Abstract
Tspan5 is a member of a subgroup of tetraspanins referred to as TspanC8. These tetraspanins directly interact with the metalloprotease ADAM10, regulate its exit from the endoplasmic reticulum and subsequent trafficking, and differentially regulate its ability to cleave various substrates and activate Notch signaling. The study of Tspan5 has been limited by the lack of good antibodies. This study provides new insights into Tspan5 using new monoclonal antibodies (mAbs), including two mAbs recognizing both Tspan5 and the highly similar tetraspanin Tspan17. Using these mAbs, we show that endogenous Tspan5 associates with ADAM10 in human cell lines and in mouse tissues where it is the most abundant, such as the brain, the lung, the kidney, or the intestine. We also uncover two TspanC8-specific motifs in the large extracellular domain of Tspan5 that are important for ADAM10 interaction and exit from the endoplasmic reticulum. One of the anti-Tspan5 mAbs does not recognize Tspan5 associated with ADAM10, providing a convenient way to measure the fraction of Tspan5 not associated with ADAM10. This fraction is minor in the cell lines tested, and it increases upon transfection of cells with TspanC8 tetraspanins such as Tspan15 or Tspan33 that inhibit Notch signaling. Finally, two antibodies inhibit ligand-induced Notch signaling, and this effect is stronger in cells depleted of the TspanC8 tetraspanin Tspan14, further indicating that Tspan5 and Tspan14 can compensate for each other in Notch signaling.
Keywords: ADAM, intracellular trafficking, metalloprotease, monoclonal antibody, Notch pathway, tetraspanin, ADAM10, Tspan5
Introduction
Tetraspanins form a family of proteins with four transmembrane domains expressed by all metazoans. These proteins possess a number of specific features, including conserved residues and a specific fold in the largest of the two extracellular domains (the large extracellular loop (LEL)5) that differentiate them from other proteins with four transmembrane domains (1–4). Identification of pathological mutations in humans and genetic approaches in the mouse or invertebrates have shown the importance of these molecules. In mammals, particular tetraspanins have, for example, been shown to play a key role in sperm-egg fusion, vision, kidney function, immunity, or muscle regeneration (1–4). A remarkable property of the most characterized tetraspanins is to associate at the cell surface with one another and with non-tetraspanin integral proteins to organize a network of interaction referred to as the “tetraspanin web” or tetraspanin-enriched microdomains. In this network, tetraspanins associate directly and specifically with a limited number of molecular partners that they connect to other tetraspanins. Well characterized primary complexes include the complexes formed by CD151 (Tspan24) and the laminin-binding integrins α3β1 and α6β1 as well as the complex formed by CD81 (Tspan28) and CD19, a co-stimulatory molecule of B lymphocytes. In addition, CD81 shares with CD9 (Tspan29) two common partners, CD9P-1/EWI-F and EWI-2, that are Ig domain proteins of unknown function (1–4).
Tspan5 is a highly conserved tetraspanin; the human, mouse, and rat proteins are completely identical and share 91, 44, and 38% identity with the closest orthologs found in Danio rerio, Drosophila melanogaster, and Caenorhabditis elegans, respectively (5–8). Tspan5 is a member of a subgroup of tetraspanins that have 8 cysteines in the LEL (others have 6 or 4 cysteines) and are consequently referred to as TspanC8 (7–10). Mammals express six of these TspanC8 tetraspanins that share a common partner, the metalloprotease ADAM10, a member of the ADAM (A Disintegrin And Metalloprotease domain) family of metalloproteases (8, 10, 11). These membrane-anchored enzymes mediate a proteolytic cleavage of various transmembrane proteins within their extracellular region, a process referred to as ectodomain shedding (12, 13). ADAM10 cleaves off the ectodomain of more than 40 transmembrane proteins, including cytokine and growth factor precursors, as well as adhesion proteins such as E- and N-cadherins (13). Notably, ADAM10-mediated cleavage of the amyloid precursor protein prevents the formation of the amyloid peptide Aβ, a major component of amyloid plaques observed in Alzheimer's disease (14). ADAM10 plays also an essential role in Notch signaling; Notch ectodomain cleavage by ADAM10 allows a second cleavage by the γ-secretase complex that results in the release of the Notch intracellular domain and its translocation to the nucleus where it acts as a transcriptional cofactor (15–18).
TspanC8 tetraspanins regulate several aspects of ADAM10. They all regulate the exit of ADAM10 from the ER and target it either to late endosomes (Tspan10, 17) or the plasma membrane (Tspan5, -14, -15, and -33) (8, 10, 11). In addition, TspanC8 tetraspanins modulate the substrate specificity of ADAM10 (19, 20). In particular, Tspan5 and Tspan14 are positive and Tspan15 and Tspan33 negative regulators of Notch signaling (8, 19). Also, of all TspanC8 tetraspanins tested, only Tspan15 was shown to regulate ADAM10-mediated cleavage of N-cadherin (11, 19, 20). These functional differences may be the result of a different action of TspanC8 on ADAM10 membrane compartmentalization (19). Alternatively, TspanC8 might direct substrate specificity by constraining ADAM10 into defined conformations (20).
In the absence of good antibodies, the study of Tspan5 and other TspanC8 has relied on the transfection of tagged molecules, with potential pitfalls arising from overexpression or the addition of a tag. Here, we report on the generation of anti-Tspan5 monoclonal antibodies and use them to investigate several aspects of Tspan5, including its expression profile, subcellular localization, and the interaction of the endogenous protein with ADAM10 and with the tetraspanin web. We also show that two of these mAbs inhibit ligand-induced Notch signaling.
Results
Generation of antibodies recognizing Tspan5
To generate anti-Tspan5 mAbs, we immunized mice twice with U2OS cells stably expressing Tspan5-GFP and twice with a Tspan5-GFP immunoprecipitate. Because the human, mouse, and rat Tspan5 molecules are completely identical, Tspan5 knock-out mice were used. Hybridomas were screened by indirect labeling of live U2OS cells stably expressing Tspan5-GFP and flow cytometry analysis. Out of more than 3000 clones tested, we isolated nine hybridomas stably secreting antibodies that stained U2OS-Tspan5 cells proportionally to the level of Tspan5-GFP expressed by the cells. Three examples are shown in Fig. 1A. As a control, the labeling by the CD81 antibody did not change according to the GFP signal, and the labeling by the anti-ADAM10 mAb 11G2 reached a plateau, as described previously (8, 19). The characteristics of these antibodies are shown in Table 1. To validate that these antibodies indeed recognize Tspan5, GFP-Tspan5 was immunoprecipitated from U2OS/Tspan5 cells using GFP trap beads after lysis in RIPA buffer (Fig. 1B). This lysis buffer is known to dissociate from tetraspanins most if not all the associated proteins. Indeed, under this condition, ADAM10 was no longer co-immunoprecipitated with Tspan5, although it is strongly co-immunoprecipitated after lysis with Brij 97 (Fig. 1B). All antibodies tested recognized by Western blotting the immunoprecipitated Tspan5 GFP (see three examples in Fig. 1B), as shown by the strong signal perfectly overlapping with the signal obtained with an anti-GFP antibody.
Figure 1.
Characterization of new anti-Tspan5 mAb. A, flow cytometry analysis of U2OS cells expressing Tspan5 GFP and stained or not with mAbs to ADAM10, CD81, or three anti-Tspan5 mAbs. B, U2OS cells expressing Tspan5 GFP were lysed in RIPA or Brij 97 lysis buffer as indicated, before immunoprecipitation (IP) of Tspan5 using GFP trap beads. The samples were analyzed by Western blotting using the indicated antibodies. Note that the interaction of Tspan5 with ADAM10 is disrupted in RIPA buffer. C, binding of mAb TS5-2 to HCT116 was analyzed by flow cytometry 3 days after transfection with a control siRNA or two siRNA targeting Tspan5. D, HCT116 cells were lysed 3 days after transfection with a control siRNA or two Tspan5 siRNAs. In the top panels, the cells were lysed directly in Laemmli buffer, before Western blotting using a combination of anti-Tspan5 mAb TS5-2 and a secondary antibody. In the bottom panel, the cells were lysed in Brij 97 buffer, and Tspan5 was immunoprecipitated using TS5-2. The presence of Tspan5 in the immunoprecipitate was determined using a combination of biotin-labeled TS-2 mAb and streptavidin. E, mouse colon CT26 cells were lysed 3 days after transfection with a control siRNA or two to Tspan5 siRNAs. The cells were lysed directly in Laemmli buffer, before Western blotting using a combination of anti-Tspan5 mAb TS5-2 and a secondary antibody.
Table 1.
Characterization of anti-Tspan5 mAbs
The binding of the mAbs to Tspan5, Tspan14, Tspan15, and Tspan33 was analyzed using U2OS cells stably expressing these molecules. The binding to the two chimeric molecules Tspan15LEL5 (in which the LEL of Tspan15 is replaced by that of Tspan5) and the reciprocal construct Tspan5LEL15 was analyzed by flow cytometry after transient transfection. The binding to Tspan10 and Tspan17 was analyzed by indirect immunofluorescence after saponin permeabilization of cells grown on coverslips. The binding to the Tspan5 mutants RDD and NIYF was analyzed by indirect immunofluorescence after Triton X-100 permeabilization of cells grown on coverslips. We did not identify the subclass of mAb 10G5, which is not an IgA, IgM, Ig1, Ig2a, Ig2b, or Ig3. ND means not determined.
| Clone | Alias | Sub class | Antibody binds to: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T5 | T10 | T14 | T15 | T17 | T33 | Ts15 LEL5 | Ts5 LEL15 | RDD | NIYF | |||
| 16B8 | TS5-2 | IgG2a | Yes | No | No | No | No | No | Yes | No | Yes | No |
| 12E1 | TS5-1r | IgG1 | Yes | No | No | No | No | No | Yes | No | Yes | No |
| 10G11 | TS5/17 | IgG2b | Yes | No | No | No | Yes | No | Yes | No | Yes | Yes |
| 8H12 | TS5-3 | IgG2a | Yes | No | No | No | No | No | Yes | No | Yes | No |
| 8B1 | IgG2a | Yes | No | No | No | No | No | Yes | No | Yes | No | |
| 13G1 | IgG2b | Yes | No | No | No | Weak | No | Yes | No | No | No | |
| 20E2 | IgG2b | Yes | No | No | No | Yes | No | Yes | No | Yes | Yes | |
| 10G5 | ND | Yes | ND | No | No | ND | No | Yes | No | ND | ND | |
The mAb TS5-2 was selected for further studies because in the initial characterization it gave one of the strongest signals in Western blotting and flow cytometry. To validate that it recognized the endogenous Tspan5, we turned to colon cancer HCT116 cells that showed the strongest surface staining among various cell lines tested (data not shown). Silencing Tspan5 in these cells by two previously validated siRNAs (19) reduced by 70–80% the staining by TS5-2 in flow cytometry experiments (Fig. 1C). In addition, the mAb TS5-2 recognized by Western blotting endogenous levels of Tspan5 in HCT116 and CT26 cells (a human and a mouse colon cancer cell line, respectively) but not in Tspan5-silenced cells (Fig. 1, D and E). Moreover, this mAb immunoprecipitated a major ∼27–34-kDa band and a fainter ∼22-kDa thin band that were recognized by Western blotting by the TS5-2 mAb (Fig. 1D). None of these bands were visualized after silencing Tspan5 further indicating that they both correspond to Tspan5 (possibly to different conformations or glycosylated forms) and that TS5-2 only recognizes Tspan5 in these cells.
Specificity of Tspan5 antibodies and demonstration that they bind to the LEL
TspanC8 tetraspanins are characterized not only by the presence of 8 cysteines in the larger extracellular domain but also by the presence of specific residues not present in other tetraspanins (8). It was therefore important to test whether these anti-Tspan5 antibodies recognized other TspanC8. In a first set of experiments, we tested by Western blotting whether these antibodies recognized Tspan14, Tspan15, or Tspan33 in the lysates of U2OS cells stably expressing GFP-tagged versions of Tspan15, Tspan14, or Tspan33 (19), or after GFP immunoprecipitation. As shown in Fig. 2A and summarized in Table 1, none of the mAbs recognized these three tetraspanins.
Figure 2.
Specificity of selected anti-Tspan5 mAbs. A, U2OS cells stably expressing or not GFP-tagged Tspan5, Tspan14, Tspan15, and Tspan33 were lysed in RIPA buffer before immunoprecipitation (IP) of the transfected protein with GFP trap beads. The recognition of the transfected tetraspanin by the different mAbs was tested by Western blotting using both the cell lysate or the GFP-trap immunoprecipitate (IP). To control for the expression of the different transfected tetraspanins, the membrane was also incubated with an anti-GFP mAb. B, HeLa cells were transiently transfected with Tspan5, Tspan17, Tspan10, or CD9 and lysed 2 days later in Laemmli buffer. To control for the expression of the different transfected tetraspanins, the membrane was also incubated with an anti-GFP mAb. C, U2OS cells were transiently transfected with plasmids coding Tspan17 or Tspan5 and immunolabeled 2 days later with mAb TS5-2 and TS5/17. Note that the mAb TS5/17 also recognizes Tspan17 but TS5-2 does not (bar: 10 μm). D, U2OS cells were transiently transfected with plasmids coding the GFP-tagged chimeric tetraspanin Ts15LEL5, in which the LEL of Tspan15 was replaced by that of Tspan5 and the reverse chimera Ts5LEL15. The panel shows flow cytometry analysis of the binding of selected anti-Tspan5 mAb to the transfected cells.
Similarly, none of these mAbs recognized Tspan17 or Tspan10 by Western blotting after transfection in HeLa cells (Fig. 2B). However, two antibodies, TS5/17 (Fig. 2) and 20E2 (Table 1), strongly stained Tspan17-transfected cells, either by flow cytometry analysis (data not shown) or immunostaining of saponin-permeabilized U2OS cells grown on coverslips (Fig. 2C and Table 1). As shown previously in HeLa cells, Tspan10 was mainly intracellular after transfection in U2OS cells. None of our antibodies recognized this tetraspanin (data not shown).
We then used Tspan5/Tspan15 chimeric molecules to determine whether our antibodies recognize the small or the large extracellular domain. As summarized in Table 1, and exemplified for three of them in Fig 2C, all antibodies recognized, as shown by flow cytometry, a chimera in which the LEL of Tspan15 was replaced with that of Tspan5 (Ts15LEL5) but not the reverse chimera (Ts5LEL15). Thus, all antibodies bind to epitopes present in the LEL of Tspan5.
Absence of major intracellular pool of Tspan5
Using flow cytometry, we have tested the surface expression of Tspan5 in ∼25 hematopoietic or non-hematopoietic cells lines. In all cell lines tested, the surface expression level of Tspan5 was low as compared with that of ADAM10 or CD81 (data not shown). This, together with the fact that some tetraspanins such as CD63 are mainly intracellular proteins (2), prompted us to compare the expression levels of endogenous Tspan5 at the cell surface and in intracellular compartments. For this purpose, the surface pool of Tspan5 was first labeled using a combination of the anti-Tspan5 mAb TS5-2 and an anti-mouse polyclonal antibody coupled to Alexa Fluor 568 (Fig. 3, top, red). In the second step, the cells were incubated, in the presence of saponin to permeabilize them, with a combination of the Tspan5 mAb and an anti-mouse polyclonal antibody coupled to Alexa Fluor 647. In this experiment, the surface pool of Tspan5 was labeled with the two secondary antibodies, whereas the internal pool was labeled only with the Alexa Fluor 647-coupled secondary antibody (Fig. 3, green). As a control for the non-specific intracellular signal, the Tspan5 mAb was replaced by a control IgG2a in the second step (Fig. 3, middle panels). As shown in Fig. 3, there was a weak intracellular signal with TS5-2 that was higher than that observed with the control IgG2a mAb, indicating the existence of an intracellular pool of Tspan5. This signal is much lower than the intracellular signal observed for CD63 (Fig. 3, right panels). Therefore, Tspan5 lacks a strong intracellular pool. Similar experiments were performed on U2OS cells (data not shown). The higher nonspecific intracellular signal observed with the IgG2a control mAb precluded determining whether there was an intracellular pool of Tspan5 in this cell line. We can conclude, however, that this cell line does not have a strong intracellular pool of Tspan5.
Figure 3.
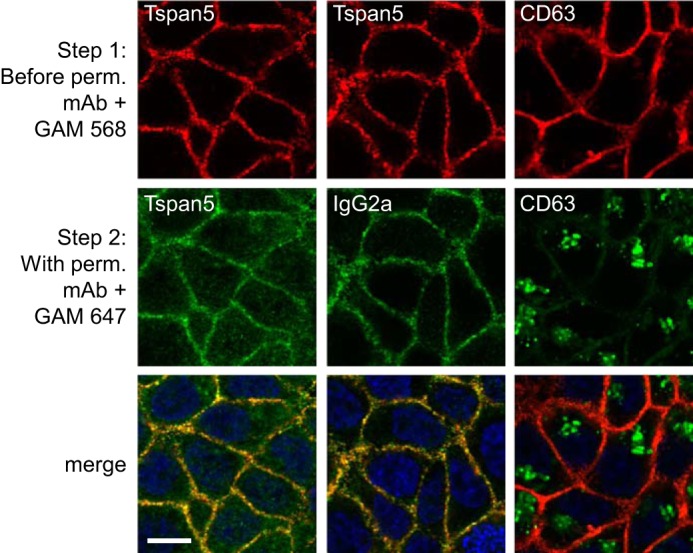
Confocal microscopy analysis of the intracellular pool of Tspan5 and CD63. HCT116 cells were grown for 2 days on coverslips before incubation with anti-Tspan5 (TS5-2) or CD63 mAb for 1 h at 4 °C. After washing and fixation with paraformaldehyde, the cells were incubated with an anti-mouse polyclonal antibody coupled to Alexa 568 to visualize the surface pool of the tetraspanin (red). In Step 2, the cells were incubated with either the same primary antibody or a control IgG2a mAb in the presence of saponin to permeabilize the cells, and subsequently with an anti-mouse polyclonal antibody coupled to Alexa 647 (green). In this experiment, the surface pool of Tspan5 or CD63 is labeled with the two secondary antibodies, whereas the internal pool is labeled only with the Alexa 647-coupled secondary antibody (green). This experiment has been performed twice with similar outcomes. Perm., permeabilization; GAM, goat anti-mouse antibody. Bar, 10 μm.
Endogenous Tspan5 interacts with ADAM10 and is a component of the tetraspanin web
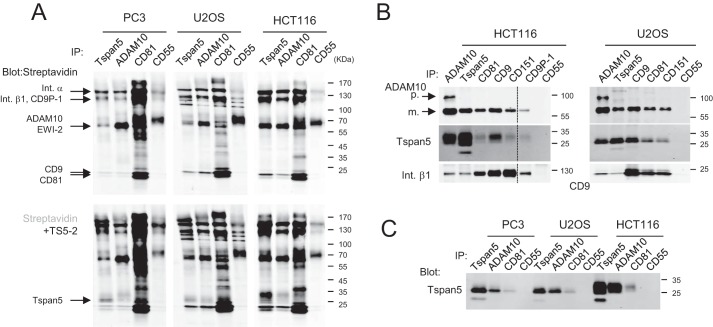
We then tested whether endogenous Tspan5 interacted with ADAM10, with other tetraspanins, and with selected components of the tetraspanin web. In a first set of experiments, several cell lines cells were surface-labeled with biotin before immunoprecipitations. All cells express Tspan5 at the mRNA level, but the most prominent TspanC8 expressed by these cells is Tspan15 in PC3 cells and Tspan14 in U2OS and HCT116 cells (8). As shown in Fig. 4A, the Tspan5 mAb precipitated in all cell types, but at different levels several biotin-labeled proteins that co-migrated with proteins co-immunoprecipitated with the prototypal tetraspanin CD81, including CD9 and CD81, or β1 integrin subunits. Although ectopically expressed Tspan5 is biotin-labeled, we did not detect Tspan5 in the Tspan5 immunoprecipitation after biotin labeling. This may be because the fraction of Tspan5 associated with ADAM10 may not be efficiently biotin-labeled (data not shown). The presence of Tspan5 in Tspan5 and ADAM10 immunoprecipitates was validated by incubating the same membrane with biotin-labeled TS5-2 mAb, yielding an additional band of 25–30 kDa corresponding to Tspan5.
Figure 4.
Endogenous Tspan5 associates with ADAM10 and other tetraspanins. PC3, U2OS, and HCT116 were lysed in Brij 97, and immunoprecipitations (IP) were performed as indicated on the top of each lane. A, surface proteins were labeled with biotin before lysis and were visualized using Alexa 680-labeled streptavidin (top). In the 2nd step, the membrane was incubated with biotin-labeled TS5-2 mAb and again with Alexa 680-labeled streptavidin to confirm the presence of Tspan5 in the immunoprecipitates and compare its molecular weight with the other proteins present in the different immunoprecipitations (bottom). B, immunoprecipitations were performed from non-labeled cells. The composition of the immunoprecipitates was analyzed by Western blotting using various biotin-labeled mAbs. The mature (m.) and proform (p.) forms of ADAM10 are indicated by arrows. C, comparison of the migration profile of Tspan5 immunoprecipitated from PC3, U2OS, and HCT116 cells. Int., integrin.
Similar experiments were performed without biotin labeling, allowing analysis of the composition of the different immunoprecipitates by Western blotting using biotin-labeled antibodies (Fig. 4B). Overall, the data show that endogenous Tspan5 co-immunoprecipitates and/or is co-immunoprecipitated with ADAM10, other tetraspanins such as CD9, CD81, and CD151, as well as the β1 integrin subunit or CD9P-1. The apparent molecular weight of Tspan5 is higher in HCT116 cells than in PC3 and U2OS cells (Fig. 4C). This is likely to be due to different glycosylation patterns, as will be addressed later.
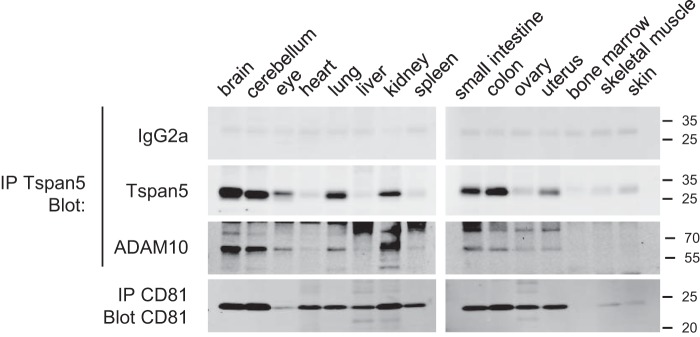
Tspan5 interacts with ADAM10 in mouse organs
Initial attempts to characterize the tissue distribution of Tspan5 by immunohistochemistry in the mouse did not succeed, probably because of the low abundance of Tspan5. As an alternative approach, various tissues were lysed in Brij 97 lysis buffer, and immunoprecipitations were performed with the mAb TS5-2 (Fig. 5). The presence of Tspan5 was determined by Western blotting using a combination of TS5-2 mAb conjugated to biotin and streptavidin. This approach revealed that the major sites of Tspan5 expression at the protein level were the brain, cerebellum, eye, kidney, lung, uterus, and intestine. The high expression of Tspan5 at the protein level in the brain, lung, and kidney is consistent with its expression at the RNA level (21). However, in this previous study Tspan5 RNA was found to be present at the same level in the liver and the heart than in the lung, which contrasts with our data because we found no expression or very little expression of Tspan5 protein in these organs. In the tissues where the expression of Tspan5 is high, including the brain, ADAM10 was co-immunoprecipitated with Tspan5.
Figure 5.
Tspan5 associates with ADAM10 in mouse organs. Mouse organs were lysed, and immunoprecipitations (IP) were performed using Tspan5 or CD81 antibodies. The presence of the target antigen was analyzed by Western blotting using biotin-labeled mAbs. Because Tspan5 co-migrates with a nonspecific band, the membranes were incubated with an irrelevant biotin-labeled mAb of the same subclass (IgG2a) before incubation with the Tspan5 mAb. The presence of ADAM10 in the Tspan5 immunoprecipitates was analyzed using a polyclonal antibody to ADAM10. This experiment has been done twice with similar outcomes.
An antibody that does not recognize Tspan5 associated with ADAM10 reveals that the majority of Tspan5 is associated with ADAM10 in U2OS and HCT116 cells
Previous studies have shown that the binding of certain anti-tetraspanin CD151 antibodies is prevented by the interaction with integrins that conceal the epitope, and as a consequence these antibodies only recognize the fraction of CD151 not associated with integrins (22–26). In our initial screening, we observed that TS5-1r failed to co-immunoprecipitate ADAM10 from U2OS-N1 cells (data not shown), suggesting that it may be such an antibody. As a first step to validate this hypothesis, immunoprecipitations in various detergents were performed using either TS5-1r or TS5-2. After cell lysis with RIPA, a condition that disrupts the interaction of ADAM10 with Tspan5, both antibodies immunoprecipitated similar amounts of Tspan5, both in HCT116 cells and U2OS cells (Fig. 6A). In contrast, after lysis with Brij 97 or digitonin, the interaction with ADAM10 was preserved, and TS5-1r precipitated only a small fraction of Tspan5. Notably, the major band co-immunoprecipitated with TS5-1r in HCT116 cells after Brij 97 or digitonin lysis had a slightly lower molecular weight than the bulk of Tspan5. Importantly, TS5-1r failed to immunoprecipitate ADAM10 under all conditions tested (except, in some experiments, for a slight amount in HCT116 cells after lysis with Brij 97, a detergent that preserves indirect interactions in the tetraspanin web), confirming our initial observation. The inability of TS5-1r to co-immunoprecipitate ADAM10 is not a consequence of the lower immunoprecipitation of Tspan5 after Brij 97 or digitonin lysis (Fig. 6A) because this mAb also failed to co-immunoprecipitate ADAM10 in U2OS cells overexpressing Tspan5, in which it efficiently immunoprecipitated this tetraspanin (Fig. 6B).
Figure 6.
The majority of Tspan5 molecules associates with ADAM10. A, HCT116 cells were lysed in the presence of Brij 97, digitonin, and RIPA buffer before immunoprecipitations (IP) with the anti-ADAM10 mAb 11G2 or the anti-Tspan5 mAbs TS5-2 and TS5-1r. The presence of Tspan5 and ADAM10 in the immunoprecipitates was visualized by immunoblotting using biotin-labeled TS5-2 and 11G2 mAbs. Note that TS5-1r does not co-immunoprecipitate ADAM10 and that its ability to immunoprecipitate Tspan5 is poor under lysis conditions preserving the interaction of Tspan5 with ADAM10. The band corresponding to the proform (p.) of ADAM10 is indicated. B, U2OS-N1 cells stably expressing GFP-tagged Tspan5 were lysed in the presence of Brij 97 or digitonin before immunoprecipitations with the anti-ADAM10 mAb 11G2 or the anti-Tspan5 mAbs TS5-2 and TS5-1r. The presence of Tspan5 and ADAM10 in the immunoprecipitates was visualized by immunoblotting using biotin-labeled TS5-2 and 11G2 mAbs. Note that TS5-1r does not co-immunoprecipitate ADAM10. C, HCT116 and U2OS cells were transfected or not with a control siRNA or an siRNA targeting ADAM10 and lysed 3 days later in Brij 97. Immunoprecipitations were performed as indicated at the top of each lane. The presence of Tspan5 and ADAM10 in the immunoprecipitates was visualized by immunoblotting using biotin-labeled TS5-2 and 11G2 mAbs. Note the change of Tspan5 molecular weight upon ADAM10 silencing and the better immunoprecipitation of Tspan5 by TS5-1r. The band corresponding to the proform (p.) of ADAM10 is indicated. D, HCT116 or U2OS-N1 cells were lysed in digitonin and were subjected to two rounds of immunoprecipitation with a control mAb or the anti-ADAM10 mAb 11G2 or the anti-Tspan5 mAb TS5-2. Each sample was then separated in two for immunoprecipitations with the anti-ADAM10 and the anti-Tspan5 mAbs. The presence of Tspan5 and ADAM10 in the immunoprecipitates was visualized by immunoblotting using biotin-labeled TS5-2 and 11G2 mAbs. Except for B, the experiments in this figure have been done three times with similar outcomes.
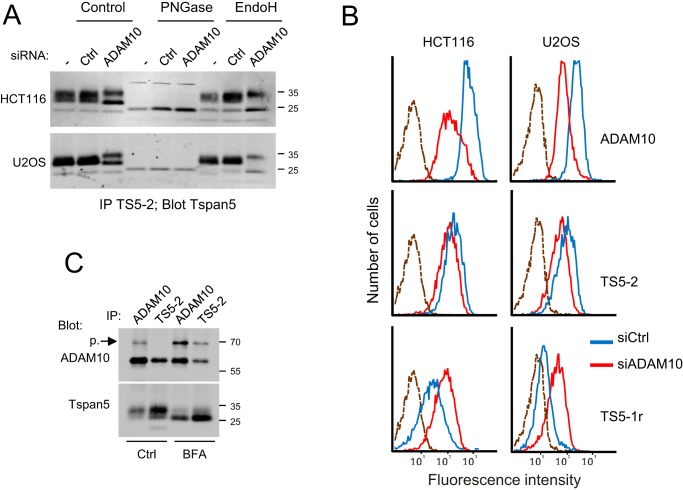
The above experiments indicated that the mAb TS5-1r efficiently immunoprecipitates Tspan5, but only under conditions disrupting the interaction with ADAM10. To validate that the low recognition of Tspan5 by TS5-1r after Brij 97 or digitonin lysis is due to the interaction with ADAM10, we next analyzed the effect of ADAM10 silencing on the ability of TS5-1r to immunoprecipitate Tspan5 after Brij 97 lysis (Fig. 6B). Surprisingly, silencing ADAM10 in HCT116 and U2OS cells with two different siRNAs (Fig. 6C, 7A) changed the migration profile of Tspan5; the main ∼27–34-kDa diffuse band disappeared, and two bands of slightly higher and lower molecular weight appeared or were reinforced. The lower molecular weight band (∼27 kDa) co-migrated with the band recognized by TS5-1r in untreated Brij 97 or digitonin-lysed cells. Both bands were similarly immunoprecipitated with mAbs TS5-1r and TS5-2 after ADAM10 silencing, showing that the low level of Tspan5 immunoprecipitation by TS5-1r in control cells is a consequence of ADAM10 expression and that ADAM10 prevents the binding of this mAb to Tspan5. As a consequence, and consistent with the finding that TS5-1r does not co-immunoprecipitate ADAM10, the fraction of Tspan5 immunoprecipitated by TS5-1r under conditions preserving the interaction with ADAM10 (∼15–20%, excluding the 22-kDa but not the 27-kDa band, Fig. 6C) corresponds to the fraction not associated with ADAM10. To confirm this finding, Tspan5 and ADAM10 were immunoprecipitated after antibody-mediated depletion of these proteins in U2OS or HCT116 cell lysates. As shown in Fig. 6D, removal of ADAM10 using the mAb 11G2 diminished by 80–85% the amount of Tspan5 present in the lysate confirming that the majority of Tspan5 is associated with ADAM10. In contrast, depletion of Tspan5 had no effect or a modest effect on the amount of ADAM10 immunoprecipitated with the mAb 11G2.
Figure 7.
ADAM10 facilitates Tspan5 exit from the ER. A, HCT116 and U2OS cells were transfected or not with a control siRNA or an siRNA targeting ADAM10 and grown for 3 days before lysis in Brij 97, and immunoprecipitations with mAb TS5-2 were performed. The immunoprecipitated proteins were treated or not with PNGase F or EndoH, as indicated before electrophoresis and immunoblotting using biotin-labeled TS5-2 mAb. Note that the lower molecular weight form of Tspan5 reinforced after ADAM10 silencing is EndoH-sensitive. B, binding of mAb TS5-2 and TS5-1r to HCT116 and U2OS cells, as well as that of the anti-ADAM10 mAb 11G2, was analyzed by flow cytometry 3 days after transfection with a control siRNA or an siRNA targeting ADAM10. Note the decrease of the binding of TS5-2 and the increase of the binding of TS5-1r. The dashed line corresponds to the labeling with only the secondary reagent. C, HCT116 cells treated or not with BFA were lysed using Brij 97 before immunoprecipitations with the anti-ADAM10 mAb 11G2 or the anti-Tspan5 mAb TS5-2 and TS5-1r. The presence of Tspan5 and ADAM10 in the immunoprecipitates was visualized by immunoblotting using biotin-labeled TS5-2 and 11G2 mAbs. Note that TS5-2 co-immunoprecipitates the proform of ADAM10 after BFA treatment and that ADAM10 co-immunoprecipitates the immature form of Tspan5. The experiments in this figure have been done twice.
ADAM10 regulates Tspan5 exit from the ER and glycosylation
The above data showed that silencing ADAM10 (Fig. 6C) changed the migration profile of Tspan5, with the main band disappearing at the profit of two bands of slightly higher and lower molecular weight (∼27 and 34 kDa). We then investigated whether these new bands corresponded to different glycoforms of Tspan5 (Fig. 7A). After digestion of Tspan5 immunoprecipitates with PNGase F, which removes all N-linked glycans, only a band of lower molecular weight was observed in all samples (∼24 kDa), both in HCT116 cells and U2OS cells. Thus, the change in Tspan5 molecular weight observed upon ADAM10 silencing corresponds to a change in glycosylation. The ∼27-kDa band observed after silencing ADAM10 was sensitive to EndoH (Fig. 7A), an enzyme that removes high mannose structures, which are trimmed in the Golgi, indicating that this band corresponds to a fraction of Tspan5 retained in the ER in the absence of ADAM10. In contrast, the higher ∼34-kDa band is not sensitive to EndoH and therefore corresponds to a fraction of ADAM10 having egressed from the ER. Because this band is efficiently immunoprecipitated with TS5-1r (Fig. 6C), it corresponds to a fraction of Tspan5 not associated with ADAM10. Thus, the interaction with ADAM10 modifies the glycosylation of Tspan5 in the Golgi.
Retention of Tspan5 in the ER in the absence of ADAM10 should be associated with a reduction of Tspan5 surface expression levels. Indeed, depletion of ADAM10 by RNA interference reduced ADAM10 surface expression levels by >70%, as determined by flow cytometry (Fig. 7B), and decreased Tspan5 expression levels (as determined by the labeling with mAb TS5-2) by ∼40%. Of note, in these experiments, the binding of TS5-1r was lower than the binding of TS5-2 in both cell lines, but it increased almost to the level observed for TS5-2 after ADAM10 silencing, further indicating that ADAM10 expression prevents TS5-1r binding to Tspan5 at the cell surface and that Tspan5 is to a large extent associated with ADAM10 at the cell surface.
Previous studies have shown that TspanC8 tetraspanins regulate the exit of ADAM10 from the ER (8, 10, 11). We now demonstrate that reciprocally ADAM10 regulates Tspan5 exit from the ER, suggesting that the formation of the complex is necessary for the ER exit of both partners. However, the Tspan5 mAbs did not immunoprecipitate the proform of ADAM10, the form of ADAM10 present in this compartment (Figs. 4 and 6). This does not exclude the possibility that they interact in the ER, because once the complex is formed, it may quickly exit the ER, and ADAM10 may be rapidly processed by pro-protein convertases in the Golgi. To determine whether the two proteins associate in the ER, immunoprecipitations were performed after treatment of cells with brefeldin A, to inhibit the egress of ADAM10 and Tspan5 to the Golgi. As shown in Fig. 7C, the band corresponding to the proform of ADAM10 was reinforced after BFA treatment in the ADAM10 immunoprecipitate, and was co-immunoprecipitated with Tspan5. Reciprocally, the ∼27-kDa band corresponding to the immature form of Tspan5 was strongly reinforced after BFA treatment and was efficiently co-immunoprecipitated with ADAM10. Thus, Tspan5 and ADAM10 interact in the ER.
Analysis of TspanC8-specific motifs in Tspan5 LEL
Because all TspanC8 tetraspanins interact with ADAM10, we hypothesized that the residues conserved in the LEL of all TspanC8 tetraspanins and not in other tetraspanins would mediate the interaction with ADAM10. Sequence analysis uncovered two TspanC8-specific motifs, present in mammalian, C. elegans, and D. melanogaster TspanC8 tetraspanins (Fig. 8A) as follows: Y(R/q/y)DDXD(L/qf/)(Q/r/k) between the two first predicted helix of the LEL (YRDDIDLQ in Tspan5, site 1; X corresponds to any residue, and lowercase letters indicate alternatives found in only one or two TspanC8); NXY(F/h) (NIYF in Tspan5, site 2), immediately before the first of the two TspanC8-specific extra cysteines. Three overlapping mutations were generated for site 1 (RDD → AAA, DID → AAA, and DLQ → AAA), whereas the three conserved residues of site 2 were changed to alanines (NIYF → AIAA).
Figure 8.
Mutations of TspanC8-specific motifs in the LEL of Tspan5 abolish the interaction with ADAM10. A, sequence alignment of the LEL of CD151 and of the different Homo sapiens (Tspan), D. melanogaster (Tsp), and C. elegans (Tsp-12) TspanC8 tetraspanins. The residues present in most tetraspanins are in pink. The residues conserved in TspanC8 are also indicated: red, >80% conservation; blue, >60% conservation; green, conservative substitutions. The two additional cysteines that are the hallmark of TspanC8 are in yellow. B, HeLa cells were transfected with GFP-tagged Tspan5 or the different mutants as indicated at the top of each lane. Two days later, the cells were lysed in Brij 97, and immunoprecipitations were performed as indicated. The presence of ADAM10, Tspan5, or the mutants in the immunoprecipitates was analyzed by Western blotting. The mature (m.) form and the proform (p.) of ADAM10 are indicated. C, HeLa cells were transfected with plasmids encoding GFP-tagged Tspan5 or the RDD and DID mutants. Two days later, the surface expression of ADAM10 and the binding of the anti-Tspan5 mAb TS5-2 to the cells were analyzed by flow cytometry. The experiments in this figure have been done twice.
The ability of GFP-tagged versions of the different mutants to interact with ADAM10 was determined after transfection in HeLa cells and immunoprecipitation with GFPtrap beads. As shown in Fig. 8B, the DID and DLQ mutants stimulated an increase in total ADAM10 expression level and co-immunoprecipitated ADAM10, whereas the RDD and NIYF mutants did not. Flow cytometry analysis indicated that the DID and DLQ mutants but not the RDD and NIYF mutants promoted an increase in surface ADAM10 expression level (Fig. 8C and data not shown). However, there was no staining of the NIYF mutant (data not shown), and only little staining of the RDD mutant with mAb TS5-2, despite GFP expression. This indicated that these two mutants do not or minimally reach the plasma membrane or alternatively were poorly recognized or not recognized by the mAbs.
This prompted us to analyze the subcellular distribution of these two mutants (Fig. 9, A and B). After transfection in U2OS or HeLa cells, both mutants showed reticulated and perinuclear labeling typical of an ER labeling. The RDD mutant was recognized by all Tspan5 mAbs except one, whereas the NIYF mutant was only recognized by the two Tspan5/17 antibodies (Fig. 9B and Table 1). Confirming the retention in the ER, the signal of the RDD and NIYF mutants strongly overlapped with the signal obtained with an mCherry-tagged sec61 construct (Fig. 9A). Similarly, mutation of YDD (corresponding to RDD in Tspan5) in Tspan15 also yielded ER retention of the mutant (Fig. 9A).
Figure 9.
Two Tspan5 mutants that do not interact with ADAM10 are retained in the endoplasmic reticulum. A, HeLa cells were co-transfected with plasmids encoding the indicated GFP-tagged Tspan5 or Tspan15 mutants and a plasmid encoding mCherry-tagged Sec61, an ER marker. The images were acquired by confocal microscopy. B, immunofluorescence microscopy analysis of HeLa cells transfected with GFP-tagged Tspan5 mutants RDD and NIYF and stained with mAbs TS5-2 and TS5/17 after Triton X-100 permeabilization. Bar: 10 μm.
TS5-2 and TS5-3 inhibit Notch signaling
We have previously shown that silencing Tspan5 in U2OS-N1 or PC3 cells reduced ligand-induced Notch signaling (8, 19). To test by an independent approach the role of Tspan5 in the regulation of Notch, we tested whether our Tspan5 mAbs could modulate this activity, using an assay in which U2OS-N1 cells are transiently transfected with a luciferase Notch reporter and co-cultured with OP9 cells expressing or not the Notch ligand DLL1. Neither TS5-1r nor TS5/17 significantly inhibited Notch signaling. In contrast, TS5-2 and TS5-3 had a modest effect, reducing Notch signaling by ∼15–20% (Fig. 10A).
Figure 10.

Effect of Tspan5 mAbs on ligand-induced Notch signaling. A, Notch activity in U2OS-N1, measured using a luciferase reporter assay. U2OS-N1 cells treated or not with the indicated mAbs were co-cultured with OP9-DLL1 cells for 20–24 h to activate Notch signaling or with parental OP9 cells to determine the basal level of luciferase production. The graph shows the mean ± S.E. of 3–7 independent experiments in duplicate. The data are expressed as a percentage of the signal observed for control U2OS-N1 cells. B, Notch activity, measured using a luciferase reporter assay, of U2OS-N1 cells treated with the indicated siRNAs. Notch was activated by incubation with OP9-DLL1 cells. The graph shows the mean ± S.E. of six independent experiments performed in duplicate. C, Notch activity, measured using a luciferase reporter assay, of U2OS-N1 cells treated with a control siRNA or an siRNA to Tspan14, and the indicated mAbs. The graph shows the mean ± S.E. of 3–6 independent experiments performed in duplicate. ***, p < 0.001; **, p < 0.01; *, p < 0.05 as compared with control.
Because both Tspan5 and Tspan14, the two major TspanC8 tetraspanins expressed by these cells, positively regulate Notch signaling, the effect of these mAb was also tested on cells depleted of Tspan14. Whereas Tspan14 alone did not significantly reduce Notch signaling, treatment of Tspan14-silenced cells with TS5-2 or TS5-3 reduced Notch signaling to the level observed after silencing both Tspan5 and Tspan14 (∼ 50% of the control, Fig. 10, B and C).
Tspan15 and Tspan33 inhibit Notch signaling by competing with endogenous TspanC8 for the interaction with ADAM10
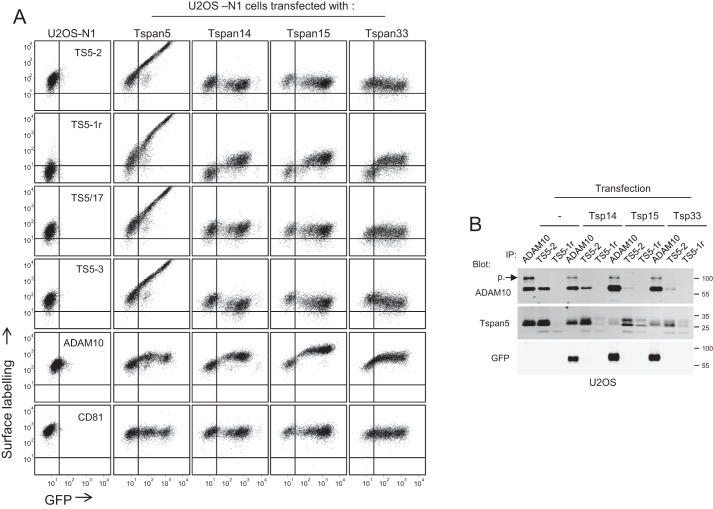
We have previously shown that expression of Tspan15 and Tspan33 in U2OS-N1 cells led to a strong reduction of ligand-induced Notch signaling (19). In the absence of antibodies to the endogenous TspanC8 tetraspanins (mainly Tspan14 and Tspan5), and because expression of Tspan15 and Tspan33 stimulated a strong increase in ADAM10 surface expression levels, it was not possible to determine whether Tspan15 and Tspan33 acted by competing with endogenous Tspan5 and Tspan14 for the association with ADAM10 or through a different mechanism. The availability of anti-Tspan5 mAbs, and notably of an antibody that recognizes Tspan5 not associated with ADAM10, has opened the way to discrimination between these two hypotheses.
As shown in Fig. 11, the staining of cells transfected with GFP-tagged Tspan14, Tspan15, or Tspan33 with the mAb TS5-1r increased proportionally to the level of GFP. However, the staining of these cells by TS5-1r remains very low as compared with the staining of cells transfected with Tspan5. In contrast, the labeling by two other Tspan5 mAbs, TS5-2 and TS5-3, slightly diminished as a function of the GFP signal, strongly suggesting a lower surface expression of Tspan5 in these cells. This diminution was more pronounced with the mAb TS5-3, and no diminution of signal was observed for TS5/17. The reason why the diminution of the staining by the three mAbs is not of the same magnitude is unknown. This could be due to the fact that some of these mAbs may have slightly different affinities for the free and ADAM10-associated forms of Tspan5.
Figure 11.
Ectopically expressed TspanC8 tetraspanins compete with endogenous Tspan5 for the association with ADAM10. A, flow cytometry analysis of U2OS cells stably expressing GFP-tagged Tspan5, Tspan14, Tspan15, or Tspan33 and stained or not with mAbs to ADAM10, CD81, or four anti-Tspan5 mAb. Note the increase in TS5-1r binding proportionally to the GFP signal and the slight decrease of the binding of TS5-2 and TS5-3. B, U2OS cells stably expressing or not GFP-tagged Tspan14, Tspan15, or Tspan33 were lysed in digitonin before immunoprecipitations (IP) with the anti-ADAM10 mAb 11G2 or the anti-Tspan5 mAbs TS5-2 and TS5-1r. The presence of Tspan5 and ADAM10 in the immunoprecipitates was visualized by immunoblotting using biotin-labeled TS5-2 and 11G2 mAbs. The band corresponding to the proform (p.) of ADAM10 is indicated. All experiments in this figure have been done three times with similar outcomes.
Because TS5-1r only recognizes Tspan5 not associated with ADAM10, the better staining of the cells expressing high levels of GFP-tagged Tspan14, Tspan15, or Tspan33 strongly suggests that these transfected tetraspanins compete with endogenous Tspan5 for the association with ADAM10. However, even if TS5-1r does not recognize these tetraspanins by Western blotting, one cannot exclude based solely on these data the possibility that TS5-1r could bind to these tetraspanins with a low affinity. To validate that the higher TS5-1r labeling indeed reveals the presence of a higher free Tspan5 fraction, we immunoprecipitated ADAM10 and Tspan5, using both TS5-2 and TS5-1r, from digitonin lysates of the different transfectants. As shown in Fig. 11B, TS5-2 immunoprecipitated less ADAM10 from all transfected cells than from parental cells, and reciprocally ADAM10 immunoprecipitated less Tspan5. This is due at least in part to the diminution of the Tspan5 level. Importantly, this was associated with a higher level of Tspan5 immunoprecipitation by TS5-1r and a change of the migration profile of Tspan5 that resembles that observed after silencing ADAM10 (Fig. 6C). Of note, not all cells express high levels of the transfected tetraspanins (Fig. 11A), which attenuates the changes observed in this experiment.
Discussion
In this paper we describe the generation of the first high quality Tspan5 mAbs and use them to get new insights into the functional implications of the Tspan5-ADAM10 complexes.
A set of anti-Tspan5 mAbs recognizing different epitopes
Because the human, mouse, and rat Tspan5 molecules are identical, we immunized Tspan5 knock-out mice to produce these antibodies. These antibodies were able to stain cells expressing minute amounts of Tspan5-GFP (see Fig. 1A), suggesting that they have a strong affinity for Tspan5. Thus the low staining of non-transfected cells actually reflects a low expression level. Analysis of the binding to chimeric Tspan5/Tspan15 chimeras indicated that all mAbs bind to epitopes present in the LEL of Tspan5. To our knowledge, all anti-tetraspanin mAbs able to bind to their target in a native conformation at the cell surface bind to an epitope present in the LEL, and none to epitopes in the small extracellular loop. This is consistent with the model of tetraspanins proposed by Seigneuret (27) in which the small extracellular loop is concealed by the LEL. A crystal structure of the full-length CD81 molecule has recently been published, but the small extracellular domain could not be resolved (28).
The mAbs produced in this study recognize at least three different epitopes. Several of these mAbs, like TS5-2, recognize Tspan5 and no other TspanC8 tetraspanins and are able to co-immunoprecipitate ADAM10. Three mAbs also recognize another TspanC8 tetraspanin, Tspan17, which is consistent with these being the two most closely related TspanC8 tetraspanins, sharing 70% identity at the amino acid level and even 79% in the large extracellular domain. Finally, we also show that one of these mAbs, TS5-1r, recognizes an epitope concealed by the interaction with ADAM10. This is supported by two lines of evidence. 1) TS5-1r never co-immunoprecipitates ADAM10 under conditions preserving this interaction (Brij 97, digitonin), in contrast to other antibodies, even after overexpression of Tspan5. 2) The immunoprecipitation of Tspan5 by TS5-1r and the binding of this mAb to the cell surface are increased following ADAM10 silencing by RNAi. TS5-1r constitutes the second example of an anti-tetraspanin mAb recognizing the “free” pool of its target tetraspanin. The first example was the CD151 mAb TS151r (followed by several other anti-CD151 mAbs), which was similarly shown to be unable to bind to CD151 in the presence of the integrin α3β1 and subsequently to require three residues involved in the interaction with the integrin for binding (22, 23).
The majority of Tspan5 is associated with ADAM10
An antibody that does not recognize the fraction of Tspan5 molecules constitutes a unique tool to evaluate the fraction of free Tspan5, i.e. not directly associated with ADAM10, and to investigate quantitative changes in the level of interaction. In this regard, we found that TS5-1r immunoprecipitated from both HCT116 and U2OS cells ∼15–20% of the amount of Tspan5 immunoprecipitated by TS5-2, consistent with immunodepletion experiments in which removal of ADAM10 from the cell lysates removed ∼85% of Tspan5. Flow cytometry analysis to compare the binding of TS5-2 and TS5-1r indicated that the majority of Tspan5 is associated with ADAM10 at the surface of a large variety of cell lines (Fig. 7B and data not shown). In the immunodepletion experiments, Tspan5 depletion had no effect or a modest effect on the amount of ADAM10 immunoprecipitated with the mAb 11G2, indicating that the majority of ADAM10 is not associated with Tspan5. We have previously demonstrated that silencing Tspan14 in HCT116 impaired the surface expression of ADAM10 (8). It is likely that in this cell line ADAM10 is mostly associated with Tspan14.
ADAM10 regulates the exit of Tspan5 from the endoplasmic reticulum and its glycosylation
We previously showed that Tspan5, like other TspanC8 tetraspanins, controlled the exit of ADAM10 from the ER (8). We now demonstrate that, reciprocally, the exit of Tspan5 from the ER is facilitated by the interaction with ADAM10. Indeed, a large fraction of Tspan5 is retained in the ER after ADAM10 silencing, as shown by acquisition of EndoH sensitivity, yielding a lower level of Tspan5 at the cell surface. Quantification by Western blotting indicated that the total amount of Tspan5 was diminished after ADAM10 silencing. There was no diminution of Tspan5 mRNA as shown by RT-qPCR (data not shown). It is possible that a fraction of ADAM10 retained in the ER has not acquired a conformation recognized by the antibodies or has been degraded through the ER-associated degradation pathway. Alternatively, the half-life of Tspan5 may be shorter when it is not associated with ADAM10.
The regulation by ADAM10 of Tspan5's exit from the ER is consistent with the finding that the majority of Tspan5 molecules is associated with ADAM10. However, two lines of evidence suggest that Tspan5 can exit from the ER without being associated with ADAM10. First, transfection of Tspan5 in U2OS cells yields a large surface pool of Tspan5 that is not associated with ADAM10 as shown by the recognition by TS5-1r (Fig. 1A). Second, a fraction of free Tspan5 is observed at the cell surface after ADAM10 silencing, as shown, again, by the increased binding of TS5-1r (Fig. 7B). This suggests that ADAM10 accelerates the exit of Tspan5 from the ER without being absolutely required. In this regard, ADAM10 is first synthesized as a proform, which undergoes a proteolytic cleavage in the Golgi by proprotein convertases (29). Our Tspan5 mAbs do not co-immunoprecipitate the ADAM10 proform, except if the egress to the Golgi is prevented by a treatment with BFA, suggesting that once Tspan5 and ADAM10 have interacted in the ER they are quickly transported to the Golgi where ADAM10 is cleaved.
There are now several examples of tetraspanins regulating the exit of their partner proteins from the ER. Besides the example of ADAM10, the best characterized example is that of CD81 regulating the trafficking of CD19 (30). In contrast, the only example of a protein that influences the exit from the ER of its partner tetraspanin is the urothelial plaque component uroplakin (UP) II, which is essential for the exit of UPIa from the ER after transfection in HEK 293 cells. In the same study, the closely related tetraspanin UPIb did not need the interaction with UPIII to leave the ER (31).
Silencing ADAM10 not only reinforced a Tspan5 band corresponding to an EndoH-sensitive form, but also a band of higher molecular weight resistant to this glycosidase, and therefore corresponding to a fraction of Tspan5 having egressed from the ER. PNGase F treatment indicated that this band corresponds to a different glycoform of Tspan5. Thus the interaction with ADAM10 modifies the glycosylation of Tspan5, and this constitutes to our knowledge the first example in which a protein regulates the glycosylation of its partner tetraspanin at a post-ER level. Again, the reverse situation has been described. For example, the glycosylation of CD19 and that of the light chain of the α3 integrin subunit are affected at a post-ER level by their interaction with CD81 and CD151, respectively (30, 32).
Essential role of TspanC8-specific residues in the LEL of TspanC8
The interaction of TspanC8 tetraspanins with ADAM10 has recently been shown to involve the extracellular domain of ADAM10. Consistent with this result, the LEL of Tspan14 was shown to contribute to the interaction with ADAM10 (20). We reasoned that because all TspanC8 tetraspanins interact with ADAM10, the residues responsible for this interaction would be conserved in the LEL of all TspanC8 tetraspanins but not in other tetraspanins. We identified two conserved motifs, located immediately after the first predicted helix of the LEL (RDD) and immediately before the first of the two TspanC8-specific extra-cysteines (NIYF), the mutation of which precludes the interaction with ADAM10. However, these two mutants were retained in the ER. This is surprising because previous mutations in the LEL of tetraspanins did not have such an effect. Indeed, it was shown that mutation of cysteines in the LEL of UPIb or in CD151, which strongly affect the conformation of this domain, replacing the LEL of CD151 by a Myc tag, or large deletions in CD81 LEL did not impair trafficking to the plasma membrane (33–35). In addition, mutation in CD81 of the residues (VVD) corresponding to RDD in Tspan5 did not affect its conformation and cell-surface expression (36).
The intracellular retention of the mutants could be due to the loss of interaction with ER proteins that facilitate its egress from the ER or, alternatively, to alterations of their conformation and recognition by chaperones that form a part of the ER quality control system (37). Our mAbs may help to differentiate between these hypotheses. The NIYF mutant was not recognized by most anti-Tspan5 mAbs except for the two anti-Tspan5/Tspan17 mAb. This may be due either to the fact that the NIYF motif is recognized by most antibodies or indicate a major change of its conformation. In contrast, the RDD Tspan5 mutant was recognized by all but one mAb, indicating no major conformation defect. However, by immunofluorescence microscopy, the staining by these antibodies was reduced in comparison with the staining of WT Tspan5 (i.e. the ratio of Tspan5 antibody labeling/GFP signal was reduced, data not shown), indicating that in the ER only a fraction of Tspan5 RDD is labeled by the mAbs. The recognition of the RDD mutant by TS5-2 was even more impaired by Western blotting, following Brij 97 lysis and GFPtrap immunoprecipitation (Fig. 8B). These data, together with the fact that a minor fraction of RDD reaches the cell surface where it is recognized by TS5-2 (probably efficiently as this surface fraction was not detectable by immunofluorescence microscopy), suggest that the RDD motif is dispensable for the correct folding of Tspan5 but facilitates and stabilizes this folding. This may be a general feature of TspanC8 because the same mutation in Tspan15 similarly causes retention of the protein in the ER.
Our data show that TspanC8-specific LEL motifs contribute to the interaction with ADAM10 and to the exit from the ER, probably by facilitating proper folding. Because previous mutations in other tetraspanins did not have such consequences, the folding and ER exit of TspanC8 might be a more complex process than that of other tetraspanins. The role of these motifs in the folding does not exclude the possibility that they could also directly contribute to the interaction of TspanC8 with ADAM10. Also, it is possible that ADAM10 facilitates the folding of Tspan5 and other TspanC8.
Tspan5, TspanC8, and Notch signaling
We previously reported that Tspan5 and Tspan14 positively regulated Notch signaling in U2OS-N1 cells (8). Surprisingly, contrasting with our previous study, silencing Tspan5 or Tspan14 separately did not significantly reduce Notch signaling. The reason for this discrepancy is unknown, and it may be due to a change in culture conditions (i.e. a different batch of FCS). However, silencing Tspan5 and Tspan14 together reduced Notch signaling by ∼50%, confirming our initial conclusion that both Tspan5 and Tspan14 positively regulate Notch signaling in U2OS-N1 cells and suggesting that Tspan5 and Tspan14 can compensate for each other in this function. The role of Tspan5 in Notch signaling is further confirmed by the finding that the mAbs TS5-2 and TS5-3 inhibit to some extent ligand-induced Notch signaling. The effect is small in untreated cells, but it reached ∼50% in cells depleted of Tspan14, consistent with the necessity of targeting both tetraspanins for a maximal effect on Notch signaling. This is consistent with in vivo data in flies showing that silencing only one of the three TspanC8 tetraspanins produced mild developmental defects, in contrast to the depletion of all three molecules (8). The inhibitory effect of TS5-2 on Notch signaling is not due to a change in ADAM10 expression (data not shown), and it is unlikely to be due to a dissociation of the Tspan5-ADAM10 complex because TS5-2 recognizes Tspan5 associated with ADAM10, in contrast to TS5-1r, which does not inhibit Notch signaling.
In this study, we provide evidence that ectopically expressed Tspan15 and Tspan33 compete with endogenous Tspan5 for the association with ADAM10. Although we could not test it, it is likely that Tspan15 and Tspan33 also compete with Tspan14. Therefore, the reduced ability of U2OS-N1 cells stably expressing Tspan15 or Tspan33 to support ligand-induced Notch signaling (19) is likely to be due to the replacement of ADAM10-Tspan5 (and ADAM10-Tspan14) complexes functional for Notch signaling by ADAM10-Tspan15 or ADAM10-Tspan33 complexes. Of note, the diminution of Tspan5-ADAM10 complexes was observed not only using a biochemical approach but also by flow cytometry, using the mAb TS5-1r that binds only to free Tspan5 molecules and permits the quantification of the free Tspan5 pool in the different transfectants at the single cell level, highlighting the usefulness of this mAb.
Conclusions
In conclusion, this study describes the production of the first mAbs against the evolutionarily conserved Tspan5 tetraspanin, and their use to characterize several aspects of this tetraspanin. These mAbs allow revealing that the inhibition of Notch signaling observed after overexpression of Tspan15 or Tspan33 is due to a competition with the endogenous Tspan5 and possibly endogenous Tspan14. They unravel some properties of Tspan5 that are unusual for tetraspanins such as the regulation of the exit from the ER by its partner protein ADAM10, or the key role of two conserved motifs in the LEL in maintaining the interaction with ADAM10 and a proper conformation, allowing the exit from the ER. Finally, the ability of some of them to inhibit Notch signaling confirms the role of Tspan5 in this process. These mAbs will be useful for further studies addressing the role of Tspan5.
Experimental procedures
Antibodies, plasmids, and mutagenesis
The mAbs directed to mouse CD81 (MT81) and CD9 (4.1F12) as well as human ADAM10 (11G2), CD9 (TS9), CD81 (TS81), CD63 (TS63), CD151 (TS151), CD9P-1 (1F11), and CD55 (12A12) have been previously described (38–41). The rabbit polyclonal antibody to the cytoplasmic domain of ADAM10 was provided by P. Saftig (11). The rabbit polyclonal anti-GFP antibody was from Santa Cruz Biotechnology. The plasmid encoding mCherry-Sec61β was a gift from Gia Voeltz (Addgene plasmid catalog no. 49155) (42). The plasmids encoding the various TspanC8 fused to GFP were previously described (8). The chimeric Tspan5/Tspan15 constructs (Ts15LEL5 in which the LEL of Tspan15 is replaced by that of Tspan5 and the reciprocal construct Ts5LEL15) and the various Tspan5 or Tspan15 mutants used in this study were generated using the overlap extension method in which two overlapping PCRs are used as a second PCR template (43). The swaps in the chimeras were made at residues conserved in Tspan5 and Tspan15. The sequences of the swap sites are Tspan5-LAFVFRNQT-Tspan15 and Tspan15-MDNYTIVAGI-Tspan5 for Ts5LEL15 and Tspan15-VALTFKDWI-Tspan5 and Tspan5-QDNLTIMAGI-Tspan15 for Ts15LEL5 (the conserved amino acid where the swap was made is shown in boldface). The siRNA targeting Tspan14 targets the sequence CUCGCUGUUGCAGAUAUUU. Its efficiency has been tested on U2OS cells stably expressing Tspan14-GFP (data not shown). The other siRNAs used in this study were previously described (19). They target the following sequences: siTspan5 #1, GACCAGCUGUAUUUCUUUA; siTspan5 #2, GCUGAUGAUUGGAACCUAA; siADAM10 #1, GGAUUAUCUUACAAUGUGG; siADAM10 #2, AGACAUUAUGAAGGAUUAU; and control siRNA, UUUGUAAUCGUCGAUACCC.
Cell culture and generation of cells expressing tetraspanins
OP9 cells expressing or not the human Notch ligand DLL-1 (OP9-DLL-1) (44) were cultured in α-minimum Eagle's medium supplemented with 10% FCS and antibiotics. The human osteosarcoma cell line U2OS expressing human Notch1 (U2OS-N1) (45) as well as the prostate carcinoma cell line PC3, the colon carcinoma cell line HCT116, the cervical carcinoma cell line HeLa and the murine colon carcinoma cell line CT26 were cultured in DMEM supplemented with 10% FCS and antibiotics. These cell lines have been previously described in terms of TspanC8 mRNA expression levels (8, 19). U2OS-N1 cells stably expressing GFP-tagged tetraspanins have also been previously described in the same studies. Transient transfection of cells was performed using FuGENE HD (Promega).
Generation of anti-Tspan5 mAbs
Tspan5 knock-out mice in a C57/Bl6 genetic background were injected intraperitoneally twice with 107 U2OS cells stably expressing Tspan5-GFP. For the two subsequent immunizations, U2OS cells expressing Tspan5-GFP were lysed in Brij 97 before immunoprecipitation of Tspan5 using GFP-trap beads (Chromotek). The beads were injected i.p., and 5 days after the last injection, the spleen cells were fused with P3X63AG8 mouse myeloma cells (5 × 107 cells) according to standard techniques and distributed into 96-well tissue culture plates. After ∼10 days, hybridoma culture supernatants were harvested and tested for the staining of U2OS cells expressing Tspan5-GFP by flow cytometry.
Flow cytometry analysis
Cells were detached with accutase, washed twice in complete DMEM, and incubated for 30 min at 4 °C with 10 μg/ml primary antibody or hybridoma supernatant. After three washings, the cells were incubated for 30 min at 4 °C with an Alexa 647-conjugated F(ab′)2 goat anti-mouse antibody The cells were analyzed using an Accuri C6 flow cytometer (BD Biosciences).
Analysis of Notch activity
This analysis was performed as described previously (45); U2OS-N1 cells were seeded at a concentration of 25,000 cells/cm2. RNA interference was performed at the time of plating using INTERFERin® (Polyplus transfection). Cells were transfected 24 h later with the CSL reporter and Renilla plasmids using FuGENE HD (Promega). 24 h later, cells were co-cultured with OP9 or OP9-DLL1 at 35,000 cells/cm2. The mAbs were added to U2OS-N1 cells 4 h before the beginning of the co-culture. The activities of firefly and Renilla luciferases were determined using a dual-luciferase reporter assay (Promega) according to the manufacturer's instructions. Statistical analysis was performed using one-way analysis of variance followed by the Dunnett's multiple comparison test.
Biotin labeling of surface proteins and immunoprecipitation
Biotin labeling of surface proteins with EZ-link Sulfo-NHS-LC-biotin (ThermoFisher Scientific) and immunoprecipitations were performed as described previously (38, 39). For immunoprecipitation, cells were lysed in a lysis buffer (30 mm Tris, pH 7.4, 150 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, protease inhibitors) supplemented with 1% digitonin or 1% Brij 97, or in RIPA buffer. After 30 min of incubation at 4 °C, the insoluble material was removed by centrifugation at 10,000 × g, and the cell lysate was precleared by addition of heat-inactivated goat serum and protein G-Sepharose beads (GE Healthcare). Proteins were then immunoprecipitated by adding 2 μg of mAb and 10 μl of protein G-Sepharose beads to 200–400 μl of the lysate or using GFP-trap beads (Chromotek). The immunoprecipitated proteins were separated by SDS-PAGE and transferred to a PVDF membrane (Amersham Biosciences). Western blotting on GFP-trap immunoprecipitates was performed using appropriate combinations of primary and fluorescent secondary antibodies. Western blotting on immunoprecipitations performed with mouse mAbs was performed using biotin-labeled antibodies and Alexa 680-labeled streptavidin. All acquisitions were performed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Brefeldin A and endoglycosidase treatment
BFA was added to the cells overnight at a concentration of 5 μg/ml before lysis and immunoprecipitation. For deglycosylation, Tspan5 immunoprecipitates were resuspended in 30 μl of 0.1% SDS and boiled for 3 min. After addition of 60 μl of H2O, the samples were distributed in 3 aliquots that were either kept undigested or incubated overnight with 0.5 unit of EndoH (Roche Applied Science) in 50 mm acetate sodium, pH 5.2, or with 1 unit of PNGase F (Promega) in 50 mm sodium phosphate buffer, pH 7.4, 1% Triton X-100. The samples were analyzed under non-reducing conditions by SDS-PAGE and Western blotting using biotin-labeled antibodies.
Immunoprecipitation of Tspan5 from mouse organs
Mice were anesthetized i.p. with 100 mg/kg ketamine and 10 mg/kg xylazine and perfused with 20 ml of PBS before harvesting various organs. The organs were covered with lysis buffer (2% Brij 97; 1 ml for 250 mg) and protease inhibitors and disrupted using a Retsch MM300 mixer mill. The samples were further diluted five times in the same buffer before immunoprecipitations as described above.
Immunofluorescence and confocal microscopy
The cells grown for 2 days on glass coverslips in complete medium were fixed for 20 min with 4% paraformaldehyde at room temperature, washed in PBS, and then incubated for 15 min in 50 mm NH4Cl in PBS. For the staining of the Tspan5 mutants in the ER, the cells were pretreated with 0.1% Triton X-100 in PBS for 2 min at 4 °C before incubation for 1 h with 10–20 μg/ml antibodies in PBS supplemented with 0.1% BSA at room temperature. In other experiments, permeabilization was achieved by adding saponin (0.1%) to the antibody solution. The binding of primary antibodies was revealed using appropriate secondary reagents. To compare the surface and intracellular pools of Tspan5 or CD63, the cells were first labeled at 4 °C in the presence of 0.1% NaN3 with specific mAb to label the surface pool of the target tetraspanin. After washing in PBS and fixation, the cells were incubated with an anti-mouse polyclonal antibody coupled to Alexa 568. The cells were washed and stained with a combination of primary mAb and an anti-mouse polyclonal antibody coupled to Alexa 647 in PBS/BSA containing 0.1% saponin to permeabilize the cells. The cells were mounted in Mowiol 4-88 (Sigma) supplemented with 1,4-diazabicyclo[2.2.2]octane (DABCO) and DAPI or Prolong Diamond (Life Technologies, Inc.) and examined with a Leica DMR fluorescence microscope equipped with a CoolSNAP HQ2 camera (Photometrics) controlled by metamorph (Molecular Devices) or a Leica SP5 confocal microscope (×63 objective, 1.4 numerical aperture, zoom 6). A Gaussian filter (Sigma X and Sigma Y = 1) has been applied using the ICY imaging software to the images of Fig. 3.
Author contributions
J. S. P. designed and performed experiments, analyzed data, and contributed to the writing of the article. M. B., E. D., and E. E. carried out experiments and analyzed data. S. C., C. B., and L. D. contributed expertise and reagents and to the writing of the article. E. R. designed the project, designed and performed experiments, analyzed data, and wrote the article.
Acknowledgments
We thank Denis Clay for expert assistance in cell sorting, and Philippe Mauduit and Stéphanie Jouannet for the generation of some constructs. We thank Christel Brou and Paul Saftig for the generous gift of reagents.
This work was supported in part by core funding from INSERM and by specific grants from the NRB-Vaincre le Cancer, Institut du Cancer et d'Immunogénétique, and the Institut National du Cancer. The authors declare that they have no conflicts of interest with the contents of this article.
- LEL
- large extracellular loop
- UP
- uroplakin
- ER
- endoplasmic reticulum
- PNGase
- peptide:N-glycosidase
- EndoH
- endoglycosidase H
- BFA
- brefeldin A.
References
- 1. Charrin S., Jouannet S., Boucheix C., and Rubinstein E. (2014) Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 [DOI] [PubMed] [Google Scholar]
- 2. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., and Rubinstein E. (2009) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 3. Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 4. Yáñez-Mó M., Barreiro O., Gordon-Alonso M., Sala-Valdés M., and Sánchez-Madrid F. (2009) Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 [DOI] [PubMed] [Google Scholar]
- 5. Serru V., Dessen P., Boucheix C., and Rubinstein E. (2000) Sequence and expression of seven new tetraspans. Biochim. Biophys. Acta 1478, 159–163 [DOI] [PubMed] [Google Scholar]
- 6. Todd S. C., Doctor V. S., and Levy S. (1998) Sequences and expression of six new members of the tetraspanin/TM4SF family. Biochim. Biophys. Acta 1399, 101–104 [DOI] [PubMed] [Google Scholar]
- 7. Huang S., Yuan S., Dong M., Su J., Yu C., Shen Y., Xie X., Yu Y., Yu X., Chen S., Zhang S., Pontarotti P., and Xu A. (2005) The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 86, 674–684 [DOI] [PubMed] [Google Scholar]
- 8. Dornier E., Coumailleau F., Ottavi J. F., Moretti J., Boucheix C., Mauduit P., Schweisguth F., and Rubinstein E. (2012) TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 199, 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seigneuret M., Delaguillaumie A., Lagaudrière-Gesbert C., and Conjeaud H. (2001) Structure of the tetraspanin main extracellular domain: a partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276, 40055–40064 [DOI] [PubMed] [Google Scholar]
- 10. Haining E. J., Yang J., Bailey R. L., Khan K., Collier R., Tsai S., Watson S. P., Frampton J., Garcia P., and Tomlinson M. G. (2012) The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J. Biol. Chem. 287, 39753–39765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prox J., Willenbrock M., Weber S., Lehmann T., Schmidt-Arras D., Schwanbeck R., Saftig P., and Schwake M. (2012) Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10. Cell. Mol. Life Sci. 69, 2919–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blobel C. P. (2005) ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 13. Saftig P., and Reiss K. (2011) The “A disintegrin and metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur. J. Cell Biol. 90, 527–535 [DOI] [PubMed] [Google Scholar]
- 14. Lichtenthaler S. F. (2011) α-Secretase in Alzheimer's disease: molecular identity, regulation and therapeutic potential. J. Neurochem. 116, 10–21 [DOI] [PubMed] [Google Scholar]
- 15. Kopan R., and Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bozkulak E. C., and Weinmaster G. (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Tetering G., van Diest P., Verlaan I., van der Wall E., Kopan R., and Vooijs M. (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284, 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Groot A. J., Habets R., Yahyanejad S., Hodin C. M., Reiss K., Saftig P., Theys J., and Vooijs M. (2014) Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol. Cell. Biol. 34, 2822–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jouannet S., Saint-Pol J., Fernandez L., Nguyen V., Charrin S., Boucheix C., Brou C., Milhiet P. E., and Rubinstein E. (2016) TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell. Mol. Life Sci. 73, 1895–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noy P. J., Yang J., Reyat J. S., Matthews A. L., Charlton A. E., Furmston J., Rogers D. A., Rainger G. E., and Tomlinson M. G. (2016) TspanC8 tetraspanins and A disintegrin and metalloprotease 10 (ADAM10) interact via their extracellular regions: evidence for distinct binding mechanisms for different TspanC8 proteins. J. Biol. Chem. 291, 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. García-Frigola C., Burgaya F., de Lecea L., and Soriano E. (2001) Pattern of expression of the tetraspanin Tspan-5 during brain development in the mouse. Mech. Dev. 106, 207–212 [DOI] [PubMed] [Google Scholar]
- 22. Serru V., Le Naour F., Billard M., Azorsa D. O., Lanza F., Boucheix C., and Rubinstein E. (1999) Selective tetraspan/integrin complexes (CD81/a4b1, CD151/a3b1, CD151/a6b1) under conditions disrupting tetraspan interactions. Biochem. J. 340, 103–111 [PMC free article] [PubMed] [Google Scholar]
- 23. Yauch R. L., Kazarov A. R., Desai B., Lee R. T., and Hemler M. E. (2000) Direct extracellular contact between integrin α3β1 and TM4SF protein CD151. J. Biol. Chem. 275, 9230–9238 [DOI] [PubMed] [Google Scholar]
- 24. Sterk L. M., Geuijen C. A., van den Berg J. G., Claessen N., Weening J. J., and Sonnenberg A. (2002) Association of the tetraspanin CD151 with the laminin-binding integrins α3β1, α6β1, α6β4 and α7β1 in cells in culture and in vivo. J. Cell Sci. 115, 1161–1173 [DOI] [PubMed] [Google Scholar]
- 25. Yamada M., Tamura Y., Sanzen N., Sato-Nishiuchi R., Hasegawa H., Ashman L. K., Rubinstein E., Yáñez-Mó M., Sánchez-Madrid F., and Sekiguchi K. (2008) Probing the interaction of tetraspanin CD151 with integrin α3β1 using a panel of monoclonal antibodies with distinct reactivities toward the CD151-integrin α3β1 complex. Biochem. J. 415, 417–427 [DOI] [PubMed] [Google Scholar]
- 26. Palmer T. D., Martínez C. H., Vasquez C., Hebron K. E., Jones-Paris C., Arnold S. A., Chan S. M., Chalasani V., Gomez-Lemus J. A., Williams A. K., Chin J. L., Giannico G. A., Ketova T., Lewis J. D., and Zijlstra A. (2014) Integrin-free tetraspanin CD151 can inhibit tumor cell motility upon clustering and is a clinical indicator of prostate cancer progression. Cancer Res. 74, 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seigneuret M. (2006) Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 90, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zimmerman B., Kelly B., McMillan B. J., Seegar T. C., Dror R. O., Kruse A. C., and Blacklow S. C. (2016) Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anders A., Gilbert S., Garten W., Postina R., and Fahrenholz F. (2001) Regulation of the α-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 15, 1837–1839 [DOI] [PubMed] [Google Scholar]
- 30. Shoham T., Rajapaksa R., Kuo C. C., Haimovich J., and Levy S. (2006) Building of the tetraspanin web: distinct structural domains of CD81 function in different cellular compartments. Mol. Cell. Biol. 26, 1373–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tu L., Sun T. T., and Kreibich G. (2002) Specific heterodimer formation is a prerequisite for uroplakins to exit from the endoplasmic reticulum. Mol. Biol. Cell 13, 4221–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baldwin G., Novitskaya V., Sadej R., Pochec E., Litynska A., Hartmann C., Williams J., Ashman L., Eble J. A., and Berditchevski F. (2008) Tetraspanin CD151 regulates glycosylation of (α)3(β)1 integrin. J. Biol. Chem. 283, 35445–35454 [DOI] [PubMed] [Google Scholar]
- 33. Berditchevski F., Gilbert E., Griffiths M. R., Fitter S., Ashman L., and Jenner S. J. (2001) Analysis of the CD151-α3β1 integrin and CD151-tetraspanin interactions by mutagenesis. J. Biol. Chem. 276, 41165–41174 [DOI] [PubMed] [Google Scholar]
- 34. Tu L., Kong X. P., Sun T. T., and Kreibich G. (2006) Integrity of all four transmembrane domains of the tetraspanin uroplakin Ib is required for its exit from the ER. J. Cell Sci. 119, 5077–5086 [DOI] [PubMed] [Google Scholar]
- 35. Homsi Y., Schloetel J. G., Scheffer K. D., Schmidt T. H., Destainville N., Florin L., and Lang T. (2014) The extracellular δ-domain is essential for the formation of CD81 tetraspanin webs. Biophys. J. 107, 100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yalaoui S., Zougbédé S., Charrin S., Silvie O., Arduise C., Farhati K., Boucheix C., Mazier D., Rubinstein E., and Froissard P. (2008) Hepatocyte permissiveness to Plasmodium infection is conveyed by a short and structurally conserved region of the CD81 large extracellular domain. PLoS Pathog. 4, e1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gidalevitz T., Stevens F., and Argon Y. (2013) Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 1833, 2410–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arduise C., Abache T., Li L., Billard M., Chabanon A., Ludwig A., Mauduit P., Boucheix C., Rubinstein E., and Le Naour F. (2008) Tetraspanins regulate ADAM10-mediated cleavage of TNF-α and epidermal growth factor. J. Immunol. 181, 7002–7013 [DOI] [PubMed] [Google Scholar]
- 39. Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., and Rubinstein E. (2001) The major CD9 and CD81 molecular partner: identification and characterization of the complexes. J. Biol. Chem. 276, 14329–14337 [DOI] [PubMed] [Google Scholar]
- 40. Charrin S., Le Naour F., Labas V., Billard M., Le Caer J. P., Emile J. F., Petit M. A., Boucheix C., and Rubinstein E. (2003) EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Silvie O., Charrin S., Billard M., Franetich J. F., Clark K. L., van Gemert G. J., Sauerwein R. W., Dautry F., Boucheix C., Mazier D., and Rubinstein E. (2006) Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci. 119, 1992–2002 [DOI] [PubMed] [Google Scholar]
- 42. Zurek N., Sparks L., and Voeltz G. (2011) Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12, 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Higuchi R., Krummel B., and Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Six E. M., Ndiaye D., Sauer G., Laâbi Y., Athman R., Cumano A., Brou C., Israël A., and Logeat F. (2004) The notch ligand Δ1 recruits Dlg1 at cell-cell contacts and regulates cell migration. J. Biol. Chem. 279, 55818–55826 [DOI] [PubMed] [Google Scholar]
- 45. Moretti J., Chastagner P., Gastaldello S., Heuss S. F., Dirac A. M., Bernards R., Masucci M. G., Israël A., and Brou C. (2010) The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol. 8, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]