Abstract
The V1R gene family comprises one of two types of putative pheromone receptors expressed in the mammalian vomeronasal organ (VNO). We searched the most recent mouse, rat, dog, chimpanzee, and human genome sequence assemblies to compile a near-complete repertoire of V1R genes for each species. Dog, human, and chimpanzee have very few intact V1Rs (8, 2, and 0, respectively) compared to more than a hundred intact V1Rs in each of the rat (106) and mouse (165) genomes. We also provide the first description of the diversity of V1R pseudogenes in these species. We identify at least 165 pseudogenes in mouse, 110 in rat, 102 in chimpanzee, 115 in human, and 54 in dog. Primate and dog pseudogenes are distributed among almost all V1R subfamilies seen in rodents, indicating that the common ancestor of these species had a diverse V1R repertoire. We find that V1R genes were subject to strikingly different fates in different species and in different subfamilies. In rodents, some subfamilies remained relatively stable or underwent roughly equivalent expansion in mouse and rat; other subfamilies expanded in one species but not the other. The small number of intact V1Rs in the dog genome is unexpected given the presumption that dogs, like rodents, have a functional VNO, and a complex system of pheromone-based behaviors. We identify an intact transient receptor potential channel 2β in the dog genome, consistent with a functional VNO in dogs. The diminished V1R repertoire in dogs raises questions about the relative contributions of V1Rs versus other candidate pheromone receptor genes in the establishment of complex pheromone systems in mammals.
The vomeronasal organ (VNO) of terrestrial vertebrates detects pheromones that evoke innate social and reproductive behaviors (Keverne 2002). Functions that are attributed to the VNO include male dominance/aggressive patterning, male sexual preference, puberty timing, and pregnancy blockage (Kaneko et al. 1980; Lloyd-Thomas and Keverne 1982; Halpern 1987; Del Punta et al. 2002; Leypold et al. 2002; Stowers et al. 2002; Halpern and Martinez-Marcos 2003). Yet the VNO is probably not the exclusive pheromone-responding system, because some pheromone-induced behaviors are not perturbed by removal of the VNO (Hudson and Distel 1986; Dorries et al. 1997; Fernandez-Fewell and Meredith 1998; Fewell and Meredith 2002).
The rodent VNO has two distinct compartments of sensory neuronal populations, each expressing a different family of G-protein-coupled, seven-transmembrane-domain putative pheromone receptors. The sensory neurons of the apical compartment of the VNO express members of the V1R gene family, which transduce signals via a coupled Gαi protein; neurons of the basal compartment express members of a second putative pheromone receptor gene family, the V2Rs, which transduce signals via a coupled Gαo protein (Dulac and Axel 1995; Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997; Pantages and Dulac 2000). Upon receptor activation, signals are relayed via a G-protein-regulated transient receptor potential (trp)ion channel (Liman et al. 1999). The trp2 gene, whose β isoform is exclusively expressed in VNO neurons (Hofmann et al. 2000), is required for VNO sensory neuronal responses (Leypold et al. 2002; Stowers et al. 2002).
Each of the Gαi neurons of the apical compartment in the VNO is thought to express only one of a large repertoire of V1Rs (Rodriguez et al. 1999). V1Rs appear to bind pheromone ligands with high affinity and specificity (Leinders-Zufall et al. 2000; Boschat et al. 2002). Therefore, singular expression of pheromone receptors enables individual VNO sensory neurons to distinguish individual chemicals in complex pheromone blends. The rodent VNO might also respond to some odorants recognized by the olfactory receptors expressed in the nose (Sam et al. 2001), and therefore these two chemosensory organs could have functional overlap.
The canine VNO, like the rodent VNO, expresses several neuronal markers that suggest it is functional, including both the Gαo and Gαi proteins important for V1R and V2R signaling, and both the GAP43 and N-CAM proteins important for neuronal synaptogenesis (Dennis et al. 2003). However, the sensory epithelium in the dog VNO is relatively thin, and the auxiliary olfactory bulb (AOB) to which VNO neurons target is relatively small (Dennis et al. 2003). Moreover, there is some evidence for reduced VNO function in several mammalian species, such as pig (Dorries et al. 1997), sheep (Cohen-Tannoudji et al. 1989), and ferret (Weiler et al. 1999), as compared to rodents. Therefore, it is not clear whether the VNO and the V1R and V2R repertoires play as extensive a role in mammalian pheromone-based behaviors as rodent studies suggest (for review, see Halpern and Martinez-Marcos 2003).
Humans and some primates seem to possess only a vestigial VNO (Trotier et al. 2000). It has been argued that the decline of the VNO in primates began shortly before the separation of hominoids and Old World monkeys, concurrent with the advent of trichromatic color vision, which could have accelerated the replacement of a chemical-based system with a vision-based system of reproductive signaling (Zhang and Webb 2003). The essential trp2 channel is a pseudogene in humans (Liman et al. 1999) and closely related primates (Zhang and Webb 2003), and the putative pheromone receptor gene families expressed in the VNO are predominantly pseudogenes in the human genome (Giorgi et al. 2000; Kouros-Mehr et al. 2001; Lane et al. 2002; Rodriguez and Mombaerts 2002), and the genomes of other closely related primates (Giorgi and Rouquier 2002; Zhang and Webb 2003). One putative functional V1R gene identified in the human genome is expressed in the main olfactory system, suggesting that the ligands of the intact human V1Rs are recognized by the nose instead of a VNO (Rodriguez et al. 2000).
The size and diversity of V1R repertoires in various species could provide insights into the relative complexity and species-specificity of VNO-mediated pheromone-based behaviors. The near-complete repertoire of mouse V1R genes was estimated previously by Rodriguez et al. (2002) and Zhang et al. (2004) to be 137 genes (plus 156 pseudogenes) and 164 genes (plus 168 pseudogenes), respectively. The intact V1R genes cluster into 12 subfamilies in a phylogenetic tree (Rodriguez et al. 2002). We described how two of these subfamilies underwent striking lineage-specific expansions since mouse-rat speciation, which could contribute to species-specific pheromone responsiveness (Lane et al. 2004).
In this study, we first set out to identify the near-complete repertoire of rat V1R genes in order to investigate the extent of species-specificity in the rodent V1R family. We find that rat has only a slightly smaller V1R gene repertoire (106 intact V1Rs) than mouse, and that this repertoire encompasses most of the subfamilies described in mouse. However, we find striking examples of lineage-specific subfamily expansions. We then extended our study to investigate the size and diversity of the V1R repertoires in the recently generated dog, chimpanzee, and human genome assemblies. The chimpanzee genome has a large repertoire of V1R pseudogenes and no apparently functional V1R genes. This massive V1R gene loss in the chimpanzee genome resembles what is observed for the human V1R repertoire (Giorgi et al. 2000; Kouros-Mehr et al. 2001; Lane et al. 2002; Rodriguez and Mombaerts 2002; herein). The most surprising observation we make in this study is that the dog V1R repertoire, like the primate repertoire, is greatly diminished. This finding is unexpected because dog, like mouse and rat, is thought to possess a functional VNO (Dennis et al. 2003), and dogs are famous for their olfactory acuity and complex social structures that would presumably demand a large, functional repertoire of pheromone receptor genes. This finding raises questions about the general importance of V1Rs, as compared to other putative pheromone receptor gene families (e.g., V2R) or odorant receptors of the nose, in the establishment of pheromone-based behaviors throughout the mammalian phylogeny.
Results and Discussion
Identification of V1R-gene and -pseudogene repertoires in five species
We mined recent sequence assemblies of the mouse, rat, dog, chimpanzee, and human genomes to identify the near-complete repertoires of V1R pheromone receptor genes in each of these species. The numbers of intact V1Rs and pseudogenes found in each genome assembly are shown in Table 1. The status as either intact or disrupted is ambiguous for a small number of atypical dog, chimpanzee, and human V1R-like sequences (see Methods). It is important to note that all V1Rs were identified in draft sequence assemblies at various stages of completion, and therefore, sequencing and assembly errors could contribute to under- or overestimation of V1R repertoire sizes.
Table 1.
Number of V1R-like sequences found in the genomes of five species
| Mouse | Rat | Dog | Chimpanzee | Human | |
|---|---|---|---|---|---|
| 364 | 220 | 65 | 116 | 117 | Total V1R-like sequences |
| Intact V1Rs | |||||
| 165 (50%) | 110 (51%) | 54 (87%) | 102 (100%) | 115 (98%) | Pseudogenes |
| 34 | 4 | 3 | 14 | 0 | Partial sequences |
| 95% | 90% | 98% | 94% | 100% | Assembly coveragea |
| 190 | 120 | 9 | 0 | 2 | Intact V1Rs extrapolated to complete genome |
Partial sequences are interrupted by assembly gaps and could be intact or pseudogenes. Complete sequence information is needed before their status can be determined. The percentages shown are intact and pseudogene proportions of all V1Rs with full-length sequence available.
See Methods.
Our survey results are in good agreement with previous studies in mouse and human. We identified 165 intact mouse V1Rs, one more than was found by Zhang et al. (2004). In the human genome, we identified two of the five intact human V1Rs found previously by Rodriguez and Mombaerts (2002); the remaining three were identified as pseudogenes (see ambiguities described in Methods). We find that rat, like mouse, has a large repertoire of intact V1Rs, and chimpanzee, like human, has a large collection of V1R pseudogenes. Two intact chimpanzee V1Rs are annotated in GenBank; we find an in-frame stop codon in both sequences. These V1Rs might be polymorphic with some alleles intact and others pseudogenes. The enormous difference in the number of intact V1Rs between rodents and primates is consistent with the fact that rodents possess a functional VNO (Leypold et al. 2002; Stowers et al. 2002), whereas the two primates probably do not (Trotier et al. 2000; Zhang and Webb 2003).
Rodent V1R repertoires are comprised of both new and old subfamilies
Our results show that mouse has a functional V1R repertoire that is ∼50% larger than its rodent relative, the rat (165 vs. 106 intact genes). A gene tree (Fig. 1) of intact V1Rs partitions into 12 major clades corresponding to the mouse subfamilies A-L described previously (Rodriguez et al. 2002) (see Methods), with only two intact mouse V1Rs outside of these clades. All but one of the 106 intact rat V1Rs also cluster within these 12 clades.
Figure 1.
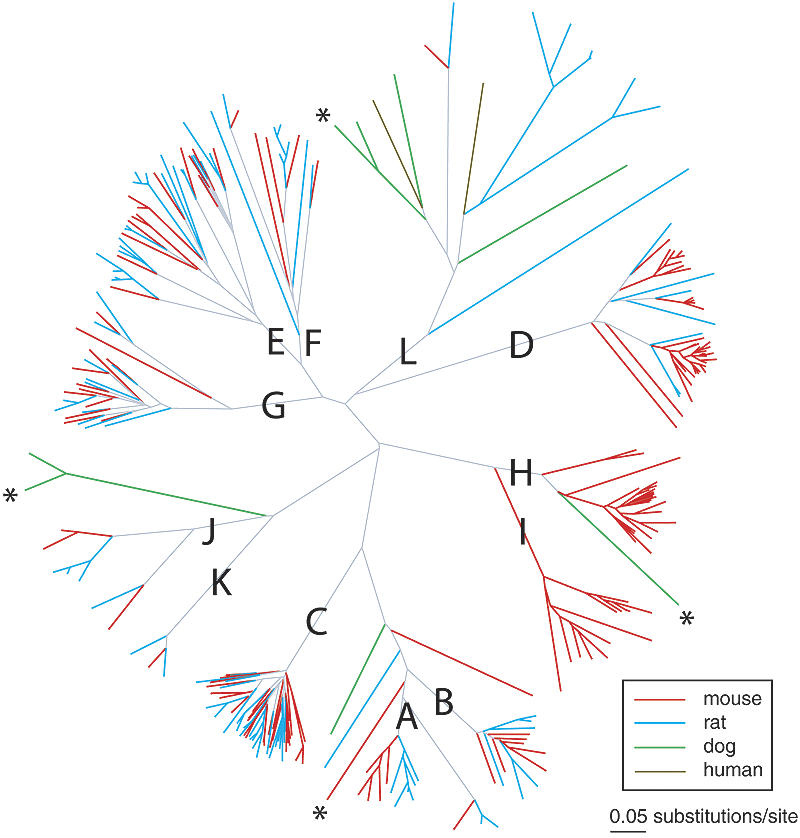
Neighbor-joining gene tree based on nucleotide alignments illustrating subfamily representation (A-L) of intact mouse (red), rat (blue), dog (green), and human (brown) V1Rs. Three dog V1Rs and one mouse V1R that could be pseudogenes (see Methods) are indicated with asterisks.
Some of the mouse/rat subfamilies delineate along species lines. For example, we previously described the species-specific divergence of the rodent A subfamily (Lane et al. 2004). That is, all of the A-subfamily mouse V1Rs group in a clade distinct from all of the rat V1Rs, consistent with duplication/divergence of this subfamily after the species split. The mouse H and I subfamilies consist of 23 and 13 intact V1Rs, respectively, and these subfamilies provide even more striking examples of species specificity, as neither clade contains an intact rat V1R. A tree that includes pseudogenes (Fig. 2) places one rat pseudogene in each of the H and I subfamilies; two additional rat pseudogenes are also I-like, but their sequences were too short to include in the tree (see Methods). Analysis of synonymous substitution levels (dS) within the H and I subfamilies range from 0.01 to 0.94 substitutions per site. Synonymous nucleotide positions can be used to estimate the level of neutral substitution between pairs of genes. Neutral substitution rates are approximately proportional to elapsed evolutionary time. Therefore, the rates of synonymous substitution (dS) indicate how long ago pairs of genes diverged. Since typical mouse-rat orthologs have median dS levels of 0.19 substitutions/site (Rat Genome Sequencing Project Consortium 2004), the H and I subfamilies therefore began expanding in the rodent ancestor (as evidenced by gene pairs with dS » 0.19 substitutions/site) and continued to expand after the mouse-rat split (dS « 0.19). Thus, the difference in the sizes of the H and I subfamilies between mouse and rat is due to both deletion in rat and post-speciation expansion in the mouse lineage. Less extreme examples of species bias include the D subfamily, which is predominantly mouse (33 of 42 intact genes), and the L subfamily, which is predominantly rat (seven of eight intact rodent genes). Finally, the isolated mouse gene below the A-B root has no rat counterpart, and the mouse counterpart of the isolated rat gene below the A-B root is a pseudogene, suggesting that species-specific loss of orthologs occurred in these two cases (Lane et al. 2004). Each of these examples illustrates possible delineations of V1R function between the two rodent species that could contribute to their ability to use pheromones to communicate within but not between species.
Figure 2.
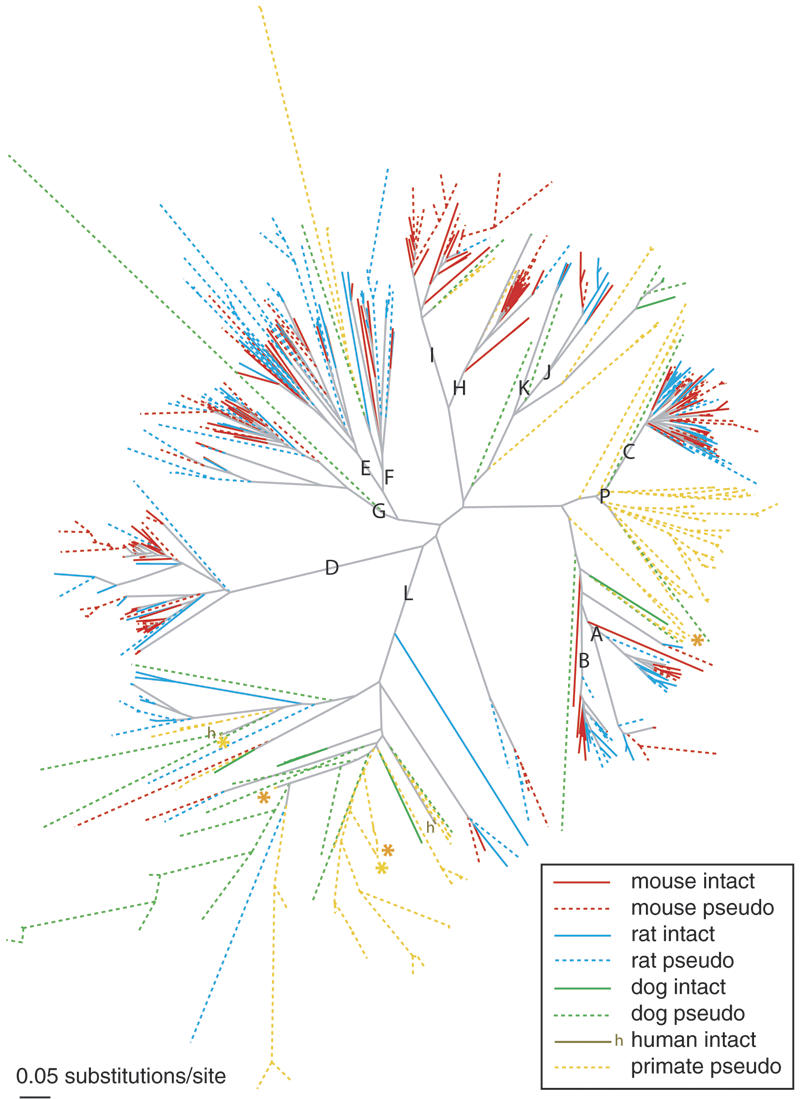
Neighbor-joining gene tree illustrating subfamily clades (A-L, P) of intact and pseudogene V1R-like sequences identified in the genomes of five mammalian species. Mouse intact (red), mouse pseudogene (red dashed), rat intact (blue), rat pseudogene (blue dashed), dog intact (green), dog pseudogene (green dashed), human intact (brown, denoted “h”), and human/chimpanzee pseudogene (yellow dashed) branches are shown. Two chimpanzee and three human pseudogenes that were previously annotated as intact V1Rs (see Methods) are indicated with yellow and orange asterisks, respectively. A total of 87 mouse, 39 rat, 27 dog, 59 chimpanzee, and 58 human V1R-like pseudogenes were excluded from this tree, because the length of confidently aligned sequence was too short (see Methods). A list of these excluded sequences, as well as their presumed subfamily (based on closest fast×34 matches), is provided in Supplemental Figure D.
In contrast, several other subfamilies (E, F, and G) consist of mouse and rat V1Rs that intermingle in the tree, and therefore do not exhibit species bias. Generally, these subfamilies did not experience extensive expansion since the mouse-rat split, and there are several examples in which unambiguous orthology of pairs of mouse and rat V1Rs is evident (e.g., one of the minor clades within the E subfamily consists of three pairs of candidate mouse-rat orthologs). These orthologous pairs could encode ancestral functions that are preserved in both species.
Dog has an unexpectedly diminished V1R repertoire
We find only eight intact V1R genes in the dog genome. This small number is unexpected, given that dogs appear to have an operational VNO (e.g., Dennis et al. 2003), have renowned olfactory acuity, and live within highly ordered pack structures that presumably require complex intraspecies signaling. The dog sequence assembly is from the boxer breed. We note that a preliminary comparison of pseudogene status between boxer and poodle indicates that all V1R pseudogenes identified in boxer are also pseudogenes in poodle (data not shown), suggesting that the decline in the dog V1R repertoire is not exclusive to the boxer breed. The small number of intact V1Rs may partly explain the thin sensory epithelium in the dog VNO, as well as the relatively small AOB of dog (Dennis et al. 2003). We wondered if dogs, like primates, might have lost functionality of the VNO-specific β isoform (Hofmann et al. 2000) of the trp2 channel. The dog trp2β appears functional from our analysis of the genome assembly, and of sequence we generated to fill an assembly gap. The dog trp2β gene's 13-exon gene structure is identical to that of its mouse ortholog, and all predicted dog exon boundaries have consensus AG/GT splice sequences. The dog and mouse predicted protein products have ∼88% amino acid identity (see Supplemental Fig. A).
These findings raise some interesting ideas. It is possible that V1Rs emerged as a dominant pheromone receptor family (perhaps exclusively) in rodents. Since we assume that dogs have a complex system of pheromonal communication, other receptor gene families expressed in the VNO (e.g., V2R) (Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997) and/or the main olfactory system (olfactory receptors) (Buck and Axel 1991) might, in fact, be more important than V1Rs for pheromone perception in most mammals. Alternatively, in the process of breeding out certain wild social behaviors (e.g., aggression and dominance) during the domestication of dogs, breeders might have depleted the dog genome of many of its pheromone receptor genes. We also note that the dog genome has far fewer total V1R sequences than the other four species (Table 2), thus it is possible that entire clusters containing intact V1Rs might have deleted during domestication. Before concluding that V1Rs grew to prominence only in rodents it will be important to investigate repertoires in undomesticated canids and other nonrodent mammalian species.
Table 2.
Coordinates of all of the V1R gene clusters in all five species
| Cluster | Flanking genes | Mouse | Rat | Dog | Chimpanzee | Human |
|---|---|---|---|---|---|---|
| AB | Txnrd3 | 6_90.182> | 4_123.974> | 20_3.615< | 2_129.650< | 3_127.608< |
| Klf15 | 6_90.892> | 4_124.958> | 20_3.346< | 2_129.394< | 3_127.343< | |
| A/B | 11 (9) | 15 (10) | ||||
| AB2 | FBLN2 | 6_91.672> | 4_125.645> | 20_6.717> | 2_14.154> | 3_13.603> |
| SLC6A6 | 6_92.172> | 4_126.158> | 20_7.416> | 2_15.058> | 3_14.476> | |
| A/B | 0 (1) | 0(1) | 1(1) | 0(2) | 0(2) | |
| L | 0(1) | |||||
| C1 | Pde1c | 6_56.140< | 4_85.187< | 14_46.859< | 6_32.731< | 7_31.556< |
| Herc3 | 6_59.045> | 4_87.981> | 32_14.842> | 3_105.880> | 4_89.919> | |
| C | 26 (23) | 23 (8) | ||||
| C2 | Prdm5 | 6_66.089> | 4_95.693> | 19_22.150> | 3_138.666< | 4_122.009< |
| II23r | 6_67.732< | 4_97.331< | 5_46.355< | 1_65.705> | 1_66.992> | |
| C | 5 (4) | 1 (0) | ||||
| D1 | U2af2 | 7_4.457> | 1_67.576< | 1_104.765< | 20_58.300> | 19_60.842> |
| Zfp28 | 7_5.726> | 1_65.997< | 1_104.145< | 20_59.305> | 19_61.741> | |
| D | 5 (8) | 8 (3)a | 0 (1) | 0 (1) | ||
| D2 | Clptm1 | 7_14.075< | 1_79.017< | 1_112.705< | 20_47.296> | 19_50.134> |
| Kcnn4 | 7_15.781> | 1_79.691> | 1_113.630> | 20_45.902< | 19_48.947< | |
| D | 10 (8) | |||||
| C | 0 (1) | |||||
| L | 0 (1) | |||||
| EFb | Has1 | 17_16.348< | 1_56.506< | 1_107.645> | 20_54.341< | 19_56.896< |
| Zfp51c | 17_19.964> | 1_59.414> | 1_106.502> | 20_55.019> | 19_57.653< | |
| PRPF31d | 7_3.065> | 1_63.968< | 1_105.761< | 20_56.755> | 19_59.295> | |
| E | 8 (6) | 15 (12) | 0 (1) | |||
| F | 5 (4) | 8 (9) | 0 (1) | 0 (2) | 0 (2) | |
| L | 0 (2) | 0 (3) | 3 (22) | 0 (11)e | 1 (11)f | |
| D | 0 (2) | 0 (3) | ||||
| ELGd | Peg3 | 7_6.057< | 1_65.593> | 1_103.799> | 20_59.565< | 19_61.999< |
| Zfp606 | 7_8.275> | 1_72.662> | 1_102.618> | 20_60.920< | 19_63.165< | |
| E | 5 (4) | 6 (9)a | ||||
| L | 1 (2) | 7 (13)a | 0 (6) | 0.2 | 1 (1) | |
| G | 12 (16) | 11 (10) | 0 (1) | |||
| D | 0 (2) | |||||
| EG | Cyp4f15 | 17_31.407> | 7_13.294> | 20_49.472< | 20_16.438< | 19_15.716< |
| X97650 | 17_32.152> | 7_16.074> | 20_55.922< | 20_8.848< | 19_8.496< | |
| E | 1 (1) | |||||
| G | 2 (12) | |||||
| HI | Zfp192 | 13_20.958< | 17_50.683> | 35_28.290> | 5_28.679> | 6_28.178> |
| Btn1a1 | 13_22.902< | 17_48.858> | 35_27.265> | 5_27.087> | 6_26.567> | |
| H | 23 (17) | 1 (1) | 0 (1) | 0 (1) | ||
| I | 13 (32) | 0 (3)g | 0 (1) | 0 (3) | 0 (3) | |
| JK | 0 (1) | 0 (1) | ||||
| JK | Trim28 | 7_8.762>h | 1_72.970< | 1_102.116< | 20_61.507> | 19_63.731> |
| Lig1 | 7_9.022> | 1_73.805> | 1_110.333> | 20_50.584< | 19_53.294< | |
| J | 3 (1) | 4 (3) | 0 (1)i | 0 (1) | 0 (1) | |
| K | 1 (1) | 2 (0) | ||||
| L | 0 (1) | 1 (0) | ||||
| LP1 | CGI-49 | 1_180.357>j | 13_95.538> | 7_44.079> | 1_227.292> | 1_242.984> |
| MGC15548 | 11_60.118< | 10_45.804< | 8_3.140< | 1_227.873< | 1_243.560< | |
| AB | 0 (2) | 0 (2) | ||||
| L | 0 (1) | 0 (1) | ||||
| P | 0 (1) | 0 (1) | ||||
| LP2 | FLJ13868 | 7_116.995< | 1_187.679< | 6_19.275< | 18_32.219< | 16_31.499< |
| Centromerek | 16_36.326 | |||||
| L | 0 (3) | 0 (5)f | ||||
| P | 0 (1) | 0 (3)f | ||||
| LP3 | GMIP | 8_68.831> | 16_20.086< | 20_46.494> | 20_20.679< | 19_19.601< |
| Centromere | 19_24.407 | |||||
| L | 0 (10)l | 0 (17)f | ||||
| E | 0 (1) | 0 (1) | ||||
| LP4 | PHKG1 | 5_127.258< | 12_27.893< | 6_3.303< | 6_57.619< | 7_55.856< |
| Centromere | 7_57.794 | |||||
| L | 0 (2)e | 0 (3) | ||||
| P | 0 (3)e | 0 (3) | ||||
| LP5 | Centromere | 7_60.794 | ||||
| TPST1 | 5_127.497> | 12_27.470< | 6_3.684> | 6_66.801> | 7_65.103> | |
| L | 0 (4)m | 0 (5) | ||||
| P | 0 (7) | 0 (9) | ||||
| Mapped in clusters | 128 (140) | 102 (99)a | 5 (35) | 0 (49)e | 1 (62)f | |
| Mapped unclustered | 5 (8) | 2 (9)a | 1 (15) | 0 (39)e | 1 (51)f | |
| Unmapped | 32 (51) | 2 (6) | 2 (7) | 0 (28) | 0 (2) |
For each V1R gene cluster, the positions (format: chromosome number_Mb position) and orientation (arrowheads) of non-V1R genes near cluster boundaries in each genome are given. All genes shown are annotated human (in capital letters) or mouse (not fully capitalized) genes. The number of intact V1Rs (number of pseudogenes plus partial sequences in brackets) from identifiable subfamilies (A-P) in each cluster is shown for each species. In cases where orthologous non-V1R genes have not remained linked, the number of V1Rs located in clusters <1 Mb from one of the non-V1R genes (bold) is shown. Clusters (shaded) are defined as a group of at least three V1Rs with <500 kb between neighboring genes (exceptions are footnoted).
The numbers shown for the rat D cluster include an isolated intact D-like V1R located ~800 kb downstream of the main D cluster; the numbers shown for the rat ELG cluster include a pair of isolated pseudogenes (one E-like and one L-like) located ~800 kb upstream of the main cluster. The totals shown at the bottom of the table count these three V1Rs within the “Mapped Unclustered” category, according to our formal cluster definition.
The EF locus contains three linked clusters in rat and two linked clusters in chimpanzee and human; the V1R totals shown reflect all three rat and both primate clusters.
Zfp51 has ambiguous orthology. A more distant, less ambiguous flanking gene (PRPF31) is downstream of the nonrodent clusters; there is a syntenic break between Zfp51 and PRPF31 in mouse.
An inversion has split the rat “ELG” cluster into three clusters on rat Chromosome 1 (two EL clusters and one LG cluster; see Supplemental Material B). The numbers shown are totals in all three rat clusters. Note that owing to these rearrangements, the rat PRPF31 and Peg3 genes no longer flank the rat EF and ELG clusters, respectively (see Supplemental Material B).
The numbers shown for the chimpanzee EF and LP4 clusters each include a pair of isolated pseudogenes located ~800 kb away from the main clusters. The totals shown at the bottom of the table count these four V1Rs within the “Mapped Unclustered” category, according to our formal cluster definition.
The numbers shown for the human EF, LP2 and LP3 clusters includes two pseudogenes (L-like), three pseudogenes (two P-like and one L-like) and one pseudogene (L-like), respectively, that are located >500 kb away from the main clusters. The totals shown at the bottom of the table count these six V1Rs within the “Mapped Unclustered” category, according to our formal cluster definition.
The rat H-like pseudogene is located as a singleton on rat Chromosome 15.
The mouse ELG and JK loci are <500 kb apart (constitute one cluster by our definition).
Two intact dog J-like genes are unmapped.
An isolated, intact AB-like V1R is located at the 182.9 position (~3 Mb away from the CGI-49 gene) on mouse chromosome 1.
The LP2, LP3, LP4, and LP5 clusters are located in gene-poor pericentromeric regions.
This locus encompasses three clusters in chimpanzee.
This locus encompasses two clusters in chimpanzee.
V1R subfamily diversity arose early in mammalian evolution
Even though present-day dog and primate genomes contain only a small number of intact V1Rs, our analyses reveal that the common ancestor of these species and rodents had a diverse repertoire of V1Rs, with representatives of many of the subfamilies that exist in rodents today. Diversification into subfamilies therefore likely occurred prior to the rodent-dog-primate split early in mammalian evolution. Intact dog V1Rs are spread among four clades (L, H, J/K, and A/B) (Fig. 1), and dog and primate pseudogenes are found in almost all major clades of the V1R tree (Fig. 2). To provide further support for this view, we measured synonymous substitution (dS) levels of intact V1Rs. Neutral sequences are expected to exhibit ∼34% substitution levels (Jukes-Cantor adjusted) along the mouse-rat lineages since the common ancestor of rodents and humans (Mouse Genome Sequencing Consortium 2002). We find that the minimum substitution levels between the closest rodent subfamilies (e.g., E vs. F, H vs. I, J vs. K) exceed 70% (Jukes-Cantor adjusted), consistent with the hypothesis that these subfamilies arose prior to the divergence of rodents from primates and dogs. The A and B subfamilies are the only exceptions, with cross-subfamily pairwise dS levels clustered around 68% (the minimum A vs. B pairwise dS is 50%). Moreover, the A and B clades in Figure 2 lack nonrodent V1Rs (nonrodent V1Rs are found below the root of these subfamily clades). These data are consistent with the diversification of the A and B subfamilies after the rodent-primate-dog splits.
The L subfamily of V1Rs is the most prominent in the primate and dog repertoires. Half of the intact dog V1Rs (four) are contained within the L clade, along with the only two intact human V1Rs. Furthermore, the L clade has the greatest representation of dog, chimpanzee, and human pseudogenes of all the clades, and therefore might have had an even more prominent role until recently. In contrast, the L subfamily is the smallest subfamily in mouse, consisting of only one intact gene (as well as six pseudogenes). The long terminal branches and diffuse topology in the L clade suggest a distant common ancestor, and a dearth of recent duplication/expansion compared to other clades. Therefore, the L subfamily might encode some of the oldest V1R functions to be fixed in ancestral mammals.
Primates have an additional large subfamily of V1R pseudogenes near the root of the rodent C clade (labeled “P” in Fig. 2). Two dog pseudogenes are also found in this section of the tree. This subfamily contains a divergent set of V1Rs (some dS pairs exceed saturated substitution levels), and therefore, the subfamily probably predates the rodent-primate split (i.e., is probably not an orthologous group to the rodent C subfamily). Therefore, this set of “P” genes has probably been lost along rodent lineages, yet might have remained prominent in primates until recently.
V1Rs are found at syntenic locations in the mouse and rat genomes
Mouse V1Rs are physically clustered at nine genomic locations (Table 2). We find that most members of each subfamily map within the same cluster as one another, indicating that the rodent V1R family expanded primarily as a result of local duplication events, as observed for many other gene families (e.g., Young et al. 2002).
We find in rat a similar number of V1R clusters, which are at syntenic genomic locations to those in mouse. We identified pairs of flanking non-V1R genes for each of these mouse clusters, and located their putative orthologs in rat (Table 2). The linkage between V1Rs and neighboring non-V1R orthologs is generally conserved between mouse and rat, and the same subfamilies are clustered together in both rodent species. The only exceptions to this conserved linkage are with clusters found on rat Chromosome 1, equivalent to the D, EF, ELG, and JK clusters on mouse Chromosomes 7 and 17. Since these species diverged, these clusters experienced two local rearrangements (one in each of the rat and mouse lineages) and one chromosomal rearrangement in mouse (see Supplemental Fig. B).
Some nonrodent V1Rs are found at syntenic locations to rodent loci, but many are dispersed
We find a small number of V1R-like sequences at many of the dog and primate locations that are equivalent to the nine mouse V1R clusters (Table 2), showing that V1Rs existed at these genomic positions in the common ancestor. However, we find many other V1R-like sequences dispersed widely throughout the dog and primate genomes; these sequences likely arose by non-local duplications. Most dispersed primate and dog V1Rs are isolated sequences, except for five clusters of L- and P-subfamily primate pseudogenes at least four of which appear to have moved to their current locations since primates diverged from the other species.
Three mouse V1R clusters do not exhibit conserved synteny in the dog and primate genomes. Yet, even in these cases, it is possible to trace the rearrangement events that occurred to account for previous syntenic relationships. The genes flanking the mouse AB cluster are still linked in the other three genomes, and while we do not identify V1Rs between these orthologs, A-like pseudogenes are on the same chromosomes in the dog, chimpanzee, and human genome (AB2 cluster in Table 2). For the other two mouse clusters (C1 and C2), the flanking genes are unlinked in the dog, human, and chimpanzee genomes, and V1Rs from the C subfamily are not evident in the dog, chimpanzee, or human genomes. However, we suggest there was a single cluster containing both P- and C-like genes in the common mammalian ancestor: The primate LP4 cluster (pericentromeric region of human Chromosome 7) is flanked by a FKBP9 gene that has a rodent homolog within 100 kb of the mouse and rat C2 loci (data not shown). Furthermore, the P pseudogenes are the most similar primate and dog relatives of the rodent C genes (Fig. 2). Members of the P and L subfamilies are also found at other pericentromeric locations in the human genome, as well as near the telomere of Chromosome 1 (Table 2). Subsequent history of a putative ancestral “CP cluster” appears complicated and probably involved several evolutionary mechanisms: loss of P-like genes in rodent and possibly all of the C-like genes in primates and dog (as noted earlier from Fig. 2); local duplications expanding the P and L (primates) and C (rodents) subfamilies; chromosomal translocations that disrupt synteny by breaking ancestral genomic segments; and segmental duplication among pericentromeric locations in primates. These events add to a growing set of examples of chromosome-remodeling events seen at clusters of highly related genes (Dehal et al. 2001) and in pericentromeric regions (She et al. 2004). This example also illustrates how elaborate evolutionary events can result in different subfamily representation at syntenic loci between species (Table 2).
The primate and dog V1R-like genes are more dispersed in their respective genomes than those of rodents. Of the 259 V1R-like sequences mapped in the dog, chimpanzee, and human genomes, only 152 (∼61%) are found within clusters (defined as a set of at least three genes spaced <500 kb apart); the median cluster size is just five genes. Even considering just the human genome (as the mapping data are less accurate in the draft dog and chimp assemblies), only 63 (∼55%) of the 115 mapped human V1R-like sequences reside in clusters. In contrast, ∼95% (267/281) of the mapped mouse and ∼95% (268/281) of the mapped rat V1R repertoire (counting both genes and pseudogenes) are located within large clusters with a median cluster size of 19 genes. In primates, chromosomal dispersal is extreme: V1R-like sequences are identified on 22 chromosomes in human and 21 chromosomes in chimpanzee compared to being found on only six chromosomes in rat and eight in mouse. Most (16 of 23) of the isolated rodent V1R-like sequences are pseudogenes. Of the six intact V1Rs mapped in dog (two intact dog V1Rs are not mapped), five map within clusters. Thus, dispersed V1R sequences are likely to be pseudogenes, with the large, scattered collection of unclustered chimpanzee and human pseudogenes representing an extreme of this trend. We previously noted an increase in the dispersal of olfactory receptor genes in human compared to mouse (Young et al. 2002). These observations might reflect selective pressures in the rodent genome to keep receptor genes in tight, easily regulated clusters, as well as a different balance between local and interchromosomal duplication mechanisms in the two genomes. These results also fuel our earlier speculation (Lane et al. 2002) that functional V1Rs could in some way depend on clustering or their native locus for their proper expression and thus might not survive as intact genes when duplicated to other locations.
Conclusions
The V1R gene family encodes pheromone receptors expressed in the mammalian VNO. The large and diverse V1R repertoire in mouse and rat, as well as striking species delineation of some subfamilies in mouse and rat, suggest that this family plays a profound role in defining species-specific social behaviors in rodents. In contrast, the dog V1R family has been decimated, with only eight remaining potentially functional V1Rs, despite an apparently functional VNO. Humans and chimpanzees appear to have suffered even more extreme deterioration of their vomeronasal organ and pheromonal signaling components, perhaps as a result of the rise of a dominant visual system. Different V1R subfamilies appear to have been dominant in each of the mouse, rat, dog, and primate lineages.
We consider three possible implications of the vastly different functional repertoire sizes in rodents and dogs: (1) V1Rs might, indeed, provide a greatly expanded range of specialized functions in rodents, or even allow the VNO to recognize additional nonpheromonal ligands. (2) The many duplication events in rodents could have produced a largely redundant and overlapping set of functions, with the extra genes making only an incremental difference in the range of encoded functions. (3) Other gene families, such as V2Rs in the VNO or olfactory receptors in the nose, might be much more important than V1Rs in pheromone perception in dogs and perhaps other mammals.
Methods
V1R gene identification
V1R-like sequences were identified using a modified version of the method used to identify olfactory receptors (Glusman et al. 2001; Young et al. 2002). First, the amino acid sequences of 49 diverse intact V1R genes were each used as queries in a sensitive TBLASTN search of five genome sequence assemblies (mouse: NCBI build 30, Feb. 2003, Mouse Sequencing Consortium and Mouse Genome Sequencing Consortium; rat: version 3.1, June 2003, Rat Genome Sequencing Consortium; human: NCBI build 33, Apr. 2003, International Human Genome Project; chimpanzee: NCBI build 1 version 1, Nov. 2003, chimpanzee: NCBI build 1 version 1, Nov. 2003, R. Waterston, pers. comm.; dog: WGSv1.0, July 2004, K. Lindblad-Toh, pers. comm. Note that because a female dog was sequenced, no Y-chromosome sequence is represented in the assembly. The diverse query set consisted of between one and three intact V1Rs from each previously described mouse subfamily (Rodriguez et al. 2002), one rat V1R from each subfamily (as identified in preliminary analyses), five human V1Rs (Rodriguez and Mombaerts 2002), and one dog V1R (Rodriguez and Mombaerts 2002). A second round of searches was performed to ensure that all dog V1R pseudogenes were identified, using as queries the eight apparently intact dog V1Rs identified in a preliminary search—only one additional dog pseudogene was identified. Second, all genomic sequences matching any of the V1R queries with an E-value of 10-5 or better were collected, along with 1 kb of flanking sequence on each side. Interspersed repeats in these ∼3-kb sequences were masked using RepeatMasker (http://repeatmasker.org), using the -nolow option to leave low-complexity regions unmasked. Third, masked sequences were compared using fast×34 (Pearson et al. 1997) to a database that included previously identified intact, full-length V1R sequences as well as a large number of non-V1R outgroups, including V2R pheromone receptors, olfactory receptors, taste receptors, rhodopsin, and representatives of many other GPCR families. Any sequence that matched a non-V1R better than a V1R or that had no good matches in this search was eliminated from further analysis. Fourth, a custom script was used to determine the most likely locations of start and stop codons, using the best fast×34-identified V1R match as guide, starting from the endpoints of sequence similarity and searching in each direction a codon at a time. Any sequences with stop codons, frameshifts, or interspersed repeats interrupting the ORF were annotated as pseudogenes. Sequences interrupted by assembly gaps were also noted. Results were tracked using a customized acedb database (http://www.acedb.org). Fifth, apparent pseudogenes were subjected to manual curation—in a handful of cases a better translation could be found, avoiding the stop codon or frameshift suggested by the closest fast×34 match. Finally, all V1R-like pseudogene sequences were manually examined; sequences that only match intact V1Rs weakly and that could not be confidently aligned to an intact V1R were discarded. We note that about the time this manuscript was submitted, Grus and Zhang (2004) reported the identification of 95 intact V1R genes (and 116 total V1R-like sequences) from the draft rat genome assembly. Our conclusions about the rodent V1R family are in close agreement with theirs, although we identify a few more intact V1Rs (106 vs. 95) and many more rat V1R-like sequences when pseudogenes are also considered (220 vs. 116).
Estimation of assembly coverage
Assembly coverage for the rat genome is a published estimate (Rat Genome Sequencing Project Consortium 2004); the human genome is considered complete. Coverage for the remaining genomes was estimated as follows. Briefly, for each species, a set of nonredundant GenBank entries that were submitted independently of any genome project (sample size >100 sequences) were compared to the assembly. The frequency with which test sequences were not represented in the genome assembly was measured (nucleotide identity threshold = 95%). This frequency of unmatched queries approximates the fraction of missing sequence content in the assembly. Similar methods have been used previously (e.g., Rat Genome Sequencing Project Consortium 2004), and full details are available upon request.
Ambiguities in pseudogene classification of some V1Rs
The eight intact genes identified in dog include three V1R-like open reading frames (ORFs) that might, in fact, be pseudogenes: (1) The 3′-most 25 bp of a V1R gene at the 106.247-Mb position on Chromosome 1, identical to GenBank accession AAM66755, is similar to a SINE repeat. (2) A sequence at the 27.476-Mb position of Chromosome 35 encodes a slightly truncated predicted protein (292 amino acids). (3) The 3′-most 170 bp of an un-mapped V1R (at the 56.471-Mb position of the contig) is similar to a SINE repeat, and the predicted protein of this gene is longer than expected. Therefore, if these three ambiguous ORFs are, in fact, pseudogenes, dogs only possess five functional V1R proteins.
The most recent chimpanzee genome assembly does not encode any V1R ORFs. Notably, two of the pseudogenes (one at the 60.395-Mb position on Chromosome 20 and the other at the 32.613-Mb position on Chromosome 18) are represented in GenBank as intact ORFs. The first has four GenBank entries, including two sequences with ORFs (AY114011.1 and AY114732.1) and two without ORFs (AY312463.1 and AY426106.1). The second encodes a stop codon at the position where the GenBank entry (AY312468) encodes an arginine. Therefore, chimpanzee (like human) could encode two intact V1Rs.
The human genome assembly encodes two V1R ORFs, although five entries for human V1Rs with ORFs are present in GenBank (V1RL1, V1RL2, V1RL3, V1RL4, and V1RL5). The V1RL1 and V1RL4 genes are intact in the assembly. However, the 5′-most 278 bp of the V1RL2 sequence are similar to an LTR-like repeat, the V1RL3 sequence in the assembly has a frameshift mutation 285 bp after the start codon, and the V1RL5 sequence in the assembly has a stop codon 150 bp after the start codon.
One of the intact mouse V1Rs (at the 90.700-Mb position on Chromosome 6) does not encode a start codon. The ORF begins with a His residue predicted to be the eighth amino acid in the nearest homolog. This sequence could make an intact V1R if it gains a start codon from an upstream exon, as seen for some olfactory receptor genes (Linardopoulou et al. 2001). Alternatively, this V1R might be a pseudogene, which would reduce our total for intact mouse V1Rs to 164.
The one mouse and three dog intact V1Rs that could be pseudogenes are marked with asterisks in Figure 1, and the three human and two chimp V1R pseudogenes that could be intact are marked with asterisks in Figure 2.
Construction of neighbor-joining trees
First, amino acid translations of all intact V1R genes were aligned under a gap-minimizing protocol. Second, predicted amino acid translations for each pseudogene were manually added to the alignment by finding the best alignment to the most similar intact representatives. Third, amino acids were back-translated to provide nucleotide alignments, and all frameshifting insertions in pseudogenes were discarded to maintain the alignment in-frame. Fourth, pseudogene sequences were trimmed so that they could be confidently aligned. Highly divergent N- and C-terminal regions were also trimmed. The resulting alignments were 675 bp long (Supplemental Fig. C). Fifth, partial sequences in which >40% of the aligned sequence was missing (because of gaps in assemblies, repeat insertions, partial gene deletions, or low confidence in alignment) were excluded, in order to ensure that all pairwise comparisons had sufficient alignment overlap to reduce spurious neighbor-joining. A complete list of pseudogenes excluded by this criterion, and their subfamily assignments, is provided in Supplemental Figure D. Aligned nucleotide sequences were imported into the PAUP phylogenetic program (Sinauer Associates), and neighbor-joining trees were constructed using Jukes-Cantor-adjusted distances. Amino acid trees give similar subfamily structure (data not shown). Preliminary trees were examined for genes that clustered in subfamily clades differing from their presumed best match (used for an initial alignment in Step 2). These genes were re-examined to ensure that their alignments were not incorrectly biased by this preliminary assignment. The trees shown in Figures 1 and 2 were colored using a custom script (E. Williams, unpubl.). Synonymous substitution levels for all V1R pairs were estimated using the SNAP program (http://www.hiv.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html); this algorithm uses the Jukes-Cantor adjustment to correct for multiple substitutions.
Subfamily assignments
Subfamily assignments for mouse V1R genes were previously described by Rodriguez et al. (2002), where inter-subfamily amino acid identities were <40%. We maintained the A/B and H/I subfamily delineations of Rodriguez et al. (2002), even though (as they point out) these groups have some inter-subfamily identities that exceed 40%. As described by Rodriguez et al. (2002), the reason for splitting the A/B and H/I subfamilies is that they form distinct clades and because their fusions would have generated subfamilies with intra-subfamily identities <40%. We assigned all new V1R genes to a subfamily based on their monophyletic grouping in the tree with previously named mouse genes. Designation of the L subfamily is exceptional, because we group L-like genes that violate the 40% amino acid cutoff threshold. Previously, only a single intact L-subfamily member was identified in mouse. For simplicity, we grouped seven L-like rat homologs into a single subfamily even though some of the V1R pairs in this group are >40% diverged. We then assigned nonrodent V1Rs to this “super subfamily” if they fell within the same clade established by the rodent L-like homologs. As a result, the L subfamily described here would consist of more than one subfamily using the original Rodriguez et al. (2002) definition. Gene branches that connected below the root of mouse subfamily clades were given descriptive subfamily names that indicate this topology (e.g., “AB” or “JK” for the numerous primate and dog V1R branches that connect below the “AB” and “JK” subfamily clades, respectively). Some partial V1R-like sequences were excluded from the tree (see Supplemental Fig. D); preliminary subfamily assignments for these excluded V1Rs were based on each sequence's best fast×34 match.
Cluster identification and synteny analysis
Clusters of V1Rs were defined as groups of at least three genes spaced <500 kb from each other. We used the UCSC Genome Browser (http://genome.ucsc.edu) to identify non-V1R genes flanking these clusters. All flanking genes selected for Table 2 satisfy one of the following three conditions to avoid ambiguity in orthology assignments: (1) the top BLAT score was >1000, and the second highest BLAT score was <500; or (2) the top BLAT score was >500, and the second highest BLAT score was <100; or (3) the top BLAT hit mapped to the same chromosomal location as top BLAT hits of adjacent genes from the reference genome.
Identification of the canine trp2β gene
The exons of the mouse trp2 gene (NM_011644) were used in a BLAT search of the dog genome assembly (http://genome.ucsc.edu). A single match is found on dog Chromosome 21. We filled a 464-bp gap in the dog assembly by sequencing a PCR product generated from dog genomic DNA (Clontech) (forward primer: AAAAGTAGGTGGGAACATGAGAAG; reverse primer: ACCCAGGACTAGCAAATGAATTAC), using big-dye terminators and standard procedures.
Acknowledgments
This work was supported by National Institutes of Health Grants R01-DC006267 to R.P.L. and R01-DC004209 to B.J.T. We thank Trygve Bakken for generating missing dog Trp2 sequence, Eleanor Williams for assistance in coloring trees, Michael Get-man for thoughtful comments on the manuscript, and Linda Buck and Nate Sutter for helpful discussions. We are grateful to the numerous sequencing centers worldwide for generating and providing access to the mouse, rat, human, chimpanzee, and dog genome assemblies.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3339905. Article published online ahead of print in January 2005.
References
- Boschat, C., Pelofi, C., Randin, O., Roppolo, D., Luscher, C., Broillet, M.-C., and Rodriguez, I. 2002. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 5: 1261-1262. [DOI] [PubMed] [Google Scholar]
- Buck, L. and Axel, R. 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175-187. [DOI] [PubMed] [Google Scholar]
- Cohen-Tannoudji, J., Lavenet, C., Locatelli, A., Tillet, Y., and Signoret, J.P. 1989. Non-involvement of the accessory olfactory system in the LH response of anoestrous ewes to male odour. J. Reprod. Fertil. 86: 135-144. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- Del Punta, K., Leinders-Zufall, T., Rodriguez, I., Jukam, D., Wysocki, C.J., Ogawa, S., Zufall, F., and Mombaerts, P. 2002. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature 419: 70-74. [DOI] [PubMed] [Google Scholar]
- Dennis, J.C., Allgier, J.G., Desouza, L.S., Eward, W.C., and Morrison, E.E. 2003. Immunohistochemistry of the canine vomeronasal organ. J. Anat. 203: 329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorries, K.M., Adkins-Regan, E., and Halpern, B.P. 1997. Sensitivity and behavioral responses to the pheromone androstenone are not mediated by the vomeronasal organ in domestic pigs. Brain Behav. Evol. 49: 53-62. [DOI] [PubMed] [Google Scholar]
- Dulac, C. and Axel, R. 1995. A novel family of genes encoding putative pheromone receptors in mammals. Cell 83: 195-206. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fewell, G.D. and Meredith, M. 1998. Olfactory contribution to fos expression during mating in inexperienced male hamsters. Chem. Sens. 23: 257-267. [DOI] [PubMed] [Google Scholar]
- Fewell, G.D. and Meredith, M. 2002. Experience facilitates vomeronasal and olfactory influence on Fos expression in medial preopticarea during pheromone exposure or mating in male hamsters. Brain Res. 941: 91-106. [DOI] [PubMed] [Google Scholar]
- Giorgi, D. and Rouquier, S. 2002. Identification of V1R-like putative pheromone receptor sequences in non-human primates. Characterization of V1R pseudogenes in marmoset, a primate species that possesses an intact vomeronasal organ. Chem. Senses 27: 529-537. [DOI] [PubMed] [Google Scholar]
- Giorgi, D., Friedman, C., Trask, B.J., and Rouquier, S. 2000. Characterization of nonfunctional V1R-like pheromone receptor sequences in human. Genome Res. 10: 1979-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman, G., Yanai, I., Rubin, I., and Lancet, D. 2001. The complete human olfactory subgenome. Genome Res. 11: 685-702. [DOI] [PubMed] [Google Scholar]
- Grus, W.E. and Zhang, J. 2004. Rapid turnover and species-specificity of vomeronasal pheromone receptor genes in mice and rats. Gene 340: 303-312. [DOI] [PubMed] [Google Scholar]
- Halpern, M. 1987. The organization and function of the vomeronasal system. Annu. Rev. Neurosci. 10: 325-362. [DOI] [PubMed] [Google Scholar]
- Halpern, M. and Martinez-Marcos, A. 2003. Structure and function of the vomeronasal system: An update. Prog. Neurobiol. 70: 245-318. [DOI] [PubMed] [Google Scholar]
- Herrada, G. and Dulac, C. 1997. A novel family of putative pheromone receptors in mammals with a topologically organized and sexually dimorphic distribution. Cell 90: 763-773. [DOI] [PubMed] [Google Scholar]
- Hofmann, T., Schaeffer, M., Schultz, G., and Gudermann, T. 2000. Cloning, expression and subcellular localization of two novel splice variants of mouse transient receptor potential channel 2. Biochem. J. 351: 115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. and Distel, H. 1986. Pheromonal release of suckling in rabbits does not depend on the vomeronasal organ. Physiol. Behav. 37: 123-128. [DOI] [PubMed] [Google Scholar]
- Kaneko, N., Debski, E.A., Wilson, M.C., and Whitten, W.K. 1980. Puberty acceleration in mice: II. Evidence that the vomeronasal organ is a receptor for the primer pheromone in male mouse urine. Biol. Reprod. 22: 873-878. [DOI] [PubMed] [Google Scholar]
- Keverne, E.B. 2002. Mammalian pheromones: From genes to behaviour. Curr. Biol. 12: R807-R809. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr, H., Pintchovski, S., Melnyk, J., Chen, Y.J., Friedman, C., Trask, B.J., and Shizuya, H. 2001. Identification of non-functional human VNO receptor genes provides evidence for vestigiality of the human VNO. Chem. Senses 26: 1167-1174. [DOI] [PubMed] [Google Scholar]
- Lane, R.P., Cutforth, T., Axel, R., Hood, L., and Trask, B.J. 2002. Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. Proc. Natl. Acad. Sci. 99: 291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, R.P., Young, J., Newman, T., and Trask, B.J. 2004. Species specificity in rodent pheromone receptor repertoires. Genome Res. 14: 603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall, T., Lane, A.P., Puche, A.C., Ma, W., Novotny, M.V., Shipley, M.T., and Zufall, F. 2000. Ultrasensitive pheromone detection by mammalian vomeronasal neurons. Nature 405: 792-796. [DOI] [PubMed] [Google Scholar]
- Leypold, B.G., Yu, C.R., Leinders-Zufall, T., Kim, M.M., Zufall, F., and Axel, R. 2002. Altered sexual and social behaviors in trp2 mutant mice. Proc. Natl. Acad. Sci. 99: 6376-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman, E.R., Corey, D.P., and Dulac, C. 1999. TRP2: A candidate transduction channel for mammalian pheromone sensory signaling. Proc. Natl. Acad. Sci. 96: 5791-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou, E., Mefford, H.C., Nguyen, O., Friedman, C., van den Engh, G., Farwell, D.G., Coltrera, M., and Trask, B.J. 2001. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum. Mol. Genet. 10: 2373-2383. [DOI] [PubMed] [Google Scholar]
- Lloyd-Thomas, A. and Keverne, E.B. 1982. Role of the brain and accessory olfactory system in the block to pregnancy in mice. Neuroscience 7: 907-913. [DOI] [PubMed] [Google Scholar]
- Matsunami, H. and Buck, L.B. 1997. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell 90: 775-784. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Pantages, E. and Dulac, C. 2000. A novel family of candidate pheromone receptors in mammals. Neuron 28: 835-845. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R., Wood, T., Zhang, Z., and Miller, W. 1997. Comparison of DNA sequences with protein sequences. Genomics 46: 24-36. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 475-476. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I. and Mombaerts, P. 2002. Novel human vomeronasal receptor-like genes reveal species-specific families. Curr. Biol. 12: R409-R411. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Feinstein, P., and Mombaerts, P. 1999. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell 97: 199-208. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Greer, C.A., Mok, M.Y., and Mombaerts, P. 2000. A putative pheromone receptor gene expressed in human olfactory mucosa. Nat. Genet. 26: 18-19. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Del Punta, K., Rothman, A., Ishii, T., and Mombaerts, P. 2002. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat. Neurosci. 5: 134-140. [DOI] [PubMed] [Google Scholar]
- Ryba, N.J. and Tirindelli, R. 1997. A new multigene family of putative pheromone receptors. Neuron 19: 371-379. [DOI] [PubMed] [Google Scholar]
- Sam, M., Vora, S., Malnic, B., Ma, W., Novotny, M.V., and Buck, L.B. 2001. Neuropharmacology: Odorants may arouse instinctive behaviours. Nature 412: 142. [DOI] [PubMed] [Google Scholar]
- She, X., Horvath, J.E., Jiang, Z., Liu, G., Furey, T.S., Christ, L., Clark, R., Graves, T., Gulden, C.L., Alkan, C., et al. 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430: 857-864. [DOI] [PubMed] [Google Scholar]
- Stowers, L., Holy, T.E., Meister, M., Dulac, C., and Koentges, G. 2002. Loss of sex discrimination and male-male aggression in mice deficient for trp2. Science 295: 1493-1500. [DOI] [PubMed] [Google Scholar]
- Trotier, D., Eloit, C., Wassef, M., Telmain, G., Bensimon, J.L., Doving, K.B., and Ferrand, J. 2000. The vomeronasal cavity in adult humans. Chem. Senses 25: 369-380. [DOI] [PubMed] [Google Scholar]
- Weiler, E., Apfelbach, R., and Farbman, A.I. 1999. The vomeronasal organ of the male ferret. Chem. Senses 24: 127-136. [DOI] [PubMed] [Google Scholar]
- Young, J.M., Friedman, C., Williams, E.M., Ross, J.A., Tonnes-Priddy, L., and Trask, B.J. 2002. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11: 535-546. [DOI] [PubMed] [Google Scholar]
- Zhang, J. and Webb, D.M. 2003. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc. Natl. Acad. Sci. 100: 8337-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Rodriguez, I., and Mombaerts, P. 2004. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics 83: 802-811. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker Web server.
- http://genome.ucsc.edu/; UCSC Genome Bioinformatics.
- http://hiv-web.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html; HIV Sequence Database: SNAP submission form.
- http://repeatmasker.org; RepeatMasker.
- http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html; BCM search launcher.
- http://www.acedb.org; acedb database.
- http://www.ncbi.nlm.nih.gov/BLAST/; NCBI BLAST.


