Abstract
To explore the role of transcriptome polymorphism in adaptation of organisms to their environment, we evaluated this parameter for the Escherichia coli/Shigella bacterial species, which is composed of well-characterized phylogenetic groups that exhibit characteristic life styles ranging from commensalism to intracellular pathogenicity. Both the genomic content and the transcriptome of 10 strains representative of the major E. coli/Shigella phylogenetic groups were evaluated using macroarrays displaying the 4290 K12-MG1655 open reading frames (ORFs). Although Shigella and enteroinvasive E. coli (EIEC) are not monophyletic, phylogenetic analysis of the binary coded (presence/absence) gene content data showed that these organisms group together due to similar patterns of undetectable K12-MG1655 genes. The variation in transcript abundance was then analyzed using a core genome of 2880 genes present in all strains, after adjusting RNA hybridization signals for DNA hybridization signals. Nonrandom changes in gene expression during the evolution of the E. coli/Shigella species were evidenced. Phylogenetic analysis of transcriptome data again showed that Shigella and EIEC strains group together in terms of gene expression, and this convergence involved groups of genes displaying biologically coherent patterns of functional divergence. Unlike the other E. coli strains evaluated, Shigella and EIEC are intracellular pathogens, and therefore face similar selective pressures. Thus, within the E. coli/Shigella species, strains exhibiting a particular life style have converged toward a specific gene expression pattern in a subset of genes common to the species, revealing the role of selection in shaping transcriptome polymorphism.
Adaptability of organisms relies on phenotypic diversity, which, in turn, is the substrate for natural selection. Phenotypic diversity can be the consequence of genetic diversity (i.e., differences) in DNA coding for structural, metabolic, or signaling proteins, but can also reflect diversity at the level of gene expression. Understanding the importance of such regulatory genetic diversity to adaptive evolution is a crucial issue in evolutionary biology. The availability of whole-genome sequences and new tools facilitating the evaluation of gene expression at the genomic level has now made it possible to identify regulatory diversity, and to assess the importance of its role in promoting adaptability.
Gene activity is controlled first and foremost at the level of transcription. Much of the control of gene activity is achieved through the interplay between regulatory proteins (trans factors) and specific DNA sequences where these transcription factors bind (cis elements) (Lemon and Tjian 2000; Lloyd et al. 2001). If polymorphism in gene expression is neutral (genetic drift), differences in the global pattern of gene expression between organisms should reflect global genomic divergence and accumulate mainly as a function of time, and therefore be linked to the organism phylogeny. The more the divergence between two organisms, the more their transcriptomes will differ, and this divergence should include variation in both cis and trans elements involved in the control of gene expression. Alternatively, if gene regulation is under adaptive selection, the transcriptome within a group of organisms should be linked to particular ecological conditions encountered by the members of that group, regardless of evolutionary distance.
To test these hypotheses, we took advantage of the Escherichia coli/Shigella single bacterial species. The evolutionary history of these organisms is well characterized, and several intraspecies phylogenetic groups have been identified (Reid et al. 2000; Escobar-Paramo et al. 2003, 2004a,b). Members of this species exhibit very different life styles, and face a wide range of selective pressures. E. coli strains are found in numerous mammals and birds, and include both commensal organisms and extracellular pathogens implicated in a variety of pathologies, including urinary tract infection, sepsis, and noninvasive diarrhea (Donnenberg 2002). In contrast, Shigella, originally was considered a distinct genus with four distinct species (Shigella dysenteriae, Shigella boydii, Shigella flexneri, and Shigella sonnei) when only phenotypic tools were available, and enteroinvasive E. coli (EIEC) are strictly human intracellular pathogens responsible for bacillary dysentery (Donnenberg 2002). These organisms are characterized by a large virulence plasmid and by several negative characters, including immobility, inability to use lactose, and absence of lysine decarboxylase activity (Parsot and Sansonetti 1996). In this study, we have analyzed the genomic content and the transcriptome of 10 strains representative of the major E. coli/Shigella phylogenetic groups, and show that transcriptome polymorphism does exist. This regulatory genetic diversity is neither randomly distributed nor linked to strain phylogeny, but correlates with the lifestyle of these organisms.
Results and Discussion
Evolutionary history of the E. coli/Shigella species
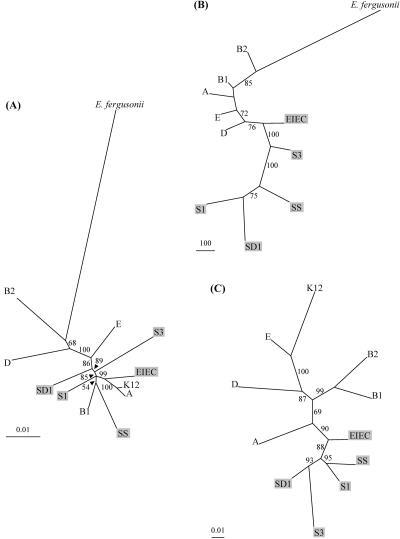
Preliminary to the analysis of the relationship between the pattern of gene expression and the phylogeny of the strains evaluated in this study, we first reconstruct a phylogenetic tree of these strains using simultaneous analysis of 11 essential genes (trpA, trpB, thrB, putP, pabB, icd, purM, polB, dnaE, dinB, and umuC) (Gerdes et al. 2003). These genes, which spanned 11,340 nucleotides, are known to exhibit low levels of horizontal gene transfer (Denamur et al. 2000; Bjedov et al. 2003). This nucleotide sequence tree yielded a well-accepted evolutionary history of the species (Fig. 1A; Pupo et al. 2000; Escobar-Paramo et al. 2003, 2004a,b). The most ancient diverged group is the B2 group, followed by the D and E groups. The B1 and A groups, as well as the Shigella groups (S1, S3, SD1, SS), emerged more recently, and in a relatively short period of time, as attested to by the short internal branches and the low bootstrap value sustaining the SS/B1 group. The EIEC appears as a sister group to the A group strains. It must be noted that Shigella/EIEC strains are clearly not monophyletic. The emergence of the most divergent E. coli group (B2) occurred ∼22 to 30 million years ago (Mya) (Lecointre et al. 1998), whereas dates ranging from 4-6 Mya (G. Lecointre and E. Denamur, pers. comm.) to <300,000 years (Pupo et al. 2000) have been proposed for the emergence of the various Shigella groups.
Figure 1.
(A) Phylogenetic tree of the 12 strains, E. fergusonii as an outgroup, and reconstructed from the DNA sequences of 11 essential genes, using Neighbor Joining procedure. The model used to estimate the pairwise distance matrix is TN93, the parameters being estimated by maximum likelihood (PHYML [Guindon and Gascuel 2003]) as follows: transition/transversion for purines = 3.28, transition/transversion for pyrimidines = 6.08, γ shape = 0.112, 10 categories, percentage of invariant = 0. Identical topologies were obtained using maximum likelihood or parsimony procedures (data not shown). (B) Phylogenetic tree of the 11 strains (K12-MG1655 being excluded), E. fergusonii as an outgroup, and obtained by parsimony procedure using PAUP4.0b (Swofford 2002) from the K12-MG1655 4290 binary-coded ORFs (“0” for undetectable sequence, “1” for sequence present at least in one copy). The tree is obtained by branch and bound algorithm, 1264 characters being parsimony informative. The length of the tree is 3584 steps, consistency index = 0.56, retention index = 0.52. (C) Unrooted tree of the transcript abundances, expressed as adjusted RNA values (see Methods), of the 2880 core genome genes for the 11 strains (E. fergusonii excluded). The tree is obtained following the Neighbor Joining algorithm applied to the euclidian distances between strains. All of the bootstrap values are obtained from 1000 replicates and are indicated only when ≥50%. Abbreviations for the strain designation are as follows: B2 (ECOR56), B1 (ECOR26), D (ECOR50), E (EDL933), A (ECOR1), K12 (K12-MG1655), EIEC (EIEC85b), SD1 (S. dysenteriae serotype 1, SD0177), SS (S. sonnei, SS92a), S1 (S. boydii serotype 10, SB1080), S3 (S. flexneri serotype 5, M90T). Shigella and EIEC strains are indicated in gray background.
Genomic DNA content
In natural bacterial isolates, the comparative analysis of genomic DNA content is an important prerequisite to the evaluation of gene expression. Prokaryotic genomes are highly plastic, and gene losses/acquisitions are frequent events (Lawrence and Ochman 1998). Thus, the absence of transcripts can reflect either the absence of the gene at the DNA level or the absence of transcription. Moreover, gene copy number can be variable among strains. For example, it has been reported that insertion sequences are expanded in Shigella as compared with E. coli (Jin et al. 2002; Wei et al. 2003). All of these differences between strains will affect the apparent transcript abundance measured by macroarrays.
We first analyzed the DNA content of each strain by determining the presence or absence of the 4290 open reading frames (ORFs) identified in K12-MG1655. As previously described for E. coli (Ochman and Jones 2000; Dobrindt et al. 2003), gene deletions/acquisitions were identified frequently. The proportion of undetectable K12-MG1655 genes in the strains evaluated by us ranged from 6% (257 genes in ECOR26 strain) to 17% (709 genes in S1 strain). Among these undetected genes, 1.4% were K12-MG1655 specific, corresponding mainly to prophages, uncharacterized ORFs, and ORFs involved in lipopolysaccharide biosynthesis (Dobrindt et al. 2003). Displaying these undetectable genes along the K12-MG1655 chromosome (Supplemental Fig. S1) shows that they are not randomly scattered, and are often clustered (1) within operons and (2) at specific locations, especially at tRNA sites, as previously reported (Dobrindt et al. 2003). Phylogenetic analysis of the binary coded (presence/absence) DNA data set shows that Shigella and EIEC strains group together (Fig. 1B), as these strains share similar patterns of undetectable K12-MG1655 genes. The comparison of the species (Fig. 1A) and genomic DNA content (Fig. 1B) trees showed incongruence (i.e., disagreement) as indicated by the proportion of unconflicting quartet at 0.66 and symmetric difference between trees at 14 (in comparison to values of 1 and 0, respectively, for identical trees). Because Shigella and EIEC strains are not monophyletic within the E. coli species (Fig. 1A), the similar pattern of undetected genes in these strains appears to be the result of convergent evolution. This convergence is also retrieved when available complete genome sequences (www.genome.wisc.edu; genome.geninfo.osaka-u.ac.jp/bacteria/o157; www.sanger.ac.uk/cgi-bin/blast/submitblast/escherichia_shigella) from another set of strains (S. dysenteriae, two S. flexneri, S. sonnei, six E. coli [two B2 group, two E group, one D group strains]) are studied by BLAST analysis (data not shown). Clearly, Shigella strains have fewer K12-MG1655 genes than the remaining E. coli strains. The mean number of undetectable genes was 621 for Shigella strains and 326 for the remaining E. coli strains. The undetectable genes in Shigella/EIEC strains appear to belong to specific gene categories, including genes coding for putative regulatory proteins (P =2.0 × 10-6 based on the two-tailed Student t-test). One hot spot of undetectable K12-MG1655 genes in Shigella/EIEC strains starts at the thrW tRNA site (located at 5.65′ on the K12-MG1655 chromosome) and overlaps the lac operon locus (Supplemental Fig. S1). This finding confirms at a genomic scale what has been previously described as “black holes” for individual genes, such as the lysine decarboxylase gene (Maurelli et al. 1998) or the lactose operon (Ito et al. 1991). Although we cannot exclude that the observed convergence in the pattern of undetectable K12-MG1655 genes is the result of genome reduction events linked to mutations become fixed by genetic drift in genes that are not maintained by selection (mutation accumulation), the role of selection (antagonistic pleiotropy) has been demonstrated for the inactivation of the lysine decarboxylase gene, cadA. When cadA was introduced into S. flexneri, virulence became attenuated, and enterotoxin activity was inhibited greatly (Maurelli et al. 1998). Cadaverine, a product of the reaction catalyzed by lysine decarboxylase, has been shown to attenuate the bacteria's ability to induce polymorphonuclear leucocytes transepithelial migration (McCormick et al. 1999) and to be the enterotoxin inhibitor (Maurelli et al. 1998).
A core genome was then defined as genes present as at least a single copy in all of the E. coli/Shigella strains studied. This conserved E. coli genetic backbone encompasses 2880 genes of the K12-MG1655 genome, a figure that is in agreement with previous estimations using the same approach (Ochman and Jones 2000; Dobrindt et al. 2003; Fukiya et al. 2004), or based on the comparison of complete genome sequences (Blattner et al. 1997; Perna et al. 2001; Jin et al. 2002; Welch et al. 2002; Wei et al. 2003; T. Le Gall and E. Denamur, pers. comm.). This core genome includes 76% (466 of 614) of the experimentally determined essential genes in K12-MG1655 (Gerdes et al. 2003), a finding that further validates our experimental approach.
Gene expression polymorphism
To accurately assess variations in gene expression, a variety of parameters have to be considered. First, when a given macroarray is hybridized with labeled DNA from K12-MG1655, dot intensities are clearly not homogeneous. Rather, intensities ranging from strong to near background are observed (Supplemental Fig. S2), and the standard deviation of all signals on an array hybridized with DNA is about half the mean of all signal intensities. This wide variability results from the intrinsic properties of the targets (the 4290 PCR-amplified ORFs spotted on the array) in the hybridization process and can introduce artifacts when quantifying transcript abundance. Indeed, if a strong signal is observed for DNA hybridization at a given ORF, a strong signal for RNA hybridization at this ORF does not necessarily indicate high transcript abundance. If two strains with low and high transcript abundance at this ORF are to be compared, the ratio between their spot intensities will not reflect their effective transcript abundance ratio. Another potential confounding variable in the interpretation of hybridization data is nucleotide divergence. In this study, macroarrays contained DNA that had been PCR amplified from the laboratory strain K12-MG1655, which belongs to the E. coli A group. Differences in the hybridization signal between isolates could be due to differential divergence between the targets (K12-MG1655 PCR-amplified ORFs) and the probes (the studied natural isolates). Indeed, nucleotide divergence between K12-MG1655 and B2 group strains is estimated to be 3.5%, whereas the divergence between K12-MG1655 and Escherichia fergusonii is about 5.5% (Escobar-Paramo et al. 2004b; data not shown). Lastly, increased transcript abundance of a gene could be due to an increase in the copy gene number, a frequent event in bacterial evolution.
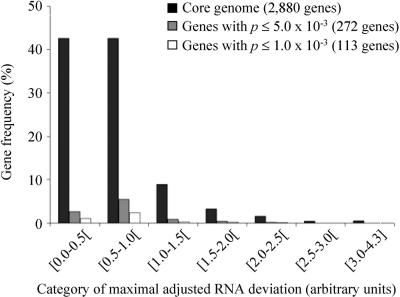
To compensate for these potential confounding factors, the RNA hybridization signal obtained for each gene was adjusted to take into account the signal obtained after DNA hybridization of the same gene from the same strain (Supplemental Fig. S3B). These values, called adjusted RNA values, ranged from -2.22 to +4.80 arbitrary units. For all strains, a Gaussian distribution of gene expression was observed (data not shown). The comparison of adjusted RNA values for a given gene between strains indicated that gene expression polymorphism does exist at the E. coli/Shigella species level. To obtain a global estimation of the extent of gene-expression polymorphism, the maximal deviation was calculated for each gene by determining the difference between the maximum and the minimum adjusted RNA values for that gene among the 11 strains studied. The distribution of these maximal deviation values showed a polymorphism peak frequency between 0.5 and 1.0 arbitrary unit for 42.6% of the genes (1228 of 2880). For 0.6% of the core genome genes (18 of 2880 genes), maximum deviation values higher than three arbitrary units were observed, 4.3 being the greatest extent of expression polymorphism (Fig. 2). This intraspecies polymorphism distribution is in agreement with that reported for strains of the yeast Saccharomyces cerevisiae (Townsend et al. 2003).
Figure 2.
Distribution of maximal adjusted RNA deviations among 11 strains of the E. coli/Shigella species for the 2880 genes of the core genome (black), the 272 genes (gray), and the 113 genes (white) that differentiate the Shigella/EIEC strains from the remaining strains at Student t-test P values ≤5.0 × 10-3 and ≤1.0 × 10-3, respectively. The maximal deviations (in arbitrary units) were calculated for each gene considering the values obtained for all of the studied strains as the difference between the maximum and the minimum adjusted RNA values. The gene frequency (%) is expressed relative to the 2880 core genome genes for the three data sets (core genome, genes with P ≤ 5.0 × 10-3 and ≤1.0 × 10-3).
Traces of selection in the gene expression pattern
To assess nonrandom changes in gene expression during the evolution of the E. coli/Shigella species, we evaluated the extent that (1) strains that face apparently identical selective pressures, (i.e., share identical life style), but belong to distinct phylogenetic groups, exhibit similar gene expression patterns, and (2) whether genes with biologically coherent functional relationships share the same pattern of expression in a group of strains.
A phylogenetic analysis of the transcriptome data was performed on the set of the core genome genes. The unrooted tree reconstructed from the adjusted RNA values of the 2880 genes common to all E. coli/Shigella strains distinguishes the Shigella/EIEC strains from the remaining strains of the species (Fig. 1C). As was observed with the analysis of genomic DNA content (Fig. 1B), Shigella/EIEC strains group together in term of gene expression. The low resolution of the branching pattern for the other strains in the genomic DNA content tree (Fig. 1B) does not permit the comparison between the two trees for these strains. In contrast, the transcriptome data tree is clearly different from the evolutionary history of the strains based on the sequence data for the 11 essential genes (Fig. 1A). Indeed, the comparison of these trees showed, here again, incongruence as indicated by the proportion of nonconflicting quartet at 0.68 and symmetric difference between trees at 18, and by the corresponding distance matrixes (the probability of incongruence is not rejected at P = 0.18 in the Mantel test). These results argue in favor of convergence in the pattern of global gene expression for Shigella/EIEC, a hallmark for adaptive evolution (Harvey and Pagel 1991). No clear grouping for the non-Shigella/EIEC strains was observed according to their lifestyles (for example, the division between commensal/pathogen) (Fig. 1C). The inclusion of more strains will be required, however, before definitive conclusions can be drawn.
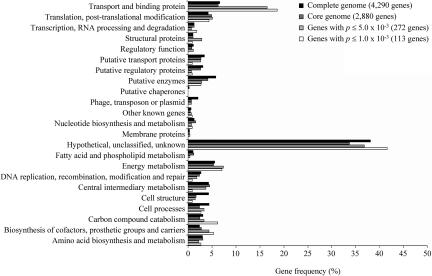
We next sought to identify genes whose expression was significantly different comparing Shigella/EIEC strains and the remaining E. coli strains using Student's t-test. Simulations were performed with sets of random values showing a Gaussian distribution to determine the P value for this test (P =1.0 × 10-3) that would give no more than 5% of false-positive results in this analysis (data not shown). At this threshold, the level of expression of 113 genes (4% of the core genome) was significantly different comparing the two groups of strains (Supplemental Table S1). These differentially expressed genes were not randomly distributed within the categories of maximal deviation of adjusted RNA values. Rather, most belonged to the 0.5-1.0 deviation category (Fig. 2, χ2 = 17.0, df = 7, P = 3.0 × 10-4). The observed differences in gene expression between the two groups of strains are thus extensive and mostly subtle, as reported for the comparison of brain transcriptomes of honey bees with distinct behaviors (Whitfield et al. 2003). The analysis of the distribution of these genes among functional categories indicated that genes coding for transporters and binding proteins were overrepresented, and 19% (21 of 113 genes) belonged to this class (χ2 = 39.0, df = 22, P =5.0 × 10-4) (Fig. 3). These findings were also observed when the Student t-test was performed using a P value of 5.0 × 10-3 for the identification of genes with significantly different expression (272 differentially expressed genes were identified under these conditions) (Figs. 2, 3). Lastly, a significant trend toward gene overexpression in the Shigella/EIEC group was observed, as 71% of the genes (80 of 113 genes) were expressed at higher levels in the Shigella/EIEC group than in the remaining E. coli strains. In contrast, only 54% of all genes in the core genome showed greater expression comparing the Shigella/EIEC group and the remaining E. coli strains. Among the 80 overexpressed genes in Shigella/EIEC strains, seven are involved in the acquisition of iron. Five (entD, entF, fepG, entE-ybdB) belong to five distinct transcriptional units coding for proteins involved in the synthesis and transport of the catecholate-type siderophore enterobactin, one (fhuC) codes for a cytoplasmic membrane component of the hydroxamate-type siderophore, and one (exbD) codes for a member of the TonB-ExbB-ExbD complex, which energizes the proton motive force required for transport across the outer membrane (Braun and Braun 2002). All of the other genes belonging to these seven transcriptional units, as well as tonB itself, showed greater expression in the Shigella/EIEC group than in the remaining E. coli, but the differences in expression did not achieve statistical significance using the P <1.0 × 10-3 threshold (Supplemental Fig. S4). These transcriptional units all belong to the Fur regulon (Salgado et al. 2001). Other overexpressed genes encoded transporter proteins for inorganic phosphate, phosphonate, nickel, potassium, amino acids, and sugars (Supplemental Table S1). Thus, the set of differentially expressed genes is not a random subset of the core genome genes, but is highly structured, an additional hallmark of adaptive evolution. Real-time quantitative RT-PCR was used as an independent method to confirm the differential regulation of some of the identified genes. The expression of 20 genes belonging to the list of 113 differentially expressed genes (Supplemental Table S1) and representing different functions and operons was analyzed in three representative strains, i.e., a noninvasive E. coli strain (EDL933), the EIEC, and a Shigella (SS). The RT-PCR data confirmed the macroarray-observed regulation in 13 cases (65%) with a significant variation between E. coli and Shigella/EIEC expression levels of 0.7-9.3 fold (Table 1). In five cases, the macroarray observed variation was retrieved by RT-PCR analysis in two of three strains. Only in two cases, the macroarray and RT-PCR data were contradictory.
Figure 3.
Distribution in main functional categories of the 4290 genes of E. coli K12-MG1655 (black), the 2880 genes of the core genome (deep gray), the 272 genes (gray), and the 113 genes (white) that differentiate the Shigella/EIEC strains from the remaining strains at Student t-test P values ≤5.0 × 10-3 and ≤1.0 × 10-3, respectively. The gene frequency (%) is expressed relative to the total number of genes in each considered data set (complete genome, core genome, genes with P ≤ 5.0 × 10-3 and ≤1.0 × 10-3).
Table 1.
Expression levels of 13 differentially expressed genes between a typical E. coli strain (EDL933) and EIEC/Shigella strains
| In vitro grown bacteria
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EDL933 (E)
|
EIEC
|
S. sonnei (SS)
|
In vivo grown bacteria
|
|||||||
| Gene | Functional group | Mean | SD | Mean | SD | Mean | SD | Ratio | P-value | S. flexneri (S3) |
| hrpB | Transcription, RNA processing and degradation | 0.83 | 0.09 | 2.22 | 0.45 | 2.61 | 0.34 | 2.9 | 5.2E-06* | 1-10 |
| fhuC | Transport and binding proteins | 0.12 | 0.03 | 0.34 | 0.06 | 0.18 | 0.01 | 2.1 | 1.1E-03 | 1-10 |
| entD | Biosynthesis of cofactors, prosthetic groups and carriers | 0.37 | 0.02 | 0.44 | 0.02 | 0.47 | 0.00 | 1.2 | 8.5E-04 | 10-100 |
| ybeA | Hypothetical, unclassified, unknown | 0.33 | 0.04 | 0.79 | 0.14 | 0.68 | 0.04 | 2.3 | 6.5E-04 | 1-10 |
| nagE | Transport and binding proteins | 2.48 | 0.68 | 9.54 | 3.21 | 6.44 | 2.31 | 3.2 | 5.5E-03 | 0.1-1 |
| fruB | Transport and binding proteins | 0.10 | 0.00 | 0.47 | 0.03 | 0.35 | 0.13 | 4.3 | 7.6E-04 | 1-10 |
| exbD | Transport and binding proteins | 0.13 | 0.02 | 0.51 | 0.04 | 0.31 | 0.05 | 3.1 | 2.1E-03 | 10-100 |
| deaD | Transcription, RNA processing and degradation | 0.59 | 0.05 | 3.20 | 0.36 | 2.82 | 1.33 | 5.1 | 6.7E-05* | 10-100 |
| bioH | Biosynthesis of cofactors, prosthetic groups and carriers | 0.36 | 0.06 | 0.93 | 0.26 | 1.01 | 0.08 | 2.7 | 1.7E-04 | 1-10 |
| yhiP | Transport and binding proteins | 3.79 | 0.40 | 21.87 | 3.65 | 12.18 | 3.64 | 4.5 | 3.4E-03 | 0.1-1 |
| uhpC | Transport and binding proteins | 0.09 | 0.03 | 0.28 | 0.05 | 0.22 | 0.03 | 2.8 | 4.0E-04 | 10-100 |
| uhpB | Transport and binding proteins | 0.01 | 0.00 | 0.15 | 0.06 | 0.12 | 0.01 | 9.3 | 5.9E-04 | 10-100 |
| yiiT | Putative regulatory proteins | 17.04 | 0.39 | 11.59 | 0.02 | 11.91 | 0.18 | 0.7 | 5.3E-04 | 0.1-1 |
Genes are listed as in Table S1 according to their physical position on the K12-MG1655 chromosome. For in vitro grown bacteria, values were normalized on dinB expression level. For each gene, ratio value corresponds to the EIEC/Shigella expression level mean to the EDL933 expression level. Student t test P values were calculated between the EDL933 and the EIEC/Shigella expression levels. (*) Genes for which RT-PCR was repeated six times. For other genes, RT-PCR was repeated three to four times. For in vivo grown bacteria, expression levels are given as a range of magnitude of the expression level ratio between Shigella strain either infecting eukaryotic cells or in vitro growing. All genes were found overexpressed in EIEC/Shigella strains except yiiT gene, which is underexpressed in these strains, as observed in the macroarray analysis. The genes involved in iron acquisition process are indicated in bold.
We explored whether the sequence divergence in the different promoter regions (as defined in Salgado et al. 2001) can predict how a gene is differentially expressed between the two groups of strains. Phylogenetic analysis of the concatenated promoters obtained from the 10 complete genome sequences (see above) of the 113 differentially expressed genes did not find any evidence for convergence, but showed a tree similar to the species tree (data not shown). These data are in agreement with a recent genetic analysis of genome-wide variation in human gene expression showing that trans-acting loci (and not cis) regulate the majority of variation in the expression levels of genes (Morley et al. 2004).
In vivo relevance
Shigella/EIEC, in contrast to the other E. coli strains studied, are obligatory intracellular pathogens, and have developed numerous strategies to survive and multiply within host cells (Parsot and Sansonetti 1996). Following initial cell invasion by a micropinocytotic process, these organisms lyse the endocytic vacuole, thereby gaining access to nutrients in the cytoplasm. Little is known about the intracellular growth environment with respect to the availability of specific nutrients, but in order to multiply intracellularly, the bacteria must compete successfully with the host for all essential nutrients, including iron. In this regard, we performed the transcriptome analysis using bacteria in log-phase growthat37°C in a nutrient-rich culture medium. Several observations suggest, however, that gene expression by such cultured bacteria shares features with that of invasive organisms in vivo. First, Shigella in log-phase growth are 10 times more invasive than organisms that have reached the stationary phase (Mounier et al. 1997), and it has been shown that genes required for invasion are expressed by Shigella and EIEC in log-phase growth at 37°C (Maurelli and Sansonetti 1988; Falconi et al. 1998; Le Gall et al. 2005). Second, the intracellular doubling time of Shigella is about 40 min (Demers et al. 1998), similar to that observed for bacteria grown in vitro in the enriched medium used by us, indicating that the growth rate of intracellular organisms is high.
In order to document the fact that the genes we found differentially expressed in log-phase bacteria grown in rich medium between Shigella/EIEC and the remaining E. coli strains are transcribed in vivo in Shigella, real-time RT-PCR was performed during infection of intestinal epithelial cell lines by the S. flexneri M90T (S3) strain. All of the 20 selected genes (Supplemental Table S2) are expressed in the early stages of infection. Reported to the number of bacteria, the ratio of expression between Shigella strain infecting either eukaryotic cells or growing in 869 ranges from 10-1 to 102 (Table 1). Interestingly, two of the five genes exhibiting the higher ratio (exbG and entD) are involved in iron metabolism.
The genetic information encoded by the 200-kb virulence plasmid is essential for the intracellular pathogenicity of Shigella/EIEC (Buchrieser et al. 2000). Nevertheless, the convergence in both the pattern of gene inactivation (Pupo et al. 2000; Escobar-Paramo et al. 2003; this work) and the pattern of gene expression in these phylogenetically distinct organisms attests to an important role for the chromosomal background in pathogenicity. Interestingly, it has been shown that inactivation of a single gene tends to increase variation in the expression of the remaining genes, accelerating the adaptation to a new phenotypic optimum (Bergman and Siegal 2003). It can be hypothesized that gene inactivation in Shigella/EIEC may have generated polymorphism in gene expression that served as a basis for subsequent selection.
Conclusions
There is now accumulating evidence that polymorphism in the pattern of gene expression is a widespread phenomenon, and can be observed in bacteria (this work), yeast (Townsend et al. 2003), mice (Schadt et al. 2003), and humans (Yan et al. 2002). It appears that multiple forces may shape this polymorphism. For example, the pattern of gene expression is under positive selection in human brain (Enard et al. 2002), in the camera eye of octopus and human (Ogura et al. 2004), in teleost fish (Oleksiak et al. 2002), and is sex dependant in Drosophila (Ranz et al. 2003). Cooper et al. (2003), studying the evolution of E. coli in glucoselimited medium, observed the convergence in gene expression in 12 independent lineages after 20,000 generations, suggesting that environment selection is a determinant of polymorphism in gene expression. Our study extends this observation to a nonlaboratory setting and on an evolutionary scale by demonstrating that a lifestyle associated with intracellular virulence can select for a particular pattern of gene expression.
Methods
Bacterial strains
A total of 10 human-isolated strains, each representative of the major E. coli/Shigella phylogenetic groups, were considered in this study, including one strain of each of the four major E. coli phylogenetic groups from the E. coli reference (ECOR) collection (Herzer et al. 1990) (A: ECOR1, B1: ECOR26, D: ECOR50, B2: ECOR56), the EDL933 strain, which is an O157:H7 enterohemoragic E. coli (EHEC) completely sequenced (Perna et al. 2001) and belonging to the minor E phylogenetic group (Escobar-Paramo et al. 2004a), one strain of each of the four major Shigella phylogenetic groups (S1: S. boydii serotype 10 [SB1080], S3: S. flexneri serotype 5 [M90T], SD1: S. dysenteriae serotype 1 [SD0177], SS: S. sonnei: [SS92a]) and one EIEC strain (EIEC85b) (Escobar-Paramo et al. 2003). ECOR strains were isolated in commensal conditions, except for the ECOR50 strain, which originated from a urinary tract infection. In addition, E. coli K12-MG1655 strain (Blattner et al. 1997) from which the ORFs were PCR amplified and spotted on the macroarrays (see below) was used as the control strain. This strain is a laboratory-adapted strain isolated in commensal conditions and belongs to the A phylogenetic group. A strain of E. fergusonii, which is the closest species to E. coli (Lawrence et al. 1991), was used as the outgroup in the phylogenetic analyses.
DNA and RNA extraction and labeling
Cells were grown in 869 medium at 37°C with constant aeration. Genomic DNA was isolated from an overnight culture (2 × 109 bacteria) using the Wizard Genomic DNA Purification Kit (Promega). A total of 500 ng of DNA were labeled by random priming (Roche Diagnostics) using [α33P]dCTP. Total RNA was isolated from cells grown at mid-log phase using RNAplus (Qbiogen) and Nucleospin RNA II kit (Macherey Nagel), which includes a DNAse treatment step. cDNA synthesis was performed from 20 μg of total RNA using random hexamer primers, Superscript II reverse transcriptase (Invitrogen) and [α33P]dCTP.
Macroarray hybridizations and data acquisition
DNA filter arrays (Panorama E. coli gene arrays) spotted with duplicate copies of each of the 4290 E. coli K12-MG1655 ORFs were obtained from Sigma-Genosys Biotechnologies and used for both DNA and RNA analyses. Hybridizations were carried out for 15-18 h at 68°C in the ExpressHyb Hybridization solution (Clontech). Each filter was then rinsed with 0.5 SSPE/0.2% SDS at room temperature for 3 min, three times, followed by three washes in the same solution at 65°C for 20 min each. The filters were exposed to a PhosphorImager screen (AGFA ADC plate MD30) for 48-60 h and scanned. Filters were stripped as described by the manufacturer and checked for efficient dehybridization. A commercial software package obtained from COSE Inc. (XDotsReader) was used to grid the phosphorimaging image and to record the pixel densities. The output data were exported to a Microsoft Excel spreadsheet for subsequent manipulations.
DNA hybridizations were repeated two and three times with different DNA preparations from M90T and K12-MG1655 strains, respectively, and then performed one time for each strain. cDNA hybridizations were repeated two to six times, each time with a different RNA preparation.
Macroarray data processing and statistical analyses
The average background signal was quantified on nonspotted zones and subtracted from the intensity obtained for each spot. Standardization was performed expressing intensity values of individual spots as a fraction of the overall intensity of the 4290 ORFs on the membrane. As an excellent correlation between the intensity of the duplicated spots was always observed, the mean of these standardized values was then used. Pairwise correlations between standardized data of each individual experiment, including new nucleic acid extraction and hybridization for a same strain, were determined to assess reproducibility. As these pairwise correlations were always higher than 0.978, values from repeated experiments were averaged and then used for the analyses detailed below.
DNA analysis
An ORF was recorded as undetectable if (1) its signal was less than or equal to the background signal, or (2) the ratio of the reference strain K12-MG1655 signal against that of the studied strain was ≥1.25. This threshold has been chosen as giving the best trade-off between sensitivity and specificity using the EDL933 hybridization experiment data compared with the complete genome sequence data (Perna et al. 2001).
RNA analysis
The RNA data, as well as the DNA data, were then normalized by transformation in a gaussian distribution (Maclean et al. 1976; Supplemental Fig. S3A). Since normalized DNA (d) and RNA (r) values were correlated (coefficients of correlation between 0.723 for E. fergusonii and 0.865 for SS) (Supplemental Fig. S3B), we defined an adjusted RNA value (r′), independent of the DNA value, as r′ =(r-f(d))/s, where f(d) was the linear regression function of r on d and s the standard deviation of the residuals.
All of the raw and processed data obtained in this study are available in tabular format at www.genome.org.
Phylogenetic analyses
DNA analysis
Phylogenetic reconstruction of the strain evolutionary history was performed by simultaneous analysis of the DNA sequences of 11 housekeeping genes (Escobar-Paramo et al. 2004b), extracted from GenBank, using Neighbor Joining (NJ) algorithm. The macroarray DNA data were binary coded (“0” for undetectable genes, “1” for genes present at least in one copy) and used to reconstruct a phylogenetic tree by parsimony, using PAUP4.0b (Swofford 2002) and branch and bound algorithm. Both trees have been rooted on E. fergusonii.
RNA analysis
The unrooted phylogenetic tree of the adjusted RNA values was obtained following the NJ algorithm, and using the euclidian distance matrix, in which the distance d(ij) between two strains i and j is the square root of the sum over all genes of the squared difference between adjusted RNA of the strains i and j.
For all trees, bootstrap proportions were calculated from 1000 iterations. Quartet method (Estabrook 1992) and symmetric difference between trees (Swofford 2002) were used for comparison between the three tree topologies (based on DNA sequences, binary-coded DNA, and adjusted RNA data sets). In addition, Mantel test (Legendre and Lapointe 2005) was used to compare the three corresponding distance matrixes.
Eukaryotic cell culture and infection
Infection of cultured epithelial cells by the S. flexneri strain M90T (S3) was performed as described (Pedron et al. 2003). Human intestinal epithelial Caco-2 cells derived from a colonic carcinoma were grown in an incubator at 37°C, 10% CO2, in Dulbecco's modified Eagle's medium supplemented with 10% decomplemented fetal calf serum, 1% nonessential amino acid, and penicillin and streptomycin at 100 U/mL and 100 μg/mL, respectively. Before infection, nonconfluent cell cultures grown in 10-cm diameter Petri dishes were washed in Dulbecco's modified Eagle's medium without serum, and incubated at 37°C for 2 h in the same medium. Bacteria harvested in exponential phase of growth in TCS medium and resuspended in Dulbecco's modified Eagle's medium were used to infect cells at a multiplicity of infection of 100 bacteria/cell. After a 15-min centrifugation at 2000 rpm, infected cells were incubated for 30 min at 37°C, washed twice in Dulbecco's modified Eagle's medium, and incubated for 150 min at 37°C in Dulbecco's modified Eagle's medium supplemented with 50 μg/mL gentamicin to kill extracellular bacteria. Infected cells were either (1) lysed in the presence of PBS containing 0.1% sodium deoxycholate and dilutions of the lysate were plated to calculate the number of intracellular bacteria; (2) fixed with ethanol and stained with Giemsa to visualize the infected cells; (3) or lysed by addition of RNAplus and processed for RNA extraction as above. A noninfected pool of cells processed as above but without infection was performed as control.
Real-time quantitative RT-PCR
The expression of a selected panel of 20 genes was studied by real-time PCR in three representative strains (EDL933, EIEC, and SS) grown in 869 medium at mid-log phase. To eliminate the effects of sequence divergence between the strains that can interfere in the assay, primers were designed in conserved regions of the 10 E. coli/Shigella complete genome sequences (see above), including the EDL933 and S. sonnei strains (Supplemental Table S2). cDNA synthesis was performed from 3 μg of total RNA using random hexamer primers, Superscript II reverse transcriptase (Invitrogen) in a final volume of 20 μL, then expanded to 200 μL at the end of the reaction. PCR was performed in a final volume of 25 μL with 5 μL of the cDNA reaction according to the manufacturer's protocol with the SYBR GREEN PCR Master Mix (Applied Biosystems) and analyzed on an ABI PRISM 7700 sequence detector (Applied Biosystems). Cycling conditions were as follows: initial step at 50°C for 2 min, followed by a denaturation at 95°C for 10 min, and 40 cycles of 95°C for 15 sec, 60°C for 60 sec. For quantification of RT-PCRs, serial dilutions of pure E. coli K12-MG1655 DNA (corresponding to 105 to 102 genome copies per 5 μL) were used. The number of copies of each transcript was then determined with the aid of the SDS 1.9 software (Applied Biosystems). DNA contamination of the RNA samples was ruled out by verifying the absence of significant signal in the real-time PCR assay performed from a RT reaction without reverse transcriptase. Normalization for all results was performed with a second quantification for a gene (dinB) expressed in the mid-log phase at a constant rate in a collection of 25 natural isolates representing the diversity of the species (this work; B. Gérard and E. Denamur, pers. comm.). Similar results were obtained when another gene (yjaD) harboring these properties was used for normalization (data not shown). Result reproducibility was first assessed on two genes (deaD and hrpB) by repeating six times the analysis from two independent RNA preparations, followed in each case by different RT reactions. Experiments were then repeated three to four times on independent RNA preparations, and the obtained values were averaged. Differences between strain expressions were analyzed using a Student t-test.
The expression level of this panel of 20 genes was also studied during the infection of cultured epithelial cells by the S. flexneri M90T (S3) strain in two independent experiments. As a control, noninfected cells were also studied. The number of intracellular bacteria used for each RNA extraction was about 4 × 107 bacteria. The absence of DNA contamination was checked as above. Each RNA preparation was analyzed twice by RT-PCR as above. A good reproducibility, assessed by small standard deviation values, was observed, and the results were averaged. The level of expression was estimated by comparing the number of copies of each transcript in in vivo-infecting condition to the number of copies in in vitro condition, normalized on the number of bacteria. Noninfected cells did not give any detectable signal by real-time PCR for all the tested genes.
Acknowledgments
We thank Claude Parsot and Olivier Tenaillon for the numerous idea exchanges that we had all over this work and Allan Hance for his valuable critical reading of the manuscript. We are grateful to Bénédicte Gérard, Agnès Bourillon, and Olivier Clermont for their help in the real-time quantitative RT-PCR. Our work was partially granted by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires-MENRT and the Fondation pour la Recherche Médicale.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: B. Gérard, G. Lecointre, and E. Denamur.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2405905.
References
- Bergman, A. and Siegal, M.L. 2003. Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549-552. [DOI] [PubMed] [Google Scholar]
- Bjedov, I., Lecointre, G., Tenaillon, O., Vaury, C., Radman, M., Taddei, F., Denamur, E., and Matic, I. 2003. Polymorphism of genes encoding SOS polymerases in natural populations of Escherichia coli. DNA Repair 2: 417-426. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. [DOI] [PubMed] [Google Scholar]
- Braun, V. and Braun, M. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529: 78-85. [DOI] [PubMed] [Google Scholar]
- Buchrieser, C., Glaser, P., Rusniok, C., Nedjari, H., D'Hauteville, H., Kunst, F., Sansonetti, P., and Parsot, C. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38: 760-771. [DOI] [PubMed] [Google Scholar]
- Cooper, T.F., Rozen, D.E., and Lenski, R.E. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. 100: 1072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers, B., Sansonetti, P.J., and Parsot, C. 1998. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 17: 2894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur, E., Lecointre, G., Darlu, P., Tenaillon, O., Acquaviva, C., Sayada, C., Sunjevaric, I., Rothstein, R., Elion, J., Taddei, F., et al. 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103: 711-721. [DOI] [PubMed] [Google Scholar]
- Dobrindt, U., Agerer, F., Michaelis, K., Janka, A., Buchrieser, C., Samuelson, M., Svanborg, C., Gottschalk, G., Karch, H., and Hacker, J. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185: 1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg. 2002. Escherichia coli. Virulence mechanisms of a versatile pathogen. Academic press, Elsevier Science, Baltimore, MD.
- Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340-343. [DOI] [PubMed] [Google Scholar]
- Escobar-Paramo, P., Giudicelli, C., Parsot, C., and Denamur, E. 2003. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J. Mol. Evol. 57: 140-148. [DOI] [PubMed] [Google Scholar]
- Escobar-Paramo, P., Clermont, O., Blanc-Potard, A.B., Bui, H., Le Bouguenec, C., and Denamur, E. 2004a. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21: 1085-1094. [DOI] [PubMed] [Google Scholar]
- Escobar-Paramo, P., Sabbagh, A., Darlu, P., Pradillon, O., Vaury, C., Denamur, E., and Lecointre, G. 2004b. Decreasing the effects of horizontal gene transfer on bacterial phylogeny: The Escherichia coli case study. Mol. Phyl. Evol. 30: 243-250. [DOI] [PubMed] [Google Scholar]
- Estabrook, G.F. 1992. Evaluating undirected positional incongruence of individual taxa between two estimates of the phylogenetic tree for a group of taxa. Syst. Biol. 41: 172-177. [Google Scholar]
- Falconi, M., Colonna, B., Prosseda, G., Micheli, G., and Gualerzi, C.O. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17: 7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukiya, S., Mizoguchi, H., Tobe, T., and Mori, H. 2004. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 186: 3911-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, S.Y., Scholle, M.D., Campbell, J.W., Balazsi, G., Ravasz, E., Daugherty, M.D., Somera, A.L., Kyrpides, N.C., Anderson, I., Gelfand, M.S., et al. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185: 5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. and Gascuel, O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696-704. [DOI] [PubMed] [Google Scholar]
- Harvey, P.H. and Pagel, M.D. 1991. The comparative method in evolutionary biology. Oxford University Press, Oxford, UK.
- Herzer, P.J., Inouye, S., Inouye, M., and Whittam, T.S. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172: 6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Kido, N., Arakawa, Y., Ohta, M., Sugiyama, T., and Kato, N. 1991. Possible mechanisms underlying the slow lactose fermentation phenotype in Shigella spp. Appl. Environ. Microbiol. 57: 2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q., Yuan, Z., Xu, J., Wang, Y., Shen, Y., Lu, W., Wang, J., Liu, H., Yang, J., Yang, F., et al. 2002. Genome sequence of Shigella flexneri 2a: Insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30: 4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J.G. and Ochman, H. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. 95: 9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J.G., Ochman, H., and Hartl, D.L. 1991. Molecular and evolutionary relationships among enteric bacteria. J. Gen. Microbiol. 137: 1911-1921. [DOI] [PubMed] [Google Scholar]
- Lecointre, G., Rachdi, L., Darlu, P., and Denamur, E. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15: 1685-1695. [DOI] [PubMed] [Google Scholar]
- Le Gall, T., Mavris, M., Martino, M.C., Bernardini, M.L., Denamur, E., and Parsot, C. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology (in press). [DOI] [PubMed]
- Legendre, P. and Lapointe, F.-J. 2005. Assessing the congruence among distance matrices: Single malt Scotch whiskies revisited. Australian and New Zealand J. Stat. (in press).
- Lemon, B. and Tjian, R. 2000. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 14: 2551-2569. [DOI] [PubMed] [Google Scholar]
- Lloyd, G., Landini, P., and Busby, S. 2001. Activation and repression of transcription initiation in bacteria. Essays Biochem. 37: 17-31. [DOI] [PubMed] [Google Scholar]
- Maclean, C.J., Morton, N.E., Elston, R.C., and Yee, S. 1976. Skewness in commingled distributions. Biometrics 32: 695-699. [PubMed] [Google Scholar]
- Maurelli, A.T. and Sansonetti, P.J. 1988. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc. Natl. Acad. Sci. 85: 2820-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli, A.T., Fernandez, R.E., Bloch, C.A., Rode, C.K., and Fasano, A. 1998. “Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. 95: 3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, B.A., Fernandez, M.I., Siber, A.M., and Maurelli, A.T. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell Microbiol. 1: 143-155. [DOI] [PubMed] [Google Scholar]
- Morley, M., Molony, C.M., Weber, T.M., Devlin, J.L., Ewens, K.G., Spielman, R.S., and Cheung, V.G. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier, J., Bahrani, F.K., and Sansonetti, P.J. 1997. Secretion of Shigella flexneri Ipa invasins on contact with epithelial cells and subsequent entry of the bacterium into cells are growth stage dependent. Infect. Immun. 65: 774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H. and Jones, I.B. 2000. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 19: 6637-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, A., Ikeo, K., and Gojobori, T. 2004. Comparative analysis of gene expression for convergent evolution of camera eye between octopus and human. Genome Res. 14: 1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiak, M.F., Churchill, G.A., and Crawford, D.L. 2002. Variation in gene expression within and among natural populations. Nat. Genet. 32: 261-266. [DOI] [PubMed] [Google Scholar]
- Parsot, C. and Sansonetti, P.J. 1996. Invasion and the pathogenesis of Shigella infections. Curr. Top. Microbiol. Immunol. 209: 25-42. [DOI] [PubMed] [Google Scholar]
- Pedron, T., Thibault, C., and Sansonetti, P.J. 2003. The invasive phenotype of Shigella flexneri directs a distinct gene expression pattern in the human intestinal epithelial cell line Caco-2. J. Biol. Chem. 278: 33878-33886. [DOI] [PubMed] [Google Scholar]
- Perna, N.T., Plunkett III, G., Burland, V., Mau, B., Glasner, J.D., Rose, D.J., Mayhew, G.F., Evans, P.S., Gregor, J., Kirkpatrick, H.A., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529-533. [DOI] [PubMed] [Google Scholar]
- Pupo, G.M., Lan, R., and Reeves, P.R. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. 97: 10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J.M., Castillo-Davis, C.I., Meiklejohn, C.D., and Hartl, D.L. 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300: 1742-1745. [DOI] [PubMed] [Google Scholar]
- Reid, S.D., Herbelin, C.J., Bumbaugh, A.C., Selander, R.K., and Whittam, T.S. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406: 64-67. [DOI] [PubMed] [Google Scholar]
- Salgado, H., Santos-Zavaleta, A., Gama-Castro, S., Millan-Zarate, D., Diaz-Peredo, E., Sanchez-Solano, F., Perez-Rueda, E., Bonavides-Martinez, C., and Collado-Vides, J. 2001. RegulonDB (version 3.2): Transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29: 72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt, E.E., Monks, S.A., Drake, T.A., Lusis, A.J., Che, N., Colinayo, V., Ruff, T.G., Milligan, S.B., Lamb, J.R., Cavet, G., et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297-302. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. 2002. PAUP* phylogenetic analyses using parsimony. Sinauer Associates, Sunderland, MA.
- Townsend, J.P., Cavalieri, D., and Hartl, D.L. 2003. Population genetic variation in genome-wide gene expression. Mol. Biol. Evol. 20: 955-963. [DOI] [PubMed] [Google Scholar]
- Wei, J., Goldberg, M.B., Burland, V., Venkatesan, M.M., Deng, W., Fournier, G., Mayhew, G.F., Plunkett III, G., Rose, D.J., Darling, A., et al. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71: 2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, R.A., Burland, V., Plunkett III, G., Redford, P., Roesch, P., Rasko, D., Buckles, E.L., Liou, S.R., Boutin, A., Hackett, J., et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. 99: 17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, C.W., Cziko, A.M., and Robinson, G.E. 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302: 296-299. [DOI] [PubMed] [Google Scholar]
- Yan, H., Yuan, W., Velculescu, V.E., Vogelstein, B., and Kinzler, K.W. 2002. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- www.genome.wisc.edu; genome.gen-info.osaka-u.ac.jp/bacteria/o157 and www.sanger.ac.uk/cgi-bin/blast/submitblast/escherichia_shigella; These Web sites provide the complete genome sequences of the ten E. coli/Shigella strains studied in silico in this work.