Abstract
Background
Antimicrobial resistant Salmonella strains are a direct threat to human health when this resistance interferes with treatment and an indirect threat when resistance can be transferred to other human pathogens. The objective of the present study was to characterize antimicrobial resistant non-typhoidal Salmonella (NTS) isolates recovered from poultry industries, including a description of genetic diversity and virulence profiles.
Results
In total of 93 Salmonella isolates shown antimicrobial resistance to one or more drugs, all isolates exhibited common resistance to streptomycin, nalidixic acid and cephalothin but no ciprofloxacin resistance. Among 26 virulence gene profiling, 12 virulence genes, invA, orgA, prgH, sopB, tolC, sipB, gatC, msgA, pagC, spiA, sifA, and sitC were found in all antimicrobial-resistant NTS isolates. In comparing the data from ERIC-PCR clusters, virulence profiles and resistance profiles, some Salmonella isolates grouped into the same cluster were found to exhibit similar virulence and resistance patterns.
Conclusions
Virulence profiling combined with ERIC-PCR offered a rapid approach to characterize antimicrobial-resistant NTS.
Keywords: Non-typhoidal Salmonella, Molecular characterization, Resistance
Background
Non-typhoidal Salmonella (NTS) are major zoonotic food-borne pathogens causing gastroenteritis worldwide. The global burden of NTS infection is estimated to be 93.8 million cases of gastroenteritis each year [30]. In Korea, total of 9,472 human cases of food and waterborne salmonellosis were detected between 1998 and 2007 [25], and NTS strains are one of the most common causes of food poisoning in humans [23]. In many countries including Korea, NTS infections are associated with the consumption of contaminated food products, especially poultry meats and eggs [22, 37].
Salmonella virulence genes are present on the bacterial chromosome, plasmids, and prophages, and Salmonella pathogenicity islands (SPIs) play important roles in adhesion, invasion, intracellular survival, systemic infection, fimbrial expression, antibiotic resistance, toxin production, and Mg2+ and iron uptake [8]. For example, genes such as invA, orgA, prgH, sipB and spaN in SPI-1 encode a type 3 secretion system 1 (T3SS-1) which allows Salmonella to invade phagocytic and non-phagocytic cells. Genes such as spiA in SPI-2 encode a type 3 secretion system 2 (T3SS-2), which allows the survival and replication of Salmonella in host cells [27]. Other chromosomal genes such as lpfA and pefA encode fimbriae-associated proteins that are important for adherence [11]. Moreover, plasmidal genes such as spvB contribute to colonization of deeper tissues, among other functions [2]. The virulence potential of Salmonella determines the differences in pathogenicity among Salmonella serotypes.
The increase in antibiotic-resistant food-borne pathogens is also a major public health problem. A high rate of antimicrobial-resistance in Salmonella strains has been reported in Korea [5, 24]. Antimicrobial resistance and virulence of Salmonella strains play an important role in systemic infections with these pathogens [14].
Pulsed-field gel electrophoresis (PFGE) is the gold standard subtyping method used to assess relatedness among Salmonella strains from different sources [28]; however it is time consuming and labor intensive [16]. DNA-based fingerprinting techniques such as enterobacterial repetitive intergenic consensus (ERIC), repetitive extragenic palindromic (REP), and BOX repeat-based (BOXAIR) PCR methods are relatively easy to perform, rapid, and sensitive in discriminating between closely related strains [12, 16]. In recent years, the ERIC-PCR fingerprinting method has been used to confirm epidemiological relationships between various isolates, and this method has shown high discriminatory power [1, 32]. The objective of the present study was to characterize antimicrobial-resistant NTS isolates recovered from poultry industries, including a description of genetic diversity and virulence profiles.
Methods
Salmonella isolates
Ninty-three Salmonella isolates showing antimicrobial resistance to one or more drugs were tested in this study. All isolates were recovered from chilled chicken carcasses (n = 25) and chillers (n = 23) in chicken slaughter houses; chilled carcasses (n = 15) and chillers (n = 8) in duck slaughter houses; egg belts (n = 2), feeders (n = 1), feces (n = 5), dust (n = 3) and egg shells (n = 8) from layer farms; and raw shell eggs (n = 2) and egg contents (n = 1) from retail markets between 2008 and 2014 (Table 1). Salmonella isolates were serotyped according to the Kauffmann-White scheme following slide agglutination testing with Salmonella-specific O and H antisera (Difco, Detroit, MI).
Table 1.
Distribution of 93 antimicrobial resistant Salmonella isolates derived from poultry industries in KoreaThe 17 chicken slaughter houses, 9 duck slaughter houses, 8 commercial layer farms and 3 retail markets were designated C1 ~ C17, D1 ~ D9, L1 ~ L8 and R1 ~ R3, respectively
| Salmonella serovars | No | Source | Place | Year | Resistance pattern (n) |
|---|---|---|---|---|---|
| S. Agona | 1 | Feces | L6 | 2014 | AMP-TE (1) |
| 1 | Egg belts | L2 | 2013 | CF-NA (1) | |
| S. Bareilly | 1 | Egg belts | L3 | 2013 | CF-NA-S (1) |
| 1 | Egg shell | L2 | 2013 | CF-NA (1) | |
| 5 | Egg shell | L3 | 2013 | S (5) | |
| 1 | Feeder | L2 | 2013 | S (1) | |
| 1 | Feces | L2 | 2013 | S (1) | |
| 1 | Feces | L3 | 2013 | S (1) | |
| S. Binza | 1 | Chilled duck carcasses | D1 | 2011 | AM-CF-S (1) |
| S. Braenderup | 1 | Egg shell | R1 | 2013 | AM (1) |
| 1 | Egg shell | R2 | 2013 | AM (1) | |
| 1 | Feces | L5 | 2013 | AM (1) | |
| 1 | Egg shell | L4 | 2013 | AM (1) | |
| 1 | Dust | L5 | 2013 | AM (1) | |
| S. Coquilhativille | 1 | Chilled chicken carcasses | C15 | 2011 | C-CF-S (1) |
| S. Enteritidis | 1 | Chicken chillers | C2 | 2008 | NA (1) |
| 1 | Chicken chillers | C3 | 2008 | NA-S-TE (1) | |
| 2 | Chicken chillers | C4 | 2008 | NA (2) | |
| 2 | Chicken chillers | C5 | 2008 | AM-NA-S (1), AM-C-NA-S-TE (1) | |
| 1 | Chilled chicken carcasses | C1 | 2008 | NA (1) | |
| 5 | Chilled chicken carcasses | C4 | 2008 | NA (5) | |
| 5 | Chilled chicken carcasses | C5 | 2008 | AM-NA-S (3), AM-CF-NA-S (2) | |
| 1 | Chicken chillers | C6 | 2011 | AM-C-CF-NA-S (1) | |
| 1 | Chicken chillers | C7 | 2011 | CF-NA (1) | |
| 1 | Chicken chillers | C8 | 2011 | CF-NA-S (1) | |
| 2 | Duck chillers | D1 | 2010 | TE (1), CF-S-TE (1) | |
| 1 | Duck chillers | D2 | 2011 | AM-C-CF-NA (1) | |
| S. Give | 1 | Chilled chicken carcasses | C14 | 2011 | AMP-TE (1) |
| S. Hadar | 2 | Duck chillers | D3 | 2008 | AM-CF-S-TE (1), AM-CF-KAN-S-TE (1) |
| 1 | Duck chillers | D9 | 2011 | CF-TE (1) | |
| 1 | Chilled duck carcasses | D1 | 2011 | CF-S-TE (1) | |
| 1 | Chilled duck carcasses | D3 | 2008 | KAN-S (1) | |
| 2 | Chilled duck carcasses | D4 | 2008 | S-TE (2) | |
| S. Hogton | 1 | Chilled duck carcasses | D4 | 2008 | KAN-S-TE (1) |
| S. Infantis | 1 | Chilled chicken carcasses | C8 | 2011 | CF-NA-S (1) |
| 1 | Egg shell | L1 | 2014 | NA (1) | |
| S. Kortrijk | 1 | Chicken chillers | C15 | 2011 | C (1) |
| S. Livingstone | 1 | Dust | L7 | 2014 | NA (1) |
| S. London | 1 | Chilled duck carcasses | D3 | 2008 | AM-CF-S-TE (1) |
| S. Malmoe | 3 | Chicken chillers | C9 | 2011 | S (2), AM-C-CAZ-CF-CTX-NA-TE (1) |
| S. Mbandaka | 2 | Chilled duck carcasses | D5 | 2010 | CF-S (1), S (1) |
| 1 | Feces | L3 | 2013 | CF (1) | |
| S. Montevideo | 1 | Chicken chillers | C1 | 2008 | NA (1) |
| 1 | Chicken chillers | C9 | 2008 | NA (1) | |
| 1 | Chicken chillers | C11 | 2011 | CF-NA (1) | |
| 1 | Chicken chillers | C12 | 2011 | CF-NA (1) | |
| 3 | Chilled chicken carcasses | C9 | 2008 | NA (3) | |
| 1 | Chilled chicken carcasses | C10 | 2011 | NA (1) | |
| S. Newbrunswick | 1 | Chilled duck carcasses | D1 | 2011 | S-TE (1) |
| S. Newport | 2 | Chicken chillers | C10 | 2011 | NA (2) |
| 2 | Chilled chicken carcasses | C10 | 2011 | AM-CF-TE (1), CF-NA (1) | |
| S. Ohio | 1 | Chilled duck carcasses | D4 | 2008 | S-TE (1) |
| S. Orion | 1 | Chilled duck carcasses | D8 | 2011 | CF-S (1) |
| S. Senftenberg | 1 | Chicken chillers | C9 | 2008 | NA (1) |
| 1 | Chicken chillers | C17 | 2011 | CF-NA-S (1) | |
| 1 | Chilled chicken carcasses | C9 | 2008 | NA (1) | |
| 1 | Chilled chicken carcasses | C13 | 2011 | NA (1) | |
| 1 | Chilled chicken carcasses | C16 | 2011 | AM (1) | |
| 1 | Dust | L8 | 2014 | GEN-NA-S (1) | |
| S. Takoradi | 2 | Chicken chillers | C9 | 2011 | AM-CF-SXT (1) |
| S. Thomson | 1 | Chilled chicken carcasses | C7 | 2011 | CF (1) |
| S. Trachau | 1 | Egg contents | R3 | 2013 | CF (1) |
| S. Typhimurium | 1 | Chilled chicken carcasses | C10 | 2011 | CTX (1) |
| 1 | Duck chillers | D1 | 2010 | CF-TE (1) | |
| 1 | Duck chillers | D3 | 2008 | S-TE (1) | |
| 1 | Chilled duck carcasses | D2 | 2011 | CF (1) | |
| 1 | Chilled duck carcasses | D6 | 2011 | S-TE (1) | |
| S. Wippra | 1 | Chilled duck carcasses | D6 | 2011 | CF-S-TE (1) |
Antimicrobial susceptibility tests
Antimicrobial susceptibility profiles of the isolates were determined by the disk diffusion method [6]. Twelve antimicrobial agents (Difco, United States) were tested at the following concentrations: gentamicin (GM, 10 μg), kanamycin (K, 30 μg), cephalothin (CF, 30 μg), cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), chloramphenicol (C, 30 μg), ciprofloxacin (CIP, 5 μg), tetracycline (TE, 30 μg), ampicillin (AM, 10 μg), streptomycin (S, 10 μg), trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg) and nalidixic acid (NA, 30 μg). An isolate was considered as multidrug-resistant (MDR) when exhibiting resistance to antimicrobials of at least three different classes [29]. Escherichia coli strain ATCC 25922 was used as a reference strain.
Analysis of virulence genes
The DNA for all analyses was extracted by the boiling method [7], and 5 μl of DNA template (approximate 60 ng) was used in each PCR reaction. Primers details are presented in Table 2. To establish the reproducibility of the experiments, PCR reactions were performed twice. The amplified PCR products were visualized by electrophoresis on 1.5% agarose gels stained with ethidium bromide (0.5 μg/ml).
Table 2.
Primers used in PCR for detection of virulence genes in antimicrobial resistant non-typhoidal Salmonella
| Primer | Broad function (gene function) | Sequence (5’ to 3’) | Referance |
|---|---|---|---|
| invA | Host recognition/invasion (type III secretion system appratus) | F-CTGGCGGTGGGTTTTGTTGTCTTCTCTATT | Skyberg et al., (2006) [34] |
| R-AGTTTCTCCCCCTCTTCATGCGTTACCC | |||
| orgA | Host recognition/invasion (type III secretion system appratus) | F-TTTTTGGCAATGCATCAGGGAACA | Skyberg et al., (2006) [34] |
| R-GGCGAAAGCGGGGACGGTATT | |||
| prgH | Host recognition/invasion (type III secretion system appratus) | F-GCCCGAGCAGCCTGAGAAGTTAGAAA | Skyberg et al., (2006) [34] |
| R-TGAAATGAGCGCCCCTTGAGCCAGTC | |||
| sopB | Host recognition/invasion (type III secretion system appratus) | F-CGGACCGGCCAGCAACAAAACAAGAAGAAG | Skyberg et al., (2006) [34] |
| R-TAGTGATGCCCGTTATGCGTGAGTGTATT | |||
| tolC | Host recognition/invasion (outer membrane channel protein) | F-TACCCAGGCGCAAAAAGAGGCTATC | Skyberg et al., (2006) [34] |
| R-CCGCGTTATCCAGGTTGTTGC | |||
| sopE | Host recognition/invasion (invasion-associated secreted protein) | F-CATAGCGCCTTTTCTTCAGG | Suez et al., (2013) [35] |
| R-ATGCCTGCTGATGTTGATTG | |||
| sseI | Host recognition/invasion (type III secretion system effector protein) | F-CGCCATCATCAGTAACCGCC | Suez et al., (2013) [35] |
| R-CTGCTGACCACATCCTCCC | |||
| ssek3 | Host recognition/invasion (type III secretion system effector protein) | F-TATCAATCTCAAATCATGG | Suez et al., (2013) [35] |
| R-CGCGTTTATATCATACGTTTGC | |||
| sspH1 | Host recognition/invasion (type III secretion system effector protein) | F-GGTCACAGGACACGTTCTACG | Suez et al., (2013) [35] |
| R-GCGCTTCTTCGTAATTTTCC | |||
| cdtB | Host recognition/invasion (toxin-like protein) | F-ACAACTGTCGCATCTCGCCCCGTCATT | Skyberg et al., (2006) [34] |
| R-CAATTTGCGTGGGTTCTGTAGGTGCGAGT | |||
| hlyE | Host recognition/invasion (hemolysis-inducing protein) | F-GCGTGATTGAAGGGAAATTG | Suez et al., (2013) [35] |
| R-CGAAAAGCGTCTTCTTACCG | |||
| lpfC | Host recognition/invasion (fimbrial protein) | F-GCCCCGCCTGAAGCCTGTGTTGC | Skyberg et al., (2006) [34] |
| R-AGGTCGCCGCTGTTTGAGGTTGGATA | |||
| pefA | Host recognition/invasion (fimbial protein) | F-TAAGCCACTGCGAAAGATGC | Suez et al., (2013) [35] |
| R-GCGTGAACTCCAAAAACCCG | |||
| tcfA | Host recognition/invasion (fimbrial protein) | F-TCGCTATGTTTGCATGTGGT | Suez et al., (2013) [35] |
| R-TTCAGGAACAGCCTCGAAGT | |||
| span | Entry into nonphagocytic cells (type III secretion system appratus) | F-AAAAGCCGTGGAATCCGTTAGTGAAGT | Skyberg et al., (2006) [34] |
| R-CAGCGCTGGGGATTACCGTTTTG | |||
| sipB | Entry into nonphagocytic cells (translocation machinery component) | F-GGACGCCGCCCGGGAAAAACTCTC | Skyberg et al., (2006) [34] |
| R-ACACTCCCGTCGCCGCCTTCACAA | |||
| spiA | Survival within macrophage (outer membrane secretory protein) | F-CCAGGGGTCGTTAGTGTATTGCGTGAGATG | Skyberg et al., (2006) [34] |
| R-CGCGTAACAAAGAACCCGTAGTGATGGATT | |||
| msgA | Survival within macrophage (macrophage survival protein) | F-GCCAGGCGCACGCGAAATCATCC | Skyberg et al., (2006) [34] |
| R-GCGACCAGCCACATATCAGCCTCTTCAAAC | |||
| pagC | Survival within macrophage (virulence membrane protein) | F-CGCCTTTTCCGTGGGGTATGC | Skyberg et al., (2006) [34] |
| R-GAAGCCGTTTATTTTTGTAGAGGAGATGTT | |||
| sodC | Survival within macrophage (periplasmic Cu/Zn superoxide dismutase) | F-ATGACACCACAGGCAAAACG | Suez et al., (2013) [35] |
| R-AGATGAACGATGCCCTGTCC | |||
| gatC | Growth within host (PTS galactitol transporter subunit IIC) | F-ATTGGTATCGGCTTCGTGGG | Suez et al., (2013) [35] |
| R-ATCCCCAGCCAGTATGAACC | |||
| spvB | Growth within host (ADP-ribosylating toxin) | F-CTATCAGCCCCGCACGGAGAGCAGTTTTTA | Skyberg et al., (2006) [34]. |
| R-GGAGGAGGCGGTGGCGGTGGCATCATA | |||
| sitC | Iron acquisition (permease) | F-CAGTATATGCTCAACGCGATGTGGGTCTCC | Skyberg et al., (2006) [34] |
| R-CGGGGCGAAAATAAAGGCTGTGATGAAC | |||
| iron | Iron acquisition (sidrophore) | F-ACTGGCACGGCTCGCTGTCGCTCTAT | Skyberg et al., (2006) [34] |
| R-CGCTTTACCGCCGTTCTGCCACTGC | |||
| sifA | Filamentous structure formation (secreted effector protein) | F-TTTGCCGAACGCGCCCCCACACG | Skyberg et al., (2006) [34] |
| R-GTTGCCTTTTCTTGCGCTTTCCACCCATCT | |||
| STM 2759 | Putative dipeptice/oligopetide/nikel ABC-type transport systems | F-ACCATTTTCACCTGGGCTCC | Suez et al., (2013) [35] |
| R-CGTTCAGGTTTTGTCGCTGG |
ERIC-PCR fingerprints analysis
Genotyping of isolates was performed by ERIC-PCR using a pair of primers (F: 5’-ATG TAA GCT CCT GGG GAT TCA C-3’; R: 5’-AAG TAA GTG ACT GGG GTG AGC G-3’) [36]. The PCR reaction was performed using a lyophilized PCR master mix according to the manufacturer’s instructions. (AccuPower PCR PreMix, Bioneer, Korea). A thermocycler (Bio-Rad, Singapore) was programmed as follows: initial denaturation at 95 °C for 7 min, followed by 30 cycles of denaturation at 90 °C for 30 s, annealing at 52 °C for 1 min and extension at 65 °C for 8 min, and a final extention step at 65 °C for 16 min [39]. A negative control consisting of the same reaction mixture without a DNA template was included in each reaction. ERIC-PCR reactions were repeated at least twice for each isolate to determine the reproducibility of banding patterns. Data were analyzed using the software package BioNumerics 7.5 (Applied Maths, Keistraat, Belgium). A similarity dendrogram was constructed by the UPGMA method with a 1% tolerance limit and 1% optimization, using the DICE similarity coefficient. Clusters were identified based on an 80% similarity cut-off [10]. The discrimination index for ERIC-PCR was calculated using Simpon’s diversity index [19].
Results
The results of ERIC-PCR, virulence gene profiling, and antimicrobial susceptibility test are summarized in Fig. 1. The 93 Salmonella isolates showed resistance to S (n = 46), NA (n = 42), CF (n = 34), AM (n = 24), TE (n = 22), C (n = 6), K (n = 3), CTX (n = 2), CAZ (n = 1), GM (n = 1), and SXT (n = 1). All isolates of Salmonella were susceptible to CIP. The 27 isolates (29%) showed multidrug resistance to more than three antibiotic classes.
Fig. 1.
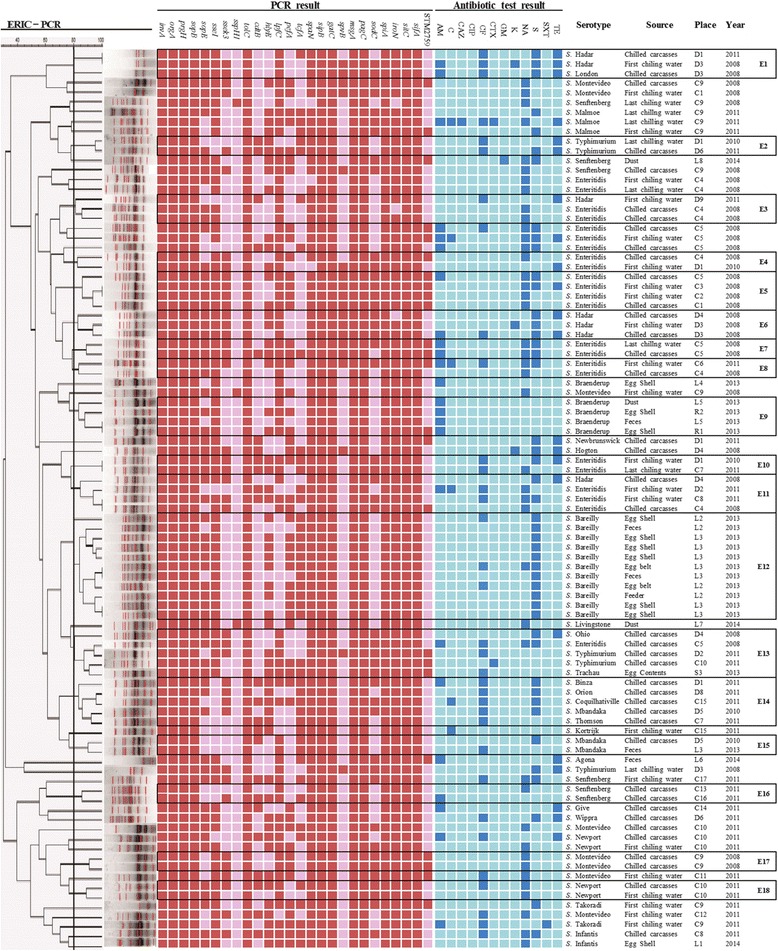
The ERIC-PCR analysis of the non-typhoidal Salmonella from Korean poultry industry is displayed using dendrograms generated by Bionumerics software. The vertical line shows the delineation level of 80%. The red color indicates the presence of the gene while the pink color indicates the absence of the gene. The dark blue color indicates resistance to the corresponding antibiotic while the light blue color indicates susceptibility. The 17 chicken slaughter houses, nine duck slaughter houses, eight commercial layer farms and three retail markets were designated C1 ~ C17, D1 ~ D9, L1 ~ L8, R1 ~ R3, respectively. AM ampicillin, C choloramphenicol, CAZ ceftazidime, CIP ciprofloxacin, CF cephalothin, CTX cefotaxime, GM gentamicin, K kanamycin, NA nalidixic acid, S streptomycin, STX trimethoprim/sulfamethoxazole, TE tetracycline
From the virulence gene profiling, 12 virulence genes, invA, orgA, prgH, sopB, tolC, sipB, gatC, msgA, pagC, spiA, sifA, and sitC were found in all isolates and almost all of the isolates were positive for spaN (97%) and iroN (97%). However, the other 12 virulence genes, sseI (83%), lpfC (76%), sopE (70%), hlyE (60%), pefA (55%), sodC (48%), tcfA (48%), ssek3 (43%), STM2759 (26%), cdtB (26%), spvB (20%) and sspH1 (3%) were variably present in the isolates. Especially, sspH1 gene was found in only 3 isolates, S. Montevideo, S. Livingstone and S. Senftenberg.
The ERIC-PCR analysis showed that 60 Salmonella isolates grouped into 18 clusters (E1 to E18) at 80% genetic similarity, whereas the remaining 33 isolates remained unclustered. The discrimination index of ERIC-PCR typing in this study was 0.974. Except for five clusters (E1, E3, E11, E13 and E14), isolates within each of the other 13 clusters belonged to the same serotype. Irrespective of their sources and places of isolation, all S. Bareilly isolates grouped into only one cluster (E12), whereas isolates of other serotypes could be found in different clusters, despite isolation from the same source and location. For example, 18 of 23 S. Enteritidis isolates grouped into eight clusters (E3, E4, E5, E7, E8, E10, E11 and E13) and the remaining five isolates of this serotype were not grouped. Two of 8 S. Montevideo isolates clustered into E17, and the remaining six isolates were not found in any cluster. Some of these Salmonella isolates representing the same serotype, sources, and location clustered separately.
Discussion
Antimicrobial-resistant Salmonella strains are a direct threat to human health when this resistance interferes with treatment and an indirect threat when resistance can be transferred to other human pathogens [13]. In Korea, S. Enteritidis, S. Montevideo, S. Typhimurium is most common serotypes in poultry slaughter houses [4, 38], and the most frequently observed Salmonella serovars in the layer farms were S. Bareilly [20].
Cephems, quinolones and aminoglycosides have been widely used in the poultry industries in Korea [18, 24]. Therefore, Salmonella isolates tested in study showed consistent resistance to S, NA, and CF. Our finding showed that 29% among 93 antimicrobial resistance isolates were MDR. All MDR isolates recovered from chicken or duck slaughter houses. However, there was no MDR isolates recovered from layer farms or eggs of retail markets. Generally, Salmonella isolated from the broilers demonstrated greater MDR compared to those isolated from layer and eggs [21].
The ability of antimicrobial resistant NTS strains to cause invasive disease can be attributed to various virulence genes, and virulotyping rapidly allows the discrimination of isolates with diverse pathogenic potential [17]. In this study the genes-invA, orgA, prgH, sopB, tolC, sipB, gatC, msgA, pagC, spiA, sifA, and sitC, located in SPI-1, SPI-2, SPI-5, SPI-11, or others, were found in all antimicrobial-resistant NTS isolates. In addition, spaN, located in SPI-1, and iroN, located in an effector were highly conserved in the isolates. Similar findings were reported in a study of samples from both sick and healthy birds [34], clinical samples, and environmental samples from poultry houses [31], and free-living birds [26].
Recently some researchers have reported that typhoid-associated virulence genes (cdtB, tcfA, and hlyE) in NTS serotypes of human and poultry origin are increasing [11, 26, 34]. NTS strains containing cdtB, tcfA, and hlyE genes were found in this study, and, to our knowledge, this is the first report to detect the presence of cdtB in the S. Hogton, S. Give, S. Newbrunswick, S. Thomson, S. Kortrijk, S. Coquilhativille, and S. Binza serotypes.
In this study, lpfC, one of three fimbrial genes, was more prevalent than the others, pefA and tcfA, which is consistent with previous studies [8, 15]. However, no S. Bareilly isolates tested in this study harbored the lpfC gene; whereas tcfA was presented in all S. Bareilly isolates, and pefA was highly conserved. Gong et al. [15] showed that the presence or absence of specific fimbrial genes in certain Salmonella serovars might have diagnostic value, as fimbrial genotypes can be used to determine certain Salmonella serotypes. Our results of fimbrial gene profiles are consistent with this report.
Molecular typing of Salmonella is vital to determining potential sources of infection and implementing effective epidemiological surveillance and control [9]. In this study, 60 out of 93 antimicrobial-resistant NTS isolates grouped into 18 ERIC-PCR clusters, and 33 isolates remained unclustered. This variability might be due to a difference in the sources of samples or in serotypes. In this study, the discrimination index of ERIC-PCR was 0.974. Based on a recommendation by Hunter et al. [19], a D value > 0.9 is desirable for good differentiation; our ability to discriminate between isolates was high. Based on these criteria, ERIC typing is useful for Salmonella typing, and our report showed that ERIC-PCR differentiated Salmonella strains indistinguishable to levels of heterogeneity of various serotypes.
The distribution of profiles among serotypes demonstrated that different serotypes showed similar fingerprinting patterns. These results are consistent with findings of Ranjbar et al. [33] who found that every Salmonella isolate had a unique fingerprinting but the serotypes were not grouped together in major branches.
In this study, the correlations among ERIC-PCR clusters, virulence profiles and resistance profiles were analyzed. We found that virulence genes and resistance profiles correlated with ERIC-PCR subtypes. Some isolates showed the same or similar virulotype or resistance pattern, irrespective of serotypes. The simultaneous presence of a resident virulence plasmid and resistance gene in the same bacterial cell has been reported in Salmonella [3]. Therefore, assessing the prevalence of virulence genes as well as the antibiotic resistance status in Salmonella serotypes would be useful to better understanding Salmonella pathogenicity.
Conclusion
In conclusion, this study provides a molecular characterization of antimicrobial-resistant NTS from poultry industries in Korea. Virulence profiling combined with ERIC-PCR may offers a rapid approach to characterize antimicrobial-resistant NTS isolates. Therefore, determination of their definitive correlations will require future studies with isolates from various source of animal.
Acknowledgements
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Agriculture, Food and Rural Affairs Research Center Support Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (716002–7).
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
JEK analyzed the samples, performed the statistical analysis and wrote the manuscript. YJL provided the basic format of the study, obtained the funding and acted as study team leader. Both authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AM
Ampicillin
- BOXAIR
BOX repeat-based
- C
Chloramphenicol
- CAZ
Ceftazidime
- CF
Cephalothin
- CIP
Ciprofloxacin
- CLSI
Clinical and Laboratory Standards Institute
- CTX
Cefotaxime
- ERIC
Enterobacterial repetitive intergenic consensus
- GM
Gentamicin
- K
Kanamycin
- MDR
Multidrug-resistant
- NA
Nalidixic acid
- NTS
Non-typhoidal Salmonella
- PFGE
Pulsed-field gel electrophoresis
- REP
Repetitive extragenic palindromic
- S
Streptomycin
- SPIs
Salmonella pathogenicity islands
- SXT
Trimethoprim/sulfamethoxazole
- T3SS-1
Type 3 secretion system 1
- T3SS-2
Type 3 secretion system 2
- TE
Tetracycline
References
- 1.Almeida F, Medeiros MI, Kich JD, Falcão JP. Virulence-associated genes, antimicrobial resistance and molecular typing of Salmonella Typhimurium strains isolated from swine from 2000–2012 in Brazil. J Appl Microbiol. 2016;120:1677–90. doi: 10.1111/jam.13110. [DOI] [PubMed] [Google Scholar]
- 2.Campioni F, Moratto Bergamini AM, Falcão JP. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol. 2012;32:254–64. doi: 10.1016/j.fm.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr Issuses Mol Biol. 2002;5:113–22. [PubMed] [Google Scholar]
- 4.Cheong HJ, Lee YJ, Hwang IS, Kee SY, Cheong HW, Song JY, Kim JM, Park YH, Jung JH, Kim WJ. Characteristics of non-typhoidal Salmonella isolates from human and broiler-chickens in southwestern Seoul, Korea. J Korean Med Sci. 2007;22:773–8. doi: 10.3346/jkms.2007.22.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chon JW, Jung H, Kuk M, Kim YJ, Seo KH, Kim SK. High occurrence of extended-spectrum β-lactamase-producing Salmonella in broiler carcasses from poultry slaughterhouses in South Korea. Foodborne Pathog Dis. 2015;12:190–6. doi: 10.1089/fpd.2014.1847. [DOI] [PubMed] [Google Scholar]
- 6.CLSI: Clinical and Laboratory Standards Institute . Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved Standard-Third edition and supplement. Wayne: CLSI; 2008. [Google Scholar]
- 7.De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L. Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl Environ Microbiol. 2003;69:3456–61. doi: 10.1128/AEM.69.6.3456-3461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elemfareji OI, Thong KL. Comparative Virulotyping of Salmonella typhi and Salmonella enteritidis. Indian J Microbiol. 2013;53:410–7. doi: 10.1007/s12088-013-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sebay NA, Gebreel HM, El-Zeedy SA, Samy AA. Genotyping of some local isolates of genus Salmonella using RAPD-PCR. World Appl Sci J. 2012;17:1377–85. [Google Scholar]
- 10.Fendri I, Ben Hassena A, Grosset N, Barkallah M, Khannous L, Chuat V, Gautier M, Gdoura R. Genetic diversity of food-isolated Salmonella strains through Pulsed Field Gel Electrophoresis (PFGE) and Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR) PLoS ONE. 2013;8:e81315. doi: 10.1371/journal.pone.0081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueiredo R, Card R, Nunes C, AbuOun M, Bagnall MC, Nunez J, Mendonça N, Anjum MF, da Silva GJ. Virulence Characterization of Salmonella enterica by a New Microarray: Detection and Evaluation of the Cytolethal Distending Toxin Gene Activity in the Unusual Host S. Typhimurium. PLoS ONE. 2015;10:e0135010. doi: 10.1371/journal.pone.0135010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foley SL, Lynne AM, Nayak R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol. 2009;9:430–40. doi: 10.1016/j.meegid.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Frye JG, Jackson CR. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front Microbiol. 2013;4:135. doi: 10.3389/fmicb.2013.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gharieb RM, Tartor YH, Khedr MH. Non-Typhoidal Salmonella in poultry meat and diarrhoeic patients: prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 2015;7:34. doi: 10.1186/s13099-015-0081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong J, Zhang J, Xu M, Zhu C, Yu Y, Liu X, Kelly P, Xu B, Wang C. Prevalence and fimbrial genotype distribution of poultry Salmonella isolates in China (2006 to 2012) Appl Environ Microbiol. 2014;80:687–93. doi: 10.1128/AEM.03223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashemi A, Baghbani-Arani F. The effective differentiation of Salmonella isolates using four PCR-based typing methods. J Appl Microbiol. 2015;118:1530–40. doi: 10.1111/jam.12805. [DOI] [PubMed] [Google Scholar]
- 17.Huehn S, La Ragione RM, Anjum M, Saunders M, Woodward MJ, Bunge C, Helmuth R, Hauser E, Guerra B, Beutlich J, Brisabois A, Peters T, Svensson L, Madajczak G, Litrup E, Imre A, Herrera-Leon S, Mevius D, Newell DG, Malorny B. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis. 2010;7:523–35. doi: 10.1089/fpd.2009.0447. [DOI] [PubMed] [Google Scholar]
- 18.Hyeon JY, Chon JW, Hwang IG, Kwak HS, Kim MS, Kim SK, Choi IS, Song CS, Park C, Seo KH. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J Food Prot. 2011;74:161–6. doi: 10.4315/0362-028X.JFP-10-327. [DOI] [PubMed] [Google Scholar]
- 19.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–6. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im MC, Jeong SJ, Kwon YK, Jeong OM, Kang MS, Lee YJ. Prevalence and characteristics of Salmonella spp. isolated from commercial layer farms in Korea. Poult Sci. 2015;94:1691–8. doi: 10.3382/ps/pev137. [DOI] [PubMed] [Google Scholar]
- 21.Iwabuchi E, Yamamoto S, Endo Y, Ochiai T, Hirai K. Prevalence of Salmonella isolates and antimicrobial resistance patterns in chicken meat throughout Japan. J Food Prot. 2011;74:270–3. doi: 10.4315/0362-028X.JFP-10-215. [DOI] [PubMed] [Google Scholar]
- 22.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998–2008. Emerg Infect Dis. 2011;19:1239–44. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim NO, Jung SM, Na HY, Chung GT, Yoo CK, Seong WK, Hong S. Enteric Bacteria Isolated from Diarrheal Patients in Korea in 2014. Osong Public Health Res Perspect. 2015;6:233–40. doi: 10.1016/j.phrp.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MS, Lim TH, Jang JH, Lee DH, Kim BY, Kwon JH, Choi SW, Noh JY, Hong YH, Lee SB, Yang SY, Lee HJ, Lee JB, Park SY, Choi IS, Song CS. Prevalence and antimicrobial resistance of Salmonella species isolated from chicken meats produced by different integrated broiler operations in Korea. Poult Sci. 2012;91:2370–5. doi: 10.3382/ps.2012-02357. [DOI] [PubMed] [Google Scholar]
- 25.Kim S. Salmonella serovars from foodborne and waterborne disease in Korea, 1998–2007: Total isolates decreasing versus rare serovars emerging. J Korean Med Sci. 2010;25:1693–9. doi: 10.3346/jkms.2010.25.12.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krawiec M, Kuczkowski M, Kruszewicz AG, Wieliczko A. Prevalence and genetic characteristics of Salmonella in free-living birds in Poland. BMC Vet Res. 2015;11:15. doi: 10.1186/s12917-015-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litrup E, Torpdahl M, Malorny B, Huehn S, Christensen H, Nielsen EM. Association between phylogeny, virulence potential and serovars of Salmonella enterica. Infect Genet Evol. 2010;10:1132–9. doi: 10.1016/j.meegid.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Lynne AM, Kaldhone P, David D, White DG, Foley SL. Characterization of antimicrobial resistance in Salmonella enterica serotype Heidelberg isolated from food animals. Foodborne Pathog Dis. 2009;6:207–15. doi: 10.1089/fpd.2008.0172. [DOI] [PubMed] [Google Scholar]
- 29.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 30.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–9. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 31.Mezal EH, Sabol A, Khan MA, Ali N, Stefanova R, Khan AA. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010. Food Microbial. 2014;38:67–74. doi: 10.1016/j.fm.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Nair A, Rawool DB, Doijad S, Poharkar K, Mohan V, Barbuddhe SB, Kolhe R, Kurkure NV, Kumar A, Malik SV, Balasaravanan T. Biofilm formation and genetic diversity of Salmonella isolates recovered from clinical, food, poultry and environmental sources. Infect Genet Evol. 2015;36:424–33. doi: 10.1016/j.meegid.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Ranjbar R, Naghoni A, Yousefi S, Ahmadi A, Jonaidi N, Panahi Y. The study of genetic relationship among third generation cephalosporin-resistant Salmonella enterica strains by ERIC-PCR. Open Microbiol J. 2013;7:142–5. doi: 10.2174/1874285801307010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skyberg JA, Logue CM, Nolan LK. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 2006;50:77–81. doi: 10.1637/7417.1. [DOI] [PubMed] [Google Scholar]
- 35.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, Agmon V, McClelland M, Rahav G, Gal-Mor O. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS ONE. 2013;8:e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–31. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo SJ, Lim HS, Lee K. Epidemiological investigation of an outbreak of salmonellosis in Gyeongju, Korea. J Prev Med Public Health. 2014;47:177–81. doi: 10.3961/jpmph.2014.47.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon RH, Cha SY, Wei B, Roh JH, Seo HS, Oh JY, Jang HK. Prevalence of Salmonella isolates and antimicrobial resistance in poultry meat from South Korea. J Food Prot. 2014;77:1579–82. doi: 10.4315/0362-028X.JFP-14-018. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Shu R, Zhao Y, Zhang Q, Xu X, Zhou G. Analysis of ERIC-PCR genomic polymorphism of Salmonella isolates from chicken slaughter line. Eur Food Res Technol. 2014;239:543–8. doi: 10.1007/s00217-014-2277-x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


