Abstract
Large-scale genetic studies are highly dependent on efficient and scalable multiplex SNP assays. In this study, we report the development of Molecular Inversion Probe technology with four-color, single array detection, applied to large-scale genotyping of up to 12,000 SNPs per reaction. While generating 38,429 SNP assays using this technology in a population of 30 trios from the Centre d'Etude Polymorphisme Humain family panel as part of the International HapMap project, we established SNP conversion rates of ∼90% with concordance rates >99.6% and completeness levels >98% for assays multiplexed up to 12,000plex levels. Furthermore, these individual metrics can be “traded off” and, by sacrificing a small fraction of the conversion rate, the accuracy can be increased to very high levels. No loss of performance is seen when scaling from 6,000plex to 12,000plex assays, strongly validating the ability of the technology to suppress cross-reactivity at high multiplex levels. The results of this study demonstrate the suitability of this technology for comprehensive association studies that use targeted SNPs in indirect linkage disequilibrium studies or that directly screen for causative mutations.
Complex human diseases are known to have a significant genetic component. Despite some important successes (Altshuler et al. 2000; Hugot et al. 2001), the elucidation of the underlying genetic determinants have proven resistant to standard methods. Linkage analysis using affected sib pairs has limited power to uncover such signals, as each individual functional variant contributes only modestly to disease risk (Risch 2000). Large-scale association studies that would allow genome-wide mapping in large collections of cases with matched controls have therefore been proposed (e.g., Kruglyak 1999; Risch 2000). Some of the barriers to adopting such strategies, including the need to establish large-case control populations (Geschwind et al. 2001; Shmulewitz et al. 2001; Cupples et al. 2003) and develop comprehensive SNP resources (dbSNP), have been overcome. In addition, the International HapMap project (The International HapMap Consortium 2003) is completing a first draft of a whole genome haplotype map in the Centre d'Etude du Polymorphisme Humain (CEU) population by the end of 2004. The Hap-Map effort will yield a broad view of the genetic architecture of human populations and allow for the efficient selection of the most informative tagging SNPs for subsequent association studies.
The remaining requirement to fully enable large-scale genetic association studies is the development of truly cost-effective and scalable SNP genotyping technologies. These methods must allow hundreds of thousands of markers to be efficiently and accurately scored in thousands of patients. The first generation of SNP genotyping technologies were based on single amplification reactions for each locus and were not appropriate for these largescale, whole-genome studies. Recent advances have assayed thousands of random SNPs using ultra high-density wafer hybridizations (Matsuzaki et al. 2004) but will not allow the advantages of the tagging SNP approach to be fully realized because these technologies will not be able to convert all the informative tagging SNPs and/or putative functional SNPs into working genotyping assays. Multiplexed genotyping technologies that allow the multiplexing of hundreds of targeted SNPs have recently been developed based on various versions of the Oligo Ligation Assay (OLA) (Grossman et al. 1994; Samiotaki et al. 1994; Oliphant et al. 2002). These technologies would still demand a fairly large infrastructure in order to process the thousands of reactions necessary per patient.
Here, we describe an advanced Molecular Inversion Probe (MIP) genotyping technology (Hardenbol et al. 2003) that combines the strengths of all of the above methods. MIP exploits the advantages of the OLA methodologies but can be more highly multiplexed due to a unique unimolecular method of action and an enzymology that combines the specificity of both ligase and polymerase enzymes. This assay enables the use of >12,000 oligo probes to simultaneously interrogate human genomic DNA and, following a single PCR, detect the results via a single universal tag DNA chip array. The application of MIP to generate a haplotype map of Chromosome 12 in CEU families (The International HapMap Consortium 2003) as part of the Human HapMap project has yielded 3,509,052 genotypes from 38,429 assays. In this study we analyze the important features of the assay performance that will enable MIP to be effectively used for many future largescale studies.
Results
MIP technology
The basic concept of MIP technology has been described previously (Hardenbol et al. 2003). Briefly, a single oligonucleotide probe with recognition sequences at each terminus is hybridized with a genomic target sequence such that it forms a circular structure, with the ends of the probe abutting. This leaves a single base gap at the location of a SNP. This gapped-duplex is then tested in four separate reactions, each with a single dNTP species present, in which successful polymerization/ligation provides allelic differentiation. The probes are subsequently released from the genomic DNA and those that have been covalently circularized in the correct allele/nucleotide reaction combinations are amplified using a “universal” PCR primer pair. Each amplified probe contains a unique tag sequence that is complementary to a sequence on the universal tag array. Tags have been selected to have a similar Tm and base composition and to be maximally orthogonal in sequence complementarity. Amplicons are fluorescently labeled and the tag sequences released from the genome homology regions using a restriction endonuclease treatment. The tags are then detected using a complementary tag array.
Multiplex tag detection
We have greatly increased the complexity of the initial MIP assay while maintaining the genotyping accuracy. A major limitation in the use of conventional nucleic acid microarrays for genotyping or gene expression is the difficulty in controlling the specificity and uniformity of hybridization of biological sequences to their complementary oligonucleotides on the array. This leads to high levels of cross-hybridization and spurious signals in complex mixtures. Therefore, previously reported multiplex SNP genotyping assays (Matsuzaki et al. 2004) that use biological sequences in detection function only for detection of a small fraction of the total possible SNP sequences in the genome.
We have used a molecular tagging (or barcoding) strategy (Shoemaker et al. 1996), to enable very highly multiplexed assays. In the current study, a set of 20,000 sequences were developed as recognition probes, with minimal sequence similarity within the set. In addition, the tags were designed with a narrow range of melting temperatures and to avoid secondary sequence structure. It was anticipated that these sequences would exhibit minimal cross-hybridization properties, while maintaining a tight dynamic range that would ensure the highest accuracy at this stage of the assay detection scheme.
The performance of the 20,000 new tags was first compared to a publicly available set of ∼16,000 tag sequences present on the Tag3 GeneChip barcode arrays (Affymetrix). First, standard MIP genotyping was performed using a 6615plex probe pool containing tags from the Tag3 set, and the assay products were analyzed by two-color detection on Tag3 arrays (see below). Specificity was evaluated by determining the number of noncomplementary (or inactive) sequences on the array (n = 9666) having signal >10% of the average signal level of the active array sequence features. On average, each signal channel had 96/9666 (=1.0%) inactive features that resulted from a >10% cross-hybridization signal.
Next, the experiment was repeated using the set of 20,000 21mers that we designed. Two arrays were designed using these sequences; one with 6,000 tags (TrueTag 5k array) and another with 12,000 tags (TrueTag 10k array). Performing the same experiment described above on a TrueTag 10k array using a probe pool containing 6072 probes led to 18 nonactive features with signals >10% of the average active signal; a total of 0.3% of all features. Thus, the new tag set exhibits a three-fold higher specificity than previous sets. This added specificity has allowed us to obtain high accuracy genotyping data at 12,000plex (see below).
Cluster analysis
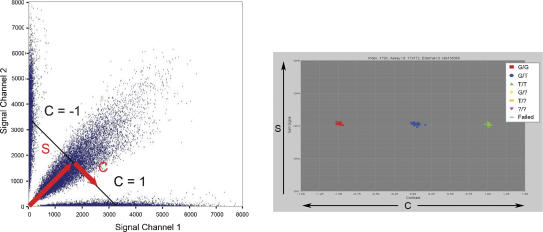
MIP data analysis has been improved by the development of a robust data clustering algorithm. In previous studies using the MIP technology, genotypes were processed from the raw data using a simple thresholding process based on the ratio of signals in the two expected allele channels, with data being rejected based on the presence of high background signals in the other channels. In this study, the new clustering algorithm declares genotypes by identifying regions of high data density that represent the three possible genotypes using an Expectation-Maximization (E-M) algorithm. This procedure allows entire data sets of thousands of markers to be analyzed without the need for any human intervention on a marker-by-marker basis. The algorithm works in the one-dimensional space of contrast between the different allele signals (Fig. 1). It assigns a probability for each data point (sample) to belong to a given cluster according to the probability distribution of the given cluster. Based on empirical observations, the shapes of the cluster probability distributions have been modeled by Gaussian distributions with non-Gaussian tails.
Figure 1.
Calling genotypes based on cluster analysis of raw data. Each SNP in a multiplex assay results in four fluorescent signal values: two for the two expected allele channels and two in background channels. Plotting the signal channels against each other (left) results in the formation of three clusters. The plot on the left shows 50,000 data points across several thousand markers. In order to decouple the overall signal of the particular data point from the contrast between the different allele signals, it is helpful to transform the data into a different space in which the sum of the signals in both channels (S) is plotted on the y-axis and the projection of the individual data point onto the line of constant S (the contrast value C) is plotted on the x-axis. The values of C range from -1 to 1 such that a value of -1 or 1 means signal in only one of the two channels while a value of 0 means equal signal in each channel. A one-dimensional E-M algorithm can then be used to find the clusters of homozygous and heterozygous calls. The colors have been automatically added by the cluster calling algorithm, which has identified the three clusters.
The overall behavior of the algorithm is controlled by parameters that are used to reject unreliable data at two different levels. The first level is to reject a given data point (sample) within the context of a given marker. Here, the main criterion is the relative probability of the data point belonging to its primary cluster versus belonging to its secondary cluster. Data points where this ratio is too low are considered ambiguous and hence not called. In addition each data point is required to have a minimum value for signal (sum of allele signals) with respect to chip noise and also with respect to nonallele signal (sum of nonallele signals).
The second level of discrimination is provided when the algorithm also rejects markers as a whole whenever a marker is deemed unreliable. The criteria here are based on the relative dispersion of the clusters, e.g., markers with loosely defined clusters are rejected. Since we do not use trio discordance (non-Mendelian inheritance) as a measure of marker reliability, we are able to use this measure as an unbiased estimate of the algorithm's overall accuracy. It is important to note that all of the fixed parameters that control the algorithm are fixed for the entire data set of x thousand markers times x hundred samples that is being fit at one time. Hence trio discordance is a reliable measure of the accuracy of the entire data set.
Multicolor detection
The current advanced MIP assay uses four-color chip detection. The methodology as originally reported used single color chip detection and four different arrays to score the four possible alleles in a 1500plex assay (Hardenbol et al. 2003). Next, an intermediate two-color detection method was developed. In that version, two fluorophores were used: one for A,C another for G,T. The A and G channels are then pooled and hybridized to one chip while the C and T channels are pooled and hybridized to a second chip. This intermediate method imparts some of the benefits of the four-color assay but necessitates two arrays.
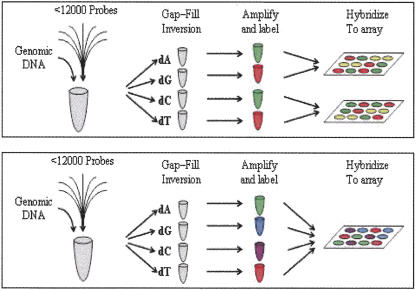
A more streamlined analysis is now enabled by labeling each of the four allele specific reactions with a spectrally distinct fluorophore (Fig. 2). These reactions can then be pooled together and hybridized to a single tag array. Fluorescent images are collected using four different filters to collect emission from each single fluorescent species such that the intensities from each of the allelic reactions can be measured from a single chip feature. In addition to significant chip cost savings, this method also has the advantage of rendering the genotype calls resistant to possible feature-to-feature variation in the arrays. A further benefit is that signal ratios between alleles, which are critical to accurate genotyping, are rendered insensitive to feature saturation that can otherwise lead to loss of linearity in response.
Figure 2.
Schematic of the MIP assay process. MIP reactions are set up adding an enzyme mix and genomic DNA to the probe pool. This mix is then split into four tubes, each receiving a distinct nucleotide species. After gap-filling and probe inversion, inverted probes are amplified using common PCR primers. These amplicons are labeled using one of two labeling processes. In the two-color labeling scheme (top), the A and C reactions are labeled with one fluorophore while the G and T reactions are labeled with a spectrally distinct fluorophore. The A and G reactions are then pooled and hybridized to one tag array while the C and T reactions pooled and hybridized to a second array. Both arrays are then scanned using a GeneChip array scanner in two spectral channels to generate four fluorescent signals for each tag. In the four-color labeling scheme (bottom), each of the four reactions is labeled with a spectrally distinct fluorophore. All four reactions are then pooled and hybridized to a single tag array which is scanned using a GeneChip AT CCD imager in four spectral bands. In both cases four images are generated containing the four allele signals for each SNP marker.
During this study, we tested both two-color and four-color assays and showed that the four-color method was superior. For all probe batches, a two-color protocol was implemented using two arrays per individual. For batch 4, a four-color protocol was also implemented using a CCD imager. Comparison of the data demonstrates that the four-color assay generates more accurate and more complete data (Table 1). Repeatability between two- and four-color data is 99.8%.
Table 1.
A summary of the genotyping for two representative batches of the HapMap study
| Batch | Detection scheme | Probes designed | Base-call parameters | Probes converted | Conversion | Repeatability | Trio accuracy | Completeness |
|---|---|---|---|---|---|---|---|---|
| 4 | 2 color | 6072 | 1 | 5469 | 90.1% | 99.5% | 99.6% | 98.0% |
| 6072 | 2 | 5127 | 84.4% | 99.6% | 99.7% | 99.3% | ||
|
|
6072
|
3
|
4706
|
77.5%
|
99.8%
|
99.9%
|
99.25%
|
|
| 4 color | 6072 | 1 | 5729 | 94.4% | 99.6% | 99.6% | 98.6% | |
| 6072 | 2 | 5416 | 89.2% | 99.6% | 99.7% | 99.7% | ||
| 6072 | 3 | 4984 | 82.1% | 99.9% | 99.96% | 99.5% | ||
| 6 | 2 color | 12234 | 1 | 10924 | 89.3% | 99.5% | 99.7% | 98.6% |
| 12234 | 2 | 10341 | 84.5% | 99.7% | 99.7% | 99.6% | ||
| 12234 | 3 | 9517 | 77.5% | 99.95% | 99.9% | 99.5% |
Batch 4 contains 6072 probes and was genotyped across the population using a two-color detection method and then repeated using a four-color detection method. Batch 6 contained 12,234 probes and was only genotyped using a two-color assay. The performance metrics are defined in the text. Results are shown for each data set with varying choices of base-calling parameters. “Parameter set 1” optimizes the conversion rate, “Parameter set 2” optimizes the completeness of the set of converted markers and “Parameter set 3” optimizes the concordance metrics as a surrogate for accuracy.
Performance metrics for genotyping
The ideal genotyping technology would identify any chosen base in the genome in all DNA samples with perfect accuracy. A series of performance metrics are now commonly applied to measure how close real data are to this ideal. “Conversion rate” is a measure of the SNPs in the genome that can be assayed and is a function of both the quality of the SNPs chosen and the technology used to score them. In addition, some SNPs can be rejected in silico, prior to probe synthesis. “Call rate” for a given marker is the percentage of DNA samples in a study whose genotype is successfully measured. For a set of converted SNPs, the percentage of total genotypes returned across all markers is defined as the “completeness” of the study. Finally, “accuracy” is the percentage of these genotypes that are correct. Ultimately, the quality of the genotype data is measured by the power that it provides in the elucidation of genetic associations. A high quality genotyping technology will have a power to find associations that closely approximates the power of the idealized technology.
We have evaluated the full set of performance metrics using this new MIP assay and produced 38,429 working probes targeting SNPs on Chromosome 12 in six batches for the HapMap project. Batches 1-3 were 6,000-8,000plex pools designed using the Tag3 tag set (Affymetrix). Batch 4 was a 6,000plex and the first set developed using the new tag set (TrueTag 5k), while Batch 6 was the first batch to be designed for over 10,000 SNPs using the expanded tag set (TrueTag 10k). As a result, these two representative probe batches were chosen to analyze the performance of the technology in detail. Each batch was genotyped on 95 samples consisting of 30 trios from the CEU collection and five repeated samples to measure repeatability. Batch 4 was repeated using the four-color detection system as described above. Table 1 summarizes the performance metrics of these two batches.
Conversion rates were high regardless of multiplex level or detection method. Here conversion rate is defined to be the fraction of probes per batch that yielded strong signals with discernable clusters to indicate the different genotypes. As mentioned previously, in silico design rates are relevant in assessing overall SNP yield. More than ∼84.7% of all SNPs passed design criteria in that they were unique in the genome. Importantly, cluster shape and signal were the only parameters that were used to select converted probes. Repeatability and concordance (see below) were not used to select for converted probes. As a result, the measures of accuracy shown below are not biased by conversion choices and represent the true accuracy of the method.
SNPs for which only one of two alleles can be detected, termed “allelic drop-outs,” are an intrinsically rare property of MIP since it uses a single probe per SNP. The allelic drop-out rate was measured to be only 0.4% of assays by comparison to publicly available data generated by other methods.
The call rate of a given probe is defined to be the number of genotypes that were unambiguously clustered divided by the total number of genotypes attempted across all individuals. Completeness is the average call rate of converted probes. Completeness is high and is again unaffected by multiplex level or detection method (Table 1).
Two measures of accuracy are possible for the data generated. The first is the data gathered from repeated samples. Such data allow random errors to be measured when the same marker gives nonconcordant data in repeated assays on the same individual. By this metric, the MIP probes performed with high accuracy. Repeatability ranges from 99.5%-99.9% across batches (Table 1).
The second measure of accuracy is trio concordance. The use of mother-father-child trios in this project was designed to allow the accuracy to be monitored by looking at the Mendelian inheritance patterns across markers (see Methods section for detailed description). This test is able to capture some forms of systematic error that cannot be estimated by repeatability. Again, the accuracy rates based on trio concordance indicate a high level of data accuracy (>99.6%). Because the criteria for assay conversion did not take into account the trio concordance rate or the repeatability rate, these measurements are good predictors of the overall accuracy in the data set.
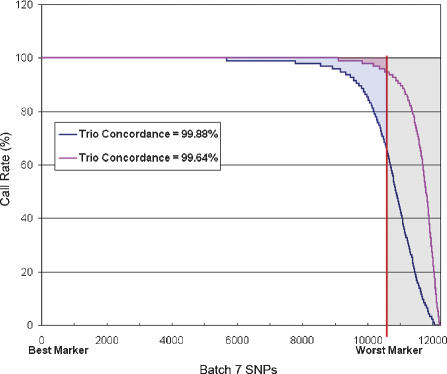
It is important to note that all of these parameters, including conversion rate, completeness, and concordance, are related through parameters that can be changed in the clustering algorithm used to assign genotypes. A permissive algorithm can result in high levels of completeness and conversion at the expense of accuracy. In general, these changes are seen at the margin between the bulk of the probes that are very complete with high accuracy and the probes that fail to call. This is shown in Figure 3. Batch 6 is shown ordered along the x-axis such that the probe with the highest call rate is plotted at the origin while the probe with the lowest call rate is at the right. The call rate for each probe across the full sample set is then shown for two different choices of cluster parameters: A stringent set, which accepts only calls very clearly in good clusters, and a more permissive set, which accepts data at the periphery of clusters. What can be seen is that the amount of missing data can be decreased at the cost of making a small number of increased errors.
Figure 3.
The effect of clustering parameters on performance metrics. In this plot, the markers for Batch 6 are ordered along the x-axis such that the marker with the highest call rate is at the origin, while the worst performing of the ∼12,000 markers is at the right. The y-axis shows the call rate for each of these markers across 95 individuals. The markers that exhibit poor call rates are called nonconverted and are shown in the gray area. The red curve shows a choice of cluster calling parameters that emphasizes high completeness by accepting calls on the periphery of clusters. More markers show very high call rates and the amount of missing data shown by the red shaded region is minimal (99.2% completeness). The overall accuracy as measured by trio concordance shows that a small number of erroneous calls are being made (99.64% concordance). If one wishes to eliminate these incorrect calls, the base caller can be tuned to be more stringent. This choice allows very high accuracy (∼99.9% trio concordance) while causing more missing data (blue shaded region). The choice of cluster calling parameters should thus be chosen according to the intended use of the data.
The optimal choice for this trade-off depends on the required use of the data. If single marker association is being used for common SNPs, it may be appropriate to choose fairly permissive clustering parameters because the gain in “usable” data quantity is more advantageous than the small number of errors that are being made. When rare markers (Kang et al. 2004) or when multimarker haplotypes are being built (Kirk and Cardon 2002), the results will be much more sensitive to genotyping errors.
Table 1 shows the effect of emphasizing conversion rate, completeness, or accuracy in choosing cluster parameters. Conversion rates of >90% are possible with accuracy rates of ∼99.6%. In this case ∼1.5%-2% of the data are “missing,” as they are too ambiguous to call. Alternatively, accuracy levels of >99.9% can be achieved by retaining only the best markers (conversion rates of 77%-82%). A compromise can be reached by optimizing a set of markers with high completeness, which results in intermediate levels of conversion and accuracy.
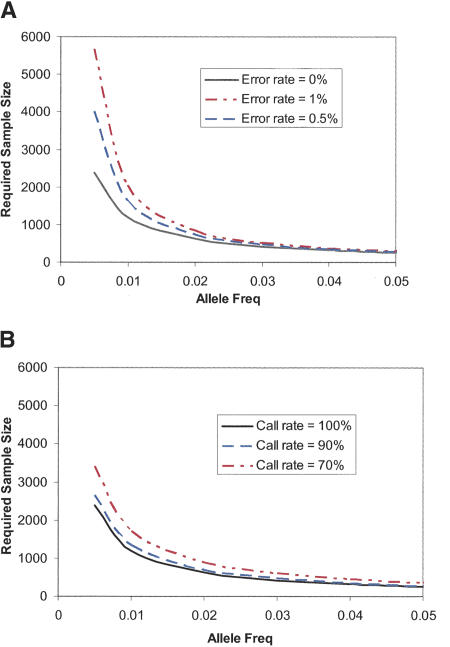
The impact of these compromises is best exemplified in the context of a model genotyping experiment. Figure 4 shows the effect of inaccurate or incomplete data from an association study for which the causative alleles are of varying frequency as shown on the x-axis. Given a genetic disease model (genetic relative risk GRR = 2 in a multiplicative model of disease) the number of patients and controls required to achieve an 80% power is plotted assuming single marker allelic tests are performed. As can be seen, common marker associations are relatively insensitive to missing data and error rates as high as 1%. On the other hand, if rare SNP markers are under study, accuracy is very important. Making an error in 1% of the data would double the population size required to find a 1% frequency marker in this model. Similarly, missing data are also more damaging when looking for signals in less frequent markers. Overall, the performance of all the converted markers in this study is sufficient to handle the full scope of association study applications with minimal loss of power relative to the ideal technology.
Figure 4.
The effect of inaccurate genotypes (A) and incomplete genotyping (B) on the number of patients required to have 80% power to find a genetic association. A genetic model has been assumed in which the relative risk of the causative allele (GRR) is two. The effect is assumed to be multiplicative. The causative allele frequency is plotted on the x-axis. The largest loss of power comes with making inaccurate calls for markers with low frequency. By contrast, incomplete data result in smaller loss of power, which is felt across the allele frequency spectrum. The data from the MIP assay are accurate enough to be used for the investigation of rare alleles without significant loss of power.
Discussion
This report of an advanced Molecular Inversion Probe technology now allows highly efficient, accurate, and low cost genotyping of targeted SNPs at levels >10,000plex. The unique features described in this study enable this technology to be scaled from 1,000plex levels to >10,000plex levels without any loss of performance in terms of accuracy completeness or conversion rate. At the same time the development of a four-color scanning solution for the tag arrays has resulted in decreased processing times and chip costs while insulating genotype calls from feature-to-feature chip noise.
This technology can be applied to complex genetic analysis in two basic ways. First, comprehensive LD mapping can be performed using a whole-genome HapMap tagging approach; second, comprehensive direct detection of potentially functional SNPs in coding and conserved regions is possible. As shown above, maintaining a high level of accuracy and completeness is critical when analyzing rare SNPs or haplotypes. The importance of being able to target specific SNPs, and to achieve a high conversion rate, is clear in the case of direct detection of causative SNPs. SNPs in functional regions tend to be rarer, leaving most without an alternative, common surrogate in high LD. This failure to convert a SNP and its concomitant loss of power to detect its genetic effect is directly proportional to the conversion rate. Hybridization-based methods whose conversion rates are significantly <50% will be unable to achieve high power to assess these SNPs. The conversion rates demonstrated in this study indicate that >85% conversion of SNPs in unique regions of the genome is possible. Furthermore, a second manufacturing and design pass has been shown to recover approximately half of the unconverted probes that failed due to failed oligo synthesis, secondary structure in the probe-tag complex, etc. (P. Hardenbol, unpubl.). With two rounds of synthesis, it should be possible to achieve conversion rates >90% and to very comprehensively analyze these SNPs.
The effect of high assay conversion rates on linkage disequilibrium mapping studies such as the HapMap is important but more subtle. The very nature of an LD mapping method presupposes a degree of redundancy in the choice of markers as some SNPs will be in high LD with each other. However, the ability to convert SNPs at a high rate does confer several advantages in large-scale HapMap approaches. First, if one cannot predict ahead of time which SNPs will fail due to a particular sequence context, a low conversion rate will increase the expense and effort required to build a map as multiple redundant SNPs will need to be attempted to find scorable tagging SNPs. This effort is onerous if several population specific maps are to be constructed. Secondly, the number of surrogates that exist for a given SNP will depend on the density of markers that one is choosing from. The first phase of the HapMap project is expected to produce an informative SNP every 5 kb. At this density, there will be a large number of SNPs that are not in LD with any others. Failure to convert these SNPs will lead to loss of power. Calculating the impact of these gaps on an association study is difficult. Larger haplotypes for which there are multiple surrogates will cover more of the genome and thus add more power than a singleton SNP in a region of high recombination, so the loss of power will not be directly proportional to the SNPs missed as in the case of direct detection. Nevertheless, it is clear that the task of building a comprehensive HapMap product is greatly simplified by a technology that can retain a high rate of assay conversion while achieving high levels of multiplexing.
Further increases in the levels of multiplexing for this assay are likely. Tag set development is proceeding and early tests on a set of 40,000 tags indicate that a similar amount of crosshybridization noise is achievable (data not shown). We believe that a set of 100,000 tags should be a realistic goal in light of recent advances in array technology. The MIP assay itself has shown no evidence of producing nonspecific probe inversion as we have moved to higher levels of multiplexing. Signal-to-noise levels have remained constant between 6,000plex and 12,000plex assays. It should be noted that the total mass of genomic DNA in the reaction is 40 times the probe amounts used in a 12,000plex reaction. As a result the probe-probe interactions that are increasing with multiplexing are still at a level far below the probe-genome interactions. It seems reasonable to assume that another order of magnitude in multiplexing should be achievable. Indeed, preliminary results using 24,000plex MIP reactions have shown highly accurate results (data not shown). The final challenge is in maintaining sufficient detection signal as one splits a single amplification reaction over increasingly large numbers of amplicons. Several means are available to address this issue including: concentration of larger PCR reactions, brighter fluorescent labels, more sensitive scanners, increased hybridization times, and increased oligo density in arrays. Taking all of these considerations into account, we believe that this technology will be able to be scaled to the level of 100,000plex in the near future.
Methods
Assay design
Batches of SNPs on Chromosome 12 were chosen according to criteria that emphasized even spacing. Once a batch of SNPs has been selected, a homology sequence is selected based on Tm optimization that is on average 40.4 bases long that is centered over the SNP and is complementary to the genome. This sequence is BLASTed against the genome to determine whether the sequence is unique in the genome, where unique is defined as an exact match to only one position. If the exact sequence appears more than once, the probe is not synthesized. This is the only filter used. Next, a tag sequence is added that is unique among the batch of assays and complementary to a feature on the detection chip system. No consideration is given to the degree of complementarity of sequences within the batch. Probes can target overlapping sequences since the genomic DNA is not saturated with hybridized probe. All probe batches were manufactured by ParAllele Bioscience using its proprietary MIP probe synthesis procedures and are commercially available (MegAllele kit, ParAllele BioScience). This process is a pooled procedure that results in a pool of up to 12,000 probes that are tested using pooled quality control procedures before being sent to Baylor College of Medicine.
Genotyping reactions
The genotyping reactions were carried out using the standard protocols recommended by the manufacturer at the Baylor College of Medicine, Human Genome Sequencing Center. MIP assays are carried out in 96-well plates using 12 individuals per plate for each of four allele channels using the MegAllele genotyping kit (ParAllele BioScience). The DNA samples were obtained from the Coriell collection (Coriell Institute) and consisted of trios from the CEU family collection. A full list of these samples is available at the HapMap Web site (www.hapmap.org). Each of the four MIP reactions was mixed with 500 ng of genomic DNA.
Two-color assay execution
After amplification and tag cleavage, the products were hybridized to two GeneChip Tag Arrays: TrueTag 5k or TrueTag 10k (Affymetrix) and stained using a two-color staining kit from ParAllele. The components were decoded by measuring the fluorescence signals at the corresponding complementary tag site on the DNA array. Four intensity values for each probe are generated using a GeneArray scanner (Affymetrix) using two arrays in two fluorescent channels (phycoerythrin and fluorescein) using the manufacturer recommended protocol (ParAllele BioScience). The A and G reactions were pooled on one chip while the C and T channels were pooled on a second chip. For each tag the two values for the expected allelic bases were compared to determine whether the sample was homozygous or heterozygous for the given SNP, and the two non-allele bases were compared to the allele bases to determine the signal-to-noise for the probe. The two non-allele bases serve as internal controls that were used to improve call rate and to reduce false calls due to missing, degraded, or noisy probes. The data from each image were tracked through a LIMS system to a probe pool and a DNA sample. This analysis was carried out using the MegAllele software package that is commercially available from ParAllele BioScience.
Four-color assay execution
The four-color data shown for Batch 4 were carried out using a four-color MegAllele assay kit from ParAllele Bioscience, using its commercially available four-color staining kit. The scanner used is a CCD camera with a broad band excitation lamp. This instrument (GeneChip AT Scanner, Molecular Devices) is commercially available for this assay from Affymetrix. Data from each chip are collected using four different filters to yield the four images that are processed in a manner identical to the data from the two chips that are used in the two-color protocol.
Data
This entire data set is now available publicly by searching Chromosome 12 at (www.hapmap.org). Trio concordances were calculated by tracking the concordance of each chromosome in a child in order to glean some information from the large numbers of markers for which at least one parent is heterozygous for a given marker. In this algorithm, a trio with two homozygous parents contributes two concordance tests (both child alleles predicted), a trio with a single heterozygous parent contributes one test (one child allele predicted), and trios with two heterozygous parents are uninformative and are not considered. Sixty-five percent of all chromosomes called in this study inform trio accuracy using this algorithm.
Acknowledgments
This work was supported by NIH grant 1U54HG02755. We wish to acknowledge the ongoing intellectual contributions of Professor Ulf Landegren and his lab to the development of the MIP technology.
Footnotes
Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.3185605.
References
- Altshuler, D., Hirschhorn, J.N., Klannemark, M., Lindgren, C.M., Vohl, M.C., Nemesh, J., Lane, C.R., Schaffner, S.F., Bolk, S., Brewer, C., et al. 2000. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 26: 76-80. [DOI] [PubMed] [Google Scholar]
- Cupples, L.A., Yang, Q., Demissie, S., Copenhafer, D., and Levy, D. 2003. Description of the Framingham Heart Study data for Genetic Analysis Workshop 13. BMC Genet. 4 Suppl 1: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind, D.H., Sowinski, J., Lord, C., Iversen, P., Shestack, J., Jones, P., Ducat, L., and Spence, S.J. 2001. The autism genetic resource exchange: A resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 69: 463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, P.D., Bloch, W., Brinson, E., Chang, C.C., Eggerding, F.A., Fung, S., Iovannisci, D.M., Woo, S., Winn-Deen, E.S., and Iovannisci, D.A. 1994. High-density multiplex detection of nucleic acid sequences: Oligonucleotide ligation assay and sequence-coded separation. Nucleic Acids Res. 22: 4527-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenbol, P., Baner, J., Jain, M., Nilsson, M., Namsaraev, E.A., Karlin-Neumann, G.A., Fakhrai-Rad, H., Ronaghi, M., Willis, T.D., Landegren, U., et al. 2003. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21: 673-678. [DOI] [PubMed] [Google Scholar]
- Hugot, J.P., Chamaillard, M., Zouali, H., Lesage, S., Cezard, J.P., Belaiche, J., Almer, S., Tysk, C., O'Morain, C.A., Gassull, M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599-603. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2003. The International HapMap Project. Nature 426: 789-796. [DOI] [PubMed] [Google Scholar]
- Kang, S.J., Gordon, D., and Finch, S.J. 2004. What SNP genotyping errors are most costly for genetic association studies? Genet. Epidemiol. 26: 132-141. [DOI] [PubMed] [Google Scholar]
- Kirk, K.M. and Cardon, L.R. 2002. The impact of genotyping error on haplotype reconstruction and frequency estimation. Eur. J. Hum. Genet. 10: 616-622. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. 1999. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22: 139-144. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, H., Loi, H., Dong, S., Tsai, Y.Y., Fang, J., Law, J., Di, X., Liu, W.M., Yang, G., Liu, G., et al. 2004. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 14: 414-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant, A., Barker, D.L., Stuelpnagel, J.R., and Chee, M.S. 2002. BeadArray technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl: 56-58, 60-61. [PubMed]
- Risch, N.J. 2000. Searching for genetic determinants in the new millennium. Nature 405: 847-856. [DOI] [PubMed] [Google Scholar]
- Samiotaki, M., Kwiatkowski, M., Parik, J., and Landegren, U. 1994. Dual-color detection of DNA sequence variants by ligase-mediated analysis. Genomics 20: 238-242. [DOI] [PubMed] [Google Scholar]
- Shmulewitz, D., Auerbach, S.B., Lehner, T., Blundell, M.L., Winick, J.D., Youngman, L.D., Skilling, V., Heath, S.C., Ott, J., Stoffel, M., et al. 2001. Epidemiology and factor analysis of obesity, type II diabetes, hypertension, and dyslipidemia (syndrome X) on the Island of Kosrae, Federated States of Micronesia. Hum. Hered. 51: 8-19. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D.D., Lashkari, D.A., Morris, D., Mittmann, M., and Davis, R.W. 1996. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 14: 450-456. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- www.hapmap.org; Chromosome 12 data, HapMap Project Web site.