Abstract
In vitro mapping studies of the MD145 norovirus (Caliciviridae) ORF1 polyprotein identified two stable cleavage products containing the viral RNA-dependent RNA polymerase (RdRp) domains: ProPol (a precursor comprised of both the proteinase and polymerase) and Pol (the mature polymerase). The goal of this study was to identify the active form (or forms) of the norovirus polymerase. The recombinant ProPol (expressed as Pro−Pol with an inactivated proteinase domain to prevent autocleavage) and recombinant Pol were purified after synthesis in bacteria and shown to be active RdRp enzymes. In addition, the mutant His-E1189A-ProPol protein (with active proteinase but with the natural ProPol cleavage site blocked) was active as an RdRp, confirming that the norovirus ProPol precursor could possess two enzymatic activities simultaneously. The effects of several UTP analogs on the RdRp activity of the norovirus and feline calicivirus Pro−Pol enzymes were compared and found to be similar. Our data suggest that the norovirus ProPol is a bifunctional enzyme during virus replication. The availability of this recombinant ProPol enzyme might prove useful in the development of antiviral drugs for control of the noroviruses associated with acute gastroenteritis.
Viruses in the genus Norovirus of the family Caliciviridae are important etiologic agents of acute gastroenteritis (12). A recent study estimates that as many as 23 million cases of norovirus-related illness occur in the United States each year (28). Noroviruses are often associated with outbreaks in settings that include hospitals, nursing homes, cruise ships, schools, and military operations (9). Studies also have suggested a significant role for these viruses as agents of pediatric gastrointestinal disease (31, 34).
Development of control strategies for the noroviruses has been hampered by the unavailability of permissive cell culture systems. However, recent advances in the understanding of norovirus biology and replication have been facilitated by the availability of recombinant proteins expressed from cloned viral genes (15). The approximately 7.6-kb polyadenylated positive-sense RNA genome of norovirus is organized into three open reading frames (ORFs) (16, 21). The first ORF (ORF1) encodes a 200-kDa polyprotein that is processed by the viral 3C-like (3CL) proteinase to release both precursors and cleavage end products (2, 13, 22, 23, 40). The second ORF (ORF2) encodes the major structural protein, VP1 (15), and the third ORF (ORF3) encodes the minor structural protein, VP2 (8). Proteolytic cleavage maps of the ORF1 polyprotein have been determined in in vitro studies for strains representing the two major genetic groups of the noroviruses that are associated with human disease, genogroup I (GI) and genogroup II (GII) (2, 3, 13, 23, 40). The proposed gene order for the norovirus ORF1 polyprotein is N-terminal protein (Nterm), NTPase, p22 (GI) or p20 (GII), VPg, proteinase (Pro), and polymerase (Pol) (2, 22, 23, 40).
The calicivirus RNA-dependent RNA polymerase (RdRp) is related to the picornavirus-like enzymes in supergroup I, represented by the picornavirus 3D polymerase (3Dpol) (20). The proteinase-polymerase (ProPol) protein of Feline calicivirus (FCV), a member of the genus Vesivirus in the Caliciviridae, was the predominant form of the RdRp observed in FCV-infected cells (44), and bacterially synthesized FCV ProPol possessed the enzymatic properties of an RdRp (46). The 58-kDa “3D-like” (3DL) polymerase of Rabbit hemorrhagic disease virus (RHDV), a member of the genus Lagovirus in the Caliciviridae, was also enzymatically active as an RdRp when produced in bacteria (25). The RHDV Pol structure has been resolved to 2.5-Å resolution and has a “right-hand” conformation that is characteristic of other RdRps of positive-strand RNA viruses (32). Both the FCV ProPol and RHDV Pol were able to synthesize RNA from a heteropolymeric template in the absence of added primer, likely by using a hairpin structure at the 3′ end of the RNA template for priming (25, 46). However, comparison of the reported activities of the recombinant FCV and RHDV polymerases indicates that the FCV ProPol was more active than RHDV Pol (46).
In vitro studies of the norovirus ORF1 polyprotein identified both a ProPol precursor protein of 76 kDa and a cleaved 57-kDa form of Pol (2, 40). The fully processed Pol form of the norovirus RdRp was recently expressed as an active enzyme in bacteria, and the three-dimensional structure of the enzyme was reported to exhibit possible unique features, such as the presence of the C-terminal region within the catalytic active-site cleft (33). In this study, we demonstrate that the ProPol precursor of the GII MD145-12 norovirus strain is an active RdRp, suggesting that the norovirus ProPol is a functional form of the RdRp during norovirus replication. The utilization of a bifunctional enzyme with both proteinase and polymerase activity may be a conserved theme in calicivirus replication.
MATERIALS AND METHODS
Viruses and materials.
Norovirus strain MD145-12 (Hu/NV/MD145-12/1987/US) was obtained from a Maryland nursing home resident with gastroenteritis (11), and the sequence of the viral genome was assigned GenBank accession no. AY032605. The FCV Urbana strain (Fe/VV/FCV/Urbana/1968/US) was characterized previously (42). The recombinant FCV ProM-Pol and poliovirus 3Dpol proteins used in this study were reported previously (10, 46). Oligonucleotide primers were purchased from Invitrogen (Carlsbad, Calif.). The nucleotide analogs (compound A, 5-iodo-UTP; compound B, pseudo-UTP; compound C, 5-methyl-UTP; compound D, 2-thio-UTP; compound E, 6-aza-UTP) were from Trilink BioTechnologies (San Diego, Calif.). The nucleoside analogs 5-iodouridine (compound A), pseudouridine (compound B), 5-methyluridine (compound C), and 2-thiouridine (compound D) were from Berry and Associates (Dexter, Mich.). Ribavirin and 6-azauridine (compound E) were from Sigma (St. Louis, Mo.) and Trilink BioTechnologies, respectively. Incorporation experiments were performed with [α-32P]UTP (400 Ci/mmol) (Amersham, Piscataway, N.J.).
Plasmids and mutagenesis.
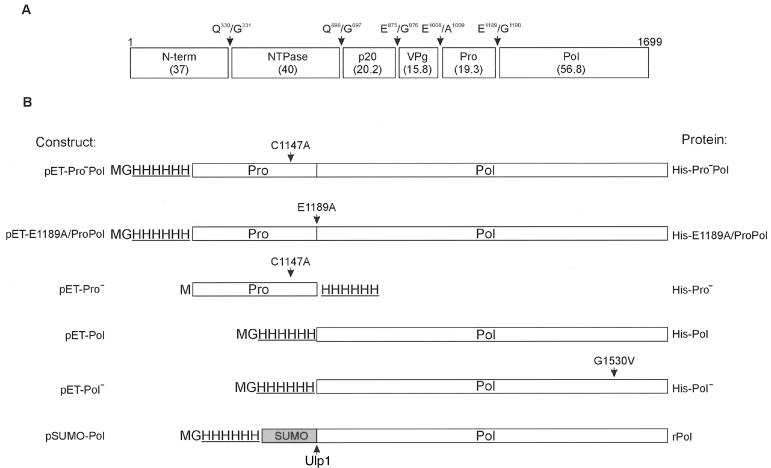
The plasmids used in this study are summarized in Fig. 1. The constructs pET-Pro, pET-Pro−Pol, and His-E1189A/ProPol were previously described (2). The plasmid cORF1 (2) was used as template for generation of the MD145 pET constructs in this study. Plasmids were maintained and selected in Escherichia coli XL1-Blue or DH5α (Invitrogen) in the presence of 30 μg of kanamycin/ml. All constructs were verified by sequence analysis.
FIG. 1.
Diagram of the plasmid constructions and recombinant proteins used in this study. (A) Map of the cleavage sites for ORF1 of the MD145-12 norovirus (2). Sequences of the dipeptide cleavage sites and their positions are indicated by arrows. The calculated molecular mass (in kilodaltons) of each protein is shown in parentheses. The designation of the cleavage products is indicated inside boxes and is adapted from the work of Liu et al. and Green et al. (12, 23). (B) Mutagenized residues are indicated above each construction by an arrow and amino acid change. The cleavage site of the protease Ulp1 is indicated below the SUMO construction by an arrow. A negative sign in superscript position indicates that the proteinase (Pro) or the polymerase (Pol) enzymatic activity was inactivated by mutagenesis. The locations of the engineered His tag (underlined) and the SUMO peptide (gray box) are indicated. Designations of the constructs and recombinant proteins are indicated on the left and the right, respectively.
The plasmid construction pET-Pol was engineered with the pET-28b vector (Novagen, Madison, Wis.). The fragment corresponding to the 3DL Pol coding sequence was amplified by PCR with the forward primer 5′-CATGCCatgGGCCATCATCATCATCATCATGGCGGTGACAGTAAGGGAACCTAC-3′, which included a NcoI site (underlined), a start codon (lowercase), and a His tag-encoding sequence (boldface), and the reverse primer 5′-AAGGAAAAAAGCGGCCGCTCACTCGACGCCATCTTCATTCAC-3′ (the NotI site is underlined) used for the cloning of ProPol into pET-28b (Novagen) (2).
Inactivation of the polymerase domain, encoded by pET-Pol, was performed by site-directed mutagenesis (QuikChange kit; Stratagene, La Jolla, Calif.) of the glycine residue of the YGDD motif to a valine at position 1530 with primer 5′-CTCCTTTTATGTCGATGATGAAATTG-3′ (nucleotide substitution is underlined) and its reverse and complementary primer. The resulting plasmid was designated pET-Pol−. Inactivation of the proteinase encoded by the plasmid pET-Pro was performed by site-directed mutagenesis, with primer C1147A as reported previously (2). The selected construct was designated pET-Pro−.
The plasmid pSUMO-Pol was engineered using the pSUMO vector (LifeSensors, Malvern, Pa.). The polymerase coding sequence was amplified by PCR with the forward primer 5′-AAGGAAAAAACGTCTCAAGGTGGCGGTGACAGTAAGGGAACCTAC-3′, which included a BsmBI site (underlined), and the reverse primer 5′-AAGGAAAAAAGCGGCCGCTCACTCGACGCCATCTTCATTCAC-3′ (the NotI site is underlined), with pET-Pol as the template. The fragment was digested with BsmBI and NotI and ligated into the BsaI and NotI sites of the pSUMO plasmid. The final plasmid (pSUMO-Pol) contained the MD145 Pol coding region fused at its N terminus to a His-tagged SUMO sequence (Fig. 1). A cleavage site for the ubiquitin-like SUMO-specific protease 1 (Ulp1) was present at the carboxyl terminus of the SUMO peptide, so that cleavage with the proteinase Ulp1 would release the precise N terminus of Pol.
Protein synthesis and purification.
Plasmids pET-Pro−Pol, pET-His-E1189A/ProPol, pET-Pol, pET-Pol−, and pET-Pro− were used to transform E. coli BL21(DE3) cells (Novagen). The experimental protocol for the protein expression was previously described (2). After elution of the protein of interest with buffer containing 10% glycerol, 10 mM β-mercaptoethanol (BME), 300 mM NaCl, 50 mM NaH2PO4 (pH 8.0), and either 60 mM imidazole (His-Pro−Pol, His-E1189A/ProPol, His-Pol, and His-Pol−) or 250 mM imidazole (His-Pro−), the recombinant proteins were dialyzed overnight at 4°C against buffer containing 150 mM KCl, 2 mM dithiothreitol (DTT), and 50 mM Tris (pH 8.0) with either 10% (His-Pro−Pol, His-Pol, His-Pol−, and His-Pro−) or 20% (His-E1189A/ProPol) glycerol. For each protein, the quantity was estimated by the Bradford method (4) by using a protein standard and Coomassie blue reagents from Pierce (Rockford, Ill.). The His-E1189A/ProPol was aliquoted, frozen on dry ice, and stored at −80°C. Recombinant His-Pro−Pol, His-Pol, His-Pol−, and His-Pro− were further purified on a 10- to 300-kDa size-exclusion column mounted on a Biologic Workstation (all from Bio-Rad Laboratories, Hercules, Calif.). One milligram of protein in 0.3 to 1 ml of buffer containing 10% glycerol, 150 mM KCl, 2 mM DTT, and 50 mM Tris (pH 8.0) was loaded onto the column at a flow rate of 0.5 ml/min. The first 5 ml of the flowthrough was discarded, and the rest was collected into 0.3-ml fractions. During the fraction collection, the presence of protein was monitored by UV absorbance at 280 nm. The purity of each protein fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the RdRp activity was assayed as described below. The fractions showing the highest RdRp activity and purest protein profiles in SDS-PAGE were pooled and dialyzed overnight at 4°C against 20% glycerol-150 mM KCl or 100 mM KCl-2 mM DTT-50 mM Tris (pH 8.0). The samples were then aliquoted and stored at −80°C.
BL21(DE3) cells transformed with pSUMO-Pol were grown at 37°C in 4 liters of NZCYM medium (Fisher, Pittsburgh, Pa.) containing 25 μg of kanamycin/ml until the optical density at 600 nm reached 0.6. The bacteria were then cooled to 25°C prior to induction with IPTG (isopropyl-β-d-thiogalactopyranoside; 500 μM) and grown for an additional 4 h. The cells were pelleted, washed once with 200 ml of 10 mM Tris (pH 8.0)-1 mM EDTA, and pelleted again. The supernatant was removed, and the bacterial pellet was frozen and stored at −70°C. Approximately 20 g of frozen cells was thawed and suspended in lysis buffer containing 100 mM potassium phosphate (pH 8.0), 0.5 mM EDTA, 20% glycerol, 1 mM BME, 1.4 μg of pepstatin A/ml, and 1.0 μg of leupeptin/ml (5 ml/g of cell paste). The cells were resuspended and lysed with a French press, followed by the addition of phenylmethylsulfonyl fluoride and NP-40 to a final concentration of 1 mM and 0.1%, respectively. The cell lysate was centrifuged at 100,000 × g for 30 min at 4°C prior to loading onto a 5-ml nickel-nitrilotriacetic acid (Ni-NTA)-agarose column at a 1-ml/min flow rate. The column was washed once with 20 ml of buffer A (50 mM Tris [pH 8.0], 20% glycerol, 1 mM BME, 500 mM NaCl, and 0.1% NP-40) and 10 ml of buffer A containing 5 mM imidazole. The His-tagged SUMO-Pol fusion protein (His-SUMO-Pol) was eluted with buffer A containing 500 mM imidazole. The His-SUMO-Pol was dialyzed against buffer A containing 50 mM NaCl and was loaded onto a 5-ml phosphocellulose column at a flow rate of 1 ml/min. The phosphocellulose column was washed to baseline with buffer A containing 50 mM NaCl, and His-SUMO-Pol was eluted with buffer A containing 200 mM NaCl. The His-SUMO-Pol was diluted fourfold with buffer A to a 50 mM NaCl final concentration and loaded onto a 1-ml Q-Sepharose column at a flow rate of 1 ml/min. The Q-Sepharose column was washed to baseline with buffer A containing 50 mM NaCl, and His-SUMO-Pol was eluted with buffer A containing 400 mM NaCl.
Five units of His-tagged Ulp1 (10 μg) (Life Sensors, Inc.) was incubated with 500 μg of His-SUMO-Pol in a final volume of 100 μl at 4°C for 5 h in buffer A containing 200 mM NaCl. The reaction mixture was passed over a Ni-NTA-agarose column to retain the His-tagged SUMO peptide and Ulp1 proteinase. The purified recombinant Pol (rPol) was collected in the flowthrough, the protein concentration was determined, and the protein was frozen and stored at −80°C.
RdRp assays.
The RdRp activity assay was performed as described by Sankar and Porter with some modifications (39). The reaction was carried out in a 15-μl mixture containing 20 μg of poly(rA) (Roche, Indianapolis, Ind.)/ml, 10 μM poly(rU)15 (Invitrogen), 1 μM [α-32P]UTP (400 Ci/mmol), 10 or 100 μM UTP, 50 mM HEPES buffer (pH 7.4), 3 mM MgCl2 or as otherwise indicated, 10 mM DTT, 60 μM ZnCl2, 200 μg of rifampin/ml, and 1 μM MD145-12 His-Pro−Pol, His-Pol, or rPol. Temperature, incubation times, salt concentrations, and amounts of the recombinant proteins and nucleotides are detailed in the figure legends. Each incorporation assay was stopped by addition of 0.5 M EDTA to a final concentration of 83 mM. Six microliters of the reaction mixture was spotted on a DE81 membrane (Whatman, Clifton, N.J.). Once dried, the disks were washed three times in 5% dibasic sodium phosphate for 10 min at room temperature. The membranes were then rinsed in absolute ethanol, and the bound radiolabeled polynucleotide was counted in liquid scintillation fluid (ICN, Costa Mesa, Calif.).
Template switching activity was examined in an RdRp assay mixture containing 4.3 μM MD145 His-Pro−Pol, 5 mM MgCl2, 10 μM poly(dT)15-poly(rA)30, and 0.67 μM [α-32P]UTP (40 Ci/mmol) (NEN). Reaction mixtures were initiated by the addition of enzyme and incubated for 3 min at 30°C. The reaction mixtures were then diluted 10-fold by addition of heparin (10 μM) and UTP (500 μM) with or without acceptor templates (100 μM) of poly(rA)30 or poly(dA)30. The reaction was allowed to proceed for 0.5, 1, 2, 3, 5, or 10 min, at which point the reaction was stopped by the addition of EDTA to a final concentration of 50 mM. Products were resolved in a 1× Tris-borate-EDTA-10% polyacrylamide gel and visualized with a PhosphorImager (Molecular Dynamics).
The transcriptase activity was examined using a natural RNA as template. In these experiments, the MluI-linearized plasmid pGLT7 was employed for synthesis of the RNA template, by using a Ribomax kit (Promega) as previously described (46). The resulting product was an 830-nucleotide (nt)-long RNA transcript, and a control RNA marker was synthesized by including [α-32P]UTP in a separate reaction. A 50-μl elongation reaction mixture included 50 mM HEPES (pH 8.0), 4 mM DTT, 3 mM MgCl2, 50 mM potassium acetate, 10 μM GTP, 10 μM ATP, 10 μM CTP, 5 μM UTP, 10 μCi of α-32P-labeled UTP, 20 μg of rifampin/ml, 40 U of RNasin (Roche), 1 μM recombinant enzyme, and 1 μg of RNA template. The reaction mixture was incubated for 1 h at 30°C. RNA was extracted with the RNAeasy kit (Qiagen) and precipitated with ethanol. The samples were then denatured in glyoxal-dimethyl sulfoxide buffer (Ambion, Inc., Austin, Tex.) for 40 min at 65°C and analyzed in a 1% agarose gel (Ambion; Northern Max-Gly kit) that was dried and exposed to X-Omat AR film (Kodak, Rochester, N.Y.). Products from the elongation assay were incubated with RNase T1 (Ambion) at 2 U/μl for 30 min at 37°C, followed by purification and analysis in a 1% agarose gel as described above.
Inhibition assays.
The norovirus His-Pro−Pol (1 μM) was incubated for 20 min at 30°C in a poly(A)-dependent UMP incorporation assay that included 100 μM UTP, 5 μCi of [α-32P]UTP (400 Ci/mmol), 3 mM MgCl2, and increasing concentrations (1, 5, 50, 100, and 200 μM) of UTP analogs. The UMP incorporation was measured as described above and compared to a control assay without nucleotide analog. For the virus growth inhibition experiments, CRFK cells were preincubated for 1 h at 37°C, in the presence of 2 mM nucleoside analogs or ribavirin in virus growth medium (VM) consisting of Eagle's minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum, amphotericin B (2.5 μg/ml), chlortetracycline (25 μg/ml), penicillin (250 U/ml), and streptomycin (250 μg/ml). The cells were washed, inoculated with FCV at a multiplicity of infection of 2, and incubated for 1 h at 37°C in VM containing 2 mM nucleoside. The inoculum was then replaced with fresh Eagle's minimal essential medium containing 2 mM nucleoside analogs without fetal bovine serum and incubated for 7 h at 37°C. A mock-treated virus control was included in all experiments. Infected cells were frozen and thawed prior to plaque titration. Cell monolayers were inoculated with serial 10-fold dilutions of virus samples and incubated for 1 h at 37°C. The inoculum was removed, and cells were overlaid with 0.8% agarose prepared in VM. Plaques were scored after 48 h, following formalin fixation of the monolayers and staining with crystal violet (Sigma).
RESULTS
Synthesis and purification of norovirus proteins.
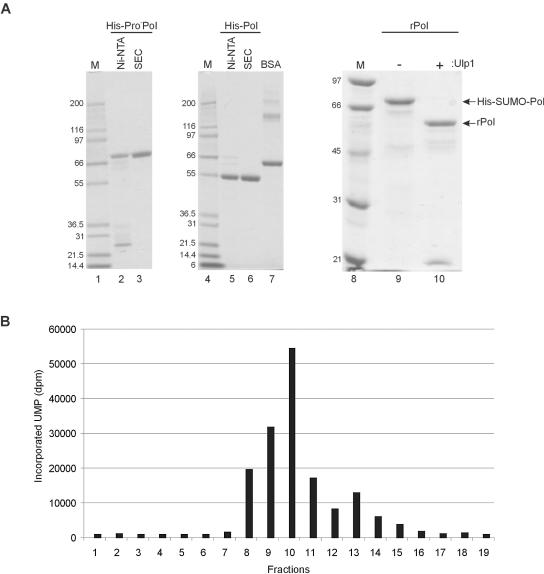
The recombinant norovirus proteins Pro−Pol, A1189A/ProPol, Pol, Pol−, and Pro− (all containing an engineered His6 tail) were synthesized in E. coli BL21(DE3) cells following IPTG induction. Following addition of the soluble material to a Ni-NTA column, the bound recombinant protein was eluted and immediately dialyzed prior to size exclusion chromatography (SEC) (His-Pro−Pol, His-Pol, His-Pol−, and His-Pro−) or storage (His-E1189A/ProPol). Analysis of all final purified recombinant proteins by SDS-PAGE was performed to determine their purity (over 90%) as shown for His-Pro−Pol and His-Pol (Fig. 2A).
FIG. 2.
Purification of recombinant MD145-12 proteins. (A) SDS-PAGE analysis of His-Pro−Pol (lanes 2 and 3), His-Pol (lanes 5 and 6), His-SUMO-Pol (lane 9), and rPol (lane 10). Purification steps (Ni-NTA binding and SEC) and Ulp1 proteinase treatment are indicated above each lane. One microgram of His-Pro−Pol and His-Pol was separated by SDS-PAGE in a 4 to 12% Bis-Tris polyacrylamide gel (Invitrogen). One microgram of bovine serum albumin (BSA; Pierce) was added as control for the protein estimation (lane 7). Two and one-half micrograms of SUMO-Pol and rPol was resolved by SDS-PAGE in a 10% polyacrylamide gel. Proteins were visualized by staining with Coomassie blue. Lanes 1, 4, and 8 contain protein molecular weight markers (weights are at left in thousands). (B) RdRp assay of His-Pro−Pol fractions obtained during SEC. One microliter of each fraction was assayed in a 15-μl reaction mixture that contained 3 mM MgCl2, 10 μM UTP, and 5 μCi of [α-32P]UTP (400 Ci/mmol) for 20 min at 30°C. For this experiment and all subsequent experiments 200 μg of rifampin/ml was added. Other components of the RdRp assay mixture are described in Materials and Methods. Fraction numbers are indicated below the graph. UMP incorporation is given in disintegrations per minute (dpm) and corresponds for each fraction to an aliquot of 5 μl spotted on a DE81 filter.
One milligram of the Ni-NTA enriched material was further purified by SEC, and 1 μl of each collected fraction was assayed for activity by using poly(A) as a homopolymeric template in an RdRp activity assay in the presence of rifampin, a potent inhibitor of bacterial DNA-dependent RNA polymerase. For His-Pro−Pol, the polymerase activity was detected in fractions 8 through 15 with maximal activity in fraction 10 (Fig. 2B). Fractions 8 through 11 were pooled, concentrated in an Amicon spin column (Millipore), dialyzed against 150 mM KCl buffer, and stored at −80°C. The same procedure (SEC and RdRp assay) was also used for the purification of His-Pol (data not shown). For His-Pro−Pol and His-Pol, no RdRp activity was detected in the absence of template poly(A) or MgCl2 in the reaction (data not shown). These experiments indicated that MD145-12 His-Pro−Pol and His-Pol were functional RdRp enzymes, whereas no activity was detected for His-Pol− and His-Pro− (data not shown).
RdRp activities of norovirus His-Pro−Pol, His-Pol, and rPol.
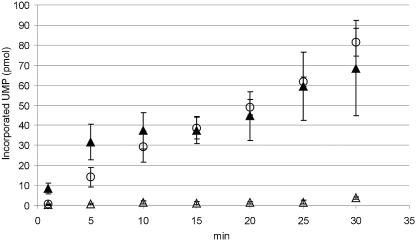
ProPol and Pol are active forms of the FCV and RHDV polymerases, respectively (25, 46). We previously proposed that the norovirus polymerase may exist in at least two forms, Pol and ProPol (2). In this study, the preliminary data suggested that both forms of the polymerase (Pol and ProPol) were enzymatically active. Experiments were performed to compare the polymerase activities of His-Pro−Pol and His-Pol (Fig. 3). His-Pro−Pol and His-Pol from three different preparations were analyzed concurrently under the same conditions. The His-Pro−Pol was consistently at least 3 (1 min) to 36 (25 min) times more active than His-Pol (Fig. 3). Calculation of the UMP incorporation rate by His-Pro−Pol at any given time point showed that the average rate was 2.4 ± 0.71 pmol/min. In contrast, the incorporation rates for Pol decreased rapidly over time (from 0.26 pmol/min at 1 min to 0.13 pmol/min at 30 min), with almost no activity detected after 5 min of incubation at 30°C. To examine whether the presence of the His tag at the amino terminus of His-Pol impaired its RdRp activity, the MD145 Pol was expressed and purified in a system (SUMO) that would generate the precise N terminus. The resulting protein, designated rPol (Fig. 2A, lane 10), was assayed under the same conditions as His-Pol and His-Pro−Pol. The incorporation rate of rPol varied from 6.1 ± 1.8 pmol/min (1 through 10 min) to 2.35 ± 0.63 pmol/min (15 through 30 min), with the latter incorporation rate comparable to that of His-Pro−Pol. These data indicate that the presence of six histidine residues at the N terminus of Pol has an inhibitory effect on its RdRp activity.
FIG. 3.
Kinetics of norovirus His-Pro−Pol, rPol, and His-Pol UMP incorporation. (A) For each time point (1, 5, 10, 15, 20, 25, and 30 min), a 15-μl reaction mixture containing His-Pro−Pol (open circles), rPol (solid triangles), or His-Pol (open triangles) at 1 μM was incubated up to 30 min at 30°C. The reaction mixture contained 100 μM UTP, 5 mM MgCl2, and 5 μCi of [α-32P]UTP. The reaction was stopped by adding 3 μl of 0.5 M EDTA, and 6 μl was spotted onto a DE81 filter. The amount of incorporated UMP is given in picomoles (ordinate). For both proteins, mean results and standard deviations of three independent experiments are shown (vertical bars).
Parameters affecting the RdRp activity of the His-Pro−Pol.
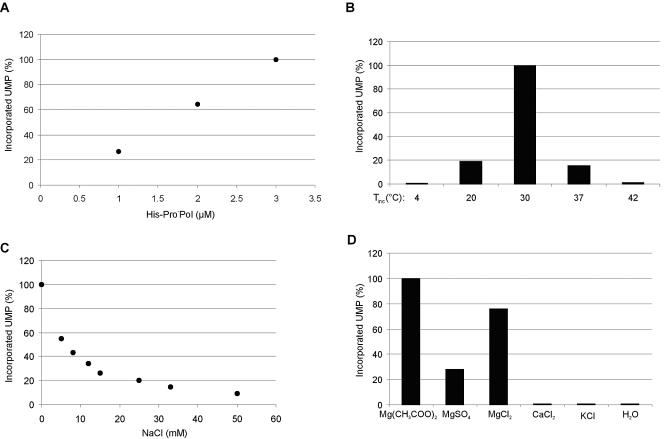
The effects of several parameters on the activity of the newly identified ProPol form of the norovirus RdRp were evaluated. The amount of incorporated UMP correlated with the quantity of His-Pro−Pol in the reaction (Fig. 4A), and the enzyme performed optimally at 30°C (Fig. 4B). The effect of NaCl on the RdRp activity of His-Pro−Pol was analyzed in the presence of increasing concentrations of salt from 0 to 50 mM. The His-Pro−Pol activity decreased in a dose-dependent manner with a 50% reduction of the RdRp activity in the presence of 6.5 mM NaCl (Fig. 4C). In comparison, 30 mM NaCl was needed to cause a 50% reduction of the RdRp activity for FCV rProM-Pol (46). The effect of various divalent cations and salts on the activity of His-Pro−Pol was examined (Fig. 4D; Table 1). Magnesium was optimal in an acetate form [Mg(CH3COO)2], and no RdRp activity was detected with CaCl2 or KCl (Fig. 4D). It was previously shown for the RHDV polymerase that magnesium could be replaced by manganese (26). We tested the RdRp activity of His-Pro−Pol in the presence of MgCl2 or MnCl2 (0.1 to 40 mM) (Table 1). The optimal concentration of MgCl2 was 5 mM (62 pmol of incorporated UMP), and that of MnCl2 was 10 mM (102 pmol of incorporated UMP), with the amount of incorporated UMP being approximately 1.7 times greater in the presence of MnCl2 than MgCl2.
FIG. 4.
Effect of various conditions on the activity of norovirus His-Pro−Pol. (A) Effect of the enzyme concentration on the amount of incorporated UMP. One, 2, and 3 μM concentrations of His-Pro−Pol were assayed in a 15-μl reaction mixture containing 3 mM MgCl2, 100 μM UTP, and 5 μCi of [α-32P]UTP. The mixture was incubated for 20 min at 30°C and stopped with 0.5 M EDTA. Six microliters of the final mixture was spotted onto a DE81 membrane for counting. Final concentrations of the recombinant polymerase are indicated below the graph. The incorporated UMP is normalized against the value of 3 μM His-Pro−Pol (100%). (B) The polymerase activity of His-Pro−Pol (1 μM) was analyzed following incubation for 30 min at various temperatures (Tinc) (indicated on abscissa). The RdRp assay was performed in a 30-μl reaction mixture which contained 10 μM UTP, 10 μCi of [α-32P]UTP, 3.3 mM MgCl2, and the other components described in Materials and Methods. The UMP incorporation rates are given in percentages of the value obtained at 30°C (100%). (C) Effect of NaCl on the His-Pro−Pol activity. The 30-μl reaction mixture was incubated for 20 min at 30°C and contained 1 μM His-Pro−Pol, 10 μM UTP, 10 μCi of [α-32P]UTP, 3.3 mM MgCl2, and increasing concentrations of NaCl (0 to 50 mM), indicated below the gel. The negative control was an RdRp assay of His-Pro−Pol without MgCl2 and NaCl. The UMP incorporation rates (ordinate) are normalized against the amount of incorporated UMP in an RdRp assay containing 3.3 MgCl2 and no NaCl (100%). (D) Effect of divalent (magnesium and calcium) and monovalent (potassium) cations on the polymerase activity. His-Pro−Pol (1 μM) was incubated at 30°C for 20 min with 3.3 mM (CH3COO)2Mg, MgSO4, MgCl2, CaCl2, KCl, or H2O (negative control) in a 15-μl reaction mixture containing 10 μM UTP and 5 μCi of [α-32P]UTP. The amount of incorporated UMP (ordinate) is normalized against values obtained with magnesium acetate (100%).
TABLE 1.
Effect of Mg2+ and Mn2+ concentrations on the norovirus Pro −Pol poly(rU) polymerase activity
| Divalent cation concn (mM) | UMP incorporated (pmol)
|
|
|---|---|---|
| Mg2+ | Mn2+ | |
| 0.1 | 0.2 | 0.8 |
| 0.5 | 1.0 | 35.4 |
| 1.5 | 9.9 | 53.8 |
| 3 | 39.9 | 87.4 |
| 5 | 61.7 | 85.1 |
| 10 | 23.8 | 102.1 |
| 20 | 4.8 | 40.2 |
| 40 | 0.3 | 15.3 |
Elongation activity of the norovirus ProPol enzyme with a heteropolymeric template.
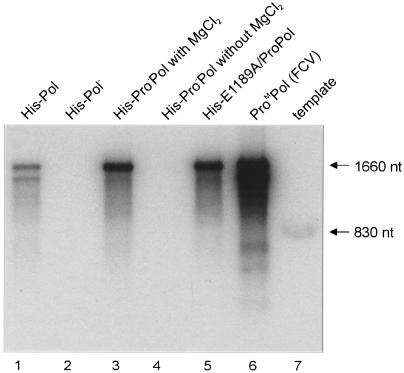
It was previously shown that FCV ProPol and RHDV Pol could synthesize RNA from an RNA heteropolymeric template (25, 26, 46). We used an 830-nt RNA transcript (46) to examine whether the norovirus His-Pro−Pol shared this property (Fig. 5). An RNA transcript of over 1,600 nt in length was observed for His-Pol, His-Pro−Pol (in the presence of MgCl2), His-E1189A-ProPol, and FCV ProM-Pol (Fig. 5, lanes 1, 3, 5, and 6, respectively). Treatment of the 1,600-nt products with RNase T1 resulted in the appearance of a template-length RNA of 830 nt (data not shown), consistent with previous studies showing the presence of a double-stranded molecule comprised of the template linked to the newly synthesized RNA by an RNA loop at the 3′ end of the template (25, 46). The amount of RNA synthesized was similar for norovirus His-Pro−Pol and His-E1189A-ProPol (Fig. 5, lanes 3 and 5) and lower for norovirus His-Pol (Fig. 5, lane 1). No product was observed with His-Pro−Pol in the absence of MgCl2 (Fig. 5, lane 4). In addition, no RNA product was synthesized by the genetically inactivated His-Pol− (Fig. 5, lane 2). We had previously reported that the ProPol precursor (His-E1189A-ProPol) examined in this experiment was an active proteinase (2). From our previous report and this study, our data indicate that the norovirus ProPol precursor protein is a bifunctional enzyme.
FIG. 5.
Synthesis of RNA from a heteropolymeric template by recombinant norovirus polymerases. Recombinant enzyme (1 μM) was incubated with 1 μg of the 830-nt green fluorescent protein RNA template for 1 h at 30°C in the presence of [α-32P]UTP; 3 mM MgCl2 was included in each reaction except that shown in lane 4 (no magnesium). The recombinant protein used in each assay is indicated above the gel (lanes 1 to 6). The synthesized RNA was purified, precipitated in ethanol, dried, heated in glyoxyl sample buffer, and analyzed in a 1% agarose gel. The gel was dried and exposed to film. In lane 7, 1 μg of the green fluorescent protein DNA template was linearized by MluI and transcribed by T7 polymerase in the presence of 10 μCi of [α-32P]UTP. The sizes of the green fluorescent protein RNA transcript (830 nt) and the RNA dimer (approximately 1,660 nt) produced by the norovirus polymerases are indicated.
Template switching activity of the norovirus ProPol.
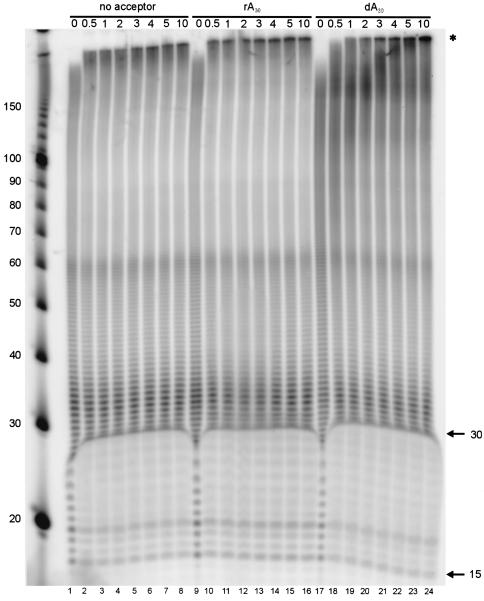
The ability of the norovirus ProPol to undergo template switching during RNA synthesis was examined as described previously for other RdRps (1, 5, 18). The norovirus His-Pro−Pol enzyme was preincubated for 3 min with preannealed poly(dT)15-poly(rA)30 complexes and a limiting amount of [α-32P]UTP to allow the formation of initiation complexes prior to the addition of heparin (to bind free His-Pro−Pol molecules) and excess UTP (1). In the presence or absence of acceptor templates (rA30 or dA30), products ranging from 15 to 60 nt were detected, consistent with those expected by polymerization from the original preannealed primer (dT15) and template (rA30) (Fig. 6). Over time, products larger than 150 nt accumulated, indicating that template switching had occurred. The template switching was most efficient in the presence of rA30 acceptor molecules, with the detection of a 200-nt product at 30 s after the addition of acceptor (Fig. 6, lane 10). The efficiency of the template switching was reduced significantly in the absence of acceptor (Fig. 6, lanes 1 through 8) or the presence of dA30 molecules (Fig. 6, lanes 17 through 24), as evidenced by the appearance of larger products at later time points. The presence of larger products in the absence of acceptor was likely due to template switching events on the original template occurring during the 3-min labeling period.
FIG. 6.
Template switching activity of MD145 His-Pro−Pol in absence or presence of RNA (rA30) and DNA (dA30) acceptor templates over a 10-min period. Reactions were initiated by mixing His-Pro−Pol (5 μM), 10 μM dT15-rA30, and [α-32P]UTP (0.67 μM), followed by incubation at 30°C for 3 min. Heparin, an excess of UTP, and either no acceptor template, rA30 acceptor template (100 μM), or dA30 acceptor template (100 μM) were then added to the reactions and incubated at 30°C. Reactions were quenched at the times (in minutes) indicated above the gel by addition of EDTA. Products were resolved by electrophoresis on a denaturing 10% polyacrylamide gel, and signals were detected using a phosphorimager. The sizes corresponding to the length of the acceptor template and primer are indicated by arrows. Larger products consistent with synthesis by template switching are indicated by an asterisk. Sizes (in nucleotides) of the DNA marker are shown on the left.
Effect of various UTP analogs on norovirus polymerase activity.
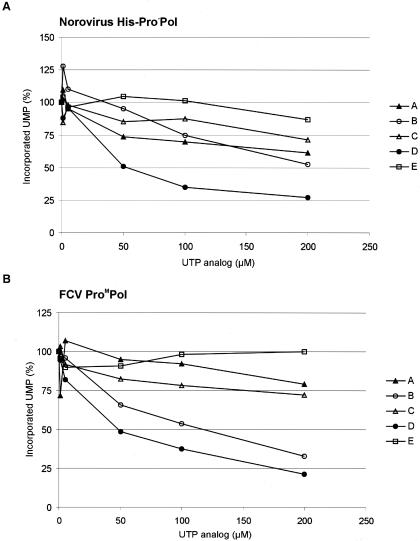
The effect of commercially available UTP analogs on the activity of the norovirus and FCV ProPol enzymes was tested (Fig. 7) in order to examine whether the calicivirus in vitro polymerase assays showed potential for drug screening applications. Five nucleoside triphosphates modified at base position 2, 5, or 6 were assayed at increasing concentrations (1 to 200 μM) in the presence of 100 μM UTP. For norovirus His-Pro−Pol, compounds A, B, C, and E at a final concentration of 200 μM resulted in norovirus polymerase activities that were 61, 51, 73, and 88% of the incorporation values of the untreated control, respectively. For FCV ProM-Pol, compounds A, B, and C at a 200 μM concentration resulted in FCV polymerase activities that were 80, 33, and 72% of the values obtained for the untreated control, respectively. Compound E at 200 μM did not affect the incorporation activity of the FCV ProM-Pol, and the inhibitory effect of compound B was greater for the FCV polymerase at a 50 or 100 μM concentration, compared to the norovirus Pro−Pol. Under these conditions, compound D was the most efficient molecule for inhibiting both the norovirus Pro−Pol and FCV ProM-Pol, resulting in activities that were 26 and 21% of the values obtained for the untreated controls, respectively.
FIG. 7.
Effect of UTP analogs on norovirus and FCV polymerase activity. (A) The norovirus His-Pro−Pol at 1 μM was incubated for 20 min at 30°C with increasing concentrations (1, 5, 50, 100, and 200 μM) of UTP analogs. The modified NTPs are 5-iodo-UTP (compound A), pseudo-UTP (compound B), 5-methyl-UTP (compound C), 2-thio-UTP (compound D), and 6-aza-UTP (compound E). The relative quantities of incorporated UMP (ordinate) are given in percentages. The data (shown as percentages of UMP incorporation) were normalized based upon the UMP incorporated by His-Pro−Pol (100%) in the absence of UTP analog. (B) The recombinant FCV ProM-Pol at 1 μM was assayed as described in the legend for panel A except that the reaction mixture was incubated for 2 min. Code names of the compounds are identical to those in panel A. The data (shown as percentages of UMP incorporation) are normalized based upon the UMP incorporated by FCV rProM-Pol (100%) in the absence of UTP analog.
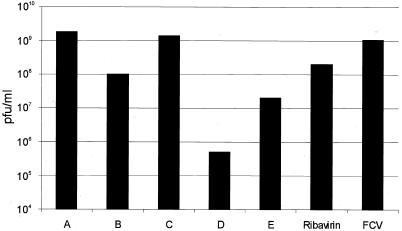
The effect of base analogs on the replication of FCV in cell culture was examined (Fig. 8). The FCV Urbana strain typically grows to titers of approximately 108 to 109 PFU/ml and exhibits a rapid cytopathic effect in cell culture. Cells were preincubated for 1 h with the nucleoside form of each compound assayed above (2 mM final concentration) and then infected with FCV. After 7 h of incubation in the presence of each nucleoside, the resulting virus titer was determined. The nucleosides 2-thiouridine (compound D) and 6-azauridine (compound E) were the most efficient in inhibiting FCV growth (4.9 × 105 and 2 × 107 PFU/ml, respectively). For pseudouridine (compound B), the virus titer was decreased by 1 log (108 PFU/ml), and virus titers observed for nucleosides 5-iodouridine (compound A) (1.89 × 109 PFU/ml) and 5-methyluridine (compound C) (1.4 × 109 PFU/ml) were similar to that of FCV without treatment. Only one compound, 2-thiouridine (compound D), showed a consistent ability to inhibit both the growth of FCV in cell culture and the in vitro enzymatic activity of the recombinant FCV and norovirus ProPol enzymes. Because ribavirin is a nucleoside analog that has been used in clinical settings for the treatment of viral disease, we tested its effect on the growth of FCV. The ribavirin caused only a minimal decrease in titer to 2 × 108 PFU/ml, even at a high concentration of 2 mM. By measuring cell viability in the presence of each base analog (2 mM), no significant cytotoxicity was observed in mock-infected CRFK cells (data not shown).
FIG. 8.
Effect of nucleoside analogs on the growth of FCV. CRFK cells were incubated with 2 mM nucleoside for 1 h prior to infection with FCV in the presence of 2 mM inhibitors. At 8 h postinfection, the effect of each analog was monitored by plaque titration assay. The nucleosides (abscissa) are 5-iodouridine (compound A), pseudouridine (compound B), 5-methyluridine (compound C), 2-thiouridine (compound D), 6-azauridine (compound E), and ribavirin. The virus titers are indicated on the left (ordinate) in PFU per milliliter (PFU/ml). FCV infection in the absence of analog was included as a control.
DISCUSSION
Norovirus-associated gastroenteritis is usually mild and self-limiting, but it can be incapacitating for a period ranging from several hours to days (12). Severe disease can occur, and recent studies have established an association between norovirus-related illness and admission to the hospital for treatment of gastroenteritis (27, 29). No vaccines or antiviral drugs are currently available, but an effective control strategy might have a significant impact on the overall disease burden. It is possible that study of calicivirus replication could facilitate the development of such strategies. In this work, we report the identification of two active forms of the norovirus polymerase: the ProPol precursor and the fully processed Pol. Further characterization of MD145 His-Pro−Pol showed that this enzyme has properties similar to those reported for FCV ProPol and RHDV Pol (25, 26, 46). From this study and our previous identification of norovirus ProPol as an active proteinase (2, 41), we propose that the norovirus ProPol is important in replication as a bifunctional enzyme with both proteinase and polymerase activities.
Noroviruses, like other caliciviruses, show genetic relatedness to the picornavirus-like positive-strand RNA viruses (20). Furthermore, the nonstructural polyprotein proteolytic processing strategy generates both precursors and products, increasing the capacity of the virus to produce several different forms of replicative enzymes with various functions (38). The picornavirus 3CD precursor (analogous to the calicivirus ProPol) is present in virus-infected cells and has been associated with multiple functions in replication (17, 35, 48). However, cleavage must ultimately occur within 3CD to release the mature 3C proteinase and 3D polymerase; 3Dpol is the only known active form of the picornavirus RdRp (38). A similar processing of the polymerase region was observed for members of the genera Lagovirus and Norovirus, in which a conserved dipeptide cleavage site between the Pro and Pol domains is recognized and cleaved by the proteinase (2, 19, 30, 47). However, the release of the fully processed Pol is not an absolute requirement for certain caliciviruses such as FCV (genus Vesivirus) in which the FCV ProPol precursor is the major form of both the RdRp and proteinase detected in infected cells (44). Although proteolytic processing was observed when the FCV ProPol precursor was overproduced in bacteria, these Pro-mediated cleavages were later shown to be nonessential for virus replication (43, 44). In this first direct comparison of the enzymatic activities of norovirus Pol and ProPol, we found that the ProPol precursor of norovirus strain MD145, like that of FCV (46), is active. The utilization of ProPol may be a shared feature among caliciviruses, regardless of the proteolytic processing strategy used by the virus during the RNA replication. It will be important to determine in future studies whether the two active forms of the norovirus polymerase identified in this study (Pol and ProPol) have different roles in replication.
An interesting observation in our study was that the presence of a histidine tag at the N terminus of the fully processed norovirus polymerase had a profound inhibitory effect on its RdRp activity. Previous studies of the RHDV and human norovirus enzymes reported the presence of two extra amino acids or a second amino acid substitution, respectively, at the N terminus of their purified recombinant products (25, 33). These enzymes, similar to the norovirus His-Pol in our study, showed activity in in vitro assays. However, it is likely that calicivirus Pol proteins may be more active when expressed as recombinant proteins with the precise N terminus as shown for poliovirus 3D (10).
The recombinant norovirus polymerase was examined for its ability to synthesize RNA from a heteropolymeric nonviral template. The two forms of norovirus RdRp could initiate synthesis from a single-stranded RNA template in the absence of an added primer. The size of the synthesized product was twice the size of the template, suggesting that a copy-back type of mechanism occurred in vitro to initiate synthesis as suggested for the recombinant RHDV and FCV RdRps (26, 46).
The FCV ProPol was previously shown to be less active (10-fold) than polio 3Dpol, and this was attributed to a lower efficiency in template switching by the FCV ProPol (46). The ability of the norovirus ProPol to undergo template switching suggests that recombination between RNA molecules could occur during norovirus RNA replication by using this mechanism. The availability of an active norovirus enzyme might facilitate the development of in vitro replication systems to study template switching events and other potential mechanisms for creating the observed genetic diversity among norovirus strains associated with epidemic gastroenteritis (14, 24).
The use of antiviral drugs is an alternative control strategy for some viruses in the absence of effective vaccines (7). We investigated whether the use of the FCV cell culture system might give insight into the identification of potential inhibitors of calicivirus replication. First, we compared several UTP analogs for their effects on the in vitro activity of the recombinant norovirus and FCV ProPol enzymes. The biochemical inhibitory profiles were similar for the two enzymes, with the compound 2-thio-UTP as the most potent inhibitor for both polymerases. The similarities in the inhibitory effects of these compounds on the FCV and norovirus ProPol enzymes suggest that the catalytic sites of these RdRps may share a similar structure, but this will require additional study. To support the results obtained using the in vitro assay, we tested the nucleoside form of each compound on the growth of FCV in cell culture. We identified one compound (2-thiouridine) that showed an ability to inhibit both the growth of FCV and the activity of recombinant ProPol enzymes in vitro. For the other compounds, there was no absolute correlation between the efficacy of the nucleotides and that of the nucleosides. The capacity for the nucleosides to be transformed into nucleoside triphosphates by the cellular machinery might explain, in part, the different inhibitory effects observed between in vitro and in vivo assays (45). Early studies reported some inhibitory effect of ribavirin on the growth of FCV (strain 255) in cell culture (37) but little therapeutic benefit in the treatment of FCV disease in cats (36). Our data evaluating the effect of ribavirin on the growth of the FCV strain Urbana in cell culture were consistent with these studies, in which the reduction of virus titer was low (0.7 log) when FCV was examined at a high multiplicity of infection. Ribavirin, which is a potent inhibitor of poliovirus replication in cell culture (6) and useful for the treatment of certain viral diseases (7), may not be an effective anticalicivirus drug. However, its efficacy for norovirus infection will require direct evaluation when replication or cell culture systems become available.
The evaluation of antiviral drugs is hampered by the lack of a norovirus cell culture system or replicon, but this study suggests that the combined data obtained from in vitro calicivirus polymerase assays and cell culture testing with FCV may provide an initiating platform for drug screening. The ability to treat or prevent acute norovirus illness should prove useful in lowering the burden from this common, and sometimes severe, disease.
Acknowledgments
We thank Tanaji Mitra (NIAID, NIH), Tauseef Butt, and John McGrath (LifeSensors, Inc.) for technical assistance. We extend our appreciation to Jerry Keith and Ellie Ehrenfeld for helpful discussions and Albert Z. Kapikian (NIAID, NIH) for support of this project.
REFERENCES
- 1.Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. [DOI] [PubMed] [Google Scholar]
- 2.Belliot, G., S. V. Sosnovtsev, T. Mitra, C. Hammer, M. Garfield, and K. Y. Green. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77:10957-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blakeney, S. J., A. Cahill, and P. A. Reilly. 2003. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology 308:216-224. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, C. P., and P. D. Nagy. 2003. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J. Virol. 77:12033-12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq, E. 2004. Antiviral drugs in current clinical use. J. Clin. Virol. 30:115-133. [DOI] [PubMed] [Google Scholar]
- 8.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):S254-S261. [DOI] [PubMed] [Google Scholar]
- 10.Gohara, D. W., C. S. Ha, S. Kumar, B. Ghosh, J. J. Arnold, T. J. Wisniewski, and C. E. Cameron. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128-138. [DOI] [PubMed] [Google Scholar]
- 11.Green, K. Y., G. Belliot, J. L. Taylor, J. Valdesuso, J. F. Lew, A. Z. Kapikian, and F. Y. Lin. 2002. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 185:133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Hardy, M. E., T. J. Crone, J. E. Brower, and K. Ettayebi. 2002. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 89:29-39. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 17.Jore, J., B. De Geus, R. J. Jackson, P. H. Pouwels, and B. E. Enger-Valk. 1988. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J. Gen. Virol. 69:1627-1636. [DOI] [PubMed] [Google Scholar]
- 18.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konig, M., H. J. Thiel, and G. Meyers. 1998. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J. Virol. 72:4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. (Erratum, 28:546.) [DOI] [PubMed] [Google Scholar]
- 21.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 22.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80:291-296. [DOI] [PubMed] [Google Scholar]
- 24.Lochridge, V. P., and M. E. Hardy. 2003. Snow Mountain virus genome sequence and virus-like particle assembly. Virus Genes 26:71-82. [DOI] [PubMed] [Google Scholar]
- 25.Lopez Vazquez, A., J. M. Martin Alonso, R. Casais, J. A. Boga, and F. Parra. 1998. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J. Virol. 72:2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez Vazquez, A. L., J. M. Martin Alonso, and F. Parra. 2001. Characterisation of the RNA-dependent RNA polymerase from rabbit hemorrhagic disease virus produced in Escherichia coli. Arch. Virol. 146:59-69. [DOI] [PubMed] [Google Scholar]
- 27.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meakins, S. M., G. K. Adak, B. A. Lopman, and S. J. O'Brien. 2003. General outbreaks of infectious intestinal disease (IID) in hospitals, England and Wales, 1992-2000. J. Hosp. Infect. 53:1-5. [DOI] [PubMed] [Google Scholar]
- 30.Meyers, G., C. Wirblich, H. J. Thiel, and J. O. Thumfart. 2000. Rabbit hemorrhagic disease virus: genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology 276:349-363. [DOI] [PubMed] [Google Scholar]
- 31.Nakata, S., S. Honma, K. K. Numata, K. Kogawa, S. Ukae, Y. Morita, N. Adachi, and S. Chiba. 2000. Members of the family Caliciviridae (Norwalk virus and Sapporo virus) are the most prevalent cause of gastroenteritis outbreaks among infants in Japan. J. Infect. Dis. 181:2029-2032. [DOI] [PubMed] [Google Scholar]
- 32.Ng, K. K., M. M. Cherney, A. L. Vazquez, A. Machin, J. M. Alonso, F. Parra, and M. N. James. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381-1387. [DOI] [PubMed] [Google Scholar]
- 33.Ng, K. K., N. Pendas-Franco, J. Rojo, J. A. Boga, A. Machin, J. M. Alonso, and F. Parra. 2004. Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638-16645. [DOI] [PubMed] [Google Scholar]
- 34.Pang, X. L., J. Joensuu, and T. Vesikari. 1999. Human calicivirus-associated sporadic gastroenteritis in Finnish children less than two years of age followed prospectively during a rotavirus vaccine trial. Pediatr. Infect. Dis. J. 18:420-426. [DOI] [PubMed] [Google Scholar]
- 35.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Povey, R. C. 1978. Effect of orally administered ribavirin on experimental feline calicivirus infection in cats. Am. J. Vet. Res. 39:1337-1341. [PubMed] [Google Scholar]
- 37.Povey, R. C. 1978. In vitro antiviral efficacy of ribavirin against feline calicivirus, feline viral rhinotracheitis virus, and canine parainfluenza virus. Am. J. Vet. Res. 39:175-178. [PubMed] [Google Scholar]
- 38.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 39.Sankar, S., and A. G. Porter. 1991. Expression, purification, and properties of recombinant encephalomyocarditis virus RNA-dependent RNA polymerase. J. Virol. 65:2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seah, E. L., J. A. Marshall, and P. J. Wright. 1999. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 73:10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seah, E. L., J. A. Marshall, and P. J. Wright. 2003. Trans activity of the norovirus Camberwell proteinase and cleavage of the N-terminal protein encoded by ORF1. J. Virol. 77:7150-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 43.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Rompay, A. R., A. Norda, K. Linden, M. Johansson, and A. Karlsson. 2001. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol. 59:1181-1186. [DOI] [PubMed] [Google Scholar]
- 46.Wei, L., J. S. Huhn, A. Mory, H. B. Pathak, S. V. Sosnovtsev, K. Y. Green, and C. E. Cameron. 2001. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J. Virol. 75:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirblich, C., M. Sibilia, M. B. Boniotti, C. Rossi, H. J. Thiel, and G. Meyers. 1995. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J. Virol. 69:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ypma-Wong, M. F., P. G. Dewalt, V. H. Johnson, J. G. Lamb, and B. L. Semler. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265-270. [DOI] [PubMed] [Google Scholar]