Abstract
Oligonucleotide microarrays provide a high-throughput method for exploring genomes. In addition to their utility for gene-expression analysis, oligonucleotide-expression arrays have also been used to perform genotyping on genomic DNA. Here, we show that in segregants from a cross between two unrelated strains of Saccharomyces cerevisiae, high-quality genotype data can also be obtained when mRNA is hybridized to an oligonucleotide-expression array. We were able to identify and genotype nearly 1000 polymorphisms at an error rate close to 3% in segregants and at an error rate of 7% in diploid strains, a performance comparable to methods using genomic DNA. In addition, we demonstrate how simultaneous genotyping and gene-expression profiling can reveal cis-regulatory variation by screening hundreds of genes for allele-specific expression. With this method, we discovered 70 ORFs with evidence for preferential expression of one allele in a diploid hybrid of two S. cerevisiae strains.
High-density oligonucleotide arrays have provided an important and versatile tool for genome-scale experimentation. In addition to their utility for measuring gene-expression levels, oligonucleotide arrays have been used to perform genotyping in a variety of genomes (Winzeler et al. 1998; Brem et al. 2002; Borevitz et al. 2003). Microarray genotyping is based on the idea that a sequence mismatch between a short oligonucleotide probe on the array and its target sequence significantly disrupts hybridization and attenuates that probe's signal. By hybridizing genomic DNA to oligonucleotide arrays and observing the relative signal strength from probes whose target sequences bear a putative polymorphic site, Winzeler et al. (1998) and Brem et al. (2002) were able to identify and genotype >3000 polymorphisms between two strains of Saccharomyces cerevisiae at error rates of 5% and <1%, respectively. Using the same approach, Borevitz et al. (2003) identified and genotyped nearly 4000 polymorphisms at a 5% error rate in the more complex Arabidopsis thaliana genome.
In this report, we build on this work by showing that polymorphic loci can be discovered and genotyped in two strains of S. cerevisiae by hybridizing mRNA to oligonucleotide arrays, allowing for simultaneous genotyping and gene-expression analysis. The marker density and genotyping error rate are comparable to those obtained when genomic DNA is hybridized to arrays. Furthermore, we show that combined genotyping and gene-expression analysis can be used to study allele-specific expression in diploid hybrids formed by mating two S. cerevisiae strains.
Detecting preferential expression of one allele in a diploid is of considerable interest in characterizing transcriptional control in a genome, because it identifies the presence of cis-regulatory polymorphisms. Several groups have recognized the importance of studying this feature of gene expression on a genome-wide scale (Knight 2004), utilizing a variety of experimental approaches, including single-base extension genotyping of RT-PCR products (Cowles et al. 2002; Yan et al. 2002), hybridization of mRNA to Affymetrix HuSNP genotyping arrays (Lo et al. 2003), and quantification of RNA polymerase II loading onto DNA (Knight et al. 2003). Thus far, the Affymetrix HuSNP array has provided the most high-throughput platform, but it is limited to genomes for which these specially designed arrays are available.
It remains to be seen which technologies for detecting allele-specific expression will be most useful in whole-genome studies. The primary advantages of the method we describe here are the large number of genes that may be interrogated and the simplicity of the experimental and analytical techniques. The method's false-positive rate suggests that it will be suitable for genome-wide identification of candidate genes, followed by selective confirmation using other techniques.
Results
In order to perform polymorphism discovery and genotyping by hybridizing mRNA (rather than genomic DNA) to an oligonucleotide array, it is necessary to distinguish between a low-intensity signal due to poor hybridization resulting from a sequence polymorphism and a low-intensity signal due to low gene expression. A simple solution to this problem is to search for cases in which one or a few probes display poor hybridization in one strain, presumably due to sequence mismatches, compared with the remaining probes in the probe set that interrogate the same ORF (Fig. 1A,B,C). In practice, this can be accomplished by determining the ratio of the observed probe intensity to the overall gene-expression level, estimated by all probes in the probe set. Another level of refinement, offered by a recently described energy model for oligonucleotide array analysis (Zhang et al. 2003), is to compare the observed probe signal with the expected probe signal, given the estimated gene expression level and thermodynamics of probe-target sequence duplex formation.
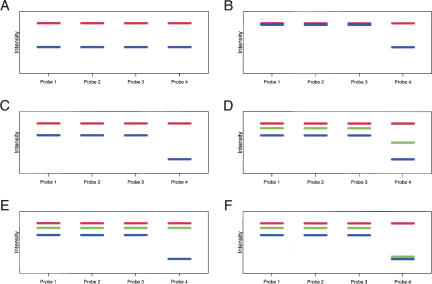
Figure 1.
Schematic illustrating the methods for genotyping and detecting allele-specific expression. (A) Routine gene-expression analysis using mRNA. Signals from all probes interrogating the same ORF are averaged to produce an estimate of the gene-expression level. In this example, the probe signals suggest that the reference strain (red) expresses this ORF at a higher level than the test strain (blue). All probes hybridize equally well in the test strain, providing no evidence of sequence mismatches between the probe and target sequence. (B) Genotyping using genomic DNA. When DNA is hybridized to arrays, the intensities in the two strains are the same, except at probes interrogating sequences where the test strain bears a polymorphism. Here, the test strain bears a polymorphism in the region interrogated by “Probe 4,” and as a result, the test strain produces a weak signal at this probe. (C) Genotyping using mRNA. The gene is expressed at a different level in the test strain than in the reference strain, but the polymorphism in the sequence interrogated by “Probe 4” is still readily detected, because the signal at this probe is substantially less than the gene-expression level as estimated using the entire probe set. (D) Equal expression of both alleles in a diploid. A diploid strain formed by mating the reference and test strains (green) is heterozygous for the sequence polymorphism at “Probe 4.” The ratio of the intensity at “Probe 4” to the remaining probes in the heterozygous diploid is equal to the average of the ratios of “Probe 4” to the remaining probes in the reference and test strains, suggesting that both alleles are expressed in equal amounts. (E) Preferential expression of the reference strain allele. Here, the ratio of the intensity at “Probe 4” to the remaining probe is the same as the ratio in the reference strain, suggesting that only the allele derived from the reference strain is expressed. (F) Preferential expression of the test strain allele.
We used this approach to detect putative polymorphisms between the S. cerevisiae strains BY4716 (BY) and RM11-1a (RM). Because the Affymetrix YGS98 microarray is designed against the genome of strain S288C, which is isogenic to BY, polymorphisms between BY and RM in a probe's target sequence are expected to disrupt hybridization of that probe to mRNA from RM. Therefore, we searched for probes in which the ratio of the observed intensity to the expected intensity was significantly greater in BY than in RM by comparing three replicates of each strain (see Methods). We identified 1049 probes at a false discovery rate (FDR) ≤ 0.05 that became candidates for use as genotyping markers based on their differential hybridization patterns in BY and RM (see Supplemental Table 1).
In order to determine whether our marker discovery algorithm was indeed detecting polymorphisms in RM, we performed BLAST searches of the marker probes against sequence reads from a recently generated whole-genome shotgun sequence of RM available from the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/). We identified alignments in RM for 1034 of the 1049 marker probes and found that 970 of these 1034 probes interrogated sequence polymorphisms in RM (a false-positive rate of 6%). In contrast, of all 30,695 probes considered in our marker discovery analysis, only 2168 of the 30,462 probes for which we found alignments interrogated polymorphic sequence. Thus, the method detected 45% of all probes interrogating polymorphic sequence, and provided a 13-fold enrichment of polymorphic probes (94% of probes identified by the method vs. 7% of all 30,695 probes). We compared this detection rate with that obtained when genomic DNA was hybridized to the arrays (Fig. 2). Considering the same 30,695 probes and utilizing the same false discovery rate and number of arrays, we were able to detect 1517 of the 2168 polymorphisms using genomic DNA. There was high concordance between the two sets of marker probes (901 of the 970 mRNA-derived markers were present in the DNA-derived set), and although the DNA-based approach provided greater sensitivity, our set of mRNA-derived markers provides coverage of the yeast genome at a 4-cM density, suggesting that these markers would still extract much of the linkage information in a data set.
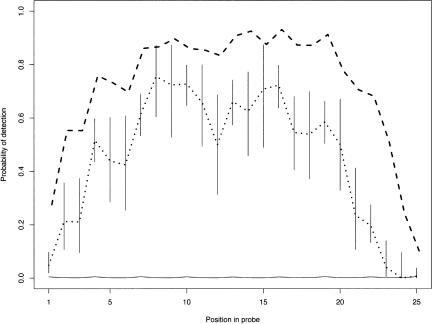
Figure 2.
Sensitivity of polymorphism discovery. The dotted line shows the probability of detecting a single base change as a function of position in the 25 base-pair probe when mRNA material is hybridized to the array. Vertical lines give the 95% confidence intervals for these probabilities. The dashed line shows the probability of detecting single base changes using genomic DNA. The solid line depicts the baseline rate of polymorphism.
We next tested our set of marker probes (including nonpolymorphic probes, to simulate the more realistic situation where a whole-genome sequence is unavailable) by comparing the mRNA-based geno-types with genomic DNA-based genotypes (Brem et al. 2002) in two segregants. Analyzing the raw mRNA-derived genotypes, we found that the genotyping error rate in the two segregants was 3.2% for the 1049 markers identified at FDR ≤ 0.05. We found that applying a simple hidden Markov model (HMM) to estimate the most likely underlying genotypes reduced the error rate to 2.7% (Fig. 3A).
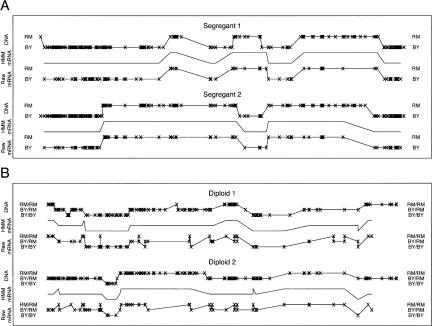
Figure 3.
Comparison of genomic DNA and mRNA-derived genotypes. (A) Chromosome IV genotypes in haploid segregants. The x's represent locations of markers. In each box, the middle line (without x's) is the most likely set of genotypes, estimated using the HMM, producing the mRNA data. (B) Chromosome XIII genotypes in diploid hybrids formed by mating two segregants.
We next extended this mRNA-based genotyping approach to the case of a diploid genome. Performing genotyping in a diploid from an mRNA-based signal is complicated by situations in which the two alleles at a heterozygous locus are expressed in unequal amounts. We therefore obtained an estimate of the observed to expected intensity ratio for the heterozygous genotype by measuring the mRNA-based genotype signal from six independent diploid hybrids formed by mating BY and RM. In general, this signal may fall anywhere between the homozygous genotypes, depending on the presence and magnitude of allele-specific expression. If this allele-specific expression is driven by a nearby polymorphism, then as long as the regulatory polymorphism remains linked to the polymorphism interrogated by the marker probe, we expect the amount of each allele expressed and the resulting observed to expected intensity ratio for the heterozygous genotype to remain constant. To test this expectation, we formed two independent diploid hybrids from haploid segregants that we previously genotyped using genomic DNA, and found that the error rate for the raw genotype calls in these diploids was 16%. After correction with a simple three-genotype HMM, the error rate of the genotype calls was reduced to 7.3% (Fig. 3B). If we only scored markers when the posterior probability of the most likely genotype was >0.9, 85% of markers were genotyped, and the error rate was reduced to 5.3%.
Given the ability to measure genotypes from mRNA, we applied this technology to measuring preferential expression of one allele in the diploid hybrids formed by mating BY and RM. In order to detect transcripts in which either the BY or RM allele was preferentially expressed, we identified probes for which the observed to expected intensity ratio in the diploid indicated a relative excess of either the BY or RM allele (Fig. 1D,E,F). We assumed a simple linear model specifying that if both alleles are equally expressed, the observed to expected intensity ratio in the diploid should be equal to the average of the ratios in BY and RM (see Methods). Analyzing all 1049 marker probes (see Supplemental Table 1 for complete results) identified by our marker discovery algorithm, which interrogated a total of 692 unique ORFs, we found that 99 probes deviated from the linear model by a maximally significant Wilcoxon-Mann-Whitney result (P ≤ 0.006). Eight of these 99 probes were among the false-positive probes detected during marker discovery whose target sequences were nonpolymorphic, while the remaining 91 probes interrogated 70 unique ORFs. Interestingly, only 11 of the 70 ORFs showed preferential expression of the RM allele. Although our linkage results suggest that the BY allele is more often up-regulated in haploid segregants (data not shown), the asymmetric nature of the probe signals most likely causes a detection bias that reduces the method's ability to reveal preferential expression of the RM allele.
To gain empirical estimates of the false-positive and false-negative rates of this method, we measured six replicates of a 1:1 mixture of BY parental mRNA and RM parental mRNA and applied our criterion for detecting allele-specific expression to these arrays under two different models (Fig. 4A). First, in order to explore the false-positive rate, we assumed the correct model for the amount of each allele expressed in the 1:1 mixture, stating that each allele is expressed at half the gene-expression level in the haploid parent. Under this scenario, probes that deviate from the linear model represent false positives that would be erroneously classified as cases of allele-specific expression. Analyzing all 1049 marker probes, we found that 26 probes, representing 24 unique ORFs, deviated from the correct model for the 1:1 mixture by a maximally significant Wilcoxon-Mann-Whitney test (P ≤ 0.006), suggesting that the nominal p-values from the rank test are somewhat anticonservative and that the true p-value may be closer to P ≤ 0.03. This analysis suggests that ∼24 of the 70 ORFs in the diploid showing evidence of allele-specific expression are likely to be false positives, indicating a false discovery rate of 0.34.
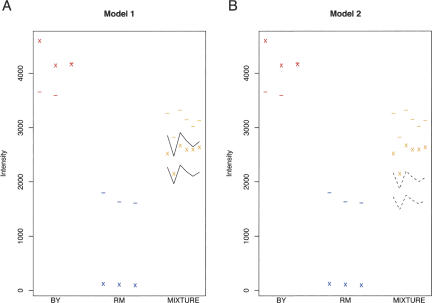
Figure 4.
Examples illustrating the use of the 1:1 mixture to estimate the false positive and sensitivity of the method for detecting allele-specific expression. In each plot, the x's represent the observed intensities (I) and the dashes represent the expected intensities (Î) for a marker probe located in FKS1. Note that the probe represents a good marker, because the I/Î ratio is much greater in BY than in RM. For this transcript, the expected probe intensities for the three BY replicates are approximately twofold higher than in the three RM replicates, suggesting that the transcript is expressed at a higher level in BY. A 1:1 mixture of total mRNA from BY and RM would therefore contain approximately two times as much of the BY FKS1 allele as the RM allele. (A) Model 1 analyzes the false-positive rate. Here, the solid lines in the 1:1 mixture represent the predicted range for the observed probe signals under a model that correctly accounts for the twofold overabundance of BY allele. The observed signals from the 1:1 mixture replicates fall within this range, providing no evidence for deviation from the model. (B) Model 2 analyzes the power. Here, the dashed lines represent the predicted range of the observed probe signals under a model assuming equal expression of both alleles. All six observed intensity signals in the 1:1 mixture fall above this range, correctly revealing that the 1:1 mixture contains more BY allele than RM allele for this transcript.
Second, in order to explore the power of the method to detect true differences in the amount of each allele present, we assumed an incorrect model, stating that each allele is expressed in equal amounts in the 1:1 mixture (Fig. 4B). Under this scenario, we determined whether our method was able to detect overabundance of either the BY or RM allele in the 1:1 mixture and thus reveal expression differences in the BY and RM parents that formed the mixture. We found that 56 probes, representing 41 unique ORFs, signaled an excess of BY allele in the 1:1 mixture. Of these 41 ORFs, 33 showed significantly higher expression in the BY parental strain at FDR ≤ 0.05. Only 16 probes, representing 13 unique ORFs, indicated excess of the RM allele in the 1:1 mixture, and 10 of these 13 ORFs showed the expected increased expression in the RM parental strain. Of all 692 ORFs considered in the analysis, 182 showed significantly greater expression in BY at FDR ≤ 0.05, while 100 showed significantly greater expression in RM. Our method detected the correct allelic expression difference in the 1:1 mixture for only 43 of the 282 ORFs with significantly different expression between the two strains, suggesting that we have low power to detect all differences in the amount of each allele present in a sample. However, the method detected 12 of 22 of the ORFs that showed significantly greater expression of the BY allele by a factor of two or greater, and eight of 22 ORFs with twofold greater expression of the RM allele, suggesting that the method has moderate power to detect large differences in the amount of each allele present.
Because the presence of allele-specific expression implies a nearby polymorphism that controls expression of the allele residing on the same DNA molecule, we expect the expression level of such a gene, when treated as a quantitative trait, to show linkage to its own locus in haploid S. cerevisiae segregants. Furthermore, we expect that if a nearby polymorphism that is linked to the BY allele acts to produce preferential expression favoring the BY allele in diploids, the BY allele should be associated with higher expression levels in the haploid segregants. In other words, detecting allele-specific expression implies not only that the expression level of the gene should link to its own locus, but also that the allele that is preferentially expressed should be associated with higher expression in haploid segregants. Using results from our linkage-mapping studies (Brem et al. 2002; Yvert et al. 2003), we found a significant enrichment for ORFs that linked to their own locus with higher expression of the predicted allele among the ORFs showing evidence of allele-specific expression (Table 1). In addition, ORFs showing evidence of allele-specific expression were less likely to show linkage to their own loci in association with elevated expression of the opposite allele.
Table 1.
Relationship between allele-specific expression results and linkage analysis results
| Type of self-linker | Type of allele-specific expression | Number of self-linkers/Total | Enrichment (or deficiency) P-value | |
|---|---|---|---|---|
| Self-link FDR ≤ 0.05 Allele-specific expression P ≤ 0.006 | BY allele upregulated | ASE-BY | 21/59 = 36% | 0.001 |
| All non-ASE-BY | 102/565 = 18% | (enrichment) | ||
| RM allele upregulated | ASE-BY | 2/59 = 3% | 0.008 | |
| All non-ASE-BY | 78/565 = 14% | (deficiency) | ||
| BY allele upregulated | ASE-RM | 0/11 = 0% | 0.10 | |
| All non-ASE-RM | 111/586 = 19% | (deficiency) | ||
| RM allele upregulated | ASE-RM | 3/11 = 27% | 0.16 | |
| All non-ASE-RM | 75/586 = 13% | (enrichment) | ||
| Self-link p ≤ 0.05 Allele-specific expression P ≤ 0.05
|
BY allele upregulated | ASE-BY | 59/159 = 37% | 4e-6 |
| All non-ASE-BY | 108/505 = 21% | (enrichment) | ||
| RM allele upregulated | ASE-BY | 15/159 = 9% | 2e-4 | |
| All non-ASE-BY | 101/505 = 20% | (deficiency) | ||
| BY allele upregulated | ASE-RM | 6/55 = 11% | 0.005 | |
| All non-ASE-RM | 145/555 = 26% | (deficiency) | ||
| RM allele upregulated
|
ASE-RM | 21/55 = 38% | 1e-4 | |
| All non-ASE-RM | 92/555 = 17% | (enrichment) |
Each cell shows the count of self-linking ORFs associated with higher expression of the BY allele (BY allele up-regulated) or higher expression of the RM allele (RM allele up-regulated), divided by the total number of ORFs showing evidence for (or against) allele-specific expression of the BY (ASE-BY) or RM allele (ASE-RM). P-values are for one-tailed tests of the hypothesis that ORFs showing allele-specific expression are enriched for self-linkages associated with higher expression of that same allele and deficient for self-linkages associated with higher expression of the other allele. Note the total number of ORFs in the denominator varies from test to test. Because a given ORF may contain multiple marker probes, each ORF is counted once as showing allele-specific expression if any of its marker probes fulfill the criterion for allele-specific expression, and once as not showing allele-specific expression if any of its marker probes fail to meet the criterion. Forming the table in this way is conservative, because unless all probes interrogating the same ORF uniformly show evidence for (or against) allele-specific expression, that ORF will contribute one count to both the rate of self-linkers (or non-self-linkers) in the allele-specific expression and the nonallele-specific expression sets, tending to make the two rates appear similar.
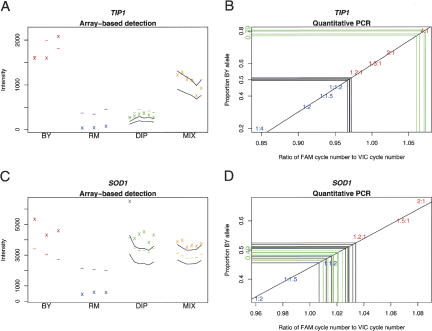
We used quantitative PCR to attempt to confirm the predicted allele-specific expression of several ORFs (see Supplemental Methods and Supplemental Table 2). Because we expect the false-positive rate of the array-based detection method to be relatively high; based on our analysis of the 1:1 mixture, we selected ORFs for confirmation using a small number of heuristics. We selected ORFs expressed at a level in the hybrid that was not far outside the range of the parents, ORFs without excessive variation in the probe signals across replicate arrays, and ORFs whose expression levels in the BY and RM haploid parents indicated an expression difference in the same direction as the predicted allele-specific expression. ORFs were chosen without regard to the linkage results, but selecting the ORFs in this way further enriched for self-linkages. Figure 5 shows two examples of ORFs analyzed in these quantitative PCR experiments. We found that five of 11 ORFs tested showed significant allele-specific expression in the direction predicted by the microarray data (CIS3, HSP150, MOG1, TIP1, and YHR032W), one ORF showed significant allele-specific expression in the direction opposite that predicted (CHO2), and five ORFs failed to show evidence of allele-specific expression (Table 2). These data suggest that the method indeed detected cases of allele-specific expression, and although a 6/11 false-positive rate is not statistically distinguishable from our expected rate of 34%, these data do suggest that the true false-positive rate may be somewhat higher than anticipated.
Figure 5.
Allele-specific expression analysis and quantitative PCR. (A,C) Array-based detection of allele-specific expression in TIP1 and SOD1, respectively. Note that the observed intensities fall above the predicted range for both TIP1 and SOD1, indicating that the observed to expected intensity ratio is more similar to BY than RM, suggesting that the BY allele is preferentially expressed. (B,D) Quantitative PCR experiments. Green lines indicate the diploid cDNA samples, and black lines indicate the diploid genomic DNA samples. Data points on the standard curve are labeled as parts BY allele: parts RM allele. Blue points represent excess RM allele, whereas red points represent excess BY allele. The estimated proportion of BY allele present in the diploid cDNA is given on the vertical axis in green.
Table 2.
Results of quantitative PCR experiments analyzing allele-specific expression
| ORF name | Predicted ASE direction | Self-linkage p-value | Estimated proportion of BY allele based on linkage data | Estimated proportion of BY allele from Q-PCR experiments | P-value for allele-specific expression from Q-PCR experiments |
|---|---|---|---|---|---|
| CIS3* | ASE-BY | 1e-6 | 0.55 | 0.55 | 0.007 |
| HSPI50* | ASE-BY | 9e-7 | 0.55 | 0.54 | 0.02 |
| MOG1* | ASE-BY | 4e-6 | 0.54 | 0.53 | 0.004 |
| TIP1* | ASE-BY | 4e-10 | 0.70 | 0.79 | 0.004 |
| YHR032W* | ASE-BY | 0.03 | 0.48 | 0.44 | 0.05 |
| ERD2 | ASE-BY | 4e-7 | 0.53 | 0.50 | 0.76 |
| ITR1 | ASE-BY | 5e-5 | 0.54 | 0.50 | 0.91 |
| OPI3 | ASE-BY | 2e-5 | 0.56 | 0.50 | 1.0 |
| SOD1 | ASE-BY | 3e-6 | 0.54 | 0.49 | 0.32 |
| SEC53 | ASE-BY | 0.27 | 0.50 | 0.51 | 0.15 |
| CHO2 | ASE-BY | 0.04 | 0.52 | 0.48 | 0.01 |
The allele predicted to show preferential expression based on the microarray analysis is shown in column 2. We calculated self-linkage p-values as described previously (Brem et al. 2002). The predicted amount of BY allele expressed based in the linkage data was determined by comparing the mean expression level in segregants bearing the BY genotype at the ORF in question to the segregants bearing the RM genotype at this locus. Single astericks indicate ORFs that showed significant allele-specific expression according to quantitative PCR in the direction predicted, thus confirming the array-based detection.
Discussion
Simultaneous genotyping and gene-expression analysis using oligonucleotide arrays will be useful for gene-mapping studies in model organisms such as S. cerevisiae. Genotyping using mRNA comes at a cost in marker density, but the advantage of the method lies in its ability to more fully exploit the genetic information in a data set through simultaneous measurement of gene-expression levels. Such an approach allows for the synthesis of gene-expression data and linkage-mapping results as a method for identifying candidate genes. It also allows for detection of allele-specific expression and for more efficient analysis of the genetics of gene expression.
Using this approach to detect allele-specific expression, we identified 70 ORFs that showed evidence of allele-specific expression in a diploid hybrid of two S. cerevisiae strains. Our quantitative PCR experiments directed at 11 of these ORFs confirmed five cases of allele-specific expression in the direction predicted by the array-based approach, while six of these ORFs failed to show allele-specific expression in the direction predicted. Our failure to detect allele-specific expression in these six ORFs confirms our suspicion that the array-based method for detecting allele-specific has a high false-positive rate. Several strategies might therefore be used to improve the detection rate. For example, for three of the six ORFs that were not experimentally confirmed, the marker probe that signaled allele-specific expression also deviated from the correct model in the 1:1 mixture. In contrast, for all five ORFs with confirmatory evidence of allele-specific expression, the marker probe fit the correct model in the 1:1 mixture analysis well. Thus, excluding probes that deviate from expectation in the 1:1 mixture could reduce the rate of false positives. In addition, synthesizing these array data with other information, such as the presence of upstream polymorphisms, might also lead to greater enrichment for true cases of allele-specific expression. Nonetheless, although a false-positive rate of 6/11 is high, analysis of an unselected set of genes (Cowles et al. 2002) led to detection of allele-specific expression in only 6% of cases. Thus, a prescreening approach such as the method we describe here that leads to a severalfold enrichment may accelerate studies of allele-specific expression and cis-regulatory variation.
The ORFs showing evidence of allele-specific expression were enriched for genes whose expression levels showed linkage to their own loci with the predicted difference in expression, suggesting that, in spite of the high false-positive rate, this method for detecting allele-specific expression provides a complementary strategy to linkage analysis for identifying cis-regulatory elements. At least two possibilities exist for the remaining ORFs whose expression levels did not show evidence of linkage to their own loci. First, some of these ORFs likely represent the false positives we expect to obtain in the method for detecting allele-specific expression. Second, more complex regulatory systems may prevent linkage analysis from detecting polymorphisms that produce allele-specific expression, either because these cis-acting polymorphisms represent minor loci having small effect sizes, or because they act in such a way as to control the rate, but not the level of gene expression. In the latter scenario, the overall gene expression level in haploids would be identical, but the allele transcribed at a faster rate in diploids would be preferentially expressed.
An important application of this mRNA-based marker discovery and genotyping analysis is in studies exploring the genetics of gene expression (Brem et al. 2002; Schadt et al. 2003; Yvert et al. 2003; Monks et al. 2004; Morley et al. 2004). We have shown that genotype data can be reliably obtained in both haploid and diploid S. cerevisiae utilizing an mRNA-based genotyping approach, making it possible to obtain both genotypes and phenotypes in a single hybridization experiment. Application of simultaneous genotyping and gene-expression analysis will accelerate gene-mapping studies exploring transcriptional regulation in model organisms, and will aid in revealing the QTLs underlying these complex phenotypes.
Methods
Expression array analysis
All statistical analyses of array data were performed using a method that estimates the expression level of a gene by modeling the energy of probe-target sequence duplex formation (Zhang et al. 2003). The expected intensity (Î) for each probe in a probe set is determined, given the estimated expression level of the gene under inspection, the population of mRNA molecules contributing to nonspecific hybridization, and the background signal for the microarray. Once nonspecific binding and background estimates are subtracted, Î represents the signal that should be obtained at a probe assuming a perfectly matched target sequence. Cases where Î dramatically overestimates the observed intensity (I) are assumed to be due to a polymorphism that compromises hybridization, and these probes are candidates for use as markers. We implemented the model described by Zhang et al. (2003) and incorporated a complete data array normalization procedure (Irizarry et al. 2003) into a C++ program that runs on the Linux platform. This program is available from the authors upon request. We also performed analyses using the RMA package (Irizarry et al. 2003) and noted similar, but slightly weaker performance in marker detection and genotyping.
Marker identification
We analyzed 30,695 probes on the Affymetrix YGS98 array (https://www.affymetrix.com/analysis/download_center.affx) that were located in transcribed ORFs, that interrogated unique sequence, and that were expressed at robust levels (expected probe intensity >100) in both BY and RM, allowing for reliable polymorphism detection and genotyping. For each probe, we calculated a t-statistic for the difference between the mean of the three IBY/ÎBY ratios and the mean of the three IRM/ÎRM ratios, setting the floor of the variance to 0.01. To estimate the null distribution, we permuted the BY and RM labels and recalculated the t-statistic for the difference in mean I/Î ratios for the two randomized groups. We assigned a p-value to each probe's observed I/Î ratio difference by counting how many of the 30,695 I/Î ratio differences were greater than the observed value, and identified significant probes using the false discovery rate (FDR) criterion (Benjamini and Hochberg 1995).
Genotyping and hidden Markov model
For the raw genotyping, we scored a haploid segregant as possessing the BY genotype if its I/Î ratio was closer to the mean of the three IBY/ÎBY ratios than to the mean of the three IRM/ÎRM ratios, and the RM genotype otherwise. For the HMM, we estimated transition probabilities by assuming recombination probabilities of 1 cM per 3 kb. We estimated emission probabilities by assuming that each genotype's I/Î ratios were normally distributed with mean equal to the mean of the three IBY/ÎBY (or IRM/ÎRM) ratios and standard deviation equal to the standard deviation of the three IBY/ÎBY (or IRM/ÎRM) ratios plus the plus the 90th percentile of all I/Î standard deviations. We assumed that any marker having Î <100 was expressed too low to produce a reliable genotype-specific signal, and we assigned such data points equal emission probabilities under all genotypes. For genotyping in the diploids, we made six measurements of BY-RM diploid hybrids and added three measurements of BY-BY diploids and three measurements of RM-RM diploids to our three BY and three RM haploid measurements. The raw and HMM genotyping calls were performed in the same way as the haploid genotyping, except that three genotype states were allowed. To determine the genotyping error rate, we scored an mRNA-derived genotype as erroneous if, and only if, it disagreed with the two most closely flanking genomic DNA-derived genotypes, and those genomic DNA-derived genotypes did not indicate a recombination event within the interval.
Measurement of allele-specific expression
The linear model states that the I/Î ratio in the BY-RM diploid IDP/ÎDP =(IBY/ÎBY +IRM/ÎRM)/2, the expected ratio under equal expression of the BY and RM alleles. Cases where IDP/ÎDP <(IBY/ÎBY + IRM/ÎRM)/2 are characteristic of preferential expression of the RM allele (since, by design, markers are probes where IRM/ÎRM is much less than IBY/ÎBY), whereas cases where IDP/ÎDP is greater are characteristic of preferential expression of the BY allele. We tested for deviations from the linear model by performing the nonparametric Wilcoxon-Mann-Whitney rank test for a difference between IDP/ÎDP and (IBY/ÎBY +IRM/ÎRM)/2, using six BY-RM diploid replicates to generate six independent IDP/ÎDP data points. To generate six independent (IBY/ÎBY +IRM/ÎRM)/2 data points, we paired each of the six IBY/ÎBY ratios from the three BY haploid replicates used in marker discovery and three BY-BY diploid replicates with a randomly chosen (without replacement) IRM/ÎRM ratio from the three RM haploid replicates and three RM-RM diploid replicates. As in our marker discovery analysis, we excluded probes having expected intensities <100.
Estimation of the false-positive rate and sensitivity using the 1:1 mixture
In the 1:1 mixture, the amount of each allele in the sample is known, the quantity of BY allele is equal to half the gene expression level in BY, and the quantity of RM allele is equal to half the gene expression level in RM. In order to estimate the false-positive rate, we modeled the I/Î ratio in the 1:1 mixture (IMIX/ÎMIX) by the average of IBY/ÎBY and IRM/ÎRM, each weighted by the expected signal at the probe, given the gene-expression level in the haploid BY and RM parents, respectively. As in our analysis of the BY-RM diploid, we performed a Wilcoxon-Mann-Whitney test using six IMIX/ÎMIX data points and six wBYIBY/ÎBY +wRMIRM/ÎRM data points, excluding probes with expected intensities <100. Here, wBY and wRM are the weights, given by the median value of ÎBY (or ÎRM) divided by the sum of the median values of ÎBY and ÎRM. We note that, although the three BY-BY diploid replicates and three RM-RM diploid replicates provide additional useful observations on IBY/ÎBY and IRM/ÎRM, they cannot be used in the estimation of the weights, since the 1:1 mixture was formed from a combination of BY and RM haploid mRNA, and in general, the gene-expression levels will not be the same in the haploid and diploid organisms. For simplicity, data from the BY-BY and RM-RM diploids are omitted from the figures.
In order to determine the sensitivity of the method, we performed the analysis on the 1:1 mixture in the same way as the diploid by assuming equal representation of each allele and attempting to detect deviations from equality. We then compared the probes deviating from this model with the BY and RM parents that formed the 1:1 mixture in order to determine whether the allele-specific expression analysis had correctly revealed an expression difference in the parent strains. To identify significant expression differences in the three BY and three RM parental replicates, we utilized the method of Efron et al. (2000). Briefly, we calculated Z = D/(S+A) for the difference in expected intensities between BY and RM for each probe. Here, Z was the mean of the differences BYi - RMi, (i = 1,2,3), S was the standard deviation of those differences, and A was the 90th percentile of all S values. We also calculated z = d/(s+a), where z was the mean of the differences BY2-BY1, BY3-BY2, RM2-RM1, and RM3-RM2, s was the standard deviation of those differences, and a was the 90th percentile of all s values. We then determined the set of probes showing significant difference in expression at an FDR ≤ 0.05 by comparing the Z values to the z values.
Acknowledgments
We thank Leah Scanlin, Hilary Coller, and Rachel Brem for helpful discussions. J.R. is supported by a Medical Scientist Training Program Grant. J.M.A. is supported by a National Science Foundation Postdoctoral Research Fellowship in Interdisciplinary Informatics. G.Y. was supported by a postdoctoral fellowship from the Human Frontier Science Program. This work was funded by the Howard Hughes Medical Institute, of which L.K. is an associate investigator. L.K. is a James S. McDonnell Centennial Fellow.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2850605.
References
- Benjamini, Y. and Hochberg, Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57: 289-300. [Google Scholar]
- Borevitz, J.O., Liang, D., Plouffe, D., Chang, H.S., Zhu, T., Weigel, D., Berry, C.C., Winzeler, E., and Chory, J. 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13: 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem, R.B., Yvert, G., Clinton, R., and Kruglyak, L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752-755. [DOI] [PubMed] [Google Scholar]
- Cowles, C.R., Hirschhorn, J.N., Altshuler, D., and Lander, E.S. 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432-437. [DOI] [PubMed] [Google Scholar]
- Efron, B., Tibshirani, R., Goss, V., and Chu, G. 2000. Microarrays and their use in a comparative experiment. Technical Report No. 213. Stanford University, Stanford, CA.
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, J.C. 2004. Allele-specific gene expression uncovered. Trends Genet. 20: 113-116. [DOI] [PubMed] [Google Scholar]
- Knight, J.C., Keating, B.J., Rockett, K.A., and Kwiatkowski, D.P. 2003. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat. Genet. 33: 469-475. [DOI] [PubMed] [Google Scholar]
- Lo, H.S., Wang, Z., Hu, Y., Yang, H.H., Gere, S., Buetow, K.H., and Lee, M.P. 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13: 1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks, S.A., Leonardson, A., Zhu, H., Cundiff, P., Pietrusiak, P., Edwards, S., Phillips, J.W., Sachs, A., and Schadt, E.E. 2004. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75: 1094-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, M., Molony, C.M., Weber, T.M., Devlin, J.L., Ewens, K.G., Spielman, R.S, and Cheung, V.G. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 733-734.15306793 [Google Scholar]
- Schadt, E.E., Monks, S.A., Drake, T.A., Lusis, A.J., Che, N., Colinayo, V., Ruff, T.G., Milligan, S.B., Lamb, J.R., Cavet, G., et al. 2003. Genetics of gene expression surveyed in maize, mouse, and man. Nature 422: 269-270. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Richards, D.R., Conway, A.R., Goldstein, A.L., Kalman, S., McCullough, M.J., McCusker, J.H., Stevens, D.A., Wodicka, L., Lockhart, D.J., et al. 1998. Direct allelic variation scanning of the yeast genome. Science 281: 1194-1197. [DOI] [PubMed] [Google Scholar]
- Yan, H., Yuan, W., Velculescu, V., Vogelstein, B., and Kinzler, K.W. 2002. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
- Yvert, G., Brem, R.B., Whittle, J., Akey, J.M., Foss, E., Smith, E.N., Mackelprang, R., and Kruglyak, L. 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57-64. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Miles, M.F., and Aldape, K.D. 2003. A model of molecular interactions on short oligonucleotide microarrays. Nat. Biotechnol. 21: 818-821. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- https://www.affymetrix.com/analysis/download_center.affx; Affymetrix Web site.
- http://www.ncbi.nlm.nih.gov/Traces/; NCBI Trace Archive.