Abstract
Warming at nearly twice the global rate, higher than average air temperatures are the new ‘normal’ for Arctic ecosystems. This rise in temperature has triggered hydrological and geochemical changes that increasingly release carbon-rich water into the coastal ocean via increased riverine discharge, coastal erosion, and the thawing of the semi-permanent permafrost ubiquitous in the region. To determine the biogeochemical impacts of terrestrially derived dissolved organic matter (tDOM) on marine ecosystems we compared the nutrient stocks and bacterial communities present under ice-covered and ice-free conditions, assessed the lability of Arctic tDOM to coastal microbial communities from the Chukchi Sea, and identified bacterial taxa that respond to rapid increases in tDOM. Once thought to be predominantly refractory, we found that ∼7% of dissolved organic carbon and ∼38% of dissolved organic nitrogen from tDOM was bioavailable to receiving marine microbial communities on short 4 – 6 day time scales. The addition of tDOM shifted bacterial community structure toward more copiotrophic taxa and away from more oligotrophic taxa. Although no single order was found to respond universally (positively or negatively) to the tDOM addition, this study identified 20 indicator species as possible sentinels for increased tDOM. These data suggest the true ecological impact of tDOM will be widespread across many bacterial taxa and that shifts in coastal microbial community composition should be anticipated.
Keywords: dissolved organic matter, bacterial diversity, Arctic, Chukchi Sea, microbial community composition, tDOM
Introduction
As much as 50% of the world’s terrestrial organic carbon (C) pool is stored in the northern hemisphere as permafrost and approximately half of that is located within the upper 3 m (Gorham, 1991; Tarnocai et al., 2003, 2009). As atmospheric temperatures rise, the once semi-impermeable layers of permafrost begin to thaw, mobilizing a portion of this C pool by increasing the active layer depth, and then increasing riverine discharge and coastal erosion (Froese et al., 2008; Schuur et al., 2008; Feng et al., 2013; Holmes et al., 2013). This change in hydrology releases water, C, and nutrients, once trapped within the ice, into the coastal ocean. The amount of terrestrially derived dissolved organic matter (tDOM) supplied to the coastal ocean is hard to constrain due to complex environmental factors including changes in climate, hydrology and vegetation (Amon et al., 2012; Kicklighter et al., 2013). However, it is estimated that mobilization of organic C has increased by 3 – 6% from 1985 to 2004 (Feng et al., 2013) and will continue to increase with a warming climate (Amon et al., 2012). Bacterial production in the coastal Arctic appears to be controlled by the bottom up supply of C (Garneau et al., 2008) and the composition of C and nitrogen (N) pools can shape microbial community structure. These microbial responses can impact higher trophic levels, biogeochemical fluxes, and climate feedbacks (McCarren et al., 2010; Vincent, 2010). We hypothesize that tDOM increases will be important to the overall productivity of the Arctic Ocean and suggest that responsive microorganisms could be important sentinels of environmental change. Therefore, understanding what fraction of tDOM is bioavailable, how coastal microbial communities will respond to increases in tDOM and what impact these changes will have on microbial community composition and nutrient cycling is paramount.
Most of the freshwater runoff (both by water volume and DOM contribution) into the coastal Arctic Ocean happens during the short-window of freshet. The term freshet describes the pulse of freshwater that occurs in spring upon the melting of winter snow and ice accumulations. On the Alaskan Arctic coast, >50% of annual freshwater river discharge occurs within a 2 week period in spring (McClelland et al., 2014). This ephemeral fluvial discharge is accompanied by tremendous loads of tDOM and inorganic nutrients (McClelland et al., 2014). As a result, Arctic rivers are highly enriched in dissolved organic C (DOC) and N (DON) (Opsahl et al., 1999; Mathis et al., 2005; Holmes et al., 2008; Frey and Mcclelland, 2009), exposing coastal microbial communities to concentrated pulses of tDOM annually. The tDOM supplied by these Arctic rivers was once thought to be highly refractory (Opsahl et al., 1999; Dittmar and Kattner, 2003; Amon and Meon, 2004; Tank et al., 2012; Xie et al., 2012). Several studies, however, have found that a portion (10 – 40%) of this riverine tDOM is bioavailable on time scales of weeks to months (Hansell et al., 2004; Holmes et al., 2008; Alling et al., 2010; Mann et al., 2012; Vonk et al., 2013). This suggests that much of the bioavailable tDOM is being degraded within estuaries and the coastal ocean before it reaches off-shore waters.
Compared to more temperate regions of the world, study sites in the coastal Arctic are rare. Much of the existing research has focused on the major Arctic rivers, and comparatively little attention has been paid to the impacts of smaller rivers. This lack of knowledge impedes our ability to predict the impact of climate change on local Arctic food webs and biogeochemical cycles (Thingstad et al., 2008). In an effort to fill this knowledge gap, this study used tDOM from the Meade River, Alaska to address two main research objectives: (1) assess the bioavailability of tDOM to microbial communities in the coastal Chukchi Sea and (2) determine how pulsed, high concentration additions of tDOM, impact microbial community composition. These objectives were achieved through a series of bioassay experiments where extracted Meade River tDOM was supplied to microorganisms collected from coastal waters of the Chukchi Sea during ice-covered (April) and open-water (August) conditions. Biological [chlorophyll a (Chl a) and bacterial community composition] and chemical [ammonium (NH4+), nitrate (NO3-), DON, DOC, and phosphate (PO43-)] changes were monitored throughout the experiments. This study provides the first evidence that some tDOM-responsive bacterial taxa could provide early indications of environmental changes in the coastal Arctic.
Materials and Methods
tDOM Collection and Extraction
Approximately 100 L of water was collected from a thermokarst draining into the Meade River, Alaska, at its mouth (70°54′39.1″N, 156°07′25.9″W), on 29 August 2010. The pH was 6.5, the salinity was 4 and the temperature was 7°C. The salinity of the samples is indicative of its proximity to the coast. The thermokarst water was collected into acid washed (10% HCl) high-density polyethylene (HDPE) carboys and transported in freeze safes (∼2 h transit) to the Barrow Arctic Research Center (BARC). The sample was filtered sequentially through pre-rinsed 5-μm polycarbonate filters, combusted (450°C for 4 h) Whatman Glass fiber filters (GF/F; nominal pore size 0.7 μm), and 0.2-μm Supor® (PALL corp) filters. A 10% HCl solution was added to the filtered water to reduce its pH to 2. Samples were stored at 4°C until being shipped in freeze safes to the Virginia Institute of Marine Science (VIMS). Data loggers (HOBO TidbiT v2) were used to monitor water temperature at 60-s intervals during both the transit to BARC and VIMS. Sample temperatures did not increase more than 0.7°C during transit.
Two different extraction methods were compared to identify the method with the highest DOC recovery prior to the large-scale extraction efforts. Sub-samples of the source water were extracted using Superlite DAX-8 resin, described by Aiken (1985) for Amberlite XAD-8 resin or PPL solid phase extraction (Dittmar et al., 2008). Both extraction methods isolate and concentrate the DOM source retaining a fraction of the DOM pool but allowing salts (including inorganic nutrients) to pass through as filtrate. The DOC recovery using the DAX-8 resin was higher than the PPL extractions (61 and 48%, respectively) for the same aqueous sample. Because of its higher recovery, Superlite DAX-8 resin (Aiken, 1985) was used to extract the DOM used in this study. Once isolated, the salinity of the DAX extracted tDOM was adjusted to 30 using a combination of baked (500°C for 4 h) sodium chloride, magnesium sulfate and sodium bicarbonate. The change in salinity causes a portion of the tDOM to precipitate out of solution, mimicking the salinity induced flocculation that occurs within estuaries and deltas. After the salinity was adjusted, the higher salinity tDOM solution was re-filtered (0.2 μm) to remove any particulates that formed through this flocculation process, thus providing a more realistic geochemical signature of the tDOM. The proportion of total riverine DOM that is removed via salinity induced flocculation ranges between 3 and 11% (Sholkovitz, 1976). We found that ∼17% of the DOC was removed during the flocculation of the extracted tDOM (humic) fraction. Prior to use in the bioassay experiments, the tDOM source was brought to the in situ temperature of the experimental waters (-1°C or 4.5°C) to ensure additions of tDOM did not shock the receiving communities.
Field Sample Collection and Bioassays
Seawater samples were collected from a site located 2.5 km northwest of Barrow, Alaska (71°20′39.6″N, 156°41′25.0″W). Whole (unfiltered) Chukchi Sea water was collected from 4 m depth in April and from a depth of 8 m in August. In April, the site was covered by 1.5 m of ice. The site was ice-free in August. Samples were collected in acid washed (10% HCl), 20 L HDPE carboys. The sampling procedures are described in detail in Baer et al. (2017). Bioassays were performed in April (26 April – 1 May 2011) and August (17 – 21 August 2011) and incubated in walk-in chambers located at the BARC in Barrow, Alaska. Temperatures were maintained at -1 °C in April and 4.5 °C in August, corresponding to ambient water temperatures at the time of collection. Both of these experiments were conducted under ambient light conditions, which was greater in August (∼50 μmol quanta m-2 s-1) than in April (4.7 μmol quanta-1 m-2 s-1), so both autotrophic and heterotrophic processes were apparent.
Pulsed DOM addition studies (Bioassays) are particularly applicable for the coastal Arctic. This is based on the well documented large pulses of high tDOM waters to the coast via Arctic rivers (Amon et al., 2012; Holmes et al., 2012, 2013; McClelland et al., 2014). While freshet represents the largest pulse of DOC to the coastal Arctic annually, the flow of DOM via rivers continues throughout the summer and can include pulses of DOM release (e.g., Holmes et al., 2008, 2012). Although DOC data in coastal waters during pulse events are limited, April DOC concentrations in the coastal Beaufort Sea range from ∼100 to 400 μmol C L-1 and August DOC concentrations range between ∼75 and 300 μmol C L-1 (Dunton and Crump, 2014). Our additions were 400 μmol C L-1 tDOM in both seasons. The same concentration and volume was used in April and August to reduce experimental variability and thus microbial response among the study seasons. Although 400 μmol C L-1 tDOM additions may be a bit high for summer incubations, with increasing atmospheric temperatures and the known C reserves contained within permafrost (Gorham, 1991; Tarnocai et al., 2003, 2009), the addition is realistic.
Bioassays were prepared by adding 1.25 L of substrate (extracted tDOM or artificial seawater in the Control treatments) to 8.75 L of whole seawater in acid washed (10% HCl), 10-L polycarbonate carboys, resulting in a 12.5% dilution of the ambient community. The addition of artificial seawater to the control ensured that the dilutions of the microbial community and ambient nutrient pools were consistent among both the control and tDOM addition treatments. This replication of dilution allowed us to specifically target the microbial response to the extracted tDOM. All bioassays were run in duplicate. Bioassays were sub-sampled for Chl a, bacterial community composition, and nutrient concentrations [DOC, total dissolved N (TDN; used to calculate DON), NO3-, nitrite (NO2-), NH4+, and PO43-], at time points 0, 1, 2, 4, and 6 days in April and 0, 1.25, 2.5, and 4 days in August. The duration of the April incubation was longer (6 days) than the August incubation (4 days) due to a shorter field opportunity created by extensive weather delays in summer. Nutrient samples were filtered through combusted (450°C for 2 h) Whatman GF/F filters (nominal pore size 0.7 μm) and stored frozen until analyzed. Bacterial abundance was measured for ambient site samples in both April and August but bacterial abundance and production measurements within the bioassays were only made during the April sampling. The August bioassay bacterial abundance samples were collected but degraded prior to analysis and complications with shipping the radioisotopes to Barrow precluded production measurements in August.
Nutrients and Dissolved Organic Matter Assessment
The colorimetric phenol-hypochlorite method was used to manually measure NH4+ concentrations in triplicate (Koroleff, 1983). Concentrations of NO3-, NO2-, and PO43- were measured using a Lachat QuikChem 8500 autoanalyzer (Parsons et al., 1984). Triplicate TDN and DOC concentrations were measured by high temperature combustion on a Shimadzu TOC-V TNM autoanalyzer (Hansell, 1993; Sharp et al., 2002, 2004). Deep-sea and low-carbon reference water from the University of Miami consensus reference material program were used as a quality control standard for DOC and TDN samples (Hansell, 1993). Concentrations of DON were determined as the difference between the TDN and dissolved inorganic N substrates; the errors from the TDN, NH4+, and NO3- + NO2- measurements were propagated to provide a standard error for DON.
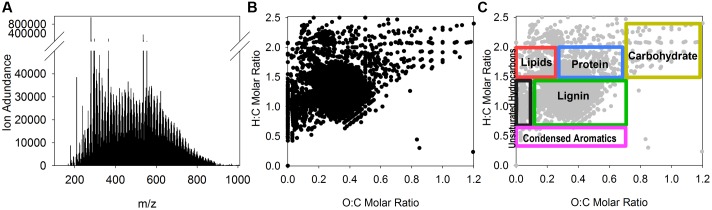
Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) was used to assess the composition of the extracted tDOM. The extracted tDOM was analyzed in positive ionization mode using a hybrid 7 Tesla linear ion trap (LTQ) FT-ICR mass spectrometer equipped with an electrospray ionization (ESI) inlet system (LTQ FT Ultra, Thermo Electron Corp., Wood Hole Oceanographic Institute Mass Spectrometer Facility). Samples were analyzed in the positive ionization mode according to methods described in Sipler et al. (2013) and data were processed and internally calibrated according to Bhatia et al. (2010). FT-ICR MS data are reported as mass-to-charge ratios (m/zs), because the data presented here represent singly charged compounds; an individual m/z generally represents a single compound. The mass range analyzed was 100 – 1000 m/z. Compound classifications were assigned based on general hydrogen to C (H:C) and oxygen to C (O:C) molar ratios. General classifications and their corresponding ranges are described in Hockaday et al. (2009) and listed in Table 1. Multiple studies have used H:C and O:C molar ratios to relate the thousands of compounds generated using FT-ICR to more common compound classifications (e.g., Kim et al., 2003; Hockaday et al., 2009; Bhatia et al., 2010; Ohno et al., 2010; Sleighter et al., 2010; Sleighter and Hatcher, 2011). These classifications provide a general understanding of composition; however, the specific ranges and nomenclature used to distinguish among these classifications continues to evolve (e.g., Kim et al., 2003; Hockaday et al., 2009; Bhatia et al., 2010; Ohno et al., 2010; Sleighter and Hatcher, 2011). For example, the terms lignin, tannin and terrestrial have all been used to describe overlapping ranges (e.g., Hockaday et al., 2009; Bhatia et al., 2010; Killberg-Thoreson et al., 2013). Here we follow the ranges and nomenclature outlined by Hockaday et al. (2009). It should also be noted that although we have provided H:C and O:C ratios for each classification, a high degree of overlap among classifications is expected (Hockaday et al., 2009). We use these general classifications as a tool to group and visualize the potential composition and thus origins of the detected compounds. The aromaticity index (AI) was also calculated for all m/zs with molecular formula assignments (Koch and Dittmar, 2006; Šantl-Temkiv et al., 2013).
Table 1.
Elemental ratios and compound classification characteristics of masses detected in extracted tDOM source using FT-ICR MS.
| Classification | H:C range | O:C range | No. of m/z | % of Total |
|---|---|---|---|---|
| Lipid-like | 1.5 to 2.0 | 0.0 to 0.3 | 494 | 9.2 |
| Protein-like | 1.5 to 2.2 | 0.3 to 0.67 | 673 | 12.6 |
| Lignin-like | 0.7 to 1.5 | 0.1 to 0.67 | 3049 | 57.1 |
| Carbohydrate-like | 1.5 to 2.4 | 0.67 to 1.2 | 143 | 2.7 |
| Unsaturated Hydrocarbon-like | 0.7 to 1.5 | 0.0 to 0.1 | 242 | 4.5 |
| Condensed Aromatics | 0.2 to 0.7 | 0.0 to 0.67 | 63 | 1.2 |
| Other/unidentified | – | – | 679 | 12.7 |
Molecular formulas were assigned to 96% (5343 m/z) of the 5543 m/zs detected. Data represent the classification, range of H:C and O:C molar ratios, the number of m/zs per classification, and proportion of total m/zs detected in positive ionization mode using Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS). Classifications described in more detail in Hockaday et al. (2009).
Biological Assessment
Concentrations of Chl a were measured fluorometrically (Parsons et al., 1984). Bacterial abundance was measured from triplicate samples taken from each incubation bottle and fixed with 4% formaldehyde. Abundance samples were stained with SYBR Green and counted on a flow cytometer (FACSCalibur, Becton-Dickinson). Fluorescing beads were used to approximate the number of cells mL-1 and at least 10,000 cells were counted per replicate. Bacterial production rates were measured via the incorporation of 3H-leucine into protein as a proxy for growth (Kirchman et al., 1985; Smith and Azam, 1992; Ducklow, 2000; Kirchman, 2001). Briefly, triplicate aliquots of 1.5 mL seawater from each treatment bottle were incubated in the dark in a water bath set to temperatures mimicking ambient temperatures for 4 h with 25 nmol L-1 of 3H-leucine (Williams et al., 2016). Incubations were stopped by rinsing each aliquot with 0.1 mL of 100% trichloroacetic (TCA). Protein was extracted by rinsing each aliquot with 1 mL of ice-cold 50% TCA and then by rinsing with 1 mL of 80% ethanol. Samples were centrifuged between each rinse. Parallel incubations for killed controls were done by adding 3H-leucine only after killing cells with 0.1 mL of 100% TCA. Activity measured in killed controls was subtracted from the sample values, eliminating any isotope adsorption to particulate protein.
Bacterial Community Composition
Whole water from the sample sites and each incubation bottle was filtered onto a 0.2-μm Supor filter and frozen at -80 °C. Before extraction, 900 μL of DNA extraction buffer (Fortunato and Crump, 2011), lysozyme (2 mg/mL final concentration) and proteinase K (0.2 mg/mL final concentration) were added to the filters. Samples were then subjected to three freeze-thaw cycles. This was followed by enzymatic lysis in a 30 min 37°C incubation and then continued lysis at 65°C for 1 – 2 h after the addition of SDS (1% final concentration). Two phenol:chloroform:isoamyl alcohol (25:24:1) extractions were then carried out to isolate nucleic acids. Nucleic acids were then precipitated using 100% isopropanol (0.6 × volume of the resulting supernatant) for 2 h up to overnight, pelleted at 13,000 rpm for 30 min, and then rinsed and re-pelleted twice with 70% ethanol before being dried in a roto-evaporator. Once dry, samples were resuspended in 250 mL of nuclease-free water.
The V4 region (515F, GTGCCAGCMGCCGCGGTAA and 806R, GGACTACHVGGGTWTCTAAT) of the 16S rRNA gene for prokaryotic composition was amplified using Earth Microbiome Project protocols1, but with only 30 total cycles. This 806R primer has a bias against SAR11 sequences (Parada et al., 2015) and thus the relative abundance of these taxa and Alphaproteobacteria in general are likely underestimated in our study. Sample libraries were sent to Argonne National Lab for 2 × 150 bp sequencing on the Illumina MiSeq platform and reads were paired using fastq-join (Aronesty, 2013). Sequences that successfully joined were quality filtered, dereplicated (derep_fulllength) and abundance sorted (sortbysize) using UPARSE v 7.0.1001 (fastq_filter) (Edgar, 2013) with an expected error rate of 0.5. Singleton sequences were removed in the latter step in order to prevent them from seeding clusters when clustering operational taxonomic units (OTU). Reads were then clustered (cluster_otus in UPARSE pipeline) at 97% similarity. Reference-based chimera filtering was performed using UPARSE (uchime) with the Gold Database2 as the reference database. Reads (including singletons) were subsequently mapped back to OTUs using UPARSE (usearch_global) and an OTU table created. Taxonomy of the representative sequences was assigned in QIIME v 1.8 (assign_taxonomy.py; Caporaso et al., 2010) using the RDP classifier trained to the Greengenes database (v. 13.83) for 16S amplicons. Any remaining singletons and OTUs occurring in only one sample, chloroplast, mitochondrial and archaeal sequences were removed in QIIME (filter_otus_from_otu_table.py). Sequences passing these quality control filters were uploaded to the NCBI Short Read Archive (SAMN06175504 to SAMN06175533) under the BioProject PRJNA310254. Samples were subsampled to 3037 sequences per sample (the minimum number of sequences per sample in the dataset) before subsequent analyses.
Statistical Analyses
We used ANOVAs (Nelson et al., 2013) and Tukey’s HSB test, which controls for multiple comparisons, to determine if an OTU significantly changed (p < 0.05) over the course of the bioassay (among any of the treatments) and the treatments and time points between which these OTUs changed. These analyses were only run on OTUs that were present in four or more samples and were abundant or became abundant during at least one of the time points. Here we define an abundant OTU as an OTU that reached > 1% of the sample at some point over the course of the bioassay. Replicates were collected from each sample bottle and were grouped by treatment and time point for these statistical analyses. We then calculated the fold-change between the relative abundance of these OTUs at 4 days and in the starting community to determine the magnitude of change in the relative abundance of these significant OTUs over the course of the experiments. Relationships between the average relative abundance of the OTUs that changed throughout the bioassays and average nutrient, Chl a and organic matter concentrations over the course of the bioassays were assessed using Spearman rank correlations. Non-metric multidimensional scaling (MDS) plots were computed in R, using metaMDS in the VEGAN package (Oksanen et al., 2016) and the stress for this ordination (0.0362) was calculated using nmds.monte in the BIOSTATS collection of R functions (Kevin McGarigal4).
Results
Site Characteristics
The thermokarst water sample contained no detectable concentrations of NH4+ or NO3-, but was highly enriched (relative to seawater) in DOC and DON (Table 2). Sixty-one percent of the thermokarst DOC and 56% of the DON pools were retained during tDOM isolation. The molar DOC:DON ratio within the thermokarst water and extracted tDOM were 24 and 26, respectively. Molecular formulas were assigned to 96% (5343 m/z) of the 5543 m/zs detected in the extracted tDOM sample by FT-ICR MS (Figure 1). Masses were detected across the entire analytical range (100–1000 Da.) but the average molecular weight of compounds detected in this sample was 590 Da. The average H:C of detected m/zs was 1.39 and the average O:C of detected m/zs was 0.33 which is similar to other lignin and tannin-rich natural DOM samples from temperate swamps (e.g., Sleighter and Hatcher, 2008; Hockaday et al., 2009; Ohno et al., 2010) and other Arctic permafrost thaw streams (Spencer et al., 2015). Based on the H:C and O:C molar ratios of the individual m/zs, the tDOM was dominated by lignin-like compounds which accounted for 57% of the m/zs for which molecular formulas could be assigned (Table 1). Of the remaining masses, protein, lipid and unsaturated hydrocarbon-like compounds represented 13, 9, and 5% of the compound pool, respectively. Approximately 13% of compounds for which molecular formulas were assigned could not be categorized under the general H:C and O:C molar ratios for known classifications (Table 1). For comparison, we also calculated the AI of the compounds for which molecular formulas could be assigned (Koch and Dittmar, 2006; Šantl-Temkiv et al., 2013). Using this approach we found that approximately 37% (1960 m/zs) of compounds could be categorized as moderately unsaturated, and aromatic and condensed aromatic each accounted for 16 and 15%, respectively. Approximately 64% of compounds that were identified as lignin-like were also identified as moderately unsaturated showing similarities between these two distinctions.
Table 2.
Biological and chemical concentrations in the tDOM source and ambient site water.
| Chl aμg L-1 | Bacterial Abundance 108 cells L-1 | Ammonium μmol N L-1 | Nitrate μmol N L-1 | DON μmol N L-1 | Phosphate μmol P L-1 | DOC μmol C L-1 | |
|---|---|---|---|---|---|---|---|
| Thermokarst | ND | ND | BD | BD | 155 ± 0 | 0.36 ± 0.01 | 3670 ± 17 |
| Extracted tDOM | ND | ND | 0.80 ± 0.52 | BD | 600 ± 14 | 0.08 ± 0.01 | 15430 ± 31 |
| April | 0.10 ± 0.01 | 2.58 ± 0.10 | 0.81 ± 0.05 | 8.41 ± 0.01 | 4 ± 0 | 1.05 ± 0.01 | 68 ± 1 |
| August | 0.37 ± 0.00 | 7.85 ± 0.40 | 0.59 ± 0.02 | 0.32 ± 0.00 | 9 ± 0 | 0.51 ± 0.01 | 94 ± 2 |
The thermokarst water sample was collected on 29 August 2010, and marine site water samples were collected on 26 April 2011 and 17 August 2011. A full survey of this system can be found in Baer et al. (2017). Numbers reported are the average of triplicate samples ± the standard deviation. BD = below detection or < 0.03 μmol N L-1. ND = not determined.
FIGURE 1.
Compound level evaluation of extracted tDOM. Fourier transform ion cyclotron resonance mass spectrometer was used to evaluate the composition of the extracted tDOM. The presented data include (A) FT-ICR mass spectra of extracted tDOM source showing the relative ion abundances of detected mass-to-charge ratios (m/zs); (B) Van Krevelen diagram showing the hydrogen to carbon (H:C) and oxygen to carbon (O:C) molar ratio distributions of masses for which molecular formulas could be assigned (96% of detected m/zs); (C) Van Krevelen diagram of the same points shown in (B) overlaid with general compound class ranges described in Hockaday et al., 2009).
Distinct chemical and biological differences in seawater were observed between April and August. Ambient Chl a and bacterial abundance concentrations were 3- to 4-fold higher in August than April (Table 2). Dissolved inorganic nutrient concentrations were higher in April than August, while DOM concentrations were lower (Table 2). The largest difference in ambient nutrient stocks occurred within the NO3- pool, which was 26-fold higher in April than in August.
Seasonal Differences in In Situ Bacterial Community Composition
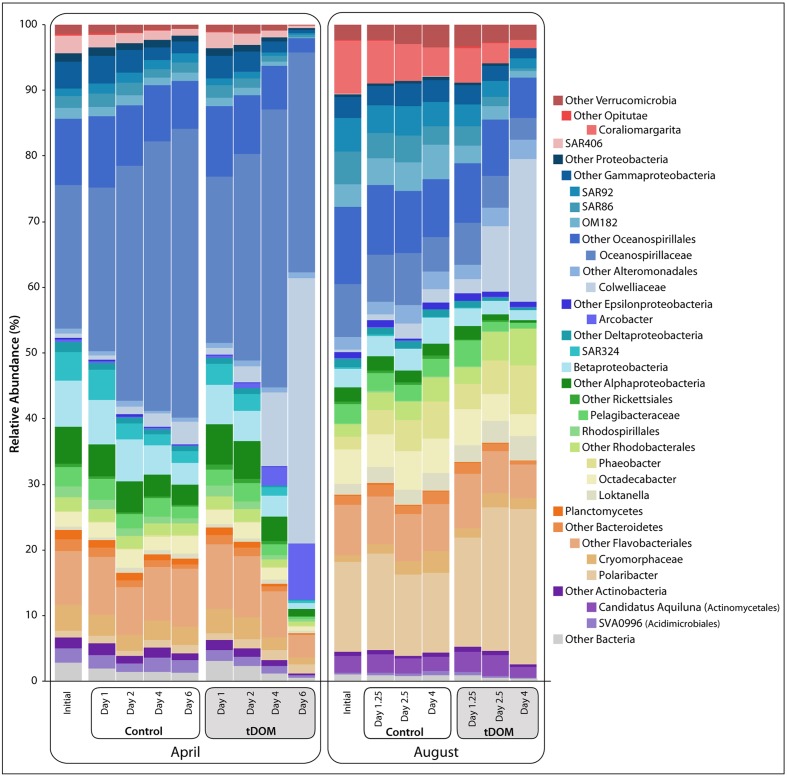
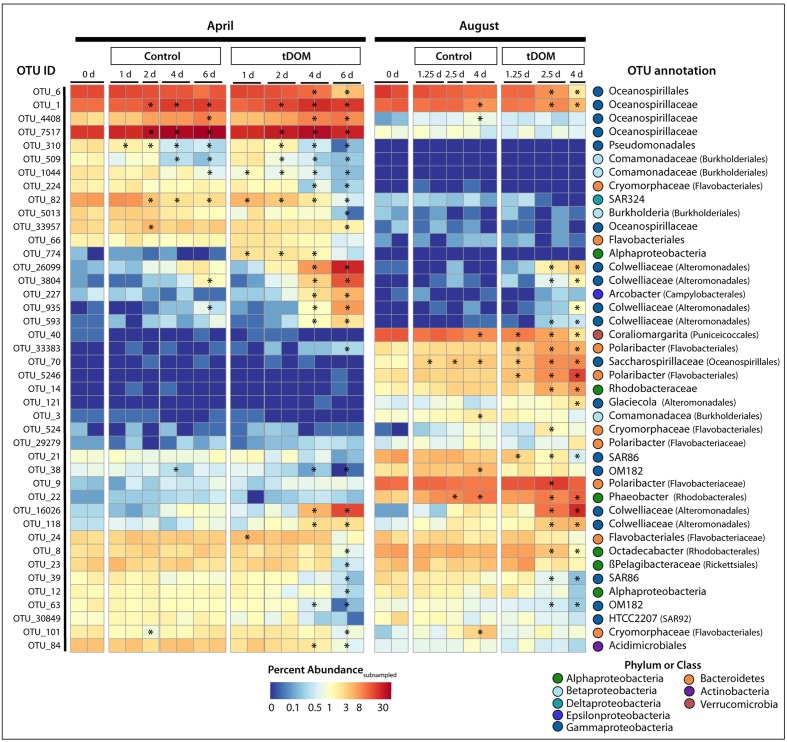
Distinct seasonal differences were observed across multiple taxonomic levels. The ambient communities used in the April and August bioassays shared fewer than 50% of their OTUs. The most abundant taxa classes were Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, and Deltaproteobacteria within the phylum Proteobacteria, as well as phyla Bacteroidetes, Verrucomicrobia, and Actinobacteria (Figure 2). Gammaproteobacteria was the dominant group in both April and August, accounting for approximately 42% and 39% of the populations, respectively. OTUs related to family Oceanospirillaceae were the largest contributors to this class and constituted approximately 22 and 8% of the April and August communities, respectively. Sequences closely related to oligotrophic marine taxa (e.g., SAR86, SAR92 and OM182; Cho and Giovannoni, 2004) constituted only 4.5% of the April population but 13.5% of the starting populations in August. Betaproteobacteria were more prevalent in April (7%) than in August (∼3%), with dominant OTUs identified within the families Comamonadaceae and Burkholderiaceae. Although the total contribution of Alphaproteobacteria was similar between the two seasons (15.7 and 16.4% in April and August, respectively), contributions from unclassified Alphaproteobacteria OTUs were relatively more abundant in April and order Rhodobacterales OTUs had higher relative abundances in August. Bacteroidetes, dominated by Polaribacter spp., comprised only 1% of the April community compared to ∼13.7% in August. Verrucomicrobia (especially Coraliomargarita sp.) was also less abundant in April (<2%) than in August (∼10%).
FIGURE 2.
Bacterial relative abundance with tDOM additions. Stacked bar chart of bacterial taxonomy throughout the course of the bioassays [Controls and treatments with additions of terrestrial-derived dissolved organic matter (tDOM)]. Taxonomy was assigned in QIIME (v. 1.8) using the RDP classifier (Caporaso et al., 2010) based on DNA sequences, randomly subsampled to 3037 sequences per sample, from the v4 region of the 16S rRNA gene. Major bacterial phyla are further broken down into more specific taxonomic ranks (class or order) to highlight the dynamics of these taxa throughout the bioassays.
Less abundant members of the starting bacterial communities, including the class Deltaproteobacteria (SAR324), and the phyla Actinobacteria (Acidimicrobiales sp., SVA0996 clade), and Marine Group A (SAR406), also exhibited distinct seasonal patterns. The proportion of SAR324, was 20-fold greater in April (4.3%) than in August (0.18%). Likewise, proportions of Acidimicrobiales were greater in April (2%) than in August (0.5%). Sequences closely related to SAR406 made up 2.7% of the April population but constituted a negligible proportion of the August community. In August, however, ∼2.5% of the Actinobacteria sequences were closely related to candidate genus “Aquiluna,” a genus that was rare in April.
Biological and Chemical Changes in Bioassays
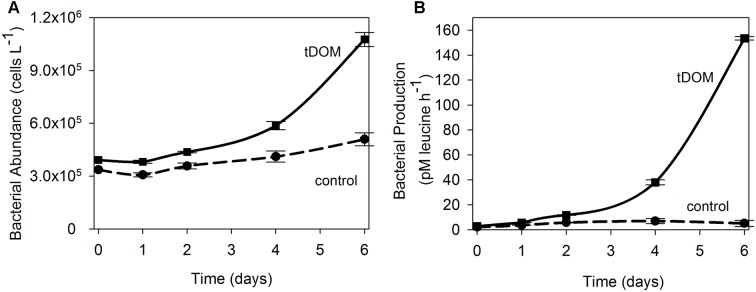
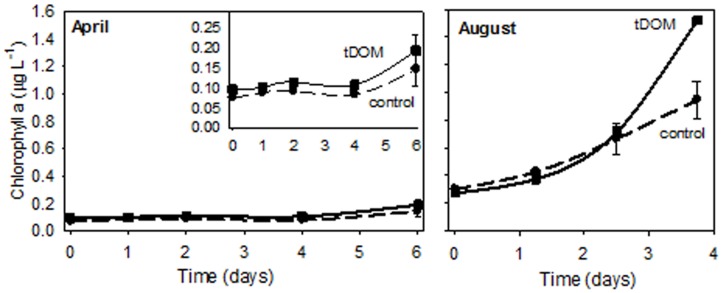
Additions of tDOM affected both ice-covered (April) and open-water (August) microbial communities. Bacterial abundance at the end of the April experiment was 3-fold higher when tDOM was added compared to the Control (Figure 3A). In April, bacterial production increased 2-fold in the Control and 55-fold in the tDOM treatment over the 6-day incubation (Figure 3B). Chl a concentration doubled in both the Control and tDOM treatments in April (Figure 4). In August, however, increases in Chl a concentration were nearly 2-fold higher (1.6 ± 0.0 μg Chl a L-1) in the tDOM treatment than in the no addition Control (0.9 ± 0.1 μg Chl a L-1; Figure 4).
FIGURE 3.
Bacterial response to tDOM additions. Time series of bacterial abundance (A) and bacterial production (B) data for the April bioassays where near coastal Arctic microbial communities were amended with terrestrially derived dissolved organic matter (tDOM). Data points are the mean (n = 2) ± half the range of duplicate samples. The tDOM treatment is depicted by the solid line and the Control treatment, which contains no additional DOM, is depicted by the dashed line.
FIGURE 4.
Chlorophyll a response to tDOM additions. Change in Chl a concentrations for the ice-covered (April) and open-water (August) bioassays. The inset in the April figure zooms in on the lower range of the Chl a scale. Control treatments are indicated by a dashed line and treatments with additions of tDOM are indicated using a solid line.
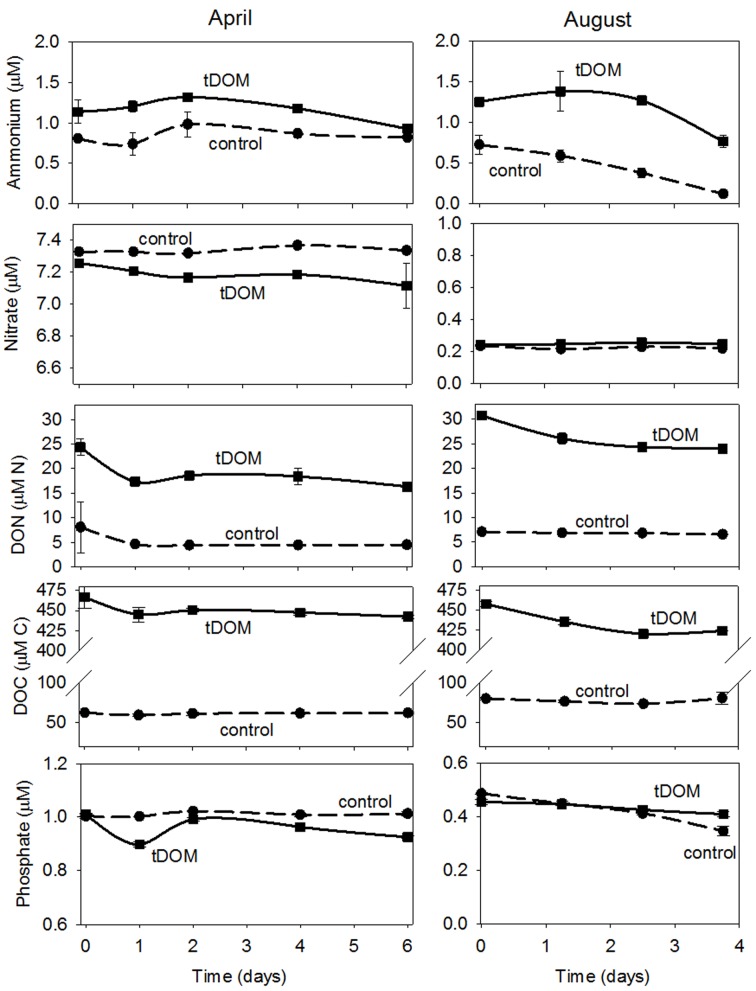
Little change in NH4+ concentration was observed in both the Control and tDOM treatments in April, but NH4+ concentrations decreased by approximately 0.5 μmol N L-1 in both the Control and tDOM treatments in August (Figure 5). Concentrations of NO3- did not change in either treatment or season, even though NO3- concentrations were an order of magnitude higher in April than August (Figure 5). DON concentrations in the tDOM treatments decreased by 7 ± 1 μmol N L-1 in April and by 8 ± 1 μmol N L-1 in August (Figure 5), compared to no change in the Control treatments. DOC concentrations in the tDOM treatments decreased by 29 ± 2 μM and 37 ± 3 μmol C L-1 in April and August, respectively (Figure 5), compared to no change in the Control treatments (Figure 5). Concentrations of PO43- decreased by 0.15 ± 0.04 μmol P L-1 in April and 0.3 ± 0.05 μmol P L-1 in August in the tDOM treatments, compared to no change in the Controls.
FIGURE 5.
Bioassay nutrient concentrations. Time series nutrient concentration data for April and August bioassays where near coastal Arctic water was amended with terrestrially derived dissolved organic matter (tDOM). Data points are the mean (n = 2) ± half the range of duplicate samples. The tDOM treatment is depicted by the solid line and the Control treatment which contains no additional DOM, is depicted by the dashed line.
Bacterial Community Response to tDOM Amendments
A clear shift in community composition was observed in response to tDOM amendments (Figure 2), resulting in decreased bacterial diversity in all tDOM treatments (Supplementary Figure S1) and indicating that tDOM selected for and against certain species and clades. Bacterial diversity also decreased in control bioassays, but to a lesser degree, suggesting that bottle effects may also play a part in bringing about the observed declines in diversity. The tDOM bioassays diverged from the Control communities after 4 days in April and after 2.5 days in August (Figure 2 and Supplementary Figure S2). A total of 138 bacterial OTUs (out of 1656) significantly (p < 0.05) increased or decreased during either April or August. Approximately 42 of these 138 OTUs (e.g., 30%) reached a relative abundance of > 1% in at least one sample, and belonged to phyla Actinobacteria, Bacteroidetes, Proteobacteria, or Verrucomicrobia (Figure 6). Sixteen of these 42 OTUs (38%) changed in either the Control or tDOM bioassays or both (Figure 6). The change in 14 of these 16 OTUs showed significant correlations (ρ > 0.5 or < –0.5 n = 16, p < 0.05) with either Chl a or inorganic nutrient concentrations indicating a response to phytoplankton growth or ambient nutrient stocks (Supplementary Figure S3). While 5 of these 16 OTUs were also correlated with DOC concentration, the correlations with Chl a were greater (ρ > 0.8 or < –0.7, p < 0.05). For 7 out of the 16 OTUs that changed in both Controls and treatments, the change in relative abundance in the tDOM bioassays was at least 2-fold higher in the tDOM treatments than in the Controls, indicating that at least some of the increase in relative abundance was due directly or indirectly to the tDOM addition.
FIGURE 6.
Change in operational taxonomic unit (OTU) relative abundance. Heatmap showing the percent abundance of OTUs identified to significantly change (adjust p-value < 0.05) in relative abundance over the course of the bioassays. Plotted are OTUs that were at least 1% of the total number of subsampled sequences in at least one sample. The OTU ID is noted to the left of the heatmap while the annotation, given as the lowest taxonomic level, is to the right of the heatmap. The bioassay (April or August), treatment (Control or tDOM) and timepoint (in days) are noted above the heatmap. Finally, for each OTU, asterisks within the heatmap denote a sample or pair of samples (duplicates) that have a relative abundance that is significantly greater or less than the Control.
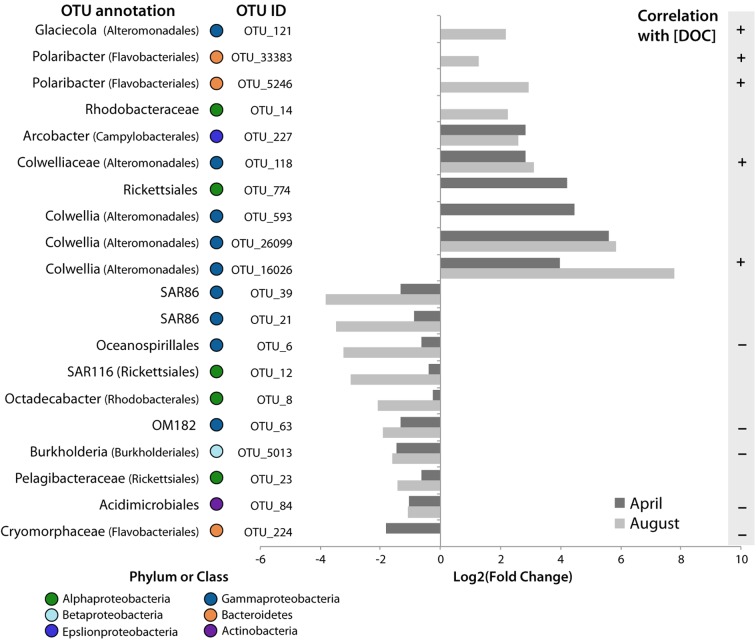
Here we highlight 20 OTUs that increased in the tDOM bioassays, but not in the Controls, and therefore, may be sentinels for increased terrestrial inputs to coastal Arctic systems (Figure 7). Specifically, 10 OTUs responded positively to the addition of tDOM and 5 of these were positively correlated with DOC concentrations (ρ > 0.56, n = 16 for each OTU, p < 0.05, Figure 7). The 10 OTUs that increased in relative abundance come from 2 bacterial phyla (Figure 7): Bacteroidetes (Polaribacter spp.) and three classes within the Proteobacteria (Gammaproteobacteria, Alphaproteobacteria, and Epsilonproteobacteria).
FIGURE 7.
Potential sentinels of increasing tDOM. Log2 of the fold change in relative abundance of OTUs that were found to significantly increase or decrease in tDOM bioassays only. The amount of change at 4 days in each bioassay is plotted. Taxonomic classification of the OTU is noted to the right of the OTU# (more detailed classifications are provided in Figure 2). The presence of a significant (p < 0.05) correlation with DOC concentration is also indicated to the right of the plot. OTU_593 and OTU_33383 also increased in the April bioassay but this change could not be calculated since it was not present at the start of the experiment in these bioassays.
Of the Gammaproteobacteria that significantly increased in the presence of tDOM, four OTUs, which increased in both April and August, were closely related to the family Colwelliaceae [OTU_118, OTU_593, OTU_26099, OTU_16026; (Figure 6)]. Three of these OTUs were closely related to the genus Colwellia (Figure 7). Another gammaproteobacterial taxa that increased in response to tDOM, but only in August, was an OTU related to Glaciecola sp. (OTU_121). While we observed a decline in some groups of marine Roseobacter clade (e.g., Octadecabacter sp.; OTU_8) in the tDOM bioassays, discussed in greater detail below, we observed an increase in other taxa, especially one related to order Rhodobacterales (OTU_14). The relative abundance of an OTU closely related to Arcobacter, an Epsilonproteobacteria, increased in response to tDOM in both seasons (Figure 7).
While the aforementioned OTUs benefited from tDOM, the relative abundance of 10 other OTUs decreased in the tDOM treatment, but not in the Controls. The relative abundance of half of these OTUs were inversely correlated with DOC concentrations (ρ < –0.54, n = 16, p < 0.05), suggesting increased DOM inputs impede the growth of these OTUs relative to other members of the community. The negatively affected OTUs included taxa within Gammaproteobacteria, Alphaproteobacteria, Bacteroidetes, Betaproteobacteria, and Actinobacteria.
Several marine clades of Gammaproteobacteria, including SAR86 (OTU_39, OTU_21), OM182 (OTU_63) and an Oceanospirillales-related OTU (OTU_6) declined significantly (p < 0.05) with the addition of tDOM. Three Alphaproteobacteria OTUs (OTUs 8, 12, and 23) showed a significant decline in relative abundance in the tDOM-amended bioassays, including an OTU belonging to the family Pelagibacteraceae (OTU_23), an Octadecabacter sp. (OTU_8), and a member of the SAR116 clade (OTU_12). Within the Bacteroidetes phylum, an OTU closely related to family Cryomorphaceae (OTU_224, with 100% identity to the Antarctic fosmid ANT39E11; Grzymski et al., 2006) was found to decline throughout the course of the April bioassay.
Discussion
Rivers, estuaries and coastal regions are particularly important for the transformation and degradation of tDOM (Raymond and Bauer, 2000; Battin et al., 2008). Approximately half of the DOC released by Arctic rivers is removed over the continental shelves (Letscher et al., 2011). This project explored the effects of increased tDOM on coastal Arctic microorganisms under ice-covered and ice-free conditions. By investigating the effect of tDOM on microbial community composition we found that additions of tDOM selected for certain species and against others, altering community composition and decreasing bacterial diversity. Increases in tDOM discharge to the coastal Arctic will change both the biogeochemistry and microbial composition of coastal regions.
Bulk Microbial Response to and Lability of tDOM
Concentrations of Chl a increased in all treatments indicating that the autotrophic communities were not nutrient limited at the time of collection. The lower initial Chl a concentration and low light level (4.7 μmol quanta-1 m-2 s-1) may have contributed to the lack of an additional response to the tDOM supplement in April. The divergence between the Chl a concentrations in the Control and in the tDOM treatments in August occurred after incubating for 3 days. Although phytoplankton may have been capable of directly using tDOM, the 3 day delay in growth may indicate that regenerated nutrients or critical microbial byproducts contributed to the observed change in Chl a. Tank et al. (2012) estimated that remineralized riverine N may support up 0.5 to 1.5 Tmol C yr-1 of primary production.
To further support the importance of remineralized nutrients in Arctic microbial growth, we found that reduced forms of N (i.e., NH4+ and DON) were more labile to the receiving community than NO3- which did not change in concentration in either treatment or season on the timescale tested, even though it was an order of magnitude higher in ambient waters in April than in August (Figure 5). This hypothesis is supported by high regeneration rates (15.2 nmol N L-1 h-1) and NH4+ uptake rates (14.1 nmol N L-1 h-1) observed in a parallel study for this specific microbial community (Baer et al., 2017). Concentrations of NH4+ decreased by approximately the same concentration (0.5 μM N) in both the Control and tDOM treatments in August, however, DON may have been remineralized into NH4+ and then quickly used by the community with no net change in concentration. Changes in nutrient concentrations are often not observed in tightly coupled regenerative systems (Sipler and Bronk, 2015).
Approximately 7 – 8% of the DOC supplied in this study was removed during the incubations. Because no change in the bulk DOC concentration was observed in the control treatments from either season it is likely that the majority of DOC lost during the incubation in the tDOM treatments was from the tDOM pool. The lability of the tDOM source to the summer community is possibly underestimated as phytoplankton growth in the tDOM treatment was observed for the final time point and plankton derived DOC production may have masked additional tDOM removal. The timing restrictions of our field season limited the duration of our incubations but it is likely, based on changes in bacterial abundance in spring and increasing Chl a concentrations at the final time point in summer, that additional degradation would occur with longer incubation times.
The 7 – 8% lability presented here is lower than riverine tDOM lability reported by other incubation studies (17 – 62%; Holmes et al., 2008; Mann et al., 2012; Vonk et al., 2013; Spencer et al., 2015). Some of this difference may be due to seasonal or regional differences in composition. For example, a study focusing on the lability DOC from Alaskan Arctic rivers showed DOC collected between July and August was ∼2 –9% labile when incubated in the dark for 90 days (Holmes et al., 2008). This proportion is similar to the 7 – 8% that we observed. Another study conducted in the Russian Arctic found that >50% of DOC in permafrost thaw streams collected in September was bioavailable to riverine bacterial communities on short 7 day time scales (Spencer et al., 2015). The distinct difference in lability over similar time scales between our study and Spencer et al. (2015) could be due to differences in the target microbial communities or DOM composition. For example, our study investigated the response of coastal marine communities while the other aforementioned studies focused on to freshwater riverine communities. The high bacterial production rates show that the community in our study was primed to use DOM but based on the complex compositional characteristics of the tDOM source many of the compounds supplied were likely less easily degraded than simple amino acids used for production estimates. Therefore it is expected that the rate of degradation decreased as the most labile fractions are removed (Cherrier et al., 1999).
Unfortunately, data showing changes in DOM composition over the incubation period for our study are not available and direct comparisons of the FT-ICR MS data between our study and Spencer et al. (2015) are difficult as samples were analyzed in different ionization modes. What we can determine from comparing these studies is that a large proportion of compounds in both sources showed elemental characteristics of being moderately unsaturated and lignin-like, and both showed district clustering of some of the compounds in the lipid/ protein-like or aliphatic region of the van Krevelen diagram (Figure 1). What cannot be determined from our current data is how many of these compounds are shared among both sites/ regions and if the same compounds are labile to both communities.
Another difference between our study and the previous studies is that we did not exclude phytoplankton from our incubations and incubated in the presence of light. The inclusion of primary production could mask the reduction of riverine DOC by supplying an additional source of DOC to a closed system but their presence also provides a more inclusive microbial response to tDOM additions by including both autotrophic and heterotrophic processes. We acknowledge that, like most bioassay studies, our conclusions are limited by the exclusion of higher trophic levels but feel that important information can be gained about potential impacts on the base of the coastal food web from our work assessing the response of the phytoplankton and bacterial community.
Seasonal Differences in In Situ Microbial Community Composition
The extreme physical and chemical differences observed between April and August likely contributed to the distinct variations between the April and August ambient microbial communities. Large seasonal changes in organic matter composition are observed throughout the Arctic (e.g., Crump et al., 2003; Connelly et al., 2015; Mann et al., 2016), and can influence the composition (Crump et al., 2003; Judd et al., 2006) and physiology of the ambient microbial community (Sipler et al., 2017). An evaluation of offshore sites found differences in community composition between seasons to be insignificant (Kirchman et al., 2010), which when combined with our results, further supports the importance of terrestrial influence (tDOM) in shaping coastal microbial communities. The observed variations in community composition are likely influenced by the dramatic seasonal changes in their physiochemical environment. Given the notable differences in the ambient microbial community compositions in April and August, it follows that the microbial response to tDOM will differ seasonally as well.
Gammaproteobacteria were the dominant phylum in both April and August. The largest contributors in April were OTUs related to the Oceanospirillaceae family. Members of this family have been shown to support phytoplankton growth in polar waters through the synthesis of cobalamin (vitamin B12) (Bertrand et al., 2015). Sequences closely related to open-ocean marine taxa (e.g., SAR86 and OM182 (Cho and Giovannoni, 2004) were more prevalent in the nutrient-poor August waters, similar to observations observed by Ghiglione et al. (2012) in Arctic and Antarctic waters.
Betaproteobacteria were more prevalent in April than in August. Members of Betaproteobacteria have been observed throughout Arctic surface waters, especially in the summer (Garneau et al., 2006; Ghiglione et al., 2012) and are often indicators of freshwater inputs. Therefore, their higher relative abundance in April, before the spring freshet, was unexpected. However, a recent study in the Beaufort Sea found that Betaproteobacteria composed up to 20% of bacterial cells sampled from ice-covered waters in February (Alonso-Sáez et al., 2014), suggesting the niche for different members of Betaproteobacteria is wider and potentially more diverse than previously thought.
Proportions of Alphaproteobacteria were similar between the two seasons. Phaeobacter sp. (Roseobacter clade), which was more dominant in August, has been shown to grow in association with phytoplankton (Brinkhoff et al., 2008). They promote algal growth through the synthesis and secretion of antibiotics and algal growth stimulants (Seyedsayamdost et al., 2011).
Bacteroidetes, dominated by Polaribacter spp., comprised ∼13.7% of the August community. Polaribacter are abundant and active members of coastal Arctic and Antarctic bacterial communities (Malmstrom et al., 2007; Ghiglione et al., 2012; Grzymski et al., 2012; Nikrad et al., 2012). Summer conditions appear to favor taxa belonging to the Polaribacter genus. For example, the proportion Polaribacter active in the uptake of simple DOC substrates is 7- to 10-fold higher in summer compared to winter (Nikrad et al., 2012). Several Polaribacter taxa have been observed to have gas vesicles (Gosink et al., 1998) and contain proteorhodopsin (González et al., 2008; Grzymski et al., 2012). Given their high relative abundances and notable activity in polar summer waters as well as their buoyancy-related adaptations, members of this taxa likely play a key role in summer C cycling in coastal Arctic ecosystems, perhaps benefiting from long days.
Verrucomicrobia (especially Coraliomargarita sp.) was another phylum that dominated August samples. The majority of Verrucomicrobia sequences in our samples belong to subdivision 4, which are observed in surface waters globally (Freitas et al., 2012). Verrucomicrobia have been shown to increase in relative abundance in response to phytoplankton-derived DOM inputs (Landa et al., 2014) and may be important polysaccharide degraders in marine environments (Martinez-Garcia et al., 2012).
Although constituting a smaller proportion of the starting communities, Deltaproteobacteria (SAR324), Actinobacteria (Acidimicrobiales sp., SVA0996 clade), and SAR406 also exhibited distinct seasonal patterns. The proportions of two taxa, SAR324 and Acidimicrobiales, that are important in winter N cycling in these waters (Connelly et al., 2014), were greater in April than in August. The higher abundance of SAR324 in April is not surprising as it is known to be a metabolically flexible taxa able to take advantage of diverse nutrient sources (Sheik et al., 2014). SAR406 was more abundant in April but negligible in August. Sequences of SAR406 have been recovered under the sea-ice east of our Barrow field site in the Beaufort Sea (Collins et al., 2010) and in surface waters elsewhere in the Arctic and Antarctic (Ghiglione et al., 2012). Both SAR324 and SAR406 tend to be present in higher abundances in deep waters, yet have also been observed in winter polar surface waters (Ghiglione et al., 2012 and this study), suggesting that their distribution is not limited to deep waters, especially in polar environments. A metagenomic assessment of Antarctic surface waters also highlighted the dominance of chemolithoautotrophic pathways characteristic of these taxa in winter surface waters (Grzymski et al., 2012). The actinobacterial sequences detected in August were closely related to candidate genus “Aquiluna,” a genus that was rare in April. Originally thought to be a freshwater genus (Hahn, 2009), sequences closely related to “ca. Aquiluna” have been recovered from marine environments, including from the Arctic (Kirchman et al., 2010; Kang et al., 2012) and have been observed to contain actinorhodopsin, suggesting the potential for photoheterotrophy by this taxa (Kang et al., 2012).
Bacterial Response to tDOM Amendments
Additions of tDOM selected for certain bacterial taxa and against others, altering community composition and decreasing bacterial diversity. Members of Actinobacteria, Bacteroidetes, Proteobacteria, and Verrucomicrobia significantly responded to the addition of tDOM. Twenty OTUs were identified as potential sentinels for increased terrestrial inputs in coastal Arctic systems (Figure 7). Polaribacter spp. within Bacteroidetes, taxa within Gammaproteobacteria (Colwelliaceae family and Glaciecola spp.), a member of the alphaproteobacterial family Rhodobacteraceae, and an Epsilonproteobacteria related to Arcobacter sp. all increased in relative abundance in the presence of tDOM (Figure 7). Many of the taxa that responded positively to the tDOM additions are common within polar waters, are known to grow in association with phytoplankton, and thrive in high nutrient environments. Common in polar waters in both spring (Malmstrom et al., 2007) and summer (Ghiglione et al., 2012; Grzymski et al., 2012), Polaribacter have been linked to the uptake and decomposition of complex polymeric DOM (Kirchman, 2002). Members of this clade are also often observed in association with polar phytoplankton blooms (Teeling et al., 2012; Landa et al., 2016). Here we extend evidence of their success to high-tDOM environments. Given their ability to thrive in the presence of phytoplankton-derived DOM or tDOM, the Polaribacter genus may be resilient to the ongoing changes in coastal Arctic waters.
The Gammaproteobacteria that significantly increased in the presence of tDOM were closely related to the family Colwelliaceae (Figure 6), specifically the genus Colwellia (Figure 7). Although this family was not very abundant in the starting communities in this study, it is clear that they may become dominant members of polar microbial communities during times of high DOM inputs. Colwellia has a diverse array of metabolic strategies aiding its survival in Arctic waters including blooming in response to phytoplankton-derived DOM (Landa et al., 2016), assimilating bicarbonate (Alonso-Sáez et al., 2010), production of extracellular enzymes capable of degrading high molecular weight compounds (Methé et al., 2005), degrading sinking organic matter (Delmont et al., 2014), and responding to tDOM inputs as described in this study. Glaciecola sp. (Gammaproteobacteria) also increased in response to tDOM. This genus is capable of growing in association with complex DOM sources. For example, this genus has been found in sea ice (Brakstad et al., 2008) and sea water contaminated with crude oil (Chronopoulou et al., 2015), appearing to be able to survive in systems with high hydrocarbon concentrations. Arctic peats naturally contain hydrocarbons (Yunker et al., 1993); therefore, it is not surprising that 16% of the DOM compounds within the tDOM source had elemental characteristics similar to Aromatics and 15% as condensed Aromatics that could have provided a source of DOM for Glaciecola sp.
Roseobacters are a metabolically diverse clade of Alphaproteobacteria (Buchan et al., 2005; Newton et al., 2010). While we observed a decline in some groups of Roseobacter (e.g., Octadecabacter sp.) in the tDOM bioassays, discussed in greater detail below, we observed an increase in other taxa, especially an OTUs related to Rhodobacterales, further highlighting the metabolic breadth of this important clade of marine bacteria. Our results suggest that some taxa within the globally important Roseobacter clade will thrive in a changing Arctic Ocean.
Arcobacter, an Epsilonproteobacteria, also increased in response to tDOM. In marine environments, Arcobacter-related sequences are often associated with metazoans (Gugliandolo et al., 2008; Fontanez et al., 2015), yet, the Arcobacter that responded to tDOM was more closely related (96% similar) to a sulfur-oxidizer (Candidatus Arcobacter sulfidicus) (Wirsen et al., 2002). Because waters between 1 and 18 m depth account for 17% of Arctic shelves (including the site where our seawater samples were collected), and because wave action is expected to increase in the coastal Arctic due to sea ice loss (Overeem et al., 2011), taxa that thrive on tDOM or near sediments (e.g., sulfur-oxidizers) may become more dominant in the water column in the future.
While several OTUs benefited from tDOM, others, including taxa within Gammaproteobacteria, Alphaproteobacteria, Bacteroidetes, Betaproteobacteria, and Actinobacteria, were negatively affected by the presence of tDOM. More than half of the OTUs that decreased in relative abundance in the tDOM bioassays were inversely correlated with DOC concentrations. This suggests that increased DOM inputs will impede the growth of these OTUs relative to other members of the community.
Several marine clades of Gammaproteobacteria that declined with the addition of tDOM have previously shown diverse responses to nutrients and DOM sources. For example, SAR86, which has been shown to have mixed responses to phytoplankton blooms (Obernosterer et al., 2011; Tada et al., 2011; Rinta-Kanto et al., 2012; Wemheuer et al., 2014; Landa et al., 2016), are found in higher concentrations in the open ocean rather than in Arctic coastal waters (Ghiglione et al., 2012). Likewise, members of the OM182 clade exhibit higher growth rates in lower C environments (Cho and Giovannoni, 2004). Given their preference for oligotrophic waters, it is not surprising that SAR86 and OM182 OTUs decreased in the tDOM bioassays. Similar to SAR86, members of the Oceanospirillales order have shown mixed responses to DOM amendments suggesting global variation in how these taxa, or ecotypes within these taxa, respond to tDOM inputs. Some studies observed a response of Oceanospirillales to both phytoplankton-derived and plant-derived organic matter (Mou et al., 2008), but other studies show that some members of this order decrease in relative abundance in response DOM amendments (Landa et al., 2013). The Oceanospirillales-related OTU in our study was negatively correlated with DOC concentration and significantly decreased in both the April and August tDOM incubations.
This study highlights the ecological complexity of Arctic microbes. As discussed above, some OTUs that are known to respond positively to phytoplankton blooms and presumably phytoplankton derived DOM decreased in relative abundance while others increased. Having access to preferred forms of DOM does not mean that the other biogeochemical conditions remain favorable enough for certain species to take advantage of the resource. High levels of DOM, and the metals that they chelate may (1) inhibit some species that benefit from the lower concentration tDOM additions or (2) species that benefit from tDOM additions may simply outcompete or actively inhibit other species neutral to tDOM additions. The underlying mechanisms that control microbial community composition are not fully understood but these mechanisms are likely much more complex than C availability alone.
Alphaproteobacteria, including OTUs belonging to the Pelagibacteraceae family, an Octadecabater sp., and a member of the SAR116 clade, also showed a significant decline in relative abundance in the tDOM amended bioassays. Pelagibacteraceae taxa specialize in the degradation of fresh, low molecular weight dissolved organic matter (Malmstrom et al., 2005; Poretsky et al., 2010), while the tDOM in this study represents a complex mixture of predominantly lignin -like compounds (Table 1), that may not favor the growth of Pelagibacteraceae or other oligotrophs. Similarly, Octadebacter sp. appear to lack the machinery needed to degrade aromatics (Newton et al., 2010), an important characteristic for utilizing many of the tDOM compounds. Paralleling our observations, SAR116 activity was observed to be low in eutrophic temperate coastal waters (Campbell et al., 2009) and ribotypes of this clade have been observed to have higher abundances in open ocean waters during highly oligotrophic periods (Morris et al., 2005).
Within the Bacteroidetes phylum, an OTU closely related to the Cryomorphaceae family (with 100% identity to the Antarctic fosmid ANT39E11; (Grzymski et al., 2006) was found to decline throughout the course of the ice-covered bioassay despite members of this family being known to thrive in high organic matter environments. Members of this taxa, as well as SAR86, the order Oceanospirillales and the Pelagibacteraceae family, have been previously shown to thrive in high DOM environments but the observed decrease in our samples highlights the importance of not generalizing an organism’s preference for high organic matter environments to all types or sources of organic matter. Even microorganisms that thrive in high DOM waters may not do well in polar waters with a high fraction of terrestrial or tundra-derived organic matter.
Although we have focused on OTUs that only responded positively or negatively to the tDOM additions, approximately half of the OTUs that changed in both treatments, increased more in the tDOM treatments than the Controls, indicating that at least a fraction of their increase in relative abundance was due directly or indirectly to the tDOM addition and not solely attributed to bottle effects. For example, commonly considered generalist taxa (Mou et al., 2008), OTUs belonging to the gammaproteobacteria family Colwelliaceae increased in both Control and tDOM bioassays in our study, but the increase in the tDOM bioassays far exceeded that in the Control bioassays (Figure 6), highlighting their potential importance in taking advantage of increasing inputs of tDOM in a changing Arctic Ocean. Because phytoplankton (Chl a) increased in the August tDOM treatments it is hard to differentiate the direct impacts of tDOM or the indirect influence of tDOM promoting phytoplankton growth/ exudates. We argue that this study addresses the overall response, direct and indirect, of target coastal microbial communities to tDOM additions. Several of the OTUs that changed in both the Control and tDOM bioassays significantly correlated with Chl a concentrations or inorganic nutrient concentrations validating their response to phytoplankton growth or ambient nutrient stocks (Supplementary Figure S3). Similarly, while alpha diversity declined in both the Control and tDOM treatments, the magnitude of these decreases was greater in tDOM treatments. This suggests that tDOM amendments enhanced selection for bacterial taxa beyond that which was observed in the controls alone indicating that this selection was not only the result of bottle effects.
Seasonal differences in how quickly microbial communities responded to tDOM were apparent, with August tDOM-amended community composition diverging from control communities more rapidly than April communities (Supplementary Figure S2). It is possible that some of these differences may be temperature driven, given that small increases in temperature can have notable effects microbial metabolism (e.g., Kirchman et al., 2005), but our experimental design does not allow for this effect to be quantified. Nevertheless, our results suggest that summertime microbial communities may be poised to respond more rapidly to pulses of tDOM than communities in early spring. Regardless of season, the addition of tDOM generally shifted bacterial communities toward more copiotrophic and away from more oligotrophic taxa. No one order was found to respond universally (positive or negative) to the tDOM addition. These data suggest that not all sources of DOM (phytoplankton DOM vs. tDOM) are created equal and generalization about the potential impact of a source (e.g., DOM) to a specific phylum, family or clade could be misleading.
Implications for a Warmer More tDOM Rich Arctic
We know that Arctic atmospheric temperatures are rising (Serreze and Francis, 2006; Screen and Simmonds, 2010), that permafrost reserves are thawing (Smith et al., 2005, 2010), and that tDOM inputs to the coastal Arctic are increasing (Fichot et al., 2013). Here we provide evidence that these increases in tDOM to coastal waters of the Arctic Ocean will likely yield a shift in microbial community composition. What remains unknown is how all of these changes will impact global C, N, and phosphorus (P) cycles.
The most obvious impact for the global C cycle is the release of carbon dioxide associated with the respiration and degradation of this abundant and increasing source of organic C. However, a less obvious consequence for the C cycle includes a potential changes in autotrophic and heterotrophic C-fixation. Several studies suggest that increased freshwater discharge will cause an overall reduction in coastal production (e.g., Balch et al., 2012, 2016; Wikner and Andersson, 2012); however, the present experiment provides evidence that some late season Arctic phytoplankton may benefit from tDOM additions through regenerated nutrients. No such benefit was observed in spring when light levels are low. Therefore, the positive Chl a response observed at the final summer time point may be indicative of a seasonal or community dependent response but that the overall impact on production may be negative.
The light absorption by the tDOM ranges into the visible spectrum, which decreases light needed for photosynthesis (Arrigo and Brown, 1996). The light attenuation associated with tDOM may be more physiologically consequential in spring when light is already limiting productivity (Sipler et al., 2017). A companion study Sipler et al. (2017) found that the addition of tDOM reduced C-fixation during a 24 h incubation. Additionally, several of the above-described Alpha- and Gammaproteobacterial taxa, including Octadecabacter and Oceanospirillales spp., that declined in abundance when tDOM was added, are important heterotrophic dissolved inorganic C-fixers (DeLorenzo et al., 2012). While other heterotrophic C-fixers, such as Colwellia sp. (Alonso-Sáez et al., 2012) or Polaribacter sp. (DeLorenzo et al., 2012), were observed to increase in response to tDOM, indicating increasing tDOM inputs may have mixed effects on heterotrophic C-fixation. Although autotrophic C-fixation dominates Arctic primary productivity, heterotrophic C-fixation may be important in seasons or regions (depth) when light is limiting (Alonso-Sáez et al., 2010; Herndl and Reinthaler, 2013).
Indirect impacts of DOM on primary production may also include promoting (Bertrand et al., 2015; Mishamandani et al., 2016) or potentially inhibiting co-existent or mutualistic bacterial species essential for phytoplankton growth. For example, reductions in specific taxonomic groups (e.g., Oceanospirillaceae sp.) may lead to reductions in bacterially synthesized compounds, including cobalamin, that are essential for phytoplankton growth (Bertrand et al., 2015). Thus, a decline in the abundance and activity of these organisms in an Arctic Ocean faced with increased inputs of tDOM may impact not only the cycling of organic C but also the sequestration of inorganic C.
Nutrient budgets will also be affected by increases in tDOM. A recent study investigated the potential for regenerated terrestrial-dissolved organic nutrients to balance Arctic Ocean inorganic nutrient budgets (Torres-Valdés et al., 2016). Although DON inputs appear insufficient in supplying enough N to offset the rather large denitrification deficit (Chang and Devol, 2009) in the Arctic Ocean, terrestrial DON and DOP sources do play an important role in Arctic and North Atlantic N and P cycles. Because microbial communities mediate the magnitude and rate of DOM regeneration, understanding how DOM and microbial pools will change in relationship to one another is critical to predicting the impacts of future environmental perturbations and trends (Kujawinski et al., 2016).
The extent and magnitude at which tDOM inputs increase in the Arctic may determine the future productivity of this sensitive ecosystem. In this interconnected system (Wassmann et al., 2011), changes in phytoplankton and bacterial community composition have enormous consequences for higher trophic levels locally and across the Arctic. Beyond the community changes we observed in response to tDOM amendments, more work is necessary to elucidate how these changes affect community metabolism, ecosystem function, and process rates important in the cycling of globally important elements.
Conclusion
Changes in heterotrophic production and microbial community composition are expected as substantial reserves of tDOM, released from once stable permafrost, become available to coastal microbial communities. Unlike sea ice melt, which has been speculated to cause short term increases in primary production, a large scale loading of a C-rich source may increase carbon dioxide release via respiration and impact the cycling of both organic and inorganic nutrients. Here we have shown that the addition of tDOM promoted certain species and inhibited others, decreased bacterial diversity, thus shifting the microbial community composition. At least 7% of the terrestrial-DOC supplied during this study was consumed by both ice-covered and ice-free microbial communities. The terrestrial-DON fraction was proportionately more labile with ∼ 38% of the supplied DON being removed during the relatively short (4 – 6 day) incubations. Through this effort we have identified 20 OTUs that significantly changed in response to the tDOM addition that may serve as sentinels for future environmental change. Our work is a step forward in identifying microbial shifts that may indicate larger future impacts. Additional studies are needed to investigate temporal and spatial trends of these species as they relate to coastal humification, to confirm their role as true sentinels of change in the Arctic.
Author Contributions
RS, DB, and TC designed the study; RS, DB, TC, and QR collected the samples; RS, CK, TC, and QR analyzed the data; RS, CK, and TC wrote the paper; all authors helped interpret the data and edited the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank M. Frischer, K. Sines, S. Baer, Z. Tait, and L. Killberg-Thoreson for their help in the field. We also thank CH2MHill, the UMIAQ science support staff and the Barrow Whaling Captains Association for their field and logistical support. We thank B. Crump for generously providing the resources for extracting DNA from and sequencing the samples involved in this project. We also acknowledge E. Kujawinski, M. Soule, K. Longnecker and the funding sources of the WHOI FT-MS Users Facility (National Science Foundation MRI program OCE-0619608 and the Gordon and Betty Moore Foundation) for assistance with MS data acquisition. This work is supported by NSF grant# ARC-0909839 to DB and ARC-0910252 to PY. This paper is Contribution No. 3638 of the Virginia Institute of Marine Science, College of William & Mary.
Funding. This work was supported by the United States National Science Foundation grants ARC-0909839 and 0910252. We also acknowledge the funding sources of the WHOI FT-MS Users Facility (National Science Foundation MRI program OCE-0619608 and the Gordon and Betty Moore Foundation) for assistance with MS data acquisition.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01018/full#supplementary-material
References
- Aiken G. R. (1985). “Isolation and concentration techniques for aquatic humic substances,” in Humic Substances in Soil, Sediment, and Water: Geochemistry, Isolation, and Characterization, eds Aiken G. R., MacKnight M. D., Wershaw R. L., MacCarthy P. (Hoboken, NJ: John Wiley and Sons; ),363–385. [Google Scholar]
- Alling V., Sanchez-Garcia L., Porcelli D., Pugach S., Vonk J. E., van Dongen B., et al. (2010). Nonconservative behavior of dissolved organic carbon across the Laptev and East Siberian seas. Glob. Biogeochem. Cycles 24 10.1029/2010gb003834 [DOI] [Google Scholar]
- Alonso-Sáez L., Galand P. E., Casamayor E. O., Pedrós-Alió C., Bertilsson S. (2010). High bicarbonate assimilation in the dark by Arctic bacteria. ISME J. 4 1581–1590. 10.1038/ismej.2010.69 [DOI] [PubMed] [Google Scholar]
- Alonso-Sáez L., Sánchez O., Gasol J. M. (2012). Bacterial uptake of low molecular weight organics in the subtropical Atlantic: Are major phylogenetic groups functionally different? Limnol. Oceanogr. 57 798–808. 10.4319/lo.2012.57.3.0798 [DOI] [Google Scholar]
- Alonso-Sáez L., Zeder M., Harding T., Pernthaler J., Lovejoy C., Bertilsson S., et al. (2014). Winter bloom of a rare betaproteobacterium in the Arctic Ocean. Front. Microbiol. 5:425 10.3389/fmicb.2014.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon R. M. W., Meon B. (2004). The biogeochemistry of dissolved organic matter and nutrients in two large Arctic estuaries and potential implications for our understanding of the Arctic Ocean system. Mar. Chem. 92 311–330. 10.1016/j.marchem.2004.06.034 [DOI] [Google Scholar]
- Amon R. M. W., Rinehart A. J., Duan S., Louchouarn P., Prokushkin A., Guggenberger G., et al. (2012). Dissolved organic matter sources in large Arctic rivers. Geochim. Cosmochim. Acta 94 217–237. 10.1016/j.gca.2012.07.015 [DOI] [Google Scholar]
- Aronesty E. (2013). Comparison of sequencing utility programs. Open Bioinform. J. 7 1–8. 10.2174/1875036201307010001 [DOI] [Google Scholar]
- Arrigo K. R., Brown C. W. (1996). Impact of chromophoric dissolved organic matter on UV inhibition of primary productivity in the sea. Mar. Ecol. Prog. Ser. 140 207–216. 10.3354/meps140207 [DOI] [Google Scholar]
- Baer S. E., Sipler R. E., Roberts Q. N., Yager P. L., Frischer M. E., Bronk D. A. (2017). Seasonal nitrogen uptake and regeneration in the western coastal Arctic. Limnol. Oceanogr. 10.1002/lno.10580 [DOI] [Google Scholar]
- Balch W., Drapeau D., Bowler B., Huntington T. (2012). Step-changes in the physical, chemical and biological characteristics of the Gulf of Maine, as documented by the GNATS time series. Mar. Ecol. Prog. Ser. 450 11–35. 10.3354/meps09555 [DOI] [Google Scholar]
- Balch W., Huntington T., Aiken G., Drapeau D., Bowler B., Lubelczyk L., et al. (2016). Toward a quantitative and empirical dissolved organic carbon budget for the Gulf of Maine, a semienclosed shelf sea. Glob. Biogeochem. Cycles 30 268–292. 10.1002/2015GB005332 [DOI] [Google Scholar]
- Battin T. J., Kaplan L. A., Findlay S., Hopkinson C. S., Marti E., Packman A. I., et al. (2008). Biophysical controls on organic carbon fluxes in fluvial networks. Nat. Geosci. 1 95–100. 10.1038/ngeo101 [DOI] [Google Scholar]
- Bertrand E. M., McCrow J. P., Moustafa A., Zheng H., McQuaid J. B., Delmont T. O., et al. (2015). Phytoplankton–bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc. Natl. Acad. Sci. U.S.A. 112 9938–9943. 10.1073/pnas.1501615112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia M. P., Das S. B., Longnecker K., Charette M. A., Kujawinski E. B. (2010). Molecular characterization of dissolved organic matter associated with the Greenland ice sheet. Geochim. Cosmochim. Acta 74 3768–3784. 10.1016/j.gca.2010.03.035 [DOI] [Google Scholar]
- Brakstad O. G., Nonstad I., Faksness L.-G., Brandvik P. J. (2008). Responses of microbial communities in Arctic sea ice after contamination by crude petroleum oil. Microb. Ecol. 55 540–552. 10.1007/s00248-007-9299-x [DOI] [PubMed] [Google Scholar]
- Brinkhoff T., Giebel H.-A., Simon M. (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189 531–539. 10.1007/s00203-008-0353-y [DOI] [PubMed] [Google Scholar]
- Buchan A., González J. M., Moran M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71 5665–5677. 10.1128/AEM.71.10.5665-5677.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B., Yu L., Straza T., Kirchman D. (2009). Temporal changes in bacterial rRNA and rRNA genes in Delaware (USA) coastal waters. Aquat. Microb. Ecol. 57 123–135. 10.3354/ame01335 [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. X., Devol A. H. (2009). Seasonal and spatial patterns of sedimentary denitrification rates in the Chukchi Sea. Deep Sea Res. Part II Top. Stud. Oceanogr. 56 1339–1350. 10.1016/j.dsr2.2008.10.024 [DOI] [Google Scholar]
- Cherrier J., Bauer J. E., Druffel E. R., Coffin R. B., Chanton J. P. (1999). Radiocarbon in marine bacteria: evidence for the ages of assimilated carbon. Limnol. Oceanogr. 44 730–736. 10.4319/lo.1999.44.3.0730 [DOI] [Google Scholar]
- Cho J.-C., Giovannoni S. J. (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70 432–440. 10.1128/AEM.70.1.432-440.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronopoulou P. M., Sanni G. O., Silas-Olu D. I., van der Meer J. R., Timmis K. N., Brussaard C. P., et al. (2015). Generalist hydrocarbon-degrading bacterial communities in the oil-polluted water column of the North Sea. Microb. Biotechnol. 8 434–447. 10.1111/1751-7915.12176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. E., Rocap G., Deming J. W. (2010). Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ. Microbiol. 12 1828–1841. 10.1111/j.1462-2920.2010.02179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly T. L., Baer S. E., Cooper J. T., Bronk D. A., Wawrik B. (2014). Urea uptake and carbon fixation by marine pelagic bacteria and archaea during the arctic summer and winter seasons. Appl. Environ. Microbiol. 80 6013–6022. 10.1128/AEM.01431-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly T. L., Mcclelland J. W., Crump B. C., Kellogg C. T., Dunton K. H. (2015). Seasonal changes in quantity and composition of suspended particulate organic matter in lagoons of the Alaskan Beaufort Sea. Mar. Ecol. Prog. Ser. 527 31–45. 10.3354/meps11207 [DOI] [Google Scholar]
- Crump B. C., Kling G. W., Bahr M., Hobbie J. E. (2003). Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69 2253–2268. 10.1128/AEM.69.4.2253-2268.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont T. O., Hammar K. M., Ducklow H. W., Yager P. L., Post A. F. (2014). Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front. Microbiol. 5:646 10.3389/fmicb.2014.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorenzo S., Bräuer S. L., Edgmont C. A., Herfort L., Tebo B. M., Zuber P. (2012). Ubiquitous dissolved inorganic carbon assimilation by marine bacteria in the Pacific Northwest coastal ocean as determined by stable isotope probing. PLoS ONE 7:e46695 10.1371/journal.pone.0046695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar T., Kattner G. (2003). The biogeochemistry of the river and shelf ecosystem of the Arctic Ocean: a review. Mar. Chem. 83 103–120. 10.1016/S0304-4203(03)00105-1 [DOI] [Google Scholar]
- Dittmar T., Koch B. P., Hertkorn N., Kattner G. (2008). A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol. Oceanogr. Methods 6 230–235. 10.4319/lom.2008.6.230 [DOI] [Google Scholar]
- Ducklow H. (2000). Bacterial production and biomass in the oceans. Microb. Ecol. Oceans 1 85–120. [Google Scholar]
- Dunton K., Crump B. (2014). Collaborative Research: Terrestrial Linkages to Microbial and Metazoan Communities in Coastal Ecosystems of the Beaufort Sea. Arlington, VA: NSF Arctic Data Center. [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Feng X., Vonk J. E., van Dongen B. E., Gustafsson Ö, Semiletov I. P., Dudarev O. V. (2013). Differential mobilization of terrestrial carbon pools in Eurasian Arctic river basins. Proc. Natl. Acad. Sci. U.S.A. 110 14168–14173. 10.1073/pnas.1307031110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichot C. G., Kaiser K., Hooker S. B., Amon R. M., Babin M., Bélanger S., et al. (2013). Pan-Arctic distributions of continental runoff in the Arctic Ocean. Sci. Rep. 3:1053 10.1038/srep01053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanez K. M., Eppley J. M., Samo T. J., Karl D. M., Delong E. F. (2015). Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front. Microbiol. 6:469 10.3389/fmicb.2015.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato C. S., Crump B. C. (2011). Bacterioplankton community variation across river to ocean environmental gradients. Microb. Ecol. 62 374–382. 10.1007/s00248-011-9805-z [DOI] [PubMed] [Google Scholar]
- Freitas S., Hatosy S., Fuhrman J. A., Huse S. M., Welch D. B., Sogin M. L., et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J. 6 1499–1505. 10.1038/ismej.2012.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey K. E., Mcclelland J. W. (2009). Impacts of permafrost degradation on arctic river biogeochemistry. Hydrol. Proc. 23 169–182. 10.1002/hyp.7196 [DOI] [Google Scholar]
- Froese D. G., Westgate J. A., Reyes A. V., Enkin R. J., Preece S. J. (2008). Ancient permafrost and a future, warmer Arctic. Science 321 1648–1648. 10.1126/science.1157525 [DOI] [PubMed] [Google Scholar]
- Garneau M. È, Roy S., Lovejoy C., Gratton Y., Vincent W. F. (2008). Seasonal dynamics of bacterial biomass and production in a coastal arctic ecosystem: Franklin Bay, western Canadian Arctic. J. Geophys. Res. Oceans 113 C07S91 10.1029/2007jc004281 [DOI] [Google Scholar]
- Garneau M. -È, Vicent W. F., Alonso-Sáez L., Gratton Y., Lovejoy C. (2006). Prokaryotic community structure and heterotrophic production in a river-influenced coastal arctic ecosystem. Aquat. Microb. Ecol. 42 27–40. 10.3354/ame042027 [DOI] [Google Scholar]
- Ghiglione J.-F., Galand P. E., Pommier T., Pedrós-Alió C., Maas E. W., Bakker K., et al. (2012). Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 109 17633–17638. 10.1073/pnas.1208160109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J. M., Fernández-Gómez B., Fernàndez-Guerra A., Gómez-Consarnau L., Sánchez O., Coll-Lladó M., et al. (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc. Natl. Acad. Sci. 105 8724–8729. 10.1073/pnas.0712027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham E. (1991). Northern peatlands: role in the carbon cycle and probable responses to climatic warming. Ecol. Appl. 1 182–195. 10.2307/1941811 [DOI] [PubMed] [Google Scholar]
- Gosink J. J., Woese C. R., Staley J. T. (1998). Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Bacteriol. 48 223–235. 10.1099/00207713-48-1-223 [DOI] [PubMed] [Google Scholar]
- Grzymski J. J., Carter B. J., Delong E. F., Feldman R. A., Ghadiri A., Murray A. E. (2006). Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl. Environ. Microbiol. 72 1532–1541. 10.1128/AEM.72.2.1532-1541.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski J. J., Riesenfeld C. S., Williams T. J., Dussaq A. M., Ducklow H., Erickson M., et al. (2012). A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 6 1901–1915. 10.1038/ismej.2012.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliandolo C., Irrera G., Lentini V., Maugeri T. (2008). Pathogenic Vibrio, Aeromonas and Arcobacter spp. associated with copepods in the Straits of Messina (Italy). Mar. Pollut. Bull. 56 600–606. 10.1016/j.marpolbul.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Hahn M. W. (2009). Description of seven candidate species affiliated with the phylum Actinobacteria, representing planktonic freshwater bacteria. Int. J. Syst. Evol. Microbiol. 59 112–117. 10.1099/ijs.0.001743-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell D. A. (1993). Results and observations from the measurement of DOC and DON in seawater using a high-temperature catalytic oxidation technique. Mar. Chem. 41 195–202. 10.1016/0304-4203(93)90119-9 [DOI] [Google Scholar]
- Hansell D. A., Kadko D., Bates N. R. (2004). Degradation of terrigenous dissolved organic carbon in the western arctic ocean. Science 304 858–861. 10.1126/science.1096175 [DOI] [PubMed] [Google Scholar]
- Herndl G. J., Reinthaler T. (2013). Microbial control of the dark end of the biological pump. Nat. Geosci. 6 718–724. 10.1038/ngeo1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockaday W. C., Purcell J. M., Marshall A. G., Baldock J. A., Hatcher P. G. (2009). Electrospray and photoionization mass spectrometry for the characterization of organic matter in natural waters: a qualitative assessment. Limnol. Oceanogr. Methods 7 81–95. 10.4319/lom.2009.7.81 [DOI] [Google Scholar]
- Holmes R. M., Coe M. T., Fiske G. J., Gurtovaya T., McClelland J. W., Shiklomanov A. I., et al. (2013). Climate change impacts on the hydrology and biogeochemistry of Arctic rivers. Clim. Change Glob. Warm. Inland Waters Impacts Mitigat. Ecosyst. Soc. 3–26. [Google Scholar]
- Holmes R. M., McClelland J. W., Peterson B. J., Tank S. E., Bulygina E., Eglinton T. I., et al. (2012). Seasonal and annual fluxes of nutrients and organic matter from large rivers to the arctic ocean and surrounding seas. Estuaries Coasts 35 369–382. 10.1007/s12237-011-9386-6 [DOI] [Google Scholar]
- Holmes R. M., Mcclelland J. W., Raymond P. A., Frazer B. B., Peterson B. J., Stieglitz M. (2008). Lability of DOC transported by alaskan rivers to the arctic ocean. Geophys. Res. Lett. 35:L03402 10.1029/2007gl032837 [DOI] [Google Scholar]
- Judd K. E., Crump B. C., Kling G. W. (2006). Variation in dissolved organic matter controls bacterial production and community composition. Ecology 87 2068–2079. 10.1890/0012-9658(2006)87[2068:VIDOMC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kang I., Lee K., Yang S.-J., Choi A., Kang D., Lee Y. K., et al. (2012). Genome sequence of “Candidatus aquiluna” sp. strain IMCC13023, a marine member of the Actinobacteria isolated from an Arctic fjord. J. Bacteriol. 194 3550–3551. 10.1128/JB.00586-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kicklighter D. W., Hayes D. J., Mcclelland J. W., Peterson B. J., Mcguire A. D., Melillo J. M. (2013). Insights and issues with simulating terrestrial DOC loading of Arctic river networks. Ecol. Appl. 23 1817–1836. 10.1890/11-1050.1 [DOI] [PubMed] [Google Scholar]
- Killberg-Thoreson L., Sipler R. E., Bronk D. A. (2013). Anthropogenic nutrient sources supplied to a Chesapeake Bay tributary support algal growth: a bioassay and high-resolution mass spectrometry approach. Estuaries Coasts 36 966–980. 10.1007/s12237-013-9604-5 [DOI] [Google Scholar]
- Kim S., Kramer R. W., Hatcher P. G. (2003). Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the van krevelen diagram. Anal. Chem. 75 5336–5344. 10.1021/ac034415p [DOI] [PubMed] [Google Scholar]
- Kirchman D. (2001). Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments. Methods Microbiol. 30 227–238. 10.1016/S0580-9517(01)30047-8 [DOI] [Google Scholar]
- Kirchman D., K’nees E., Hodson R. (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D. L. (2002). The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39 91–100. 10.1016/s0168-6496(01)00206-9 [DOI] [PubMed] [Google Scholar]
- Kirchman D. L., Cottrell M. T., Lovejoy C. (2010). The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12 1132–1143. 10.1111/j.1462-2920.2010.02154.x [DOI] [PubMed] [Google Scholar]
- Kirchman D. L., Malmstrom R. R., Cottrell M. T. (2005). Control of bacterial growth by temperature and organic matter in the Western Arctic. Deep Sea Res. Part II: Top. Stud. Oceanogr. 52 3386–3395. 10.1016/j.dsr2.2005.09.005 [DOI] [Google Scholar]
- Koch B., Dittmar T. (2006). From mass to structure: an aromaticity index for high-resolution mass data of natural organic matter. Rapid Commun. Mass Spectrom. 20 926–932. 10.1002/rcm.2386 [DOI] [Google Scholar]
- Koroleff F. (1983). “Simultaneous oxidation of nitrogen and phosphorus compounds by persulfate,” in Methods of Seawater Analysis, eds Grasshoff K., Eberhardt M., Kremling K. (Weinheim: Verlag Chemie; ). [Google Scholar]
- Kujawinski E. B., Longnecker K., Barott K. L., Weber R. J. M., Kido Soule M. C. (2016). Microbial community structure affects marine dissolved organic matter composition. Front. Mar. Sci. 3:45 10.3389/fmars.2016.00045 [DOI] [Google Scholar]
- Landa M., Blain S., Christaki U., Monchy S., Obernosterer I. (2016). Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J. 10 39–50. 10.1038/ismej.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa M., Cottrell M. T., Kirchman D. L., Blain S., Obernosterer I. (2013). Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquat. Microb. Ecol. 69 157–168. 10.3354/ame01632 [DOI] [Google Scholar]
- Landa M., Cottrell M. T., Kirchman D. L., Kaiser K., Medeiros P. M., Tremblay L., et al. (2014). Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ. Microbiol. 16 1668–1681. 10.1111/1462-2920.12242 [DOI] [PubMed] [Google Scholar]
- Letscher R. T., Hansell D. A., Kadko D. (2011). Rapid removal of terrigenous dissolved organic carbon over the Eurasian shelves of the Arctic Ocean. Mar. Chem. 123 78–87. 10.1016/j.marchem.2010.10.002 [DOI] [Google Scholar]
- Malmstrom R. R., Cottrell M. T., Elifantz H., Kirchman D. L. (2005). Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl. Environ. Microbiol. 71 2979–2986. 10.1128/AEM.71.6.2979-2986.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom R. R., Straza T. R., Cottrell M. T., Kirchman D. L. (2007). Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aquat. Microb. Ecol. 47 45–55. 10.3354/ame047045 [DOI] [Google Scholar]
- Mann P., Spencer R. G. M., Hernes P. J., Six J., Aiken G. R., Tank S. E., et al. (2016). Pan-arctic trends in terrestrial dissolved organic matter from optical measurements. Front. Earth Sci. 4:25 10.3389/feart.2016.00025 [DOI] [Google Scholar]
- Mann P. J., Davydova A., Zimov N., Spencer R. G. M., Davydov S., Bulygina E., et al. (2012). Controls on the composition and lability of dissolved organic matter in Siberia’s Kolyma River basin. J. Geophys. Res. Biogeosci. 117:G01028 10.1029/2011JG001798 [DOI] [Google Scholar]
- Martinez-Garcia M., Brazel D. M., Swan B. K., Arnosti C., Chain P. S., Reitenga K. G., et al. (2012). Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS ONE 7:e35314 10.1371/journal.pone.0035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis J. T., Hansell D. A., Bates N. R. (2005). Strong hydrographic controls on spatial and seasonal variability of dissolved organic carbon in the Chukchi Sea. Deep Sea Res. Part II Top. Stud. Oceanogr. 52 3245–3258. 10.1016/j.dsr2.2005.10.002 [DOI] [Google Scholar]
- McCarren J., Becker J. W., Repeta D. J., Shi Y., Young C. R., Malmstrom R. R., et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U.S.A. 107 16420–16427. 10.1073/pnas.1010732107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland J. W., Townsend-Small A., Holmes R. M., Pan F., Stieglitz M., Khosh M., et al. (2014). River export of nutrients and organic matter from the North Slope of Alaska to the Beaufort Sea. Water Resour. Res. 50 1823–1839. 10.1002/2013WR014722 [DOI] [Google Scholar]
- Methé B. A., Nelson K. E., Deming J. W., Momen B., Melamud E., Zhang X. (2005). The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U.S.A. 102 10913–10918. 10.1073/pnas.0504766102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishamandani S., Gutierrez T., Berry D., Aitken M. D. (2016). Response of the bacterial community associated with a cosmopolitan marine diatom to crude oil shows a preference for the biodegradation of aromatic hydrocarbons. Environ. Microbiol. 18 1817–1833. 10.1111/1462-2920.12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. M., Vergin K. L., Cho J.-C., Rappé M. S., Carlson C. A., Giovannoni S. J. (2005). Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol. Oceanogr. 50 1687–1696. 10.4319/lo.2005.50.5.1687 [DOI] [Google Scholar]
- Mou X., Sun S., Edwards R. A., Hodson R. E., Moran M. A. (2008). Bacterial carbon processing by generalist species in the coastal ocean. Nature 451 708–711. 10.1038/nature06513 [DOI] [PubMed] [Google Scholar]
- Nelson C. E., Goldberg S. J., Kelly, Hass A. F., Smith J. E., Rohwer F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7 962–979. 10.1038/ismej.2012.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R. J., Griffin L. E., Bowles K. M., Meile C., Gifford S., Givens C. E., et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J. 4 784–798. 10.1038/ismej.2009.150 [DOI] [PubMed] [Google Scholar]
- Nikrad M. P., Cottrell M., Kirchman D. (2012). Abundance and single-cell activity of heterotrophic bacterial groups in the western Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 78 2402–2409. 10.1128/AEM.07130-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer I., Catala P., Lebaron P., West N. J. (2011). Distinct bacterial groups contribute to carbon cycling during a naturally iron fertilized phytoplankton bloom in the Southern Ocean. Limnol. Oceanogr. 56 2391–2401. 10.4319/lo.2011.56.6.2391 [DOI] [Google Scholar]
- Ohno T., He Z., Sleighter R. L., Honeycutt C. W., Hatcher P. G. (2010). Ultrahigh resolution mass spectrometry and indicator species analysis to identify marker components of soil-and plant biomass-derived organic matter fractions. Environ. Sci. Technol. 44 8594–8600. 10.1021/es101089t [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2016). Vegan: Community Ecology Package. Available at: http://cran.rproject.org/web/packages [Google Scholar]
- Opsahl S., Benner R., Amon R. M. W. (1999). Major flux of terrigenous dissolved organic matter through the arctic ocean. Limnol. Oceanogr. 44 2017–2023. 10.4319/lo.1999.44.8.2017 [DOI] [Google Scholar]
- Overeem I., Anderson R. S., Wobus C. W., Clow G. D., Urban F. E., Matell N. (2011). Sea ice loss enhances wave action at the Arctic coast. Geophys. Res. Lett. 38:L17503 10.1029/2011GL048681 [DOI] [Google Scholar]
- Parada A. E., Needham D. M., Fuhrman J. A. (2015). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18 1403–1414. 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- Parsons T. R., Maita Y., Lalli C. (1984). A manual of Chemical and Biological Methods for Seawater Analysis. Oxford: Pergamon Press. [Google Scholar]
- Poretsky R. S., Sun S., Mou X., Moran M. A. (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12 616–627. 10.1111/j.1462-2920.2009.02102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond P. A., Bauer J. E. (2000). Bacterial consumption of DOC during transport through a temperate estuary. Aquat. Microb. Ecol. 22 1–12. 10.3354/ame022001 [DOI] [Google Scholar]
- Rinta-Kanto J. M., Sun S., Sharma S., Kiene R. P., Moran M. A. (2012). Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 14 228–239. 10.1111/j.1462-2920.2011.02602.x [DOI] [PubMed] [Google Scholar]
- Šantl-Temkiv T., Finster K., Dittmar T., Hansen B. M., Thyrhaug R., Nielsen N. W., et al. (2013). Hailstones: a window into the microbial and chemical inventory of a storm cloud. PLoS ONE 8:e53550 10.1371/journal.pone.0053550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuur E. A. G., Bockenheim J., Canadell J. P., Euskirchen E., Field C. B., Goryachkin S. V., et al. (2008). Vulnerability of permafrost carbon to climate change: implications for the global carbon cycle. BioScience 58 701–714. 10.1641/B580807 [DOI] [Google Scholar]
- Screen J. A., Simmonds I. (2010). The central role of diminishing sea ice in recent Arctic temperature amplification. Nature 464 1334–1337. 10.1038/nature09051 [DOI] [PubMed] [Google Scholar]
- Serreze M. C., Francis J. A. (2006). The Arctic amplification debate. Climatic Change 76 241–264. 10.1007/s10584-005-9017-y [DOI] [Google Scholar]
- Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J. (2011). The jekyll-and-hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3 331–335. 10.1038/nchem.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. H., Beauregard A. Y., Burdige D., Cauwet G., Curless S. E., Lauck R., et al. (2004). A direct instrument comparison for measurement of total dissolved nitrogen in seawater. Mar. Chem. 84 181–193. 10.1016/j.marchem.2003.07.003 [DOI] [Google Scholar]
- Sharp J. H., Carlson C. A., Peltzer E. T., Castle-Ward D. M., Savidge K. B., Rinker K. R. (2002). Final dissolved organic carbon broad community intercalibration and preliminary use of DOC reference materials. Mar. Chem. 77 239–253. 10.1016/S0304-4203(02)00002-6 [DOI] [Google Scholar]
- Sheik C. S., Jain S., Dick G. J. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16 304–317. 10.1111/1462-2920.12165 [DOI] [PubMed] [Google Scholar]
- Sholkovitz E. R. (1976). Flocculation of dissolved organic and inorganic matter during the mixing of river water and seawater. Geochim. Cosmochim. Acta 40 831–845. 10.1016/0016-7037(76)90035-1 [DOI] [Google Scholar]
- Sipler R. E., Baer S. E., Connelly T. L., Frischer M. E., Roberts Q. N., Yager P. L., et al. (2017). Chemical and photophysiological impact of terrestrially-derived dissolved organic matter on nitrate uptake in the coastal western Arctic. Limnol. Oceanogr. 10.1002/lno.10541 [DOI] [Google Scholar]
- Sipler R. E., Bronk D. A. (2015). “Dynamics of marine dissolved organic nitrogen,” in Biogeochemistry of Marine Dissolved Organic Matter, eds Hansell D. A., Carlson C. A. (Cambridge, MA: Academic Press; ), 127–232. 10.1016/B978-0-12-405940-5.00004-2 [DOI] [Google Scholar]
- Sipler R. E., Bronk D. A., Seitzinger S. P., Lauck R. J., McGuinness L. R., Kirkpatrick G. J., et al. (2013). Trichodesmium-derived dissolved organic matter is a source of nitrogen capable of supporting the growth of toxic red tide Karenia brevis. Mar. Ecol. Prog. Ser. 483 31–45. 10.3354/meps10258 [DOI] [Google Scholar]
- Sleighter R. L., Hatcher P. G. (2008). Molecular characterization of dissolved organic matter (DOM) along a river to ocean transect of the lower Chesapeake Bay by ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 110 140–152. 10.1016/j.marchem.2008.04.008 [DOI] [Google Scholar]
- Sleighter R. L., Hatcher P. G. (2011). Fourier Transform Mass Spectrometry for the Molecular Level Characterization of Natural Organic Matter: Instrument Capabilities, Applications, and Limitations. Rijeka: Intech. [Google Scholar]
- Sleighter R. L., Liu Z., Xue J., Hatcher P. G. (2010). Multivariate statistical approaches for the characterization of dissolved organic matter analyzed by ultrahigh resolution mass spectrometry. Environ. Sci. Technol. 44 7576–7582. 10.1021/es1002204 [DOI] [PubMed] [Google Scholar]
- Smith D. C., Azam F. (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6 107–114. [Google Scholar]
- Smith L. C., Sheng Y., Macdonald G., Hinzman L. (2005). Disappearing arctic lakes. Science 308 1429–1429. 10.1126/science.1108142 [DOI] [PubMed] [Google Scholar]
- Smith S., Romanovsky V. E., Lewkowicz A. G., Burn C. R., Allard M., Clow G. D., et al. (2010). Thermal state of permafrost in North America: a contribution to the international polar year. Permafrost Periglacial Process. 21 117–135. 10.1002/ppp.690 [DOI] [Google Scholar]
- Spencer R. G., Mann P. J., Dittmar T., Eglinton T. I., McIntyre C., Holmes R. M., et al. (2015). Detecting the signature of permafrost thaw in Arctic rivers. Geophys. Res. Lett. 42 2830–2835. 10.1002/2015GL063498 [DOI] [Google Scholar]
- Tada Y., Taniguchi A., Nagao I., Miki T., Uematsu M., Tsuda A., et al. (2011). Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl. Environ. Microbiol. 77 4055–4065. 10.1128/AEM.02952-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank S., Manizza M., Holmes R., Mcclelland J., Peterson B. (2012). The processing and impact of dissolved riverine nitrogen in the arctic ocean. Estuaries Coast. 35 401–415. 10.1007/s12237-011-9417-3 [DOI] [Google Scholar]
- Tarnocai C., Canadell J., Schuur E., Kuhry P., Mazhitova G., Zimov S. (2009). Soil organic carbon pools in the northern circumpolar permafrost region. Globa.l Biogeochem. Cycles 23:GB2023 10.1029/2008GB003327 [DOI] [Google Scholar]
- Tarnocai C., Kimble J., Broll G. (2003). Determining carbon stocks in cryosols using the Northern and Mid Latitudes soil database. Swets Zeitlinger Lisse 2 1129–1134. [Google Scholar]
- Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336 608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Thingstad T. F., Bellerby R. G. J., Bratbak G., Børsheim K. Y., Egge J. K., Heldal M., et al. (2008). Counterintuitive carbon-to-nutrient coupling in an Arctic pelagic ecosystem. Nature 455 387–390. 10.1038/nature07235 [DOI] [PubMed] [Google Scholar]
- Torres-Valdés S., Tsubouchi T., Davey E., Yashayaev I., Bacon S. (2016). Relevance of dissolved organic nutrients for the Arctic Ocean nutrient budget. Geophys. Res. Lett. 43 6418–6426. 10.1002/2016GL069245 [DOI] [Google Scholar]
- Vincent W. F. (2010). Microbial ecosystem responses to rapid climate change in the Arctic. ISME J. 4 1087–1090. 10.1038/ismej.2010.108 [DOI] [PubMed] [Google Scholar]
- Vonk J. E., Mann P. J., Davydov S., Davydova A., Spencer R. G. M., Schade J., et al. (2013). High biolability of ancient permafrost carbon upon thaw. Geophys. Res. Lett. 40 2689–2693. 10.1038/ncomms8856 [DOI] [Google Scholar]
- Wassmann P., Duarte C. M., Agusti S., Sejr M. K. (2011). Footprints of climate change in the Arctic marine ecosystem. Glob. Change Biol. 17 1235–1249. 10.1111/gcb.13152 [DOI] [Google Scholar]
- Wemheuer B., Güllert S., Billerbeck S., Giebel H. A., Voget S., Simon M., et al. (2014). Impact of a phytoplankton bloom on the diversity of the active bacterial community in the southern North Sea as revealed by metatranscriptomic approaches. FEMS Microbiol. Ecol. 87 378–389. 10.1111/1574-6941.12230 [DOI] [PubMed] [Google Scholar]
- Wikner J., Andersson A. (2012). Increased freshwater discharge shifts the trophic balance in the coastal zone of the northern Baltic Sea. Glob. Chang. Biol. 18 2509–2519. 10.1111/j.1365-2486.2012.02718.x [DOI] [Google Scholar]
- Williams C. M., Dupont A. M., Loevenich J., Post A. F., Dinasquet J., Yager P. L., et al. (2016). Pelagic microbial heterotrophy in response to a highly productive bloom of Phaeocystis antarctica in the Amundsen Sea Polynya, Antarctica. Elem. Sci. Anth. 4:02 10.12952/journal.elementa.000102 [DOI] [Google Scholar]
- Wirsen C. O., Sievert S. M., Cavanaugh C. M., Molyneaux S. J., Ahmad A., Taylor L. T., et al. (2002). Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68 316–325. 10.1128/AEM.68.1.316-325.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Bélanger S., Song G., Benner R., Taalba A., Blais M., et al. (2012). Photoproduction of ammonium in the southeastern Beaufort Sea and its biogeochemical implications. Biogeosciences 9 3047–3061. 10.5194/bg-9-3047-2012 [DOI] [Google Scholar]
- Yunker M. B., Macdonald R. W., Cretney W. J., Fowler B. R., Mclaughlin F. A. (1993). Alkane, terpene and polycyclic aromatic hydrocarbon geochemistry of the Mackenzie River and Mackenzie shelf: riverine contributions to Beaufort Sea coastal sediment. Geochim. Cosmochim. Acta 57 3041–3061. 10.1016/0016-7037(93)90292-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.