Abstract
Divalent ions fulfill essential cellular roles and are required for virulence by certain bacteria. Free intracellular Mg2+ can approach 5 mm, but at this level Mn2+, Ni2+, or Co2+ can be growth-inhibitory, and magnesium fluoride is toxic. To maintain ion homeostasis, many bacteria have evolved ion sensors embedded in the 5′-leader sequences of mRNAs encoding ion uptake or efflux channels. Here, we review current insights into these “metalloriboswitches,” emphasizing ion-specific binding by structured RNA aptamers and associated conformational changes in downstream signal sequences. This riboswitch-effector interplay produces a layer of gene regulatory feedback that has elicited interest as an antibacterial target.
Keywords: crystallography, ion channel, metal homeostasis, Salmonella enterica, transcription regulation, riboswitch
Introduction
Riboswitches are highly structured RNA motifs often located in the 5′-leader sequences of bacterial mRNAs (1–3). A riboswitch recognizes its cognate effector by a conserved aptamer domain, which elicits conformational changes in a downstream expression platform that alter transcription termination, translation initiation, message stability, or alternative splicing of the associated transcript (2, 4, 5). Because riboswitches evolved distinct molecular determinants to bind specific effectors, they have garnered interest not only for their elegant feedback mechanisms, but also for their potential to serve as novel drug targets. A prime example is the flavin mononucleotide (FMN) riboswitch, which can be inhibited by natural or synthetic ligand analogs that blunt the essential biosynthesis of riboflavin in bacteria (6–10). At present, nearly 40 riboswitch classes that recognize >25 chemically distinct effectors have been described (11), providing many opportunities to interrupt key anabolic pathways. Conversely, a growing cohort of riboswitches activate genes involved in remediation of cellular toxins, such as S-adenosylhomocysteine (12), azaaromatics (13), guanidine (14), Mg2+ (15), transition metals (16–18), or magnesium fluoride (19). Here, we discuss the latter three classes of riboswitches that evolved elegant but distinct RNA folds to recognize specific divalent cations, leading to gene-regulatory changes that maintain cellular homeostasis. In some cases, metalloriboswitch affinity and cooperativity can rival that of proteins. Many excellent riboswitch reviews provide broad overviews of the field (2, 3, 11). As such, we focus here on the handful of known metalloriboswitches with an emphasis on their discovery, validation, specificity, and gene regulation, as well as their potential to serve as drug targets.
Mg2+-I riboswitches are non-specific sensors that regulate Mg2+ import pumps
Mg2+ is the most abundant intracellular, multivalent ion with free concentrations in bacteria ranging from 1 to 5 mm (20). Although numerous enzymes require Mg2+ for catalysis, the ion can also fulfill structural roles by neutralizing charged DNA or RNA backbones or acting as a nexus for functional group coordination in RNA tertiary structure (21, 22). The Escherichia coli 70S ribosome has ∼170 coordinated Mg2+ ions (23), and the lack of Mg2+ in growth media leads to subunit dissolution and degradation (20). As a rule, bacteria employ a broad range of metallosensors and transporters to attain metal ion homeostasis (24). Some bacteria link Mg2+ concentration to virulence genes to coordinate gene expression in cation-deficient host cells, such as macrophages (25). Salmonella enterica has three Mg2+ importers: the widespread CorA channel and P-type ATPases MgtA and MgtB (26, 27). Recent salmonellosis outbreaks caused by the Heidelberg serovar are alarming due to the multidrug resistance of this organism (28, 29). In this and many other Gram-negative bacteria, mgtA expression is controlled by an upstream Mg2+-sensing riboswitch (known as the Mg2+-II class (11)) that regulates transcription via rho-dependent and RNase E degradation mechanisms (30–32). An additional level of control appears to be exerted by a proline-rich open reading frame inside the mgtA riboswitch that works in concert with Mg2+-sensing to increase mgtA mRNA levels when Mg2+ is low (33, 34). Additional studies are needed to clarify the details of regulation, which appears to be present in only a handful of γ-proteobacteria (11). A second class of Mg2+-sensing riboswitch known as the Mg2+-I (or M-box) is more broadly distributed in Gram-positive and some Gram-negative bacteria, where it is controls a variety of genes (15, 25), including the widespread Mg2+-uptake channel mgtE (27). In Aeromonas hydrophila, mgtE mutations impair swarming and biofilm formation (35), which are important aspects of its pathogenicity. The defined Mg2+-I aptamer domain and readily discernible expression platform make it a model system for structure and function analysis, which provides a benchmark for additional comparisons herein.
The aptamer of the Mg2+-I riboswitch is not selective for Mg2+, unlike other metal-sensing riboswitches (see below), as shown by the variety of divalent ions that promote compaction of its fold and favor formation of a downstream intrinsic terminator hairpin within the expression platform (15, 36). In this respect, Mg2+-I riboswitches likely function by sensing the most prevalent intracellular ion, Mg2+, which requires only modest affinity and selectivity, as corroborated by EC50 values of 0.4 mm for Mg2+ or Ca2+ and 0.13 mm for Mn2+ (36). Thus, Mg2+-I riboswitches promote homeostasis but are outperformed by regulatory metalloproteins in terms of selectivity (16). To investigate the metal-binding determinants, structures of the Mg2+-I riboswitch were determined in Mg2+- and Mn2+-bound states (15, 36). The global architecture features three parallel helical domains comprising P1-P2-P6, P3-P4, and P5 (Fig. 1a). Tertiary interactions are stabilized by six Mg2+ ions that promote compaction as shown biochemically (37). Four key Mg2+ sites (I–IV) reside at the P1-P2-P5 interface, whereas sites V and VI are in the P4 internal loop. The former metal constellation (core 1) contributes to interhelical tertiary stability, whereas the latter (core 2) appears to orient the P4 loop to facilitate long-range contacts (36). Biochemical evidence also supports additional Mg2+ binding, but these sites (core 3) were visible only in structures obtained from crystals grown from 5 mm Mn2+, which was used as a proxy for Mg2+. Importantly, only core 1 ions were present in all structures, including crystal forms harboring multiple Mg2+-I riboswitch copies per asymmetric unit. Moreover, sites I–III exhibited the highest phosphorothioate interference (37) with superimposable Mg2+ and Mn2+ coordination in pairwise structural comparisons (36). As such, it is worthwhile to describe these strong binding sites with a focus on how their tertiary contacts at these locations relate to gene regulatory conformations.
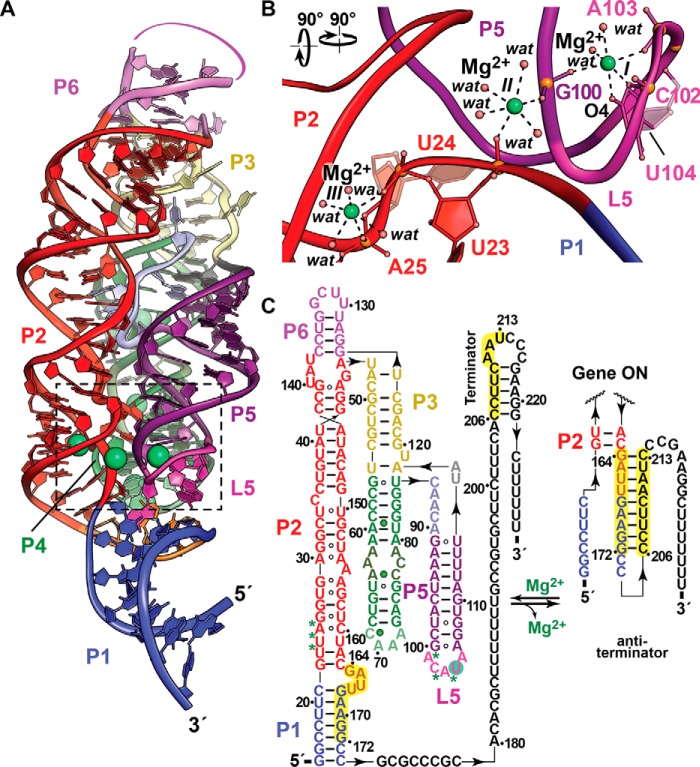
Figure 1.
Tertiary fold and ion coordination by the Mg2+-I riboswitch in the Mg2+-bound state. a, ribbon diagram of the B. subtilis Mg2+-I riboswitch co-crystal structure determined at 2.7 Å resolution (Protein Data Bank entry 2QBZ). The riboswitch fold contains three parallel helical domains, P1-P2-P6, P3-P4, and P5, wherein the former two segments coaxially stack. Four coordinated Mg2+ ions are depicted as green spheres at the interface of helices P1, P2, and P5 (foreground) and in P4 (background). b, close-up view of three Mg2+-binding sites (I–III, boxed in a). The metal ions at each site coordinate in an octahedral manner. The site I Mg2+ is coordinated by the non-bridging phosphate oxygens of G100, C102, and A103. A fourth ligand is contributed by the O4 keto moiety of U104 with two inner-shell water molecules completing the coordination sphere. Site II uses non-bridging oxygens from G100 and U23 with the remaining coordination sphere completed by waters. Site III uses two non-bridging oxygens from U24 and A25, as well as four inner-sphere waters. c, secondary structure depicting changes in conformational states that regulate transcription. Paired regions are colored as in a with backbone binding contributions to Mg2+ from sites I–III depicted as green asterisks; additional Mg2+ sites in P4 are depicted as green spheres. In the gene-off state, Mg2+ binding at sites I–III promotes a conformation that destabilizes anti-terminator base pairing between the P1-P2 bulged loop (164–172) and the 3′-tail (206–213), favoring intrinsic-terminator hairpin formation that attenuates mgtE transcription, reducing Mg2+ uptake by the MgtE channel. Conversely, low Mg2+ favors RNA polymerase transcriptional read-through via an anti-terminator helix in place of P1. The full-length message leads to MgtE translation with increased Mg2+ import into the cell.
The site I Mg2+ is coordinated by non-bridging phosphate oxygens at the transition of P5 into L5 with a fourth ligand contributed by the U104 base (Fig. 1b). Site II also uses non-bridging oxygens from P5 and P2 with the remaining coordination sphere completed by waters. Likewise, site III comprises two non-bridging oxygens from P2 and four waters. Shared coordination groups, such as the phosphate of G100, provide a plausible basis for cooperativity between Mg2+-binding sites (Hill coefficient 4.3) (36), which confers a sharp “digital” response to the effector over a narrow concentration range.
Gene regulation by the Mg2+-I riboswitch is predicated on Mg2+-mediated condensation of the RNA fold (Fig. 1a) that favors sequestration of an anti-terminator sequence (164–172) located between P1 and P2 (Fig. 1c). This effector-bound conformation promotes an alternate, intrinsic terminator harboring the 3′-anti-terminator sequence (206–213), thus blocking transcription of the downstream mgtE gene, leading to attenuated Mg2+ uptake by a diminished population of MgtE channels. Conversely, depleted cellular Mg2+ alters the interface between P1-P2 and P5-L5 (sites I–III), promoting formation of an anti-terminator hairpin (Fig. 1c) that favors full-length mgtE transcription, resulting in Mg2+ import. The presence of distinct Mg2+-sensing riboswitches in human pathogens suggests that these RNA elements could be targets for antibacterials, especially because the associated Mg2+ transport genes are linked to virulence (discussed below).
Orphan riboswitch yybP-ykoY senses Mn2+ to activate intake pumps
Manganese is an essential trace element that plays key cellular roles, including facilitating the catalytic activity of various ribonucleotide reductases and superoxide dismutases expressed under low iron conditions or H2O2 stress (38). Bacteria exhibit varied Mn2+ requirements that reflect survival adaptations to specific microenvironments. E. coli can tolerate up to 20 mm Mn2+ (39) but maintains cellular levels at ∼1 μm (24), whereas Bacillus subtilis has cytosolic levels of ∼10 μm (40). For some bacteria, Mn2+ is toxic because in various enzymes it replaces Fe2+, which is not tolerated functionally (38). Conversely, Borrelia and Lactobacillus are members of small bacterial cohorts that use Mn2+ with preference over iron (41, 42), a strategy that bypasses host efforts to restrict iron levels, at least in the former case. Likewise, mutations in the macrophage-specific natural resistance-associated macrophage protein 1 (Nramp1) transporter–a Mn2+, Fe2+, and Zn2+ uptake antiporter–led to diminished host resistance against Salmonella, Leishmania, and Mycobacterium (43), possibly resulting from a reduced capacity to remove divalent cations from bacteria-containing vacuoles in the host (44). Conversely, bacterial virulence can be attenuated by mutation of Nramp1 homologs of the pathogen, linking virulence to Mn2+ uptake. Salmonella uses Mn2+ transporters MntH (an Nramp1 homolog) and SitABCD (an ATP-binding cassette protein) (45), and a typhimurium serovar lacking both is avirulent (46). The mntH and sitABCD genes are repressed by the trans-acting Fur and MntR proteins in high Mn2+, although an additional cis-acting Mn2+-responsive riboswitch is hypothesized to reside in the 5′-leader of Salmonella mntH genes (47). This element requires further validation, but it ostensibly favors anti-terminator stem formation at low Mn2+ concentrations, leading to MntH expression and Mn2+ uptake. Exporters of Mn2+ are also required by bacteria to attain homeostasis. The finding that the mntP (formerly yebN) encodes Mn2+ efflux pumps, and is controlled by the Mn2+-responsive Fur and MntR proteins (48–50), provided insight into a longstanding riboswitch mystery. The ubiquitous yybP-ykoY RNA motif, classified for years as an “orphan” riboswitch for lack of a known effector (51, 52), was identified in the 5′-leaders of mntP genes, suggesting that Mn2+ was the long-sought ligand (17, 18). Indeed, independent labs validated the yybP-ykoY or Mn2+ riboswitch as an Mn2+ sensor that controls expression of a P-type uptake pump (17, 18). This provided a firm basis to understand how a metalloriboswitch discriminates between Mg2+ and Mn2+, leading to cellular resistance against the toxic effects of a transition metal.
A structure of the Mn2+ riboswitch revealed a four-way helical junction comprising tandem coaxially stacked helices (Fig. 2a). Mn2+-induced fold compaction based on chemical modification showed reduced loop flexibility at L1 and L3 and the base of P1 (17) with reactivity changes at U87 and A88 producing a KD, app value for Mn2+ of 27 ± 6 μm. Chemical mapping also corroborated the structure, wherein two divalent ion-binding sites localize in bulged loops L1 and L3 to compose a prominent tertiary docking interface at the helical junction (Fig. 2, a and b). At site I, Mn2+ is coordinated by the N7 moiety of A41 and non-bridging phosphate oxygens from L3 and L1. The nearby site II pocket binds Mg2+ using only oxygen atoms contributed by non-bridging groups of L1 and L3, as well as a single water molecule, consistent with the recalcitrance of Mg(H2O)62+ to release its inner-sphere waters (53). Regarding the basis of Mn2+ selectivity at site I, it is notable that Mn2+ exhibits a more polarizable (softer) character in high-coordination-number environments, whereas Mg2+ is oxophilic (harder) with less preference for nitrogen (53). Nonetheless, Mn2+ functionally substitutes for Mg2+ in many enzymes, and octahedral Mg2+ will coordinate nitrogen (53). Consistent with these findings, riboswitch crystals prepared from 2.5 mm Mn2+ incorporated Mn2+ at both binding sites (17), but only site I was occupied when the Mn2+ concentration was reduced to 100 μm. Although biochemical cooperativity was not analyzed, interdependence of the ion-binding sites is likely based on shared coordination of non-bridging phosphate oxygens (Fig. 2b). As anticipated, 41A→U and 41A→G mutations are less responsive to Mn2+ and are accompanied by enhanced L3 flexibility, consistent with rearrangement of the coordination pocket. It has been noted for proteins that insertion of a single nitrogen into a coordination sphere can exclude Mg2+ and favor Mn2+ (53). As such, it is reasonable that N7 of conserved A41 is a major selectivity determinant for Mn2+ in the Mn2+ riboswitch, which could account for the ability of Mn2+ to bind even in the presence of relatively high Mg2+ ion levels in the cytosol.
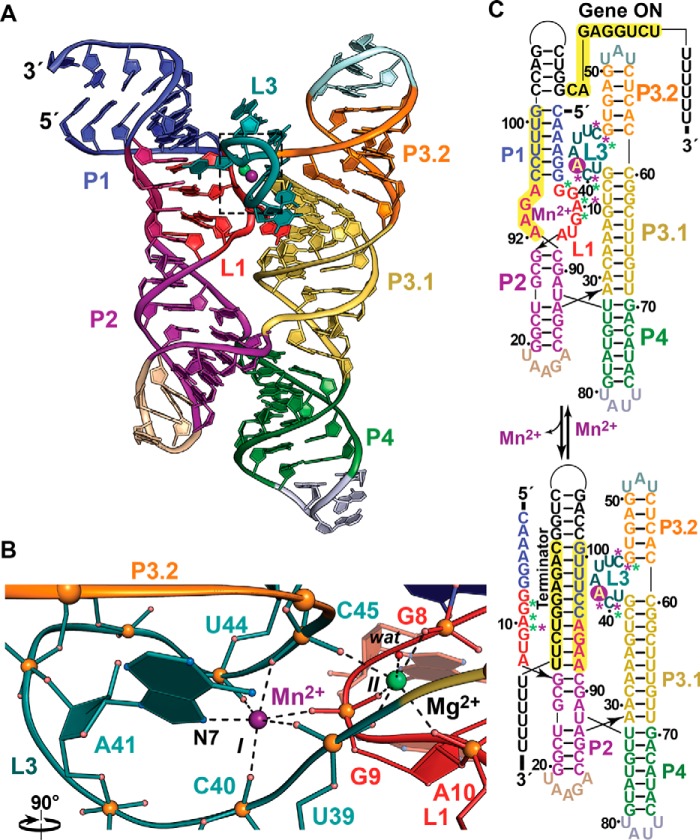
Figure 2.
Tertiary fold and ion coordination by the Mn2+ riboswitch bound to Mn2+ and Mg2+. a, ribbon diagram of the Lactococcus lactis Mn2+ riboswitch co-crystal structure determined at 2.85 Å resolution (Protein Data Bank entry 4Y1I) showing a four-way helical junction containing parallel, coaxially stacked helices P1-P2 and P3-P4. Divalent ion recognition occurs at the helical junction between loops L1 and L3, where Mn2+ (site I, magenta) and Mg2+ (site II, green) bind. b, close-up view of the octahedral divalent ion coordination at sites I and II (boxed in a). Mn2+ is observed at site I and is preferred over Mg2+ based on the N7 group of the A41 base. Non-bridging phosphate oxygens are contributed from U39, C40, U44, and C45 of L3, as well as G9 of L1. Site II is coordinated by non-bridging oxygens of G8, G9, and A10 of L1 and U39 and C45 of L3; an inner-sphere water molecule completes the coordination shell. Each non-bridging phosphate oxygen of G9 contributes to ion binding at sites I and II, providing a basis for cooperative binding. c, secondary structure diagrams depicting changes in Mn2+ riboswitch conformational states that regulate transcription. Backbone contributions to Mn2+ and Mg2+ binding are depicted as magenta and green asterisks; Mn2+ coordination at N7 of A41 is depicted as a magenta circle. In the gene-on state, Mn2+ and Mg2+ binding at sites I and II promote a conformation that facilitates base pairing of an anti-terminator hairpin above P1, favoring transcription of the mntP (yoaB) efflux pump that confers Mn2+ resistance. Conversely, low Mn2+ levels favor formation of an intrinsic terminator hairpin at the expense of the P1 helix and the L1-L3 ion-binding sites. A premature mntP (yoaB) message leads to reduced expression of the associated Mn2+ efflux channel.
Gene regulation by Mn2+ riboswitches occurs by two distinct mechanisms involving transcriptional and translational control (17, 18). Structural analysis of the E. coli riboswitch showed that site I collapses in the absence of Mn2+, whereas site II remains intact and occupied by Mg2+ (17). However, Mg2+ alone did not stabilize the L1-L3 binding interface in chemical-modification experiments, which has implications for formation of mutually exclusive expression platform conformations that regulate transcription. At high Mn2+ concentrations, the L1-L3 interface is well ordered (Fig. 2b), giving rise to a downstream transcription anti-terminator hairpin following P1 (Fig. 2c, top). This conformation favors polymerase read-through, leading to full-length transcripts encoding the MntP (YoaB) efflux pump that confers Mn2+ resistance. Conversely, the L3 sensor loop melts in the Mn2+-free state, favoring intrinsic terminator formation (Fig. 2c, bottom) that leads to premature mntP termination. Similar conformational changes are envisioned for translational control, although the details of these changes are unclear at present.
NiCo riboswitches confer heavy metal resistance by sensing Ni2+ or Co2+
Heavy metals (i.e. ρ > 5 g/cm3 (39)) such as copper and silver have long been known to possess antimicrobial properties (54). Even essential metals at sufficiently high concentrations can be toxic, as is the case for trace nutrients Ni2+ and Co2+. These ions can be incorporated into Fe-S proteins, leading to lethal effects that necessitate strict cellular control over their uptake (39, 55). E. coli senses Ni2+ and Co2+ as repellants (56) and maintains intracellular levels in the low- to sub-μm range (57). In minimal media, growth of E. coli is inhibited at 8 μm Ni2+, and 160 μm Co2+ arrests S. enterica (58, 59). Whereas Co2+ is used for carbon rearrangements in the context of coenzyme B12, Ni2+ is needed for enzymes such as hydrogenase and urease (24). Once inside the periplasm, ABC transporters and NiCo permeases are the major modes of bringing Ni2+ and Co2+ into the bacterial cell (24). The non-specific CorA transporter also facilitates cytosolic accumulation (27). To attain homeostasis, bacteria have cadmium-zinc-cobalt (czc) genes that encode efflux pumps and cation diffusion facilitators such as CzcD. Like the Mg2+-I and Mn2+ riboswitches, comparative sequence analysis revealed a structured RNA motif upstream of czcD genes in multiple bacterial species (16). Isolated czc RNA motifs showed in vitro KD, app values of 5.6 and 12 μm for Co2+ and Ni2+, with five metal-responsive regions in the four-way helical junction core. Comparatively weaker binding (KD, app of 220 μm) was seen for Mn2+.
The czc motif or “NiCo” riboswitch must selectivity bind Co2+ or Ni2+ amid much higher intracellular Mg2+ concentrations. To elucidate the basis of affinity and recognition of these ions, a riboswitch structure was determined in the presence of Co2+ (16). Like the Mg2+-I and Mn2+ riboswitches, the NiCo structure lacks pseudoknot interactions and exhibits close packing, whereby four helices coaxially stack in pairs (Fig. 3a). Three co-planar Co2+-binding sites (I–III) reside at the interface between L2-3 and L4-1 with a fourth site (IV) at the base of P2. Like the Mn2+ riboswitch motif, Co2+ or Ni2+ selectivity is conferred by the purine N7 moiety, with sites I–III each coordinating two imines (Fig. 3b). Hill coefficients of 2.0 and 1.6 were measured for Co2+ and Ni2+, using the Clostridium botulinum NiCo riboswitch. The structure nicely explains such positive cooperativity in that sites I and II coordinate the N7 and 2′-OH groups from opposing L4-1 and L2-3 nucleotides that knit the junction together. In this manner, the G46 N7 group coordinates site II, and its 2′-OH coordinates site I; conversely, the N7 group of G87 coordinates site I, and its 2′-OH coordinates sites II (Fig. 3b). As such, Co2+ binding at one site stabilizes coordination at the adjoining site. N7-deaza substitutions at G87 and G88 underscore the importance of N7 coordination. The former mutation cannot support Co2+-dependent stabilization at G46, indicating that sites I and II are indeed linked. In contrast, N7-deaza G87 did not affect site III, consistent with its distal location from site I. At present, site IV appears to provide stabilization of P1-P2, with the site exhibiting a significantly lower anomalous signal than sites I–III (16).
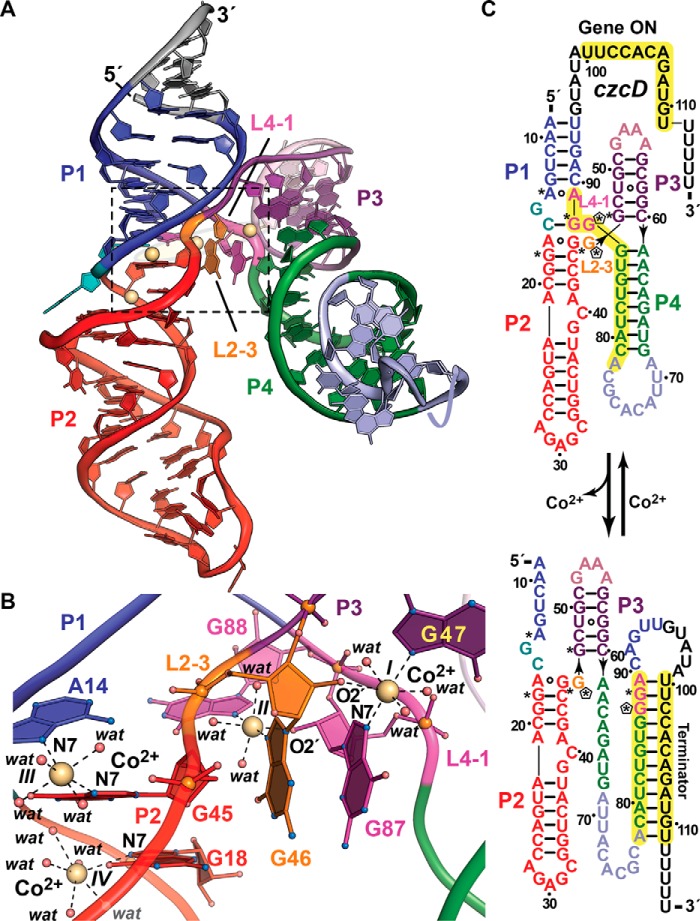
Figure 3.
Tertiary fold and ion coordination by the NiCo riboswitch bound to Co2+. a, ribbon diagram of the Erysipelotrichaceae bacterium riboswitch co-crystal structure determined at 2.64 Å resolution (Protein Data Bank entry 4RUM). The overall architecture comprises tandem, coaxially stacked P1-P2 and P3-P4 helices offset by 30° and joined by a central four-way helical junction. Like the Mg2+-I and Mn2+ riboswitches, divalent ion sensing occurs at the junction interface. Here, four Co2+ sites (I–IV, yellow spheres) bind between loops L2–3 and L4–1. b, close-up view of the divalent ion-binding positions (boxed in a). Sites I–III interact directly with bases G47 and G87, G46 and G88, and A14 and G45, which are conserved among most czc homologs. G46 and G87 each contribute to coordination at sites I and II via purine N7 and ribose hydroxyl groups that form the basis of cooperativity during Co2+ or Ni2+ binding. In contrast to the Mg2+-I and Mn2+ riboswitches that make use of non-bridging phosphate oxygens to coordinate Mg2+ or Mn2+, NiCo riboswitch coordination utilizes one or two N7 moieties, 2′-OH groups, and water molecules to bind Co2+ and presumably Ni2+. c, secondary-structure diagram depicting effector-dependent conformational changes that regulate transcription; asterisks indicate sites of purine N7 Co2+ coordination in b, and pentagons show coordination from ribose 2′-OH groups. Transcriptional regulation by the NiCo riboswitch parallels that of the Mn2+ riboswitch because Co2+ or Ni2+ sensing favors an RNA fold that allows RNA polymerase to read through the transcript, leading to a full-length message that confers heavy-atom resistance when translated. In the effector-free state, P4 remodels to form a terminator hairpin leading to premature transcription termination, potentially resulting in Co2+ or Ni2+ accumulation in the cell.
Inspection of the NiCo riboswitch expression platform suggests the formation of two mutually exclusive conformations. At low Co2+ or Ni2+ concentrations, an intrinsic terminator hairpin is favored, whereas elevated ion levels support polymerase read-through leading to czcD expression (Fig. 3c). This mechanism was confirmed by in vitro transcription assays, which yielded full-length products only in the presence of cognate effector ions. Regulation was validated in vivo when a representative czc motif from Clostridium scindens was placed upstream of a putative cation efflux gene. Whereas levels of the associated mRNA increased with increasing Ni2+, a similar analysis using non-cognate ions did not affect downstream transcript levels (16). Overall, the cooperative binding and unique mode of metal recognition by the NiCo riboswitch impart high selectivity, enabling feedback to minute changes in Co2+ or Ni2+ in a milieu of competing cellular ions to confer heavy-metal resistance. Notably, NiCo riboswitches are present in Listeria monocytogenes, a common foodborne pathogen (60) that is of concern because of the identification of multi-antibiotic resistance isolates in “ready-to-eat”' foods (61).
Fluoride riboswitches require Mg2+ to sense and regulate export of a cellular toxin
Fluorine is the most electronegative and reactive element and is found predominantly in the biosphere as the fluoride ion (F−). Although a trace element, fluorine is ubiquitous with levels ranging from 1.2 ppm (mg/liter) in seawater to 200 ppm in some soils (62, 63). High levels of F− are toxic to bacteria, fungi, and animals (62, 64, 65), and in many regions F− is added at 0.7 ppm to municipal water supplies as an anticaries agent (66). The beneficial and toxic effects of F− arise from similarities in its charge and radius to hydroxide (67). The distinct antimicrobial properties of F− are derived from its affinity for divalent cations. F− is toxic to enolase because it stabilizes a bis-MgF-PO4 complex that mimics the reaction intermediate (68–70). MgF3− and AlF3 complexes also form trigonal-bipyramidal mimics of phosphoryl-transfer transition states (71) to inhibit catalysis by this essential class of enzymes.
Chronic contact with F− contributed to the evolution of fluoride toxicity-resistance factors that stimulate expression of genes to ameliorate intracellular F− levels, including transporters (19, 72–74). The observation that many bacteria and archaea use fluoride-responsive RNAs to regulate such genes suggests that F−-sensing riboswitches arose as part of an ancient strategy to achieve F− resistance (19, 75). Such riboswitches were first identified as conserved, non-coding motifs upstream of genes such as crcB, which encodes a fluoride-specific channel (19, 72, 73), as well as metabolic enzymes such as enolase (19, 76). Biochemical experiments on the ∼80-nucleotide crcB 5′-leader RNA confirmed a 1:1 binding stoichiometry for F− only in the presence of Mg2+ (KD, app ∼60–135 μm) (19, 77), with no apparent binding to Cl−, Br−, I−, or the infused gases CO and NO. Mutations that alter conserved core sequences or secondary structures of the riboswitch adversely impact F− binding, as expected for molecular determinants that evolved to bind a specific effector (78). Fusion of the Bacillus cereus 5′-leader of crcB to a lacZ reporter in B. subtilis revealed F−-dependent β-galactosidase activity consistent with its modulation of an intrinsic transcription terminator. Evidence for translational control by an alternative expression platform came from a crcB riboswitch from Pseudomonas syringae that lies upstream of eriC genes. This riboswitch was used to control lacZ in an E. coli crcB knock-out strain that is ∼200-fold more sensitive to F− than WT (19). This work demonstrated a role for CrcB in reducing cellular F− levels and provided insight into eriC function and selectivity. Specifically, EriCF proteins rescued growth of E. coli crcB knock-out cells under high F− conditions, suggesting that these genes are functionally homologous (19). Accordingly, it is now established that EriCF proteins are fluoride-selective antiporters (19, 74).
To elucidate the mode of F− sensing and the underlying basis for gene regulation, the structure of a fluoride riboswitch was determined (77). The structure confirmed that the conserved aptamer folds as an HLout pseudoknot (Fig. 4a) (79). Unexpectedly, a constellation of five backbone phosphates at the topological confluence of P2, L1, and L3 chelate three Mg2+ ions crucial for F− sensing. These cations form the vertices of a nearly equilateral triangle with a central cavity that is ideal for F− coordination (radius 1.30 Å) (Fig. 4b), while selectively excluding larger halides such as Cl− (radius 1.81 Å) (80). Similar tri-Mg2+ coordination of F− has been observed previously in proteins (81, 82).
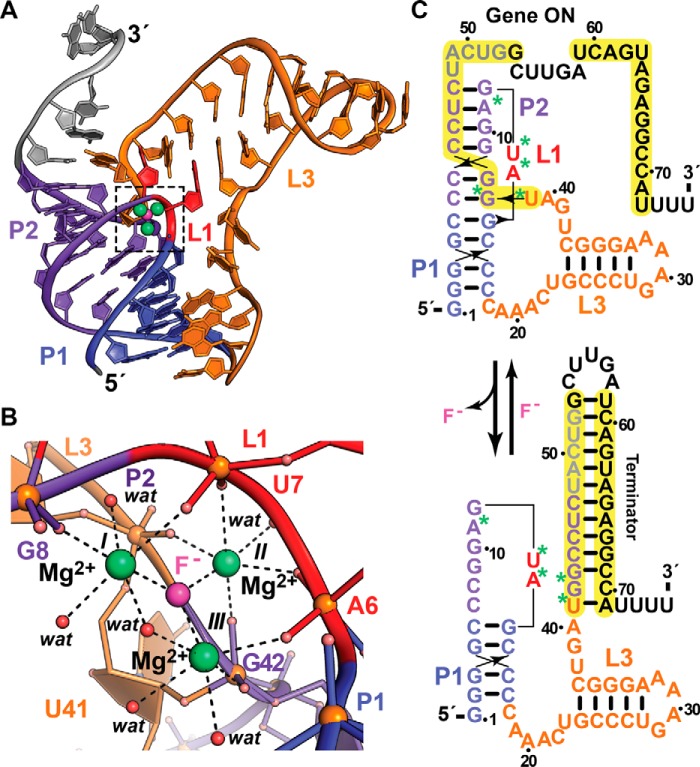
Figure 4.
Tertiary fold and ion coordination by the fluoride riboswitch in complex with magnesium fluoride. a, ribbon diagram of the T. petrophila riboswitch co-crystal structure determined at 2.3 Å resolution (Protein Data Bank entry 4ENC). The riboswitch folds as an HLout pseudoknot wherein F− ion (pink) sensing occurs by three Mg2+ ions (green) coordinated between loops L1 and L3 that mediate long-range contacts between coaxially stacked helices P1 and P2; loop L2 = 0. b, close-up view of the ion-sensing pocket (boxed in a), revealing a trigonal constellation of octahedrally coordinated Mg2+ ions, I–III, which share a central F− ion. Like the Mg2+-I and Mn2+ riboswitch, non-bridging phosphate oxygens provide coordination sites for Mg2+ with inner-sphere water molecules completing the coordination sphere. c, secondary-structure diagram depicting effector-dependent conformational changes that regulate transcription; asterisks indicate the sites of backbone oxygen coordination to Mg2+ in b. In the presence of fluoride, the 5′-end of a terminator hairpin is buried by the pseudoknot, leading to transcription of a full-length message and translation of a CrcB channel that exports F− to confer resistance. In the absence of fluoride, there is no need for F− export, and the intrinsic terminator hairpin is favored, leading to the gene being maintained in an off state.
The gene regulatory functions of Thermotoga petrophila and B. cereus fluoride riboswitches likely entail modulation of an intrinsic terminator that forms at low F− concentration (Fig. 4c), ostensibly reducing CrcB levels. NMR data corroborate F−-dependent compaction of the T. petrophila crcB motif with interconversion of the effector free state into a compact form (77). Mapping the Mg2+ coordination sites onto the riboswitch structure suggests that structurally disparate regions in L1, P2, and L3 may be pre-organized early in transcription to coordinate F− and Mg2+, which promotes complete folding of P2 before the 5′-end of the terminator helix folds into a hairpin that arrests transcription. An alternative expression platform has been identified in the F− riboswitch from P. syringae in which a Shine-Dalgarno sequence in the 3′-tail is sequestered from the 16S rRNA of the ribosome under low F− levels (19), thus attenuating translation initiation required for EriCF synthesis.
Prospects for therapeutic targeting and discovery of new metalloriboswitches
A handful of riboswitches are known to bind metabolite analogs that alter gene expression and bacterial growth (6–9, 83, 84). Although metalloriboswitches do not bind organic ligands, they are expected to be susceptible to small molecules that target conserved binding pockets (85). Such small molecules will likely hit other members of the same class, and any escape mutant will impede normal ion binding and gene regulation. High-throughput and fragment-based screening, as well as structure-guided methods, have yielded compounds that target metabolite-sensing riboswitches (6–9, 83, 86, 87), and these approaches will likely succeed for metalloriboswitches. Small molecules that lock the Mg2+-I riboswitch into a compact gene-off state (e.g. Fig. 1a) would attenuate MgtE expression, depriving bacteria of Mg2+. Deletion of mgtE in B. subtilis yielded a defect requiring >25 mm Mg2+ for growth; by contrast, mgtA deletion–controlled by some Mg2+-II riboswitches–had no effect (88).
As for the Mn2+ and NiCo riboswitches, these regulators are important when iron is scarce or H2O2 is present, such as inside a host cell. Therein, iron enzymes of the pathogen can lose activity, and Mn2+ import by MntH becomes essential (45, 89). Stabilizing the mntH riboswitch in a gene-off state could reduce virulence, although the details of this conformation are unclear. Similarly, small molecules that fix the mntP riboswitch in a gene-on state (Fig. 2a) would also lower Mn2+ levels. Importantly, heavy metals can enter bacteria by non-specific import that requires regulated export for homeostasis (90). Whereas the czcD gene confers Co2+ and Ni2+ resistance, its deletion in Streptococcus pneumoniae led to increased Zn2+ sensitivity (IC50 of ∼250 μm) (91). Zn2+ varies in human tissues from 20 μm in serum to 230 μm in lung (92), and high levels can impair iron enzymes by mismetallation (93). It is conceivable that trapping the NiCo riboswitch in a gene-off state (Fig. 3c) would impair growth under certain environmental conditions (91).
Deletion of crcB increases bacterial sensitivity to F− (19), but certain compounds are also known to enhance F− sensitivity (94–96). This work supports the prospect of targeting F− riboswitches in concert with supplemental F− (75). Such molecules would be selected to promote gene-off states (Fig. 4c), blocking F− export. Beyond bacteria, CrcB homologs confer F− resistance in fungi and yeast (73) but are absent in mammals. In general, F− riboswitches are missing in eukaryotes (11), supporting prospects for antibacterial development. Overall, the metalloriboswitch field would benefit greatly by placing greater emphasis on analysis in the context of infectious disease models.
The abundance of representatives for various riboswitch classes obeys a power-law relationship that suggests ∼1000 new classes remain to be discovered (11). In terms of new metalloriboswitches, it is reasonable to consider that magnesium, calcium, manganese, iron, cobalt, copper, and zinc are essential for most life forms, whereas vanadium, nickel, and tungsten are present only in bacteria (97). Each metal represents a likely effector, but it is reasonable to expect new riboswitches will sense the known ions described here. Such aptamers might possess unique sequences or use known scaffolds with variations to alter specificity (98). For example, the Mg2+-I riboswitch might be co-opted to bind softer metals by replacing its oxygen ligands with nitrogen. Conversely, adding an equatorial oxygen ligand could favor pentagonal bipyramidal coordination of Ca2+, which is maintained at 100–300 nm in bacteria (99). Perhaps the most likely effector is iron because it is the fourth most abundant element in the earth's crust and is essential for most life. A precedent exists for small RNA regulation of iron-storage proteins in bacteria (100). Overall, the discovery of new and possibly rare metalloriboswitches will require traditional bioinformatic methods along with targeted searches (30). All evidence suggests that the number of metal-sensing RNAs will continue to grow (11) and that this niche of genetic control is worth further consideration on the path to restoring a waning antimicrobial armamentarium.
Footnotes
This work was supported in part by National Institutes of Health Grant GM063162 from NIGMS (to J. E. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Breaker R. R. (2011) Prospects for riboswitch discovery and analysis. Mol. Cell 43, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Serganov A., and Nudler E. (2013) A decade of riboswitches. Cell 152, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sherwood A. V., and Henkin T. M. (2016) Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374 [DOI] [PubMed] [Google Scholar]
- 4. Breaker R. R. (2012) Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 4, a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roth A., and Breaker R. R. (2009) The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blount K. F., Megyola C., Plummer M., Osterman D., O'Connell T., Aristoff P., Quinn C., Chrusciel R. A., Poel T. J., Schostarez H. J., Stewart C. A., Walker D. P., Wuts P. G., and Breaker R. R. (2015) Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob. Agents Chemother. 59, 5736–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howe J. A., Wang H., Fischmann T. O., Balibar C. J., Xiao L., Galgoci A. M., Malinverni J. C., Mayhood T., Villafania A., Nahvi A., Murgolo N., Barbieri C. M., Mann P. A., Carr D., Xia E., et al. (2015) Selective small-molecule inhibition of an RNA structural element. Nature 526, 672–677 [DOI] [PubMed] [Google Scholar]
- 8. Howe J. A., Xiao L., Fischmann T. O., Wang H., Tang H., Villafania A., Zhang R., Barbieri C. M., and Roemer T. (2016) Atomic resolution mechanistic studies of ribocil: a highly selective unnatural ligand mimic of the E. coli FMN riboswitch. RNA Biol. 13, 946–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee E. R., Blount K. F., and Breaker R. R. (2009) Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 6, 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ott E., Stolz J., Lehmann M., and Mack M. (2009) The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 6, 276–280 [DOI] [PubMed] [Google Scholar]
- 11. McCown P. J., Corbino K. A., Stav S., Sherlock M. E., and Breaker R. R. (2017) Riboswitch diversity and distribution. RNA 23, 10.1261/rna.061234.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J. X., Lee E. R., Morales D. R., Lim J., and Breaker R. R. (2008) Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell 29, 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li S., Hwang X. Y., Stav S., and Breaker R. R. (2016) The yjdF riboswitch candidate regulates gene expression by binding diverse azaaromatic compounds. RNA 22, 530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Breaker R. R., Atilho R. M., Malkowski S. N., Nelson J. W., and Sherlock M. E. (2017) The biology of free guanidine as revealed by riboswitches. Biochemistry 56, 345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dann C. E. 3rd, Wakeman C. A., Sieling C. L., Baker S. C., Irnov I., and Winkler W. C. (2007) Structure and mechanism of a metal-sensing regulatory RNA. Cell 130, 878–892 [DOI] [PubMed] [Google Scholar]
- 16. Furukawa K., Ramesh A., Zhou Z., Weinberg Z., Vallery T., Winkler W. C., and Breaker R. R. (2015) Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol. Cell 57, 1088–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Price I. R., Gaballa A., Ding F., Helmann J. D., and Ke A. (2015) Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol. Cell 57, 1110–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dambach M., Sandoval M., Updegrove T. B., Anantharaman V., Aravind L., Waters L. S., and Storz G. (2015) The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol. Cell 57, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baker J. L., Sudarsan N., Weinberg Z., Roth A., Stockbridge R. B., and Breaker R. R. (2012) Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335, 233–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nierhaus K. H. (2014) Mg2+, K+, and the ribosome. J. Bacteriol. 196, 3817–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Draper D. E., Grilley D., and Soto A. M. (2005) Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34, 221–243 [DOI] [PubMed] [Google Scholar]
- 22. Sigel R. K., and Pyle A. M. (2007) Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 107, 97–113 [DOI] [PubMed] [Google Scholar]
- 23. Schuwirth B. S., Borovinskaya M. A., Hau C. W., Zhang W., Vila-Sanjurjo A., Holton J. M., and Cate J. H. (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 [DOI] [PubMed] [Google Scholar]
- 24. Ma Z., Jacobsen F. E., and Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramesh A., and Winkler W. C. (2010) Magnesium-sensing riboswitches in bacteria. RNA Biol. 7, 77–83 [DOI] [PubMed] [Google Scholar]
- 26. Ibarra J. A., and Steele-Mortimer O. (2009) Salmonella–the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11, 1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moomaw A. S., and Maguire M. E. (2008) The unique nature of Mg2+ channels. Physiology 23, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Centers for Disease Control and Prevention (2014) Multistate outbreak of multidrug-resistant Salmonella Heidelberg infections linked to foster farms brand chicken (final update). http://www.cdc.gov/salmonella/heidelberg-10-13/
- 29. Rothrock M. J. Jr., Ingram K. D., Gamble J., Guard J., Cicconi-Hogan K. M., Hinton A. Jr., and Hiett K. L. (2015) The characterization of Salmonella enterica serotypes isolated from the scalder tank water of a commercial poultry processing plant: recovery of a multidrug-resistant Heidelberg strain. Poult. Sci. 94, 467–472 [DOI] [PubMed] [Google Scholar]
- 30. Cromie M. J., Shi Y., Latifi T., and Groisman E. A. (2006) An RNA sensor for intracellular Mg2+. Cell 125, 71–84 [DOI] [PubMed] [Google Scholar]
- 31. Hollands K., Proshkin S., Sklyarova S., Epshtein V., Mironov A., Nudler E., and Groisman E. A. (2012) Riboswitch control of rho-dependent transcription termination. Proc. Natl. Acad. Sci. U.S.A. 109, 5376–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spinelli S. V., Pontel L. B., García Véscovi E., and Soncini F. C. (2008) Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol. Lett. 280, 226–234 [DOI] [PubMed] [Google Scholar]
- 33. Park S. Y., Cromie M. J., Lee E. J., and Groisman E. A. (2010) A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao G., Kong W., Weatherspoon-Griffin N., Clark-Curtiss J., and Shi Y. (2011) Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J. 30, 1485–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merino S., Gavín R., Altarriba M., Izquierdo L., Maguire M. E., and Tomás J. M. (2001) The MgtE Mg2+ transport protein is involved in Aeromonas hydrophila adherence. FEMS Microbiol. Lett. 198, 189–195 [DOI] [PubMed] [Google Scholar]
- 36. Ramesh A., Wakeman C. A., and Winkler W. C. (2011) Insights into metalloregulation by M-box riboswitch RNAs via structural analysis of manganese-bound complexes. J. Mol. Biol. 407, 556–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wakeman C. A., Ramesh A., and Winkler W. C. (2009) Multiple metal-binding cores are required for metalloregulation by M-box riboswitch RNAs. J. Mol. Biol. 392, 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin J. E., Waters L. S., Storz G., and Imlay J. A. (2015) The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet. 11, e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nies D. H. (1999) Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750 [DOI] [PubMed] [Google Scholar]
- 40. Ma Z., Faulkner M. J., and Helmann J. D. (2012) Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Mol. Microbiol. 86, 1144–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Archibald F. S., and Duong M. N. (1984) Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 158, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Posey J. E., and Gherardini F. C. (2000) Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653 [DOI] [PubMed] [Google Scholar]
- 43. Papp-Wallace K. M., and Maguire M. E. (2006) Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60, 187–209 [DOI] [PubMed] [Google Scholar]
- 44. Jabado N., Jankowski A., Dougaparsad S., Picard V., Grinstein S., and Gros P. (2000) Natural resistance to intracellular infections: Natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192, 1237–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kehres D. G., Zaharik M. L., Finlay B. B., and Maguire M. E. (2000) The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36, 1085–1100 [DOI] [PubMed] [Google Scholar]
- 46. Zaharik M. L., Cullen V. L., Fung A. M., Libby S. J., Kujat Choy S. L., Coburn B., Kehres D. G., Maguire M. E., Fang F. C., and Finlay B. B. (2004) The Salmonella enterica serovar typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1G169 murine typhoid model. Infect. Immun. 72, 5522–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi Y., Zhao G., and Kong W. (2014) Genetic analysis of riboswitch-mediated transcriptional regulation responding to Mn2+ in Salmonella. J. Biol. Chem. 289, 11353–11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li C., Tao J., Mao D., and He C. (2011) A novel manganese efflux system, YebN, is required for virulence by Xanthomonas oryzae pv. oryzae. PLoS ONE 6, e21983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waters L. S., Sandoval M., and Storz G. (2011) The Escherichia coli mntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J. Bacteriol. 193, 5887–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Veyrier F. J., Boneca I. G., Cellier M. F., and Taha M. K. (2011) A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog. 7, e1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barrick J. E., Corbino K. A., Winkler W. C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I., Wickiser J. K., and Breaker R. R. (2004) New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 101, 6421–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meyer M. M., Hammond M. C., Salinas Y., Roth A., Sudarsan N., and Breaker R. R. (2011) Challenges of ligand identification for riboswitch candidates. RNA Biol. 8, 5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bock C. W., Katz A. K., Markham G. D., and Glusker J. P. (1999) Manganese as a replacement for magnesium and zinc: Functional comparison of the divalent ions. J. Am. Chem. Soc. 121, 7360–7372 [Google Scholar]
- 54. Lemire J. A., Harrison J. J., and Turner R. J. (2013) Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384 [DOI] [PubMed] [Google Scholar]
- 55. Nies D. H. (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339 [DOI] [PubMed] [Google Scholar]
- 56. Parkinson J. S., and Houts S. E. (1982) Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Outten C. E., and O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 [DOI] [PubMed] [Google Scholar]
- 58. Macomber L., Elsey S. P., and Hausinger R. P. (2011) Fructose-1,6-bisphosphate aldolase (class II) is the primary site of nickel toxicity in Escherichia coli. Mol. Microbiol. 82, 1291–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barras F., and Fontecave M. (2011) Cobalt stress in Escherichia coli and Salmonella enterica: molecular bases for toxicity and resistance. Metallomics 3, 1130–1134 [DOI] [PubMed] [Google Scholar]
- 60. Centers for Disease Control and Prevention. (2016) Listeria outbreaks. http://www.cdc.gov/listeria/outbreaks/
- 61. Lim S. Y., Yap K. P., and Thong K. L. (2016) Comparative genomics analyses revealed two virulent Listeria monocytogenes strains isolated from ready-to-eat food. Gut Pathog. 8, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kanduti D., Sterbenk P., and Artnik B. (2016) Fluoride: a review of use and effects on health. Mater. Sociomed. 28, 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adler P., Armstrong W. D., Bell M. E., Bhussry B. R., Buttner W., Cremer H.-D., Demole V., Ericcson Y., Gedalia I., Hodge H. C., Jenkins G. N., Jolly S. S., Largent E. J., Leone N. C., Ludwig T. G., et al. (1970) Fluorides and human health. World Health Organization Monograph Series, No. 59, pp. 1–115, Geneva, Switzerland [Google Scholar]
- 64. Arthus M., and Gavelle J. (1903) Effect of sodium fluoride at 1% on a yeast. Comptes Rendus Des Seances De La Societe De Biologie Et De Ses Filiales 55, 1481–1483 [Google Scholar]
- 65. Marquis R. E., Clock S. A., and Mota-Meira M. (2003) Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev. 26, 493–510 [DOI] [PubMed] [Google Scholar]
- 66. Abrams S., Beltrán-Aguilar E., Martinez-Mier E. A., Kumar J., Slade G. D., and Gooch B. (2015) Water fluoridation: safety, effectiveness and value in oral health: a symposium at the 2014 annual meeting of the American and Canadian associations for dental research. J. Can. Dent. Assoc. 80, f16. [PubMed] [Google Scholar]
- 67. Garcia M. G., and Borgnino L. (2015) in Fluorine: Chemistry, Analysis, Function and Effects (Preedy V. R., ed), pp. 1–21, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 68. Warburg O., and Christian W. (1942) Isolation and crystallisation of the fermenting process of enolase. Biochemische Zeitschrift 310, 384–421 [Google Scholar]
- 69. Qin J., Chai G., Brewer J. M., Lovelace L. L., and Lebioda L. (2006) Fluoride inhibition of enolase: crystal structure and thermodynamics. Biochemistry 45, 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Reed G. H., Poyner R. R., Larsen T. M., Wedekind J. E., and Rayment I. (1996) Structural and mechanistic studies of enolase. Curr. Opin. Struct. Biol. 6, 736–743 [DOI] [PubMed] [Google Scholar]
- 71. Jin Y., Richards N. G., Waltho J. P., and Blackburn G. M. (2017) Metal fluorides as analogs for studies on phosphoryl transfer enzymes. Angew. Chem. Int. Ed. Engl. 56, 4110–4128 [DOI] [PubMed] [Google Scholar]
- 72. Stockbridge R. B., Robertson J. L., Kolmakova-Partensky L., and Miller C. (2013) A family of fluoride-specific ion channels with dual-topology architecture. eLife 2, e01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li S., Smith K. D., Davis J. H., Gordon P. B., Breaker R. R., and Strobel S. A. (2013) Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 19018–19023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stockbridge R. B., Lim H. H., Otten R., Williams C., Shane T., Weinberg Z., and Miller C. (2012) Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc. Natl. Acad. Sci. U.S.A. 109, 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Breaker R. R. (2012) New insight on the response of bacteria to fluoride. Caries Res. 46, 78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weinberg Z., Wang J. X., Bogue J., Yang J., Corbino K., Moy R. H., and Breaker R. R. (2010) Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 11, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ren A., Rajashankar K. R., and Patel D. J. (2012) Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486, 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Montange R. K., and Batey R. T. (2008) Riboswitches: Emerging themes in RNA structure and function. Annu. Rev. Biophys. 37, 117–133 [DOI] [PubMed] [Google Scholar]
- 79. Peselis A., and Serganov A. (2014) Structure and function of pseudoknots involved in gene expression control. Wiley Interdiscip. Rev. RNA 5, 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shannon R. D. (1976) Revised effective ionic-radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Cryst. A 32, 751–767 [Google Scholar]
- 81. Thorsell A. G., Persson C., Gräslund S., Hammarström M., Busam R. D., and Hallberg B. M. (2009) Crystal structure of human diphosphoinositol phosphatase 1. Proteins 77, 242–246 [DOI] [PubMed] [Google Scholar]
- 82. Fabrichniy I. P., Lehtiö L., Tammenkoski M., Zyryanov A. B., Oksanen E., Baykov A. A., Lahti R., and Goldman A. (2007) A trimetal site and substrate distortion in a family II inorganic pyrophosphatase. J. Biol. Chem. 282, 1422–1431 [DOI] [PubMed] [Google Scholar]
- 83. Sudarsan N., Cohen-Chalamish S., Nakamura S., Emilsson G. M., and Breaker R. R. (2005) Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem. Biol. 12, 1325–1335 [DOI] [PubMed] [Google Scholar]
- 84. Blount K. F., and Breaker R. R. (2006) Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24, 1558–1564 [DOI] [PubMed] [Google Scholar]
- 85. Breaker R. R. (2009) Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 4, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mulhbacher J., Brouillette E., Allard M., Fortier L. C., Malouin F., and Lafontaine D. A. (2010) Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 6, e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Matzner D., and Mayer G. (2015) (Dis)similar analogues of riboswitch metabolites as antibacterial lead compounds. J. Med. Chem. 58, 3275–3286 [DOI] [PubMed] [Google Scholar]
- 88. Wakeman C. A., Goodson J. R., Zacharia V. M., and Winkler W. C. (2014) Assessment of the requirements for magnesium transporters in Bacillus subtilis. J. Bacteriol. 196, 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Anjem A., Varghese S., and Imlay J. A. (2009) Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72, 844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nies D. H., and Silver S. (1995) Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14, 186–199 [DOI] [PubMed] [Google Scholar]
- 91. Kloosterman T. G., van der Kooi-Pol M. M., Bijlsma J. J., and Kuipers O. P. (2007) The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 65, 1049–1063 [DOI] [PubMed] [Google Scholar]
- 92. Versieck J. (1985) Trace elements in human body fluids and tissues. Crit. Rev. Clin. Lab. Sci. 22, 97–184 [DOI] [PubMed] [Google Scholar]
- 93. Gu M., and Imlay J. A. (2013) Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol. Microbiol. 89, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nelson J. W., Plummer M. S., Blount K. F., Ames T. D., and Breaker R. R. (2015) Small molecule fluoride toxicity agonists. Chem. Biol. 22, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li S., and Breaker R. R. (2012) Fluoride enhances the activity of fungicides that destabilize cell membranes. Bioorg. Med. Chem. Lett. 22, 3317–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nelson J. W., Zhou Z., and Breaker R. R. (2014) Gramicidin D enhances the antibacterial activity of fluoride. Bioorg. Med. Chem. Lett. 24, 2969–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Maret W. (2016) The metals in the biological periodic system of the elements: concepts and conjectures. Int. J. Mol. Sci. 17, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Weinberg Z., Nelson J. W., Lünse C. E., Sherlock M. E., and Breaker R. R. (2017) Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc. Natl. Acad. Sci. U.S.A. 114, E2077–E2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dominguez D. C. (2004) Calcium signalling in bacteria. Mol. Microbiol. 54, 291–297 [DOI] [PubMed] [Google Scholar]
- 100. Massé E., and Gottesman S. (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]