Abstract
The entomopoxvirus from Amsacta moorei (AmEPV) contains none of the commonly recognized vertebrate poxvirus apoptotic suppressor genes. However, AmEPV carries a single inhibitor of apoptosis (iap) gene (AMViap) not present in vertebrate poxviruses. The AMViap gene was active when coexpressed with the Drosophila proapoptotic gene hid in Ld652 cells and can rescue cells from apoptosis as shown by increased number of surviving cells and reduced levels of caspase-3-like activity. We also showed that expression of the AMViap gene rescued polyhedron production in Autographa californica M nucleopolyhedrovirus (AcMNPV)Δp35-infected Sf9 cells during an otherwise abortive infection induced by apoptosis. Surprisingly, deletion of the AMViap gene from the AmEPV genome led to only a modest (10-fold) loss of virion production in infected Ld652 cells, indicating that the AMViap gene is nonessential for virus replication under these conditions. However, infection of Ld652 cells by AmEPV lacking a functional iap gene led to a more rapid induction of cytotoxicity and increased levels of caspase-3-like activity. Similar results were observed and were more pronounced in infected Sf9 and S2 cells. The purified AMVIAP protein also inhibits the enzymatic activities of human caspase-9 and caspase-3 in vitro. Our results indicate that while the AMViap gene was active in controlling apoptosis through the intrinsic pathway, the virus likely encodes additional proteins that also regulate apoptosis.
Apoptosis, or programmed cell death, is a well-conserved and integral process necessary for normal organism development which serves to remove unwanted, damaged, mutated, or infected cells (43, 54, 70). Apoptosis can be initiated by both external and internal stimuli such as UV-induced DNA damage, oncogenic transformation, drugs such as actinomycin D, virus infection, and a variety of extracellular signals (68, 71). These various stimuli lead to the activation of either the intrinsic or extrinsic apoptotic pathway (7). The extrinsic pathway is triggered by the binding of external (death) ligands to their cognate (death) receptors as exemplified by members of the tumor necrosis factor (TNF) superfamily. Receptor-ligand engagement then allows transmission of external signals into the cell. The intrinsic pathway is initiated by signals originating within the cell from a series of death-triggering genes, which in insect systems include the hid, grim, and reaper genes of Drosophila and in mammalian cells the Smac/Diablo, GSPT1, and Omi/HtrA2 genes (39, 68). The apoptotic killing of cells by these death-inducing genes can be blocked and regulated by apoptotic suppressor genes, including members of the inhibitor of apoptosis (iap) gene superfamily (26, 59).
Both intrinsic and extrinsic apoptotic pathways result in the formation of a cytosolic protein complex that activates a family of aspartic acid-specific cysteine proteases (caspases). Caspases are a diverse family of proteases, and it is through the action of active caspases that apoptotic death occurs. Caspases are divided into “initiator” (caspases 2, 8, 9, and 10) and “executioner” or “terminal” caspases (caspases 3 and 7) (35, 54). Once activated, the initiator caspases are responsible for activating the executioner caspases by proteolytic cleavage. The activated executioner caspases are then responsible for the proteolytic cleavage and degradation of a broad spectrum of cellular targets, eventually leading to apoptosis that is characterized by changes in the cells such as shrinkage, membrane blebbing, chromatin condensation, formation of the apoptosome, and DNA fragmentation (54, 71).
Virus infections frequently lead to the induction of apoptosis in host cells. Apoptosis in infected cells can be considered as a host defense mechanism which ultimately results in reduction of virus production (7, 43). However, viruses have developed strategies to block apoptosis allowing more time for progeny virus to be produced (7, 11, 27, 48). One of the first apoptotic suppressor genes, p35, was discovered in Autographa californica M nucleopolyhedrovirus (AcMNPV). The p35 gene is required for virus growth since deletion of p35 from AcMNPV blocks virus production (13). The P35 protein is believed to function in the terminal stages of apoptosis as a caspase inhibitor that acts to block the lethal effects of terminal caspase activation and prevent cell death. In addition to P35, baculoviruses also encode at least two other types of apoptotic suppressor proteins, P49 and IAP (12, 17, 41, 73). The P49 protein, produced by the Spodoptera littoralis nucleopolyhedrosis virus, inhibits the activation of initiator caspases, including human caspase-9 and the other P35-sensitive initiator caspases (17, 46, 73). Thus, P49 functions to inhibit apoptosis by blocking activity of the initiator caspases that serves to prevent activation of the downstream effector caspases.
Viral iap genes were first described from the Orgyia pseudotsugata M nucleopolyhedrovirus (OpMNPV), the Op-iap gene, and Cydia pomonella granulosis virus (CpGV), the Cp-iap (8, 15). Structurally, IAPs are characterized by the presence of two signature motifs: the so-called baculovirus IAP repeats (BIRs) and a RING domain (16, 59). RING domains are a specialized form of zinc finger involved in protein-protein interactions. IAP proteins generally act to block apoptosis by interacting through the BIR domain to block the activity of a variety of proapoptotic proteins such as REAPER, HID, and GRIM in insect cells and SMAC/DIABLO in vertebrate cells (34, 63). Op-IAP has been shown to bind to HID, REAPER, and GRIM; to down-regulate Sf-caspase-X; and therefore to inhibit apoptosis induced by various proapoptotic inducers (34, 63). The IAP family of proteins is evolutionarily well conserved, and most IAP proteins appear to be able to function across different species. Unlike p35 and p49 genes, which have only been identified in a few baculoviruses, iap genes have been found widely distributed in most if not all baculoviruses, as well as in eukaryotes, including mammals (16).
The Poxviridae comprise a large family of double-stranded DNA-containing viruses, which, unlike baculoviruses, develop in the cytoplasm and include viruses of both vertebrates (Chordopoxvirinae) and invertebrates (Entomopoxvirinae). The vertebrate poxviruses are known to encode a number of proteins that regulate apoptosis, including the serine proteinase inhibitor (serpin), SPI-2/CrmA. The crmA/SPI-2-encoded serpin blocks apoptosis by direct inhibition of initiator caspases, including caspases 1, 8, and 10 (69), as well as granzyme B (40, 47, 58). However, no serpin genes have been identified within the two sequenced insect poxvirus genomes, the entomopoxviruses from Amsacta moorei (AmEPV) (5) and Melanoplus sanguinipes (MsEPV) (1). Furthermore, examination of the genomic sequences suggests none of the many other vertebrate poxvirus apoptosis suppressors exist within AmEPV.
AmEPV, a group B (β) entomopoxvirus, has been reported to infect agriculturally important pests, such as Estigmene acrea (24) and Lymantria dispar (5). The 232-kb AmEPV genome was recently sequenced (5) and appears to contain a single iap gene, AMViap (AMV021). As the only candidate apoptotic suppressor gene, it is likely that the single AmEPV iap gene homolog has a role in controlling apoptosis. In this paper, we show that the AMViap gene is active, inhibits apoptosis as expected, and represents one mechanism by which AmEPV can control cell viability.
MATERIALS AND METHODS
Cells and viruses.
IPLB-LD-652 (Ld652), Sf9, and S2 insect cell lines were maintained at 28°C in Sf900 II SFM medium supplemented with 10% fetal bovine serum (FBS), 50 U of penicillin G, and 50 μg of streptomycin per ml (Invitrogen). Wild-type (WT) AmEPV (25) and all recombinant viruses were grown in Ld652 cells (23). WT AcMNPV was produced in Sf9 cells, whereas AcMNPV lacking the p35 gene (vAcΔp35) (courtesy of Paul Friesen at University of Wisconsin) was grown in TN368 cells. For preparation of virus stocks, Sf9 and TN368 cell lines were maintained in TC-100 medium plus 10% FBS (Invitrogen) (28).
Virus infections.
Ld652 cells were plated at 106 cells/well in six-well plates (Corning) for 2 h at 28°C. The growth medium in each well was then removed and replaced with 1 ml of fresh Sf900 II SFM medium plus 10% FBS containing virus at the multiplicity of infection (MOI) indicated in the text, and the monolayer was gently rocked for 2 h at room temperature. The plates were then placed at 28°C for another 2 h, after which the inoculum was removed and replaced with another 1 ml of fresh growth medium. At various times after infection, the combined cells and supernatant were collected. Cell lysis was achieved by four quick-freeze-thaw cycles. Cell debris was removed by low-speed centrifugation (1,000 × g), and the titers of the resulting supernatants were determined on Ld652 cells at 28°C with a standard plaque assay as previously described (37).
Plasmid constructs.
The following two primers were used to amplify the AMViap gene from WT AmEPV genomic DNA: 5′-TATGGATCCTATGATGGATGACATT-3′ and 5′-GGCTCGAGATCTTAATATATTGTTAAGG-3′. The PCR products were sequenced and cloned into the pIE1-4 vector (Novagen), using the unique BamHI and XhoI sites (underlined) to create the pIE1AMViap plasmid. The enhanced green fluorescent protein (gfp) and Op-iap (courtesy of Lois K. Miller at the University of Georgia) genes were also cloned into the pIE1-4 vector using the restriction sites NotI for construction of pIE1gfp, and BamHI/XhoI for construction of pIE1Opiap (Fig. 1). Plasmids containing the p35 gene and hid gene were obtained from Lei Zhou, University of Florida, and have been cloned into the pIE1-4 vector using NotI/NruI sites (pIE1p35) and BamHI/NruI sites (pIE1hid) (Fig. 1A), respectively.
FIG. 1.
Plasmids used for transfections and gene replacements. (A) Expression plasmids. The AMViap gene was cloned into the pIE1-4 vector and designated as pIE1AMViap. Plasmids pIE1Opiap, pIE1p35, pIE1hid, and pIE1gfp were created similarly. (B) The plasmid pDU20gfp was used to insert gfp into the sph gene locus of AmEPV. Expression of the gfp gene is mediated by the sph promoter. (C) The plasmid pBSAmΔiap was generated by inserting the lacZ gene, under the control of cowpox virus ATI promoter, into a plasmid containing the AMViap gene and 1 kb of upstream and downstream flanking sequences. The plasmid backbone was pBluescript KS(+). The lacZ gene disrupts and replaces a portion of the AMViap.
The plasmid pDU20gfp, used to replace the spheroidin (sph) gene with gfp, was constructed from the plasmid pDU20lacZ as follows. The plasmid pDU20lacZ contains the sph gene and 1-kb flanking sequences into which a portion of the sph gene was replaced by the lacZ gene (36). The lacZ gene was replaced with the gfp gene under the control of the sph promoter (6) to create the pDU20gfp plasmid (Fig. 1B).
The plasmid pBSAmΔiap (Fig. 1C) used to inactivate the AMViap gene was constructed as follows. A 3.8-kb DNA fragment, containing the AMViap gene with 1-kb flanking sequences on each side, was amplified from WT AmEPV viral DNA using VENT DNA polymerase (NE BioLab) with the following primers: 5′-CTGCATATATATCTAGAAGATTTAG-3′ and 5′-CACCCTCGAGACCATAATAATATGAGTC-3′. The PCR fragment was cloned into the pBluescript KS II+ vector (Stratagene) using PstI and XhoI restriction sites (underlined). A portion (637 bp) of the AMViap gene was then removed through a modified PCR-based deletion method (21) using the following two primers: 5′-TCGATATCTAGCAAATGATTC-3′ and 5′-GAAAAGCTTCTATATCGATGGA-3′. A lacZ gene cassette containing the cowpox virus early ATI promoter (36) was then inserted to replace the 637 bp of the AMViap gene.
Construction of recombinant AmEPV viruses.
Standard transfection/gene replacement techniques were used to construct AmEPV recombinant viruses. Ld652 cells, seeded at 106 cells/well in a six-well plate, were infected with AmEPV at an MOI of 0.5. The infected cells were incubated at 28°C for 18 h and then cotransfected with an appropriate transplacement plasmid, pBSAmΔiap or pDU20gfp (2 μg per transfection), with 8 μl of Cellfectin (Invitrogen) per transfection as specified by the manufacturer. WT AmEPV was used as the parental virus to generate AmEPV(sph−::gfp+) following transfection with pDU20gfp. For simplicity, we refer to this virus as vAmΔsph/gfp. The vAmΔsph/gfp recombinant virus was identified by occlusion body-negative, GFP-positive plaques. The vAmΔsph/gfp was then used as the parental virus for generation of AmEPV(sph−::gfp+)(iap−::lacZ+) recombinant virus following transfection of the plasmid pBSAmΔiap. For simplicity, we refer to this virus as vAmgfp/Δiap/lacZ. The vAmgfp/Δiap/lacZ virus was identified by occlusion body-negative, β-galactosidase-positive, GFP-positive plaques. A total of three to five rounds of plaquing were required to isolate a pure AmEPV recombinant virus.
All constructs were verified by PCR analysis using recombinant virus genomic DNA as template. The viral genomic DNA was isolated from Ld652 cells infected with AmEPV recombinants, using the DNeasy tissue kit (QIAGEN). To verify the vAmΔsph/gfp virus, the following two primers, 5′-CCATCAGCCATATGTTTAAT-3′ and 5′-AGTACTGGTATTAATTTAACATAT-3′, were used for amplification, which is predicted to produce a 1.5-kb PCR product from vAmΔsph/gfp virus. To identify vAmgfp/Δiap/lacZ, the same pair of primers that were previously used to amplify the AMViap open reading frame (ORF) from the AmEPV genome is predicted to generate a 4-kb PCR product from the vAmgfp/Δiap/lacZ recombinant, whereas PCR amplification of parental vAmΔsph/gfp is predicted to generate a 1-kb PCR product. All PCR products were subsequently sequenced.
Inhibition of hid-induced apoptosis as measured by GFP expression.
Briefly, 70 to 80% confluent Ld652 cells were seeded in six-well plates (106 cells/well). Cellfectin was used to transfect cells, using the Drosophila proapoptotic gene hid (0.5 μg of pIE1hid/transfection), a gfp-marked gene (2 μg of pIE1gfp/transfection). The concentrations of pIE1hid and pIE1gfp were constant in all transfections. The cells were also transfected with various amounts of a third plasmid comprising the potential apoptotic suppressor genes (p35, Op-iap, or AMViap) to be tested. Various ratios of hid to a given apoptotic suppressor gene were used that ranged from 1:1, 1:2, 1:4, 1:8, and 1:16 up to 1:32. For example, at a ratio of pIE1hid to pIE1AMViap of 1:4, there were 0.5 μg of pIE1hid plus 2 μg of pIE1AMViap and 2 μg of pIE1gfp per transfection. The transfection conditions were those described by the manufacturer with some slight modifications. After mixing 8 μl of Cellfectin with the required amount of DNA at the designated ratios in 200 μl of Sf900 II SFM medium, the DNA-Cellfectin mixture was incubated for 45 min at room temperature with occasional mixing. The mixture was then added to the Ld652 cells seeded as described above, in a final volume of 1 ml of Sf900 II SFM medium. After rocking for 15 min at room temperature, the Ld652 cells were further incubated at 28°C for 6 h. The DNA-Cellfectin mixture was then replaced by 1 ml of Sf900 II SFM medium plus 10% FBS and incubated at 28°C until harvested at 48 h after transfection. Cells were photographed at 48 h following transfection with a Zeiss inverted microscope. Rescue was indicated by GFP-positive surviving cells.
Polyhedron formation.
Polyhedron formation was evaluated visually in apoptosis-sensitive Sf9 cells transfected with apoptotic suppressor genes and then infected with vAcΔp35, as previously described by Zoog et al. (72). Briefly, uninfected Sf9 cells were first transfected with 8 μl of Cellfectin containing 2 μg of one of the following plasmids: pIE1p35, pIE1Opiap, or pIE1AMViap. At 16 h after transfection, cells were infected with vAcΔp35 (0.5 PFU/cell) and rocked for 1 h at room temperature. The infected cells were then incubated at 28°C for another 2 h, after which the inoculum was removed and replaced with 1 ml of fresh Sf900 II SFM medium plus 10% FBS. The infected cells were then incubated at 28°C until 96 h postinfection (hpi). Photographs were taken with a Zeiss inverted microscope. Rescue from apoptosis was indicated by polyhedron formation.
Recombinant protein expression and purification.
The human XIAP and AMViap genes were cloned into pGEX-4T3 (Pharmacia) vector as described previously (6). Overnight cultures of pGEX-4T3, pGEX-XIAP, or pGEX-AmIAP plasmid-containing cells were diluted 1:10 in fresh medium and allowed to grow for 1 h prior to induction. Zn acetate (50 μM) was then added to the cultures along with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cultures were induced at 28°C for 2.5 h. Bacteria were pelleted and resuspended in STE buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM EDTA) containing protease inhibitors (10 μM aprotinin, 100 μM pepstatin A, 10 μM leupeptin, 100 μM phenylmethlysulfonyl fluoride). Dithiothreitol (DTT) was added to 1 mM, and the pellet was vortexed before adding 20-mg/ml lysozyme (Roche). The suspension was then incubated on ice for 15 min. A total of 1/50 the volume of 10% taurocholic acid (Calbiochem) was added along with 0.1 mM DNase (Sigma) and 0.1 mM MgCl2, and the suspension was incubated on ice for 10 min, sonicated for 20 s three times with a probe sonicator with an amplitude of 20%, and then centrifuged at 14,000 × g for 15 min. The supernatant was filtered through cheesecloth, and 1/50 the volume of 20% Triton X-100 (Sigma) was added. Glutathione-Sepharose bead slurry (Amersham) was added to the lysate, and this mixture was incubated at 4°C for 1 h on a rotating platform. Bead-protein complexes were collected and washed four times in NETN buffer (2 M Tris-HCl, pH 8.0, 4 M NaCl, 20% Triton X-100, 0.5 M EDTA). Recombinant glutathione S-transferase (GST) fusion proteins were eluted in 50 mM Tris 8.0-5 mM reduced glutathione, dialyzed for 48 h against three changes of caspase assay buffer: 20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 100 mM NaCl, 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1 mM EDTA, 10% sucrose, and 10 mM DTT. Protein quantity and quality were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using bovine serum albumin standards to estimate protein concentrations. Recombinant proteins were used immediately in caspase inhibition assays.
Assays for caspase-3-like activity.
Ld652, Sf9, and S2 cells were infected in suspension with AmEPV or an AmEPV recombinant for 1 h at room temperature with occasional mixing. The cells were then plated in a six-well plate at 1 × 106 cells/well for Ld652, 3 × 106 cells/well for Sf9, and 1 × 107 cells/well for S2 cells and incubated at 28°C for additional 2 h. The inocula were then removed and replaced with 1 ml of Sf900 II SFM medium plus 10% FBS and harvested at different times as indicated in the figures. At 48 hpi, photographs of the cells were also taken with a Zeiss microscope.
Protein samples were prepared from the combined 1 ml of cell-supernatant mixture by four quick-freeze-thaw cycles following the addition of 100 μl of caspase buffer to a final concentration of a mixture of 100 mM HEPES, pH 7.5, 2 mM EDTA, 0.1% CHAPS, and 1 mM DTT. Cell debris was then removed by centrifugation at 10,000 × g for 10 min. In a 96-well black plate, 50-μl protein samples were then mixed with 150 μl of reaction buffer (100 mM HEPES [pH 7.5], 10% sucrose, 0.1% CHAPS, 10 mM DTT) containing 14 μM fluorogenic tetrapeptide substrate Ac-DEVD-AMC [acetyl-Asp-Val-Ala-Asp-(amino-4-methyl coumarin)] (Sigma). Cleavage of the peptide substrate was monitored by emission at 460 nm following excitation at 380 nm in a Tecan SpectraFluor microplate reader to detect activity as indicated by release of free amino methyl coumarin. Values are the averages of triplicate assays and are reported as the rate of product formation obtained from the linear portion of the reaction curves, expressed as the fluorescence unit (FSU) increase per second in the number of cells indicated (45).
Caspase-9 and caspase-3 inhibition assays.
Caspase-9 reactions were performed in 96-well plates containing caspase reaction buffer supplemented with 0.4 mM Ac-LEHD-AMC (Biomol) and 200 U of recombinant active caspase-9 (100 U/μl; Biomol) in a total volume of 100 μl. Reactions were initiated by addition of caspase 9 and carried out in the presence of GST (5.2 μg; 2 μM), GST-XIAP (4.1 μg; 500 nM final concentration), GST-AmIAP (2.9 μg; 500 nM), or a 10-fold excess of GST-AmIAP (29.0 μg; 5 μM). Caspase-3 inhibition assays were carried out in a total volume of 100 μl containing caspase reaction buffer supplemented with 0.4 mM Ac-DEVD-AMC (Biomol) by adding 200 U of recombinant caspase-3 (Biomol; 100 U/μl) in the absence of recombinant IAPs or with GST-XIAP (4.1 μg; 500 nM final concentration), GST-AmIAP (2.9 μg; 500 nM), or GST alone (5.2 μg; 2 μM). Fluorescence emission (490 nm) was measured at 1-min intervals for 120 min in a BMG Polarstar Galaxy plate reader using an excitation wavelength of 400 nm.
RESULTS
Features of the AMViap gene and its potential encoded protein product.
Analysis of the genome of AmEPV indicates a single ORF (AMV021), 819 bp in length, located at 24,548 to 23,757 bp from the left end of the genome, which based on sequence, appears to be an iap gene (AMViap) (5). The AMViap gene encodes a putative 264-amino-acid protein with a predicted molecular mass of 30,544 Da. The AMViap ORF is preceded by a potential poxvirus consensus early promoter motif, TGAAAAATAA, located 31 bp upstream of the initiating methionine codon (nucleotides −32 to −41).
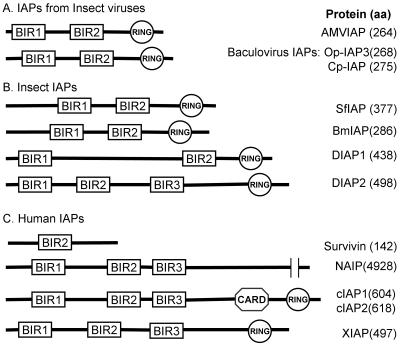
IAPs are widespread in nature and have been found in viruses, insects, birds, and mammals. Typically, IAPs contain one or more so-called Cys/His BIR domains (CX2C16HX6-8C) as well as a carboxyl-terminal RING finger motif (C3HC4), both of which are required for activity to suppress apoptosis (Fig. 2). The AMVIAP contains two BIR domains (BIR1 and BIR2) and a RING finger motif. The AMVIAP BIR1 and BIR2 domains consist of 70 and 71 amino aids, respectively, and are separated by 27 amino acids. Although not all proteins that contain BIR domains inhibit apoptosis, the BIR domains have been implicated in the binding to and subsequent inhibition of caspase activation (16). The AMVIAP RING domain consists of 34 amino acids and is separated from the BIR2 motif by 41 amino acids. The RING finger domain of DIAP1 has been shown to be involved in ubiquitin-mediated protein degradation (50).
FIG. 2.
Comparison of AMVIAP domain structure with other IAPs. The relative positions of BIR and RING domains of the AMVIAP are compared to IAPs from insect viruses (A); insect hosts, including Drosophila IAP 1 and 2 (DIAP1 and DIAP2), Spodoptera frugiperda IAP (Sf-IAP) (30), and Bombyx mori IAP (Bm-IAP) (31) (B); and human sources, including NAIP (39), c-IAP1/HIAP-2 (49, 67), c-IAP2/HIAP-1 (49, 67), XIAP/hILP (18), and survivin (3) (C). aa, amino acids. All BIR domains shown contain the conserved cystine and histidine residues of the CX2C16HX6-8C motif as well as the C3HC4 backbone motif of the RING domain. The sequences were aligned with the SMART program (52).
The AMVIAP is 47 and 45% identical to the Cp-IAP and the Op-IAP, respectively (8, 15). Based on phylogenetic analysis, the AMVIAP protein has been shown to fall into a distinct cluster, grouping with a number of functional IAPs that include the Drosophila DIAP1 protein, several baculovirus IAPs (Op-IAP3 and Cp-IAP3), and several IAPs from Lepidoptera, including Sf-IAP from Spodoptera frugiperda and Bm-IAP from Bombyx mori (30-32). Based on these comparisons, we anticipate that the AMViap gene will be active and function as an inhibitor of apoptosis.
Rescue of Drosophila hid gene-induced apoptosis by the AMViap gene.
The Drosophila proapoptotic genes hid, grim, and reaper are known inducers of apoptosis based on both in vivo genetic studies in Drosophila as well as in vitro transfection studies of both insect and mammalian cell lines (10, 38, 60, 66). Based on previous research, transfection of Ld652 cells with hid is predicted to lead to apoptosis (60, 66). To evaluate the apoptotic suppressor activity of AMViap, we cotransfected Ld652 cells with pIE1hid plus a pIE1p35- or pIE1iap-containing plasmid at different ratios, as well as a fixed amount of a pIE1gfp-containing plasmid DNA (2 μg/transfection) as an internal marker gene for viable cells. In this assay, the expression of a functional apoptotic suppressor gene is predicted to block apoptosis induced by hid. This rescue is indicated by expression of GFP from a cotransfected plasmid in surviving cells. Cells cotransfected with the hid and gfp genes but no apoptotic suppressor gene would be expected to undergo apoptosis, and expression of GFP would not be seen. In parallel experiments, we have evaluated the levels of caspase-3-like activity in those transfected cells. Caspase-3 activation can be conveniently measured by cleavage of the fluorescent substrate DEVD-AMC. In apoptotic cells, one would expect the levels of caspase-3-like activity to be increased relative to control cells.
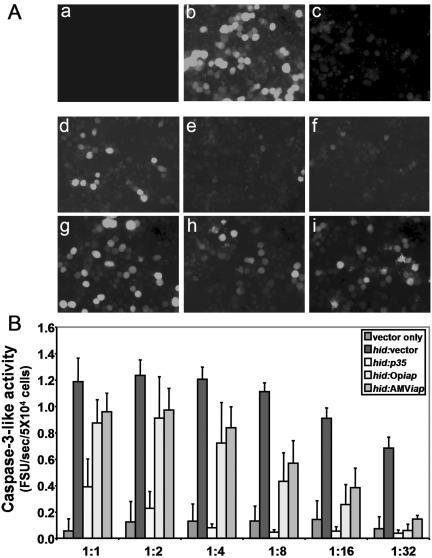
We designed our experiments to employ vectors expressing the p35 and Op-iap genes as positive controls, empty pIE1 vector plasmid as a negative control, and the AMViap gene as the gene to be tested. All genes were cloned into the same pIE1-4 vector, which places control of expression of the genes under the baculovirus ie1 promoter (Novogen) (Fig. 1A). Transfection of vector and pIE1gfp plasmid DNA into Ld652 cells, based on the generation of GFP-positive cells, indicated that about 60 to 70% of the cells were transfected (Fig. 3A, panel b). Under these conditions, there were only background levels of caspase-3-like activity, ranging from 0.1 to 0.2 FSU/5 × 104 cells (Fig. 3B). When hid was cotransfected with gfp in Ld652 cells, apoptosis was induced as evidenced by a significant reduction in the number of GFP-positive cells (Fig. 3A, panel c) and increased levels of caspase-3-like activity (Fig. 3B). Regardless of the hid/empty vector ratio, less than 1% of the cells were GFP positive (Fig. 3A, panel c). Although the levels of caspase activity were slightly dependent on the hid/gfp ratio, the levels of caspase-3-like activity were considerably higher than those of the empty vector control and ranged from 0.8 up to 1.2 FSU/5 × 104 cells (Fig. 3B).
FIG. 3.
Rescue of cells from hid-induced apoptosis by p35 and iap genes. Transfection with hid induces apoptosis and prevents expression of GFP from a second cotransfected plasmid. (A) The rescue of hid-induced apoptosis in Ld652 cells was shown as the rescue of GFP expression in cells after transfection with mock (a), (b) vector-vector (b), hid-vector (c), hid-p35 (1:1) (d), hid-Opiap (1:1) (e), hid-AMViap (1:1) (f), hid- p35 (1:8) (g), hid-Opiap (1:8) (h), hid-AMViap (1:8) (i). All samples were also cotransfected with 2 μg of the indicator pIE1gfp, 0.5 μg of pIE1hid, and either 4 μg of pIE1 vector or pIE1 plasmids containing apoptotic suppressor genes at the ratio indicated for each transfection. Photos were taken at 48 h after transfection in a Zeiss inverted phase-contrast microscope (magnification, ×20). (B) Caspase-3-like activity in Ld652 cells as measured by Ac-DEVD-AMC cleavage. Cells were cotransfected with 2 μg of pIEgfp as the indicator plasmid, 0.5 μg of pIE1hid, and a third plasmid, either pIE1-4 vector or pIE1-containing one of the apoptotic suppressor genes (p35, Op-iap, or AMViap) at different ratios of hid to the third plasmid, ranging from 1:1 up to 1:32. Relative value units of caspase activity were expressed as the FSU per second in 5 × 104 cells.
We then attempted to block hid-induced apoptosis by cotransfection with p35, Op-iap, or AMViap genes. The p35-positive control gene rescued hid-induced apoptosis at ratios as low as a 1:1 hid/p35 ratio, with over 50% GFP-positive cells observed (Fig. 3A, panels d and g) and a 75% reduction of caspase-3-like activity compared to that seen in the absence of p35 (Fig. 3B). Rescue was also observed with both the AMViap and Op-iap genes (Fig. 3A, panels h and i) but only at higher levels of the rescuing iap gene DNA (Fig. 3B) with concomitant reduction in the level of caspase-3-like activity. When these iaps were cotransfected with hid in a 1:1 ratio, little rescue of the hid-induced apoptosis was observed as the number of GFP-positive cells was low (<5%) (Fig. 3A, panels e and f) and the caspase levels were still high (Fig. 3B). However, at hid/iap ratios of 1:8, the rescue became apparent and the number of GFP-positive cells increased up to 30 to 40% (Fig. 3A, panels h and i) and the levels of caspase-3-like activity dropped 60% compared to cells transfected in the absence of either of these iap genes (Fig. 3B). At ratios of 1:16 and 1:32, the level of rescue by Op-iap and AMViap was comparable to that observed with the p35 gene in terms of the number of GFP-positive cells (data not shown) and the reduction of caspase-3-like activity to background levels (Fig. 3B). Therefore, it appears that while all three apoptotic suppressor genes can prevent hid-induced apoptosis, the p35 gene is the most efficient, and that the Op-iap and AMViap genes, while functional and equivalent to each other, are less efficient than the p35 gene.
AMViap restores polyhedron formation in vAcΔp35-infected Sf9 cells.
Infection of Sf9 cells by AcMNPV lacking the p35 gene (vAcΔp35) leads to rapid induction of apoptosis, resulting in an abortive infection and lack of polyhedron formation. Apoptosis can be blocked and virus replication and polyhedron formation can be rescued, if infection is preceded by expression of an apoptotic suppressor gene such as p35 or an iap gene (13, 73). We tested the ability of AMViap to rescue polyhedron formation in vAcΔp35-infected Sf9 cells. Cells were transfected with ”empty“ pIE1 plasmid, pIE1p35, pIE1Opiap or pIE1AMViap. The transfected cells were then infected with vAcΔp35, and polyhedron formation was assessed at 96 hpi.
In WT AcMNPV-infected Sf9 cells, polyhedron formation was readily visible (Fig. 4B). However, in vAcΔp35-infected cells previously transfected with empty vector, apoptosis was induced and no polyhedra were formed (Fig. 4C). However, if vAcΔp35-infected cells were transfected with a plasmid expressing either p35 or Op-iap, apoptosis was sufficiently delayed to allow polyhedron formation (Fig. 4D and E). Similarly, transfection of pIE1AMViap also blocked premature cell death and rescued the formation of polyhedra, albeit less efficiently as many fewer polyhedra were noted compared to rescue by either the p35 or Op-iap gene (Fig. 4F). Therefore, AMViap does function to block apoptosis under these conditions; however, rescue was less efficient than rescue by either p35 or Op-iap.
FIG. 4.
Polyhedron formation in vAcΔp35-infected Sf9 cells. Sf9 cells were transfected with pIE1-4 (A), pIE1AMViap (B), pIE1-4 (C), pIE1p35 (D), pIE1Op-iap (E), or pIE1AMViap (F) and then mock infected (A) or infected with WT AcMNPV (B) or with vAcΔp35 (C to F). Polyhedra were observed in cells transfected with apoptotic suppressor genes and are indicated by the arrows.
Properties of AmEPV lacking a functional iap gene (vAmgfp/Δiap/lacZ).
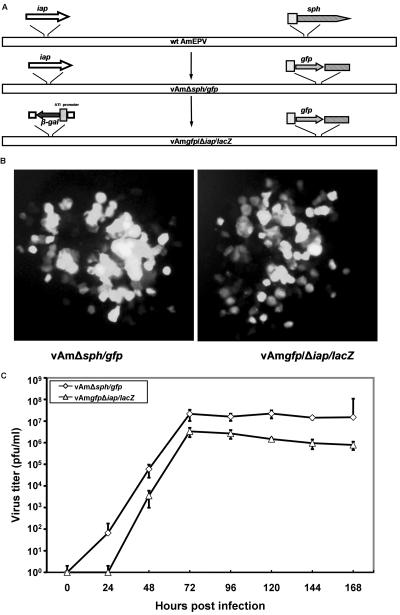
Our evidence suggests that AMViap can act to inhibit apoptosis. One of the key questions is whether the AMViap gene functions in this capacity during an AmEPV infection and is essential for AmEPV growth. In order to address this question, we produced an AmEPV recombinant virus in which the nonessential sph gene was disrupted by the gfp gene under control of the strong, late sph promoter (36). This recombinant AmEPV (vAmΔsph/gfp) is both sph− and gfp+, allowing easy identification of this virus via GFP-positive plaques (Fig. 5A). The iap gene of this virus was then disrupted to create vAmgfp/Δiap/lacZ by insertion of the lacZ gene under control of the strong, late cowpox virus ATI promoter (36). The final resultant virus lacks a functional iap gene and is both lacZ+ and gfp+ (Fig. 5A).
FIG. 5.
Growth and spread of vAmΔsph/gfp and vAmgfp/Δiap/lacZ in infected Ld652 cells. (A) Diagrams of the construction of vAmΔsph/gfp and vAmgfp/Δiap/lacZ recombinant viruses. (B) Plaque comparison of vAmΔsph/gfp and vAmgfp/Δiap/lacZ in infected Ld652 cells 6 days postinfection (magnification, ×20). (C) Time course of virus replication in Ld652 cells. Ld652 cells were infected with vAmΔsph/gfp or vAmgfp/Δiap/lacZ at an MOI of 0.01, and samples were removed at the times indicated. Cells were lysed and virus growth was evaluated by plaque assay in Ld652 cells.
The fact that we had little difficulty creating vAmgfp/Δiap/lacZ and that the virus grew well in Ld652 cells suggested that a functional iap gene was not necessary for growth in these cells. We have compared the growth of vAmΔsph/gfp and vAmgfp/Δiap/lacZ viruses in Ld652 cells following low-multiplicity infections, conditions that measure both growth and spread of the virus. Infected cultures were harvested daily, and the titer of progeny virus was determined at different time points. Plaques expressing GFP could be observed for either virus by 3 days postinfection. While the plaques of both viruses were very similar, the vAmgfp/Δiap/lacZ virus appeared to generate slightly less fluorescence and perhaps slightly smaller plaques. However, these differences were subtle, and there were very few observable differences between the two viruses (Fig. 5B).
A more quantitative comparison of the growth of vAmΔsph/gfp and vAmgfp/Δiap/lacZ viruses shows that significant growth of both viruses was evident by 48 hpi, with some delay in vAmgfp/Δiap/lacZ production, and both viruses reached a plateau by 72 hpi. The titers of vAmΔsph/gfp virus approached 107 PFU/ml, whereas the titers of vAmgfp/Δiap/lacZ were 106 PFU/ml, about 10-fold lower than the parental vAmΔsph/gfp virus (Fig. 5C). These results suggest that while the iap gene is not absolutely essential for growth in cell culture, virus yields are slightly impaired in the absence of a functional iap gene.
Induction of apoptosis in cells infected with vAmΔsph/gfp and vAmgfp/Δiap/lacZ.
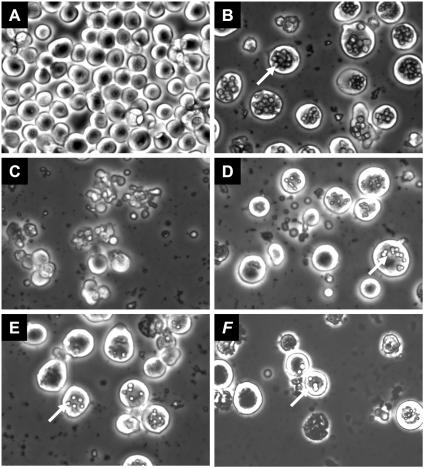
While Ld652 cells are fully permissive for AmEPV growth, yields of virus from Sf9 cells are considerably lower and no virus is produced from infected Drosophila S2 cells even though virus enters these cells quite well (Q. Li and R. W. Moyer, unpublished results). We evaluated the effect of the AMViap gene deletion in the context of recombinant virus-induced apoptosis in each of these cell lines. A similar assay using both permissive and nonpermissive cells has been previously reported in a study on the recombinant AcMNPV virus (p49+ p35− iap−) (73). Monolayers of Ld652, Sf9, or S2 cells were infected with vAmΔsph/gfp or vAmgfp/Δiap/lacZ at an MOI of 15. The rate and extent of cytoxicity induced by vAmgfp/Δiap/lacZ differ among the three cell lines. Microscopic examination revealed that clear signs of cellular cytotoxicity were readily noticeable by 24 hpi and were clearly observed at 48 hpi in S2 and Sf9 cells infected with vAmgfp/Δiap/lacZ (Fig. 6C and F). Far less cytotoxicity was observed in vAmΔsph/gfp-infected Ld652 cells (Fig. 6B and E). Infection of S2 cells by vAmgfp/Δiap/lacZ results in high levels of cellular destruction with a >80% decrease in the adherent cell population compared with little cell loss in S2 cells infected with vAmΔsph/gfp. Infection of Sf9 cells with vAmgfp/Δiap/lacZ also resulted in cellular damage, with loss of about 30% of the overall cell population, far less than that observed for S2 cells. Again, infection of Sf9 cells with vAmΔsph/gfp led to even less cytotoxicity. Finally, Ld652 cells infected with vAmgfp/Δiap/lacZ led to relatively little cytotoxicity and little observable loss of the adherent cell population (Fig. 6I). It appears that the presence of the AMViap gene aids in the preservation of cellular integrity and lessens infected cell cytotoxicity in a cell line-dependent fashion.
FIG. 6.
Cytotoxicity in cell lines infected with vAmΔsph/gfp or vAmgfp/Δiap/lacZ. Different levels of cellular cytotoxicity were observed in S2, Sf9, and Ld652 cells infected with vAmΔsph/gfp or vAmgfp/Δiap/lacZ at an MOI of 15. Photographs were taken at 48 hpi, using a Zeiss inverted microscope at ×20.
We have also measured the induction of caspase-3-like activity in S2, Sf9, and Ld652 cells following infection. In vAmΔsph/gfp-infected S2 cells, only low levels of caspase-3-like activity were induced. In vAmgfp/Δiap/lacZ-infected S2 cells, the level of caspase-3-like activity was higher and could be observed as early as 6 hpi and increased only slightly thereafter up to 48 hpi (Fig. 7A). In Sf9 cells, while there is little induction of caspase-3-like activity in cells infected with vAmΔsph/gfp, infection with vAmgfp/Δiap/lacZ leads to a steady induction of caspase-3-like activity throughout the 48-h period of infection (Fig. 7B). In Ld652 cells, the patterns of caspase induction were similar to those observed in S2 cells; however, vAmgfp/Δiap/lacZ induced much lower levels of caspase-3-like activity than the levels observed in S2 cells (Fig. 7B). It appears that some cells (Ld652) are relatively resistant to AmEPV-induced caspase-3-like activity and apoptosis. In other cells, particularly S2 cells, but to some extent Sf9 cells, there is more intrinsic susceptibility to apoptosis following infection and there is significant protection against the induction of caspase-3-like activity afforded by the AMViap gene. These results suggest that the AMViap gene can inhibit apoptosis within the context of viral infection and that induction of apoptosis by the virus is cell line specific.
FIG. 7.
Induction of caspase-3-like activity in cell lines infected with vAmΔsph/gfp or vAmgfp/Δiap/lacZ. Caspase-3-like activity was examined in protein samples comprising combined supernatants and cell pellets at the times indicated. Caspase-3-like activity was measured by the cleavage of Ac-DEVD-AMC. S2 (A), Sf9 (B), and Ld652 (C) cells were infected with vAmΔsph/gfp or vAmgfp/Δiap/lacZ at an MOI of 15.
AMVIAP protein inhibits human caspase-9 and caspase-3 activities in vitro.
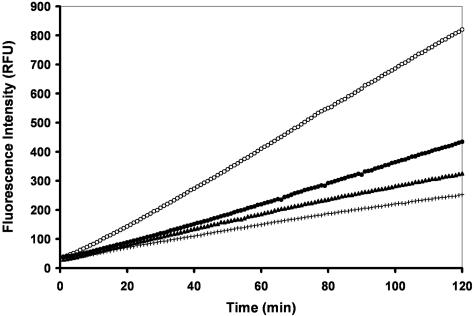
To better understand the mechanism of AMVIAP-mediated apoptosis inhibition, we performed in vitro assays to determine if AMVIAP can directly inhibit caspase activity. The AMViap and human xiap ORFs were cloned into the pGEX-4T3 expression vector. Both AMVIAP and XIAP proteins were expressed as GST fusion proteins and purified on glutathione-Sepharose beads. The purified AMVIAP and XIAP proteins were then assessed for their ability to inhibit the activity of purified caspase-9 in vitro by using the fluorogenic tetrapeptide substrate LEHD-AMC. As previously reported, XIAP effectively inhibited caspase-9 activity (Fig. 8). AMVIAP also inhibited caspase-9 activity, albeit to a lesser degree than XIAP. Even a 10-fold molar excess of AMVIAP did not inhibit caspase-9 activity to the same extent as that of XIAP (Fig. 8).
FIG. 8.
In vitro inhibition of caspase-9 activity by AMVIAP. Human caspase-9 activity was assayed in vitro with Ac-LEHD-AMC as a substrate. Caspase-9 was mixed with either GST at 2 μM (○), XIAP at 500 nM (+), and AMVIAP at 500 nM (•) or 5,000 nM (5 μM) (▴). Activity was estimated by LEHD-AMC cleavage. Each data series represents fluorescence emission (490 nm) at 1-min intervals for 120 min.
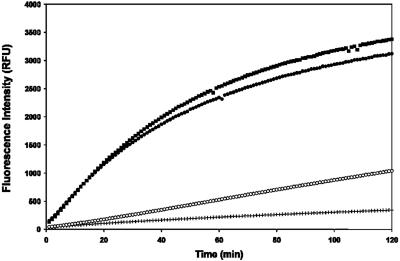
We also examined the ability of AMVIAP to inhibit the purified caspase-3 activity in vitro as measured by cleavage of the fluorogenic tetrapeptide substrate DEVD-AMC. XIAP was again used as a positive control, since XIAP effectively inhibits caspase-3 activity in vitro as reported previously and presented in Fig. 9. Our data suggest that AMVIAP also inhibited caspase-3 activity comparable to the inhibition by XIAP (Fig. 9).
FIG. 9.
In vitro inhibition of caspase-3 activity by AMVIAP. Human caspase-9 activity was assayed in vitro with Ac-DEVD-AMC as the substrate. Caspase-3 was mixed with either, in the absence of recombinant IAPs (▪), GST at 2 μM (•), GST-XIAP at 4.1 μg (500 nM final) (+), or GST-AMVIAP at 2.9 μg (500 nM) (○). Activity was estimated by DEVD-AMC cleavage. Each data series represents fluorescence emission (490 nm) at 1-min intervals for 120 min.
Therefore, AMVIAP may inhibit apoptosis through direct inhibition of either the insect host initiator caspase equivalent to caspase-9 or the terminal caspases of the intrinsic pathway.
DISCUSSION
Collectively, all our results indicate that the AMViap gene is active in controlling apoptosis, as predicted based on the sequence analysis. The AMViap gene functions in a manner similar to the well-characterized Op-iap gene (42, 61, 63). Both the AMViap gene and Op-iap gene were somewhat less effective than the p35 gene in mediating rescue from hid-induced apoptosis, as more copies of both iap genes were required to give levels of inhibition of apoptosis comparable to that seen for p35. In addition to blocking apoptosis induced by the Drosophila proapoptotic hid gene (Fig. 3), AMViap also inhibited apoptosis induced by the Drosophila proapoptotic genes grim and reaper and chemical inducers of apoptosis such as actinomycin D and staurosporine (Li and Moyer, unpublished).
In mammalian cells, IAPs bind to Smac/DIABLO through their BIR domains (26, 59). Human XIAP binds to Smac/DIABLO through a peptide-binding groove on the surface of BIR3 that is important for IAP function (64). Similarly, Drosophila HID, GRIM, and REAPER bind to DIAP1 directly through a groove on the surface of its BIR2 (65), and HID binds to the BIR2 domain of viral Op-IAP via an analogous structural feature (63). AMVIAP may interact with Hid in a manner similar to the HID-BIR2 interaction in Op-IAP and to the Smac/DIABLO binding to BIR3 of XIAP.
Human XIAP also inhibits caspase-9 activity through an interaction involving the Smac binding groove and the ATPF interaction motif exposed on the small subunit of caspase-9 following autocatalysis. Additional contacts between XIAP BIR3 and caspase-9 induce a conformational change that maintains caspase-9 in an inactive configuration (55). AMVIAP, like XIAP, shows some activity against caspase-9, indicating that AMVIAP might inhibit apoptosis in a fashion similar to that of XIAP by inhibition of an initiator caspase in the insect host (Fig. 8). Similar results have also been shown for Cp-IAP and the S. frugiperda IAP (SfIAP), viral and insect IAPs, respectively, both of which can specifically inhibit mammalian caspase-9 (30). Op-IAP also inhibits the activation of the insect initiator caspase, the Sf-caspase-X (34, 41). Although we have demonstrated that AMVIAP can inhibit the equivalent initiator caspase in the mammalian intrinsic pathways, the interaction appears to be weaker than XIAP, suggesting that AMVIAP may act on other components of the insect apoptosis pathways—for example, the terminal caspases.
AMVIAP also appears to inhibit caspase-3 and hence might be expected to inhibit the equivalent terminal caspase in the insect host. XIAP is also a potent inhibitor of caspase-3 and -7 in vitro (16). This XIAP-mediated inhibition of caspase-3 occurs via the occupation of the active site of caspase-3 or -7 by the linker segment of XIAP, which in turn serves to block the substrate entry and hence the enzymatic activity (55). We are currently exploring potential interactions between AMVIAP and proapoptotic proteins such as HID, GRIM, and REAPER and various other insect caspases.
Apoptotic suppressor genes are critical for baculovirus replication in certain cell lines. In AcMNPV, deletion of the p35 gene severely decreases virus yield by 1,000-fold in both cultured cells (28) and insect larvae (14), with no formation of virus containing polyhedra. Inactivation of the Op-iap gene in OpMNPV induces apoptosis in 98% of the infected Ld652 cells and virus replication is blocked (42). We were therefore surprised that deletion of the iap gene from the AmEPV genome did not affect plaque growth in Ld652 cells and had only minimal effects on virus yield (Fig. 5B and C). One possibility is that there are additional AmEPV genes that control apoptosis, despite the lack of the commonly recognized apoptotic suppressor genes found in vertebrate poxviruses. An alternative possibility is that, in Ld652 cells, apoptosis is not induced in response to AmEPV infection, thus rendering AMViap nonessential under these conditions.
It is interesting to compare the known strategies of the vertebrate and insect poxviruses for control of apoptosis. Of the completely sequenced insect poxviruses, AmEPV carries one iap gene (5) and MsEPV carries two: Ms248 (iap-1) and Ms242 (iap-2) (1). In contrast, none of the sequenced vertebrate poxviruses carries iap gene(s). Vertebrate poxviruses do carry a plethora of apoptosis regulators and interfere with the onset of apoptosis by a variety of mechanisms (53, 58). The extrinsic apoptotic pathway appears to be the preferred target of these viral strategies. The serpin crmA/SPI-2 inhibits apoptosis by blocking caspase-8 (45) and granzyme B-mediated apoptosis (47). The viral DED-containing proteins (vFLIPs), such as the molluscum contagiosum proteins MC159 and MC160, bind to FADD and pro-caspase-8 to inhibit transduction of death receptor-mediated apoptosis signals (29, 56). The two protein kinase inhibitors encoded by orthopoxviruses, the double-stranded RNA-binding protein E3L and the eIF2α homolog K3L of vaccinia virus, also control activation of caspase-8 (22, 33). Two unrelated viral RING finger proteins, P28 of ectromelia virus and N1R of Shope fibroma virus are able to block UV-induced apoptosis (9). The TNF receptor homolog M-T2 gene from myxoma virus also regulates apoptosis through binding to TNF-α and inhibiting the TNF-α-induced receptor signaling (51). The glutathione peroxidase from molluscum contagiosum virus (MC66) protects cells against the cytotoxic effects of hydrogen peroxide induced by TNF to prevent cells from undergoing apoptosis (57). However, M011L from myxoma virus (19, 20) and the comparable protein F1L of vaccinia virus (62) inhibit apoptosis through the intrinsic, rather than extrinsic, pathway. M011L inhibits the transduction of death signals via the mitochondrial checkpoint and prevents the loss of inner mitochondrial membrane potential associated with apoptosis. Several other apoptotic suppressor genes, such as the endoplasmic reticulum-resident protein encoded by the M-T4 gene and the ML005L/R ankyrin repeat protein encoded by myxoma virus, have also been shown to protect infected cells from apoptosis (4, 44). The Bcl-2 homolog has only been identified in fowlpox virus (FPV039) through genome sequencing, and its exact function is still unknown (2).
Given the large number of apoptosis regulatory proteins encoded by vertebrate poxviruses and the minimal effects on virus growth noted upon deletion of the sole functional iap gene from AmEPV, it is likely there are a number of as-yet-undiscovered AmEPV genes which control apoptosis.
Acknowledgments
We thank Lei Zhou (University of Florida) for providing the pIE1-4 vector, the pIE1p35 and pIE1hid plasmids, and the Drosophila S2 cell line. We also thank Paul Friesen at University of Wisconsin for his generous help and for providing numerous reagents, including the WT AcMNPV(E2) and vAcΔp35 viruses, together with the TN368 cells. We also thank Elliot J. Lefkowitz for a preliminary analysis of the AmEPV genome for apoptosis-controlling genes. Numerous discussions with members of our laboratory have been greatly appreciated.
This research is supported by NIH grant 5R01AI046479-04.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrosini, G., C. Adida, and D. C. Altieri. 1997. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 3:917-921. [DOI] [PubMed] [Google Scholar]
- 4.Barry, M., S. Hnatiuk, K. Mossman, S. F. Lee, L. Boshkov, and G. McFadden. 1997. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology 239:360-377. [DOI] [PubMed] [Google Scholar]
- 5.Bawden, A. L., K. J. Glassberg, J. Diggans, R. Shaw, W. Farmerie, and R. W. Moyer. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120-139. [DOI] [PubMed] [Google Scholar]
- 6.Becker, M. N., W. B. Greenleaf, D. A. Ostrov, and R. W. Moyer. 2004. Amsacta moorei entomopoxvirus expresses an active superoxide dismutase. J. Virol. 78:10265-10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benedict, C. A., P. S. Norris, and C. F. Ware. 2002. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 3:1013-1018. [DOI] [PubMed] [Google Scholar]
- 8.Birnbaum, M. J., R. J. Clem, and L. K. Miller. 1994. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 68:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brick, D. J., R. D. Burke, A. A. Minkley, and C. Upton. 2000. Ectromelia virus virulence factor p28 acts upstream of caspase-3 in response to UV light-induced apoptosis. J. Gen. Virol. 81:1087-1097. [DOI] [PubMed] [Google Scholar]
- 10.Chai, J. J., N. Yan, J. R. Huh, J. W. Wu, W. Y. Li, B. A. Hay, and Y. G. Shi. 2003. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat. Struct. Biol. 10:892-898. [DOI] [PubMed] [Google Scholar]
- 11.Clarke, T. E., and R. J. Clem. 2003. Insect defenses against virus infection: the role of apoptosis. Int. Rev. Immunol. 22:401-424. [DOI] [PubMed] [Google Scholar]
- 12.Clem, R. J. 2001. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ. 8:137-143. [DOI] [PubMed] [Google Scholar]
- 13.Clem, R. J., and L. K. Miller. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 14:5212-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clem, R. J., M. Robson, and L. K. Miller. 1994. Influence of infection route on the infectivity of baculovirus mutants lacking the apoptosis-inhibiting gene p35 and the adjacent gene p94. J. Virol. 68:6759-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crook, N. E., R. J. Clem, and L. K. Miller. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67:2168-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deveraux, Q. L., and J. C. Reed. 1999. IAP family proteins—suppressors of apoptosis. Genes Dev. 13:239-252. [DOI] [PubMed] [Google Scholar]
- 17.Du, Q., D. Lehavi, O. Faktor, Y. Qi, and N. Chejanovsky. 1999. Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J. Virol. 73:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duckett, C. S., V. E. Nava, R. W. Gedrich, R. J. Clem, J. L. VanDongen, M. C. Gilfillan, H. Shiels, J. M. Hardwick, and C. B. Thompson. 1996. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 15:2685-2694. [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, H., M. Barry, S. F. Lee, X. J. Sun, K. Graham, J. Stone, R. C. Bleackley, and G. McFadden. 2000. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 191:1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett, H., M. Barry, X. Sun, S. F. Lee, C. Frantz, L. G. Berthiaume, G. McFadden, and R. C. Bleackley. 2002. The myxoma poxvirus protein, M11L, prevents apoptosis by direct interaction with the mitochondrial permeability transition pore. J. Exp. Med. 196:1127-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher, C. L., and G. K. Pei. 1997. Modification of a PCR-based site-directed mutagenesis method. BioTechniques 23:570-571 [DOI] [PubMed] [Google Scholar]
- 22.Gil, J., M. Esteban, and D. Roth. 2000. In vivo regulation of the dsRNA-dependent protein kinase PKR by the cellular glycoprotein P67. Biochemistry 39:16016-16025. [DOI] [PubMed] [Google Scholar]
- 23.Goodwin, R. H., J. R. Adams, and M. Shapiro. 1990. Replication of the entomopoxvirus from Amsacta moorei in serum-free cultures of a Gypsy moth cell line. J. Invertebr. Pathol. 56:190-205. [Google Scholar]
- 24.Hall, R. L., and W. F. Hink. 1990. Physical mapping and field inversion gel-electrophoresis of Amsacta moorei entomopoxvirus DNA. Arch. Virol. 110:77-90. [DOI] [PubMed] [Google Scholar]
- 25.Hall, R. L., and R. W. Moyer. 1991. Identification, cloning, and sequencing of a fragment of Amsacta moorei entomopoxvirus DNA containing the spheroidin gene and three vaccinia virus-related open reading frames. J. Virol. 65:6516-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hay, B. A. 2000. Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ. 7:1045-1056. [DOI] [PubMed] [Google Scholar]
- 27.Hay, S., and G. Kannourakis. 2002. A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83:1547-1564. [DOI] [PubMed] [Google Scholar]
- 28.Hershberger, P. A., J. A. Dickson, and P. D. Friesen. 1992. Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol. 66:5525-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, S. M., C. Vincenz, M. Buller, and V. M. Dixit. 1997. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J. Biol. Chem. 272:9621-9624. [DOI] [PubMed] [Google Scholar]
- 30.Huang, Q., Q. L. Deveraux, S. Maeda, G. S. Salvesen, H. R. Stennicke, B. D. Hammock, and J. C. Reed. 2000. Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl. Acad. Sci. USA 97:1427-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, Q. H., Q. L. Deveraux, S. Maeda, H. R. Stennicke, B. D. Hammock, and J. C. Reed. 2001. Cloning and characterization of an inhibitor of apoptosis protein (IAP) from Bombyx mori. Biochim. Biophys. Acta Mol. Cell Res. 1499:191-198. [DOI] [PubMed] [Google Scholar]
- 32.Hughes, A. L. 2002. Evolution of inhibitors of apoptosis in baculoviruses and their insect hosts. Infect. Genet. Evol. 2:3-10. [DOI] [PubMed] [Google Scholar]
- 33.Kibler, K. V., T. Shors, K. B. Perkins, C. C. Zeman, M. P. Banaszak, J. Biesterfeldt, J. O. Langland, and B. L. Jacobs. 1997. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J. Virol. 71:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaCount, D. J., S. F. Hanson, C. L. Schneider, and P. D. Friesen. 2000. Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J. Biol. Chem. 275:15657-15664. [DOI] [PubMed] [Google Scholar]
- 35.LeBlanc, A. C. 2003. Natural cellular inhibitors of caspases. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 27:215-229. [DOI] [PubMed] [Google Scholar]
- 36.Li, Y., R. L. Hall, and R. W. Moyer. 1997. Transient, nonlethal expression of genes in vertebrate cells by recombinant entomopoxviruses. J. Virol. 71:9557-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, Y., R. L. Hall, S. L. Yuan, and R. W. Moyer. 1998. High-level expression of Amsacta moorei entamopoxvirus spheroidin depends on sequences within the gene. J. Gen. Virol. 79:613-622. [DOI] [PubMed] [Google Scholar]
- 38.Lisi, S., I. Mazzon, and K. White. 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154:669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liston, P., W. G. Fong, and R. G. Korneluk. 2003. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene 22:8568-8580. [DOI] [PubMed] [Google Scholar]
- 40.Macen, J. L., R. S. Garner, P. Y. Musy, M. A. Brooks, P. C. Turner, R. W. Moyer, G. McFadden, and R. C. Bleackley. 1996. Differential inhibition of the Fas- and granule-mediated cytolysis pathways by the orthopoxvirus cytokine response modifier A/SPI-2 and SPI-1 protein. Proc. Natl. Acad. Sci. USA 93:9108-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manji, G. A., and P. D. Friesen. 2001. Apoptosis in motion. An apical, P35-insensitive caspase mediates programmed cell death in insect cells. J. Biol. Chem. 276:16704-16710. [DOI] [PubMed] [Google Scholar]
- 42.Means, J. C., I. Muro, and R. J. Clem. 2003. Silencing of the baculovirus Op-iap3 gene by RNA interference reveals that it is required for prevention of apoptosis during Orgyia pseudotsugata M nucleopolyhedrovirus infection of Ld652Y cells. J. Virol. 77:4481-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, L. K. 1997. Baculovirus interaction with host apoptotic pathways. J. Cell Physiol. 173:178-182. [DOI] [PubMed] [Google Scholar]
- 44.Mossman, K., S. F. Lee, M. Barry, L. Boshkov, and G. McFadden. 1996. Disruption of M-T5, a novel myxoma virus gene member of the poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J. Virol. 70:4394-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nathaniel, R., A. L. MacNeill, P. C. Turner, Y.-X. Wang, and R. W. Moyer. 2004. Cowpox virus CrmA, Myxoma virus SERP2 and baculovirus P35 are not functionally interchangeable caspase inhibitors in poxvirus infections. J. Gen. Virol. 85:1267-1278. [DOI] [PubMed] [Google Scholar]
- 46.Pei, Z. F., G. Reske, Q. H. Huang, B. D. Hammock, Y. P. Qi, and N. Chejanovsky. 2002. Characterization of the apoptosis suppressor protein P49 from the Spodoptera littoralis nucleopolyhedrovirus. J. Biol. Chem. 277:48677-48684. [DOI] [PubMed] [Google Scholar]
- 47.Quan, L. T., A. Caputo, R. C. Bleackley, D. J. Pickup, and G. S. Salvesen. 1995. Granzyme-B is inhibited by the cowpox virus serpin cytokine response modifier-A. J. Biol. Chem. 270:10377-10379. [DOI] [PubMed] [Google Scholar]
- 48.Ray, C. A., and D. J. Pickup. 1996. The mode of death of pig kidney cells infected with cowpox virus is governed by the expression of the crmA gene. Virology 217:384-391. [DOI] [PubMed] [Google Scholar]
- 49.Roy, N., Q. L. Deveraux, R. Takahashi, O. Zhou, G. Ambrosini, D. Altieri, G. S. Salvesen, and J. C. Reed. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. Blood 90:2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryoo, H. D., A. Bergmann, H. Gonen, A. Ciechanover, and H. Steller. 2002. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4:432-438. [DOI] [PubMed] [Google Scholar]
- 51.Schreiber, M., and G. McFadden. 1996. Mutational analysis of the ligand-binding domain of M-T2 protein, the tumor necrosis factor receptor homologue of myxoma virus. J. Immunol. 157:4486-4495. [PubMed] [Google Scholar]
- 52.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 54.Shi, Y. G. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9:459-470. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y. G. 2004. Caspase activation: revisiting the induced proximity model. Cell 117:855-858. [DOI] [PubMed] [Google Scholar]
- 56.Shisler, J. L., and B. Moss. 2001. Molluscum contagiosum virus inhibitors of apoptosis: the MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology 282:14-25. [DOI] [PubMed] [Google Scholar]
- 57.Shisler, J. L., T. G. Senkevich, M. J. Berry, and B. Moss. 1998. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science 279:102-105. [DOI] [PubMed] [Google Scholar]
- 58.Turner, P. C., and R. W. Moyer. 1998. Control of apoptosis by poxviruses. Semin. Virol. 8:453-469. [Google Scholar]
- 59.Verhagen, A. M., E. J. Coulson, and D. L. Vaux. 5 July 2001, posting date. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2:reviews3009.1-reviews3009.10. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vucic, D., W. J. Kaiser, A. J. Harvey, and L. K. Miller. 1997. Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl. Acad. Sci. USA 94:10183-10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vucic, D., W. J. Kaiser, and L. K. Miller. 1998. A mutational analysis of the baculovirus inhibitor of apoptosis Op-IAP. J. Biol. Chem. 273:33915-33921. [DOI] [PubMed] [Google Scholar]
- 62.Wasilenko, S. T., T. L. Stewart, A. F. A. Meyers, and M. Barry. 2003. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. USA 100:14345-14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright, C. W., and R. J. Clem. 2002. Sequence requirements for Hid binding and apoptosis regulation in the baculovirus inhibitor of apoptosis Op-IAP. Hid binds Op-IAP in a manner similar to Smac binding of XIAP. J. Biol. Chem. 277:2454-2462. [DOI] [PubMed] [Google Scholar]
- 64.Wu, G., J. J. Chai, T. L. Suber, J. W. Wu, C. Y. Du, X. D. Wang, and Y. G. Shi. 2000. Structural basis of IAP recognition by Smac/DIABLO. Nature 408:1008-1012. [DOI] [PubMed] [Google Scholar]
- 65.Wu, J. W., A. E. Cocina, J. J. Chai, B. A. Hay, and Y. G. Shi. 2001. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell 8:95-104. [DOI] [PubMed] [Google Scholar]
- 66.Yoo, S. J., J. R. Huh, I. Muro, H. Yu, L. J. Wang, S. L. Wang, R. M. R. Feldman, R. J. Clem, H. A. J. Muller, and B. A. Hay. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4:416-424. [DOI] [PubMed] [Google Scholar]
- 67.Young, S. S., P. Liston, J. Y. Xuan, C. McRoberts, C. A. Lefebvre, and R. G. Korneluk. 1999. Genomic organization and physical map of the human inhibitors of apoptosis: HIAP1 and HIAP2. Mamm. Genome 10:44-48. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, L., and H. Steller. 2003. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev. Cell 4:599-605. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, Q., and G. S. Salvesen. 2000. Viral caspase inhibitors CrmA and p35. Apoptosis 322:143-154. [DOI] [PubMed] [Google Scholar]
- 70.Zimmermann, K. C., C. Bonzon, and D. R. Green. 2001. The machinery of programmed cell death. Pharmacol. Ther. 92:57-70. [DOI] [PubMed] [Google Scholar]
- 71.Zimmermann, K. C., and D. R. Green. 2001. How cells die: apoptosis pathways. J. Allergy Clin. Immunol. 108:S99-S103. [DOI] [PubMed] [Google Scholar]
- 72.Zoog, S. J., J. Bertin, and P. D. Friesen. 1999. Caspase inhibition by baculovirus P35 requires interaction between the reactive site loop and the beta-sheet core. J. Biol. Chem. 274:25995-26002. [DOI] [PubMed] [Google Scholar]
- 73.Zoog, S. J., J. J. Schiller, J. A. Wetter, N. Chejanovsky, and P. D. Friesen. 2002. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. EMBO J. 21:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]