Abstract
Excess Phosphorus (P) in agriculture is causing serious environmental problems like eutrophication of lakes and rivers. Unlike the enormous information available for phosphate starvation response (P0), very few information is available for the effect of excess phosphate Pi on plants. Characterization of Excess Phosphate Response (EPiR) is essential for designing strategies to increase phosphate accumulation and tolerance. We show a significant modulation in the root developmental plasticity under the increasing supply of excess Pi. An excess supply of 20 mM Pi (P20) produces a shallow root system architecture (RSA), reduces primary root growth, root apical meristem size, and meristematic activity in Arabidopsis. The inhibition of primary root growth and development is indeterminate in nature and caused by the decrease in number of meristematic cortical cells due to EPiR. Significant changes occurred in metal nutrients level due to excess Pi supply. A comparative microarray investigation of the EPiR response reveals a modulation in ethylene biosynthesis and signaling, metal ions deficiency response, and root development related genes. We used ethylene-insensitive or sensitive mutants to provide more evidence for ethylene-mediated signaling. A new role of EPiR in regulating the developmental responses of plants mediated by ethylene has been demonstrated.
Introduction
Phosphate (Pi) availability is often limited in the soil, because of its low mobility, thus creating a major problem in various regions of the world. Several studies have undertaken to characterize plant responses under Pi starvation condition1–4. On the other hand in the developed industrialized world, the excessive application of Pi based chemical fertilizers and phosphate-rich animal manures often results in the accumulation of Pi in the top soil5–8. It leaches into surface water runoffs posing serious environmental concerns like eutrophication5–11. Chesapeake Bay region Maryland, Okeechobee Basin region of Florida (USA) and Sussex County, Delaware (USA) were rated as excessive in phosphorus8, 12, 13. The maximum inorganic P content reaches up to the order of 2610–7343 mg kg−1 in the soil around the world14–16. However, the availability of P varies greatly in different soils of the different regions. In Mehlich 3 soil of New York state (USA), total inorganic phosphorus level reached as high as 7343 mg P kg−1, but the resin-extractable P (available phosphorus) was found up to 2330 mg P kg−1 15. In the same soil, the content of dilute acid Pi pool increased up to 4815 mg kg−1 15. In a recent report, swine manure amended paddy soil profile showed an orthophosphate level up to 2610 mg kg−1 with 80% extraction efficiency16. Moreover, this situation may increase as Tillman et al.17 predicted that the most natural ecosystems will be converted into agriculture land by the year of 2050 owing to the increase in demand for food. To mitigate the adverse effect of agriculture expansion, we need to explore modern biotechnological tools to increase phosphate acquisition efficiency of crop plants which require less input of fertilizer.
In our earlier studies, we have evaluated the potential of Lolium multiflorum cultivars for remediation of excess phosphorus8, 10, 11. Studies conducted to test the efficacy of the plant-based remediation supports the use of this technology; however, the current P uptake rates in plants are not sufficient enough to tackle the problem8, 9. There is a need to significantly increase the P uptake and accumulation efficiency in plant to achieve the goal9. Genetic engineering may hold the promise for developing P-hyperaccumulator plants. However, no study has been taken up to investigate the plant responses under a wide range of P regime in detail. Furthermore, unlike the well-documented mechanism of heavy metal toxicity18, there is no information available about the toxicity of phosphorus in plants except few reports showing Fe or Zn deficiency under high P condition19, 20. Therefore, to design an effective strategy for P acquisition, accumulation, and tolerance, we need to understand and characterize the plant responses under excess phosphate conditions.
Several adaptive responses to Pi starvation have been established; for example, attenuation of primary root growth, increase in the lateral root density, and accumulation of anthocyanin21–24. Several transcriptomic studies have been carried out in plants to characterize the molecular response to phosphate deficiency or sufficiency using the ATH1 microarray chips25–28. Unlike these enormous investigations on phosphate (Pi) starvation, very few studies have been conducted to characterize plant responses under excess phosphate10, 29. A comprehensive morphophysiological study coupled with high-throughput gene expression technology to characterize the effect of varying concentrations of excess Pi on plants is still lacking. Thus we have designed experiments to augment the present understanding of the excess phosphate response (EPiR) in the root and shoot tissues of Arabidopsis. We also intend to unravel the underlying signaling mechanism that regulates growth and development of plant during excess phosphate treatment.
Here, we showed concentration dependent changes in root system architecture (RSA), total soluble Pi content, and metal content (P, Fe, Zn, Ca, Mn). The global transcriptomic profiling reveals that the excess Pi supply induces an ethylene-mediated response which may be responsible for the alteration of the phenotype. We further provide evidence for the involvement of ethylene by using ethylene-insensitive mutants (etr1–3, ein4, ein2-T) and constitutive ethylene sensitive mutant (ctr1–1).
Results
Excess Pi supply affects shoot morphophysiological traits
The morphophysiological traits of Arabidopsis were examined after 7 days of growth under P0 (0 mM), P1.25 (1.25 mM), P2.5 (2.5 mM), P5 (5.0 mM), P10 (10 mM) and P20 (20 mM) Pi concentrations (Figs 1 and 2). Treatments P1.25, P2.5, P5 and P10 exhibited a larger shoot area similar to each other (Fig. 1b). P20 showed a significant lower shoot area compared to P1.25 only (Fig. 1b). The P0 treatment showed a leaf area significantly smaller than all other treatments except P20. The shoot to root ratio increased significantly at the P20 as compared to P10, P5, and P2.5 treatments. It indicates that the shoot growth was not as much compromised as root at the high P20 treatment (Fig. 1c). The physiologically available total soluble Pi content peaked at P2.5 but later dropped to a plateau, at treatments P5, P10, and P20 (Fig. 1d). Anthocyanin content is considered to be a specific marker for the Pi deficiency response and was found to be highest at P0 and P20 (Fig. 1e). The expression level of Pht1 family transporters significantly repressed under the excess concentrations of Pi (Fig. 1f, Supplementary Fig. S1). Overall the shoot growth was promoted at P1.25 and adversely affected at P0 and P20 treatments.
Figure 1.
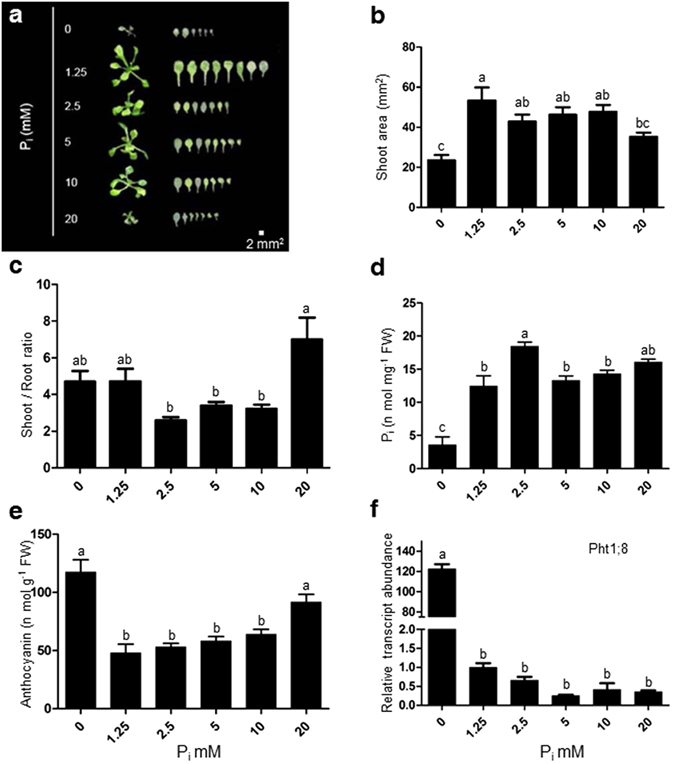
Supply of different concentrations of excess phosphate (Pi) modulates morphophysiological traits of the shoot. WT seedlings were initially grown hydroponically in 0.5X MS media for 5 days thereafter subjected to different Pi supplies (0, 1.25, 2.5, 5, 10, 20 mM) and grown for 7 days. Seedlings were spread on the agar Petri-plates for measurement of traits (a,b). Data are presented for shoot area (b), shoot to root ratio (c), total soluble Pi content (d), anthocyanin content (e), the expression level of Pht1;8 in shoot (f). Values are means ± SE, n = 21 (b,c), n = 3 consists of 30–70 mg shoot tissue in each assay (d, and e), n = 3 consists of 50 mg shoot tissue for each replicate (f). Mean Bars with different alpha letter differ significantly (P ≤ 0.05) according to analysis of variance (1X-ANOVA) with Tukey’s Multiple Comparison Posttest.
Figure 2.
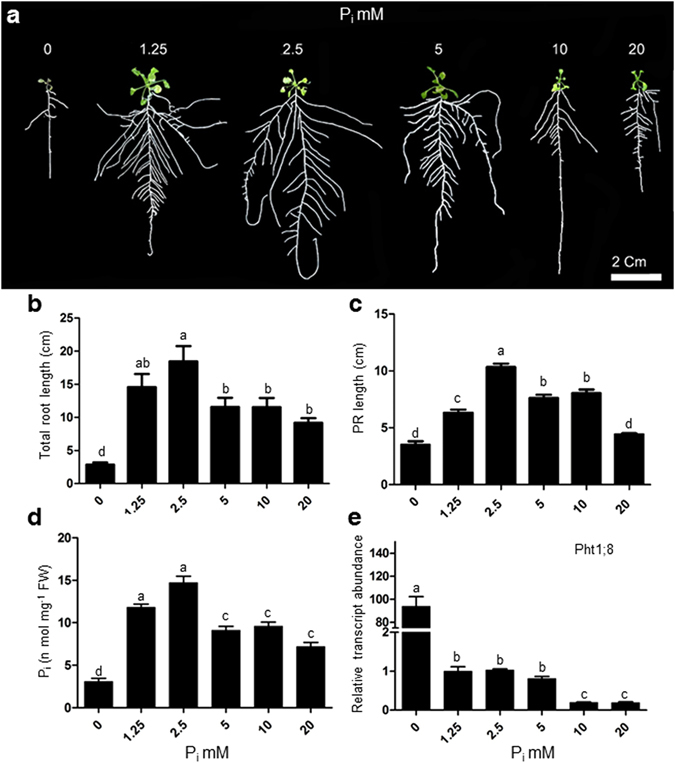
Supply of different concentrations of excess phosphate (Pi) modulates different morphophysiological parameters of root. WT seedlings were grown as described in the caption to Fig. 1. Data are presented for Total root length (b), Primary root length (c), total soluble Pi content (d), Expression level of Pht1;8 (e), Values are means ± SE, and n = 21 (b,c) or n = 3 consists of 20–50 mg root tissue in each assay (d). n = 3 consists of 50 mg root tissue for each replicate (e). Mean Bars with different alpha letter differ significantly (P ≤ 0.05) according to analysis of variance (1X-ANOVA) with Tukey’s Multiple Comparison Posttest.
Excess Pi supply affects morphophysiological traits of root
The excess Pi treatment (P20) produced a striking root phenotype exhibiting shorter, shallow and less branched RSA (Fig. 2a). Total root length and primary root length increased till P2.5 and then decreased under subsequent concentrations with a maximum decrease at P20 (Fig. 2b,c). The root growth showed a correlation with total soluble Pi content in the root that was highest at P2.5 thereafter dropped to a plateau at treatments P5, P10, with a maximum decrease at P20 treatment (Fig. 2d). The soluble Pi content of shoot was higher as compared to root under different Pi treatments. It may be due to the complete repression of Pi uptake transporters expression like Pht1 family members in root (Fig. 2e, Supplementary Fig. S1); whereas, the expression of PHO1, a phosphate xylem loading transporter, was found to be increased at excess Pi concentrations (Supplementary Fig. S2).
Excess Pi supply modulates the number and length of higher order lateral roots
The length of the branching zone (BZPR) increased till treatment P2.5 thereafter decreased at P5 and P10 and then showed a sharp decline at P20 (Fig. 3a). The number of primary lateral roots (1° LR) did not significantly change from P1.25 to P10 but later significantly decreased at P20 (Fig. 3b). The primary lateral root density did not change significantly at any of the concentration of treatments (P0 to P20) (Fig. 3c). The average 1° LR length increased till the treatment of P2.5 thereafter decreased at subsequent treatments but maintained a level found similar to P1.25 (Fig. 3d). The 2° LR density significantly decreased at P5, and P10 treatments as compared to P1.25 and rest of the treatments did not show any significant difference (Fig. 3e). Average 2° LR length significantly increased at P2.5 as compared to treatments P0 and P20 and remaining there was no significant difference among the treatments (Fig. 3f). Altogether these results show the initial Pi concentrations till 2.5 mM Pi promotes the root growth but subsequent concentrations of excess phosphate (10 or 20 mM) showed an adverse effect on RSA traits.
Figure 3.
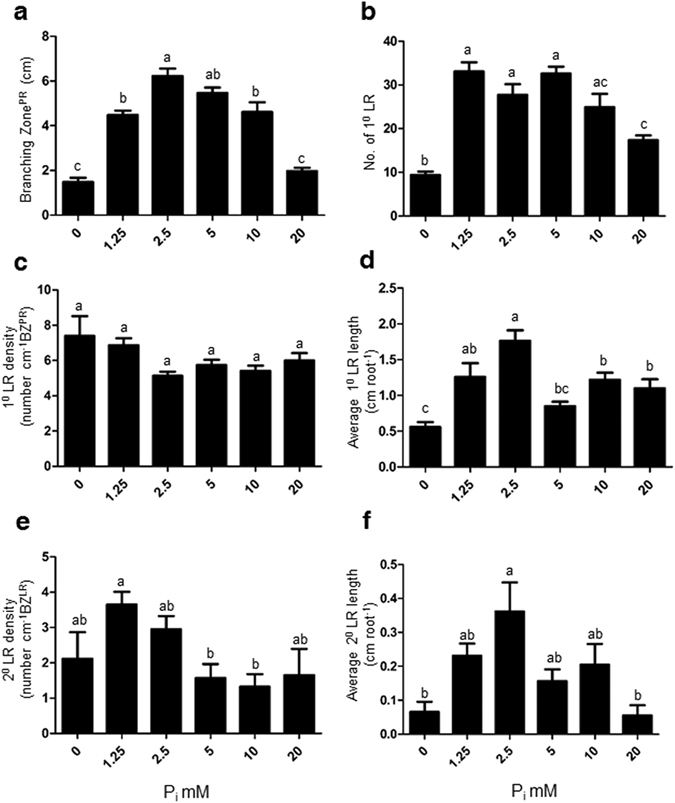
Supply of different concentrations of excess phosphate (Pi) modulates different traits of root system architecture. WT seedlings were grown as described in the caption to Fig. 1. Individual seedlings were carefully pulled out from the mesh and spread on the agar Petri-plates for documentation of different traits (a–f). Data are presented for Branching zone (a), number of first order lateral root (1° LR) (b), first order lateral root density (1° LR density) (c), average first order lateral root length (1° LR length) (d), Second order lateral root density (2° LR density) (e), average second order lateral root length (2° LR length) (f). Values are means ± SE, and n = 21 (a–f). Mean Bars with different alpha letter differ significantly (P0 ≤ 0.05) according to analysis of variance (1X-ANOVA) with Tukey’s Multiple Comparison Posttest.
Excess Pi supply inhibits primary root growth by attenuating meristematic activity
Earlier reports demonstrated a loss of apical meristem activity of primary root during Pi deprivation which results in a determinate developmental shift30, 31. Primary root tip of seedlings grown under the treatment of P1.25, P2.5, and P5, showed a high staining of CycB1;1::uidA activity; however, a significant decline in staining was observed under P10 and P20, treatment respectively (Fig. 4a). It suggests that the higher concentrations of Pi supply trigger the loss of meristematic activity. The next question is whether this developmental shift is determinate or indeterminate. To answer this, we transferred the plants grown in P1.25, P20 and P0 medium to P1.25 medium and allowed them to grow further for 7 days. Interestingly, the length of primary root (P20, P1.25) increased nearly 2-fold at 14 day relative to earlier time point (7 day); whereas the length of plants grown in P0 did not gain any significant increase under the same conditions (Fig. 4b,c). Similar parallel experiment was setup with CycB1;1::uidA transgenic plants to record the meristematic activity. Plants replenished into P1.25, P20 showed GUS staining, and plants replenished into P0 did not show any GUS staining (Fig. 4c). It suggests that inhibition of primary root growth by P20 is developmentally indeterminate in nature.
Figure 4.
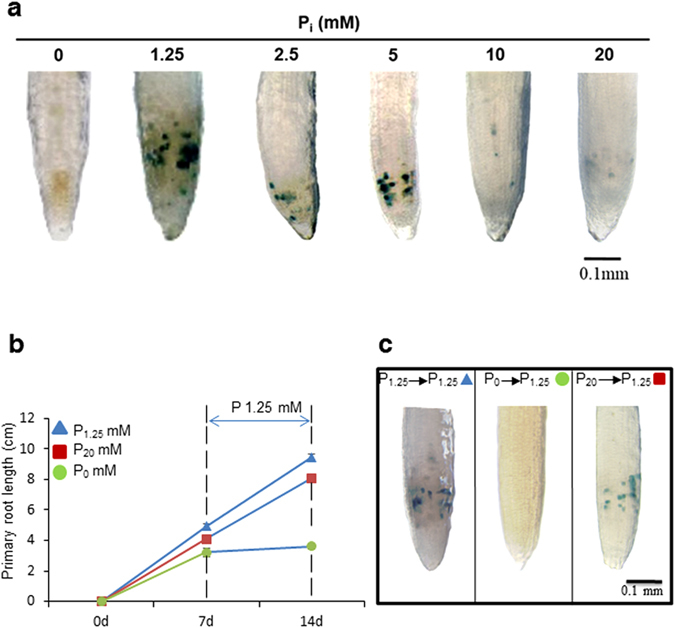
Excess Pi treatment attenuates the meristematic activity of primary root indeterminately. Histochemical GUS staining of primary root tips of CycB1;1:uidA transgenic seedlings grown as described in the legend to Fig. 1 (a). WT Arabidopsis seedlings were grown on P1.25, P0, and P20 media for 7 days as described earlier, thereafter replenished with P1.25 media and growth monitored after 7 days (b). A similar experiment was performed with transgenic seedlings of CycB1;1:uidA and documented for histochemical GUS staining (c). Pictures are representative of 10–12 seedlings. Values are means ± SE; n = 21 (b).
Evaluation of metal nutrients contents under excess Pi supply
We estimated the level of P, Fe, Zn, Ca, and Mn in Arabidopsis seedlings grown under excess Pi using ICP-OES. A concentration dependent increase was observed in P content till P5; thereafter a steady level was observed at P10 and P20 (Fig. 5a). In contrast, the maximum level of Fe was found at P0 that rapidly depleted in the presence of Pi until P2.5 and then maintained a steady level at subsequent concentrations (Fig. 5b). Zn content did not change significantly at increasing concentration of Pi till P5, thereafter only a significant decrease was observed at P10 and P20 (Fig. 5c). The content of Ca and Mn peaked at P2.5 then begin to drop at the subsequent treatments (Fig. 5d). Overall, the higher concentrations of Pi adversely affect the metal nutrients contents.
Figure 5.
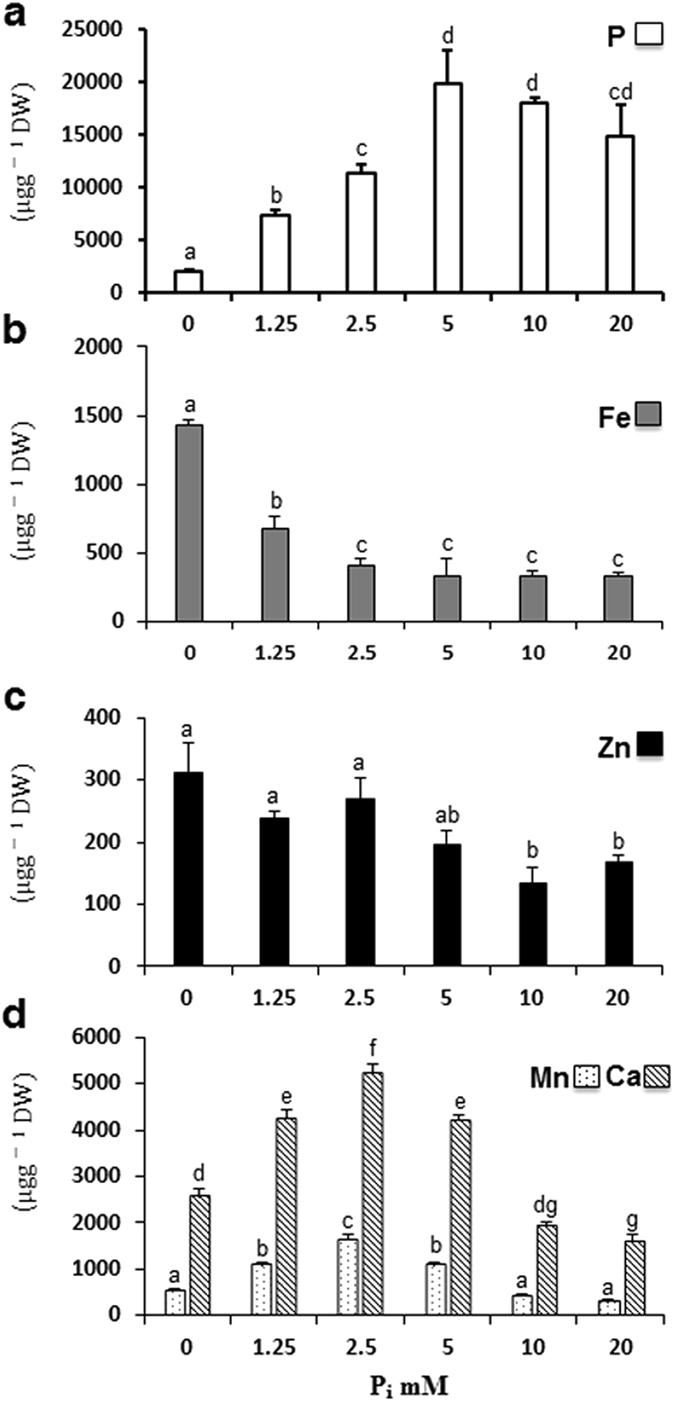
Metal nutrients analysis of Arabidopsis seedlings grown on varying concentrations of Pi hydroponically as described in the legend to Fig. 1. Data are presented as phosphorus content (a), iron content (b), zinc content (c), manganese and calcium content (d). Values are means ± SE; n = 3 replicates of 25 mg of dry weight of seedlings tissue. Mean Bars with different alpha letter differ significantly (P ≤ 0.05) according to analysis of variance (1X-ANOVA) with Tukey’s Multiple Comparison Posttest.
Transcriptomic comparison of excess phosphate response (EPiR) with phosphate starvation response (PiSR)
We comparatively studied plant transcriptome under P20 and P0 supply. A novel unbiased exon ST array chip from Affymetrix microarray technology was used to hybridize the RNA isolated from root and shoot tissues of Arabidopsis plants treated with P20, P0 and, P1.25 (control). This array uses the entire transcript to measure the expression of a gene thereby generates a precise and unbiased gene expression data (Supplementary Dataset File 1). Principal component analysis revealed the differential responses between root and shoot tissues across the treatments (Supplementary Fig. S3). Treatment components, P20 and P1.25, were relatively close to each other, indicating some similarities in their transcriptomic profiles (Supplementary Fig. S3). We obtained a total number of 31 and 2490 differentially regulated transcripts due to treatment P20 and P0, respectively in root (Fig. 6a). In shoot, a total number of 151 and 3603 transcripts were found to be differentially regulated due to treatment P20 and P0, respectively (Fig. 6b). We found a greater number of differentially regulated genes in shoots compared to root. Shoot serves as a sink organ for phosphate and exhibits a systemic adaptive response under varying Pi availability. Since our emphasis on excess phosphate response, we annotated in detail those transcripts which were relevant to EPiR. In root, only 9 genes were co-regulated by P20, P0 treatments (Supplementary Dataset File 2). The flowering promoting factor 1 (FPF1) is expressed in response to gibberellin stimulus and implicated in root growth inhibition32. The NRT1.8 is a low-affinity nitrogen transporter removes N from xylem tissue33. Nitrogen uptake is reported to be regulated by varying P availability34. FRD3 (Ferric Reductase Defective 3) is an antiporter/efflux transporter encodes a member of MATE (Multidrug and toxin efflux) is expressed in roots but not in shoots35. Expansin A17, a member of the α-expansin gene family, causes loosening, and extension of plant cell walls and is involved in an ethylene mediated signaling pathway36. There were 22 transcripts uniquely regulated by P20 treatment in root (Supplementary Dataset File 2). These transcripts represent a significant enriched GO term, transition metal ion binding indicating metal nutrient deficiency response (Supplementary Dataset File 3, Supplementary Fig. S4). Nicotianamine synthase 2 (NAS2) is responsible for the synthesis of nicotianamine, involved in iron sensing and transport37. Ferric reduction oxidase 5, is reported to participate in iron reduction to facilitate the transport38. An unknown, defensin-like protein 203 was found to be highly expressed (33-fold) possessing a GASA-like (Gibberellin-regulated protein) domain in polypeptide sequence. We also found a novel upregulated calmodulin binding protein (FDR 0.25), an MLO14-like protein that possesses a 7-transmembrane domain similar to G-Protein Coupled Receptor (GPCR), known to express during seedling stage in developing primary root tip39 (Supplementary Dataset File 1).
Figure 6.
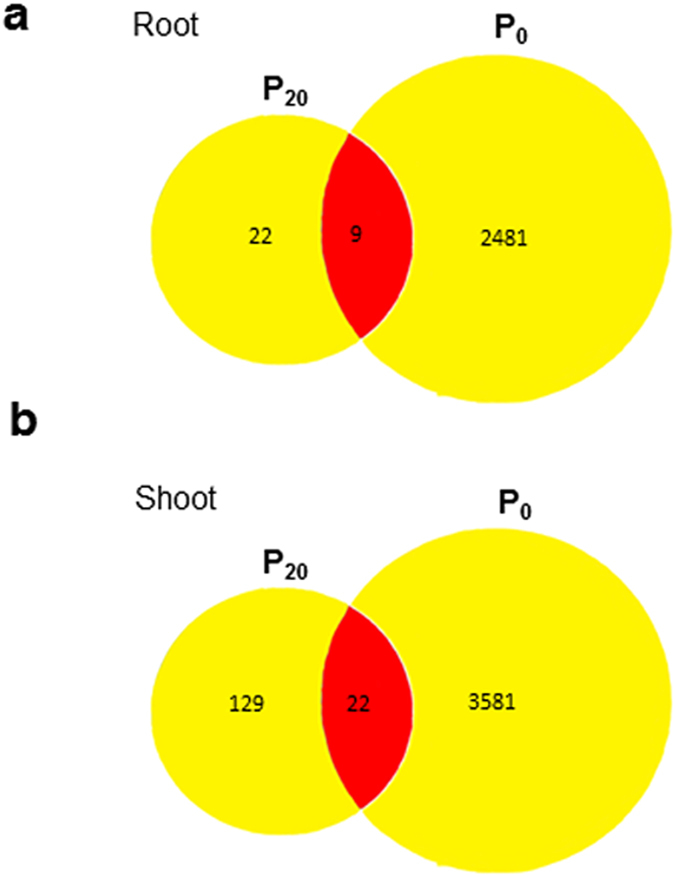
Differentially regulated genes, due to treatments P20 (20 mM Pi) or P0 (no Pi) relative to control (1.25 mM Pi) in root and shoot. Venn diagrams show the total number of uniquely and commonly regulated genes due to treatments P0 and P20 in root (a) and shoot (b).
In shoot, 22 transcripts were commonly regulated by both the treatments (Supplementary Dataset File 4). Three different peroxidase genes related to oxidative stress were found. Exordium (EXO//Phosphate-Responsive Family Protein), is a brassinosteroid stimulus-response gene, localized in the cell wall, was found to be upregulated40. These transcripts were associated with GO terms like lipid biosynthetic process and hydrolase activity (Supplementary Dataset File 3, Supplementary Fig. S5). Two other important genes (FDR ≤ 0.28) co-regulated were ACS7 and MDR4 (Supplementary Dataset File 1). ACS7 is an important member of ethylene biosynthetic pathway. MDR4 is an auxin influx transporter mediates basipetal transport of auxin in hypocotyl and root tip41. A number of 129 transcripts were uniquely regulated in shoot, and some of the important ones are described here. The AtACS11 gene, involved in ethylene biosynthesis, was found to be up-regulated. Four ethylene responsive factors namely ERF003, ERF012, ERF022, and ERF055, were found to be expressed. ATPMEPCRB is a probable pectin esterase inhibitor was found to be expressed. A recent report demonstrates that the pectin content plays an active role in solubilizing phosphate in the cell wall42. These transcripts represent enriched GO terms such as root system development, mitochondrial organization, protein targeting, RNA methylation, nucleotide metabolic process and response to metal ion (Supplementary Dataset File 3, Supplementary Dataset File 4, Supplementary Fig. S6).
To obtain a picture of the whole plant as a system, we combined root and shoot transcripts differentially expressed under excess phosphate (P20) regime and performed GO enrichment analysis. We observed three distinct biological processes enriched such as response to ethylene stimulus, root growth and development and response to metal nutrients (Fig. 7, Supplementary Fig. S7, Supplementary Dataset File 3). It provides evidence that the plant as a whole induces ethylene mediated signaling when incubated under excess phosphate regime. Altogether, these results indicate the underlying signaling mechanism which involves ethylene during excess phosphate response.
Figure 7.
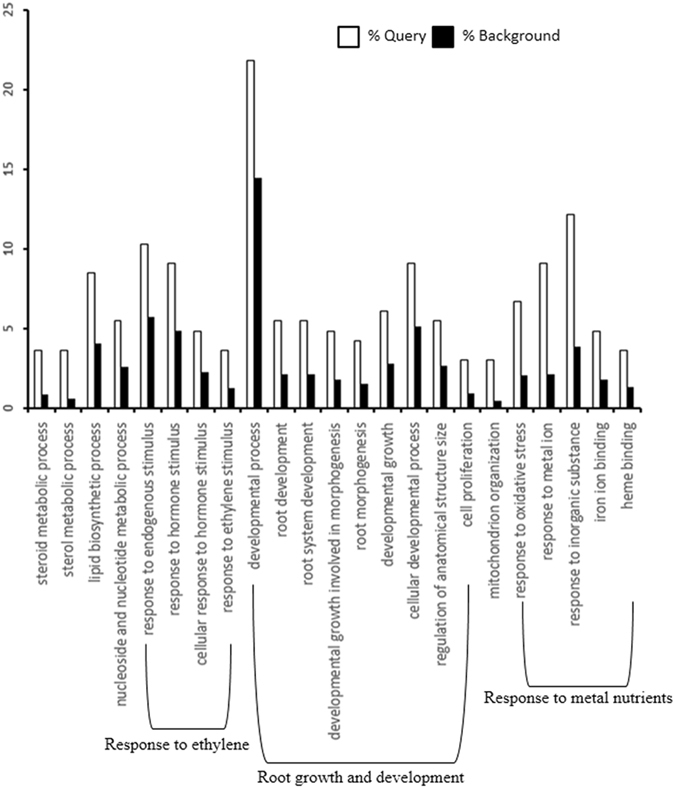
Singular Enrichment Analysis (SEA) for differentially expressed genes of root + shoot due to P20 (20 mM) treatment. It shows the change in ethylene, root growth and development, and response to metal nutrients related processes. The analysis was performed using the agriGO analysis tool under advanced mode, Fisher test with P < 0.05 and minimum 5 mapping entries. The white bar shows the fraction of query genes, and the black bar shows a fraction of genes in background (whole genome).
Validation of microarray expression data through qRT-PCR
Sixteen key genes were selected for validation of microarray data on the basis of metal ion deficiency response, ethylene mediated signaling, and root development (Supplementary Fig. S8). The expression of selected genes is more or less consistent with the microarray results. In general, treatment specific expression of genes is more consistent with the microarray results compared to tissue-specific expression. The iron deficiency response has been validated in root under P20; whereas, some members (NAS2, FRO5) showed deficiency response in shoot even under P0. It may be due to the variation in availability of different metal cations among the root and shoot. Ethylene biosynthesis and signaling related responses were validated; although, root and shoot were more or less equally responsive to both Pi treatments except ERF003. ERF003 showed a differential expression specifically under P20 in both tissues. Root development related genes also followed a similar expression trend with the array result; however, MLO14 was distinctly induced in root by P20. DEFL203 followed more or less similar trend as MLO14. Two oxidative stress related genes showed a high expression in root and shoot tissues under P20. Differential expression was confirmed for a SPX domain-containing protein that has been implicated in the Pi starvation response4. Overall, the expression of these genes coincides with array results.
Ethylene insensitive mutants show a better developmental response under excess phosphate
To obtain more evidence for ethylene mediated signaling, we quantified shoot area and traits of RSA in ethylene-insensitive mutants etr1–3, ein4, ein2-T 43, 44, and a sensitive ethylene mutant ctr1–1 45 under control (P1.25) and excess Pi condition (P20) (Fig. 8). Shoot area decreased significantly by 28% in WT under P20. In the ethylene-insensitive mutants, no significant difference was observed in shoot area due to P20 treatment (Fig. 8b). Total root length of WT decreased significantly by 27% due to P20; whereas, it increased significantly by 29% in ein2-T or increased slightly in other insensitive mutants (Fig. 8c). Inhibition of primary root length was higher in WT than in ethylene-insensitive mutants etr1–3, ein4, ein2-T, (Fig. 8d). There was no significant difference in the phenotype of ethylene sensitive mutant ctr1–1 except a significant increase in 1° LR density due to P20 (Fig. 8e). Altogether ethylene-insensitive mutants exhibited a better growth and development at P20 indicating some involvement of ethylene signaling.
Figure 8.
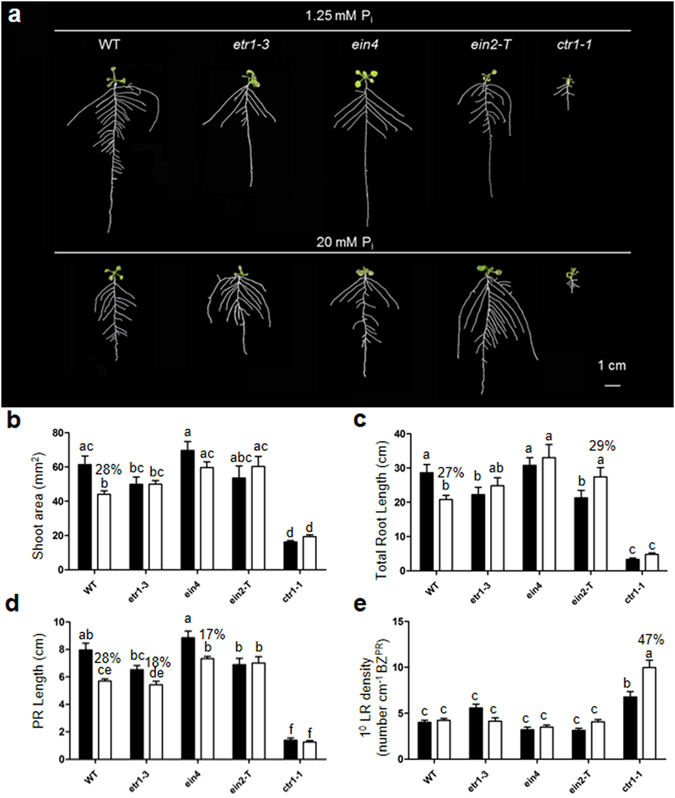
Root system architecture of wild type, ethylene-insensitive mutants (etr1–3, ein4, ein2-T) and sensitive mutants (ctr1–1) under 1.25 mM Pi and 20 mM Pi. Wild-type (WT) and ethylene mutants were grown as described in the caption to Fig. 1. Individual seedlings were spread as described in the caption to Fig. 3 for documentation of different traits (b–e). Data are presented for shoot area (b), total root length (c), primary root length (d), and 1° lateral root density (e). Black and white bar represent treatments 1.25 mM and 20 mM, respectively. Values are means ± SE n = 15. Mean bars with different alpha letter differ significantly (P ≤ 0.05–0.001) according to two-way ANOVA with Bonferroni correction posttest comparisons.
Excess phosphate response reduces the root apical meristem size by decreasing the cell number
To determine the nature of the primary root length reduction, we analyzed the size of root apical meristem (RAM) (Fig. 9a)46. RAM size decreased significantly in WT; whereas, it significantly increased in ein2-T or showed no change in other mutants due to P20 (Fig. 9b, Supplementary Fig. S9). No significant change in RAM size of ctr1–1 mutant was observed due to P20 (Supplementary Fig. S10). We also analyzed the relative change in RAM size by taking the ratio at P20 to the P1.25. The relative increase in RAM size due to P20 treatment was significantly higher for ethylene-insensitive mutants as compared to WT (Fig. 9c). Taken together, it indicates the participation of ethylene mediated signaling during excess Pi treatment.
Figure 9.
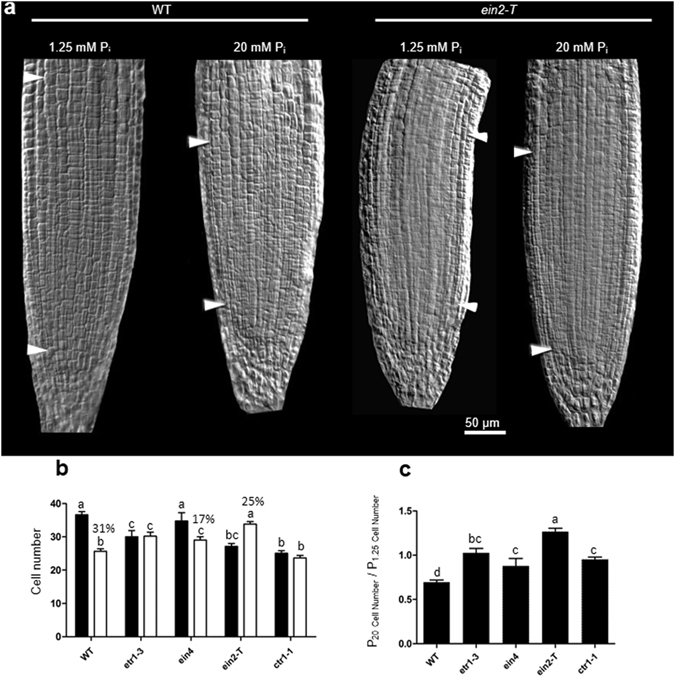
Excess phosphate supply reduced root apical meristem size of WT but not in ethylene- insensitive mutants. WT and ethylene insensitive (etr1–3, ein4, ein2-T) or sensitive mutant (ctr1–1) were grown as described in the caption to Fig. 1. A 5 mm section from the tip of primary root was cut and mounted on a slide with diluted Visikol in 10% glycerol, and root cells were visualized under the DIC filter. Root meristem size was measured by counting the number of meristematic cortex cells between the quiescent center (QC) and the first elongated cell. White arrows mark the QC and first elongated cortex cells in a representative picture of WT and ein2-T mutant (a). Data are presented for the number of cortical cells (b), and the ratio of RAM size (cell number) of plants grown at P20 to P1.25 conditions, as shown in (c). Values are means ± SE, and n = 15 (b,c). Bars with different alpha letter differ significantly (P ≤ 0.05–0.001) according to two-way ANOVA (b), and one-way ANOVA (c) with Bonferroni’s posttest.
To summarize the important findings of this study, we proposed a schematic model which shows that the excess phosphate treatment induces the ethylene response may be directly or by altering the nutrient level and attenuates primary root growth by reducing the cell number in root apical meristem (Fig. 10).
Figure 10.
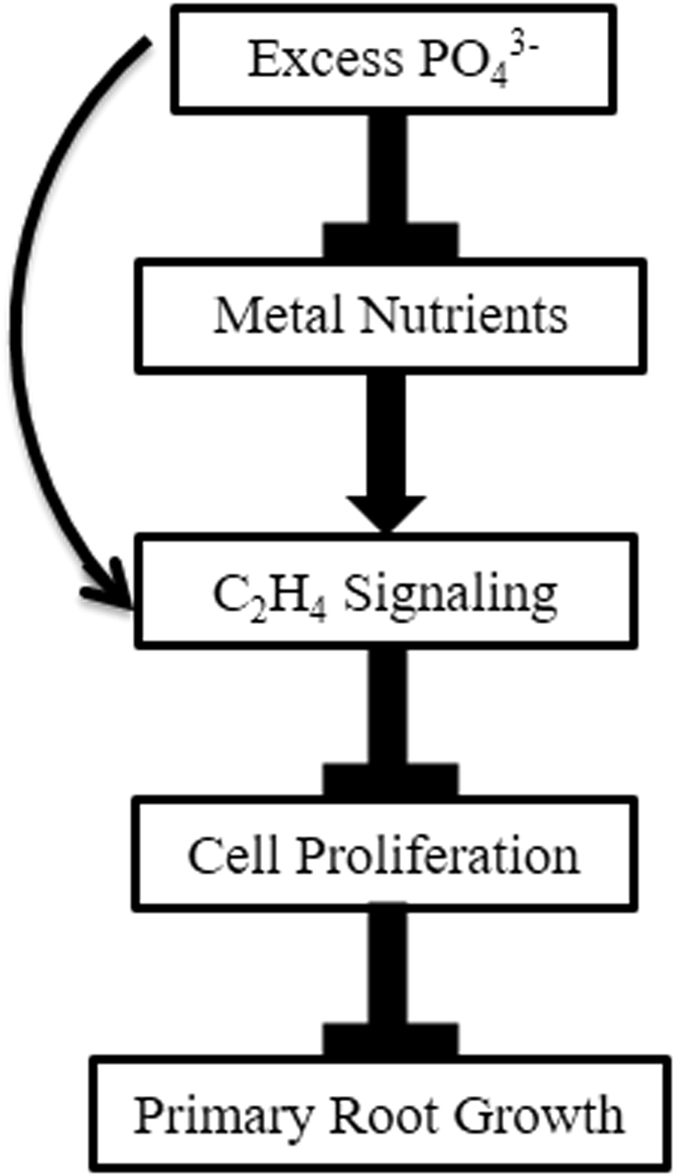
A common minimum consensus model displaying underlying signaling occurred during Excess Phosphate Response (EPiR). Excess phosphate treatment triggers the ethylene response may be directly or by altering nutrient profile and negatively regulates primary root growth by reducing cell proliferation in root apical meristem.
Discussion
Despite a rapid change in agronomic condition, causes environmental problems17, excess phosphate response in plants has not been elucidated in detail. Earlier reports showed that the high P condition in soil creates Fe and Zn deficiency in plants19, 20, 47, 48. In our earlier studies, we have attempted to identify genes modulated in crop plants like sunflower in excess Pi condition but obtained a little information29. In the present study, using morphophysiological, biochemical, genomics and genetic approaches we have characterized the excess phosphate response in Arabidopsis that indicates an ethylene mediated response.
We have shown that how varying concentrations of Pi could be sensed and rendered into phenotypic alterations. Different morphometric traits suggest that root morphology is more sensitive than shoot morphology by the supply of excess Pi (Figs 1 and 2). The total soluble Pi content was higher in the shoot under the excess Pi treatment points as compared to root (Figs 1d and 2d). The lesser level of soluble Pi in root may be linked to the shutdown of phosphate uptake transporters (Fig. 2e, Supplementary Fig. S1). At the same time, the mobilization of Pi from root to shoot keeps occurring because the expression of PHO1 was high (Supplementary Fig. S2a) therefore, leading to a low Pi level in the root (Supplementary Fig. S2b). The higher anthocyanin content of seedlings grown at P20, which was comparable to the level found in P0, supports this notion (Fig. 1e).
We observed a dramatic change in RSA by supplying varying concentrations of Pi (Fig. 2a). The primary root length increased initially and later decreased under increasing concentrations of Pi (Fig. 2c). This is in agreement with a study in which primary root length was shown to be inhibited by higher treatment concentrations of Pi at 10 and 25 mM23. The number of lateral roots remained steady till P5, then slightly decreased at P10 and with a significant decrease at P20 (Fig. 3b). This is consistent with an earlier study showing a sharp decline in the number of lateral roots at 10 and 25 mM Pi 23. Similarly, they showed a sharp decrease in L.R. density between 1 and 25 mM Pi 23. However, we did not observe any difference in L.R. density under P0, P1.25, and P20 (Fig. 3c). There are several contrasting reports on how Pi starvation-induced changes in number, length, and density of lateral roots. For example, an increase in L.R. density was reported under P0 condition21, 23. In contrast, a decrease in L.R. density under P0 conditions was also reported24, 49, 50. These variations may be due to the use of different growth conditions such as light, sucrose, incubation time51, 52. Also, the concentration of Pi 23, 24 and, the use of the various gelling media possessing varying levels of Pi contamination may attribute to these variations52, 53. Ethylene is known to reduce primary root length under Pi-sufficient condition54–56. Ethylene was also reported to be involved in lateral root spacing and to negatively regulate the number of lateral roots while increasing the length of existing lateral roots56. Interestingly, we have observed attenuation of primary root length, reduction in the number of lateral roots, and no alteration in the length of lateral roots under excess P10 and P20 condition.
We observed a significant decline in CycB1;1:uidA staining in the primary root tip grown at P10 and P20 (Fig. 4a). More than one factor may be responsible for these results. One of the factors may be that a depletion of total physiologically available Pi which plays a critical role in the growth and development of root (Fig. 2d). Other regulating factors may be linked hormonal regulation. For example, indoleacetic acid (IAA) treatment inhibits primary root growth without affecting the meristematic activity, but since we observed a decline in the meristematic activity, auxin-mediated inhibition of primary root growth might be ruled out. Another hormone factor is ethylene which not only negatively-regulates primary root growth but may also be involved in repressing the Cyclin B1 activity46. Moreover, the inhibition of primary root growth and meristematic activity due to P20 was indeterminate thus developmentally distinct from the inhibition caused by phosphate starvation (P0) (Fig. 4b and c).
The nutrient composition of P, Fe, Zn, Mn and Ca plays a crucial role in determining RSA plasticity24, 53. Earlier studies reported that the micronutrients deficiency in plants caused by high P condition19, 20, 47, 48. We observed a maximum reduction in primary root under P0 and P20 however, developmental program were different. It is known that the loss of root meristematic activity under P0 condition is due to the elevated Fe level as it could produce hydroxyl radicals leading to the death of meristematic cells47, 57, 58 (Fig. 5b). Under P20, Fe level is relatively low, despite we observed a reduction in primary root length and root meristematic activity (Fig. 5b). It may be due to the depletion of other essential nutrients (Zn, Mn, Ca) in the seedlings (Fig. 5c,d). The low Fe content could also be a reason for an indeterminate reduction in root meristematic activity under P20 (Fig. 5b). Zn deficiency reduces the primary root length and enhances the lateral root number, but meristematic activity is not affected53. Ca deficiency also causes a decline in primary root elongation24. Taken together, it is plausible that the alteration in the level of nutrients elicits the hormonal responses that control the RSA plasticity59. Although, the specific coordination mechanisms involved in root development are not known.
The use of the Arabidopsis ST array offers access to an extensive number (28403) of transcripts relative to ATH1 array (22810 probes) and highlights the advantage of using 1.0 ST array chips. Eighteen tissue and treatment specific arrays were analyzed with the goal of creating a reference dataset to gain novel insight into the excess phosphate response (EPiR). We presented a comparative gene expression Venn diagram by distributing differentially regulated genes under EPiR, PiSR and both for root and shoot tissues as shown in Fig. 6. The majority of the differentially expressed genes and associated enriched GO terms under excess phosphate point towards ethylene mediated signaling (Fig. 7).
The gene AtACS11 and AtACS7 which is an isoform of 1-aminocyclopropane -1-carboxylate synthase (ACS) provides the rate-limiting step in ethylene biosynthesis were found to be upregulated60. We obtained four members of ERFs, expressed uniquely in the P20 treatment in the shoot (Supplementary Dataset File 4). AtERF003 and AtERF012 each have a repressor motif called EAR (ERF-associated amphiphilic repression) at the C-terminal. This motif is responsible for the repression of transcription of several target genes61, 62 and controls the major phenotypic changes in plant growth and development63. However, direct evidence is lacking for the function of AtERF003. Another gene Expansin A17, reported to be involved in ethylene signaling, was upregulated by P20 treatment and downregulated by P0 36. Upregulation of several ethylene-related genes and their GO enrichment analysis suggest that ethylene at large mediating the excess phosphate response.
Nicotianamine synthase (NAS2), Ferric Reduction Oxidase 5 (FRO5), and FRD3 (Ferric Reductase Defective 3) were upregulated in root by P20 (Supplementary Dataset File 2). The expression of these genes is regulated by ethylene and was reported to be expressed under Fe, Zn and Cu deficiency35, 37, 38, 64 (Supplementary Dataset File 2). Some members of the peroxidase gene family were found to be differentially regulated by P20 and may be an indication of oxidative stress caused by metal deficiency (Supplementary Dataset File 2). Additionally, the GO enrichment analysis in root tissue resulted in GO terms like transition metal ion binding, response to metal ion, and heme binding (Supplementary Fig. 4). It suggests a metal ion deficiency occurring under excess phosphate condition.
We identified several enriched GO terms associated with root system development mainly for transcripts regulated in shoot (Supplementary Fig 6, Supplementary Dataset File 3). It indicates the important systemic role played by shoot regulating RSA under varying phosphate regime. Several previously known genes for controlling the RSA were identified. We found an upregulation of MLO 14 like protein in root, a recent study of MLO mutant revealed its involvement in polar auxin transport which is one of the components jointly responsible for an aberrant root phenotype65. A channeling protein, Annexin (ANNAT7) is reported to be involved in lateral root development by modulating the Ca2+ ion gradient66, 67. A defensin-like protein, DEFL203, was expressed at its maximum level in the P20 treatment. Defensins are small secreted peptides having diverse biological roles and may be involved in growth and development68. DEFL203 possesses a GARE element in its promoter region and a GASA-like domain in its peptide sequence that has a role in meristematic cell division activity69. The AtERF003 might play a major role in restructuring the root system. It is known to be involved in ethylene and/or gibberellic acid mediated signaling pathways70. The direct role of ERF in restructuring the root system architecture has been demonstrated in a study, in which the overexpression of AtERF070 leads to decrease in primary root length and lateral root number71.
Root system architecture is a complex trait as its development depends upon the hormonal interplay (ethylene, auxins, gibberellins, brassinosteroids and many more) and their interaction with the environment72. Since we obtained a set of genes which directly or indirectly points towards ethylene and signaling, we tested the hypothesis of ethylene by examining ethylene insensitive or sensitive mutants. Interestingly we observed a better growth phenotype in ethylene-insensitive mutants relative to WT under excess Pi condition (Fig. 8a). Constitutive ethylene sensitive mutant ctr1–1 produces no significant change in phenotype presumably due to exhibiting a full response at control treatment in our experimental condition thus only a slight alteration in RSA occurred due to P20. Shoot area shows a better development in ethylene-insensitive mutants (Fig. 8b). There are reports which show the role of ethylene in negative regulation of shoot growth and development73. Primary root shows a relatively better growth phenotype in ethylene-insensitive mutants (Fig. 8d). We observed some variation in phenotype of etr1–3, ein4, and ein2-T relative to WT at control condition (P1.25) (Fig. 8). It may be due to the variation in responses depending upon the degree of ethylene insensitivity. For example, ein4 mutant does not exhibit a strong insensitivity to ethylene rather behaves similar to WT74. Ethylene-sensitive mutant ctr1–1 shows lesser growth relative to WT under normal growth condition75. We observed a significant increase in the 1° lateral root density of ctr1–1 due to P20. This increase could be attributed to the increase in the number of lateral roots in a highly short primary root length (Fig. 8a).
Ethylene reduces root apical meristem (RAM) size and meristem activity46. Similarly, we observed a significant reduction in RAM size of WT due to P20 and relatively a larger meristem size in ethylene-insensitive mutants (Fig. 9a b and c). This provides insight into how the primary root length is decreased under excess Pi (P20). We have also shown that the P20 treatment represses the activity of a cell cycle marker Cyclin B1, which is consistent with these RAM size results (Fig. 4a).
In conclusion, we provide comprehensive reference datasets of plant responses under excess and deficient Pi regimes. We have demonstrated that the ethylene mediates the negative regulation of plant growth and development like attenuation of primary root length, and decrease in root apical meristem size under excess phosphate. Our study sheds light on the intricate relationship of P, metal ions, ethylene and their effect on plant growth and development. This study not only helps in dissecting the mechanism of plant response to excess Pi but also paves the way to identify the pathways and additional novel genes involved in phosphorus homeostasis and accumulation. Although we characterized excess Pi responses, furthermore experiments may be required to test the specificity of these responses to other high level nutrient conditions.
Methods
Plant materials, growth, and treatments
Wild-type and mutants of Arabidopsis thaliana used in the study were in the Columbia (Col) background. Mutant seeds of etr1–3 (CS3070), ein4 (CS8053), were obtained from ABRC (Ohio). Seeds of ein2-T and ctr1–1 genotypes were kindly provided by Rishikesh Bhalerao (Umea Plant Science Center, Sweden). Seeds were surface sterilized and suspended in water and incubated 2 days at 4 °C for stratification. Seeds were germinated and grown in 0.5X MS basal medium with vitamins (PhytoTechnology laboratory) + 1.5% (w/v) Sucrose medium on a polypropylene mesh hydroponically for 5 days (Supplementary Fig. S11)52. After 5 days, the seedlings were transferred into modified MS nutrient media containing 2.0 mM NH4NO3, 1.9 mM KNO3, 0.15 mM MgSO4·7H2O, 0.1 mM MnSO4·H2O, 3.0 µM ZnSO4·7H2O, 0.1 µM CuSO4·5H2O, 0.3 mM CaCl2.2H2O, 5.0 µM KI, 0.1 µM CoCl2·6H2O, 0.1 mM FeSO4·7H2O, 0.1 mM Na2EDTA.2H2O, 1.25 mM KH2PO4, 100 µM H3BO3, 1 µM Na2MoO4·2H2O, 1.5% sucrose, 1.25 mM MES, pH 5.7 adjusted with 0.1 M MES (pH 6.1), and grown for 7 days. For P0 (0 mM) treatment, KH2PO4 was replaced with 0.62 mM K2SO4. For excess phosphate treatments, the concentration of KH2PO4 was increased in modified MS medium (2.5, 5.0, 10.0, 20.0 mM). We chose the KH2PO4 salt because potassium is considered as a suitable ion that did not affect the plant growth and development76–78. Plants were grown under standard growth condition as 16 h light /8 h dark photoperiod, 120 µmol m−2 s−1 light intensity, 60–70% humidity, at 23 °C in a growth chamber. Nutrient solutions were changed on alternate days.
Examination of root system architecture and shoot area
Seedlings were spread gently on agar (1.2%) Petri-plates using art brush. Leaves were dissected and flattened on agar plates. Petri-plates were photographed for the documentation of phenotypic traits. The root length and shoot area were measured using the ImageJ program.
Anthocyanin measurement
Shoot tissue of Arabidopsis seedlings was separated and ground into powder using liquid N2. Around 50–100 mg tissue was used to extract the anthocyanin following the method described by Neff and Chory79.
Estimation of total soluble Pi content
Root and shoot tissues were separated, rinsed with distilled water, blotted dry, and ground to fine powder using liquid N2. The estimation was carried out using the standard method31.
Histochemical GUS staining
The GUS staining of Arabidopsis genotype CycB1;1:uidA 80 seedlings was performed as per the standard protocol. The pictures of the stained seedlings were taken in the bright field with a Leica M16 stereomicroscope with an axial carrier using Automontage software (Syncroscopy-Frederick, Maryland USA).
ICP-OES analysis
ICP-OES analysis was performed as described earlier in our study53.
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from frozen root and shoot tissues of 12 days old plant using the method described earlier81. The cDNA synthesis and quantitative real-time PCR were carried out as described by Shukla et al.81. The primer sequences are listed in Supplementary Table S1. The qRT-PCR experiments were repeated two times independently, and in each experiment, three technical replicates were used.
Transcriptome analysis
Affymetrix GeneChip® ara ST 1.0 GeneChips (data submitted to GEO accession number GSE66925) were used to carry out the microarray experiments following the instructions of the manufacturer (Affymetrix, USA). Three independent biological experiments were carried out for control (1.25 mM Pi) and experimental conditions (0 mM, 20 mM Pi) separately for root and shoot tissue of Arabidopsis. The feature intensity (.cel) files were normalized at gene level using RMA algorithms provided in the software Partek Genomics Suite 6.6, St. Louis, MO, USA. To identify statistically significant differentially expressed genes, a two-way ANOVA model was applied to the intensity data. A step-up false discovery rate (FDR) corrected p-value was included for every p-value calculated, and different contrasts were generated. The list of differentially expressed genes (DEGs) was generated by filtering the data at p < 0.1 (FDR) and >2-fold change (P0/P1.25) and >1-fold change (P20/P1.25) in expression value.
GO enrichment analysis of differentially regulated genes
The singular enrichment analysis (SEA) (http://bioinfo.cau.edu.cn/agriGO/index.php) was performed on differentially regulated genes. The TAIR AGI IDs were subjected to agriGO analysis tool using the supported species mode. Arabidopsis TAIR10 database was used as a background. The hieratical graphs and GO enrichment analysis table of genes were constructed, using a Fisher test at P-value 0.05–0.01, with 5 minimum number of mapping entries with complete GO gene ontology. The GO abundance chart was prepared manually by selecting the biologically relevant GO terms.
Analysis of root apical meristem (RAM)
The mutants and WT seedlings were grown and treated with 1.25 mM Pi and 20 mM Pi as described in earlier section. The size of the root apical meristem (RAM) was determined by following the protocol of Duan et al.82. Instead of using Chloral hydrate, we used VisikolTm (Visikol Inc.) for clearing the root section. Visikol was diluted 200 times in 10% glycerol and applied to the root tissue. The number of cells counted between the quiescent cells and to the first elongated cells of the meristem was visualized with Carl Zeiss Axioplan 2 Imaging fluorescent microscope at 20X magnification using Differential Interference Contrast (DIC) filter.
Statistics
Each experiment was repeated 2–3 times independently. Statistical significance of mean values was determined by analysis of variance (1X-, 2X-ANOVA) with Tukey’s Multiple Comparison Test or Bonferroni correction post-test were used for comparing all pairs of data sets.
Electronic supplementary material
Acknowledgements
U.S. Department of Agriculture (Grant 58-6406-1-017) is acknowledged for supporting this research. We acknowledge the partial support by National Institutes of Health, KBRIN grant P20GM103436, P30GM106396. WKU Biotechnology Centre is acknowledged for providing the instrument facilities. We are thankful to Prof. Rishikesh Bhalerao, UPSC, Sweden for his generous gift of mutant seeds. We thank Dr. K.G. Raghothama, Purdue University, USA and Dr. Ajay Jain, NRCPB, India for providing CyclinB1 genetic material. Dr. John Andersland is acknowledged for his help during microscopy. We thank Sabine Waigal and Dr. Eric Rouchka for performing Microarray hybridization and providing primary expression data at University of Louisville Genomics Core Facility.
Author Contributions
S.V.S. and D.S. conceived the project and designed the experiments. D.S. performed the experiments and analyzed the data. D.S. and C.A.R. performed the transcriptome analysis. S.V.S. supervised the experiments and contributed intellectually to all aspects of research; D.S. wrote the manuscript with the contribution of all the authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03061-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Devesh Shukla, Email: devesh.nbri@gmail.com.
Shivendra V. Sahi, Email: shiv.sahi@wku.edu
References
- 1.Bari R, Datt Pant B, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define aphosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 3.Chiou TJ, Lin SI. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011;62:185–206. doi: 10.1146/annurev-arplant-042110-103849. [DOI] [PubMed] [Google Scholar]
- 4.Puga MI, et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpley A, Tunney H. Phosphorus research strategies to meet agricultural and environmental challenges of the 21st century. J. Environ. Qual. 2000;29:176–181. doi: 10.2134/jeq2000.00472425002900010022x. [DOI] [Google Scholar]
- 6.Maguire, R.O., Chardon, W.J. & Simard, R. R. Assessing potential environmental impacts of soil P by soil testing in Phosphorus: agriculture and the environment (eds Sims, J.T. & Sharpley, A.N.) 145–180 (ASA-CSSA-SSSA, 2005).
- 7.Foy, R. H. The return of the phosphorus paradigm: agricultural phosphorus and eutrophication in Phosphorus:agriculture and the environment (eds Sims, J.T. & Sharpley, A.N.) 911–939 (ASA-CSSA-SSSA, 2005).
- 8.Sharma, N. C. & Sahi, S. V. Excess soil phosphorus: accelerated P transfer, water quality deterioration, and sustainable remediation strategies in Improving crop resistance to abiotic stress (eds Tuteja, N., Gill, S.S., Tiburcio, A.F. & Tuteja, R.) 165-191 (Wiley-VCH, 2012).
- 9.Novak JM, Chan SK. Development of P-hyperaccumulator plant strategies to remediate soils with excess P concentrations. Crit. Rev. Plant Sci. 2002;21:493–509. doi: 10.1080/0735-260291044331. [DOI] [Google Scholar]
- 10.Sharma NC, Sahi SV. Characterization of phosphate accumulation in Lolium multiflorum for remediation of phosphorus-enriched soils. Environ. Sci. Technol. 2005;39:5475–5480. doi: 10.1021/es050198t. [DOI] [PubMed] [Google Scholar]
- 11.Sharma NC, Starnes DL, Sahi SV. Phytoextraction of excess soil phosphorus. Environ. Pollut. 2007;146:120–127. doi: 10.1016/j.envpol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Graetz DA, Nair VD. Fate of phosphorus in Florida Spodosols contaminated with cattle manure. Ecol. Eng. 1995;5:163–181. doi: 10.1016/0925-8574(95)00023-2. [DOI] [Google Scholar]
- 13.Nair VD, Graetz DA, Reddy KR. Dairy manure influences on phosphorusretention capacity of Spodosols. J. Environ. Qual. 1998;27:522–527. doi: 10.2134/jeq1998.00472425002700030007x. [DOI] [Google Scholar]
- 14.Frossard E, Condron LM, Oberson A, Sinaj S, Fardeau JC. Processes governing phosphorus availability in temperate soils. J. Environ. Qual. 2000;29:15–23. doi: 10.2134/jeq2000.00472425002900010003x. [DOI] [Google Scholar]
- 15.Lehmann J, et al. Long-term dynamics of phosphorus forms and retention in manure-amended soils. Environ. Sci. Technol. 2005;39:6672–6680. doi: 10.1021/es047997g. [DOI] [PubMed] [Google Scholar]
- 16.Liang X, et al. Composition of phosphorus species and phosphatase activities in a paddy soil treated with manure at varying rates. Agr. Ecosyst. Environ. 2017;237:173–181. doi: 10.1016/j.agee.2016.12.033. [DOI] [Google Scholar]
- 17.Tilman D, et al. Forecasting agriculturally driven global environmental change. Science. 2001;292:281–284. doi: 10.1126/science.1057544. [DOI] [PubMed] [Google Scholar]
- 18.Shukla, D., Trivedi, P. K., Nath, P. & Tuteja, N. Metallothioneins and phytochelatins: role and perspectives in heavy metal(loid)s stress tolerance in crop plants in Abiotic stress response in plants (eds Tuteja, N. & Gill, S.S.) 237–264 (Wiley-VCH, 2016).
- 19.Webb MJ, Loneragan JF. Zinc translocation to wheat roots and its implications for a phosphorus/zinc interaction in wheat plants 1. J. Plant Nut. 1990;13:1499–1512. doi: 10.1080/01904169009364171. [DOI] [Google Scholar]
- 20.Gianquinto G, Abu-Rayyan A, Di Tola L, Piccotino D, Pezzarossa B. Interaction effects of phosphorus and zinc on photosynthesis, growth and yield of dwarf bean grown in two environments. Plant Soil. 2000;220:219–228. doi: 10.1023/A:1004705008101. [DOI] [Google Scholar]
- 21.Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002;29:751–760. doi: 10.1046/j.1365-313X.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- 22.Lynch JP, Brown KM. Top soil foraging: an architectural adaptation of plants to low phosphorus availability. Plant Soil. 2001;237:225–237. doi: 10.1023/A:1013324727040. [DOI] [Google Scholar]
- 23.López-Bucio J. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruber BD, Giehl RF, Friedel S, Von Wirén N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013;163:161–179. doi: 10.1104/pp.113.218453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morcuende R, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 26.Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2007;143:156–171. doi: 10.1104/pp.106.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thibaud MC, et al. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010;64:775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- 28.Lan P, Li W, Schmidt W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteom. 2012;11:1156–1166. doi: 10.1074/mcp.M112.020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padmanabhan P, Sahi SV. Suppression subtractive hybridization reveals differential gene expression in sunflower grown in high P. Plant Physiol. Biochem. 2011;49:584–591. doi: 10.1016/j.plaphy.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 30.Nacry P, et al. A role for auxin redistribution in the response of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain A, et al. Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 2007;144:232–247. doi: 10.1104/pp.106.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu ML, et al. FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep. 2005;24:79–85. doi: 10.1007/s00299-004-0906-8. [DOI] [PubMed] [Google Scholar]
- 33.Li JY, et al. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell. 2010;22:1633–1646. doi: 10.1105/tpc.110.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prodhan M, et al. Tight control of nitrate acquisition in a plant species that evolved in an extremely phosphorus‐impoverished environment. Plant Cell Environ. 2016 doi: 10.1111/pce.12853. [DOI] [PubMed] [Google Scholar]
- 35.Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144:197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen X, et al. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 2014;86:303–317. doi: 10.1007/s11103-014-0230-9. [DOI] [PubMed] [Google Scholar]
- 37.Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J, Bauer P. Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell. 2012;24:2380–2400. doi: 10.1105/tpc.112.099077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong J, Connolly EL. Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant Sci. 2009;176:709–714. doi: 10.1016/j.plantsci.2009.02.011. [DOI] [Google Scholar]
- 39.Kim MC, et al. MLO, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 2002;277:19304–19314. doi: 10.1074/jbc.M108478200. [DOI] [PubMed] [Google Scholar]
- 40.Schröder F, Lisso J, Lange P, Müssig C. The extracellular EXO protein mediates cell expansion in Arabidopsis leaves. BMC Plant Biol. 2009;9:20. doi: 10.1186/1471-2229-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santelia D, et al. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 2005;579:5399–5406. doi: 10.1016/j.febslet.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 42.Zhu XF, et al. Pectin enhances rice (Oryza sativa) root phosphorus remobilization. J. Exp. Bot. 2015;66:1017–1024. doi: 10.1093/jxb/eru461. [DOI] [PubMed] [Google Scholar]
- 43.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;25:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 44.Fischer U, et al. Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr. Biol. 2006;16:2143–2149. doi: 10.1016/j.cub.2006.08.091. [DOI] [PubMed] [Google Scholar]
- 45.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 46.Street IH, et al. Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 2015;169:338–350. doi: 10.1104/pp.15.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 2008;147:1181–1191. doi: 10.1104/pp.108.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ova EA, Kutman UB, Ozturk L, Cakmak I. High phosphorus supply reduced zinc concentration of wheat in native soil but not in autoclaved soil or nutrient solution. Plant Soil. 2015;393:147–162. doi: 10.1007/s11104-015-2483-8. [DOI] [Google Scholar]
- 49.Williamson LC, Ribrioux S, Fitter AH, Leyser O. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 2001;126:875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Ghazi Y, et al. Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signaling. Plant Cell Environ. 2003;26:1053–1066. doi: 10.1046/j.1365-3040.2003.01030.x. [DOI] [Google Scholar]
- 51.Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 2006;29:115–125. doi: 10.1111/j.1365-3040.2005.01405.x. [DOI] [PubMed] [Google Scholar]
- 52.Jain A, et al. Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiol. 2009;150:1033–1049. doi: 10.1104/pp.109.136184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A, Sinilal B, Dhandapani G, Meagher RB, Sahi SV. Effects of deficiency and excess of zinc on morphophysiological traits and spatiotemporal regulation of zinc-responsive genes reveal incidence of cross talk between micro-and macronutrients. Environ. Sci. Technol. 2013;47:5327–5335. doi: 10.1021/es400113y. [DOI] [PubMed] [Google Scholar]
- 54.Borch K, Bouma TJ, Lynch JP, Brown KM. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 1999;22:425–431. doi: 10.1046/j.1365-3040.1999.00405.x. [DOI] [Google Scholar]
- 55.Ma Z, Baskin TI, Brown KM, Lynch JP. Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol. 2003;131:1381–1390. doi: 10.1104/pp.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roldan, M., Dinh, P., Leung, S. & McManus, M.T. Ethylene and the responses of plants to phosphate deficiency. AoB Plants5, plt013, doi:10.1093/aobpla/plt013 (2013).
- 57.Svistoonff S, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- 58.Zheng LQ, et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 2009;151:262–274. doi: 10.1104/pp.109.141051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubio V, et al. Plant hormones and nutrient signaling. Plant Mol. Biol. 2009;69:361–373. doi: 10.1007/s11103-008-9380-y. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;10:5–14. doi: 10.1186/1471-2229-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/tpc.13.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiratsu K, Matsui K, Koyama T, Ohme‐Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 63.Upadhyay RK, et al. SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 2013;64:3237–3247. doi: 10.1093/jxb/ert162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koen E, et al. Arabidopsis thaliana nicotianamine synthase 4 is required for proper response to iron deficiency and to cadmium exposure. Plant Sci. 2013;209:1–11. doi: 10.1016/j.plantsci.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Bidzinski P, et al. Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant Cell Environ. 2014;37:2738–2753. doi: 10.1111/pce.12353. [DOI] [PubMed] [Google Scholar]
- 66.Vellosillo T, et al. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell. 2007;19:831–846. doi: 10.1105/tpc.106.046052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kodavali PK. Structural and functional characterization of annexin 1 from Medicago truncatula. Plant Physiol. Biochem. 2013;73:56–62. doi: 10.1016/j.plaphy.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 68.Stotz HU, Thomson J, Wang Y. Plant defensins: defense, development and application. Plant Signal. Behave. 2009;4:1010–1012. doi: 10.4161/psb.4.11.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998;36:871–883. doi: 10.1023/A:1005938624418. [DOI] [PubMed] [Google Scholar]
- 70.Heyndrickx KS, V de Velde J, Wang C, Weigel D, Vandepoele KA. Functional and Evolutionary Perspective on Transcription Factor Binding in Arabidopsis thaliana. Plant Cell. 2014;26:3894–3910. doi: 10.1105/tpc.114.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramaiah M, Jain A, Raghothama KG. Ethylene Response Factor070 regulates root development and phosphate starvation-mediated responses. Plant Physiol. 2014;164:1484–1498. doi: 10.1104/pp.113.231183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung JKH, McCouch S. Getting to the roots of it: genetic and hormonal control of root architecture. Fron. Plant Sci. 2013 doi: 10.3389/fpls.2013.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkinson S, Davies WJ. Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ. 2010;33:510–525. doi: 10.1111/j.1365-3040.2009.02052.x. [DOI] [PubMed] [Google Scholar]
- 74.Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 75.Jung JY, Shin R, Schachtman DP. Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell. 2009;21:607–621. doi: 10.1105/tpc.108.063099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernstein L. Effects of salinity and sodicity on plant growth. Annu. Rev. Phytopathol. 1975;13:295–312. doi: 10.1146/annurev.py.13.090175.001455. [DOI] [Google Scholar]
- 77.Xiong, L. & Zhu J. Salt tolerance in The Arabidopsis book 1, e0048, doi:10.1199/tab.0048 (2002). [DOI] [PMC free article] [PubMed]
- 78.Hu R, et al. Overexpression of PP2A‐C5 that encodes the catalytic subunit 5 of protein phosphatase 2A in Arabidopsis confers better root and shoot development under salt conditions. Plant Cell Environ. 2016 doi: 10.1111/pce.12837. [DOI] [PubMed] [Google Scholar]
- 79.Neff MM, Chory J. Genetic Interactions between Phytochrome A, Phytochrome B, and Cryptochrome 1 during Arabidopsis Development. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colón‐Carmona A, You R, Haimovitch‐Gal T, Doerner P. Spatio‐temporal analysis of mitotic activity with a labile cyclin–GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 81.Shukla D, Krishnamurthy S, Sahi SV. Genome-wide transcriptome analysis reveals ABA mediated response in Arabidopsis during gold (AuCl4-) treatment. Front. Plant Sci. 2014 doi: 10.3389/fpls.2014.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duan, L., Sebastian, J. & Dinneny, J. R. Salt-stress regulation of root system growth and architecture in Arabidopsis seedlings in Plant cell expansion: methods and protocols, methods in molecular biology (ed. Estevez, J.M.) 105–122 (Springer, 2015). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


