Abstract
Inversions breaking the 1041 bp int1h-1 or the 9.5-kb int22h-1 sequence of the F8 gene cause hemophilia A in 1/30,000 males. These inversions are due to homologous recombination between the above sequences and their inverted copies on the same DNA molecule, respectively, int1h-2 and int22h-2 or int22h-3. We find that (1) int1h and int22h duplicated more than 25 million years ago; (2) the identity of the copies (>99%) of these sequences in humans and other primates is due to gene conversion; (3) gene conversion is most frequent in the internal regions of int22h; (4) breakpoints of int22h-related inversions also tend to involve the internal regions of int22h; (5) sequence variations in a sample of human X chromosomes defined eight haplotypes of int22h-1 and 27 of int22h-2 plus int22h-3; (6) the latter two sequences, which lie, respectively, 500 and 600 kb telomeric to int22h-1 are five-fold more identical when in cis than when in trans, thus suggesting that gene conversion may be predominantly intrachromosomal; (7) int1h, int22h, and flanking sequences evolved at a rate of about 0.1% substitutions per million years during the divergence between humans and other primates, except for int1h during the human-chimpanzee divergence, when its rate of evolution was significantly lower. This is reminiscent of the slower evolution of palindrome arms in the male specific regions of the Y chromosome and we propose, as an explanation, that intrachromosomal gene conversion and cosegregation of the duplicated regions favors retention of the ancestral sequence and thus reduces the evolution rate.
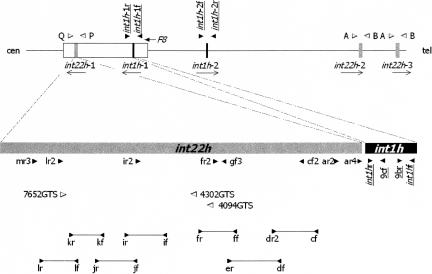
One in 30,000 males is born with an inversion breaking the factor VIII gene (F8) and causing severe hemophilia A. In 90% of these patients the break is in intron 22 of F8 and affects a 9.5-kb sequence called int22h-1 (Fig. 1) which is also present in inverted orientation 500 kb (int22h-2) and 600 kb (int22h-3) more telomerically (Naylor et al. 1992, 1993, 1995; Lakich et al. 1993; P.M. Green, N. Waseem, R.D. Bagnall, and F. Giannelli, unpubl.).
Figure 1.
Map of int1h and int22h duplicons. (Top) F8 gene (clear box), with arrow indicating direction of transcription, plus sequence of int1h (black boxes) and int22h (gray boxes) with arrows indicating orientation. Clear and solid arrow heads indicate location and orientation of primers flanking int22h copies and int1h copies, respectively (for primer sequences see Liu et al. 1998 and Bagnall et al. 2002). (Bottom) Int22h and int1h showing location and orientation of primers listed in Supplemental Table 3. These were primers required for completing the sequence of the chimpanzee's int22h sequences (solid arrow heads), or for allele-specific PCR in int22h (clear arrow heads), or for amplification of DNA for FSPCCM analysis (lines terminating with arrow heads). Other primers for sequencing int22h were from Naylor et al. (1995) (not shown). Primers for sequencing int1h (solid arrow heads and underlined names) are from Bagnall et al. (2002).
In the remaining 10% of patients the break is in intron 1 and affects a 1041-bp sequence called int1h-1 (Fig. 1; GenBank accession no. AY619998), which is duplicated in inverted orientation 140 kb more telomerically (int1h-2; GenBank accession no. AY781298) (Bagnall et al. 2002; P.M. Green, N. Waseem, R.D. Bagnall, and F. Giannelli, unpubl.).
The above inversions result from frequently recurring homologous recombination between the above sequences in the F8 gene and their more telomeric copies on the same DNA molecule (Naylor et al. 1995; Bagnall et al. 2002).
The sequences of both the int22h and int1h copies are >99.9% identical; hence they either duplicated very recently or are undergoing concerted evolution.
Aradhya et al. (2002) showed that a probe containing part of int22h hybridized to three DNA segments in blots of chimpanzee and gorilla DNA and to two segments in blots of orangutan and pygmy chimpanzee DNA, thus suggesting that duplication of int22h might not be recent. Furthermore, analysis of the hemophilia A mutation in the Chapel Hill colony of hemophilic dogs (Lozier et al. 2002) provides evidence of two copies of the F8A gene, which is a segment of int22h. Thus, at least for int22h, the hypothesis of recent duplication seems less likely than concerted evolution. In this article we demonstrate that both int1h and int22h have duplicated at least 25 million years ago (Mya) and that gene conversion is the process responsible for high copy identity. Our results also suggest how intrachromosomal gene conversion may sometimes appear to have a bias in favor of restoration to the ancestral sequence.
Results
Int1h
DNA from a male chimpanzee (Pan troglodytes), African Green monkey (Cercopithecus aethiops), and Rhesus monkey (Macaca mulatta) was used as a template for PCR reactions that amplify human int1h-1 and int1h-2. These reactions yielded sequences homologous to int1h-1 and int1h-2 from all the above DNAs (Supplemental Fig. 1; GenBank accession nos. AY781299-AY781304), thus showing that the duplication of int1h has occurred before the split between the human and Rhesus or African Green monkey and hence more than 25 Mya (Purvis 1995). Interspecies comparisons of int1h (Table 1 and Supplemental Fig. 1) and flanking sequences (Table 2 and Supplemental Fig. 2) showed rates of evolution close to 0.1% base substitutions per million years for all regions and during each interspecies divergence except for int1h during human-chimpanzee divergence when a significantly lower rate of evolution was observed (Table 2). This intriguing exception will be discussed later. However, in contrast with the interspecies comparisons, the two int1h copies of each species showed much greater similarity than could be expected after 25 Myr of separate existence as they showed only 1-, 0-, 6-, and 9-nucleotide (nt) differences and hence divergences of 0.096%, 0%, 0.57%, and 0.86% in humans, chimpanzee, African Green, and Rhesus monkey, respectively (Table 1 and Supplemental Fig. 1).
Table 1.
Divergence between int1h repeats of humans and nonhuman primates
| Int1h nucleotide | 5 | 44 | 62 | 107 | 108 | 153 | 159 | 178 | 190 | 233 | 248 | 296 | 310 | 339 | 354 | 438 | 450 | 476 | 488 | 521 | 540 | 549 | 561 | 563 | 592 | 608 | 611 | 653 | 665 | 679 | 687 | 726 | 733 | 769 | 789 | 790 | 798 | 816 | 823 | 834 | 855 | 856 | 880 | 930 | 944 | 950 | 960 | 975 | 982 | 991 | 1000 | 1001 | 1002 | 1008 | 1011 | 1023 | 1024 | 1028 | 1031 | 1038 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human int1h-1 | T | C | T | G | T | G | C | A | G | C | G | C | A | C | T | G | T | C | G | A | G | C | T | C | C | A | G | C | T | C | C | C | C | C | A | G | C | G | C | T | C | G | A | G | T | C | G | A | C | G | C | A | A | A | G | G | T | C | A | A |
| human int1h-2 | T | C | T | G | T | G | C | A | G | C | G | C | A | C | T | G | T | C | G | A | G | C | T | C | C | A | G | C | T | C | C | C | C | C | A | G | C | G | C | T | C | G | A | G | T | C | G | A | C | G | C | A | A | C | G | G | T | C | A | A |
| chimpanzee int1h-1 | T | C | T | G | T | G | C | A | G | C | G | C | A | C | T | G | T | C | G | A | G | C | T | C | T | A | G | C | T | C | C | C | C | C | A | G | C | G | C | T | C | G | A | G | T | C | G | A | C | G | C | A | A | A | G | G | T | C | A | A |
| chimpanzee int1h-2 | T | C | T | G | T | G | C | A | G | C | G | C | A | C | T | G | T | C | G | A | G | C | T | C | T | A | G | C | T | C | C | C | C | C | A | G | C | G | C | T | C | G | A | G | T | C | G | A | C | G | C | A | A | A | G | G | T | C | A | A |
| AGM int1h-1 | A | T | T | A | C | G | T | G | A | T | T | T | G | T | C | A | A | T | A | G | C | T | C | G | T | A | T | T | C | A | C | T | T | T | C | T | G | C | T | C | C | A | C | C | T | T | A | G | G | C | C | G | G | G | G | G | A | A | G | A |
| AGM int1h-2 | T | T | T | A | C | G | T | G | A | T | T | T | G | T | C | A | A | T | A | G | C | T | C | G | T | A | T | T | C | A | C | T | T | T | C | T | G | C | T | C | C | A | C | C | T | T | A | G | G | C | T | G | G | A | G | A | T | A | A | A |
| Rhesus int1h-1 | A | T | G | A | C | A | C | G | A | T | T | C | G | T | C | A | T | C | A | G | C | T | C | G | T | C | T | T | C | G | T | T | C | T | C | T | G | G | T | C | T | A | C | C | G | T | A | G | G | C | C | A | G | G | A | G | T | A | G | A |
| Rhesus int1h-2 | T | T | T | A | C | G | C | G | A | T | T | C | G | T | C | A | T | C | A | G | C | T | C | G | T | C | T | T | C | G | T | T | C | T | C | T | G | G | T | C | T | A | C | C | G | T | A | G | G | C | C | G | G | A | G | A | T | A | A | C |
Int1h residue number is shown on top.
Table 2.
Frequency of base substitutions observed in int1 h and flanking sequences of different pairs of species
|
Int1 h
|
Flanking sequence
|
||||||
|---|---|---|---|---|---|---|---|
| Species pairs | d | nt | pro | d | nt | pro | P value |
| H + C | 3 | 2082 | 0.00144 | 51 | 6941 | 0.00735 | <0.001* |
| H + A | 99 | 2080 | 0.0476 | 200 | 3516 | 0.0568 | 0.131 |
| H + R | 96 | 2082 | 0.0461 | 233 | 4409 | 0.0528 | 0.239 |
| C + A | 96 | 2080 | 0.0461 | 185 | 3465 | 0.0534 | 0.260 |
| C + R | 93 | 2082 | 0.0447 | 218 | 4366 | 0.0499 | 0.390 |
| A + R | 29 | 2092 | 0.0138 | 59 | 3516 | 0.0168 | 0.460 |
(H) human, (C) chimpanzee, (A) African Green monkey, (R) Rhesus monkey; (d) number of divergences, (nt) number of aligned nucleotides excluding sites of insertion/deletion mutations, (pro) proportion of divergence; P value is result of Normal test of equal proportions involving int1 h and flanking sequences; *statistically significant difference.
To see if this high intraspecies identity was the result of gene conversion, the test developed by Balding et al. (1992) was used. This test compares the duplicated sequences in pairs of species, considering in particular the frequency of instances where the two species differ by the same base substitution at the same site of both repeats. These instances, called “co-doubles,” are expected to be rare in independently evolving duplicates, whereas they can be readily produced, in each species, by gene conversion as this process can copy a mutation of one duplicate into the other of the same species. The Balding et al. (1992) test provided strong evidence of gene conversion in all species comparisons, as the null hypothesis of independent evolution was shown to be improbable, P < 0.001 for human + chimpanzee and even more improbable P « 0.001 for all other pairs, namely, human + African Green monkey, human + Rhesus monkey, chimpanzee + African Green monkey, chimpanzee + Rhesus monkey, and African Green + Rhesus monkey.
The distribution of co-doubles along the int1h sequence was not uniform (Supplemental Table 1), as a modest excess of co-doubles relative to Poisson expectation was observed in the central region (nt 521-624) and also toward the ends of the duplicates that are farthest apart, although the last 30 nt of these ends appear also rich in singles (Supplemental Fig. 1 and Supplemental Table 1).
Human int1h sequence variation was analyzed by examining 57 X chromosomes and this showed that nt 1008 was always A in int1h-1 and C in int1h-2, while nt 698 of int1h-2 was G in 9 and C in 48 X chromosomes (Bagnall et al. 2002).
Int22h
Naylor et al. (1995) reported the sequence of int22h-1, its flanking regions, and the regions flanking either of both int22h-2 and int22h-3. They also detected some differences between int22h-1 and int22h-2 plus int22h-3; however, they could not independently sequence int22h-2 and int22h-3 because clones containing only one or the other of these repeats had not yet been identified. We used such clones (Hassock 2000) to sequence the two distal int22h copies and also resequenced int22h-1 from a British male as the sequence published by Naylor et al. (1995) contained 31 undefined residues. The revised sequence of int22h-1 and the sequences of int22h-2 and int22h-3 are available at GenBank accession nos. AY619999, AY620000, and AY620001, respectively.
Long-range PCR experiments on primate male DNA showed that human sequence primers flanking int22h-1 and either of both int22h-2 and int22h-3 readily amplified the corresponding sequences of chimpanzee DNA. The PCR reaction that amplifies int22h-2 and int22h-3 yielded products that had no site showing the presence of two different nucleotides. Therefore, because the results of Aradhya et al. (2002) had shown that the chimpanzee has three copies of int22h, we assume that, in the chimpanzee we examined, int22h-2 and int22h-3 were identical. The African Green monkey DNA did not sustain the amplification of the full int22h sequence, but when a primer internal plus one external to int22h-1 were used simultaneously, a 6.5-kb product was obtained consisting of 1 kb homologous to the human sequence flanking int22h-1 on the telomeric side followed by 5.5 kb homologous to nt 1-5553 of human int22h (GenBank accession no. AY781307). Similarly, a 6.6-kb product was obtained with a primer external to both telomeric int22h sequences and one internal. This PCR product was homologous to human int22h from nt 1261 to its telomeric end and terminated with 40 bp of sequence homologous to the sequence flanking either of the two human telomeric int22h copies on their telomeric side (GenBank accession no. AY781308). However nucleotides homologous to human 6027-7842 were absent in African Green monkey, presumably due to a deletion/insertion event.
These data provide further proof that duplication of int22h predates the human-African Green monkey split.
The sequence of the human and chimpanzee's int22h copies were compared (Table 3 and Supplemental Fig. 3). The chimpanzee's int22h differed from the human by nine insertions/deletions, of which two affected only int22h-1, replacement of human nt 7854-8549 with an inverted copy of nt 7866-7911 and the tandem duplication of nt 8755-8802 (GenBank accession nos. AY781305 and AY781306). These insertions/deletions reduce the length of int22h common to the human and chimpanzee DNA samples to 8854 nt.
Table 3.
Divergence between int22h repeats of humans and chimpanzee
| Int22h residue | 8 | 49 | 193 | 459 | 472 | 683 | 742 | 1113 | 1319 | 1437 | 1450 | 1602 | 1701 | 2086 | 2133 | 2170 | 2478 | 2520 | 2798 | 2820 | 2834 | 3156 | 3223 | 3546 | 3777 | 3867 | 4094 | 4098 | 4099 | 4157 | 4178 | 4247 | 4257 | 4302 | 4303 | 4331 | 4340 | 4350 | 4353 | 4360 | 4364 | 4378 | 4414 | 4610 | 4612 | 4640 | 4759 | 4769 | 4776 | 4827 | 4859 | 4860 | 4869 | 4870 | 4943 | 5040 | 5076 | 5150 | 5259 | 5346 | 5431 | 5470 | 5640 | 5674 | 5725 | 5748 | 5785 | 5845 | 5876 | 5880 | 5909 | 5971 | 6017 | 6083 | 6157 | 6233 | 6275 | 6373 | 6420 | 6428 | 6456 | 6499 | 6565 | 6685 | 6702 | 6718 | 6976 | 7145 | 7192 | 7245 | 7278 | 7411 | 7609 | 7638 | 7652 | 7667 | 7733 | 7765 | 7779 | 7830 | 8579 | 8726 | 8731 | 8742 | 8746 | 8854 | 9085 | 9217 | 9243 | 9479 | 9502 | 9503 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human h-1 | G | T | T | a | G | C | T | T | G | A | G | A | T | G | T | T | A | A | A | C | T | C | A | G | G | T | a | G | G | C | T | G | T | C | G | C | a | C | C | C | G | G | A | C | G | G | G | C | G | T | C | A | G | A | A | T | g | G | C | C | C | A | C | C | A | A | C | G | C | A | A | G | G | G | G | C | T | C | C | C | C | A | C | G | G | T | C | C | G | C | C | G | G | T | a | A | A | A | C | C | A | T | C | C | A | A | C | G | G | T | A | C |
| Human h-2 + 3 | G | c | T | G | G | C | T | T | G | A | G | A | T | G | T | T | A | A | A | C | T | C | A | G | G | T | G | G | G | C | T | G | T | g | G | C | G | C | C | C | G | G | A | C | G | G | G | C | G | T | C | A | G | A | A | T | A | G | C | C | C | A | C | C | A | A | C | G | C | A | A | G | G | G | G | C | T | C | C | C | C | A | C | G | G | T | C | C | G | C | C | G | G | T | G | A | A | A | C | C | A | T | C | C | A | A | C | G | G | T | A | C |
| Chimp h-1 | a | T | T | G | a | A | C | C | T | G | T | C | C | C | C | G | G | G | C | T | C | T | G | A | A | C | G | T | A | G | G | A | C | C | T | T | G | T | g | T | c | C | G | G | C | A | A | G | T | C | G | G | A | G | G | G | A | C | T | G | T | G | T | T | G | G | G | C | G | G | G | T | T | A | A | G | C | T | G | G | T | G | T | A | A | G | T | T | c | A | C | T | A | T | G | G | T | T | t | t | g | g | t | C | A | A | G | G | t | T | g | t |
| Chimp h-2 + 3 | G | T | c | G | G | A | C | C | T | G | T | C | C | C | C | G | G | G | C | T | C | T | G | A | A | C | G | T | A | G | G | A | C | C | T | T | G | T | t | T | G | C | G | G | C | A | A | G | T | C | G | G | A | G | G | G | A | C | T | G | T | G | T | T | G | G | G | C | G | G | G | T | T | A | A | G | C | T | G | G | T | G | T | A | A | G | T | T | G | A | t | T | A | c | G | G | T | T | C | C | A | T | C | t | g | g | G | c | G | c | A | C |
Int22h residue number is shown on top. Base divergences representing co-doubles are highlighted gray while singles and doubles are in bold lower case. Base difference between human int22h-2 (C) and int22h-3 (T) at residue 4350 is indicated by showing the C of int22h-2 in bold uppercase.
This alignment of the human and chimpanzee's sequences showed 102 base substitutions between the int22h-1 sequences, 95 between the int22h-2, and 94 between the int22h-3 (Table 3). This indicated an evolution rate of about 0.1% base substitutions per million years of divergence. In contrast, human int22h-1 differed from int22h-2 and int22h-3 at 12 and 13 sites, respectively, while the latter two int22h sequences differed at a single site. Similarly, the chimpanzee's int22h-1 differed at 24 sites from both int22h-2 and int22h-3, as the latter two appeared to be identical.
The int22h-1 and int22h-2 or int22h-3 sequences of humans and chimpanzee were then examined using Balding et al's (1992) test of gene conversion after disregarding the variation at nt 4350 between the human int22h-2 and int22h-3. The vast majority of base differences were found to be co-doubles, and the Balding test provided very strong evidence of gene conversion as the probability of the null hypothesis of independent evolution of the int22h copies was extremely small (P « 0.001). The distribution of co-doubles along the int22h repeat was nonhomogeneous because when the sequence was divided into 19 segments of 466 bp, the frequency of co-doubles departed significantly from Poisson expectation in several segments (Supplemental Table 2). An excess of co-doubles was observed over a wide region comprising nt 4198 to 6532, while co-doubles were relatively rare near to the ends of the duplicated sequence. These results suggest a broad peak of gene conversion events in the central region (nt 4195-6524) and a dearth of gene conversion at the periphery (nt 1-466 and 8581-9512) of the human and chimpanzee's int22h sequence alignment.
The alignment of the human int22h copies with the sequence we obtained from the African Green monkey (see Supplemental Fig. 4) gives data in keeping with the results of the human-chimpanzee comparison because it shows 616 base substitutions in 11,835 pairs of nucleotides or a divergence of 5.2%, equivalent to an evolution rate of 0.1% per million years of divergence. In contrast, the sequence available on the int22h duplicates of the African Green monkey showed only 29 base substitutions in 4468 nucleotide pairs or a divergence of 0.064%. Furthermore, the base differences between the int22h duplicates of humans and African Green monkey are mostly in the form of co-doubles (i.e., 209 co-doubles, 31 singles and two doubles). Thus, during the human-African Green monkey divergence, int22h has evolved at a rate similar to that observed during the human-chimpanzee split and the high similarity in the segment of the int22h duplicates that we were able to study in African Green monkey is due to gene conversion.
The variation of the int22h sequence among humans was examined by investigating 19 normal British males and 16 hemophilia A patients with inversions involving int22h (i.e., six inversions due to recombination of int22h-1 with int22h-2 and 10 due to recombination of int22h-1 with int22h-3). The int22h-1 sequences of the 19 control males showed eight different haplotypes, defined by the association of alleles at 12 variable sites (Table 4). Int22h-2 and int22h-3, which cannot be individually amplified, were identical in six control males and different at a single site in nine. In the remaining four control males more differences were found and in order to distinguish the haplotypes of the two sequences it was necessary to use allele-specific primers so as to examine the association of nucleotides at the sites of divergence (Table 5A). In the hemophilia A patients the unrecombined distal sequence (either int22h-3 or int22h-2) can be specifically amplified and its haplotype can be directly determined (Table 5B,C). The 54 distal int22h sequences examined showed 27 different haplotypes (Table 6), which fall clearly into two groups: haplotypes 1-16 and haplotypes 18-27. These are distinguished by the presence or absence of a nonancestral allele (i.e., divergent from chimpanzee) at nt 49, 1007, 1477, 1567, and 4619 andaGorAatnt 8509. Haplotype 17 appears more similar to the 18-27 group but does not differ from the 1-16 group at nt 1567 and 8509. Human int22h-1 differs from int22h-2 and int22h-3 at three sites (nt 3, 10, and 12) that appear nonpolymorphic (Supplemental Fig. 3) and at a further 23 sites that are polymorphic among the individuals we have examined (Tables 4 and 6).
Table 4.
Haplotypes of int22h-1
| 459 | 4094 | 4302 | 4340 | 4341 | 4350 | 4619 | 5076 | 5880 | 7652 | 8564 | 8878 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chimpanzee h1 | G | G | C | G | G | T | T | A | G | A | A | G | int22h-1 haplotype number | |
| Control individual
|
C1 h1 | A | A | A | C | G | 1 | |||||||
| C2 h1 | A | A | A | C | G | 1 | ||||||||
| C3 h1 | A | A | A | C | G | 1 | ||||||||
| C4 h1 | A | A | A | C | G | 1 | ||||||||
| C5 h1 | A | A | A | C | G | 1 | ||||||||
| C6 h1 | A | A | A | C | G | 1 | ||||||||
| C7 h1 | A | A | A | C | G | 1 | ||||||||
| C8 h1 | A | A | A | C | G | 1 | ||||||||
| C9 h1 | A | A | C | G | 2 | |||||||||
| C10 h1 | A | A | C | G | G | 3 | ||||||||
| C11 h1 | A | A | C | G | G | 4 | ||||||||
| C12 h1 | A | G | C | 5 | ||||||||||
| C13 h1 | A | G | C | T | 6 | |||||||||
| C14 h1 | G | C | G | 7 | ||||||||||
| C15 h1 | G | C | G | 7 | ||||||||||
| C16 h1 | G | C | G | 7 | ||||||||||
| C17 h1 | G | C | G | A | G | 8 | ||||||||
| C18 h1 | G | C | G | A | G | 8 | ||||||||
| C19 h1 | G | C | G | A | G | 8 |
Int22h-1 human nucleotide number at sites of polymorphism is shown at the top. Chimpanzee nucleotides at these sites are shown in bold. Empty spaces are identical to ancestral (chimpanzee). Novel alleles are in upper case.
Table 5.
Int22h-2 and int22h-3 polymorphic sites and sites of divergence from int22h-1
| 3
|
10
|
12
|
49
|
459
|
880
|
1007
|
1168
|
1477
|
1567
|
4069
|
4094
|
4302
|
4340
|
4341
|
4350
|
4619
|
5076
|
5880
|
7652
|
8444
|
8449
|
8509
|
8564
|
8878
|
8802-8850
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chimpanzee h2/h3 | A | T | T | T | G | G | G | A | C | A | C | G | C | G | G | T | T | A | G | A | — | — | — | A | G | Y | |
| A. Control individuals | C1 h2/3 | C | A | G | C | A | C | A | G | N | |||||||||||||||||
| C1 h2/3 | C | G | G | T | A | N | |||||||||||||||||||||
| C2 h2/3 | A | G | G | G | C | G | G | T | G | A | Y | ||||||||||||||||
| C2 h2/3 | A | A | G | G | C | G | G | T | G | A | Y | ||||||||||||||||
| C3 h2/3 | C | A | G | A | G | N | |||||||||||||||||||||
| C3 h2/3 | C | G | G | A | G | N | |||||||||||||||||||||
| C4 h2/3 | C | G | C | G | A | G | N | ||||||||||||||||||||
| C4 h2/3 | C | G | C | A | A | G | N | ||||||||||||||||||||
| C5 h2/3 | C | G | A | N | |||||||||||||||||||||||
| C5 h2/3 | C | G | A | N | |||||||||||||||||||||||
| C6 h2/3 | C | A | G | C | A | G | N | ||||||||||||||||||||
| C6 h2/3 | C | A | G | C | A | G | N | ||||||||||||||||||||
| C7 h2/3 | A | A | G | G | G | C | G | A | Y | ||||||||||||||||||
| C7 h2/3 | G | A | G | G | G | C | G | A | Y | ||||||||||||||||||
| C8 h2/3 | C | A | G | A | N | ||||||||||||||||||||||
| C8 h2/3 | C | G | G | A | N | ||||||||||||||||||||||
| C9 h2/3 | C | G | A | G | N | ||||||||||||||||||||||
| C9 h2/3 | C | G | A | G | N | ||||||||||||||||||||||
| C10 h2/3 | T | A | G | G | C | G | A | N | |||||||||||||||||||
| C10 h2/3 | C | G | C | G | C | A | G | Y | |||||||||||||||||||
| C11 h2/3 | C | C | A | C | A | G | N | ||||||||||||||||||||
| C11 h2/3 | C | G | G | T | A | G | N | ||||||||||||||||||||
| C12 h2/3 | A | G | G | G | C | G | T | A | N | ||||||||||||||||||
| C12 h2/3 | A | G | G | G | C | G | C | A | N | ||||||||||||||||||
| C13 h2/3 | C | A | G | A | G | C | A | N | |||||||||||||||||||
| C13 h2/3 | C | G | G | G | A | T | G | N | |||||||||||||||||||
| C14 h2/3 | A | G | G | G | C | G | G | A | Y | ||||||||||||||||||
| C14 h2/3 | A | G | G | G | C | G | A | A | Y | ||||||||||||||||||
| C15 h2/3 | A | G | G | G | G | C | G | G | A | Y | |||||||||||||||||
| C15 h2/3 | A | G | G | C | G | C | G | G | A | Y | |||||||||||||||||
| C16 h2/3 | C | A | G | C | A | G | N | ||||||||||||||||||||
| C16 h2/3 | C | A | G | C | A | G | N | ||||||||||||||||||||
| C17 h2/3 | A | G | G | G | C | G | T | A | Y | ||||||||||||||||||
| C17 h2/3 | A | G | G | G | C | G | T | A | Y | ||||||||||||||||||
| C18 h2/3 | C | G | G | G | N | ||||||||||||||||||||||
| C18 h2/3 | C | G | A | G | N | ||||||||||||||||||||||
| C19 h2/3 | C | G | A | G | N | ||||||||||||||||||||||
| C19 h2/3 | C | G | A | G | N | ||||||||||||||||||||||
| B. Proximal inversions | a h3 | C | G | C | G | N | |||||||||||||||||||||
| b h3 | A | G | G | G | C | G | T | A | Y | ||||||||||||||||||
| c h3 | C | A | G | C | G | A | G | N | |||||||||||||||||||
| d h3 | A | G | G | G | C | G | T | A | Y | ||||||||||||||||||
| e h3 | C | G | C | G | N | ||||||||||||||||||||||
| f h3 | A | G | G | G | C | G | T | A | Y | ||||||||||||||||||
| C. Distal inversions
|
UKA181 h2 | C | G | C | A | G | N | ||||||||||||||||||||
| UKA36 h2 | C | G | C | G | A | G | N | ||||||||||||||||||||
| UKA502 h2 | C | G | C | A | G | N | |||||||||||||||||||||
| UKA57 h2 | C | G | C | G | N | ||||||||||||||||||||||
| UKA657 h2 | C | G | A | G | N | ||||||||||||||||||||||
| UKA667 h2 | C | G | G | A | G | N | |||||||||||||||||||||
| UKA695 h2 | C | G | C | A | G | N | |||||||||||||||||||||
| UKA697 h2 | C | C | A | G | N | ||||||||||||||||||||||
| UKA708 h2 | C | G | C | G | N | ||||||||||||||||||||||
| UKA744 h2 | C | G | A | G | N |
Int22h-2 and int22h-3 polymorphic sites and sites of divergence from int22h-1 are shown at top. Chimpanzee nucleotides at these sites are in bold. Empty spaces are identical to ancestral (chimpanzee) sequence or to bases found in reference human DNA and absent in chimpanzee (italics). Novel sequence variations are in upper case. Column 8802-8850 corresponds to presence (Y) or absence (N) of polymorphic tandem duplication. (A) Data from normal individuals (i.e., C1 to C19); each h2/3 row represents one or the other of the distal repeats of the controls. (B) Data from hemophilia A patients with inversions resulting from recombination of int22h-1 with int22h-2. (C) Data from hemophilia A patients with inversions resulting from recombination of int22h-1 with int22h-3. a-f and UKA numbers are patients′ codes; h2 and h3 refer to int22h-2 and int22h-3, respectively.
Table 6.
Haplotypes of two distal int22h sequences
| 49
|
880
|
1007
|
1168
|
1477
|
1567
|
4069
|
4094
|
4302
|
4340
|
4341
|
4350
|
4619
|
5076
|
5880
|
7652
|
8444
|
8449
|
8509
|
8802-8850
|
No. observed
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chimpanzee | T | G | G | A | C | A | C | G | C | G | G | T | T | A | G | A | — | — | — | Y | 1 | |
| Haplotype number
|
1 | C | G | A | C | A | G | N | 4 | |||||||||||||
| 2 | C | G | A | G | N | 9 | ||||||||||||||||
| 3 | C | G | C | A | G | N | 5 | |||||||||||||||
| 4 | C | G | G | N | 1 | |||||||||||||||||
| 5 | C | G | C | G | N | 3 | ||||||||||||||||
| 6 | C | G | C | G | N | 1 | ||||||||||||||||
| 7 | C | A | C | A | G | N | 1 | |||||||||||||||
| 8 | C | C | A | G | N | 1 | ||||||||||||||||
| 9 | C | A | G | A | N | 1 | ||||||||||||||||
| 10 | C | A | G | C | A | N | 1 | |||||||||||||||
| 11 | C | A | G | A | G | N | 2 | |||||||||||||||
| 12 | C | A | G | C | A | G | N | 4 | ||||||||||||||
| 13 | C | A | G | C | G | A | G | N | 1 | |||||||||||||
| 14 | C | G | T | G | N | 1 | ||||||||||||||||
| 15 | C | G | C | G | A | G | N | 2 | ||||||||||||||
| 16 | C | G | G | A | G | N | 1 | |||||||||||||||
| 17 | A | G | G | C | N | 1 | ||||||||||||||||
| 18 | A | G | G | G | C | G | A | Y | 2 | |||||||||||||
| 19 | A | A | G | G | G | C | G | A | Y | 1 | ||||||||||||
| 20 | A | G | G | G | G | C | G | G | A | Y | 1 | |||||||||||
| 21 | A | G | G | G | C | G | G | A | Y | 1 | ||||||||||||
| 22 | A | G | G | G | C | G | G | A | Y | 1 | ||||||||||||
| 23 | A | G | G | G | C | G | A | N | 1 | |||||||||||||
| 24 | A | G | G | G | C | G | T | A | N | 1 | ||||||||||||
| 25 | A | G | G | G | C | G | T | A | Y | 5 | ||||||||||||
| 26 | A | G | G | C | G | G | T | G | A | Y | 1 | |||||||||||
| 27 | A | G | G | G | C | G | G | T | G | A | Y | 1 |
Distal int22h human nucleotide number at sites of polymorphism is on the top. Empty spaces indicate that base is identical to that of chimpanzee's sequence or to bases found in the reference human DNA and absent in chimpanzee (italics). Novel alleles are in upper case. Column 8802-8850 shows presence (Y) or absence (N) of polymorphic tandem duplication.
The two distal int22h sequences on the same X chromosome (in cis) usually showed the same haplotype, and the average number of differences between these sequences was 1.32 (95% confidence interval obtained from bootstrapping with individuals resampled was 0.68-2.11). This is fivefold smaller than the number of differences between all possible pairs formed by the unrecombined int22h-2 and int22h-3 sequences specifically amplified from the inversion patients, which was 6.67 (95% confidence interval obtained from balanced bootstrap, with 6 int22h-2 and 10 int22h-3 chosen for each sample, was 3.63-9.60). Similarly the average number of differences between all possible pairs formed by one or the other of the distal int22h sequences of one control male with one or the other distal int22h of the remaining 18 controls was 5.98 (95% confidence interval 3.82-6.79). Both these two average numbers of differences are significantly greater than the differences between distal int22h sequences on the same chromosome as shown by the nonoverlapping confidence intervals.
It is clear from Tables 4 and 6 that the nonancestral alleles at nt 4094, 4302, 4340, 4341, 4350, 4619, 5076, 5880, and 7652 of human int22h-1 and the other two int22h copies may represent co-doubles when considered in relation to the chimpanzees' sequences. Thus the above variants may reveal gene conversion events that occurred during the history of the int22h sequences examined.
Suggestions of gene conversion associated with recombination were obtained from the analysis of the 16 hemophilia A patients mentioned above and one of seven patients with the inversion involving int1h. In this latter patient the nonancestral nt C observed at site 1008 of only int1h-2 in all 57 control human X chromosomes analyzed (Bagnall et al. 2002) was found in both recombined int1h sequences, thus suggesting gene conversion of the region containing this nucleotide. Analysis of the patients with the int22h-related inversion was more complex. Since the haplotypes of the int22h sequences prior to the inversion were not known, the events that occurred during the recombination event causing the inversion could only be indirectly reconstructed. To do this all combinations of known haplotypes of the relevant sequences (either int22h-1 plus int22h-2 or int22h-1 plus int22h-3) were considered for each patient and then probable combinations were selected with the help of the following criteria:
That the distal int22h of each likely combination should be as close a match as possible to the patient's unrecombined distal int22h, because of the close similarity of these sequences observed in control X chromosomes.
That the combination of haplotypes was capable of yielding the recombined int22h sequences seen in the patient either directly or only with the help of gene conversion as the observation of de novo mutations arising during the recombination event was considered unlikely.
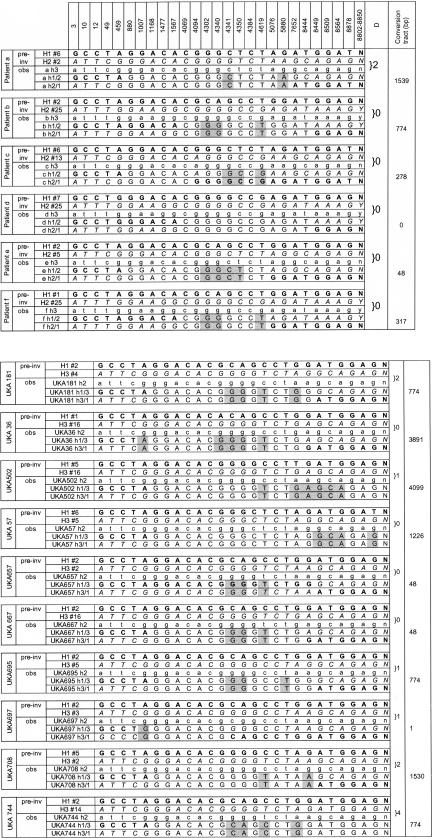
The vast majority of haplotype combinations thus selected were found to require gene conversion in order to account for the haplotypes of the patients' recombined int22h sequences. The length of the gene conversion tract, of course, varied according to the combination considered and, for the purpose of illustration, Figure 2 shows for each patient the combination of haplotypes that best fits the above two selection criteria and requires the shortest gene conversion tract to account for the patient's recombined int22h sequences. The gene conversion events proposed in Figure 2 are usually in the region between nt 4094 and 5880, which lies within the region of int22h, showing a broad peak of co-doubles. This is the region thought to have experienced most gene conversion events during the evolution of int22h in humans and chimpanzees.
Figure 2.
Observed and possible pre-inversion int22h haplotypes for patients with int22h-related inversions. For each patient a set of proposed pre-inversion haplotypes (pre-inv) are shown. The haplotype of the proposed pre-inversion proximal and distal int22h sequence are in bold and italics, respectively, and the haplotype number is indicated (e.g., H1#2 is haplotype 2 of int22h-1 and H3#23 is haplotype 23 of distal int22h). The observed haplotypes (obs) show the non-recombined int22h in lower case and the recombined haplotypes. The distribution of bold and italic characters in the latter two haplotypes shows how the pre-inversion haplotypes have rearranged. Standard capital letters indicate noninformative sites. Proposed gene conversions are highlighted gray. h1/3 and h1/2 are the recombined sequences at proximal location while h2/1 and h3/1 are those at distal location. Column “D” indicates number of differences between the patient's non-recombined int22h-2 or int22h-3 sequence and the distal int22h haplotype chosen to represent the pre-inversion distal int22h. The distance between the first and last allele that requires a gene conversion event to account for the observed recombined sequences is the gene conversion tract (last column).
Discussion
The results presented above show that int1h as well as int22h duplicated more than 25 Mya. During species divergence these sequences evolved at substitution rates close to those expected for noncoding DNA (Ebersberger et al. 2002), i.e., about 0.1% per million years except for int1h during the human-chimpanzee split. However, the copies of int1h and int22h within each species analyzed are more than 99% identical. This suggests that they undergo a process of intraspecies homogenization and that their identity is not due to functional constraints strong enough to result in marked suppression of interspecies divergence. In fact no essential function has so far been assigned to either int1h or int22h. However, the latter contains the intronless F8A gene that encodes a protein found to coprecipitate with huntingtin (Levinson et al. 1992; Peters and Ross 2001). This coding sequence represents 1.1 kb of int22h and shows less human-chimpanzee divergence than the rest of int22h (0.45% vs. 1.3%; P < 0.01). Nevertheless, in humans the loss of one F8A gene appears to be well tolerated, as deletions of the F8 gene resulting in loss of int22h-1 do not cause other phenotypes than hemophilia A (Casula et al. 1990).
The process causing homogenization of the duplicated int1h and int22h sequences appears to be gene conversion, as our results demonstrate that in most regions of int1h and int22h all interspecies divergences are in the form of co-doubles. In these regions gene conversion events must have been frequent enough to allow either restoration of the ancestral sequence or duplication of any mutation that occurred in one copy into the (or one) other copy.
The distribution of co-doubles in int22h shows that gene conversion between int22h-1 and the two distal int22h sequences is most frequent in the central region and most rare near the ends. The overlap between the region of int22h with the greatest co-double density and the region of putative gene conversion in hemophilia A inversion patients suggests that the same region of int22h is a focus for recombination as well as gene conversion not associated with recombination (“pure” gene conversion) between int22h-1 and the distal int22h. This is in keeping with the results of the direct analysis of meiotic crossover hotspots in human sperm, which were found also to be active sites of pure gene conversion (Jeffreys and May 2004). However, the concerted evolution of the int1h duplicates and of int22h-1 relative to the two distal int22h sequences is completely unrelated to sequence interactions resulting in recombination because these are expected to yield unviable mutations, namely inversions found to cause severe hemophilia A, if the recombinations are between duplicates on the same DNA molecule, or dicentric and acentric fragments, if recombinations are between duplicates not on the same DNA molecules (i.e., intersister chromatids, interhomologous chromatids, interhomologous chromosomes).
We found that human int22h-2 and int22h-3 sequences are fivefold more similar to each other when in cis than when in trans (i.e., on different chromosomes). This is interesting for two reasons. First, this is because it shows that the fivefold excess of inversions caused by recombination of int22h-1 with int22h-3 relative to those caused by recombination of int22h-1 with int22h-2 among patients with hemophilia A (Antonarakis et al. 1995) must be explained by factors other than differences in sequence similarity between int22h duplicates; for example, the effect of chromatin structure on recombination that has been noted in yeast (Paques and Haber 1999). Second, this is because the above finding suggests that pure gene conversion, at least between int22h-2 and int22h-3, is more often intrachromosomal than interchromosomal as only the former homogenizes sequences in cis without affecting sequences in trans.
In fact intrachromosomal gene conversion may help to explain the significant deficit of divergences we observed between human and chimpanzee int1h (see Table 2) if we assume that int1h-1 and int1h-2 are tightly linked in humans and chimpanzee. This assumption is likely to be correct at least in humans, where these sequences are only 140 kb apart. In chimpanzee the distance between the two int1h sequences is likely to be similar to that found in humans but firm data are not yet available as gaps remain in the sequence of this region of the chimpanzee's X chromosome. We argue that, in the presence of tight linkage, intrachromosomal gene conversion should favor the retention of the ancestral sequence because mutations duplicated by gene conversion and cosegregating will be lost from the population through genetic drift at a rate similar to that of single mutations. Thus the duplication of an int1h mutation by intrachromosomal gene conversion will not adequately compensate the alternative event, resulting in conversion of the mutation back to the ancestral sequence. Clearly if int1h in humans and chimpanzees tend to retain the sequence of the common ancestor they should show a reduced rate of interspecies divergence. Of course, interchromosomal gene conversion such as conversion between sequences on homologous chromosomes or chromatids should not contribute to the above effect because the copies of the mutations duplicated by these types of gene conversion do not cosegregate.
Intrachromosomal gene conversion and cosegregation of converted sequences may also explain Rozen et al.'s (2003) observation that the human-chimpanzee sequence divergence is significantly lower in the arms of palindromes than in other male specific Y chromosome regions. The male specific part of the Y chromosome contains 5.7 Mb of euchromatin, showing an imperfect palindromic structure (Skaletsky et al. 2003). The arms of these palindromes measure from 9 kb to 1.45 Mb and the between arm spacers are 2-170 kb long. The arms have sequence identity greater than 99%, and this is thought to result from gene conversions, which probably occur at a rate of at least 600 nt per Y chromosome per generation and retard the evolutionary decay of testis-specific genes located in the arms of palindromes (Rozen et al. 2003). Thus in the Y chromosome gene conversion acquires an important functional role.
In contrast to the above data on the int1h duplicates, which are expected to be fairly tightly linked, and those on the arms of the palindromes of the Y chromosome, which are absolutely linked (Rozen et al. 2003) the int22h sequences of humans and chimpanzee show a substitution rate per million years (0.115%) appropriate for noncoding DNA and not significantly different (P = 0.071) from that of 4818 bp of sequence flanking int22h-1 (base substitution rate = 0.0809% per million years; see Supplemental Fig. 5). As int22h-1 is 500 and 600 kb away from int22h-2 and int22h-3, respectively, recombination between int22h-1 and the two distal int22h sequences may so reduce cosegregation of regions that experienced intrachromosmal gene conversion as to prevent the bias in favor of the retention of the ancestral sequence and hence, the suppression of human-chimpanzee interspecies divergence.
When the first report of gene conversion in higher eukaryotes (Slightom et al. 1980) was followed by many other examples (Liebhaber et al. 1981; Bentley and Rabbitts 1983; Mellor et al. 1983; Michelson and Orkin 1983; Weiss et al. 1983; Hardison and Margot 1984; Stoeckert et al. 1984) Powers and Smithies (1986) asked whether pure gene conversion represented a distinct process from recombination. This question has not yet been fully answered, but as the number of duplicated sequences known to undergo concerted evolution increases, the pervasive effect of gene conversion on the genome becomes clearer.
Here we have shown how pure gene conversion has maintained the identity of two different duplicated sequences of the X chromosome that predispose to inversions breaking the F8 gene and causing hemophilia A. This gene conversion is of an intensity reminiscent of the palindromic regions of the Y chromosome and thus suggests that gene conversion may be an important common factor in the concerted evolution of intrachromosomally duplicated sequences. Some of our results suggest a prevalence of intrachromosomal gene conversion events and we propose that when these involve closely linked regions they favor the preservation of the ancestral sequence and thus reduce the rate of evolutionary change in the region involved.
Methods
DNA was extracted using standard procedures (Miller et al. 1988) from (1) 10 mL of blood donated (with informed consent) by 19 normal British males and 25 males with hemophilia A (9 with the int1h related inversion, 6 with inversions due to recombination of int22h-1 with int22h-2 and 10 with inversions due to recombination of int22h-1 with int22h-3); (2) cultured fibroblasts (Coriell Cell Repositories, cell strain GM03452) from a male chimpanzee; (3) brain from a male African Green monkey; and (4) peripheral lymphocytes of a male Rhesus monkey. LLNL human clones U100A9, containing int22h-2, and U214E7, containing int22h-3, were obtained from the MRC UK HGMP Resource Centre and DNA extracted according to the supplier's protocol.
PCR amplification of int1h duplicates and flanking regions was performed as previously described (Bagnall et al. 2002).
Int22h duplicates were PCR amplified from 100 ng of genomic DNA or 10 ng of clone DNA using 1 μL 10× Expand Long PCR polymerase buffer (Roche Diagnostics), 0.5 mM of each dATP, dCTP, and dTTP, 0.25 mM dGTP, 0.25 mM deaza dGTP (Roche Diagnostics), 7.5% DMSO, 50 ng of each oligonucleotide primer, and 1 U Expand Long PCR DNA polymerase (Roche Diagnostics). Ten cycles of PCR were performed (94°C 30 sec, 68°C 12 min) and were immediately followed by 20 further cycles of PCR (94°C 30 sec, 68°C 12 min plus 20 sec per cycle). Primer sequences for amplification of human and chimpanzee int22h sequences were as previously described (Liu et al. 1998). African green monkey int22h-1 and distal int22h segments were amplified using, respectively, primer pairs P (Liu et al. 1998) plus IR and CF (Supplemental Table 3) plus B (Liu et al. 1998).
Allele-specific PCR of distal int22h sequences was performed using the products of the PCR directed by primers A and B of Liu et al. (1998). The reactions comprised 1 μL of PCR product, 1 μL 10× Expand Long PCR polymerase buffer, 0.5 mM of each dATP, dCTP, and dTTP, 0.25 mM dGTP, 0.25 mM deaza dGTP, 7.5% DMSO, 50 ng of each oligonucleotide primer, and 1 U Expand Long PCR DNA polymerase. Thirty cycles of PCR were performed (94°C 30 sec, 68°C 12 min). Allele-specific PCR primer sequences are listed in Supplemental Table 3.
Sequencing of int1h and int22h duplicates and flanking regions was performed according to the manufacturer's instructions using the BigDye v3.1 dye terminator kit (ABI Perkin-Elmer) on 4-μL aliquots of PCR product incubated with 2 μL ExoSAP-It (USB Bioproducts) at 37°C for 15 min followed by heating at 80°C for 15 min. The products of the sequencing reactions were analyzed on an ABI 3100 DNA sequencer. Primers for sequencing int1h and int22h have been previously described (Naylor et al. 1995; Bagnall et al. 2002), and additional primers required for sequencing the chimpanzee int22h copies are shown in Supplemental Table 3.
To analyze variation of the int22h sequences among normal males and hemophilia A patients, int22h-1 and the distal int22h duplicates were amplified. Int22h nucleotides 1-1100, 4100-5100, and 8400-9512 were sequenced directly from PCR products as described above whereas nucleotides 1100-4100 and 5100-8400 were analyzed by fluorescent solid phase chemical cleavage of mismatches (FSPCCM) as follows: DNA for FSPCCM was prepared by initially amplifying, with 10 cycles of PCR, int22h-1 or both distal duplicates. A 1-μL aliquot of the primary PCR was further amplified using 2.5 μL 10× Amplitaq reaction buffer (Perkin-Elmer), 1.5 mM MgSO4, 200 ng of each nested oligonucleotide primer, 0.5 mM of each dNTP, and 2.5 U Amplitaq DNA polymerase (Perkin-Elmer). Thirty cycles of PCR were performed at 94°C for 30 sec, 65°C for 30 sec, and 72°C for 2 min. The seven primer pairs for amplification of mismatch target sequences are listed in Supplemental Table 3. Identical biotinylated fluorescent mismatch probes were amplified directly from a human clone (U100A9), which contains int22h-2, using 25 PCR cycles. Probe sequences were purified from a 1% agarose gel using GeneClean (Bio101) according to the manufacturer's instructions. FSPCCM analysis was performed as previously described (Waseem et al. 1999).
Recombined int22h-1, recombined distal int22h, and unrecombined int22h sequences were specifically amplified from inversion patient DNA using, respectively, primer pairs PB, AQ, and AB (Liu et al. 1998). Int22h nucleotides 1-1100, 4100-5100, and 8400-9512 were sequenced directly from PCR products as described above. Constant regions comprising nucleotides 1100-4100 and 5100-8400 were not analyzed in inversion patients.
The presence of gene conversion was tested using the method of Balding et al. (1992) for synonymous sites in codons, as the calculations developed there for fourfold degenerate sites are applicable to introns where a constant rate of mutation across the region and no selection is expected.
Uniformity of the occurrence of co-double sites and other types of sites (doubles, which differ from co-doubles because the divergent bases are not the same in both repeats, and singles, where a divergence occurs in only one of the repeats) was assessed via the Poisson approximation to the binomial distribution. The overall probability of a type (co-double, double, single) was estimated by the observed proportion of that type for the entire region. The region was then divided into smaller regions of length m and the Poisson probability of observing k sites of the given type out of m potential sites was calculated.
Significance of two divergence rates was determined via a Normal test of proportions (implemented in Minitab v14, Minitab Inc.) Confidence intervals for pairwise average differences were obtained by the method of bootstrapping (e.g., Manly, 1997) using 1000 samples.
Acknowledgments
We thank the hemophilia patients and the following hemophilia centers: Royal Bournemouth Hospital; Bristol Royal Infirmary; Arthur Bloom Centre, University Hospital of Wales, Cardiff; Royal Hospital for Sick Children, Yorkhill, Glasgow; Royal Post-graduate Medical School, Hammersmith Hospital, London; Lewisham Hospital; Churchill Hospital, Oxford; St Mary's General Hospital, Portsmouth; The Royal Free Hospital, London; The Royal London Hospital, for donation and collection of blood samples and for DNA with proximal type int22h-related inversions, Dr. J.D. Elsworth (supported by the St. Kitts Biomedical Research Foundation) for supplying African Green monkey brain tissue and Lesley Bergmeier at King's College London for supplying Rhesus monkey peripheral blood lymphocytes. This work was supported by the UK Medical Research Council.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY619998-AY620001 and AY781298-AY781308.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2946205.
References
- Antonarakis, S.E., Rossiter, J.P., Young, M., Horst, J., de Moerloose, P., Sommer, S.S., Ketterling, R.P., Kazazian Jr., H.H., Negrier, C., Vinciguerra, C., et al. 1995. Factor VIII gene inversions in severe hemophilia A: Results of an international consortium study. Blood 86: 2206-2212. [PubMed] [Google Scholar]
- Aradhya, S., Woffendin, H., Bonnen, P., Heiss, N.S., Yamagata, T., Esposito, T., Bardaro, T., Poustka, A., D'Urso, M., Kenwrick, S., et al. 2002. Physical and genetic characterization reveals a pseudogene, an evolutionary junction and unstable loci in distal Xq28. Genomics 79: 31-40. [DOI] [PubMed] [Google Scholar]
- Bagnall, R.D., Waseem, N., Green, P.M., and Giannelli, F. 2002. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood 99: 168-174. [DOI] [PubMed] [Google Scholar]
- Balding, D.J., Nichols, R.A., and Hunt, D.M. 1992. Detecting gene conversion: Primate visual pigment genes. Proc. R. Soc. Lond. B Biol. Sci. 249: 275-280. [DOI] [PubMed] [Google Scholar]
- Bentley, D.L. and Rabbitts, T.H. 1983. Evolution of immunoglobulin V genes: Evidence indicating that recently duplicated human V κ sequences have diverged by gene conversion. Cell 32: 181-189. [DOI] [PubMed] [Google Scholar]
- Casula, L., Murru, S., Pecorara, M., Ristaldi, M.S., Restagno, G., Mancuso, G., Morfini, M., De Biasi, R., Baudo, F., Carbonara, A., et al. 1990. Recurrent mutations and three novel rearrangements in the factor VIII gene of hemophilia A patients of Italian descent. Blood 75: 662-670. [PubMed] [Google Scholar]
- Ebersberger, I., Metzler, D., Schwarz, C., and Paabo, S. 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R.C. and Margot, J.B. 1984. Rabbit globin pseudogene ψ β 2 is a hybrid of δ- and β-globin gene sequences. Mol. Biol. Evol. 1: 302-316. [DOI] [PubMed] [Google Scholar]
- Hassock, S. 2000. “Physical and transcriptional mapping in the distal Xq28 region of the human X chromosome.” Chapter 4: Identification and mapping of transcripts in distal Xq28, pp. 133-162. Ph.D. thesis. King's College, London University.
- Jeffreys, A.J. and May, C.A. 2004. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36: 151-156. [DOI] [PubMed] [Google Scholar]
- Lakich, D., Kazazian, H.H., Antonarakis, S.E., and Gitschier, J. 1993. Inversions disrupting the factor VIII gene are a common cause of severe hemophilia A. Nat. Genet. 5: 236-241. [DOI] [PubMed] [Google Scholar]
- Levinson, B., Bermingham, J.R., Metzenberg, A., Kenwrick, S., Chapman, V., and Gitschier, J. 1992. Sequence of the human factor VIII-associated gene is conserved in mouse. Genomics 13: 862-865. [DOI] [PubMed] [Google Scholar]
- Liebhaber, S.A., Goossens, M., and Kan, Y.W. 1981. Homology and concerted evolution at the α 1 and α 2 loci of human α-globin. Nature 290: 26-29. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Nozari, G., and Sommer, S.S. 1998. Single-tube polymerase chain reaction for rapid diagnosis of the inversion hotspot of mutation in hemophilia A. Blood 92: 1458-1459. [PubMed] [Google Scholar]
- Lozier, J.N., Dutra, A., Pak, E., Zhou, N., Zheng, Z., Nichols, T.C., Bellinger, D.A., Read, M., and Morgan, R.A. 2002. The Chapel Hill hemophilia A dog colony exhibits a factor VIII gene inversion. Proc. Natl. Acad. Sci. 99: 12991-12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly, B.F.J. 1997. Randomization, bootstrap and Monte Carlo. In Methods in Biology 2nd ed., pp. 34-67. Chapman & Hall, London.
- Mellor, A.L., Weiss, E.H., Ramachandran, K., and Flavell, R.A. 1983. A potential donor gene for the bm1 gene conversion event in the C57BL mouse. Nature 306: 792-795. [DOI] [PubMed] [Google Scholar]
- Michelson, A.M. and Orkin, S.H. 1983. Boundaries of gene conversion within the duplicated human α-globin genes. Concerted evolution by segmental recombination. J. Biol. Chem. 258: 15245-15254. [PubMed] [Google Scholar]
- Miller, S.A., Dykes, D.D., and Polesky, H.F. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor, J.A., Green, P.M., Rizza, C.R., and Giannelli, F. 1992. Factor VIII gene explains all cases of hemophilia A. Lancet 340: 1066-1067. [DOI] [PubMed] [Google Scholar]
- Naylor, J., Brinke, A., Hassock, S., Green, P.M., and Giannelli, F. 1993. Characteristic mRNA abnormality found in half the patients with severe hemophilia A is due to large DNA inversions. Hum. Mol. Genet. 2: 1773-1778. [DOI] [PubMed] [Google Scholar]
- Naylor, J.A., Buck, D., Green, P., Williamson, H., Bentley, D., and Giannelli, F. 1995. Investigation of the factor VIII intron 22 repeated region (int22h) and the associated inversion junctions. Hum. Mol. Genet. 4.: 1217-1224. [DOI] [PubMed] [Google Scholar]
- Paques, F. and Haber, J.E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, M.F. and Ross, C.A. 2001. Isolation of a 40-kDa Huntingtin-associated protein. J. Biol. Chem. 276: 3188-3194. [DOI] [PubMed] [Google Scholar]
- Powers, P.A. and Smithies, O. 1986. Short gene conversions in the human fetal globin gene region: A by-product of chromosome pairing during meiosis? Genetics 112: 343-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis, A. 1995. A composite estimate of primate phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348: 405-421. [DOI] [PubMed] [Google Scholar]
- Rozen, S., Skaletsky, H., Marszalek, J.D., Minx, P.J., Cordum, H.S., Waterston, R.H., Wilson, R.K., and Page, D.C. 2003. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423: 873-876. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825-837. [DOI] [PubMed] [Google Scholar]
- Slightom, J.L., Blechl, A.E., and Smithies, O. 1980. Human fetal G γ- and A γ-globin genes: Complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell 21: 627-638. [DOI] [PubMed] [Google Scholar]
- Stoeckert, C.J., Collins, F.S., and Weissman, S.M. 1984. Human fetal globin DNA sequences suggest novel conversion event. Nucleic Acids Res. 12: 4469-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem, N.H., Bagnall, R.D., Green, P.M., and Giannelli, F. 1999. Start of UK confidential hemophilia A database: Analysis of 142 patients by solid phase fluorescent chemical cleavage of mismatch. Hemophilia Centres. Thromb. Haemost. 81: 900-905. [PubMed] [Google Scholar]
- Weiss, E., Golden, L., Zakut, R., Mellor, A., Fahrner, K., Kvist, S., and Flavell, R.A. 1983. The DNA sequence of the H-2kb gene: Evidence for gene conversion as a mechanism for the generation of polymorphism in histocompatibilty antigens. EMBO J. 2: 453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]