Abstract
Dendritic spines are heterogeneous and exist with various morphologies. Altered spine morphology might underlie the cognitive deficits in neurodevelopmental disorders such as autism, but how different subtypes of dendritic spines are selectively maintained along development is still poorly understood. Spine maturation requires spontaneous activity of N-methyl-d-aspartate (NMDA) receptor and local dendritic protein synthesis. STRN4 (also called zinedin) belongs to the striatin family of scaffold proteins, and some of the potential striatin-interacting proteins are encoded by autism risk genes. Although previous studies have demonstrated their localization in dendritic spines, the function of various striatin family members in the neuron remains unknown. Here, we demonstrate that Strn4 mRNA is present in neuronal dendrites, and the local expression of STRN4 protein depends on NMDA receptor activation. Notably, STRN4 is preferentially expressed in mushroom spines, and STRN4 specifically maintains mushroom spines but not thin spines and filopodia through interaction with the phosphatase PP2A. Our findings have therefore unraveled the local expression of STRN4 as a novel mechanism for the control of dendritic spine morphology.
Keywords: dendritic spine; N-methyl-d-aspartate receptor (NMDA receptor, NMDAR); scaffold protein; signal transduction; synapse
Introduction
Dendritic spines are protrusions on the dendritic arbors of the postsynaptic neuron where the majority of excitatory neurotransmission occurs. The morphology of dendritic spines is heterogeneous and can be classified into three main categories: the short stubby spines; the thin spines with elongated necks and small heads; and the mushroom-shaped spines with larger heads (1, 2). In addition, there are long and thin filopodia, which are thought to represent either the precursors of dendritic spines during synaptogenesis (3) or unstable spines that are undergoing turnover in the adult brain (4). The various morphologies of dendritic spines are believed to confer different structural and functional properties of the synapse, including the spine turnover rate, compartmentalization of Ca2+ signal, and the size of the postsynaptic density (5, 6). Moreover, altered spine morphology is associated with various neurodevelopmental disorders such as Fragile-X syndrome, Rett syndrome, and Angelman syndrome, which are monogenetic disorders that share significant phenotypic overlap with autism spectrum disorder (ASD)3 (7–10). These studies therefore underscore the importance of the proper control of dendritic spine morphology in neurons for normal cognitive function.
The morphology of dendritic spines is tightly regulated by spontaneous glutamatergic neurotransmission and neuronal activity (11). Blockade of action potential or ionotropic glutamate receptors in dissociated hippocampal neurons or hippocampal slices changes the motility, stability, and morphology of dendritic spines (12–15). The importance of synaptic activity in refining neuronal connectivity in vivo was also demonstrated by intravital imaging of neurons in live animals, where sensory experience affects the turnover of dendritic spines in the cerebral cortex (16). Interestingly, the two types of ionotropic glutamate receptors, namely α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-d-aspartate (NMDA) receptors, have distinct roles in the regulation of spine morphogenesis. Whereas pharmacological blockade of AMPA receptor affects dendritic spine maintenance and results in spine loss (13, 17, 18), inhibition of NMDA receptor changes the morphology to long and thin filopodia without affecting the total spine number (13). Because different subtypes of dendritic spines can be regulated independently (19), it is plausible that spontaneous activation of NMDA receptor might selectively promote the maintenance of mushroom spines as well as suppress the formation of thin spines and filopodia. Nonetheless, despite numerous studies elucidating the mechanisms that underlie dendritic spine morphogenesis, little is known about the molecular basis of how dendritic spines with different morphologies are selectively maintained and eliminated.
The presence of polyribosomes and a specific subset of mRNA transcripts near dendritic spines allows local protein synthesis to occur rapidly in response to synaptic activities. Multiple lines of evidence suggests that dendritic protein synthesis could play a crucial role in the regulation of spine morphology and maturation. For example, an increased number of filopodia is observed in neurons lacking the RNA-binding protein FMRP, which is associated with impaired dendritic RNA transport and aberrant local mRNA translation (20, 21). Depletion of eEF2 kinase or manipulation of the mechanistic target of rapamycin (mTOR) pathway, which regulates dendritic protein synthesis, also impairs spine maintenance and maturation (15, 22, 23). Notably, neurons carrying the mutated 3′-untranslated region of the Bdnf gene and hence the deficit in dendritic targeting of the Bdnf mRNA exhibit abnormal spine density and morphology (24, 25), suggesting that the dendritic synthesis of specific mRNAs is a key process to control spine morphology. Early studies have identified ∼350 mRNA transcripts in neuronal processes (26–28), but a recent study using RNA sequencing has indicated the presence of a much more extensive transcriptome in the hippocampal neuropil, with 2,250 mRNA transcripts being identified (29). Importantly, many of these mRNAs encode proteins that have unknown function in neuron. Characterizing these dendritic mRNA transcripts, which may undergo translation in response to synaptic activity, should potentially identify novel mechanisms in the activity-dependent control of dendritic spine morphology.
One of the transcripts in the hippocampal neuropil identified by Cajigas et al. (29) was the mRNA encoding STRN4 (also called zinedin). STRN4 is a multidomain scaffold protein that belongs to the striatin family, which also includes STRN1 (also called striatin) and STRN3 (also called SG2NA). The striatin family is characterized by the presence of four protein-protein interaction domains, namely the caveolin-binding domain, coiled-coil domain, Ca2+-calmodulin (CaM)-binding domain, and tryptophan-aspartate (WD)-repeat domain (30). All three striatin family members are predominantly expressed in the central nervous system and are present in dendritic spines (31, 32). Many interacting partners of the striatin scaffold proteins have been identified. Unlike many synaptic scaffold proteins that bind to ion channels and other scaffolding networks, the interacting proteins of the striatin family identified thus far are mostly signaling proteins, including multiple kinases such as members of the germinal center kinase family (MST3, MST4, and YSK1), NCK-interacting kinase (NIK), and TRAF2- and NCK-interacting kinase (TNIK) (33). The protein complex, named striatin-interacting phosphatase and kinase (STRIPAK), might therefore act as a signaling hub at the plasma membrane to regulate multiple cellular functions, rather than providing a structural scaffold in organizing the postsynaptic density. The STRIPAK complex also contains the phosphatase PP2A, and it has been suggested that the presence of PP2A maintains the activity of the various kinases within the complex under control by direct dephosphorylation (33, 34). Notably, despite information on their subcellular localization and interacting proteins, the function of striatin family proteins in neuron remains unknown. Dysfunction of some of the STRIPAK components, including MST3 and cortactin-binding protein 2 (CTTNBP2), are linked to ASD (35, 36). Given the association between altered spine morphology and ASD, we hypothesize that STRN4 might represent a novel key protein in the control of spine morphology, and impaired function of the STRIPAK complex might contribute to the altered neuronal connectivity in autism. Here, we provide evidence for a new regulatory mechanism for spine morphology that involves local dendritic expression of STRN4 driven by the NMDA receptor.
Results
Spontaneous NMDA receptor activity locally regulates the expression of STRN4 in dendritic spine
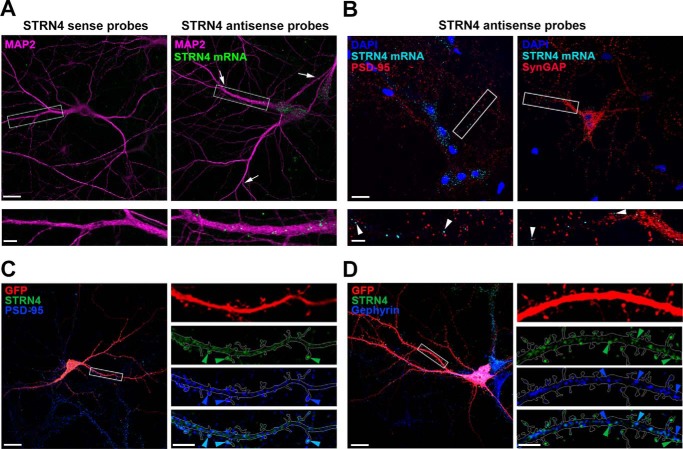
As a first step to understand the function of STRN4 in synapse development, we examine the expression of Strn4 mRNA and protein in the neuron as well as their regulation by synaptic activity. A previous large-scale transcriptome study revealed that Strn4 mRNA is one of the transcripts detected in the hippocampal neuropil (29). We verified the dendritic localization of Strn4 mRNA in dissociated hippocampal neurons by high-resolution and sensitive fluorescence in situ hybridization (FISH) as described by previous studies (29, 37). Using a set of antisense oligonucleotide probes targeting STRN4 in FISH followed by immunocytochemistry with antibody against the somatodendritic marker MAP2, we found that Strn4 mRNA existed as distinct puncta that were detected not only in the cell soma but also in distal dendrites (Fig. 1A). The puncta on dendrites were absent when neurons were hybridized with the sense STRN4 probe set, indicating specificity of the in situ hybridization signal. The Strn4 mRNA puncta were not co-localized with the excitatory postsynaptic proteins PSD-95 or SynGAP, but some of them were situated close to (∼1 μm) excitatory synapses (Fig. 1B). This is consistent with the notion that dendritic mRNA granules are not located in the heads of dendritic spines or synapses, but instead are present in the dendrites or at the base of the spines to dynamically survey individual synapses (38, 39).
Figure 1.
Strn4 mRNA is dendritically localized, and STRN4 protein is mainly present at excitatory synapses but not inhibitory synapses. A, in situ hybridization was performed on dissociated rat hippocampal neurons (17 DIV) using nucleotide probes targeting STRN4 (STRN4 antisense probes). Discrete Strn4 mRNA puncta (green) were observed in MAP2-positive dendrites (magenta); some of the puncta (arrows) were localized ∼60–80 μm away from the cell body. The green puncta were absent when neurons were hybridized with the sense probes. B, hippocampal neurons (17 DIV) were subjected to Strn4 mRNA in situ hybridization followed by PSD-95 or SynGAP immunofluorescence staining. Some Strn4 mRNA granules (cyan, arrowheads) were in close proximity, but not precisely co-localized, to PSD-95 or SynGAP puncta at excitatory synapses (red). C and D, hippocampal neurons (21 DIV) expressing GFP (red) were co-stained with STRN4 protein (green) and the excitatory postsynaptic protein PSD-95 (blue) or the inhibitory postsynaptic protein gephyrin (blue). Merged images are shown in the bottom right panels, and examples of co-localized puncta are shown (light blue arrowheads). C, co-localization of the STRN4 puncta (green arrowheads) with PSD-95 (blue arrowheads) on dendritic spines. D, many STRN4 puncta (green arrowheads) in the dendritic shaft were distinct from the gephyrin puncta (blue arrowheads). Scale bars, 20 μm (upper panels in A and B and left panels in C and D) or 5 μm (lower panels in A and B and right panels in C and D).
Next, the subcellular localization of STRN4 protein in hippocampal neuron was examined. Specificity of the STRN4 antibody in immunocytochemistry was validated by RNAi-mediated knockdown of STRN4 (Fig. 4, B and C, and see below). Excitatory and inhibitory synapses are preferentially present in dendritic spines and dendritic shaft, respectively. STRN4 was previously reported to be localized at excitatory synapses (31, 32, 35), but it is not clear whether it is preferentially expressed at excitatory synapses or present at both excitatory and inhibitory synapses. We found that STRN4 existed as discrete puncta in both the dendritic shaft and dendritic spines. The majority (78.1 ± 4.1%; 50 STRN4 puncta from three dendrites) of the spine-associated STRN4 puncta were co-localized with the excitatory postsynaptic protein PSD-95 (Fig. 1C). In contrast, relatively few (24.4 ± 5.6%; 46 puncta from two dendrites) of the non-spine-associated STRN4 puncta in the dendritic shaft were co-localized with the inhibitory postsynaptic scaffold protein gephyrin (Fig. 1D). These findings therefore indicate that STRN4 is mostly expressed at excitatory synapses.
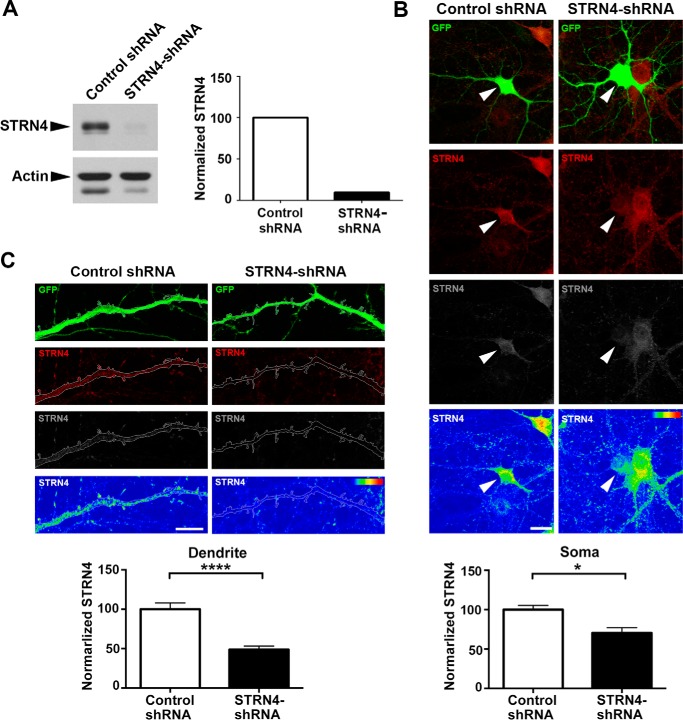
Figure 4.
shRNA-mediated knockdown of STRN4 expression in neuron. A, STRN4 shRNA (or control shRNA) was transfected into rat cortical neurons by nucleofection, and lysate was collected at 5 DIV for Western blot analysis. The intensity of STRN4 was normalized with that of actin. B and C, immunofluorescence staining of STRN4 in hippocampal neurons co-transfected with GFP and the STRN4 shRNA (or control shRNA). Fluorescence intensity of STRN4 (red) was indicated in the heat maps (lower panels). STRN4 immunoreactivity was significantly reduced in both the soma (arrowheads in B) and the dendrites (C) of the GFP-positive, STRN4-shRNA-transfected neurons, as compared with the control shRNA-transfected neurons (eight dendrites from five neurons were quantified for each condition). Data are mean ± S.E.; *, p < 0.05; ****, p < 0.0001; Student's t test. Scale bars, 20 μm (B) and 10 μm (C).
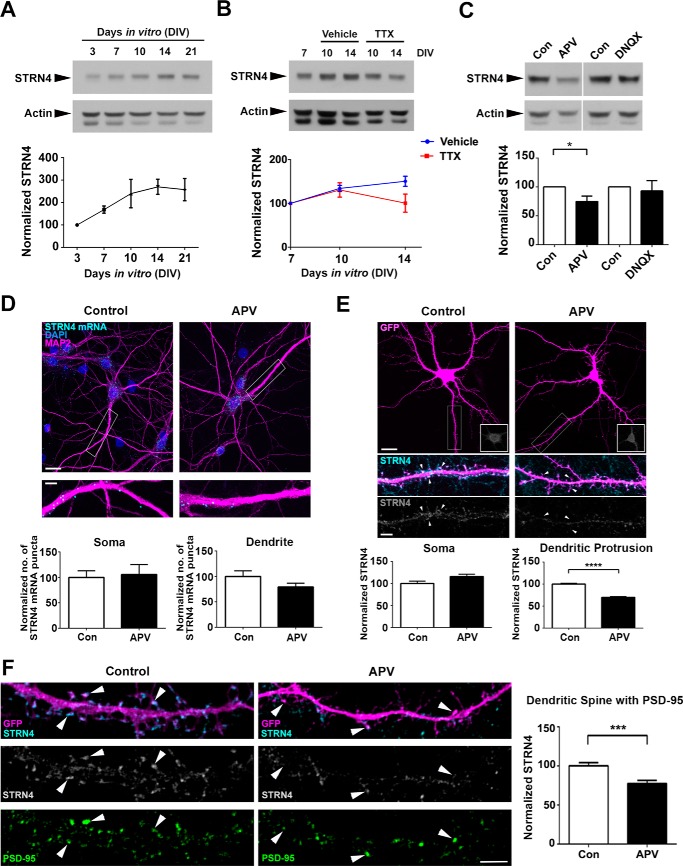
Expression of proteins that are encoded by dendritic mRNAs often depends on synaptic activity (40). We therefore determined how STRN4 protein expression is regulated in the hippocampal neuron. Consistent with our hypothesis that STRN4 regulates dendritic spine maturation, the expression of STRN4 protein was up-regulated between 7 and 14 days in vitro (DIV) (Fig. 2A), a period when there is a drastic change from thin spines to mushroom spines (41). Interestingly, the up-regulation of STRN4 expression from 7 to 14 DIV was abolished by blocking the spontaneous action potential with tetrodotoxin (TTX) (Fig. 2B), indicating that the developmental increase in STRN4 expression depends on neuronal activity.
Figure 2.
STRN4 expression at excitatory synapses depends on spontaneous NMDA receptor activity. A, Western blot analysis showing that the expression of STRN4 protein increased along maturation of hippocampal neurons. The intensity of STRN4 was normalized with that of actin. Results were pooled from three independent experiments. B, developmental up-regulation of STRN4 expression at 14 DIV was blocked by TTX (2 μm). Results were pooled from four independent experiments. C, hippocampal neurons (16 DIV) were treated with vehicle (Con), the NMDA receptor antagonist APV (100 μm), or the AMPA and kainate receptor antagonist DNQX (20 μm) for 48 h, and STRN4 protein expression was examined by Western blotting. Results were pooled from four independent experiments. D, Strn4 mRNA level was examined in dissociated rat hippocampal neurons (18 DIV) by in situ hybridization following treatment with vehicle (control) or APV (100 μm) for 48 h. The number of Strn4 mRNA (cyan) granules in the cell soma or dendrites was not significantly different between the two conditions (n = 17); p > 0.05, Student's t test. E, representative images of GFP-transfected hippocampal neurons treated with vehicle (control) or APV (100 μm) at 16 DIV for 48 h, followed by immunofluorescence staining using STRN4 antibody. STRN4 intensity (cyan or gray) in dendritic protrusions (arrowheads) and cell soma (insets) were quantified. APV treatment specifically reduced STRN4 fluorescence intensity in dendritic protrusions but not the cell soma (15 dendrites from 7 neurons were quantified for each condition). F, double immunofluorescence staining of GFP-transfected hippocampal neurons by STRN4 and PSD-95 antibodies indicated that the synaptic STRN4 was significantly reduced by APV (100 μm for 48 h) in dendritic spines containing PSD-95 clusters (arrowheads). (9 dendrites from 6 to 7 neurons were quantified for each condition.) Data are mean ± S.E. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001; Student's t test. Scale bars, 20 μm (upper panels in D and E) or 5 μm (lower panels in D–F).
Firing of action potential triggers glutamate release from excitatory neurons. The findings that STRN4 expression depends on spontaneous neuronal activity prompted us to ask whether activation of ionotropic glutamate receptors are required for STRN4 expression. We found that selective blockade of NMDA receptor by APV significantly reduced STRN4 protein expression in hippocampal neurons, whereas the AMPA and kainate receptor antagonist 6,7-dinitroquinoxaline-2,3-dione (DNQX) did not affect STRN4 expression (Fig. 2C). The regulation of STRN4 expression by the NMDA receptor occurred at the protein rather than the mRNA level, because APV did not significantly affect the Strn4 mRNA expression revealed by either in situ hybridization or semi-quantitative RT-PCR (Fig. 2D and data not shown).
Next, we ask whether the NMDA receptor-dependent expression of STRN4 protein occurs in a cell-wide or compartment-specific manner by performing immunofluorescence staining of hippocampal neurons after blockade of NMDA receptors. Notably, compared with the vehicle control, APV treatment reduced STRN4 expression specifically in the dendritic protrusions but not the cell soma (Fig. 2E). Double immunofluorescence staining with PSD-95 antibody verified the reduction of synaptic STRN4 expression after APV treatment (Fig. 2F). Taken together, these findings indicate that the expression of STRN4 is regulated locally at excitatory synapses by spontaneous activation of the NMDA receptor.
STRN4 is expressed preferentially in mushroom and stubby spines
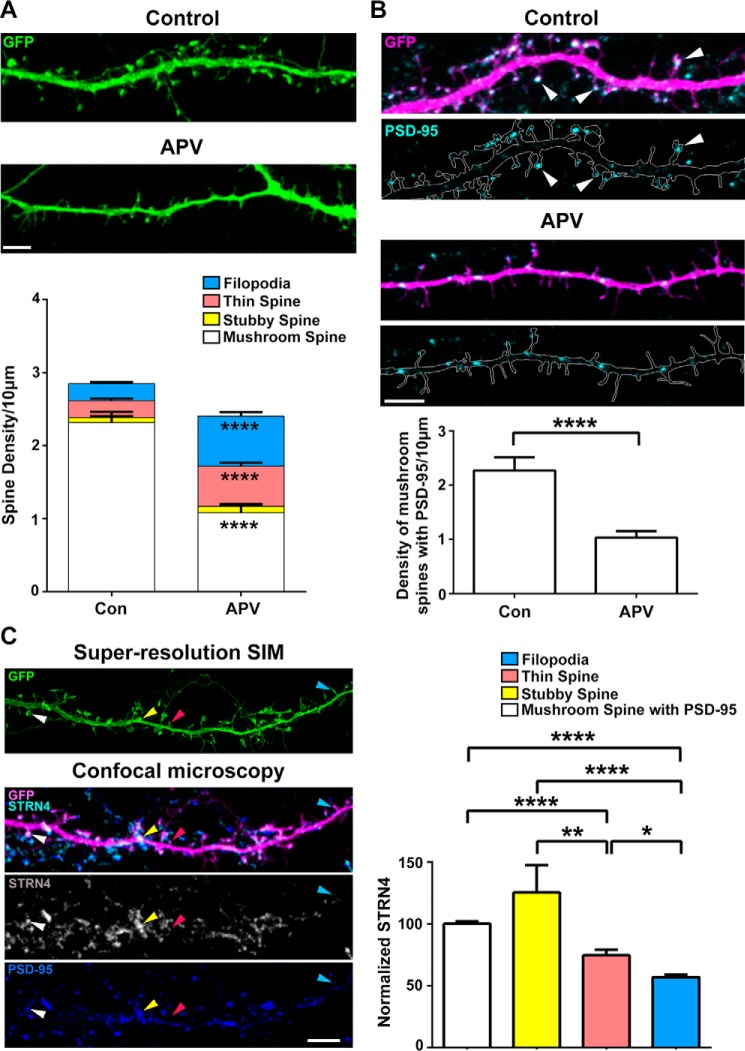
Activation of NMDA receptors, but not AMPA receptors, controls dendritic spine morphology (13, 41). We confirmed that in our culture condition, blocking NMDA receptor activity of hippocampal neurons for 48 h by APV resulted in similar change of spine morphology from mushroom spines toward thin spines and filopodia (Fig. 3A and supplemental Fig. S1). APV treatment also reduced the number of mushroom spines that contained PSD-95 clusters at their spine heads (Fig. 3B). Given the dependence of its local expression on the NMDA receptor, we postulate that STRN4 might represent one of the key players in regulating spine morphology by being expressed preferentially and hence selectively maintaining the PSD-95-positive mushroom spines. Although the presence of the striatin family of scaffold proteins in dendritic spines has been previously demonstrated, its relative abundance in different types of dendritic spines has not been explored. To better define the various spine types, the images of dendritic spines were acquired using Super-resolution Structured Illumination Microscopy (SR-SIM), which has about 2-fold higher axial resolution than confocal microscopy (42) and is suitable for visualization of cellular structures around 200 nm, such as the neck of a dendritic spine (see supplemental Fig. S2) (43). We found that STRN4 expression differed in distinct subtypes of dendritic spines, with the highest STRN4 immunoreactivity present in stubby spines and the spine heads of mushroom-shaped spines that contained PSD-95 clusters, whereas STRN4 immunoreactivity was significantly reduced in thin spines and filopodia (Fig. 3C). The expression level of STRN4 therefore correlates with spine morphology, and the down-regulation of STRN4 upon NMDA receptor blockade is correlated with the selective loss of mushroom spines where STRN4 is abundant.
Figure 3.
STRN4 is preferentially expressed in mushroom and stubby spines. A, representative confocal images of GFP-transfected hippocampal neurons treated with vehicle or APV (100 μm) at 16 DIV for 48 h. APV treatment significantly reduced the density of mushroom spines, and concomitantly increased the density of thin spines and filopodia. Results were pooled from two experiments; 35 dendrites from 17 neurons were quantified for each condition. Data are mean ± S.E.; ****, p < 0.0001; Student's t test. B, PSD-95 immunofluorescence staining on GFP-transfected hippocampal neurons treated with vehicle or APV (100 μm) at 16 DIV for 48 h, indicating the loss of PSD-95-containing mushroom spines after APV treatment. Results were pooled from two independent experiments (28 dendrites from 16–17 neurons were quantified for each condition). Data are mean ± S.E.; ****, p < 0.0001; Student's t test. C, representative images taken by Super-resolution SIM and confocal microscopy of a hippocampal neuronal dendrite (18 DIV) expressing GFP (green or magenta) and co-stained by STRN4 and PSD-95 antibodies. Examples of mushroom (white arrowheads), stubby (yellow arrowheads), thin (red arrowheads) spines and filopodia (blue arrowheads) are indicated. The average signal intensity of STRN4 was significantly higher in mushroom and stubby spines than thin spines and filopodia. Results were pooled from two independent experiments (880 spines from 27 dendrites of 15 neurons were quantified). Data are mean ± S.E.; *, p < 0.05; **, p < 0.01; ****, p < 0.0001; one-way ANOVA, Tukey's multiple comparisons test. Scale bars, 5 μm.
Knockdown of STRN4 results in the selective loss of mushroom spines
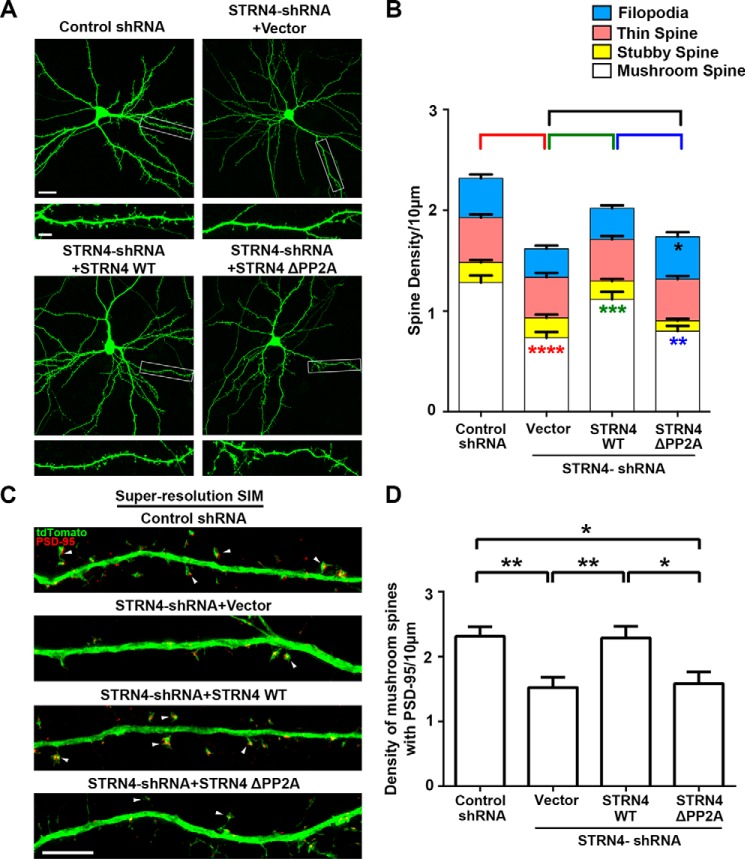
To directly address whether STRN4 is indeed crucial for regulating spine morphology, we created the short hairpin RNA (shRNA) construct that specifically targeted STRN4. The knockdown efficiency of the STRN4 shRNA was confirmed by both Western blot analysis and immunocytochemistry (Fig. 4). Next, we examined the density of various subtypes of spines after transfecting the STRN4 shRNA or control shRNA together with enhanced GFP construct into dissociated hippocampal neurons. Compared with the control shRNA, introduction of the STRN4 shRNA triggered significant reduction of the density of mushroom spines. Importantly, the density of stubby spines, thin spines, and filopodia was not affected (Fig. 5, A and B, and supplemental Fig. S3). To confirm the loss of mushroom spines that contain excitatory synapses, hippocampal neurons were co-transfected with the expression construct of PSD-95 intrabody to label the endogenous PSD-95 clusters (44), followed by SIM imaging. Knockdown of STRN4 significantly reduced the number of PSD-95-containing mushroom spines (Fig. 7, C and D). To rule out the potential off-target effect, we performed a rescue experiment using the STRN4 construct, which carried silent mutations at the region where the shRNA was designed and therefore became RNAi-resistant. We found that the loss of mushroom spines and PSD-95 clusters upon knockdown of STRN4 could be reversed by co-expression of the RNAi-resistant STRN4, indicating that STRN4 is required for maintaining proper spine morphology (Fig. 7, A–D, and supplemental Fig. S5).
Figure 5.
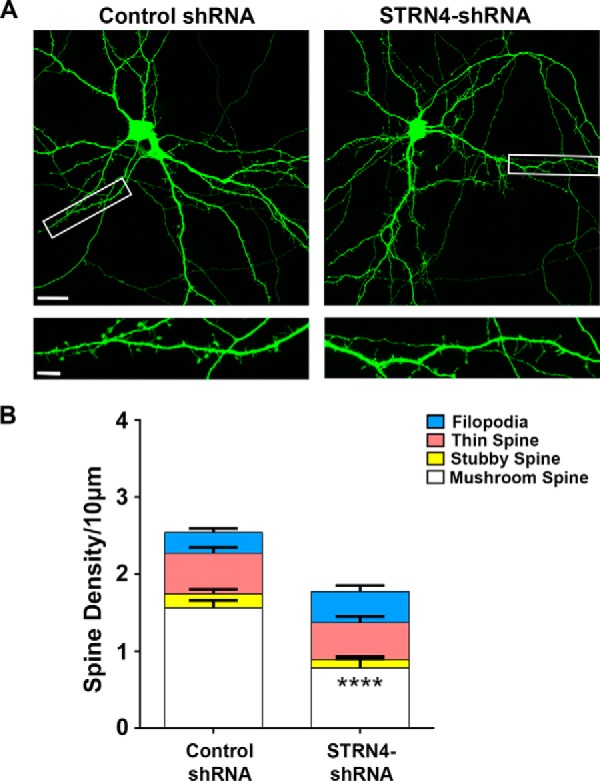
Knockdown of STRN4 results in the specific loss of mushroom spines. A, hippocampal neurons (15 DIV) were co-transfected with GFP and the STRN4 shRNA or control shRNA. Cells were fixed and stained with GFP antibody at 17 DIV. Representative images are shown. B, knockdown of STRN4 significantly reduced the density of mushroom spines, but not the stubby spines, thin spines, and filopodia. 15–16 dendrites from six neurons were quantified for each condition. Data are mean ± S.E.; ****, p < 0.0001, Student's t test (see also supplemental Fig. S3). Scale bars, 20 μm (upper) and 5 μm (lower).
Figure 7.
Function of STRN4 in dendritic spine maturation requires interaction with PP2A. A, hippocampal neurons (16 DIV) were co-transfected with GFP and control shRNA, STRN4 shRNA with or without the RNAi-resistant wild-type, or the PP2A-binding-deficient (ΔPP2A) STRN4 mutant construct. Cells were fixed and stained with GFP antibody at 18 DIV. Representative confocal images are shown. Scale bars, 20 μm (upper) and 5 μm (lower). B, quantification revealed that only the wild type (WT) but not the STRN4-ΔPP2A mutant could reverse the loss of mushroom spines induced by STRN4-shRNA. Introduction of the STRN-ΔPP2A mutant also led to increased density of filopodia. Results were pooled from three independent experiments. 71–79 dendrites from 33 to 39 neurons in each group were quantified (see also supplemental Fig. S5). Red asterisks denoted the difference in mushroom spine density between control shRNA and STRN4-shRNA+ vector; green asterisks denoted the difference in mushroom spine density between STRN4-shRNA+ vector and STRN4-shRNA+ STRN4-WT. Blue asterisk denoted difference in mushroom spine density between STRN4-shRNA+ STRN4-WT and STRN4-shRNA+ STRN4-ΔPP2A. Black asterisk denoted difference in filopodia density between STRN4-shRNA+ vector and STRN4-shRNA+ STRN4-ΔPP2A. C, hippocampal neurons (16 or 17 DIV) were co-transfected with tdTomato and PSD-95 intrabody together with control shRNA, STRN4 shRNA with or without the RNAi-resistant wild-type or PP2A-binding-deficient (ΔPP2A) STRN4 mutant construct. Cells were fixed and stained with RFP and GFP antibodies 2 days post-transfection to visualize tdTomato and PSD-95 intrabody, respectively. Representative images acquired by super-resolution SIM showing mushroom spines that contained PSD-95 clusters in the spine heads (arrowheads). Scale bar, 5 μm. D, density of PSD-95-positive mushroom spines were quantified. Wild-type STRN4, but not the STRN4-ΔPP2A mutant, rescued the spine loss induced by STRN4-shRNA. Results were pooled from two independent experiments (34–37 dendrites from 25 to 28 neurons in each group were quantified). Data are mean ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; one-way ANOVA, Tukey's multiple comparisons test.
STRN4 regulates the maintenance of mushroom spines by interacting with PP2A
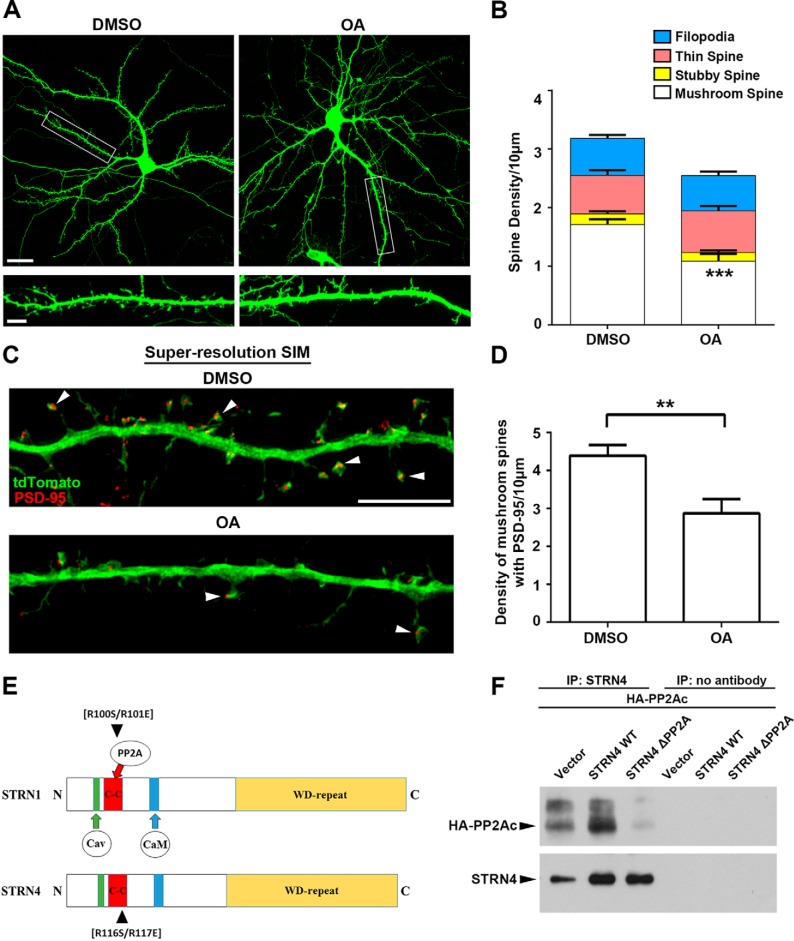
STRN4 is one of the components of the STRIPAK protein complex, which contains multiple kinases and the phosphatase PP2A (33). Biochemical studies have shown that PP2A can inactivate the different kinases by dephosphorylation, and striatin proteins can act as the PP2A regulatory subunit to regulate the activity or substrate specificity of the phosphatase (34, 45). Compared with the other phosphatases such as PP1 and PP2B (calcineurin), the role of PP2A in synapse development is not clear. We first demonstrated that, similar to the knockdown of STRN4, inhibition of PP2A by okadaic acid triggered the selective loss of mushroom spines without affecting the other three types of spines (Fig. 6, A and B, and supplemental Fig. S4). Likewise, the number of mushroom spines containing PSD-95 clusters was also reduced by okadaic acid treatment (Fig. 6, C and D). We therefore propose that STRN4 regulates spine morphology by acting as the PP2A regulatory subunit. To address whether STRN4 function indeed depends on PP2A, we created a PP2A-binding-deficient mutant of STRN4 based on the previous identification of Arg-100/101 in the coil-coiled domain of STRN1 as the critical amino acid residues for the binding to PP2A (34). Amino acid sequence alignment revealed that the two critical arginine residues in STRN1 corresponded to Arg-116/117 of STRN4 (Fig. 6E). We found that STRN4 interacted with the catalytic subunit of PP2A, and this interaction was abolished by the STRN4 mutant that harbored the R116S/R117E mutations (Fig. 6F). Next, RNAi-resistant wild-type or the PP2A-binding-deficient R116S/R117E mutant was introduced together with STRN4-shRNA into hippocampal neurons, and the effect on spine morphology of the transfected neurons was examined. Although co-expression of wild-type STRN4 reversed the loss of mushroom spines induced by the shRNA, the PP2A-binding-deficient STRN4 mutant failed to rescue the spine defects. Furthermore, compared with wild-type STRN4, co-expression of the PP2A-binding-deficient STRN4 mutant resulted in the formation of more filopodia (Fig. 7, A and B). The reduction of PSD-95-containing mushroom spines triggered by STRN4-shRNA was also rescued by wild-type STRN4 but not the PP2A-binding-deficient mutant (Fig. 7, C and D). Taken together, our findings collectively indicate that the tight regulation of PP2A by STRN4 in the STRIPAK complex is critical to govern dendritic spine morphology.
Figure 6.
PP2A inhibition affects spine morphology. A, hippocampal neurons (13 DIV) were transfected with GFP. Cells were treated with DMSO (vehicle control) or okadaic acid (OA, 30 nm) for 4 h at 18 DIV before fixation and immunostained with GFP antibody. Representative images taken by confocal microscopy are shown. Scale bars, 20 μm (upper) or 5 μm (lower). B, inhibition of PP2A by okadaic acid significantly reduced the density of mushroom spines, but not the stubby spines, thin spines, and filopodia. (15 dendrites from 9 neurons in each group were quantified.) C, hippocampal neurons (13 DIV) were transfected with tdTomato and PSD-95 intrabody and treated with DMSO (vehicle control) or okadaic acid (30 nm) for 4 h at 18 DIV before fixation. Neurons were immunostained by RFP and GFP antibodies to visualize the tdTomato and PSD-95, respectively. Representative images acquired by SR-SIM showing mushroom spines that contained PSD-95 clusters in the spine heads (arrowheads). D, okadaic acid treatment significantly reduced the density of PSD-95-positive mushroom spines. Results were pooled from two independent experiments (21–22 dendrites from 16 to 19 neurons for each group were quantified). Scale bar, 5 μm. E, schematic diagram illustrating the conserved domains between STRN1 and STRN4, and the position of the two arginine residues that are important for binding to PP2A. F, plasmid construct of HA-tagged PP2A catalytic subunit α isoform (PP2AC) was co-transfected with wild-type STRN4 or STRN4-R116S/R117E double mutant (ΔPP2A) into HEK-293T cells. Much less STRN4-ΔPP2A was co-immunoprecipitated with PP2AC compared with the wild-type STRN4. Data are mean ± S.E.; **, p < 0.01; ***, p < 0.001, Student's t test.
Discussion
Although the importance of spontaneous synaptic activity in the maintenance and maturation of dendritic spines has been well established, the underlying molecular mechanism is not well understood. Moreover, it is not clear why different subtypes of glutamate ionotropic receptors have distinct roles in spine morphogenesis. One plausible explanation is that activation of the NMDA receptor but not the other ionotropic glutamate receptors promotes the expression of specific proteins that mediate the morphological changes of dendritic spines. Nonetheless, very few activity-induced proteins that regulate spine morphology have actually been characterized. Here, we found that the expression of STRN4 is specifically regulated by spontaneous activity of NMDA but not AMPA receptors, and STRN4 is preferentially expressed in mushroom and stubby spines. We further demonstrated that down-regulation of STRN4 partially mimicked the spine phenotypes after blockade of NMDA receptors, i.e. the selective loss of mushroom spines containing PSD-95 clusters, whereas the thin spines and filopodia where STRN4 expression is low is not affected by the STRN4 knockdown. Our study therefore provides new insights into the mechanism by which spontaneous NMDA receptor activity selectively maintains different subtypes of dendritic spines.
The down-regulation of STRN4 after blockade of spontaneous NMDA receptor activity is not cell-wide but occurs locally at dendritic spines. This is not attributed to a redistribution of STRN4 from dendritic spines to the dendritic shaft because we did not observe a corresponding increase in STRN4 immunoreactivity in the dendritic shaft after APV treatment (data not shown). The total Strn4 mRNA expression is not changed after APV treatment, suggesting that the effect is at the post-transcriptional level. One possibility is that APV treatment results in the reduction of local dendritic translation of STRN4. We found that the Strn4 mRNA is localized in proximal and distal dendrites of hippocampal neurons. A previous transcriptomic study has also identified Strn4 mRNA as a putative cargo of FMRP (46), a well characterized RNA-binding protein that transports mRNA to distal dendrites and regulates their local translation. It is noteworthy that there is a trend of reduced Strn4 mRNA in the dendrite, but not cell soma, after APV treatment (Fig. 2D). Blockade of the NMDA receptor might therefore reduce both the dendritic transport of Strn4 mRNA and the translation process at the synapse, which together account for the reduced local expression of STRN4 in dendritic spines. The specific enrichment of STRN4 in mushroom spines might also be attributed to STRN4 local translation, as mushroom spines may possess greater translation capacity than thin spines and filopodia.
How does STRN4 promote the maintenance of mushroom spines? Many components of the STRIPAK complex have been identified, and one of them is the phosphatase PP2A. Despite an early study showing the presence of PP2A at the synapse (47), the physiological role of PP2A in synapse development and function is not well understood. It is difficult to dissect the synaptic function of PP2A because of its numerous and diverse substrates and its dependence on various regulatory subunits (48). It is possible to elucidate the mechanism of PP2A at the synapse by focusing on specific regulatory subunits (49). We have demonstrated that PP2A interacts with STRN4. More importantly, a rescue experiment using PP2A-binding-deficient mutant indicates that PP2A is essential for STRN4 function in the control of spine morphology. Our findings will therefore facilitate further delineation of the synaptic role of PP2A by focusing on its action on different components of the STRIPAK complex in the regulation of spine morphogenesis. For example, the F-actin-binding protein cortactin, which binds to CTNNBP2 of the STRIPAK complex, is a putative PP2A substrate (35, 50). It is possible that STRN4 recruits PP2A to dephosphorylate cortactin and locally modulates actin cytoskeleton in mushroom spines. Alternatively, PP2A can directly dephosphorylate and negatively regulate multiple kinases, such as MST3, MST4, and YSK1, in the STRIPAK complex (34, 51). Given that hyperactivity of kinases such as Cdk5 and ERK is detrimental to synaptic function and connectivity (52, 53), STRN4 might control spine morphology through balancing the activities of various kinases within the STRIPAK complex at the synapse. Interestingly, increased dephosphorylation of ERK by PP2A was observed in FMRP knock-out neurons (54). Because Strn4 mRNA is a putative cargo of FMRP, it would be interesting to further investigate whether the transport and translation of Strn4 mRNA is dysregulated in neurons lacking FMRP, and how might it contribute to the immature spine phenotypes of the neurons.
Emerging studies suggest a link between altered synaptic connections and autism (55). CTTNBP2 is encoded by autism-risk gene, and it binds to and recruits STRN4 and other striatin family scaffold proteins to dendritic spines (35). One of the kinases in the STRIPAK complex is MST3, which is required for spine maturation by phosphorylation and activation of another kinase TAOK2 (36), a protein affected in ASD (56). STRN4 might therefore play a key role in orchestrating the local regulation of various signaling proteins for the proper spine development and maintenance, and it is possible that disruption of STRN4 function might be related to altered synaptic connectivity in autism. In this regard, it is noteworthy that the human STRN4 gene is located in chromosome 19q13.32, where deletion has been observed in ASD (57). Further investigation on the potential link between copy number variants of the Strn4 gene and ASD is warranted.
Experimental procedures
Antibodies, chemicals, and DNA constructs
Antibodies against rat STRN4 (NeuroMab), human STRN4 (Abcam), GFP (Invitrogen or Aves Labs), MAP2 (Sigma), actin (Sigma), influenza hemagglutinin (HA) protein (Cell Signaling), SynGAP (Thermo Fisher Scientific), PSD-95 (Thermo Fisher Scientific and Cell Signaling), RFP (Rockland), gephyrin (Synaptic Systems), and Alexa-conjugated secondary antibodies (Invitrogen), were purchased commercially. Horseradish peroxidase-conjugated goat anti-rabbit IgG or anti-mouse IgG were purchased from Cell Signaling. dl-APV and DNQX were purchased from TOCRIS, and TTX was from Abcam. The PSD-95 intrabody construct pCAG_PSD-95.FingR-eGFP-CCR5TC was purchased from Addgene (plasmid no. 46295) (44).
For knockdown of STRN4, a 19-nucleotide sequence (5′-CAGCGAGTACTGTTACAGT-3′) derived from the rat STRN4 nucleotide sequence was used to create the shRNA construct after subcloning into the pSUPER vector (Oligoengine). Full-length human STRN4 cDNA was amplified by PCR using the plasmid pF1KB3518 from Kazusa Genome Technologies as template, which contains the insert of the full-length human STRN4-coding region (transcript variant 2). All PCRs in this study were performed using high-fidelity Pfu DNA polymerase (Agilent Technologies, Inc.). Primers used were forward 5′-CGGCCAGGTACCGCCACCATGATGGAGGAGCGAGCGGC-3′ and reverse 5′-GTTGCACTCGAGTCATACGAAGACCTTGGCCAG-3′. The PCR product was then digested by KpnI and XhoI (New England Biolabs) and subsequently subcloned into the pcDNA3 vector. For making the RNAi-resistant human STRN4 (RNAir STRN4) expression construct, site-directed mutagenesis was performed using primers designed by the QuikChange Primer Design: forward 5′-GCTATGGGCAGCAACAGTGAATATTGCTATAGCGGCGGGGCAGAT-3′; and reverse 5′-ATCTGCCCCGCCGCTATAGCAATATTCACTGTTGCTGCCCATAGC-3′. The PCR product was digested by DpnI (New England Biolabs) in a 37 °C water bath for 3 h before transformation into Escherichia coli competent cells. The human STRN4 mutant R116S/R117E (STRN4-ΔPP2A) construct was created by site-directed mutagenesis using the primers 5′-GAATCTAAAGACGGACCTGGTGAGCGAGATCAAGATGCTAGAGTATGCG-3′ and reverse 5′-CGCATACTCTAGCATCTTGATCTCGCTCACCAGGTCCGTCTTTAGATTC-3′. The expression construct of rat PP2A catalytic subunit α isoform with HA tag at the amino terminus (HA-PP2Ac) was made by PCR using cDNA of rat hippocampal neurons as template. The restriction sites of KpnI and XhoI were introduced into primers forward 5′-CGGCCAGGTACCGCCACCATGTATCCATATGATGTTCCAGATTATGCTGACGAGAAGTTGTTCACCAAG-3′ and reverse 5′-GTTGCACTCGAGTTACAGGAAGTAGTCTGGGGTACGACGAGTGACATGTGGCTCGCC-3′. The resulting PCR product was subcloned into pcDNA3 vector between the KpnI and XhoI sites. The nucleotide sequence of each expression construct was verified by Sanger sequencing.
Primary cell culture and transfection
Primary hippocampal neurons and cortical neurons were prepared from embryonic day 18–19 embryos of Sprague-Dawley rats according to a previous study (58). Hippocampal neurons were cultured on 18-mm coverslips or 35-mm dishes coated with 1 mg/ml of poly-d-lysine (P0899, Sigma) at high density (1.4 × 105 cells per coverslip for dendritic spine analysis; 4–5 × 105 cells per 35-mm dish for biochemical analysis) or low density (0.4 × 105 cells per coverslip for immunocytochemistry), in Neurobasal medium supplemented with 2% B27 and 0.25% l-glutamine (Invitrogen). Cortical neurons were plated on 35-mm dishes (1 × 106 cells per dish) coated with 0.1 mg/ml poly-d-lysine and cultured with Neurobasal medium supplemented with 2% B27 and 0.5% l-glutamine. Cells were transfected with different plasmids using calcium phosphate precipitation as described previously (58). Electroporation of control shRNA or STRN4 shRNA into cortical neurons was carried out using the Neon nucleofector transfection system (Thermo Fisher Scientific), in which a total of 1 × 106 suspension cells were electroporated in each reaction with the parameters of 1600-V pulse voltage and 20-ms pulse width. After electroporation, cells were plated on 35-mm dishes and cultured for 5 days before Western blot analysis.
Pharmacological treatment of neurons, Western blot analysis, and immunoprecipitation
To examine the expression of STRN4, hippocampal neurons cultured on 35-mm dishes were lysed at 3, 7, 10, 14, and 21 DIV. To determine the effects of neuronal activity on STRN4 expression at different culture duration, hippocampal neurons were treated with TTX (2 μm) at 7 DIV and lysed at 10 or 14 DIV or were treated with APV (100 μm) or DNQX (20 μm) at 15–16 DIV and lysed after 48 h. Neurons were lysed by RIPA containing various protease and phosphatase inhibitors (10 μg/ml soybean trypsin inhibitor, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 2 μg/ml antipain, 30 nm okadaic acid, 5 mm benzamidine, 1 mm sodium orthovanadate, 1 mm PMSF, 1 mm sodium fluoride, 100 mm β-glycerophosphate). The lysate was incubated in a cold room with rocking for 45 min, and the cell debris was cleared by centrifugation at 13,000 rpm for 10 min at 4 °C. The protein extract was boiled in sample buffer (5× sample buffer: 300 mm Tris-HCl buffer, pH 6.8, 10% (w/v) SDS, 25% (v/v) β-mercaptoethanol, 50% (v/v) glycerol, 0.05% (w/v) bromphenol blue) for 5 min, separated by SDS-PAGE, and transferred onto PVDF membranes, followed by blocking with 5% skim milk in TBS with 0.1% Tween 20 (TBST) for 1 h at room temperature. The membrane was incubated in a cold room with primary antibody diluted in TBST containing 5% BSA overnight. After washing three times with TBST, membranes were incubated for 1 h at room temperature with HRP-conjugated secondary antibody diluted in 5% skim milk in TBST. The HRP signal was detected by ECL (Thermo Fisher Scientific) and quantified by densitometry using Photoshop software.
To investigate the interaction between STRN4 and HA-PP2Ac, HEK-293T cells cultured on 100-mm dishes with 80% confluence were transfected with various plasmids using Lipofectamine (Thermo Fisher Scientific) according to instructions from the manufacturer. Twenty four hours after transfection, the cells were washed by cold D-PBS and then washing buffer (135 mm NaCl, 10% glycerol, 20 mm Tris, pH 8) and lysed by Tris lysis buffer (135 mm NaCl, 1% Nonidet P-40, 10% glycerol, 20 mm Tris, pH 8) plus various protease and phosphatase inhibitors. Lysate was incubated in a cold room with rocking for 45 min and cleared from cell debris by centrifugation at 13,000 rpm for 10 min at 4 °C. Equal amount of lysate (1 mg) was incubated with STRN4 antibody (1:100, Abcam) in a cold room with rocking overnight. Immunoprecipitate was obtained after incubation with protein A-Sepharose beads (GE Healthcare) for 1 h in a cold room with rocking. Beads were washed four times with Tris lysis buffer containing various protease and phosphatase inhibitors, and proteins were eluted by boiling in sample buffer for 6 min. The eluate was collected by centrifugation at 13,000 rpm for 1 min at 4 °C and then subjected to SDS-PAGE and Western blot analysis.
Semi-quantitative RT-PCR
Hippocampal neurons cultured on 35-mm dishes were treated with APV (100 μm) at 15 DIV for 48 h, lysed by TRIzol (Invitrogen), and then subjected to RNA extraction using RNeasy mini kit (Qiagen) according to instructions from manufacturer. Total RNA was transcribed into cDNA by RT-Superscript kit (Invitrogen), and the cDNA was used as template for PCR. The primers used were as follows: STRN4 forward 5′-GAATCAGGGGGAGAAGAAGG-3′ and STRN4 reverse 5′-CATCCTCATCGCTGTCTTC-3′; GAPDH forward 5′-CCCTTCATTGACCTCAACTA-3′ and GAPDH reverse 5′-CCAAAGTTGTCATGGATGAC-3′. PCR products were amplified using a different number of cycles and analyzed on agarose gel. Images were taken by Gel Doc 1000 (Bio-Rad), and the band intensity was quantified by Photoshop software.
In situ hybridization
The subcellular localization of Strn4 mRNA in hippocampal neurons was examined using the ViewRNA ISH Cell Assay kit from Affymetrix. A set of sense and antisense probes was designed against rat STRN4 (NM_001107480.2) by Affymetrix. This assay was performed as described in the Affymetrix manual with the following modification. The protease QS treatment was eliminated to preserve the protein markers for subsequent immunocytochemistry. After completion of the in situ hybridization, cells were washed with PBS and incubated in blocking buffer (10% goat serum in PBS) for 30 min, followed by incubation at 4 °C overnight with MAP2 (1:500), PSD-95 (Thermo Fisher Scientific, 1:200) or SynGAP (1:200) antibody diluted in blocking buffer. After washing by PBS three times, cells were incubated for 1 h with Alexa-conjugated secondary antibody (1:1000) diluted in blocking buffer, washed by PBS three times, and mounted with gold antifade reagent (Life Technologies, Inc.). To compare the Strn4 mRNA level in neurons treated with vehicle or APV, the number of Strn4 mRNA puncta was manually counted in the cell body and dendrites of neurons. The total number of puncta in the cell body was then divided by the area of the soma measured by the MetaMorph software to calculate the number of puncta per unit area. The average number of Strn4 mRNA puncta in dendrites was determined after dividing the number of dendritic mRNA puncta by the number of dendritic intersections at 50 μm away from the center of the cell body.
Immunofluorescence staining, image acquisition, and quantitative analysis
Cells were fixed by 4% paraformaldehyde, 4% sucrose in PBS for 15 min at room temperature. After washing with PBS, cells were incubated with blocking buffer (0.4% Triton X-100 (v/v) and 1% BSA (w/v)) for 45 min at room temperature and incubated with primary antibody (GFP, 1:500; RFP, 1:500; STRN4 (NeuroMab) 1:100; PSD95 (Cell Signaling) 1:100; gephyrin, 1:100) in blocking buffer at 4 °C overnight. Cells were washed three times with washing buffer (0.02% Triton X-100 and 0.25% BSA in D-PBS), incubated with Alexa-conjugated secondary antibody (1:1000 diluted in 0.02% Triton X-100 and 1% BSA in PBS) at room temperature for 1 h, followed by washing twice in washing buffer and once by D-PBS, and mounted with hydromount medium (National Diagnostics). To stain GFP-transfected neurons for confocal microscopy imaging, cells were incubated with GFP antibody (1:2000) in GDB buffer at 4 °C overnight. After washing three times with wash buffer (20 mm phosphate buffer and 0.5 m NaCl), neurons were incubated with Alexa-488-conjugated secondary antibodies (1:2000 diluted in GDB buffer) at room temperature for 1 h, then washed three times by the washing buffer, and mounted with hydromount medium.
Carl Zeiss LSM 700 or 780 confocal microscopes installed with Zen digital imaging software were used to acquire fluorescent images at a resolution of 1024 × 1024 pixels and a pinhole of 1 airy unit for each channel. For fluorescence signal intensity analysis and Strn4 mRNA in situ hybridization, LSM 780 was used to acquire Z-stack images using a ×40 oil-immersion objective (NA_1.40) with the following parameters: ×1 optical zoom, averaged two times, scan speed 6–8, 0.40-μm interval with 16-bit dynamic range. The images from the same experiment were captured using identical acquisition settings, except for the GFP or tdTomato (RFP) staining, which served to visualize dendritic arbors and spines. The staining intensity was quantified by MetaMorph software. For the analysis of dendritic spine morphology of GFP-transfected hippocampal neurons, Z-stack images were taken by LSM 700 using a ×63 oil-immersion objective (NA_1.40) with the following parameters: 1 AU pinhole, ×0.5 optical zoom, averaged two times, scan speed 6–8, 0.35-μm or 0.40-μm interval. For SR-SIM, which can achieve a resolution beyond theoretical limits of resolution (200 nm) for light microscopy (43), images were taken by Zeiss Elyra S1 SIM installed with Zen 2.3 imaging software with the following parameters: 1024 × 1024-pixel frame size, ×63 oil-immersion objective (NA_1.40), 3 rotations grating, 5 phases, 1.0 optical zoom, averaging 1 time, optimal Z interval set by software for Z-stack mode. Segments (60–75 μm for images taken by LSM 700 and LSM 780 (confocal microscopy), 35–50 μm for images taken by SIM) of two to three basal dendrites or secondary apical dendrites of a transfected neuron were analyzed from maximum-projected images.
To classify dendritic spines, the length (L), head width (H), and neck width (N) of each individual spine were measured manually using the MetaMorph software. Based on the criteria described by Harris et al. (59), spines were quantitatively classified into mushroom, stubby, or thin spines using (i) the head to neck diameter ratio (H/N), which has a value considerably greater than 1 for mushroom spine to reflect its characteristic bulbous head and narrow neck (60); and (ii) the length to neck ratio (L/N), which is used to distinguish between the short stubby spines and the long thin spines (61). Accordingly, mushroom spines were defined as those having H/N ≥1.5; stubby spines were defined as those with H/N ≤1 and L/N ≤1; thin spines had the ratio of 1 ≤ H/N <1.5 and 1.5≤ L/N ≤3. The cutoff value of 1.5 for the H/N ratio has been used in other studies (18, 60, 62, 63). Filopodia were defined as those with the ratio of H/N <1.2 and L/N >3 based on the study by Grutzendler et al. (64). Those spines that did not fit into any of the criteria described above were excluded for quantitative analysis. Because the criteria described by Harris et al. (59) are qualitative, some variations in the H/N and L/N cutoff values exist among different studies. To determine whether these variabilities in cutoff ratios would affect our conclusions, additional analyses of spine morphology using alternative H/N and L/N values of 1.7 or 2.0 based on previous studies (65, 66) were performed and are shown in supplemental Figs. S1A, S3A, S4A, and S5A. Mushroom spines were defined as having H/N ≥1.7, and thin spines had the ratio of 1≤ H/N <1.7 and 1.7 ≤ L/N ≤ 3; in supplemental Figs. S1B, S3B, S4B, and S5B, mushroom spines were defined as having H/N ≥ 2, and thin spines had the ratio of 1≤ H/N <2 and 2≤ L/N≤ 3. The quantification of the density of different spine types in every experiment was performed blinded.
Statistical analysis
Data are represented as mean ± S.E. in quantitative analysis. Statistical analysis was performed by Student's t test or ANOVA followed by Tukey post hoc test. Statistical significance was defined as p < 0.05.
Author contributions
K. O. L. supervised the project. K. O. L., L. L., and L. H.-Y. L. designed the experiments. L. H.-Y. L., L. L., and Q. L. performed the experiments. L. L. and L. H.-Y. L. analyzed the data. K. O. L. wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to Nancy Ip (Hong Kong University of Science and Technology) for the STRN4 shRNA construct, Dan Wang (iCeMS of Kyoto University) for the helpful advice on in situ hybridization, and members of the Lai laboratory for technical assistance and helpful discussions. We also thank the Imaging Core Facility of the Faculty of Medicine at the University of Hong Kong for assistance with image acquisition and analysis.
This work was supported by Research Grants Council of Hong Kong General Research Fund 16100814 and 17135816, Early Career Scheme Grant 27119715, and the University of Hong Kong Seed Funding Programme for Basic Research Grants 201407159004 and 201511159170. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5.
- ASD
- autism spectrum disorder
- DNQX
- 6,7-dinitroquinoxaline-2,3-dione
- ANOVA
- analysis of variance
- STRIPAK
- striatin interacting phosphatase and kinase
- RFP
- red fluorescent protein
- DIV
- days in vitro
- TTX
- tetrodotoxin
- SR-SIM
- super-resolution structured illumination microscopy
- H
- head
- N
- neck
- L
- length
- APV
- 2-amino-5-phosphopentanoic acid.
References
- 1. Bourne J. N., and Harris K. M. (2008) Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 31, 47–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yuste R., and Bonhoeffer T. (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 5, 24–34 [DOI] [PubMed] [Google Scholar]
- 3. Ziv N. E., and Smith S. J. (1996) Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17, 91–102 [DOI] [PubMed] [Google Scholar]
- 4. Nimchinsky E. A., Sabatini B. L., and Svoboda K. (2002) Structure and function of dendritic spines. Annu. Rev. Physiol. 64, 313–353 [DOI] [PubMed] [Google Scholar]
- 5. Bourne J., and Harris K. M. (2007) Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 17, 381–386 [DOI] [PubMed] [Google Scholar]
- 6. Holtmaat A. J., Trachtenberg J. T., Wilbrecht L., Shepherd G. M., Zhang X., Knott G. W., and Svoboda K. (2005) Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45, 279–291 [DOI] [PubMed] [Google Scholar]
- 7. Irwin S. A., Galvez R., and Greenough W. T. (2000) Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex 10, 1038–1044 [DOI] [PubMed] [Google Scholar]
- 8. Kim H., Kunz P. A., Mooney R., Philpot B. D., and Smith S. L. (2016) Maternal loss of Ube3a impairs experience-driven dendritic spine maintenance in the developing visual cortex. J. Neurosci. 36, 4888–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Penzes P., Cahill M. E., Jones K. A., VanLeeuwen J.-E., and Woolfrey K. M. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Phillips M., and Pozzo-Miller L. (2015) Dendritic spine dysgenesis in autism related disorders. Neurosci. Lett. 601, 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lai K.-O., and Ip N. Y. (2013) Structural plasticity of dendritic spines: The underlying mechanisms and its dysregulation in brain disorders. Biochim. Biophys. Acta 1832, 2257–2263 [DOI] [PubMed] [Google Scholar]
- 12. Lai K.-O., Liang Z., Fei E., Huang H., and Ip N. Y. (2015) Cyclin-dependent kinase 5 (Cdk5)-dependent phosphorylation of p70 ribosomal S6 kinase 1 (S6K) is required for dendritic spine morphogenesis. J. Biol. Chem. 290, 14637–14646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKinney R. A., Capogna M., Dürr R., Gähwiler B. H., and Thompson S. M. (1999) Miniature synaptic events maintain dendritic spines via AMPA receptor activation. Nat. Neurosci. 2, 44–49 [DOI] [PubMed] [Google Scholar]
- 14. Papa M., and Segal M. (1996) Morphological plasticity in dendritic spines of cultured hippocampal neurons. Neuroscience 71, 1005–1011 [DOI] [PubMed] [Google Scholar]
- 15. Verpelli C., Piccoli G., Zibetti C., Zanchi A., Gardoni F., Huang K., Brambilla D., Di Luca M., Battaglioli E., and Sala C. (2010) Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J. Neurosci. 30, 5830–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rochefort N. L., and Konnerth A. (2012) Dendritic spines: from structure to in vivo function. EMBO Rep. 13, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kang M.-G., Guo Y., and Huganir R. L. (2009) AMPA receptor and GEF-H1/Lfc complex regulates dendritic spine development through RhoA signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 106, 3549–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang Z., Zhan Y., Shen Y., Wong C. C., Yates J. R. 3rd, Plattner F., Lai K.-O., and Ip N. Y. (2016) The pseudokinase CaMKv is required for the activity-dependent maintenance of dendritic spines. Nat. Commun. 7, 13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spiga S., Talani G., Mulas G., Licheri V., Fois G. R., Muggironi G., Masala N., Cannizzaro C., Biggio G., Sanna E., and Diana M. (2014) Hampered long-term depression and thin spine loss in the nucleus accumbens of ethanol-dependent rats. Proc. Natl. Acad. Sci. U.S.A. 111, E3745–E3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dictenberg J. B., Swanger S. A., Antar L. N., Singer R. H., and Bassell G. J. (2008) A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muddashetty R. S., Nalavadi V. C., Gross C., Yao X., Xing L., Laur O., Warren S. T., and Bassell G. J. (2011) Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 42, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhattacharya A., Kaphzan H., Alvarez-Dieppa A. C., Murphy J. P., Pierre P., and Klann E. (2012) Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron 76, 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tavazoie S. F., Alvarez V. A., Ridenour D. A., Kwiatkowski D. J., and Sabatini B. L. (2005) Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat. Neurosci. 8, 1727–1734 [DOI] [PubMed] [Google Scholar]
- 24. An J. J., Gharami K., Liao G.-Y., Woo N. H., Lau A. G., Vanevski F., Torre E. R., Jones K. R., Feng Y., Lu B., and Xu B. (2008) Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orefice L. L., Waterhouse E. G., Partridge J. G., Lalchandani R. R., Vicini S., and Xu B. (2013) Distinct roles for somatically and dendritically synthesized brain-derived neurotrophic factor in morphogenesis of dendritic spines. J. Neurosci. 33, 11618–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T.-M., Chi M. C., Chong J., et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 [DOI] [PubMed] [Google Scholar]
- 27. Poon M. M., Choi S.-H., Jamieson C. A., Geschwind D. H., and Martin K. C. (2006) Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J. Neurosci. 26, 13390–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhong J., Zhang T., and Bloch L. M. (2006) Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cajigas I. J., Tushev G., Will T. J., tom Dieck S., Fuerst N., and Schuman E. M. (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benoist M., Gaillard S., and Castets F. (2006) The striatin family: a new signaling platform in dendritic spines. J. Physiol. Paris 99, 146–153 [DOI] [PubMed] [Google Scholar]
- 31. Benoist M., Baude A., Tasmadjian A., Dargent B., Kessler J. P., and Castets F. (2008) Distribution of zinedin in the rat brain. J. Neurochem. 106, 969–977 [DOI] [PubMed] [Google Scholar]
- 32. Gaillard S., Bailly Y., Benoist M., Rakitina T., Kessler J. P., Fronzaroli-Molinières L., Dargent B., and Castets F. (2006) Targeting of proteins of the striatin family to dendritic spines: role of the coiled-coil domain. Traffic 7, 74–84 [DOI] [PubMed] [Google Scholar]
- 33. Hwang J., and Pallas D. C. (2014) STRIPAK complexes: structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 47, 118–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gordon J., Hwang J., Carrier K. J., Jones C. A., Kern Q. L., Moreno C. S., Karas R. H., and Pallas D. C. (2011) Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 12, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y.-K., Chen C.-Y., Hu H.-T., and Hsueh Y.-P. (2012) CTTNBP2, but not CTTNBP2NL, regulates dendritic spinogenesis and synaptic distribution of the striatin-PP2A complex. Mol. Biol. Cell 23, 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ultanir S. K., Yadav S., Hertz N. T., Oses-Prieto J. A., Claxton S., Burlingame A. L., Shokat K. M., Jan L. Y., and Jan Y.-N. (2014) MST3 kinase phosphorylates TAO1/2 to enable myosin Va function in promoting spine synapse development. Neuron 84, 968–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taylor A. M., Berchtold N. C., Perreau V. M., Tu C. H., Li Jeon N., and Cotman C. W. (2009) Axonal mRNA in uninjured and regenerating cortical mammalian axons. J. Neurosci. 29, 4697–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Doyle M., and Kiebler M. A. (2011) Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30, 3540–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoon Y. J., Wu B., Buxbaum A. R., Das S., Tsai A., English B. P., Grimm J. B., Lavis L. D., and Singer R. H. (2016) Glutamate-induced RNA localization and translation in neurons. Proc. Natl. Acad. Sci. U.S.A. 113, E6877–E6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Butko M. T., Savas J. N., Friedman B., Delahunty C., Ebner F., Yates J. R. 3rd, and Tsien R. Y. (2013) In vivo quantitative proteomics of somatosensory cortical synapses shows which protein levels are modulated by sensory deprivation. Proc. Natl. Acad. Sci. U.S.A. 110, E726–E735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Papa M., Bundman M. C., Greenberger V., and Segal M. (1995) Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J. Neurosci. 15, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gustafsson M. G. (2000) Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 [DOI] [PubMed] [Google Scholar]
- 43. Schouten M., De Luca G. M., Alatriste González D. K., de Jong B. E., Timmermans W., Xiong H., Krugers H., Manders E. M., and Fitzsimons C. P. (2014) Imaging dendritic spines of rat primary hippocampal neurons using structured illumination microscopy. J. Vis. Exp. 10.3791/51276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gross G. G., Junge J. A., Mora R. J., Kwon H.-B., Olson C. A., Takahashi T. T., Liman E. R., Ellis-Davies G. C., McGee A. W., Sabatini B. L., Roberts R. W., and Arnold D. B. (2013) Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moreno C. S., Park S., Nelson K., Ashby D., Hubalek F., Lane W. S., and Pallas D. C. (2000) WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275, 5257–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Darnell J. C., Van Driesche S. J., Zhang C., Hung K. Y., Mele A., Fraser C. E., Stone E. F., Chen C., Fak J. J., Chi S. W., Licatalosi D. D., Richter J. D., and Darnell R. B. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strack S., Barban M. A., Wadzinski B. E., and Colbran R. J. (1997) Differential inactivation of postsynaptic density-associated and soluble Ca2+/calmodulin-dependent protein kinase II by protein phosphatases 1 and 2A. J. Neurochem. 68, 2119–2128 [DOI] [PubMed] [Google Scholar]
- 48. Janssens V., Longin S., and Goris J. (2008) PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem. Sci. 33, 113–121 [DOI] [PubMed] [Google Scholar]
- 49. Liu X., Huai J., Endle H., Schlüter L., Fan W., Li Y., Richers S., Yurugi H., Rajalingam K., Ji H., Cheng H., Rister B., Horta G., Baumgart J., Berger H., Laube G., Schmitt U., Schmeisser M. J., Boeckers T. M., Tenzer S., Vlachos A., Deller T., Nitsch R., and Vogt J. (2016) PRG-1 regulates synaptic plasticity via intracellular PP2A/β1-integrin signaling. Dev. Cell 38, 275–290 [DOI] [PubMed] [Google Scholar]
- 50. Chen Y.-K., and Hsueh Y.-P. (2012) Cortactin-binding protein 2 modulates the mobility of cortactin and regulates dendritic spine formation and maintenance. J. Neurosci. 32, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pombo C. M., Bonventre J. V., Molnar A., Kyriakis J., and Force T. (1996) Activation of a human Ste20-like kinase by oxidant stress defines a novel stress response pathway. EMBO J. 15, 4537–4546 [PMC free article] [PubMed] [Google Scholar]
- 52. Lai K.-O., and Ip N. Y. (2009) Recent advances in understanding the roles of Cdk5 in synaptic plasticity. Biochim. Biophys. Acta 1792, 741–745 [DOI] [PubMed] [Google Scholar]
- 53. Yang K., Cao F., Sheikh A. M., Malik M., Wen G., Wei H., Ted Brown W., and Li X. (2013) Up-regulation of Ras/Raf/ERK1/2 signaling impairs cultured neuronal cell migration, neurogenesis, synapse formation, and dendritic spine development. Brain Struct. Funct. 218, 669–682 [DOI] [PubMed] [Google Scholar]
- 54. Kim S. H., Markham J. A., Weiler I. J., and Greenough W. T. (2008) Aberrant early-phase ERK inactivation impedes neuronal function in fragile X syndrome. Proc. Natl. Acad. Sci. U.S.A. 105, 4429–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Walsh C. A., Morrow E. M., and Rubenstein J. L. (2008) Autism and brain development. Cell 135, 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. de Anda F. C., Rosario A. L., Durak O., Tran T., Gräff J., Meletis K., Rei D., Soda T., Madabhushi R., Ginty D. D., Kolodkin A. L., and Tsai L.-H. (2012) Autism spectrum disorder susceptibility gene TAOK2 affects basal dendrite formation in the neocortex. (Report). Nat. Neurosci 15, 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaminsky E. B., Kaul V., Paschall J., Church D. M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J. G., Warren S. T., Richard G., Compton J. G., Fuller A. E., Gliem T. J., Huang S., et al. (2011) An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 13, 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lai K.-O., Wong A. S., Cheung M. C., Xu P., Liang Z., Lok K. C., Xie H., Palko M. E., Yung W. H., Tessarollo L., Cheung Z. H., and Ip N. Y. (2012) TrkB phosphorylation by Cdk5 is required for activity-dependent structural plasticity and spatial memory. Nat. Neurosci. 15, 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harris K. M., Jensen F. E., and Tsao B. (1992) Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 12, 2685–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zagrebelsky M., Holz A., Dechant G., Barde Y.-A., Bonhoeffer T., and Korte M. (2005) The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J. Neurosci. 25, 9989–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McNair K., Spike R., Guilding C., Prendergast G. C., Stone T. W., Cobb S. R., and Morris B. J. (2010) A role for RhoB in synaptic plasticity and the regulation of neuronal morphology. J. Neurosci. 30, 3508–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bär J., Kobler O., van Bommel B., and Mikhaylova M. (2016) Periodic F-actin structures shape the neck of dendritic spines. Sci. Rep. 6, 37136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weyer S. W., Zagrebelsky M., Herrmann U., Hick M., Ganss L., Gobbert J., Gruber M., Altmann C., Korte M., Deller T., and Müller U. C. (2014) Comparative analysis of single and combined APP/APLP knockouts reveals reduced spine density in APP-KO mice that is prevented by APPsα expression. Acta Neuropathol. Commun. 2, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Grutzendler J., Kasthuri N., and Gan W.-B. (2002) Long-term dendritic spine stability in the adult cortex. Nature 420, 812–816 [DOI] [PubMed] [Google Scholar]
- 65. Gardoni F., Saraceno C., Malinverno M., Marcello E., Verpelli C., Sala C., and Di Luca M. (2012) The neuropeptide PACAP38 induces dendritic spine remodeling through ADAM10-N-cadherin signaling pathway. J. Cell Sci. 125, 1401–1406 [DOI] [PubMed] [Google Scholar]
- 66. Spires T. L., Molnár Z., Kind P. C., Cordery P. M., Upton A. L., Blakemore C., and Hannan A. J. (2005) Activity-dependent regulation of synapse and dendritic spine morphology in developing barrel cortex requires phospholipase C-β1 signalling. Cereb. Cortex 15, 385–393 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.