Abstract
The 26S proteasome is a large cellular assembly that mediates the selective degradation of proteins in the nucleus and cytosol and is an established target for anticancer therapeutics. Protein substrates are typically targeted to the proteasome through modification with a polyubiquitin chain, which can be recognized by several proteasome-associated ubiquitin receptors. One of these receptors, RPN13/ADRM1, is recruited to the proteasome through direct interaction with the large scaffolding protein RPN2 within the 19S regulatory particle. To better understand the interactions between RPN13, RPN2, and ubiquitin, we used human proteins to map the RPN13-binding epitope to the C-terminal 14 residues of RPN2, which, like ubiquitin, binds the N-terminal pleckstrin-like receptor of ubiquitin (PRU) domain of RPN13. We also report the crystal structures of the RPN13 PRU domain in complex with peptides corresponding to the RPN2 C terminus and ubiquitin. Through mutational analysis, we validated the RPN2-binding interface revealed by our structures and quantified binding interactions with surface plasmon resonance and fluorescence polarization. In contrast to a previous report, we find that RPN13 binds ubiquitin with an affinity similar to that of other proteasome-associated ubiquitin receptors and that RPN2, ubiquitin, and the deubiquitylase UCH37 bind to RPN13 with independent energetics. These findings provide a detailed characterization of interactions that are important for proteasome function, indicate ubiquitin affinities that are consistent with the role of RPN13 as a proteasomal ubiquitin receptor, and have major implications for the development of novel anticancer therapeutics.
Keywords: crystallography, proteasome, protein-protein interaction, surface plasmon resonance (SPR), ubiquitin, RPN13, RPN2, fluorescence polarization
Introduction
The 26S proteasome, which selectively degrades targeted protein substrates in the cytosol and nucleus, is important for many cellular functions, including protein quality control, DNA repair, cell-cycle regulation, signal transduction, and antigen presentation (1). Structurally, the proteasome is an ∼2.5-MDa complex composed of a central 20S core particle (CP),4 which houses the proteolytic sites, capped on one or both ends by the 19S regulatory particle (RP), which possesses subunits responsible for the recognition, processing, and translocation of targeted protein substrates into the CP for degradation (2). Within the RP, polyubiquitin chains can be bound by two stably associated subunits, RPN10 and RPN13/ADRM1, and by several transiently associated proteasome-interacting proteins such as RAD23 (HHR23A/B in humans), DSK2 (PLIC2 in humans), and DDI1, which are recruited to the proteasome by the large scaffolding subunit RPN1 (3–7). Recent advances in cryo-EM technology have resulted in several near-atomic resolution reconstructions of proteasome complexes, which have provided an invaluable tool for better understanding the mechanisms underlying proteasome function (8–17). Curiously, reconstructions of proteasomes from both Saccharomyces cerevisiae (PDB codes 4CR2, 4CR3, and 4CR4) (12) and Homo sapiens (PDB code 5L4K) (16) reveal that the 19S ubiquitin receptors occupy disparate locations, with RPN1, RPN10, and RPN13 being separated by >70 Å from each other in the absence of the proteasome substrate. Further analysis of the substrate-bound and translocation states of yeast proteasomes reveals only minor changes in relative position of these subunits (12). Unlike the transiently associated ubiquitin receptors and RPN10, which bind ubiquitin via ubiquitin-associated domains or ubiquitin-interaction motifs, RPN13 utilizes its N-terminal pleckstrin-like receptor of ubiquitin (PRU) domain to bind ubiquitin (18, 19). This same PRU domain also binds the RP scaffolding subunit RPN2, which recruits RPN13 to the proteasome (18–22).
Interestingly, it has been reported that RPN13 from S. cerevisiae binds ubiquitin with a KD of 65 μm but that the human RPN13 PRU domain binds ubiquitin over 200 times tighter (KD = 0.3 μm) (19). Furthermore, full-length human RPN13 was reported to bind ubiquitin roughly 25-fold more weakly than the PRU domain alone (KD = 8 μm) and that autoinhibition was relieved by binding of a C-terminal peptide of RPN2 (23). The high affinity reported for these interactions is in sharp contrast to the relatively weak affinities seen in other characterized ubiquitin receptors (24). Moreover, these data pose a challenge for understanding proteasome mechanisms because the high abundance of monomeric ubiquitin and ubiquitin conjugates that are not degraded by the proteasome would be expected to occupy, and hence block, a high-affinity receptor on the proteasome (25).
To better understand interactions between RPN13, RPN2, and ubiquitin, we have mapped the minimal RPN13-binding segment of RPN2 to residues 940–953, and we determined a crystal structure of this RPN2 peptide in a ternary complex with residues 19–132 of human RPN13, which constitute its N-terminal PRU domain (RPN13PRU), and ubiquitin. We also report quantification of both RPN13-RPN2 interactions and RPN13-ubiquitin interactions, which, in contrast to previous studies, reveal that the PRU domain of RPN13 binds ubiquitin with an affinity similar to that of other ubiquitin receptors and that the RPN13-ubiquitin interaction is not substantially modulated by binding of RPN2.
Results
Mapping the RPN13-binding domain of RPN2
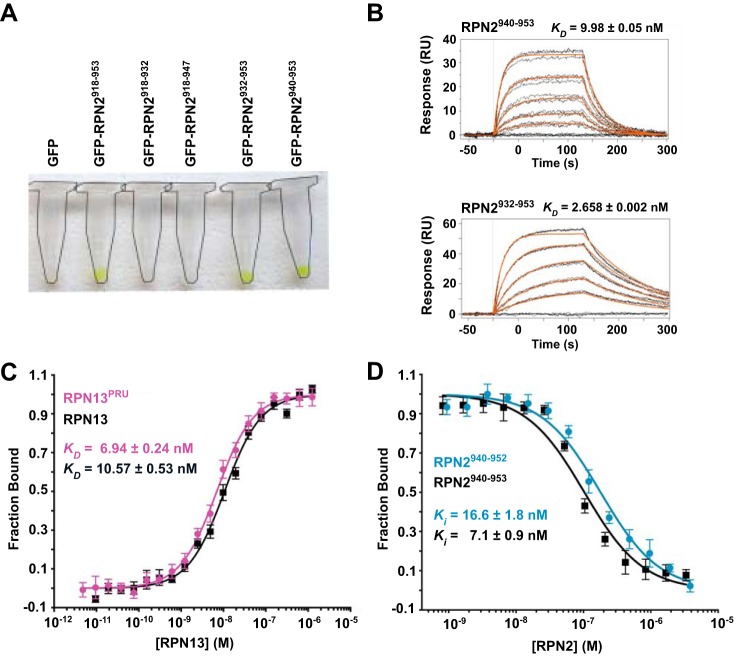
Using human protein constructs, the RPN13-binding region of RPN2 was mapped by affinity co-purification. Guided by an earlier report that the C-terminal 52 residues of RPN2 (RPN2(902–953)) facilitate binding of the RPN13PRU domain (18), we expressed in bacteria a series of constructs of N-terminally His-tagged GFP fused to C-terminal peptides of RPN2. Following co-lysis with bacteria expressing GST-tagged RPN13PRU, complex formation was visually detected by retention of the GFP-RPN2 constructs after sequential purification on nickel-chelate and glutathione resins (Fig. 1A). The shortest construct tested that exhibited binding in this assay contained the last 14 residues of RPN2, RPN2(940–953). No retention of GFP was observed using constructs consisting of RPN2 residues 918–932 or 918–947, indicating that the last six residues are required for binding.
Figure 1.
Examination of the RPN2-RPN13 interaction. A, GFP retention assays used to visualize stable association of various RPN2 peptides with RPN13. Complex formation is visualized by retention of green GFP-RPN2 on the resin. Microcentrifuge tubes outlined for clarity. B, representative SPR sensorgrams illustrating full-length RPN13 binding to RPN2 peptides corresponding to residues 940–953 (top) and 932–953 (bottom). C, representative fluorescence polarization binding curves of full-length RPN13 (black) and RPN13PRU (magenta) binding to fluorescently labeled RPN2(940–952). D, representative fluorescence polarization competition curves of RPN2(940–952) (blue) and RPN2(940–953) (black) competing with fluorescently labeled RPN2(940–952) for binding of RPN13PRU.
The RPN13-RPN2 interaction was quantified using both surface plasmon resonance (SPR) and fluorescence polarization (FP). For SPR analyses, GST-RPN13 was purified and captured on a biosensor chip coated with anti-GST antibody, and GFP-RPN2 constructs were assayed for binding. Consistent with the observations from affinity co-purification, GFP-RPN2(940–953) bound tightly with a KD of 9.98 ± 0.05 nm, whereas the slightly longer RPN2(932–953) construct bound slightly tighter, with a KD of 2.658 ± 0.002 nm (Fig. 1B). Further extensions of the RPN2 fragment did not increase affinity further. For fluorescence polarization studies, a peptide comprising residues 940–952 of RPN2 (RPN2(940–952)) was synthesized and labeled C-terminally with fluorescein. In agreement with the SPR data, FP studies indicate KD values of 10.57 ± 0.53 nm for the RPN13-RPN2(940–952) interaction and 6.94 ± 0.24 nm for the RPN13PRU-RPN2(940–952) interaction (Fig. 1C), illustrating that the RPN2 peptide binds to full-length RPN13 and RPN13PRU with similar affinities (within 2-fold). In addition, we performed a binding competition assay using unlabeled peptides corresponding to residues 940–952 (Ki = 16.6 ± 1.8 nm) and 940–953 (Ki = 7.1 ± 0.9 nm) of RPN2 to validate that neither the fluorophore nor Asp-953 of RPN2 contribute to the binding energy (Fig. 1D). Combined, these data indicate that RPN2(940–952) contains almost the entire RPN13 binding determinant, with a small contribution provided by residues 932–939, and are consistent with a recent report indicating that the PRU domain of human RPN13 binds a slightly longer RPN2 construct, RPN2(918–953), with a KD of 12.3 nm (26).
Crystal structure of RPN2-RPN13-ubiquitin complex
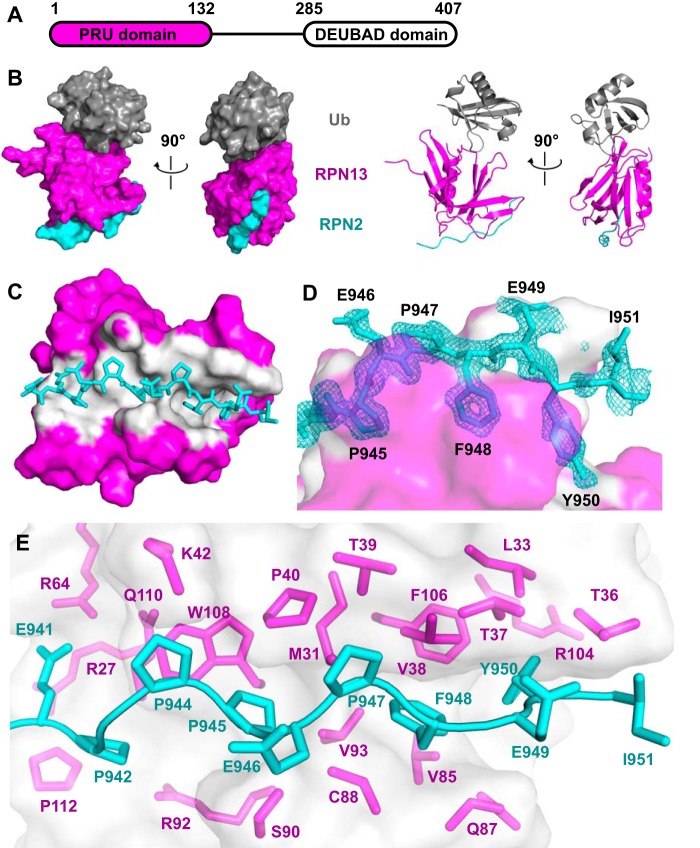
To visualize the binding interactions of RPN13PRU (Fig. 2A) with RPN2 and ubiquitin, we crystallized a ternary RPN13PRU-RPN2(940–953)-ubiquitin complex. This structure was determined by molecular replacement using the murine RPN13PRU domain (PDB code 2R2Y) (18) and ubiquitin (PDB code 1CMX) (27) as search models and was refined against 1.45 Å data to Rwork/Rfree values of 0.140/0.175 (Fig. 2B and Table 1). The two complexes within the asymmetric unit superimpose closely with an r.m.s.d. of 2.07 Å over all atoms. The main chain is clearly defined for RPN2 residues 940–950, RPN13 residues 20–130, and ubiquitin residues 1–72 and reveals that the ubiquitin and RPN2 binding surfaces are on opposite sides of the RPN13PRU domain (Fig. 2B). The largest deviations between the two asymmetric complexes are at the protein termini and in the Thr-7–Lys-11 loop of ubiquitin, where the Cα atoms of Gly-10 and Lys-11 in the two complexes deviate by 5.1 and 2.0 Å, respectively.
Figure 2.
Structure of the RPN13-RPN2-ubiquitin complex. A, schematic of human RPN13 domain architecture. B, two orthogonal views of the ternary complex of RPN13PRU (magenta), RPN2(940–953) (cyan), and ubiquitin (gray) in space-filling and ribbon representations. C, overview of the RPN13-RPN2 interface. Surface of RPN13 residues within 3.5 Å of RPN2, white. Ubiquitin removed for clarity. D, detailed view of RPN2 residues (Pro-945, Phe-948, and Tyr-950) within the binding groove of RPN13. RPN13 colored as in C. RPN2 residues labeled in black. Fo − Fc omit map (cyan mesh, 2× r.m.s.d.) within 1.6 Å of RPN2 peptide. The map was calculated by removing RPN2 peptides from the model, introducing random shifts to all atoms in remaining chains, and performing a single round of refinement. E, detailed view of RPN13 residues at RPN13-RPN2 interface. RPN13 residues within 4 Å of RPN2, magenta. RPN2 residues, cyan.
Table 1.
Data collection and refinement statistics
| Data collection | RPN13(19–132)-RPN2(940–953)-Ub | RPN13(19–132)-RPN2(932–953)-Ub |
|---|---|---|
| Space group | P21 | P31 |
| Cell dimensions | ||
| a, b, c (Å) | 36.13, 96.10, 57.61 | 100.85, 100.85, 37.57 |
| α, β, γ (°) | 90.0, 96.4, 90.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.00–1.42 (1.47–1.42) | 35.00–2.00 (2.03–2.00) |
| Rsym (%) | 6.6 (34.9) | 9.8 (62.8) |
| σI | 24.8 (5.2) | 16.0 (2.5) |
| Completeness (%) | 98 (93) | 100 (100) |
| Redundancy | 7.6 (7.0) | 4.7 (4.6) |
| Refinement | ||
| Resolution (Å) | 36.80–1.42 (1.44–1.42) | 33.01–2.00 (2.07–2.00) |
| No. of reflections | 72,405 (2,526) | 28,875 (2,748) |
| Rwork/Rfree | 0.140 (0.131)/0.175 (0.195) | 0.146 (0.246)/0.182 (0.253) |
| No. of atoms | ||
| Protein | 3,643 | 3,244 |
| Water | 510 | 81 |
| B-Factors (Å2) | ||
| Protein | 23 | 31 |
| Water | 38 | 31 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.008 | 0.009 |
| Bond angles (°) | 0.984 | 0.887 |
We also crystallized a complex with a longer peptide, RPN2(932–953), which SPR data indicate binds with slightly higher affinity (Fig. 1B). This structure was also determined by molecular replacement and was refined against 2.0 Å data to Rwork/Rfree values of 0.146/0.182. Crystals of the RPN13PRU-RPN2(932–953)-ubiquitin complex belong to a different space group than the RPN2(940–953) complex (Table 1), yet they still contain two complexes within the asymmetric unit. Importantly, each of the complexes from the two structures are essentially identical to one another, superimposing with r.m.s.d. values between 1.19 and 2.98 Å over all atoms. Consistent with the small difference in RPN13 binding affinity between RPN2(932–953) and RPN2(940–953), RPN2 residues 932–939 lack electron density, except for the backbone of Glu-939, which is visible in only one of the two complexes in the asymmetric unit of the RPN2(932–953) complex. Although no specific contacts are seen between Glu-939 and RPN13, and the Glu-939 side chain lacks defined density, this residue is in the vicinity of the similarly flexible RPN13 side chains of Arg-27 and Lys-113. Model building suggests that the charged atoms could approach as close as 2.6 Å in unstrained conformations, and these potential charge interactions likely explain the ∼3.5-fold tighter binding observed when this residue is included in the SPR-binding experiments. Because of the higher resolution of the RPN13PRU-RPN2(940–953)-ubiquitin structure, all figures and calculations will be based on that model unless otherwise noted.
RPN13-RPN2 interaction
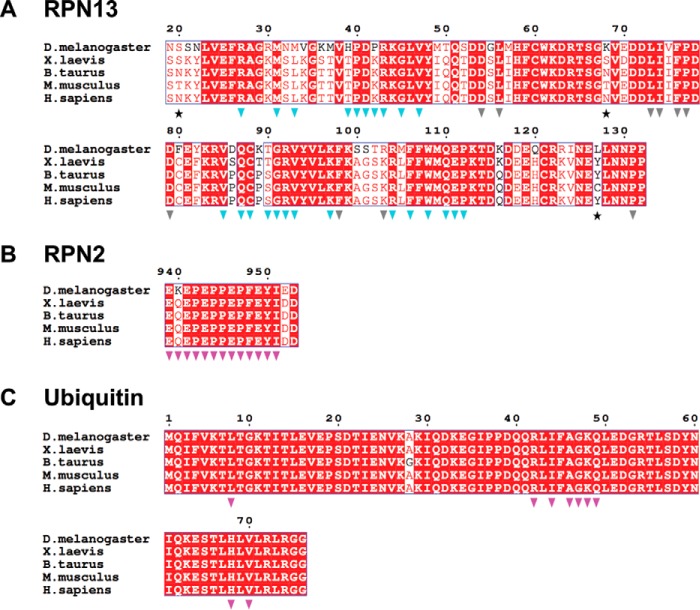
The RPN2 peptide adopts an extended conformation to bind in a surface groove on RPN13, with an average total buried surface area of ∼1,600 Å2 over the four complexes in the asymmetric units of both crystal structures (Fig. 2C). The most striking feature of this interface is the packing of RPN2 residues Pro-945, Phe-948, and Tyr-950 into a deep and largely hydrophobic binding pocket in RPN13 (Fig. 2D). As expected, the buried surface on RPN13 overlaps with residues previously implicated in binding (18, 23, 26). RPN2 residues Pro-942–Pro-947 form a polyproline II helix that is followed by residues Phe-948 to Tyr-950, which form main-chain to main-chain hydrogen bonds with RPN13 residues Thr-36 and Val-38 that extend an anti-parallel β-sheet. Residue Ile-951, which is only visible in one complex, extends away from RPN13, and it makes no significant intermolecular contacts. The RPN13-RPN2 interface is predominantly hydrophobic and includes the side chains of Glu-941, Pro-942, Pro-944, Pro-945, Pro-947, Phe-948, Glu-949, and Tyr-950 of RPN2 and the side chains of Arg-27, Met-31, Leu-33, Thr-36–Pro-40, Lys-42, Arg-64, Val-85, Gln-87, Cys-88, Ser-90, Arg-92, Val-93, Arg-104, Phe-106, Trp-108, Gln-110, and Pro-112 of RPN13 (Fig. 2E). At the center of the interface, the RPN2 Glu-946 main chain forms a hydrogen bond with the conserved RPN13 Ser-90 side chain. Additional electrostatic interactions flanking the groove include salt bridges between RPN2 Glu-941 and RPN13 residues Arg-64 and Arg-27, and hydrogen bonding between the side chain of RPN2 Glu-949 and the side chains of RPN13 Thr-36 and Thr-37. Importantly, both the RPN2-contacting residues on RPN13 (Fig. 3A, blue triangles) and the RPN2 peptide itself (Fig. 3B) are highly conserved throughout eukaryotes, indicating that details of this interaction are likely to be preserved in other species.
Figure 3.
Sequence alignments of RPN13, RPN2, and ubiquitin from higher eukaryotes. A, RPN13 alignments. Stars indicate residues not conserved between mouse and human. Blue triangles indicate residues at the RPN13-RPN2 interface. Gray triangles indicate residues at the RPN13-ubiquitin interface. B, RPN2 sequence alignment. Magenta triangles indicate residues at the RPN13-RPN2 interface. C, ubiquitin sequence alignment; magenta triangles indicate residues at the RPN13-ubiquitin interface.
We determined the importance of specific residues at the crystallographic interface by quantifying the ability of alanine point mutants within the RPN2(940–952) peptide to compete with fluorescently labeled wild-type peptide (Table 2). Consistent with our structural models, mutation of the buried interface residues Pro-945, Phe-948, and Tyr-Y950 (Fig. 2D) each result in a substantial decrease in binding affinity as reflected by their Ki values. Specifically, P945A and Y950A point mutations each result in an ∼100-fold reduction in binding affinity, whereas an F948A mutation results in an ∼1,000-fold reduction in binding affinity relative to the wild-type peptide. These studies also reveal an ∼35-fold decrease in binding affinity for an E949A mutant as well as an ∼18-fold reduction in binding affinity for an E941A mutant, both of which are consistent with loss of their respective interactions with RPN13. As expected, individual mutation of Pro-942, Pro-944, Pro-947, and Ile-951 only slightly decrease the binding affinity of the peptide (Table 2). We hypothesize that the modest 1.7–2.9-fold effect observed for each of the proline to alanine mutations likely results from the entropic penalty imposed by reduction of backbone conformational restraints rather than changes in contact between RPN2 and RPN13. In further support of these models, it was recently reported that a transgenically expressed peptide corresponding to the RPN2 C terminus fails to displace RPN13 from the proteasome in 293T cells upon introduction of either 1) an F948R point mutation, 2) a Y950D/I951D double mutation, or 3) by truncating residues Phe-948–Asp-953 (26). In the case of the Y950D/I951D double mutant, our data further suggest that dissociation results exclusively from the Y950D mutation.
Table 2.
FP analysis of RPN2 point mutants
| Construct | Peptide sequence | Ki (nm) |
|---|---|---|
| WT | PQEPEPPEPFEYID | 16.6 ± 1.8 |
| E941A | PQAPEPPEPFEYID | 291.1 ± 21.4 |
| P942A | PQEAEPPEPFEYID | 48.0 ± 3.2 |
| P944A | PQEPEAPEPFEYID | 39.0 ± 4.0 |
| P945A | PQEPEPAEPFEYID | 1,410 ± 100 |
| P947A | PQEPEPPEAFEYID | 28.9 ± 3.4 |
| F948A | PQEPEPPEPAEYID | 18,760 ± 960 |
| E949A | PQEPEPPEPFAYID | 590.5 ± 83.5 |
| Y950A | PQEPEPPEPFEAID | 1,480 ± 90 |
| I951A | PQEPEPPEPFEYAD | 43.1 ± 3.1 |
RPN13-ubiquitin interaction
In our structures, the ubiquitin molecules (residues 1–72) closely superimpose with multiple previously reported ubiquitin structures (27–31), with r.m.s.d. values ranging from 1.24 to 1.84 Å over all atoms. Similarly, the RPN13PRU domain in our complex agrees well with structures of the isolated RPN13PRU domains from H. sapiens (PDB code 5IRS) (32), Mus musculus (PDB code 2Z59) (18), and S. cerevisiae (PDB code 2Z4D) (30), aligning over all atoms with r.m.s.d. values of 1.97, 2.13, and 5.18 Å, respectively. The largest deviation observed between crystal structures of unbound human RPN13PRU (PDB code 5IRS) (32) and RPN2-bound RPN13PRU is a shift of ∼6.5 Å in residues 31–39, which lie at the RPN13-RPN2 interface.
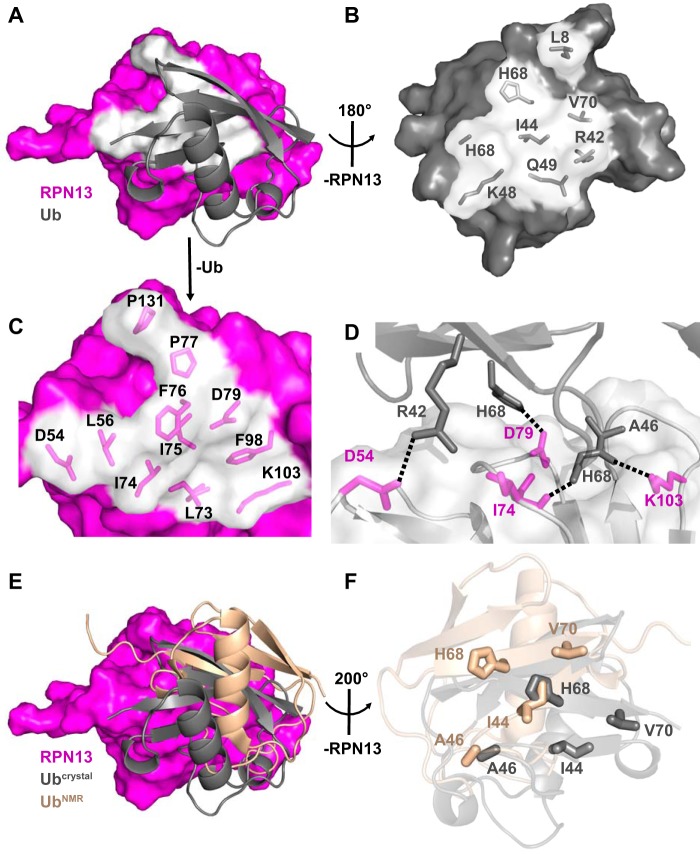
The RPN13PRU-ubiquitin interface (Fig. 4A) comprises mostly hydrophobic contacts (Fig. 4, B and C) but also features three intermolecular hydrogen bonds and one intermolecular salt bridge (Fig. 4D). The interface buries between 786 and 937 Å2 in the four crystallographically independent models and includes packing of the ubiquitin surface residue Ile-44 (Fig. 4B), a feature conserved in most characterized ubiquitin interactions (24). Notably, with the absolute conservation of each of the ubiquitin residues (Fig. 3C, magenta triangles) and many of the RPN13 residues (Fig. 3A, gray triangles) at the ubiquitin interface, including Leu-56, Leu-73, Ile-74, Phe-76, and Phe-98 (Fig. 4C), it is probable that the interface is conserved between species.
Figure 4.
RPN13-ubiquitin interaction. A, overview of the RPN13-ubiquitin interface. RPN13 residues within 4 Å of ubiquitin, white. B, view of ubiquitin residues at RPN13 interface upon ∼180° rotation around y axis of Fig. 3A. RPN13 removed for clarity. C, zoomed view of RPN13 residues at ubiquitin interface as visualized in A. Ubiquitin removed for clarity. D, electrostatic interactions at the RPN13-ubiquitin interface. E, disparity in ubiquitin positioning between crystallographic (gray) and NMR docking (wheat, PDB code 2Z59) models. F, differences in positioning of key residues (shown as sticks) between crystallographic (gray) and NMR docking (wheat) models upon alignment on RPN13.
Comparison of the four crystallographically independent ternary complexes in our structures indicates the existence of two areas of flexibility at the RPN13-ubiquitin interface. The first is in the N-terminal loop of ubiquitin, consisting of residues Thr-7 through Lys-11, where residue Leu-8 assumes multiple conformations while interacting with a hydrophobic patch created by RPN13 residues Leu-56, Phe-76, and Pro-77. The second is in a loop of RPN13 composed of residues Lys-99 through Lys-103, where Ala-100 Cα is 3.7 Å from Phe-45 of ubiquitin in complexes containing RPN2(932–953) and 5.6 Å distant in complexes with RPN2(940–953). Despite these differences, alignment of each of the four crystallographically independent ubiquitin molecules reveals r.m.s.d. values of 0.95–1.38 Å over all atoms, thereby implying that the RPN13-ubiquitin interaction observed in the crystal structures accurately represents a range of structures that would be highly populated in solution.
Comparison with a previously reported RPN13-ubiquitin model
Our crystallographically determined RPN13-ubiquitin interface resembles a previously reported model based on the docking of ubiquitin onto a crystal structure of the murine RPN13PRU domain using experimentally determined NOE restraints (PDB code 2Z59) (18), although several differences are observed. When the two structures are superimposed on RPN13, the ubiquitin molecules differ by a center-of-mass translation of 8.4 Å and a rotation of 30° (Fig. 4E). This discrepancy is surprising given that only three residues (Asn-20, Asn-68, and Tyr-127) vary between the human and mouse proteins used in the two studies, none of which lie at the RPN13-ubiquitin interface (Fig. 3A, stars). Moreover, structures of the human and mouse RPN13PRU superimpose with an average r.m.s.d. of only 1.58 Å, whereas the ubiquitin molecules from each complex overlay with an average r.m.s.d. of only 0.84 Å over all pairs of Cα atoms. The disparities seen between the two RPN13PRU-ubiquitin models can be traced to the packing of several key residues (Fig. 4F), including ubiquitin Ile-44, His-68, and Val-70, which result in a substantial difference in buried surface area at the RPN13-ubiquitin interface. Importantly, the finding of ∼1,260 Å2 of buried surface area at the RPN13-ubiquitin interface in the NMR model, which is larger than many reported ubiquitin interactions, prompted the rationale that this underlies a relativity tight binding affinity (19). In contrast, our models reveal a buried surface area of ∼860 Å2 at the RPN13PRU-ubiquitin interface.
RPN13-ubiquitin binding affinity
The difference in buried surface area between RPN13PRU and ubiquitin in these models prompted us to independently quantify the human RPN13-ubiquitin interaction. To this end, we used RPN13PRU and ubiquitin-fluorescein (Boston Biochem) in an FP assay. Even up to a concentration of 1.25 μm RPN13PRU, which is the maximum attainable before protein begins to aggregate, we observed no changes in polarization, indicating the interaction is much weaker than previously reported. To validate this finding, we utilized the independent approach of SPR, which indicates that RPN13PRU binds ubiquitin with a KD of 90.6 ± 0.6 μm in HBS Buffer (Table 3), an affinity that is ∼300-fold weaker than previously reported for the human RPN13PRU domain but similar to the 65 μm affinity reported for the S. cerevisiae RPN13-ubiquitin interaction (19) and the low to mid-micromolar affinity reported for most other characterized ubiquitin interactions (24). To determine whether differences in solution conditions could explain the discordant results, we repeated the SPR-binding experiments with reduced salt to replicate the ionic strength condition used previously (19). Under these conditions (LSPB Buffer, Table 3), the affinity increased 3-fold (KD = 31.5 ± 0.3 μm) but is still more than 2 orders of magnitude weaker than the previously reported dissociation constant. We also evaluated the binding affinity of Lys-48-linked di-ubiquitin (K48-Ub2) to RPN13PRU in HBS buffer, and we found that although K48-Ub2 bound with 2–3-fold higher affinity than monomeric ubiquitin (KD = 37.4 ± 0.2 μm, Table 3), its affinity is still more than 400-fold weaker than the 90 nm value reported earlier (19). These results are consistent with a recent isothermal titration calorimetry study showing the human RPN13PRU domain binds K48-Ub2 with a KD of 29 ± 2 μm (32).
Table 3.
SPR analysis of RPN13-ubiquitin and RPN13-K48-Ub2 interactions
| Ligand | Analyte | Observed KD (μm) in specified buffers |
|||
|---|---|---|---|---|---|
| HBS-TBP | HBS-TBP + RPN2 | LSPB-DBP | LSPB-DBP + RPN2 | ||
| GST-RPN13 | Ub | 109.2 ± 0.6 | 109.0 ± 0.7 | 39.8 ± 0.4 | 38.3 ± 0.4 |
| GST-RPN13(19–132) | Ub | 90.7 ± 0.6 | 93.3 ± 0.5 | 31.5 ± 0.3 | 28.9 ± 0.3 |
| GST-RPN13 | K48-Ub2 | 46.3 ± 0.2 | 68.5 ± 0.4 | 20.0 ± 0.2 | 22.6 ± 0.2 |
| GST-RPN13(19–130 | K48-Ub2 | 37.4 ± 0.2 | 64.1 ± 0.4 | 14.1 ± 0.1 | 18.9 ± 0.2 |
Lack of cooperativity in RPN2 and ubiquitin binding
Based upon the results of RPN13-ubiquitin binding studies, we decided to independently evaluate the report that ubiquitin binding by full-length RPN13 is antagonized by the C-terminal deubiquitylase adaptor domain of RPN13 (Fig. 2A) and that this effect can be relieved if RPN13 is also bound to RPN2 (23). This published model was attractive as it suggested a mechanism to ensure that the very tight ubiquitin-binding affinity reported for the isolated human RPN13PRU domain was masked until RPN13 was properly assembled in the 26S proteasome. In contrast, however, our SPR data show that full-length RPN13 binds ubiquitin or K48-Ub2 with only minor decreases in affinity compared with the isolated PRU domain (Table 3). To assay for the reported energetic coupling of RPN2 and ubiquitin binding, we analyzed binding in the presence of a saturating concentration of RPN2 by adding GFP-RPN2(932–953) to the SPR running buffer at a concentration of 100 nm, which is ∼40 times greater than the KD of the RPN2(932–953)-RPN13 interaction. The presence of RPN2(932–953) did not alter the affinity of monomeric ubiquitin for RPN13 or RPN13PRU beyond experimental error (Table 3). Interestingly, although RPN2(932–953) did induce some change in the affinity of K48-Ub2 toward RPN13 and RPN13PRU under both high- and low-salt conditions, these effects are small, and unlike previous reports there are decreases in binding affinity (23). We therefore conclude that binding of RPN13 to RPN2 and ubiquitin are energetically uncoupled, which is consistent with our observation that RPN13 does not undergo substantial conformational changes upon binding RPN2 and ubiquitin.
RPN13 binding of UCH37 and RPN2 are not energetically coupled
The RPN13 deubiquitylase adaptor domain, which is tethered to the PRU domain by a flexible linker of ∼150 residues in the human protein (Fig. 2A), recruits UCH37 to the proteasome and activates it in many fungal species and higher eukaryotes (20, 22, 31, 33, 34). For this reason, we questioned whether binding of UCH37 and RPN2 to RPN13 is energetically coupled. As in previous SPR experiments, we immobilized GST-RPN13 and flowed purified UCH37 or GFP-RPN2(932–953) as analytes, either on their own or in the presence of 100 nm of the other binding partner. In this series of experiments, we find that UCH37 binds ∼3-fold more tightly to RPN13 than does RPN2, with KD values of 1.7 nm for the RPN13-UCH37 interaction and 5.5 nm for the RPN13-RPN2 interaction (Table 4). Importantly, we also find that binding affinities are unaltered by the presence of the additional binding partner. These data indicate that UCH37 and RPN2 bind to RPN13 independently, and in both cases the tight binding constants are consistent with stable association in vivo (20, 33).
Table 4.
SPR analysis of RPN13 interactions with RPN2 and UCH37
| Ligand | kon (×105) | koff (×10−4) | KD (nm) |
|---|---|---|---|
| UCH37 | 1.5 ± 0.1 | 2.3 ± 0.6 | 1.7 ± 0.5 |
| UCH37 + RPN2(932–953) | 1.8 ± 0.1 | 3.2 ± 0.1 | 1.7 ± 0.1 |
| RPN2(932–953) | 9.7 ± 0.7 | 52 ± 3 | 5.4 ± 0.2 |
| RPN2(932–953) + UCH37 | 13.3 ± 0.1 | 73 ± 4 | 5.5 ± 0.4 |
Discussion
The proteasome 19S RP possesses subunits that are responsible for the recruitment, processing, and delivery of ubiquitylated protein substrates to the CP for degradation. Two of these subunits, RPN10 and RPN13, function as ubiquitin receptors that appear to be largely functionally redundant (35–38) but occupy disparate locations within the RP. In previous studies, it was reported that recognition of ubiquitin by RPN13 was modulated by its proteasomal recruiting partner, RPN2, which formed the basis for an attractive model whereby the ubiquitin receptor activity of RPN13 was autoinhibited until it was properly assembled in the proteasome. Moreover, RPN13 was reported to bind ubiquitin moieties with binding affinities that are orders of magnitude tighter than would be expected for a typical ubiquitin receptor (24).
In contrast to these studies, we have shown via two different techniques that measure two distinct phenomena (SPR and FP) that the binding affinities of RPN13 and RPN13PRU for ubiquitin are much weaker than reported by Husnjak et al. (19) and are instead more typical of characterized ubiquitin interactions (24). One possible explanation for the disparate data is that intrinsic tryptophan fluorescence (IWF), which was used in previous studies, reports only on the local environment of tryptophans, and thus changes in fluorescence can be difficult to attribute to a specific phenomenon. Analysis of our crystal structures reveals two major concerns for the use of IWF in this system. The first is that the two tryptophans in RPN13, Trp-61 and Trp-108, do not appear to be good reporters for ubiquitin association because they are not proximal to the RPN13-ubiquitin interface, lying ∼12 and ∼20 Å away from the closest ubiquitin atom, respectively. Second, structural overlap of unbound RPN13 (PDB code 5IRS) with our ubiquitin-bound RPN13 structures does not reveal a perturbation that seems likely to explain the quenching of fluorescence upon binding of ubiquitin. These observations question the reliability of IWF as a method to accurately describe these interactions. The inference of allosteric regulation of the RPN13-ubiquitin interaction by RPN2 (23) also depended upon use of IWF. Therefore, we believe that our SPR data more accurately represent the RPN13-RPN2-ubiquitin interactions and argue against an allosteric regulatory mechanism in the binding of ubiquitin to RPN13. Overall, the work presented here is consistent with the established role of RPN13 as a ubiquitin receptor, which requires that binding be reversible to allow efficient release of polyubiquitin after substrate delivery, and to avoid indiscriminate targeting of all ubiquitylated proteins and of free ubiquitin, whose estimated concentration in the cell is ∼10 μm (39).
Despite the low nanomolar affinity described for the RPN13-RPN2 interaction, RPN13 is found in sub-stoichiometric amounts in purified 26S proteasomes (8, 23). This may result from either variable cell-specific expression levels of RPN13 or through modulation of binding via post-translational modification of either RPN2 (40) or RPN13 itself (41). Further investigation into the factors that influence recruitment of RPN13 to the 26S proteasome will provide insight into the functional relevance of RPN13 as a ubiquitin receptor and to its regulation of the recruitment and activation of the deubiquitylase UCH37.
The proteasome is an established target for the treatment of multiple myeloma, with three Food and Drug Administration-approved inhibitors of the CP proteolytic sites, bortezomib (Velcade®, Millenium Pharmaceuticals), ixazomib (Ninlaro®, Millenium Pharmaceuticals), and carfilzomib (Kyprolis®, Onyx Pharmaceuticals) currently in clinical use. The problem of resistance to these drugs (42–45) places emphasis on developing new anticancer drugs to target alternative sites within the proteasome, including the deubiquitylases (46) and, more recently, RPN13 (47–50).
In humans, RPN13 resides within a recurrent amplicon in chromosome band 20q13, whose amplification has been implicated in early stages of cancer development (51) and was identified as one of the most commonly overexpressed genes in solid tumors (52). Moreover, increased levels of RNA and protein levels of RPN13 have been directly correlated to cell proliferation and metastatic potential in hepatocellular carcinoma (53), and to stage, time to recurrence, and metastatic potential in ovarian cancer (50), suggesting that RPN13 is intimately involved in pathways related to both oncogenesis and metastasis. Correlated with this potential, RPN13 knockdown studies have revealed significant decreases of cell proliferation in ovarian, liver, head and neck, and gastric and colorectal cancer cell lines (50, 51, 53–57).
In 2013, the small, electrophilic compound RA190 was reported to bind RPN13 and displayed selective toxicity against cancer cells in both cell culture and mouse xenograft models (47). More recently, the discovery of a peptoid-based ligand-targeting RPN13 was reported, which was shown to act synergistically with the proteasome inhibitor bortezomib to elicit cell death in several multiple myeloma cell lines (48). Despite revealing the efficacy of these molecules, the mechanisms by which these small molecules elicit cell death are still unknown. One provocative observation from our structural models is the presence of RPN13 Cys-88 at the base of the RPN2-binding groove, which was reported as the site in RPN13 to which RA190 covalently binds via Michael addition reaction (47). Based on our structures, we anticipate that this modification would greatly destabilize the RPN13-RPN2 interaction, although treatment with RA190 is reported to not displace RPN13 from the proteasome in 293T cells (47). Our structural and biochemical observations now make understanding the biochemical consequences of RA190 treatment and the potential therapeutic impact of disrupting the RPN13-RPN2 interaction a high priority for future research efforts.
Experimental procedures
Protein expression and purification
All expression plasmids have been deposited to the Addgene plasmid repository (plasmid IDs: 61937, 73741–73749, and 73913). Recombinant RPN13 and RPN2 proteins were expressed in BL21 (DE3) codon+ (RIL) Escherichia coli cells (Stratagene) in autoinduction media, ZYP-5052 (58), at 37 °C to an A600 of ∼1.0 and then transferred to 19 °C for 20 h. For purification of complexes, cultures of cells expressing GST-RPN13 and His6-GFP-RPN2 were mixed in a 1:1 ratio prior to harvesting by centrifugation and storing at −80 °C. Pellets were resuspended in lysis buffer (20 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10 mm imidazole) supplemented with 0.5% Triton X-100 and protease inhibitors (Sigma). Lysis by sonication was followed by clarification by centrifugation. RPN13-RPN2 complexes were purified by the following steps: 1) Ni-NTA affinity chromatography; 2) binding to glutathione-Sepharose (GS) resin; 3) on-column HRV3C protease cleavage to remove GST and GFP from RPN13 and RPN2, respectively; 4) anionic exchange chromatography; and 5) gel filtration chromatography.
Specifically, clarified lysate was bound to Ni-NTA resin (Qiagen), washed with 20 CV of lysis buffer, and eluted with 5 CV of nickel elution buffer (lysis buffer + 240 mm imidazole). EDTA and DTT were added to a final concentration of 1 mm in the eluted protein fraction, which was then incubated with pre-equilibrated GS (GE Healthcare) for 1 h. After washing the resin with 10 CV of GS-BIND buffer (20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 1 mm DTT, 1 mm EDTA), bound protein was incubated with HRV3C protease in 5 CV of GS-BIND buffer at 4 °C overnight before elution. The eluate was dialyzed into buffer QA (20 mm Tris-HCl, pH 7.5, 10 mm imidazole), loaded directly onto a HiTrap Q HP column (5 ml, GE Healthcare), washed with 10 CV of QA, and eluted with a gradient of 0–400 mm NaCl in QA over 20 CV. Protein and/or protein complexes eluted from the Q column were concentrated (3,000 MWCO Vivaspin® centrifugal concentrator) and loaded onto an SD200 gel filtration column (GE Healthcare) equilibrated with HBS-TE (10 mm HEPES-HCl, pH 7.4, 150 mm NaCl, 1 mm TCEP, 1 mm EDTA) for final purification. Mass spectrometry was used to confirm the presence of the RPN2 peptides 932-C and 940-C in purified samples as they are not visible by Coomassie staining of SDS-polyacrylamide gels.
The chromatographic steps used for purification of GST-RPN13 (expressed with an N-terminal His6 tag) and GFP-RPN2 constructs for binding assays were as follows: 1) Ni-NTA affinity chromatography; 2) dialysis and His6 affinity tag removal by TEV protease; 3) anionic exchange chromatography; 4) Ni-NTA affinity chromatography; and 5) gel filtration chromatography.
Ni-NTA affinity purification of individual constructs was performed as described above. Eluted proteins were treated with EDTA (1 mm), DTT (1 mm), and TEV protease (0.01 mg/ml final concentration) to remove the His6 tag intrinsic to the pET151 vector and dialyzed at 4 °C overnight (20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 1 mm DTT, 1 mm EDTA). The protein solution was applied to a HiTrap Q HP column (5 ml, GE Healthcare) and washed and eluted as described previously. The fractions containing the desired protein were pooled and applied again to Ni-NTA resin to remove uncleaved protein and the histidine-tagged TEV protease. The flow-through was concentrated (10,000 MWCO Vivaspin® centrifugal concentrator), loaded onto an SD200 gel filtration column, and eluted with HBS-TE. Purified protein was concentrated and stored in HBS-TE + 10% glycerol at −20 °C.
Recombinant ubiquitin was expressed in Rosetta 2 pLysS cells and purified as described previously (59). Lys-48-linked di-ubiquitin was a gift from Robert E. Cohen (Colorado State University).
Peptide synthesis
Peptides were synthesized on Protein Technologies, Inc., Prelude or PreludeX instruments using standard Fmoc chemistry, at 32-μmol scale on TentaGel R RAM resin (Rapp Polymere, 0.18 mmol/g). Fmoc deprotection employed three additions of 20% piperidine in DMF (2 ml for 3 min). Standard coupling conditions were 1.3:1.3:1 of 200 mm Fmoc-protected amino acid in NMP, 198 mm HATU in DMF, 600 mm NMM in DMF (4–5× resin density) for 15 min. Amino acid couplings on the PreludeX were performed at 40 °C. Peptides were cleaved for 2.5 h with 95:2.5:2.5 of TFA/water/TIS and then precipitated and washed thoroughly with ether. Fluorescein was installed on a non-native C-terminal lysine using 5(6)-carboxyfluorescein (Acros Organics). Peptides were dissolved in water/acetonitrile and lyophilized for long-term storage and reconstituted in minimal FP Assay Buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA) for assays. Peptide concentrations were determined by either A492 (fluorescein-labeled peptides), A280 (unlabeled peptides possessing aromatic residues), or by FTIR (unlabeled peptide lacking aromatic residues) using a Millipore Direct Detect Spectrometer. For consistency, all synthesized peptides contain a non-native N-terminal proline that was present in SPR and crystallography experiments due to cleavage from the engineered internal HRV3C protease cleavage site.
Crystallization and structure determination
Purified RPN13-RPN2 complexes were mixed in a 1:1.5 ratio with ubiquitin and concentrated to ∼17 mg/ml (E280 = 20,935 cm−1 m−1). Crystals were grown by sitting drop vapor diffusion. RPN13PRU-RPN2(932–953)-ubiquitin crystals grew at 20 °C from 2-μl drops comprising a 1:1 mixture of protein and reservoir solutions (0.1 m sodium acetate, pH 4.6, 22.5% PEG-3350). Crystals of the RPN13PRU-RPN2(940–953)-ubiquitin complex grew at 4 °C from a 0.6-μl drop in a 1:2 ratio of protein and reservoir solutions (0.1 m citric acid, pH 4.6, 20% PEG-6000). In both cases, crystals were cryoprotected by brief immersion in a solution of the reservoir made up with 20% ethylene glycol and cooled by plunging into liquid nitrogen.
Data for the RPN13PRU-RPN2(932–953)-ubiquitin and RPN13PRU-RPN2(940–953)-ubiquitin complexes were collected at the Stanford Synchrotron Radiation Lightsource and the National Synchrotron Light Source, respectively, and processed using HKL2000 (60). Phases were determined by molecular replacement using PHASER (61) with the murine RPN13PRU domain (PDB code 2R2Y) (18) and ubiquitin (PDB code 1CMX) (27) as search models. Model building and refinement were performed using Coot (62) and PHENIX (63). Data for the RPN13PRU-RPN2(932–953)-ubiquitin complex were perfectly twinned and were refined using the twin law (−h, −k, l).
Model geometries were analyzed by MolProbity (64) within PHENIX. For the RPN13PRU-RPN2(940–953)-ubiquitin model, 100.0% of residues have favorable backbone dihedrals. For the RPN13PRU-RPN2(932–953)-ubiquitin model, 95.1% of residues have favorable backbone dihedrals, whereas 4.9% fall into allowed regions.
Coordinates and structure factor amplitudes have been deposited at the PDB under the accession ID codes 5V1Y (RPN13PRU-RPN2(940–953)-ubiquitin) and 5V1Z (RPN13PRU-RPN2(932–953)-ubiquitin).
Buried surface area calculations and figures of molecular structures were generated using PyMOL (65). Specifically, buried surface area (BSA) was calculated by Equation 1,
| (Eq. 1) |
where A indicates total surface area of the individual protein or complex.
Biosensor binding assays
Anti-GST antibody (GE Healthcare) was immobilized on all four flow cells of a CM5 sensor chip in a Biacore2000 instrument (GE Healthcare) using standard amine coupling chemistry. A single flow cell was used to immobilize GST-RPN13PRU (400 response units) on the anti-GST surface, two flow cells for GST-RPN13 (300–600 response units), and the fourth flow cell for GST (250 response units). Binding of GFP-RPN2 and UCH37 constructs was tested in HBS-TBP (10 mm Tris, pH 7.4, 150 mm NaCl, 1.0 mm TCEP, 0.1 mg/ml BSA, 0.005% polyoxyethylene sorbitan monolaurate (P20)) and 100 nm GFP-RPN2(918–953) or UCH37, when applicable, with a flow rate of 50 μl/min in a 2- or 3-fold dilution series using the highest concentration for each sample as follows: RPN2(932–953) (32 nm), RPN2(940–953) (32 nm), GFP (10 μm), and UCH37 (32 nm). Experiments evaluating GFP-RPN2 binding required no regeneration step because RPN2 completely dissociates within 20 min. However, UCH37 did not completely dissociate after 60 min, and the chip was regenerated with regeneration buffer of HBS-TBP plus 850 mm NaCl and 0.05% SDS. The binding of ubiquitin was measured under three buffer conditions: HBS-TBP, HBS-TBP supplemented with 100 nm GFP-RPN2(918–953), and LSPB-TBP (20 mm NaH2PO4, pH 6.5, 30 mm NaCl, 4.0 mm DTT, 0.1 mg/ml BSA, 0.005% polyoxyethylene sorbitan monolaurate (P20)). Binding of ubiquitin was tested in a 1.5-fold dilution series with the highest concentration of 500 μm. Sensor data were processed using ScrubberPro6 (BioLogic Software) and globally fit to a 1:1 interaction model, using a kinetic analysis for RPN2 binding and equilibrium analysis for ubiquitin.
Fluorescence polarization
Fluorescence polarization assays were performed on a Biotek Synergy Neo HTS Multi-Mode Microplate Reader using 485/528 nm excitation/emission wavelengths. For binding studies, a dilution series of purified RPN13 or RPN13PRU was made in FP Assay Buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mg/ml casein, 0.005% Tween 20) in the presence of 100 pm fluorescein-labeled RPN2 peptide or 30 nm Ub-fluorescein (Boston Biochem) at room temperature. For binding competition assays, a dilution series of unlabeled peptides was made in FP Assay Buffer with 50 nm RPN13PRU and 100 pm fluorescein-labeled RPN2(940–952) peptide. Reaction equilibrium was reached within the time frame of experimental setup for all assays, so additional incubation times were not required. Dissociation constants (KD values) and IC50 values were calculated using GraphPad Prism 6 (GraphPad Software) by fitting raw polarization data to Equations 2 and 3.
| (Eq. 2) |
and
| (Eq. 3) |
In the case of the F948A mutant, where the lower baseline was not reached due to the weak IC50, FPmin was manually constrained to the polarization value of the peptide-only control. IC50 values were converted to Ki values using Equation 4 (66).
| (Eq. 4) |
Sequence alignments
Sequence alignments were performed using the Clustal Omega server provided by EMBL-EBI (67, 68). Highlighting of conserved residues and denotations of residues involved in protein-protein interactions were applied through the ESPript 3.0 web server (69, 70).
Author contributions
C. P. H. and T. Y. conceived the study. C. P. H., R. T. V., and C. W. H. designed experiments and wrote the paper. R. T. V. and C. W. H. conducted the experiments. H. R. made important contributions to X-ray crystallography experiments.
Acknowledgments
We thank Frank Whitby, Heidi Schubert, Erik Kish-Trier, Kath Ferrell, Binita Shakya, Ira Baci, and Matt Sdano for technical assistance. We also thank Mark Peterson for peptide synthesis and Bob Cohen (Colorado State University) for generous gifts and helpful comments on the manuscript. Portions of this research were performed at the National Synchrotron Light Source (NSLS) and the Stanford Synchrotron Radiation Lightsource (SSRL). The NSLS is funded by National Institutes of Health Grant P41RR012408 from the NCRR. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by National Institutes of Health Grant P41RR001209 from NIGMS and the NCRR. Use of the NSLS and SSRL is supported by the Department of Energy Office of Basic Energy Sciences (SSRL Contract No. DE-AC02-76SF00515). NSLS operations are also supported by the National Institutes of Health. Mass spectrometry validation of purified proteins was performed by the University of Utah Mass Spectrometry and Proteomics Core Facility, which is supported by National Institutes of Health Grant P30CA042014 from the NCI. Sequencing of constructs used in this work was performed at the DNA Sequencing Core Facility, University of Utah.
This work was supported in part by National Institutes of Health Grants R01 GM059135 and R21 CA191929 (to C. P. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5V1Y and 5V1Z) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CP
- core particle
- SPR
- surface plasmon resonance
- FP
- fluorescence polarization
- RP
- regulatory particle
- PRU
- pleckstrin-like receptor of ubiquitin
- IWF
- intrinsic tryptophan fluorescence
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- Ni-NTA
- nickel-nitrilotriacetic acid
- TCEP
- tris(2-carboxyethyl)phosphine
- GS
- glutathione-Sepharose
- Ub
- Ubiquitin
- TEV
- tobacco etch virus
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- DMF
- N,N-dimethylformamide
- CV
- column volume.
References
- 1. Pickart C. M., and Cohen R. E. (2004) Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 2. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenzweig R., Bronner V., Zhang D., Fushman D., and Glickman M. H. (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 287, 14659–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomez T. A., Kolawa N., Gee M., Sweredoski M. J., and Deshaies R. J. (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fatimababy A. S., Lin Y. L., Usharani R., Radjacommare R., Wang H. T., Tsai H. L., Lee Y., and Fu H. (2010) Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 277, 796–816 [DOI] [PubMed] [Google Scholar]
- 6. Saeki Y., Sone T., Toh-e A., Yokosawa H. (2002) Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem. Biophys. Res. Commun. 296, 813–819 [DOI] [PubMed] [Google Scholar]
- 7. Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., and Finley D. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 8. da Fonseca P. C., He J., and Morris E. P. (2012) Molecular model of the human 26S proteasome. Mol. Cell 46, 54–66 [DOI] [PubMed] [Google Scholar]
- 9. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., and Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., and Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sakata E., Bohn S., Mihalache O., Kiss P., Beck F., Nagy I., Nickell S., Tanaka K., Saeki Y., Förster F., and Baumeister W. (2012) Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Unverdorben P., Beck F., Śledź P., Schweitzer A., Pfeifer G., Plitzko J. M., Baumeister W., and Förster F. (2014) Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 111, 5544–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aufderheide A., Beck F., Stengel F., Hartwig M., Schweitzer A., Pfeifer G., Goldberg A. L., Sakata E., Baumeister W., and Förster F. (2015) Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 112, 8626–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Śledź P., Unverdorben P., Beck F., Pfeifer G., Schweitzer A., Förster F., and Baumeister W. (2013) Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 7264–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beck F., Unverdorben P., Bohn S., Schweitzer A., Pfeifer G., Sakata E., Nickell S., Plitzko J. M., Villa E., Baumeister W., and Förster F. (2012) Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 109, 14870–14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schweitzer A., Aufderheide A., Rudack T., Beck F., Pfeifer G., Plitzko J. M., Sakata E., Schulten K., Förster F., and Baumeister W. (2016) Structure of the human 26S proteasome at a resolution of 3.9 A. Proc. Natl. Acad. Sci. U.S.A. 113, 7816–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang X., Luan B., Wu J., and Shi Y. (2016) An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 23, 778–785 [DOI] [PubMed] [Google Scholar]
- 18. Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., and Groll M. (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., and Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., and Murata S. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., and Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., and Cohen R. E. (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]
- 23. Chen X., Lee B. H., Finley D., and Walters K. J. (2010) Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell 38, 404–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hicke L., Schubert H. L., and Hill C. P. (2005) Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 25. Mukhopadhyay D., and Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 26. Lu X., Liu F., Durham S. E., Tarasov S. G., and Walters K. J. (2015) A high affinity hRpn2-derived peptide that displaces human Rpn13 from proteasome in 293T cells. PLoS One 10, e0140518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnston S. C., Riddle S. M., Cohen R. E., and Hill C. P. (1999) Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 18, 3877–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Q., Young P., and Walters K. J. (2005) Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J. Mol. Biol. 348, 727–739 [DOI] [PubMed] [Google Scholar]
- 29. Boudreaux D. A., Maiti T. K., Davies C. W., and Das C. (2010) Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc. Natl. Acad. Sci. U.S.A. 107, 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang D., Raasi S., and Fushman D. (2008) Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 377, 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. VanderLinden R. T., Hemmis C. W., Schmitt B., Ndoja A., Whitby F. G., Robinson H., Cohen R. E., Yao T., and Hill C. P. (2015) Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol. Cell 57, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X., Randles L., Shi K., Tarasov S. G., Aihara H., and Walters K. J. (2016) Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 reveal distinct binding mechanisms between substrate receptors and shuttle factors of the proteasome. Structure 24, 1257–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., and Goldberg A. L. (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sahtoe D. D., van Dijk W. J., El Oualid F., Ekkebus R., Ovaa H., and Sixma T. K. (2015) Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol. Cell 57, 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamazaki J., Sasaki K., Kawahara H., Hisanaga S., Tanaka K., and Murata S. (2007) Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell. Biol. 27, 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamazaki J., Hirayama S., and Murata S. (2015) Redundant roles of Rpn10 and Rpn13 in recognition of ubiquitinated proteins and cellular homeostasis. PLoS Genet. 11, e1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szlanka T., Haracska L., Kiss I., Deák P., Kurucz E., Andó I., Virágh E., and Udvardy A. (2003) Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J. Cell Sci. 116, 1023–1033 [DOI] [PubMed] [Google Scholar]
- 38. Al-Shami A., Jhaver K. G., Vogel P., Wilkins C., Humphries J., Davis J. J., Xu N., Potter D. G., Gerhardt B., Mullinax R., Shirley C. R., Anderson S. J., and Oravecz T. (2010) Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS One 5, e13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haas A. L., and Bright P. M. (1985) The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J. Biol. Chem. 260, 12464–12473 [PubMed] [Google Scholar]
- 40. Ryu H., Gygi S. P., Azuma Y., Arnaoutov A., and Dasso M. (2014) SUMOylation of Psmd1 controls Adrm1 interaction with the proteasome. Cell Rep. 7, 1842–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Besche H. C., Sha Z., Kukushkin N. V., Peth A., Hock E. M., Kim W., Gygi S., Gutierrez J. A., Liao H., Dick L., and Goldberg A. L. (2014) Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 33, 1159–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oerlemans R., Franke N. E., Assaraf Y. G., Cloos J., van Zantwijk I., Berkers C. R., Scheffer G. L., Debipersad K., Vojtekova K., Lemos C., van der Heijden J. W., Ylstra B., Peters G. J., Kaspers G. L., Dijkmans B. A., et al. (2008) Molecular basis of bortezomib resistance: proteasome subunit β5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood 112, 2489–2499 [DOI] [PubMed] [Google Scholar]
- 43. Kumar S., and Rajkumar S. V. (2008) Many facets of bortezomib resistance/susceptibility. Blood 112, 2177–2178 [DOI] [PubMed] [Google Scholar]
- 44. Lü S., and Wang J. (2013) The resistance mechanisms of proteasome inhibitor bortezomib. Biomark. Res. 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kortuem K. M., and Stewart A. K. (2013) Carfilzomib. Blood 121, 893–897 [DOI] [PubMed] [Google Scholar]
- 46. D'Arcy P., Wang X., and Linder S. (2015) Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54 [DOI] [PubMed] [Google Scholar]
- 47. Anchoori R. K., Karanam B., Peng S., Wang J. W., Jiang R., Tanno T., Orlowski R. Z., Matsui W., Zhao M., Rudek M. A., Hung C. F., Chen X., Walters K. J., and Roden R. B. (2013) A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24, 791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trader D. J., Simanski S., and Kodadek T. (2015) A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells. J. Am. Chem. Soc. 137, 6312–6319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song Y., Ray A., Li S., Das D. S., Tai Y. T., Carrasco R. D., Chauhan D., and Anderson K. C. (2016) Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia 30, 1877–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fejzo M. S., Anderson L., von Euw E. M., Kalous O., Avliyakulov N. K., Haykinson M. J., Konecny G. E., Finn R. S., and Slamon D. J. (2013) Amplification target ADRM1: role as an oncogene and therapeutic target for ovarian cancer. Int. J. Mol. Sci. 14, 3094–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fejzo M. S., Dering J., Ginther C., Anderson L., Ramos L., Walsh C., Karlan B., and Slamon D. J. (2008) Comprehensive analysis of 20q13 genes in ovarian cancer identifies ADRM1 as amplification target. Genes Chromosomes Cancer 47, 873–883 [DOI] [PubMed] [Google Scholar]
- 52. Pilarsky C., Wenzig M., Specht T., Saeger H. D., and Grützmann R. (2004) Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia 6, 744–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang X., Miao X., Wen Y., Hu J., Dai W., and Yin B. (2012) A possible connection between adhesion regulating molecule 1 overexpression and nuclear factor kappa B activity in hepatocarcinogenesis. Oncol. Rep. 28, 283–290 [DOI] [PubMed] [Google Scholar]
- 54. Fejzo M. S., Anderson L., Chen H. W., Anghel A., Zhuo J., Anchoori R., Roden R., and Slamon D. J. (2015) ADRM1-amplified metastasis gene in gastric cancer. Genes Chromosomes Cancer 54, 506–515 [DOI] [PubMed] [Google Scholar]
- 55. Zheng X., Guo Y., Chen Y., Chen M., Lin Z., Wu Y., and Chen Y. (2015) Knockdown of adhesion-regulating molecule 1 inhibits proliferation in HL60 cells. Acta Haematol. 134, 88–100 [DOI] [PubMed] [Google Scholar]
- 56. Chen W., Hu X. T., Shi Q. L., Zhang F. B., and He C. (2009) Knockdown of the novel proteasome subunit Adrm1 located on the 20q13 amplicon inhibits colorectal cancer cell migration, survival and tumorigenicity. Oncol. Rep. 21, 531–537 [PubMed] [Google Scholar]
- 57. Jang S. H., Park J. W., Kim H. R., Seong J. K., and Kim H. K. (2014) ADRM1 gene amplification is a candidate driver for metastatic gastric cancers. Clin. Exp. Metastasis 31, 727–733 [DOI] [PubMed] [Google Scholar]
- 58. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 59. Pickart C. M., and Raasi S. (2005) Controlled synthesis of polyubiquitin chains. Methods Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 60. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 61. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., and Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 62. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen V. B., Arendall W. B. 3rd., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. DeLano W. L. (2015) The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC, New York [Google Scholar]
- 66. Nikolovska-Coleska Z., Wang R., Fang X., Pan H., Tomita Y., Li P., Roller P. P., Krajewski K., Saito N. G., Stuckey J. A., and Wang S. (2004) Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 332, 261–273 [DOI] [PubMed] [Google Scholar]
- 67. McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., Cowley A. P., and Lopez R. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gouet P., Robert X., and Courcelle E. (2003) ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]