is a simple duplication that originated from chromosome 11, as other modules with similar nucleotide identities are scattered more frequently to other pericentromeric locations. An interchromosomal duplication unit solely present on chromosomes 14 and 22 shows 99.4% nucleotide identity (Bailey et al. 2002). Intrachromosomal duplications appear to occur very rarely in the pericentromeric region of the human Y chromosome. This is in sharp contrast to the ampliconic sequence classes present in the MSY euchromatin of Yq11 (Skaletsky et al. 2003).
Segmental duplications have emerged during the past 30 million years of primate evolution and spread to a large number of pericentromeric and subtelomeric regions. Yet, despite the common evolutionary history of the modern sex chromosomes, we have not detected paralogous sequences on the human X chromosome. Because the centromeric/euchromatic boundaries and the subtelomeric regions of the human X have been completely sequenced, we think that we can exclude that the human X has acquired such duplications. The reason for this phenomenon remains elusive. As reviewed by Eichler et al. (2004), She et al. (2004), and Rudd and Willard (2004), gaps exist in the pericentric heterochromatic/euchromatic transition regions, and recent studies show that additional material is uncovered in these pericentric transition regions of nearly all human chromosomes. There are only quantitative differences between different chromosomes, including the Y. Our work shows in detail that part of the pericentromeric region of the Y chromosome consists of a mosaic of interchromosomal segmental duplications, indicating that this region is typical of autosomal pericentromeric regions.
With the exception of the DUXY1-4 genes, segmental duplications of Yq11-paralogous sequences have been involved in the multiplication of all genic sequences in this region. Among them, we detected two genes with intact ORFs (transcripts 7 and 17a), five genes with partial exon-intron structure, eight degenerated processed pseudogenes, and two EST clusters with an unidentified ORF. Members of one gene family could only be detected by direct homology comparison using the nr database of NCBI, as their genomic environment does not reflect the characteristics of a segmental duplication. The fate of the duplicated genic sequences is consistent with the birth-and-death model of gene evolution. Beyond that, the existence of genes possessing an intact ORF in Yq11-pericentromeric-paralogous segments supports the hypothesis that these duplications have an important evolutionary impact on functional change (Nei et al. 1997).
The homeobox-containing DUX gene family cluster in pericentromeric Yq11 is the first completely sequenced cluster of DUX genes in the human genome. The first members of the DUX gene family were identified on distal 4q (Ding et al. 1998), and paralogous gene clusters have been mapped to chromosomes 10, 13, 14, 15, 21, and 22 in the meantime. Proof of active transcription of the DUX genes has been provided for six family members (DUX1-5, -10). Similar to DUX2, none of the four Y-chromosomal DUX copies preserves a complete second homeodomain. Nevertheless, the high nucleotide sequence identities of these gene candidates warrants expression profiling to determine whether the Y-specific DUX copies adopted a distinct tissue-specific expression pattern. This is especially intriguing for DUXY1, as its predicted carboxy terminus presents striking differences from all other members, although the homeodomain I is extremely highly conserved. It will be interesting to find out if this carboxy terminus is of particular importance to the putative protein. One of the DUX gene family members, DUX4, has been suggested to play a role in facioscapulohumeral muscular dystrophy (FSHD) (Gabriëls et al. 1999).
By investigating the FKSG72-74 gene family and the tektin A1-homologous genes, two interesting features contributing to the continuing process of gene duplication and divergence were observed: First, the development of a possible functional FKSG72-74 gene family member (transcript 17a) from a degenerated retrotransposed pseudogene of ARP3β (transcript 17), and second, the integration of part of the distal end of a degenerated processed pseudogene FLJ00310 (transcript 8) into exon 1 of the tektin A1-homologous genes. A similar observation of exon usage in reverse transcription orientation was presented by Bailey et al. (2002). Actively transcribed retrotransposed pseudogenes might furthermore be involved in the regulation of their functional progenitors (Hirotsune et al. 2003). As the human genome sequences of pericentromeric regions will eventually approach completion, it will be interesting to find out whether these genic alterations are intrinsic evolutionary features of paralogous sequence blocks.
All potential genes described here are embedded in regions with extensive homologies to each other. Such highly duplicated regions with a locally increased number of recombination-promoting CAGGG repeats are prone to chromosomal rearrangements (Ji et al. 2000; Samonte and Eichler 2002; Stankiewicz and Lupski 2002). These rearrangements might lead to an abnormal phenotype by disruption or dosage alteration of the involved genes. Therefore, investigation of the eight potential genes as candidates for human disease mapped to paralogous sequences of nonhomologous chromosomes might shed new light on human disorders. Especially, the search for the yet unknown stature (GCY) (Smith et al. 1985; Ogata and Matsuo 1992) and gonadoblastoma locus on the Y chromosome (GBY) (Page 1987; Salo et al. 1995; Tsuchiya et al. 1995) might profit from the putative gene sequences provided in this study.
Methods
Generation and analysis of pericentromeric Y-specific STSs
Y-specific STSs termed SKY1, 2, 5, 6, 7 were derived from YAC, BAC, and PAC end sequences or from clone-internal sequences amplified by various combinations of Alu primers (Kirsch et al. 2002).
Subjects and PCR analysis
Subjects were unrelated healthy male individuals belonging to various Central European and Mediterranean populations. Additional samples from the Middle East and Far East were also included. Genomic DNA samples were extracted from peripheral blood leukocytes using standard protocols. PCR cycling conditions and analysis were as described by Kirsch et al. (2002). Primer sequences (5′-3′) for DUXY are as follows: DUXYfor, CCGACACCTTCGGACAG CAC; DUXYrev, GTGGTCTGGGATCC GGTGAC.
Isolation and sequencing of chromosome Y clones
Large-insert genomic PAC and BAC clones were identified through screening of PCR pools from the RPCI1,3-5 and RPCI11 male human genomic libraries provided by the German Resource Center (RZPD). From a total of 20 positive clones, four were selected to form the minimum tiling path for sequencing, namely RP1-85D24 (AC140113), RP11-131M06 (AC134878), RP11-886I11 (AC134882), and RP11-295P22 (AC134879). Genomic sequencing was carried out by conventional high-throughput sequencing techniques. Finished sequences from overlapping clones were assembled into one contiguous sequence of 554,625 bp.
Detection of segmental duplications
We used whole-genome assembly comparison (WGAC; Bailey et al. 2001) to detect segmental duplications within the contiguous sequence. This method aims to identify large alignments without being affected by intervening large deletions and/or insertions. We compared the entire 554-kb segment to the July 2003 human genome assembly. Sequences submitted to GenBank after this target date were analyzed separately. Briefly, common repeat elements were identified and removed from the sequence so as to leave putatively unique DNA. Global BLAST comparisons identified nonredundant duplications. Repeats were reinserted into the sequence and the alignment ends were fine-tuned to optimize the definition of duplication boundaries. Global alignments were generated using ALIGN (Myers and Miller 1988). Representation of single alignments with large gaps (up to 10 kb) was achieved by merging the statistics for global alignments. Merged alignments of ≥1 kb and ≥90% identity were passed on for further analysis. Two hundred interchromosomal and five intrachromosomal alignments with mean lengths of 23,355 bp and 5857 bp, respectively, were detected. Four intrachromosomal duplications were exclusively restricted to satellite repeats and not further investigated. The graphical alignment viewer PARASIGHT (J.A. Bailey, unpubl.) was used to generate diagrams of pairwise alignments and other sequence features. The evolutionary genetic distance for multiple substitutions was corrected using a two-parameter model (Kimura 1980).
Fluorescence in situ hybridization (FISH)
FISH analysis of chromosomal metaphase spreads derived from lymphocytes was performed from two different human males. Prior to FISH, the slides were treated with RNase followed by pepsin digestion as described (Ried et al. 1992). FISH followed the method described by Schempp et al. (1995). Chromosome in situ suppression was applied to the clones from the Human Male Genome PAC library (RPCI1) and from the BAC library (RPCI11) (RPCI1-85D24, RPCI11-131M06, RPCI11-886I11, RPCI11-295P22). The probes D4Z1 and D9Z1 (Appligene Oncor) were used as a marker for the centromere of chromosome 4 and for the centromere and heterochromatic region of chromosome 9. After FISH the slides were counterstained with DAPI (0.14 μg/mL) and mounted in Vectashield (Vector Laboratories). Preparations were evaluated using a Zeiss Axiophot epifluorescence microscope equipped with single-bandpass filters for excitation of red, green, and blue (Chroma Technologies). During exposures, only excitation filters were changed, allowing for pixel-shift-free image recording. Images of high magnification and resolution were obtained using a black-and-white CCD camera (Photometrics Kodak KAF 1400; Kodak) connected to the Axiophot. Camera control and digital image acquisition involved the use of an Apple Macintosh Quadra 950 computer.
Potential gene content and module definition
After in silico detection and manual correction, a BLAST analysis of the nonredundant part of the 554-kb sequence versus the full-length NCBI Locus Link/reference sequence (RefSeq) plus UniGene and TIGR human transcripts was carried out. Transcripts with high homologies to the Y-chromosomal sequence were selected for further analysis. Transcripts derived from specific autosomal regions that differed in exon composition were analyzed in detail. A total of 24 genic sequences were extracted from the Y-specific sequence, and the most likely allelic loci of the transcript in the human genome were identified using BLASTN. Exon-intron structures were delineated by Sim4. The underlying genomic sequence of the allelic loci was RepeatMasked and re-blasted against the 554-kb segment to characterize its modular structure.
Note added in proof
According to the current human genome assembly (NCBI Build 35, May 2004), the sequenced portion of the Y chromosome must be inserted at position 12.15 Mb of the human Y chromosome (in between the proximal block of satellite 3 sequences and the assembled contig NT_011875). The genomic clone RP11-322K23 connects our contig to NT_011875.
Acknowledgments
The human genomic PAC and BAC libraries used in this work were constructed at the RPCI in Buffalo, NY. Clones isolated from these libraries were purchased from the same institution. Satellite probes p22hom48.4, pHY10, pKFC68, pKFC52, pKFC11, pKFC37, and pKFC43 were kindly provided by Chris Tyler-Smith (Oxford). This work was supported by a grant from Pharmacia AB, Stockholm and the Deutsche Forschungsgesellschaft (Ra 380/10-1).
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3302705. Article published online ahead of print in January 2004.
References
- Arfin, S.M., Cirullo, R.E., Arredondo-Vega, F.X., and Smith, M. 1983. Assignment of structural gene for asparagine synthetase to human chromosome 7. Somatic Cell Genet. 9: 517-531. [DOI] [PubMed] [Google Scholar]
- Avarello, R., Pedicini, A., Caiulo, A., Zuffardi, O., and Fraccaro, M. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum. Genet. 89: 24-49. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 11: 1005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Viggiano, L., Misceo, D., Horvath, J.E., Archidiacono, N., Schwartz, S., Rocchi, M., and Eichler, E.E. 2002. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am J. Hum. Genet. 70: 83-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini, A., Ried, T., Shridhar, V., Ogura, K., D'Aiuto, L., Rocchi, M., and Ward, D.C. 1993. An alphoid DNA sequence conserved in all human and great ape chromosomes: Evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum. Genet. 90: 577-583. [DOI] [PubMed] [Google Scholar]
- Beckers, M., Gabriels, J., van der Maarel, S., De Vriese, A., Frants, R.R., Collen, D., and Belayew, A. 2001. Active genes in junk DNA? Characterization of DUX genes embedded within 3.3 kb repeated elements. Gene 264: 51-57. [DOI] [PubMed] [Google Scholar]
- Cooper, K.F., Fisher, R.B., and Tyler-Smith, C. 1992. Structure of the pericentric long arm region of the human Y chromosome. J. Mol. Biol. 228: 421-432. [DOI] [PubMed] [Google Scholar]
- Ding, H., Beckers, M.C., Plaisance, S., Marynen, P., Collen, D., and Belayew, A. 1998. Characterization of a double homeodomain protein (DUX1) encoded by a cDNA homologous to 3.3 kb dispersed repeated elements. Hum. Mol. Genet. 7: 1681-1694. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Lu, F., Shen, Y., Antonacci, R., Jurecic, V., Doggett, N.A., Moyzis, R.K., Baldini, A., Gibbs, R.A., and Nelson, D.L. 1996. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum. Mol. Genet. 5: 899-912. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Budarf, M.L., Rocchi, M., Deaven, L.L., Doggett, N.A., Baldini, A., Nelson, D.L., and Mohrenweiser, H.W. 1997. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum. Mol. Genet. 6: 991-1002. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Clark, R.A., and She, X. 2004. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat. Rev. Genet. 5: 345-354. [DOI] [PubMed] [Google Scholar]
- Gabriëls, J., Beckers, M.C., Ding, H., De Vriese, A., Plaisance, S., van der Maarel, S.M., Padberg, G.W., Frants, R.R., Hewitt, J.E., Collen, D., et al. 1999. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236: 25-32. [DOI] [PubMed] [Google Scholar]
- Grange, T., de Sa, C.M., Oddos, J., and Pictet, R. 1987. Human mRNA polyadenylate binding protein evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 15: 4771-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune, S., Yoshida, N., Chen, A., Garrett, L., Sugiyama, F., Takahashi, S., Yagami, K., Wynshaw-Boris, A., and Yoshiki, A. 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91-96. [DOI] [PubMed] [Google Scholar]
- Horvath, J.E., Viggiano, L., Loftus, B.J., Adams, M.D., Archidiacono, N., Rocchi, M., and Eichler, E.E. 2000a. Molecular structure and evolution of an α satellite/non-α satellite junction at 16p11. Hum. Mol. Genet. 9: 113-123. [DOI] [PubMed] [Google Scholar]
- Horvath, J.E., Schwartz, S., and Eichler, E.E. 2000b. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., Eichler, E.E., Schwartz, S., and Nicholls, R.D. 2000. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 10: 597-610. [DOI] [PubMed] [Google Scholar]
- Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111—120. [DOI] [PubMed] [Google Scholar]
- Kirsch, S., Weiss, B., Kleiman, S., Roberts, K., Pryor, J., Milunsky, A., Ferlin, A., Foresta, C., Matthijs, G., and Rappold, G.A. 2002. Localisation of the Y chromosome stature gene to a 700 kb interval in close proximity to the centromere. J. Med. Genet. 39: 507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijten, M., Wang, Y., Smith, B.T., Westerveld, A., Smink, L.J., Dunham, I., Roe, B.A., and Hulsebos, T.J. 2000. Mechanism of spreading of the highly related neurofibromatosis type 1 (NF1) pseudogenes on chromosomes 2, 14 and 22. Eur. J. Hum. Genet. 8: 209-214. [DOI] [PubMed] [Google Scholar]
- Lyle, R., Wright, T.J., Clark, L.N., and Hewitt, J.E. 1995. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 28: 389—397. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M. and Gould, K.L. 1999. The Arp2/3 complex: A multifunctional actin organizer. Curr. Opin. Cell Biol. 11: 117-121. [DOI] [PubMed] [Google Scholar]
- Matsuoka, S., Huang, M., and Elledge, S.J. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893-1897. [DOI] [PubMed] [Google Scholar]
- Myers, E.W. and Miller, W. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4: 11-17. [DOI] [PubMed] [Google Scholar]
- Nei, M., Gu, X., and Sitnikova, T. 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. 94: 7799-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander, J.M., Amos, L.A., and Linck, R.W. 1992. Primary structure of tektin A1: Comparison with intermediate-filament proteins and a model for its association with tubulin. Proc. Natl. Acad. Sci. 89: 6567—6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, T. and Matsuo, N. 1992. Comparison of adult height between patients with XX and XY gonadal dysgenesis: Support for a Y specific growth gene(s). J. Med. Genet. 29: 539-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, D.C. 1987. Hypothesis: A Y-chromosomal gene causes gonadoblastoma in dysgenetic gonads. Development. (Suppl.) 101: 151-155. [DOI] [PubMed] [Google Scholar]
- Reymond, A., Camargo, A.A., Deutsch, S., Stevenson, B.J., Parmigiani, R.B., Ucla, C., Bettoni, F., Rossier, C., Lyle, R., Guipponi, M., et al. 2002. Nineteen additional unpredicted transcripts from human chromosome 21. Genomics 79: 824-832. [DOI] [PubMed] [Google Scholar]
- Ried, T., Lengauer, C., Cremer, T., Wiegant, J., Raap, A.K., van der Ploeg, M., Groitl, P., and Lipp, M. 1992. Specific metaphase and interphase detection of the breakpoint region in 8q24 of Burkitt lymphoma cells by triple-color fluorescence in situ hybridization. Genes Chromosomes Cancer 4: 69-74. [DOI] [PubMed] [Google Scholar]
- Rudd, M.K. and Willard, H.F. 2004. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 20: 529-533. [DOI] [PubMed] [Google Scholar]
- Salo, P., Kaariainen, H., Petrovic, V., Peltomaki, P., Page, D.C., and de la Chapelle, A. 1995. Molecular mapping of the putative gonadoblastoma locus on the Y chromosome. Genes Chromosomes Cancer 14: 210-214. [DOI] [PubMed] [Google Scholar]
- Samonte, R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Schempp, W., Binkele, A., Arnemann, J., Glaser, B., Ma, K., Taylor, K., Toder, R., Wolfe, J., Zeitler, S., and Chandley, A.C. 1995. Comparative mapping of YRRM- and TSPY-related cosmids in man and hominoid apes. Chromosome Res. 3: 227-234. [DOI] [PubMed] [Google Scholar]
- Shaikh, T.H., Kurahashi, H., Saitta, S.C., O'Hare, A.M., Hu, P., Roe, B.A., Driscoll, D.A., McDonald-McGinn, D.M., Zackai, E.H., Budarf, M.L., et al. 2000. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 9: 489-501. [DOI] [PubMed] [Google Scholar]
- She, X., Horvath J.E., Jiang, Z., Liu, G., Furey, T.S., Christ, L., Clark, R., Graves, T., Gulden, C.L., Alkan, C., et al. 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430: 857-864. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825-837. [DOI] [PubMed] [Google Scholar]
- Smith, D.W., Marokus, R., and Graham Jr., J.M. 1985. Tentative evidence of Y-linked statural gene(s). Clin. Pediatr. 24: 189-192. [DOI] [PubMed] [Google Scholar]
- Stankiewicz, P. and Lupski, J.R. 2002. Molecular-evolutionary mechanisms for genomic disorders. Curr. Opin. Genet. Dev. 12: 312-319. [DOI] [PubMed] [Google Scholar]
- Tilford, C.A., Kuroda-Kawaguchi, T., Skaletsky, H., Rozem, S., Brown, L.G., Rosenberg, M., McPherson J.D., Wylie, K., Sekhon, M., Kucaba, T.A., et al. 2001. A physical map of the human Y chromosome. Nature 409: 943-945. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, K., Reijo, R., Page, D.C., and Disteche, C.M. 1995. Gonadoblastoma: Molecular definition of the susceptibility region on the Y chromosome. Am. J. Hum. Genet. 57: 1400-1407. [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith, C. 1987. Structure of repeated sequences in the centromeric region of the human Y chromosome. Development (Suppl.) 101: 93-100. [PubMed] [Google Scholar]