Abstract
Several RNA interference (RNAi)-based functional genomic projects have been performed in Caenorhabditis elegans to identify genes required during embryogenesis. These studies have demonstrated that the ovary is enriched for transcripts essential for the first cell divisions. However, comparing RNAi results suggests that many genes involved in embryogenesis have yet to be identified, especially those eliciting partially penetrant phenotypes. To discover additional genes required for C. elegans embryonic development, we tested by RNAi 1123 ORFeome clones selected to represent ovary-enriched genes not associated with an embryonic phenotype. We discovered 155 new ovary-enriched genes with roles during embryogenesis, of which 69% show partial penetrance lethality. Time-lapse microscopy revealed specific phenotypes during early embryogenesis for genes giving rise to high penetrance lethality. Together with previous studies, we now have evidence that 1843 C. elegans genes have roles in embryogenesis, and that many more remain to be found. Using all available RNAi phenotypic data for the ovary-enriched genes, we re-examined the distribution of genes by chromosomal location, functional class, ovary enrichment, and conservation and found that trends are driven almost exclusively by genes eliciting high-penetrance phenotypes. Furthermore, we discovered a striking direct relationship between phylogenetic distribution and the penetrance level of embryonic lethality elicited by RNAi.
A continuing challenge in contemporary biology is to extract meaningful functional data from the genome sequence. High-throughput functional genomics projects are currently ongoing in several model systems to characterize gene function on multiple levels, including gene expression patterns (Spellman et al. 1998; Reinke et al. 2000, 2004; Jiang et al. 2001; Arbeitman et al. 2002; Baugh et al. 2003), protein-protein interactions (Uetz et al. 2000; Walhout et al. 2000; Ito et al. 2001; Giot et al. 2003; Li et al. 2004), and protein structures (Chance et al. 2002; Goh et al. 2003). Phenotypic data complement these studies and are necessary to make the connection between predicted biochemical function and biological role in living cells. The development of RNA interference (RNAi) (Guo and Kemphues 1995; Fire et al. 1998) has made possible large-scale in vivo gene analyses in a functional genomic context in Caenorhabditis elegans (Gönczy et al. 2000; Piano et al. 2000, 2002; Hanazawa et al. 2001; Maeda et al. 2001; Kamath et al. 2003; Simmer et al. 2003; Rual et al. 2004) and in cell culture systems from Drosophila melanogaster and mammals (Somma et al. 2002; Kiger et al. 2003; Berns et al. 2004; Boutros et al. 2004; Paddison et al. 2004). RNAi studies enable loss-of-function analyses to be performed on genes that had not been uncovered previously by mutagenesis, thus potentially allowing phenotypic characterization of every gene in the genome. Data from different types of functional genomics projects can be combined to make inferences about gene function and protein networks on a large scale (Marcotte et al. 1999; Ge et al. 2001; Boulton et al. 2002; von Mering et al. 2002; Walhout et al. 2002).
C. elegans embryogenesis is a powerful in vivo model to study gene function in the context of animal development. Its germline is highly sensitive to RNAi, facilitating the detection of genes with essential functions in the embryo. The set of genes known to be required during embryogenesis is enriched for components of pathways that drive cell proliferation and other fundamental processes, and these genes tend to be highly conserved across species (Gönczy et al. 2000; Piano et al. 2000, 2002; Kamath et al. 2003). Thus defining a comprehensive set of genes needed for C. elegans embryogenesis is an important step toward gaining general insights into mechanisms underlying animal development. Having in hand a complete list of genes required for fertility and embryogenesis is important both to systems biologists who study large networks and to classical biologists who study in detail the genes that control a single process.
Despite the unprecedented scale of in vivo functional data obtained using RNAi, there are technical challenges to RNAi-based phenotypic analysis. In C. elegans, variables that can affect RNAi include the double-stranded (ds)RNA template source, the delivery method, and scoring criteria (for review, see Piano and Gunsalus 2002). Different studies have varied in their use of dsRNAs derived from genome amplicons that include intronic sequences (Gönczy et al. 2000; Piano et al. 2002; Kamath et al. 2003; Simmer et al. 2003) or from cDNA clones composed exclusively of exonic sequences (Piano et al. 2000; Hanazawa et al. 2001; Maeda et al. 2001; Rual et al. 2004). Delivery of dsRNA has been accomplished by injection (Guo and Kemphues 1995; Fire et al. 1998), by feeding animals on bacterial strains expressing dsRNA (Timmons and Fire 1998), and by soaking them in a solution containing dsRNA (Tabara et al. 1998). Despite these various approaches and the inherent complexities of phenotypic analyses, independent large-scale RNAi studies have shown a relatively high reproducibility rate (Piano and Gunsalus 2002; Simmer et al. 2003). However, just as a genome must be sequenced at several-fold depth to achieve sufficient coverage and accuracy, comparisons between different RNAi studies also suggest that multiple passes of genome-wide RNAi will be necessary to obtain a comprehensive view of the in vivo functions of each gene in C. elegans.
Previous RNAi studies in C. elegans have shown that the set of genes expressed at higher levels in the ovary, relative to the soma or male germline, is enriched for genes required for embryogenesis (Piano et al. 2002). Using a partial C. elegans microarray, Reinke et al. (2000) identified 766 genes enriched in the ovary, and RNAi analysis revealed that at least 43% of these genes are required for embryogenesis (Piano et al. 2002). Recently, using a C. elegans genome-wide microarray containing about 90% of predicted genes, a total of 3015 ovary-enriched genes were identified (Reinke et al. 2004). However, only ∼28% of genes tested from this set have been found to be required for embryogenesis in any published RNAi study to date, suggesting that hundreds more remain to be identified (Reinke et al. 2004).
To identify new genes required for embryogenesis, we analyzed 1011 ovary-enriched genes represented in the ORFeome. Over 85% of these had been assayed by RNAi in prior studies and not shown embryonic lethality. This group of genes may thus be enriched for genes whose embryonic requirement, if any, is more difficult to detect by RNAi. We used a combination of repeated RNAi by soaking and injection, and here report the identification of 192 genes involved in egg production or embryogenesis, 155 of which have no previously reported requirement in these stages.
A majority of the genes in the ovary-enriched set have now been studied by RNAi by at least three independent studies, making it the most deeply studied set of genes available in C. elegans. Using the combined data from all previous studies of this set, we now include the dimension of penetrance in our analyses of phenotypic trends among the ovary-enriched genes. We observe that trends with regard to sequence similarity, expression level, and chromosomal location are predominantly due to genes with highly penetrant embryonic lethality or sterility. We also observe a relationship between the pattern of conservation across phyla and the penetrance level of embryonic lethality elicited by RNAi.
Results and Discussion
Evaluation of RNAi results
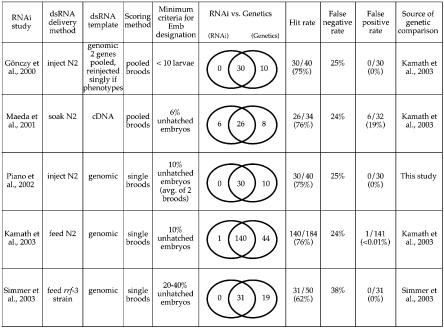
To explore the large-scale RNAi data, we evaluated results from five RNAi studies in C. elegans (Gönczy et al. 2000; Piano et al. 2002; Maeda et al. 2001; Kamath et al. 2003; Simmer et al. 2003) with respect to their ability to detect sterile or embryonic lethal phenotypes. Comparisons of RNAi-induced phenotypes with phenotypes arising from genetic mutation suggest that large-scale RNAi studies performed in the wild-type genetic background have a detection rate of ∼75%, or a false-negative rate of ∼25%, for maternal sterility or embryonic lethality (Fig. 1). False positive rates, estimated from the number of genes with RNAi-induced phenotypes that are not reported by genetics, were negligible in RNAi studies that used feeding (Kamath et al. 2003; Simmer et al. 2003) or injection (Gönczy et al. 2000; Piano et al. 2002) as a delivery method. In contrast, of the 32 embryonic lethal or sterile genes represented by mutant alleles from the large-scale soaking study (Maeda et al. 2001), six were not reported to be embryonic lethal or sterile by genetics (Kamath et al. 2003). One interpretation of this observation is that RNAi by soaking gives rise to more false positives than does feeding or injection. Another possibility is that the soaking protocol causes physiological changes that influence the effects of reducing gene function on fecundity or embryonic development. However, our own data (see below) suggest a third possibility: that comparisons to genetic analyses include cases where the genetic results were not performed using null alleles or did not report on embryonic lethality simply because the focus of the study was not the germline or embryo. This is more likely to be the case when the genetic null allele shows only partially penetrant sterility (reduced brood size) or embryonic lethality (few unhatched embryos).
Figure 1.
Summary of large-scale RNAi studies. First five columns: For each published RNAi study, the dsRNA delivery method, the template used for generation of dsRNA, the scoring method, and the minimum criteria for scoring are indicated (for reviews of large-scale RNAi projects, see Piano and Gunsalus 2002 and Sugimoto 2004). The scoring method refers to whether individual broods or composites of two or more broods were scored to obtain the percentage of unhatched embryos. The scoring criteria indicate the minimum level of lethality that must be observed in order to score a gene as embryonic lethal. For the Gönczy study, three injected hermaphrodites were placed on a single plate and allowed to lay eggs for 48 h; plates with fewer than 10 hatched larvae were scored as embryonic lethal. Because over 100 eggs are commonly produced by three animals during this time period, in this study the cutoff for scoring lethality was ∼90% unhatched embryos. The Simmer study used the rrf-3 strain, which shows some embryonic lethality and sterility in the absence of RNAi; thus their cutoff for scoring lethality was more stringent than most of the other studies. Last five columns: Comparison of RNAi phenotypes with phenotypes from genetic mutations for genes from each RNAi study that are represented by characterized mutant alleles. Venn diagrams indicate genes identified as Ste or Emb by RNAi only (left; potential false positives), by both RNAi and genetics (overlap), or by genetic analysis only (right; potential false negatives). We define the “hit rate” as the number of Ste or Emb genes identified by RNAi relative to the number of genes with known sterile or lethal genetic alleles. We define the “false positive rate” as the number of genes with Ste or Emb phenotypes by RNAi that do not give rise to sterility or lethality according to genetic analysis, divided by the total number of RNAi-lethal genes represented by genetic alleles.
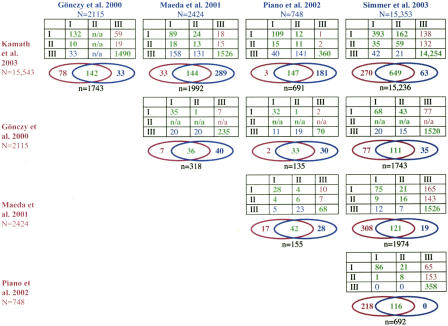
An alternative way to examine the efficacy of various RNAi approaches is to compare results from different large-scale RNAi studies directly with each other (Fig. 2). Among genes that give rise to embryonic lethality, studies agreed more often when the penetrance level was high (Class I, 80%-100% embryonic lethality or maternal sterile) than when the penetrance was partial (Class II, 6%-79% embryonic lethality), as judged by the relative number of genes placed into the same versus different penetrance bins. Studies thus appeared more closely matched in their ability to detect genes giving rise to highly penetrant phenotypes and exhibited more variability in reporting partially penetrant lethality, which can be partly explained by scoring protocols (for review, see Sugimoto 2004; Piano and Gunsalus 2002). Focusing on the three studies with the most similar scoring methods and conducted in the same genetic background (Piano et al. 2002, “injection”; Maeda et al. 2001, “soaking”; and Kamath et al. 2003, “feeding”), we observe trends not revealed by comparisons with genetics. In direct comparisons between RNAi studies, injection and soaking show similar levels of detection among genes tested by both methods; however when compared to feeding, soaking and injection show more than twice the number of genes required for embryogenesis or oogenesis (Fig. 2). The magnitude of the differences cannot be fully explained by false positives in both the soaking and injection studies, since in comparisons with genetics the injection study had no detectable false positives and soaking had at most 19%. These differences are most parsimoniously explained by a higher level of false negative results in the feeding study that are not revealed by the comparisons with genetics because genes with partially penetrant phenotypes are likely underrepresented among the characterized genetic loci.
Figure 2.
Comparison of results of large-scale RNAi studies. Each table shows the comparison of results between two RNAi studies, subdivided by gross penetrance levels. I, Class I penetrance (sterile and ≥80% embryonic lethal); II, Class II penetrance (6%-79% embryonic lethal); III, Class III (no sterility or embryonic lethality detected). Numbers in green indicate similar results by both studies; purple and blue numbers represent genes with sterility or lethality according to one study but not the other (blue: detected only by the study listed at the head of the panel column; purple: detected only by the study at the head of the panel row). For each study the total number of unique genes (excluding alternatively spliced forms) assayed (N) is indicated. Venn diagrams summarize the data in adjacent tables and show the total numbers of genes with sterile or embryonic lethal phenotypes detected by both (green) or only one study (blue, purple). The total number of overlapping genes analyzed by both studies (n) is indicated below each Venn diagram. Results from Simmer et al. (2003) that were reported as not reproducible were omitted from this analysis; all other wild-type results from this study were inferred (see Methods). No Class II results are indicated for Gönczy et al. (2000) (“n/a”, not applicable) because that study effectively scored only high-penetrance lethality. Figure 1 provides further information about how each of these studies was conducted.
Since the comparisons suggest a difference in the strength of the RNAi effect elicited by the different methods, we might expect any such differences to be reflected in the penetrance levels of phenotypes reported. Indeed, when the same gene is analyzed by different studies, we observe a consistent trend in which soaking and injection elicit equivalent or stronger phenotypes than feeding, as judged by the relative penetrance of embryonic lethal phenotypes. For example, 67% of the 46 genes reported as Class II by feeding are classified in either the same or higher penetrance category based on soaking, whereas 22% of the 168 Class II genes reported by soaking are in an equal or higher class based on feeding (Fig. 2). Similarly, 93% of the 28 genes in Class II by feeding are in the same or higher penetrance category in the injection results, whereas 14% of the 164 Class II genes by injection are in an equivalent or higher penetrance category by feeding.
A stronger RNAi effect might also be expected to elicit an earlier defect relative to the time of dsRNA delivery. In all studies analyzed, the RNAi treatment was initiated at the young adult stage, so the earliest observable defect in all studies was sterility of the treated animal, followed by embryonic defects among the progeny, and finally postembryonic phenotypes in the progeny. We therefore asked whether any genes displayed sterility or embryonic lethality in one study but only postembryonic defects that were not also accompanied by earlier defects in another study (Supplemental Fig. S1). Comparing genes studied by both feeding and soaking, the total number of genes showing only post-embryonic defects is similar: 128 by feeding and 125 by soaking. However, of the 128 genes that showed only post-embryonic phenotypes in the feeding study, the soaking study found 38 (30%) to be embryonic lethal or sterile. In contrast, of the 125 genes found to have post-embryonic defects alone from the soaking study, only 10 (8%) were found to show embryonic lethality or sterility when tested by feeding. The trend is even more pronounced when comparing feeding and injection (Supplemental Fig. S1). In addition to revealing technical aspects of RNAi analyses, these comparisons indicate that pleiotropy, or the reuse of the same cellular components in different developmental contexts, is a prevalent theme in the biology of this organism.
Since the largest studies in C. elegans have been carried out using feeding, the combined data indicate that many genes required for embryogenesis remain to be found, and using RNAi by either soaking or injection could enhance the ability to find them. In addition, the data suggest that comparing phenotypes arising from genetic mutation with those elicited by RNAi does not realistically assess RNAi hit rates among genes with partially penetrant embryonic lethality. We conclude from this analysis that all RNAi delivery methods can detect high-penetrance embryonic lethal or maternal sterile genes with similar efficiency, and that RNAi by soaking or injection appear to be more efficacious in discovering genes that give rise to partial-penetrance embryonic lethality.
Selection of the target gene set and RNAi results
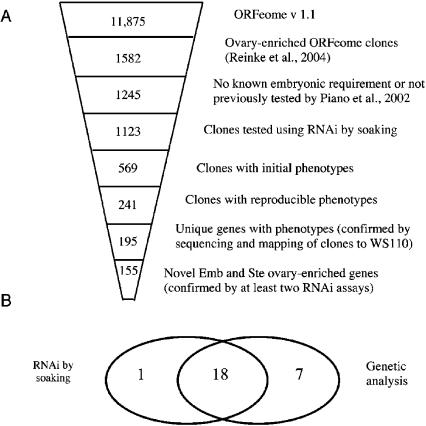
The above analysis suggests that additional genes required for embryogenesis in C. elegans remain to be found by RNAi. To test this prediction we focused on ovary-enriched genes (Reinke et al. 2004) found in the ORFeome collection (Reboul et al. 2003). Since our goal was to find new genes required for embryogenesis, we specifically excluded clones corresponding to known embryonic lethal genes at the time we initiated this study and those we had previously tested by injection (Fig. 3). Thirty-one of the clones we originally selected for analysis correspond to genes that are currently known to be required for egg production or embryonic viability from other, independent RNAi studies. The remaining 1092 clones fall into three classes, based on data available from WormBase (WS110) and the high-confidence (reproducible) data from a study using a mutant strain (Simmer et al. 2003): the majority (834) represent genes that displayed no phenotype in previous RNAi assays, 172 correspond to genes previously reported to show only post-embryonic phenotypes by RNAi, and 86 represent genes that have no reported RNAi assay.
Figure 3.
Pipeline for selection of ORFeome clones and summary of results from RNAi by soaking. (A) ORFeome v. 1.1 contains 11,875 clones. Of these, 1582 represent ovary-enriched genes. We chose the 1245 clones from this set representing genes for which no embryonic requirement had been established by RNAi by any study and which we had not tested previously. From these we tested 1123 clones and found 241 that showed a high-confidence Emb or Ste phenotype. After removing clones for which sequence could not be verified and mapping the clones to gene models from WormBase (WS110), we identified 195 unique genes (excluding alternative splice forms) with high-confidence Emb or Ste phenotypes, 155 of which are new and ovary-enriched. (B) We compared phenotypes for the 26 genes we tested that were also represented by characterized mutant alleles. For 18 of these, both RNAi and genetic analysis revealed an Emb or Ste phenotype. Seven were identified only by genetics, and one was identified only by RNAi.
We optimized the RNAi by soaking protocol and validated that we could obtain results comparable to those from singlegene injections using a control gene set of 83 genes (data not shown). In addition we tested our RNAi protocol using GFP-specific dsRNA and found negligible levels of embryonic lethality (1.2%, n = 1572). In a first round of RNAi assays applied to the test set, we found a total of 569 clones that gave rise to sterility of the RNAi-treated animal (Ste) and/or embryonic lethality (Emb); 241 of these were retained as Emb or Ste upon retesting. Sequencing verified the expected identity of 201 of the 241 clones; the remaining 40 were discarded from further analysis. By mapping these clones to genes we identified 195 independent genes that gave rise to high-confidence Emb and/or Ste phenotypes, of which 158 are identified here for the first time (Fig. 3, Supplemental Table S1). Three of these were removed from the final analyses because they currently map to genes not in the ovary-enriched list (Supplemental Table S1).
We estimated the likelihood of false positive results by comparing our phenotypes to those derived from genetic analyses (Fig. 3B, Supplemental Table S2). Nineteen genes that we found to be maternal sterile or embryonic lethal (cdk-5, mex-5, mdt-6, air-2, mom-5, hrp-1, aph-1, egl-18, dpy-30, ife-3, mom-4, aph-2, sqv-7, sqv-5, ooc-3, spn-4, lrs-2, csc-1, and lin-3) were represented by characterized mutant alleles. For this analysis we excluded genes represented by mutant alleles that were not well characterized (listed in Supplemental Table S2). Null or severe hypomorphic alleles of all but one of these genes also give rise to embryonic lethality or maternal sterility. Our single putative false positive is cdk-5. We conclude that our false positive rate is low (1/19) and that the majority of the genes we uncovered here have a role in embryogenesis.
To gauge our false-negative rate we also performed comparisons with published reports from genetic analyses. Of the 25 total genes we studied that also give rise to embryonic lethal or maternal sterile phenotypes upon genetic mutation, we identified 18 (72%) and failed to report seven (hus-1, mom-1, inx-3, cup-5, puf-8, sqv-1, and lin-41). Notably, four of the genes we missed showed lethality in one round of assays only, and were thus classified as low-confidence lethals (Supplemental Table S2). Thus, although our selection criteria might be expected to preferentially include genes that are harder to identify using RNAi, our false-negative rate based on genetic comparisons was comparable to those of previous studies (Fig. 1).
Independent RNAi studies found that 25 of the genes we tested elicit embryonic lethal or maternal sterile phenotypes upon RNAi. We detected 17 of these, of which 14 were detected reproducibly. In order to minimize false positives, we chose to be conservative in our assignment of phenotypes; thus we did not report embryonic lethality in our final analysis for any of the low-confidence lethal results. Our comparisons suggest that this practice elevated our false-negative rate and also indicate that we have not yet identified all the genes required for embryogenesis in our sample set.
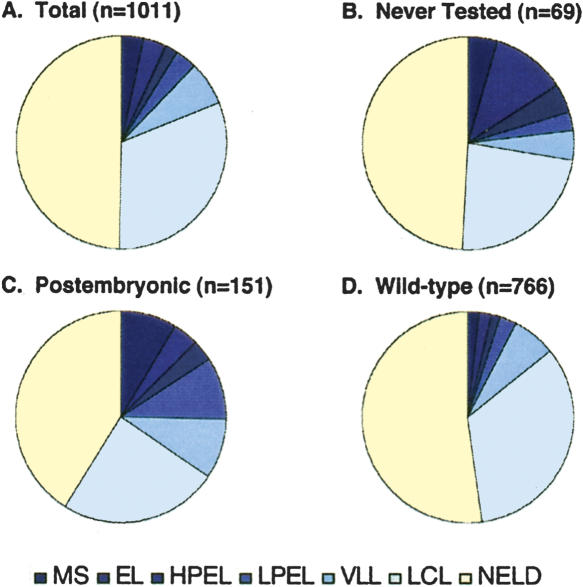
Based on our comparisons of published RNAi studies and the gene selection strategy we used in this study, we would expect to recover a nonrandom distribution of Emb or Ste genes among the different subsets of genes we assayed by RNAi. The distribution of phenotypes among the different subsets is consistent with this expectation (Fig. 4). First, a significantly higher overall rate of Emb and Ste phenotypes was detected in the gene sets that either had never been tested by RNAi (19/69 or 28%; Fig. 4B) or had been tested and found to give rise to post-embryonic phenotypes only (52/151 or 34%; Fig. 4C), versus the set of genes that previously failed to give rise to any detectable phenotype (107/766 or 14%; Fig. 4D). Furthermore, 58% (11/19) of the Emb and Ste genes found in the set that had never been tested were in the highest penetrance classes (MS and EL), whereas only 30% (47/159) of Emb or Ste genes from among those that had been tested before but had not previously shown embryonic lethality fell into these classes. These results are consistent with the idea that many of the highest-penetrance embryonic lethal genes had already been removed from the set we analyzed. In addition, the data show that genes with known post-embryonic RNAi phenotypes are much more likely to display embryonic lethality upon retesting than genes with no reported RNAi phenotypes from prior assays.
Figure 4.
Results of RNAi by soaking. (A) All ovary-enriched genes tested in this study. (B) Genes not previously tested by RNAi. (C) Genes that displayed only post-embryonic defects in previous RNAi studies. (D) Genes previously tested by RNAi for which no phenotype has been reported. MS, maternal sterile, three broods with ≤10 progeny; EL, embryonic lethal, 80%-100% embryonic lethality; HPEL, high-penetrance embryonic lethality, 50%-80%; LPEL, low-penetrance embryonic lethality, 20%-50%; VLL, very low lethality, 5%-20%; LCL, low-confidence lethality; NELD, no embryonic lethality detected.
The 158 new genes we have identified with roles in embryogenesis or egg production augment by ∼9% our knowledge of the genes required for these stages of development in C. elegans. The early embryo is particularly well suited to study cell biological defects generated by the RNAi treatments and can help reveal specific roles in cell division. To further characterize the effects on embryogenesis, we repeated RNAi by injection or soaking for the 39 genes whose RNAi analysis elicited the highest-penetrance embryonic lethality. We studied the early embryonic defects by time-lapse digital Nomarski microscopy from the one-cell stage to the eight-cell stage (we obtained, on average, eight time-lapse recordings per gene, available as Quicktime movies though www.RNAi.org). Among these are five genes (csc-1, mom-5, mex-5, air-2, spn-4) whose role in early embryogenesis was already known, and we recovered the known phenotypes in our assays. For the 34 genes that were newly identified here or were not previously studied in the early embryo, we analyzed detailed phenotypes during the first 50 min of development and scored them as previously described (Piano et al. 2002; Supplemental Table S4). The range of defects observed is similar to that of previous reports (Gönczy et al. 2000, Piano et al. 2000, 2002; Zipperlen et al. 2001), and results in the identification of new genes required for specific early embryonic events such as completion of meiosis, nuclear movements, and mitosis.
Among the genes identified as being required for embryogenesis, we identified four genes (C47D12.8, F14B4.2, F54F2.8, and F35F11.1) whose human homologs have roles in disease (ERCC4, HK2, PXF, and HRPT2, respectively). We recorded early embryonic phenotypes for C47D12.8 and F35F11.1 (the others showed partial-penetrance lethality). Both of these genes are homologs of human genes whose mutations are associated with tumors. Interestingly, RNAi of F35F11.1 (the HPRT2 homolog) causes defects in the proper formation of polar bodies, pronuclei, and mitotic nuclei, indicating that this gene is important for the proper completion of both meiosis and mitosis (see Supplemental Table S4 and associated supplementary recordings). Although HRPT2 was demonstrated to function as a tumor suppressor in parathyroid carcinoma (Carpten et al. 2002), HRPT2'sroleinthe cell remains essentially unstudied in mammals. Our phenotypic analysis suggests that this gene product is important for microtubule-based functions in the early embryo, providing a functional link between its cellular activity and its role in tumorigenesis.
How many genes are required for embryogenesis in C. elegans?
These data allow us to refine our predictions of how many genes are required for embryogenesis or egg production in C. elegans. Based on the results of this study, and considering our estimated false-negative rate, we can predict that about 45% of ovary-enriched genes are required for embryogenesis. From these data we project that a total of ∼1300 genes in the ovary-enriched fraction are required for embryogenesis or oogenesis. Currently, the ovary-enriched genes contain about half the total number of genes that are known to be required for embryogenesis or oogenesis. Therefore, by extrapolation, we predict that a lower estimate for the total number of genes required for these processes in C. elegans is ∼2600 genome-wide. So far, considering no false positives, a little over two-thirds (1843) have been identified through the multiple large-scale RNAi studies. The ovary-enriched genes are the most extensively tested set of genes in the genome, yet only about three-quarters (956) of the predicted total have been found so far. We anticipate that further in-depth analyses across the entire genome will reveal many more genes that play a role in embryonic development. Our data also suggest that the majority of these as yet undiscovered genes will give rise to a partially penetrant embryonic lethal phenotype.
Functional trends for the ovary-enriched genes
The ovary-enriched genes have been heavily studied by RNAi: of the 3015 genes in WormBase (WS110) that fit the criteria for oogenic germline-enrichment, 2944 (98%) have been tested by RNAi at least once, 2793 (93%) have been tested at least twice, and 2201 (73%) have been tested at least three times. Using the strongest reported phenotype for each gene, we characterized patterns of gene function among ovary-enriched genes.
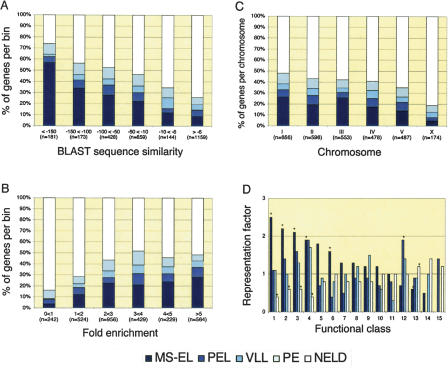
When we examined the combined data with respect to penetrance, we found that functional trends are driven mostly by genes giving rise to the highest-penetrance phenotypes. The ovary-enriched gene set displays a positive and statistically significant correlation between essential gene function in the embryo and both sequence conservation (Fig. 5A) and fold enrichment in the ovary (Fig. 5B); however, these trends are dominated by the highest-penetrance phenotypic class (“MS-EL”). Previous studies have observed that the X chromosome contains significantly fewer than expected genes required for oogenesis or embryogenesis (Piano et al. 2000, 2002; Maeda et al. 2001; Kamath et al. 2003), even taking into account the paucity of X-linked genes in the ovary-enriched set (Reinke et al. 2004). When we examined the distribution of phenotypes on different chromosomes with respect to penetrance (Fig. 5C), we observed that this trend is also restricted to genes with highly penetrant phenotypes.
Figure 5.
Phenotypic trends from a compendium of RNAi data for the ovary-enriched gene set. (A) Positive correlation between penetrance category and sequence conservation with D. melanogaster. A similar trend was observed in comparisons with human (data not shown). The highest-penetrance group, MS-EL, is significantly (P < 0.001) overrepresented in all bins containing genes with a BLAST e-value < 1e-50 and underrepresented among genes with an e-value > 1e-6. The converse is true for NELD. Binned e-value ranges on the x-axis are indicated by e-value exponents; sequence similarity decreases from left to right. (B) Positive correlation between penetrance category and fold enrichment in the oogenic germline. The MS-EL group is significantly (P < 0.001) overrepresented among genes with ≥5-fold enrichment and significantly underrepresented among genes <2-fold enriched; the converse is true for the NELD category. Trends in A and B are similar to observations from a smaller set of ovary-enriched genes (Piano et al. 2002). (C) Ovary-enriched genes on the X chromosome are significantly less likely to encode essential gene functions. Emb and Ste genes on the whole are significantly underrepresented on the X chromosome (P < 0.001). Among separate phenotypic categories, only the MS-EL class is significantly underrepresented on the X chromosome (P < 0.001). (D) Functional classes by phenotypic penetrance category. Functional classes (from Kamath et al. 2003): 1, RNA synthesis; 2, DNA synthesis and repair/cell cycle; 3, cellular architecture/collagen; 4, protein synthesis; 5, proteases; 6, protein degradation; 7, RNA binding; 8, signal transduction; 9, chromatin regulation; 10, transcription factor; 11, nucleic acid binding; 12, metabolism; 13, unknown; 14, neuronal; 15, small molecule transport. One functional class with fewer than 10 members in the ovary-enriched set (retroviral- and transposon-derived) is not shown. *, significant (P < 0.001) over- or underrepresentation. For all panels, significance was calculated using the hypergeometric probability test. Phenotypic categories are as in Figure 3, with the addition of PE (postembryonic phenotypes only) in panels A-C; HPEL and LPEL are shown combined (“PEL”); LCL genes from this study are included in the NELD category.
We also examined patterns of functional annotations with respect to penetrance (Fig. 5D). In the highest penetrance class, we observed a highly significant (P < 0.001) overrepresentation of genes involved in fundamental cellular processes (RNA and protein synthesis; DNA synthesis and repair/cell cycle, and cellular architecture) that was mirrored by a significant depletion among genes with no known requirement in the embryo. Overrepresentation of these functional classes was also observed among genes with nonviable phenotypes in a genome-wide RNAi survey (Kamath et al. 2003). However, including the new dimension of penetrance here offers a level of resolution that reveals a more subtle pattern in the data: the proportion of genes involved in these processes rises as the penetrance of embryonic lethality increases. Thus there is a positive correlation between penetrance and enrichment in these functional classes. The only other statistically significant deviation is an overrepresentation of partially penetrant genes with metabolic functions, potentially reflecting some level of redundancy in embryonic metabolic pathways. Among genes that display only post-embryonic phenotypes, no functional classes are significantly over- or underrepresented (P < 0.01 for all classes, data not shown). One way to interpret these trends is that genes classified as playing a role in fundamental cellular processes are more likely to be indispensable for embryogenesis than genes classified in more regulatory roles.
Penetrance level and conservation across phyla
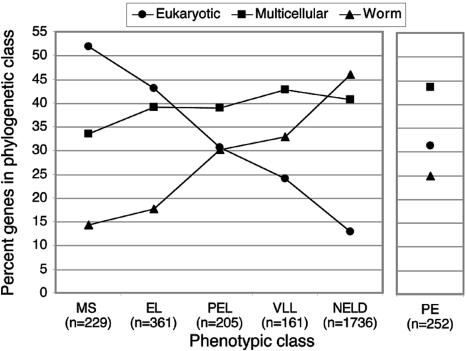
To explore these observations further, we asked whether there was a correlation between the pattern of sequence conservation across phyla and the penetrance of the embryonic lethal phenotype elicited by RNAi, and we found a striking trend (Fig. 6). We observed a highly significant (P < 0.001) enrichment in the highest penetrance classes (MS, EL) for genes that are conserved among all eukaryotes, and a corresponding depletion for genes with no identifiable homolog in other species, whereas the reverse is true for phenotypic classes with no reported phenotypes (LCL, NELD). Genes giving rise to partial-penetrance embryonic lethality show an intermediate distribution between these two extremes. Interestingly, genes that give rise to only postembryonic phenotypes display a similar phylogenetic distribution to the partial-penetrance genes. Thus there is a positive correlation between increasing phenotypic penetrance and the proportion of genes conserved across all eukaryotes; conversely, there is a negative correlation between penetrance and the proportion of worm-specific genes. This trend suggests that there is a relationship between the age of a gene and the degree to which its function is required for embryogenesis.
Figure 6.
Phenotypic trends in the ovary-enriched genes across different phyla. Genes were classified as “All Eukaryotic” if a homolog was identified in yeast, as “Multicellular” if a homolog was identified in either fly, human, and/or Arabidopsis (only a handful of genes were shared between C. elegans and Arabidopsis but not fly or human) and was not found in yeast, or as “Worm” if no homolog was identified in any of these species. Eukaryotic genes are significantly (P < 0.001, hypergeometric probability test) overrepresented in MS and EL phenotypic categories and significantly underrepresented in the NELD category. Conversely, worm-specific genes are significantly overrepresented in the NELD category and significantly underrepresented in MS and EL categories.
A priori, we suspected that penetrance designations derived from RNAi assays may not be particularly informative, as RNAi does not always deplete gene function completely and thus may not necessarily phenocopy null phenotypes. However, our analyses revealed strong trends among the different penetrance classes. This suggests that penetrance reported by RNAi can genuinely reflect differences in gene function when animals are observed under laboratory conditions, particularly when a gene has been assayed multiple times by different groups. The group of genes showing the strongest penetrance not only contains a higher proportion of genes involved in a few critical cellular processes, but is also significantly enriched for genes that are highly conserved in other species—a trend that is not observed among other, less penetrant phenotypic categories. Together these observations suggest that highly conserved genes carrying out indispensable functions occupy critical positions in gene networks that are essential to cell survival at any stage of development, whereas less-conserved genes may contribute more peripheral, or modulatory, functions that serve to adapt these core modules to specific cellular contexts and provide plasticity to genetic networks. The observation that the proportion of eukaryotic genes (the most ancient phylogenetic class) increases with increasing penetrance and that the proportion of worm-specific genes (the most recently evolved phylogenetic class) decreases with increasing penetrance suggests that the importance of a given gene's function can change over time. We speculate that as new genes arose over history, some have gradually become fixed in more crucial roles in the networks underlying biological processes. Another way to express this is to say that those genes that are retained over very long evolutionary times are those that have taken on indispensable roles. This suggests the possibility of a general architectural motif in genetic networks, in which genes entrenched in critical positions consist largely of older components, whereas newer functions added later provide additional layers of regulation that lend robustness or adaptive functions to these core networks. Combined analysis of phenotypic, gene expression, and protein interaction data from C. elegans will help resolve questions about how gene networks are adapted to different cellular contexts, and will reveal underlying architectural principles of metazoan development.
Methods
Generation of dsRNA
ORFeome clones were amplified by PCR using T7 primers. Amplified products were obtained for 1123 of the 1245 ORFeome clones successfully cherry-picked for this study. PCR products were used as templates for in vitro transcription by T7 RNA polymerase in a 50-μL reaction. All RNA products were verified on 1% agarose gels, then precipitated by adding 5 μL sodium acetate (pH 5.2), 2 μL Pellet Paint (Novagen), and 100 μL ethanol. The dsRNA was stored in this manner at -20°C for a maximum of 1 wk. All molecular biology procedures and soaking were performed in 96-well plates.
RNAi by soaking
N2 Bristol worms were synchronized using 0.8% hypochlorite solution, and L1 larvae were hatched without food in M9 medium before being plated to NGM media seeded with OP50. Animals were grown at 25°C for 40-44 h until the L4 stage. To prepare the soak, dsRNAs were spun down, washed with 70% ethanol, dried completely, and resuspended in 4 μL soaking buffer (0.25X Mg++-free M9, 3mM spermidine, 0.05% gelatin). L4 larvae were washed from plates using M9 and allowed to crawl on unseeded plates 15 min; this step was repeated. After the last incubation on unseeded plates, worms were washed three times in M9 and twice in soaking buffer. Then, 10-30 worms in 1 μL of soaking buffer were added to each tube of resuspended dsRNA. Worms were incubated for 24 h at 20°C in dsRNA and then plated to NGM seeded with OP50 to recover for 24 h at 20°C. For each gene tested, five worms were placed onto individual egg laying plates and incubated for 24 h at 20°C. Treated animals were removed from egg-laying wells, and plates were placed at 25°C for 15-18 h to allow eggs to hatch. We then counted the number of eggs and the number of larvae present in each of the five replicate broods for each gene tested.
Penetrance designations
Genes that reproducibly (i.e., observed in at least two broods) gave rise to 20%-50%, 50%-80%, or 80%-100% embryonic lethality were designated as LPEL (“low partial embryonic lethal”), HPEL (“high partial embryonic lethal”), or EL (“embryonic lethal”), respectively. Genes were termed VLL (“very low lethal”)if at least two broods showed ≥5% embryonic lethality or if at least one brood showed ≥10% embryonic lethality. Genes were termed MS (“maternal sterile”) if at least three broods yielded ≤10 progeny, whether accompanied by embryonic lethality or not. We called a gene NELD (“no embryonic lethality detected”) if the minimum criteria for VLL were not satisfied and a minimum of four broods was scored. Genes that did not show phenotypes upon retesting were termed LCL (low-confidence lethals) and grouped with the NELD class for all bioinformatic analyses. Those that showed higher penetrance phenotypes upon retesting were moved to the higher penetrance class.
Selection of ORFeome clones and mapping to C. elegans gene products
Ovary-enriched ORFeome clones were selected by matching WormPep95 (Harris et al. 2003) with C. elegans ORFeome Version 1.1 (Reboul et al. 2003) and a list of microarray assays designated as “oogenic germline-enriched” according to the selection criteria defined in Reinke et al. (2004). The intersection of these two sets identified 1582 ovary-enriched ORFeome clones, which were then filtered using all RNAi results available in WormBase (WS95) and classified into four categories: (1) clones representing genes not previously assayed by RNAi; or (2-4) previously assayed and falling into one of three phenotypic classes: (2) WT (“wild-type”), (3) postembryonic only, or (4) embryonic lethal. We selected all genes from sets 1-3 that we had not previously assayed by injection (Piano et al. 2002) for characterization in this study (see Fig. 3).
After completing the RNAi analysis, we remapped this set of ORFeome clones and the PCR products used to define the ovary-enriched gene set to the latest WormBase freeze (WS110). The set of 1083 ORFeome clones for which we reported RNAi results currently maps to 1011 ovary-enriched and 16 non-ovary-enriched genes (not including alternative splice forms). Only the ovary-enriched fraction was used in global analyses of ovary-enriched gene function. A total of 53 genes were independently assayed by two or more different ORFeome clones; for 14 of these, replicate assays yielded results in different penetrance classes. RNAi results for all clones assayed are reported in Supplemental Table S1.
Some ORFeome clones overlap two adjacent genes. This indicates a discrepancy between the current gene models in WormBase and sequence evidence from these ORFeome clones; each of these cases was reviewed at WormBase, and the gene models were left unchanged due to discrepancies between EST data from cDNA clones and evidence based on the ORFeome clones. For clones that extensively overlapped two open reading frames, when the RNAi assay was negative (LCL or NELD), we reported this result for each gene. In the few rare cases where a clone overlapped one ORF extensively and another by <200 base pairs, we considered only the former gene to have been reliably assayed and conservatively reported no RNAi result for that clone in association with the second gene. Wherever we had independent confirmation of such assays from clones that spanned only one of the two adjacent transcription units, the results were always consistent with these policies.
Time-lapse imaging
Time-lapse digital recordings were made as described (Piano et al. 2002). At least three independent RNAi-affected worms from clones giving rise to ≥80% lethality were dissected to collect the early embryos ∼36-48 h after dsRNA delivery, and were mounted on a 2% agarose pad. Movies were made with a Leica microscope equipped with a 40× lens and differential interference contrast (DIC) optics. We used digital acquisition through a Cohu CCD camera driven by macros written for Openlab (Improvision) to record 300 frames at 10-sec intervals. QuickTime Pro 4.0 (Apple Computers) was used to compress all movies (medium, JPEG B compression) and prepare them for internet streaming. All results from this study, including digital movies, are accessible through http://www.RNAi.org and http://www.wormbase.org.
Bioinformatic analysis
To facilitate the bioinformatic analyses, we built a MySQL database containing information about genes, RNAi experiments, expression studies, sequence similarities, and other functional genomics data. Data processing and informatics analyses were performed using Perl scripts and/or SQL queries to this database. Data sources included C. elegans “Predicted_gene” and “RNAi” objects from WormBase (WS110), as well as supplementary material from publications (e.g. Simmer et al. 2003), prepublication expression data (Reinke et al. 2004), multispecies sequence data from genome centers, results of in-house BLAST analyses (described below), and RNAi data collected in this study. All RNAi results contained in this database are searchable through www.RNAi.org. RNAi data collected in this study were reported to WormBase. Functional categories were assigned using the classification system from Kamath et al. (2003).
Reciprocal whole-proteome BLASTP analysis (Altschul et al. 1997) was performed using an in-house BLAST pipeline on complete predicted protein sequence sets from Saccharomyces cerevisiae (SGD; Cherry et al. 1998; updated Aug. 8, 2003; 5889 sequences), C. elegans (WormPep100; C. elegans Sequencing Consortium 1998; Harris et al. 2003; 21,383 unique sequences)], D. melanogaster (Release 3.1; Celniker et al. 2002; Flybase Consortium 2003; 18,498 sequences), Homo sapiens (IPI v2.22; http://www.ebi.ac.uk/IPI/, 29,539 sequences), and Arabidopsis thaliana (TAIR Release 4.0; Garcia-Hernandez et al. 2002; 28,786 sequences). Homologous genes between species were identified using a simple reciprocal best-hit (e-value < 1e-6) criterion. Summarized results of this analysis were transferred to the above MySQL database. Off-target genes were considered potentially inhibited by RNAi if they shared contiguous blocks of 20 nucleotides totaling at least 60 nucleotides of sequence with the primary gene target.
For comparative analyses and characterization of phenotypic trends among ovary-enriched genes, all RNAi results were mapped to genes in WormBase (WS110) using the chromosome coordinates of the reagents used for each assay. Genes were placed into exclusive categories according to the most severe phenotype observed by any group. Our comparative analyses included only those phenotypic results from Simmer et al. (2003) that were reproducible [listed in Simmer et al. 2003, Supplemental Tables 1A, 1B; nonreproducible results (Supplemental Table 1C) were excluded from this comparison]. In addition, because assays from that study with a wild-type result were not reported in WormBase, we inferred them for the remainder of genes assayed by Kamath et al. (2003), since both studies used the same set of reagents. In all, we included results for 15,353 genes (excluding alternative splice forms) assayed by Simmer et al. (2003).
Acknowledgments
We thank Casey Roehrig, Danny Shapiro, Tony Tao, and Krishna Vijayendran for technical assistance; Wanchen Yueh, Tong Hao, and Philippe Lamesch for help with data processing; WormBase curators for reviewing gene models; Dr. Julie Ahringer for critically reviewing our manuscript; Dr. Jane Hubbard for useful insights; and anonymous reviewers who provided useful comments that improved our manuscript. This work was supported by grants from NSF (DBI-0137617, to K.C.G.) and NIH(HG003019-01, to F.P.). A.J.S. is currently funded by the NCI CPFP. The ORFeome project is supported by a grant from NCI (7R33CA81658-02, to M.V.).
Footnotes
[Supplemental material is available online at www.genome.org and RNAiDB (www.RNAi.org). Results from this study are also available through WormBase (www.wormbase.org).]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3194805.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman, M.N., Furlong, E.E., Imam, F., Johnson, E., Null, B.H., Baker, B.S., Krasnow, M.A., Scott, M.P., Davis, R.W., and White, K.P. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270-2275. [DOI] [PubMed] [Google Scholar]
- Baugh, L.R., Hill, A.A., Slonim, D.K., Brown, E.L., and Hunter, C.P. 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889-900. [DOI] [PubMed] [Google Scholar]
- Berns, K., Hijmans, E.M., Mullenders, J., Brummelkamp, T.R., Velds, A., Heimerikx, M., Kerkhoven, R.M., Madiredjo, M., Nijkamp, W., Weigelt, B., et al. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431-437. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127-131. [DOI] [PubMed] [Google Scholar]
- Boutros, M., Kiger, A.A., Armknecht, S., Kerr, K., Hild, M., Koch, B., Haas, S.A., Consortium, H.F., Paro, R., and Perrimon, N. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832-835. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Carpten, J.D., Robbins, C.M., Villablanca, A., Forsberg, L., Presciuttini, S., Bailey-Wilson, J., Simonds, W.F., Gillanders, E.M., Kennedy, A.M., Chen, J.D., et al. 2002. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 32: 584-588. [DOI] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: research0079.1-0079.14. [DOI] [PMC free article] [PubMed]
- Chance, M.R., Bresnick, A.R., Burley, S.K., Jiang, J.S., Lima, C.D., Sali, A., Almo, S.C., Bonanno, J.B., Buglino, J.A., Boulton, S., et al. 2002. Structural genomics: A pipeline for providing structures for the biologist. Protein Sci. 11: 723-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., Adler, C., Ball, C., Chervitz, S.A., Dwight, S.S., Hester, E.T., Jia, Y., Juvik, G., Roe, T., Schroeder, M., et al. 1998. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 26: 73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez, M., Berardini, T.Z., Chen, G., Crist, D., Doyle, A., Huala, E., Knee, E., Lambrecht, M., Miller, N., Mueller, L.A., et al. 2002. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genomics 2: 239-253. [DOI] [PubMed] [Google Scholar]
- Ge, H., Liu, Z., Church, G.M., and Vidal, M. 2001. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29: 482-486. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727-1736. [DOI] [PubMed] [Google Scholar]
- Goh, C.S., Lan, N., Echols, N., Douglas, S.M., Milburn, D., Bertone, P., Xiao, R., Ma, L.C., Zheng, D., Wunderlich, Z., et al. 2003. SPINE 2: A system for collaborative structural proteomics within a federated database framework. Nucleic Acids Res. 31: 2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- Guo, S. and Kemphues, K.J. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611-620. [DOI] [PubMed] [Google Scholar]
- Hanazawa, M., Mochii, M., Ueno, N., Kohara, Y., and Iino, Y. 2001. Use of cDNA subtraction reveals genes required for germ-line development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 98: 8686-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T.W., Lee, R., Schwarz, E., Bradnam, K., Lawson, D., Chen, W., Blasier, D., Kenny, E., Cunningham, F., Kishore, R., et al. 2003. WormBase: A cross-species database for comparative genomics. Nucleic Acids Res. 31: 133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, M., Ryu, J., Kiraly, M., Duke, K., Reinke, V., and Kim, S.K. 2001. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 98: 218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kiger, A., Baum, B., Jones, S., Jones, M., Coulson, A., Echeverri, C. and Perrimon, N. 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2: 27-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11: 171-176. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Thompson, M.J., Yeates, T.O., and Eisenberg, D. 1999. A combined algorithm for genome-wide prediction of protein function. Nature 402: 83-86. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J., Silva, J.M., Conklin, D.S., Schlabach, M., Li, M., Aruleba, S., Balija, V., O'Shaughnessy, A., Gnoj, L., Scobie, K., et al. 2004. A resource for large-scale RNA-interference-based screens in mammals. Nature 428: 427-431. [DOI] [PubMed] [Google Scholar]
- Piano, F. and Gunsalus, K.C. 2002. RNAi-based functional genomics in Caenorhabditis elegans. Curr. Genomics 3: 68-81. [Google Scholar]
- Piano, F., Schetter, A.J., Mangone, M., Stein, L., and Kemphues, K.J. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10: 1619-1622. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetter, A.J., Morton, D.G., Gunsalus, K.C., Reinke, V., Kim, S.K., and Kemphues, K.J. 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12: 1959-1964. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Reinke, V., Smith, H.E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S.J., Davis, E.B., Scherer, S., Ward, S., et al. 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6: 605-616. [DOI] [PubMed] [Google Scholar]
- Reinke, V., Gil, I.S., Ward, S., and Kazmer, K. 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311-323. [DOI] [PubMed] [Google Scholar]
- Rual, J.-F., Ceron, J., Koreth, J., Hao, T., Nicot, A.-S., Hirozane-Kishikawa, T., Vandenhaute, J., Orkin, S.H., Hill, D.E., van den Heuvel, S., et al. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-Based RNAi library. Genome Res. 14: 2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., Moorman, C., Van Der Linden, A.M., Kuijk, E., Van Den Berghe, P.V., Kamath, R., Fraser, A.G., Ahringer, J., and Plasterk, R.H. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLOS Biol. 1: 77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma, M.P., Fasulo, B., Cenci, G., Cundari, E., and Gatti, M. 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13: 2448-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, A. 2004. High-throughput RNAi in Caenorhabditis elegans: Genome-wide screens and functional genomics. Differentiation 72: 81-91. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Grishok, A., and Mello, C.C. 1998. RNAi in C. elegans: Soaking in the genome sequence. Science 282: 430-431. [DOI] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417: 399-403. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Reboul, J., Shtanko, O., Bertin, N., Vaglio, P., Ge, H., Lee, H., Doucette-Stamm, L., Gunsalus, K.C., Schetter, A.J., et al. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12: 1952-1958. [DOI] [PubMed] [Google Scholar]
- Zipperlen, P., Fraser, A.G., Kamath, R.S., Martinez-Campos, M., and Ahringer, J. 2001. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20: 3984-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.rnai.org; Web portal to database of RNAi data from C. elegans.
- http://www.wormbase.org; Web portal to model organism database for C. elegans and related nematodes.
- http://www.ebi.ac.uk/IPI/; Provides access to database of non-redundant sets of cross-referenced human, mouse, and rat proteins from UniProt, RefSeq, and Ensembl.