Abstract
Coxsackievirus B3 (CVB3) is the most common causal agent of viral myocarditis, but existing drug therapies are of limited value. Application of small interfering RNA (siRNA) in knockdown of gene expression is an emerging technology in antiviral gene therapy. To investigate whether RNA interference (RNAi) can protect against CVB3 infection, we evaluated the effects of RNAi on viral replication in HeLa cells and murine cardiomyocytes by using five CVB3-specific siRNAs targeting distinct regions of the viral genome. The most effective one is siRNA-4, targeting the viral protease 2A, achieving a 92% inhibition of CVB3 replication. The specific RNAi effects could last at least 48 h, and cell viability assay revealed that 90% of siRNA-4-pretreated cells were still alive and lacked detectable viral protein expression 48 h postinfection. Moreover, administration of siRNAs after viral infection could also effectively inhibit viral replication, indicating its therapeutic potential. Further evaluation by combination found that no enhanced inhibitory effects were observed when siRNA-4 was cotransfected with each of the other four candidates. In mutational analysis of the mechanisms of siRNA action, we found that siRNA functions by targeting the positive strand of virus and requires a perfect sequence match in the central region of the target, but mismatches were more tolerated near the 3′ end than the 5′ end of the antisense strand. These findings reveal an effective target for CVB3 silencing and provide a new possibility for antiviral intervention.
Coxsackievirus B3 (CVB3) is a member of the genus Enterovirus, which is within the family Picornaviridae. Epidemiological studies indicated that nearly 50% of North American clinical myocarditis cases are attributable to picornaviral infection, with the CVB3 serogroup making up the most significant portion of such infections (23). The peak age group in which CVB-induced myocarditis occurs is young adults, primarily between 20 and 39 years of age (47). Although most CVB3-caused diseases are mild, some acute infections are severe and lethal. Clinically, CVB3 infections are associated with different forms of subacute, acute, and chronic myocarditis (38, 47). The infections may cause cardiac arrhythmias and acute heart failure, while in some cases the myocardial inflammation may persist chronically and progress to dilated cardiomyopathy (5, 26, 32, 39), requiring heart transplantation, or to death.
The CVB3 genome is a positive single-stranded RNA molecule. It is ∼7.4 kb in length and has a single open reading frame, which is flanked by 5′- and 3′-untranslated regions (UTR). The 5′UTR contains a highly structured internal ribosome entry site that directs viral translation initiation (30, 49). The 3′UTR contains three stem-loops followed by a poly(A) tail. The genomic RNA can serve as a template for viral RNA transcription to synthesize more copies of positive genomic RNA through a negative-strand intermediate. It can also be employed directly as an mRNA template for translation of a single polyprotein that is posttranslationally processed primarily by CVB3-endoded proteases 2A and 3C to produce individual structural and nonstructural proteins. The nonstructural proteins, particularly the RNA-dependent RNA polymerase 3D, are responsible for viral RNA replication, which takes place with rapid kinetics in the small membranous vesicles of cytoplasm. The entire replication cycle of CVB3 from entry of the host cell to release of progeny virus takes approximately 6 to 8 h. Although the life cycle of CVB3 appears clear, there is no specific drug available to inhibit this viral replication.
Small interfering RNAs (siRNAs) are short, double-stranded RNA (dsRNA) molecules that can target mRNA of a specific sequence for degradation via a cellular process known as RNA interference (RNAi) (1, 12, 16, 43). RNAi is an evolutionarily conserved phenomenon of posttranscriptional gene silencing that has been described for plants, invertebrates, and vertebrates (10, 42). In this process, dsRNA is cleaved into siRNA of 21 to 28 nucleotides (nt) by an RNAseIII-like enzyme known as Dicer, followed by incorporation of siRNA into an RNA-induced silencing complex (RISC) that recognizes and cleaves the target sequence (11). In mammals, however, dsRNAs longer than 30 nt can induce a nonspecific interferon response and, in turn, lead to the shutdown of a number of gene expressions. This limitation of application in mammals has been overcome by introduction of synthetic siRNA. They are short enough to bypass general dsRNA-induced nonspecific interferon response (28, 34) and thus provided a powerful reverse genetic approach to develop siRNA in gene functional study and antiviral drug development. To date, several laboratories have demonstrated that siRNA can be used as powerful antiviral agents for different viral infection, such as poliovirus (18), influenza A virus (17, 33), respiratory syncytial virus (3), hepatitis B (21, 50), C (27, 37, 41, 46, 51), and D virus (6), human immunodeficiency virus type 1 (HIV-1) (4, 7, 9, 24, 25, 29, 35, 36), and West Nile virus (33). Therefore, there has been considerable interest in the development of siRNA as a possible treatment for CVB3-induced heart diseases.
In this study, we examined the effects of RNAi on CVB3 replication using five siRNAs targeting different regions of CVB3 genomic RNA. We demonstrated that three of the five candidates exerted potent antiviral abilities in HeLa cells and cardiomyocytes. Among them, the most effective siRNA is the one targeting the viral protease 2A coding region. Furthermore, mutational analysis of the specific interactions between the siRNA and its target sequences revealed that the antisense strand of the siRNA plays a critical role in the specific targeting of siRNA on viral mRNA, and a single nucleotide mutation at the center or near the 5′ end of the antisense strand of siRNA can eliminate its antiviral activity.
MATERIALS AND METHODS
siRNAs.
All siRNA sequences were designed according to the manufacturer's recommendations and were subjected to a BLAST search of the National Center for Biotechnology Information's expressed sequence tag library to ensure that they targeted only the desired genes. Five CVB3-specific double-stranded siRNAs and two controls (21-mer) were synthesized by QIAGEN-Xeragon, and the siRNAs used for mutational analysis of antiviral activity were synthesized by Dharmacon. The final concentration of all siRNA was 40 μM in provided buffer. Sequences of all siRNAs are shown in Table 1.
TABLE 1.
Sequences and locations of siRNAs used to target the CVB3 RNA
| siRNA | Target sequence (5′-3′) | Location in CVB3 RNAa |
|---|---|---|
| 1 | GTAACACACACCGATCAAC | nt 115-133, 5′UTR |
| 2 | TACAGCAAAATGGGAGCTC | nt 733-751, AUG start codon region |
| 3 | GCATGTCAAAGCGTGGATA | nt 3171-3189, VP1 |
| 4 | GGTCCAAGAGAGTGAATAC | nt 3543-3561, 2A |
| 5 | GACTAAGGACCTAACAAAG | nt 6312-6330, 3D |
| Control | TTCTCCGAACGTGTCACGT | Irrelevant siRNA |
| Lamin-A/C | CTGGACTTCCAGAAGAACA | nt 608-630, coding regionb |
Virus, cell culture, infection, and transfection.
CVB3 was produced from a full-length cDNA clone (provided by Reinhard Kandolf, University of Tubingen, Germany) and amplified in HeLa cells (American Type Culture Collection) by transfection. Virus titer was routinely determined at the beginning of the experiment by plaque assay. HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 100 μg of penicillin-streptomycin/ml (Invitrogen). The HL-1 cell line, a cardiac muscle cell line established from an AT-1 mouse atrial cardiomyocyte tumor lineage, was a gift from William C. Claycomb (8). The cells were maintained in Claycomb medium (JRH Biosciences) supplemented with 10% fetal bovine serum (JRH Bioscience), 100 μg of penicillin-streptomycin/ml, 0.1 mM norepinephrine (Sigma), and 2 mM l-glutamine (Invitrogen).
The transfection of siRNAs was performed under optimal conditions. Briefly, 2 × 105 cells were grown at 37°C overnight. When cells reached 50 to 60% confluency, they were washed and overlaid with transfection complexes containing siRNAs and Oligofectamine (Invitrogen) overnight. Following transfection, cells were washed and infected with CVB3 at the indicated multiplicity of infection (MOI) for 1 h. The cells were then overlaid with complete medium and were incubated at 37°C in 5% CO2. At different time points postinfection, supernatants and cell lysates were collected and stored in a −80°C freezer. For therapeutic experiments, cells were infected with virus at the indicated MOI for 1 h and then were transfected with siRNAs.
Western blot.
Western blotting was performed by standard protocols as previously described (45). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then were transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk containing 0.1% Tween 20 for 1 h. The blots were probed with primary mouse antibody against CVB3 capsid protein VP1 (DAKO) or β-actin (Sigma) for 1 h, followed by incubation with horseradish peroxidase-conjugated secondary antibody. Finally, VP1 and β-actin expression were detected by ECL reagents (Amersham).
Viral plaque assay.
The virus titer was determined by plaque assay as described previously (52). Briefly, HeLa cells were seeded into 6-well plates (8 × 105 cells/well) and incubated at 37°C for 20 h. When cell confluency reached approximately 90%, cells were washed with phosphate-buffered saline and then overlaid with 500 μl of diluted supernatants. The cells were incubated at 37°C for 60 min, and the supernatants were removed. Finally, cells were overlaid with 2 ml of sterilized soft Bacto-agar—minimal essential medium. The cells were incubated at 37°C for 72 h, fixed with Carnoy's fixative for 30 min, and then stained with 1% crystal violet. The plaques were counted, and the amount of virus (PFU/milliliter) was calculated.
In situ hybridization.
Intracellular viral RNA was detected by in situ hybridization as previously described (31). Fixed cells were hybridized with digoxigenin-labeled CVB3 antisense riboprobes prepared by in vitro transcription. Hybridized positive signals were visualized using an alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) and the color substrate Vector Red (Vector Laboratories).
Cell viability assay.
Cell viability was measured by using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS) assay kit (Promega) according to the manufacturer's instructions. Cells were incubated with MTS solution for 2 h, and the absorbance was measured at 492 nm using an enzyme-linked immunosorbent assay (ELISA) reader. The absorbances of sham-infected cells were defined as the values of 100% survival, and the remaining data, including that for siRNA-treated, nontreated, and control cells, were converted to the ratio of the sham-infected sample. Morphological changes of cells following CVB3 infection were evaluated by phase-contrast microscopy.
Genotypic analysis of the siRNA-4 target region of CVB3.
Viral RNA was isolated from culture supernatant as described previously (52). Reverse transcription (RT) was conducted according to the manufacturer's instructions (Invitrogen) using 30 μl of RNA and 1 μl of 3 μM hexamer primer, followed by PCR to amplify CVB3 cDNA (the 2A region; nt 3423 to 3864). The PCR mixture contained 10 μl of RT products and 1 μl of 15 μM sense and antisense primers, and the reaction was run for 35 cycles with standard parameters. The PCR products representing the 2A region were analyzed with a 0.8% agarose gel and were purified with a QIAquick gel extraction kit (QIAGEN). DNA sequencing was preformed by the Biotechnology Laboratory, University of British Columbia.
RESULTS
Design of siRNA sequences specific for CVB3.
To test which region of the CVB3 genome is the most effective site for siRNA targeting, we selected sequences at different locations that may have important functions in viral replication. These regions include the 5′UTR, start codon, viral capsid protein VP1, viral protease 2A, and RNA-dependent RNA polymerase 3D. A total of five siRNAs targeting these regions were designed (Table 1) following published selection criteria (13, 14), except for siRNA-2, which targets the start codon region of CVB3. A BLAST search did not find complementation of these siRNAs with human genes. In addition, the sequences of lamin A/C siRNA (siRNA-L) (12) and irrelevant siRNA (siRNA-C) were synthesized by QIAGEN and used as controls.
Inhibition of CVB3 replication in HeLa cells.
The HeLa cell line is a widely used in vitro system for studying CVB3 replication. To verify that chemically synthetic siRNAs could efficiently silence gene expression in this cellular environment, siRNA-L was chosen as a validated model, as silencing of lamin A/C by siRNA-L has been reported previously (12). siRNA-L or siRNA-C was transfected into HeLa cells under optimal conditions. Two days posttransfection, cell lysates were collected and examined for lamin A/C protein levels by Western blotting. The lamin A/C level was barely detectable in siRNA-L-transfected samples, whereas the irrelevant siRNA-C had no effect on lamin A/C expression (data not shown). This result confirmed that transfection of synthetic siRNAs into HeLa cells is an effective approach for evaluation of the activity of siRNA in silencing gene expression in this cellular environment.
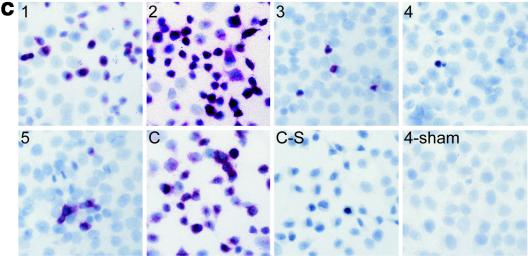
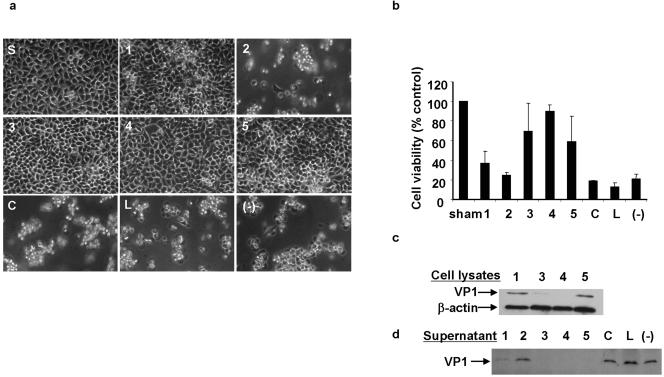
We next tested whether CVB3 replication could be inhibited by introducing various CVB3-specific siRNAs. HeLa cells were transfected with siRNAs at a final concentration of 300 nM overnight and were subsequently infected with CVB3 at an MOI of 10 for 8 h. CVB3-specific viral protein VP1 expression in cell lysates and infectious viral particles in the supernatants were analyzed by Western blotting and viral plaque assay, respectively. As shown in Fig. 1a, viral protein VP1 expression dramatically decreased in cells treated with siRNA-4, −3, or −5, but it did not decrease in cells treated with siRNA-1, siRNA-2, siRNA-C, or siRNA-L or in mock-transfected cells. Compared to the mock-transfected control, VP1 expression in siRNA-1- to -5-treated samples decreased 31, 18, 85, 92, and 65%, respectively, while siRNA-C- and -L-treated samples had no change. To evaluate the inhibition of infectious viral particle production, we performed plaque assays to detect the virus titer in the supernatants. Data demonstrated that virus titer of all CVB3-specific siRNAs except siRNA-2 decreased around 2 log10 compared to that of supernatant treated with control siRNAs or left untreated (Fig. 1b). The effects of RNAi on viral replication were further assessed by in situ hybridization. Consistent with the above results, the cells treated with CVB3-specific siRNA showed smaller amounts of positive cells and weaker signal intensity than the controls (Fig. 1c), indicating that siRNAs specifically inhibited viral replication in HeLa cells and that their antiviral activities can be ranked, in order, as siRNA-4 > siRNA-3 > siRNA-5 > siRNA-1 > siRNA-2. Considering the clinical setting, siRNAs were further used to treat wild-type CVB3. As expected, the result obtained was the same as that obtained from cDNA-derived virus (data not shown).
FIG. 1.
CVB3-specific siRNAs inhibit CVB3 replication in HeLa cells. (a) Western blot analysis of CVB3 capsid protein VP1. HeLa cells were transfected with siRNAs at a final concentration of 300 nM using Oligofectamine and then were infected with CVB3 at an MOI of 10. At 8 hpi, cell lysates were collected for VP1 detection by Western blot and supernatants were used for detecting infectious viral particles by plaque assay. The VP1 expression levels were quantified by densitometry and were normalized to the level of β-actin, which served as a loading control. The ratios of VP1 to β-actin were calculated and are expressed in the graph. (−), mock transfection. (b) Plaque assays of infectious viral particles. The assay was conducted on HeLa cell monolayers as described in Materials and Methods. Data are presented as log10 values of virus titer. (c) In situ hybridization of CVB3 RNA. After treatments and infections in chamber slides, CVB3 positive-strand RNAs were detected by in situ hybridization using antisense riboprobes (red). Cell nuclei were counterstained with hematoxylin (blue). Images 1 to 5 and C (control) represent different siRNAs treatments. Two negative controls were used: cells transfected with siRNA-C and infected with CVB3 were detected by sense probes (C-S), and cells transfected with siRNA-4 but sham infected with DMEM were detected with antisense probes (4-sham). Magnification, ×200. Data shown are from one of two independent experiments.
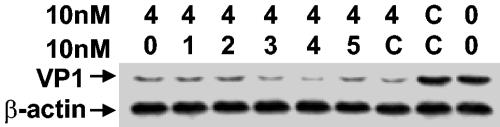
To further determine the potency of siRNAs, we performed experiments to study the effect of concentration of siRNA on its antiviral activity. We selected siRNA-4 as a candidate for this purpose. HeLa cells were transfected with a graded amount of siRNA-4 and were followed by CVB3 infection. Data presented in Fig. 2a and b demonstrated that as the amount of siRNA-4 increased, virus titer in the supernatants and VP1 protein in the cells correspondently decreased, even at concentrations as low as 10 nM. Compared to the levels of three controls, an approximate 2 log10 decrease in virus particle production was detected in these cultures treated with different concentrations of siRNA-4. These results indicate that siRNA-4 is a highly efficient antiviral agent and that it exerts a potent anti-CVB3 ability in a dose-dependent manner. To clarify the time course of RNAi, cells were pretreated with siRNA-4 or siRNA-C, followed by virus infection. At different times points postinfection, cell lysates and the supernatants were collected for detecting VP1 expression levels and virus titers. We hardly detected the difference between siRNA-4 and -C in the supernatants at 2 and 4 h postinfection (hpi). This may be because the first virus replication cycle had not finished yet, as a single replication cycle of CVB3 requires approximately 6 h (Fig. 2c). However, at 6 and 8 hpi, fewer virus particles and lower VP1 levels were observed in siRNA-4-treated samples than in the control-treated cultures (Fig. 2c and d).
FIG. 2.
siRNA-4 interferes with CVB3 replication in HeLa cells. (a and b) Dose-dependent inhibition of CVB3 production by siRNA-4. HeLa cells were transfected with siRNA-4 at a final concentration as indicated and followed by infection with a CVB3 MOI of 10 for 8 h. Supernatants were used for viral plaque assay (a), and cell lysates were collected for viral VP1 detection by Western blotting (b). β-Actin served as the loading control. (c and d) Time course of inhibition of CVB3 replication by siRNA-4. Cells were transfected with siRNA-4 at a final concentration of 300 nM and followed by CVB3 infection at an MOI of 10. Supernatants and cell lysates were collected at the indicated time points for plaque assay (c) and Western blotting (d), respectively. Data shown are representatives of two independent experiments.
Considering clinical application of siRNAs in viral myocarditis, siRNAs must be able to effectively inhibit an ongoing virus infection. To further evaluate its therapeutic potential, HeLa cells were infected with CVB3 at an MOI of 0.01 for 1 h and then were transfected with siRNA-4 or siRNA-C. Viral protein VP1 expression and virus titer were analyzed at 40 hpi As shown in Fig. 3a and b, virus titer as well as viral protein VP1 expression was significantly reduced by siRNA-4 treatment. Thus, administration of siRNAs after viral infection can also inhibit viral replication effectively.
FIG. 3.
siRNA-4 inhibits ongoing CVB3 replication in HeLa cells. Cells were infected with CVB3 at an MOI of 0.01 for 1 h and then were transfected with siRNA-4 at a final concentration of 300 nM. Forty hours after infection, supernatants and cell lysates were collected for detection of virus titer by plaque assay (a) and VP1 by Western blotting (b), respectively. Data shown are representative of two independent experiments.
We next evaluated the long-term effects of various CVB3-specific siRNAs on CVB3 replication. After transfection of the cells with each of the five siRNAs or controls, cytopathic effects were evaluated by phase-contrast microscopy. Figure 4a shows that mock-transfected cells and control cells were more susceptible to CVB3 infection. Approximately 80% of cells were killed by virus, whereas cells treated with CVB3-specific siRNAs except siRNA-2 were more resistant to infection and the majority of the cells were still alive. These observations were further validated and quantified by MTS assay, as shown in Fig. 4b. Notably, in the siRNA-4-treated cells, 90% of cells were alive compared to levels of sham-infected controls. The viral capsid protein VP1 was also detected by using cell lysates and supernatants. Figure 4c and d demonstrate that VP1 expression was undetectable in siRNA-3- or siRNA-4-treated samples. The anti-CVB3 abilities of different siRNAs in long-term (48 h) experiments were similar to those in the short-term (8 h) treatments (Fig. 2), which are ranked according to their potentials, i.e., siRNA-4 is the strongest one, followed by siRNA-3, -5, -1, and -2.
FIG. 4.
siRNAs protect cells against CVB3-induced cytopathic effects. (a) Morphological changes of HeLa cells following infection. Cells were transfected with each siRNA at a final concentration of 300 nM and then infected with CVB3 at an MOI of 0.01. Cell morphology was observed under a phase-contrast microscope at 48 hpi (magnification, ×100). S, sham infected; (−), mock transfected. (b) MTS cell viability assay. The assay was performed as described in Materials and Methods. Cell viability of each sample was expressed relative to that of the sham-infected control, which was defined as 100% survival. Values shown here are means ± standard deviations of three independent experiments. P < 0.005. (c and d) Western blot analysis of CVB3 VP1 in the cell lysates (c) and supernatants (d). Note that cells transfected with siRNA-2 or control, as well as mock-transfected cells, were dead after CVB3 infection for 48 h. Thus, no intact cells were remaining 48 hpi for preparing cell lysates used for Western blot analysis (c). Data shown are representatives of three independent experiments.
Inhibition of CVB3 replication in cardiomyocytes.
Because CVB3 is a cardiotropic virus and commonly induces viral myocarditis, we further evaluated these siRNAs in the HL-1 mouse cardiomyocyte cell line. Using optimal transfection conditions, HL-1 cells were first transfected with siRNAs and then infected with CVB3 at an MOI of 10. Forty-eight hours postinfection, the antiviral activities of siRNAs were evaluated by Western blotting and plaque assay. The inhibition pattern of the siRNAs in HL-1 cells (Fig. 5a and b) was similar to that obtained with HeLa cells, demonstrating that siRNA-4 exhibited the greatest potency in inhibiting CVB3 replication, followed by the other three siRNAs. However, no significant reduction of virus titer was observed in siRNA-2-treated cardiomyocytes compared to that of the controls.
FIG. 5.
CVB3-specific siRNAs inhibit CVB3 replication in HL-1 cells. Cells were transfected with each siRNA at a final concentration of 300 nM by the Oligofectamine method overnight and then were infected with CVB3 at an MOI of 10. At 48 hpi, supernatants were used for detecting virus titer by plaque assay (a), and cell lysates were collected for VP1 detection by Western blot (b). β-Actin was used as the loading control. Data shown are representatives of two independent experiments.
Effects of mismatches on antiviral activity of siRNA.
Because CVB3 RNA replication occurs through a dsRNA intermediate, the issue arises of which strand of the viral RNA is the site of sequence complementation with siRNA and which strand of the siRNA guides its targeting to the viral genome. To address this issue, we performed site-directed mutagenesis at the center of the sense and/or antisense strands of siRNA-4 (Fig. 6a) and then evaluated its antiviral activity in HeLa cells. Figure 6b and c demonstrated that one nucleotide mutation in the middle of the antisense strand (siRNA-4mAS) or of both strands of siRNA (siRNA-4mSAS) could eliminate RNAi completely as they lost their specific antiviral activity, whereas the corresponding mutation on the sense strand (siRNA-4mS) did not interfere with the RNAi. This suggests that the positive strand of viral RNA is the target of siRNA, and the central sequence match between the antisense strand of siRNA and the viral positive strand is critical for RNAi. To further test whether mismatches at the ends are better tolerated than mismatches at the center, siRNAs with a 1-nt mutation near either the 5′ end (siRNA-4-5′mAS) or the 3′ end (siRNA-4-3′mAS) of the antisense strand of siRNA-4 was applied to inhibit CVB3 replication (Fig. 6a). Compared to siRNA-4, siRNA-4-5′mAS exhibited a small degree of silencing, while siRNA-4-3′mAS still maintained similar inhibitory ability, implying that the 5′ sequence of antisense siRNA is more critical for target RNA recognition than the 3′-end sequence (Fig. 6d).
FIG. 6.
Effects of mismatches on antiviral activity of siRNA. (a) Sequences of the mutated siRNA-4. The targeting sequence for siRNA-4 is listed at the top, with the center base boxed. The sequences of wild-type and mutated siRNAs are shown, with mismatched nucleotides underlined. (b and c) siRNA-4mAS with one point mutation in the center of the antisense strand failed to inhibit virus replication, which was detected by viral plaque assay (b) and Western blotting (c). (d) siRNA-4 with one nucleotide mismatch near the 5′ end of the antisense strand but not with one near the 3′ end partially reduced its antiviral activity. HeLa cells were transfected with wild-type and mutated siRNA-4 at a final concentration of 300 nM, followed by CVB3 infection at an MOI of 10 for 8 h. Virus titer in the supernatants and VP1 protein expression in the cell lysates were analyzed by plaque assay (b) and Western blot (c and d). β-Actin served as the loading control. Data shown are representatives of two independent experiments.
No enhanced inhibitory effects on CVB3 replication by cotransfection of two siRNAs.
The enhanced antiviral effect of multiple siRNAs has been reported in HIV-1 infection (25). To test whether cotransfection of cells with a combination of two specific siRNAs targeting different regions of CVB3 RNA could increase the antiviral effect in our system, two siRNAs were cotransfected into the HeLa cells and followed by evaluation of the antiviral effects. Because we found that 10 and 20 nM concentrations of siRNA-4 were within the linear correlation between the dose and inhibitory effect, we selected these two concentrations for evaluation by using combinations containing siRNA-4 and each of the four remaining CVB3-specific siRNAs. The viral replication was measured by detection of VP1 protein production. As shown in Fig. 7, siRNA-4 at a final concentration of 20 nM exerted a higher inhibitory capacity than any other combination of 10 nM siRNA-4 with 10 nM siRNA-1, -2, -3, -5, or -C, suggesting that there is no additive or synergistic effect on anti-CVB3 activity between siRNA-4 and any other individual siRNA tested.
FIG. 7.
Effect of combined siRNAs on antiviral activity. HeLa cells were cotransfected with siRNA-4 and each of the other four siRNAs at a common final concentration as indicated, followed by CVB3 infection at an MOI of 10 for 8 h. Cell lysates and supernatants in cultures were used for detecting viral protein expression by Western blot. β-Actin served as the loading control. Note that no enhanced inhibitory effect on CVB3 infection was observed by cotransfection of each siRNA with siRNA-4. Data shown are representatives of two independent experiments.
No escape mutants generated upon siRNA treatment.
Emergence of escape viral mutants has been reported in previous studies following siRNA or short hairpin RNA treatment (4, 18). To determine whether mutated CVB3 would occur to evade siRNA treatment, virus was passaged on newly siRNA-4- or siRNA-C-transfected cells for four generations. Viral RNA was extracted from culture supernatant, and the siRNA-4 target region was PCR amplified and sequenced. The data indicate that the sequence of the 2A region, even up to the fourth generation, was identical to the original viral cDNA (data not shown).
DISCUSSION
Initially, when we selected the targeting sequences within the CVB3 RNA, the 5′UTR and the initiation codon regions were chosen for potential weaker target controls because the UTR-binding proteins and/or translation initiation complexes may interfere with binding of siRNA and the RISC in these regions (http://www.rockefeller.edu/labheads/tuschl/sirna.html). As expected, cells pretreated with siRNA-1 and -2 targeting the 5′UTR and initiation codon region demonstrated poorer antiviral capability than those pretreated with siRNA-3, -4, and -5 targeting the coding regions. However, several studies have demonstrated excellent protection against various virus infections by specific siRNAs targeting the 5′UTR (24, 51). These studies suggest that whether the binding of protein factors at certain sites of the 5′UTR has a negative or positive effect on siRNA function may depend on the local sequence characteristics. For certain regions, protein complex binding may change the higher ordered structure of the target sequence and, therefore, facilitate the access of the siRNA to the target sequence. On the other hand, if the siRNA and the protein factor compete for the same target, which is the regulatory sequence of gene expression, the binding of the protein factors may inhibit the RNAi activity of siRNA.
Among the siRNAs that we tested, siRNA-4, which is directed against the viral protease 2A region, was the most effective one, followed by siRNA-3, -5, -1, and -2. These results may be due to different positional accessibility caused by steric hindrance by a secondary or tertiary structure and/or protein binding. In this regard, there are inconsistent reports. Several studies focusing on the relationship between secondary structure and siRNA effects showed that the secondary structure at least in part influences the efficiency of siRNAs (22, 44, 51). Conversely, it has been reported that the secondary structure of the target mRNA does not appear to have a strong effect on gene silencing (22, 48). Whether secondary structure plays a role in siRNA machinery binding is still debated. However, we believe that binding of cellular proteins on the target sites or the secondary structure of the mRNA may affect, at least in part, the efficacies of siRNAs in the cells. Although we are not clear on the higher ordered structure of the siRNA-4 targeting sequence in the cellular environment, we believe that it possesses a more accessible conformation for siRNA. This may also be related to the thermodynamic instability of the dsRNA intermediate at this specific locus during CVB3 transcription, which is evidenced by this segment lacking any continuous two repeating bases at the middle region (Table 1), or this region may be favorable for binding of RISC required for siRNA function. In addition to the secondary structure and protein binding, the inefficacy of siRNA-2 may also be due to the sequence containing three continuous cytosines, which may hyperstack and therefore form agglomerates that potentially interfere in the silencing mechanism. Similarly, the siRNA containing five continuous guanosines targeted on the 5′UTR of hepatitis C virus did not show inhibitory ability on virus replication (51). Therefore, avoiding more than three Gs or Cs in the siRNA may exclude this problem. Interestingly, the sequence covering the initiation codon region has been reported to be an effective target for gene knockdown by an antisense deoxynucleotide (AS-DON) agent (45). Our previous study of antiviral activity of AS-DONs also confirmed this in anti-CVB3 replication (45, 52). This inconsistent data regarding siRNA targeting the same region may be due to the distinct mechanism of action for AS-DON. A commonly exploited antisense mechanism is RNase H-dependent degradation of the targeted RNA through recognition of a DNA-RNA heteroduplex (28), while siRNAs bind to targeted RNA by Watson-Crick base pairing and induce site-specific cleavage of the RNAs by a specific unknown RNase. This suggests that the best target sequences for AS-DON may not be the best candidate sites for siRNAs. Due to the different targeting sites used for siRNA and AS-DON, it is hard to compare their inhibitory effects on CVB3 replication. Overall, under optimal conditions it seems that CVB3-specific siRNAs are more effective than AS-ODNs in terms of potency, efficacy, and duration, which is consistent with other studies (2, 19, 48).
Sequence specificity of siRNA is very stringent, as single base pair mismatches between the siRNA and its target sequence dramatically reduce the silencing capability (3, 12, 14). However, there are different reports on this issue with different experimental systems. A detailed siRNA functional anatomy analysis revealed that RNAi required a perfect match between cellular mRNA and the antisense strand of siRNA, but several mutations in the sense strand of siRNAs did not eliminate the gene silencing (14, 20). On the other hand, in another report an siRNA with two nucleotide mismatches in the central region still had partial inhibitory activity (51). The experiments reported here used a positive single-stranded RNA virus which can produce a dsRNA intermediate during replication. This raises the possibility that siRNA may target positive, negative, or both strands of virus RNA. To clarify this question, we perform evaluations using a series of siRNA-4 mutants containing point mutations within the sense and/or antisense strands at different locations. The data suggest that only one point mutation in the middle of the antisense strand could eliminate the anti-CVB3 activity, whereas the corresponding mutation on the sense strand did not interfere with the viral replication, suggesting that the negative-strand RNAs produced during viral replication are not the direct target of siRNA. This result might be explained by the fact that the replicating negative strands of virus only exist as a double-stranded form in the vesicles (15), thus, they are less likely to be accessible to siRNAs. Conversely, the positive strand is the recognition site for RNAi, which forms complementary base pairs with the antisense strand of the siRNA. This conclusion is not only drawn from our study using positive single-stranded RNA virus but also has been reported recently for a negative single-stranded influenza virus in which the siRNA targets the mRNA of virus during replication (17). For the point mutation closest to the 5′ or 3′ end of the antisense siRNA, it is likely that the mismatch near the 5′ end has more negative effects on gene silencing than that near the 3′ end, which is consistent with a previous report (40). However, the molecular mechanism of this phenomenon needs to be further studied.
Cotransfection of cells with two or more siRNAs targeting different sites on HIV-1 coreceptor CXCR4 mRNA has been reported to result in enhanced gene silencing compared to that of each single siRNA (25). This could be explained by specific binding of certain siRNAs that may change the secondary structure of RNA and result in more accessible sites for other siRNA molecules. However, as with a previous report (22), we did not observe enhancement effects when using any combinations of two agents, including siRNA-4, in our system. These particular siRNAs probably could not affect the secondary structure of the targets or open more space to other siRNAs, or the amount of siRNA-associated proteins was limited for silencing rather than target accessibility. In general, the reasons for the discrepancy between the studies may be due to differences in mRNA targets and the evaluation methods. For antiviral evaluation, although the underlying mechanism of the enhanced gene silencing with multiple specific siRNAs is not clear, cotransfection with multiple siRNAs may benefit long-term treatment, as mutated virus variants may be produced following infection to escape from protection by siRNA (3, 4, 18).
To investigate whether escaping CVB3 mutants were generated following siRNA-4 treatment, a series of passages of CVB3 were challenged with fresh siRNA-4. However, we did not detect any mutants in this study. The discrepancy between previous reports (4, 18) and our result could be due to the fact that (i) exposure time to siRNA was not long enough compared to that of a previous HIV study, which showed emergence of mutation at the target site at 25 days posttreatment (4); and (ii) a different targeting region was used, as the siRNA-4 targeting sequence is a critical site for CVB3, because mutations in this area would markedly impair the fitness of the virus. Therefore, highly conserved regions should be used as targets for siRNA design to limit the occurrence of escape mutants.
In summary, this in vitro study is the first step to demonstrate that siRNA technology is a very promising approach to antiviral gene therapy. The very strong anti-CVB3 activity of siRNA-4 has indicated attractive new directions for further investigation of the underlying mechanism and the development of siRNA-4 as a prophylaxis and therapy for CVB3 infection.
Acknowledgments
We thank Reinhard Kandolf, University of Tubingen, Germany, and William C. Claycomb (Louisiana State University) for generously providing us with CVB3 cDNA and HL-1 cells, respectively. We also thank Bruce McManus for his critical discussion on experimental design and data analysis. Special thanks go to Agripina Suarez for her help with in situ hybridization and to Brian Wong, Elizabeth Walker, Zongshu Luo, and Jingchun Zhang for their technical assistance and manuscript preparation.
This work was supported by grants from the Canadian Institutes of Health Research (MOP-14068) and the Heart and Stroke Foundation of British Columbia and Yukon (20R20002).
REFERENCES
- 1.Baulcombe, D. 2002. RNA silencing. Curr. Biol. 12:R82—R84. [DOI] [PubMed] [Google Scholar]
- 2.Bertrand, J. R., M. Pottier, A. Vekris, P. Opolon, A. Maksimenko, and C. Malvy. 2002. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun. 296:1000-1004. [DOI] [PubMed] [Google Scholar]
- 3.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boden, D., O. Pusch, F. Lee, L. Tucker, and B. Ramratnam. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77:11531-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cetta, F., and V. V. Michels. 1995. The autoimmune basis of dilated cardiomyopathy. Ann. Med. 27:169-173. [DOI] [PubMed] [Google Scholar]
- 6.Chang, J., and J. M. Taylor. 2003. Susceptibility of human hepatitis delta virus RNAs to small interfering RNA action. J. Virol. 77:9728-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, Y. L., H. Cao, J. M. Jacque, M. Stevenson, and T. M. Rana. 2004. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1). J. Virol. 78:2517-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claycomb, W. C., N. A. Lanson, Jr., B. S. Stallworth, D. B. Egeland, J. B. Delcarpio, A. Bahinski, and N. J. Izzo, Jr. 1998. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 95:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 2002. RNA interference: antiviral defense and genetic tool. Nat. Immunol. 3:597-599. [DOI] [PubMed] [Google Scholar]
- 11.Dorsett, Y., and T. Tuschl. 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3:318-329. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, B. N., D. M. Knipe, P. M. Howley, and D. E. Griffin. 2001. Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 17.Ge, Q., M. T. McManus, T. Nguyen, C. H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. USA 100:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 19.Grunweller, A., E. Wyszko, B. Bieber, R. Jahnel, V. A. Erdmann, and J. Kurreck. 2003. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 31:3185-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamada, M., T. Ohtsuka, R. Kawaida, M. Koizumi, K. Morita, H. Furukawa, T. Imanishi, M. Miyagishi, and K. Taira. 2002. Effects on RNA interference in gene expression (RNAi) in cultured mammalian cells of mismatches and the introduction of chemical modifications at the 3′-ends of siRNAs. Antisense Nucleic Acid Drug Dev. 12:301-309. [DOI] [PubMed] [Google Scholar]
- 21.Hamasaki, K., K. Nakao, K. Matsumoto, T. Ichikawa, H. Ishikawa, and K. Eguchi. 2003. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 543:51-54. [DOI] [PubMed] [Google Scholar]
- 22.Holen, T., M. Amarzguioui, M. T. Wiiger, E. Babaie, and H. Prydz. 2002. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 30:1757-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, S. A., C. J. Gauntt, and P. Sakkinen. 1998. Enteroviruses and myocarditis: viral pathogenesis through replication, cytokine induction, and immunopathogenicity. Adv. Virus Res. 51:35-80. [DOI] [PubMed] [Google Scholar]
- 24.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji, J., M. Wernli, T. Klimkait, and P. Erb. 2003. Enhanced gene silencing by the application of multiple specific small interfering RNAs. FEBS Lett. 552:247-252. [DOI] [PubMed] [Google Scholar]
- 26.Kandolf, R. 1993. Molecular biology of viral heart disease. Herz 18:238-244. [PubMed] [Google Scholar]
- 27.Kronke, J., R. Kittler, F. Buchholz, M. P. Windisch, T. Pietschmann, R. Bartenschlager, and M. Frese. 2004. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, M. T., G. A. Coburn, M. O. McClure, and B. R. Cullen. 2003. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol. 77:11964-11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Z., C. M. Carthy, P. Cheung, L. Bohunek, J. E. Wilson, B. M. McManus, and D. Yang. 1999. Structural and functional analysis of the 5′ untranslated region of coxsackievirus B3 RNA: in vivo translational and infectivity studies of full-length mutants. Virology 265:206-217. [DOI] [PubMed] [Google Scholar]
- 31.Luo, H., J. Zhang, C. Cheung, A. Suarez, B. M. McManus, and D. Yang. 2003. Proteasome inhibition reduces coxsackievirus B3 replication in murine cardiomyocytes. Am. J. Pathol. 163:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martino, T. A., P. Liu, and M. J. Sole. 1994. Viral infection and the pathogenesis of dilated cardiomyopathy. Circ. Res. 74:182-188. [DOI] [PubMed] [Google Scholar]
- 33.McCown, M., M. S. Diamond, and A. Pekosz. 2003. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses. Virology 313:514-524. [DOI] [PubMed] [Google Scholar]
- 34.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 35.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 36.Park, W. S., M. Hayafune, N. Miyano-Kurosaki, and H. Takaku. 2003. Specific HIV-1 env gene silencing by small interfering RNAs in human peripheral blood mononuclear cells. Gene Ther. 10:2046-2050. [DOI] [PubMed] [Google Scholar]
- 37.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyes, M. P., and A. M. Lerner. 1985. Coxsackievirus myocarditis-with special reference to acute and chronic effects. Prog. Cardiovasc. Dis. 27:373-394. [DOI] [PubMed] [Google Scholar]
- 39.Rose, N. R., A. Herskowitz, and D. A. Neumann. 1993. Autoimmunity in myocarditis: models and mechanisms. Clin. Immunol. Immunopathol. 68:95-99. [DOI] [PubMed] [Google Scholar]
- 40.Saxena, S., Z. O. Jonsson, and A. Dutta. 2003. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 278:44312-44319. [DOI] [PubMed] [Google Scholar]
- 41.Seo, M. Y., S. Abrignani, M. Houghton, and J. H. Han. 2003. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J. Virol. 77:810-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants-defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 43.Vaucheret, H., C. Beclin, and M. Fagard. 2001. Post-transcriptional gene silencing in plants. J. Cell Sci. 114:3083-3091. [DOI] [PubMed] [Google Scholar]
- 44.Vickers, T. A., S. Koo, C. F. Bennett, S. T. Crooke, N. M. Dean, and B. F. Baker. 2003. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 278:7108-7118. [DOI] [PubMed] [Google Scholar]
- 45.Wang, A., P. K. Cheung, H. Zhang, C. M. Carthy, L. Bohunek, J. E. Wilson, B. M. McManus, and D. Yang. 2001. Specific inhibition of coxsackievirus B3 translation and replication by phosphorothioate antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 45:1043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, J. A., S. Jayasena, A. Khvorova, S. Sabatinos, I. G. Rodrigue-Gervais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA 100:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodruff, J. F. 1980. Viral myocarditis. A review. Am. J. Pathol. 101:425-484. [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, Y., H. Y. Zhang, D. Thormeyer, O. Larsson, Q. Du, J. Elmen, C. Wahlestedt, and Z. Liang. 2003. Effective small interfering RNAs and phosphorothioate antisense DNAs have different preferences for target sites in the luciferase mRNAs. Biochem. Biophys. Res. Commun. 306:712-717. [DOI] [PubMed] [Google Scholar]
- 49.Yang, D., J. E. Wilson, D. R. Anderson, L. Bohunek, C. Cordeiro, R. Kandolf, and B. M. McManus. 1997. In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5′ untranslated region of coxsackievirus B3: potential targets for antiviral action of antisense oligomers. Virology 228:63-73. [DOI] [PubMed] [Google Scholar]
- 50.Ying, C., E. De Clercq, and J. Neyts. 2003. Selective inhibition of hepatitis B virus replication by RNA interference. Biochem. Biophys. Res. Commun. 309:482-484. [DOI] [PubMed] [Google Scholar]
- 51.Yokota, T., N. Sakamoto, N. Enomoto, Y. Tanabe, M. Miyagishi, S. Maekawa, L. Yi, M. Kurosaki, K. Taira, M. Watanabe, and H. Mizusawa. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 4:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan, J., P. K. Cheung, H. Zhang, D. Chau, B. Yanagawa, C. Cheung, H. Luo, Y. Wang, A. Suarez, B. M. McManus, and D. Yang. 2004. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab. Investig. 84:703-714. [DOI] [PubMed] [Google Scholar]