Abstract
A low-activity variant of endoplasmic reticulum aminopeptidase 1 (ERAP1), Hap10, is associated with the autoinflammatory disorder Behçet's disease (BD) in epistasis with HLA-B*51, which is the main risk factor for this disorder. The role of Hap10 in BD pathogenesis is unknown. We sought to define the effects of Hap10 on the HLA-B*51 peptidome and to distinguish these effects from those due to HLA-B*51 polymorphisms unrelated to disease. The peptidome of the BD-associated HLA-B*51:08 subtype expressed in a Hap10-positive cell line was isolated, characterized by mass spectrometry, and compared with the HLA-B*51:01 peptidome from cells expressing more active ERAP1 allotypes. We additionally performed synthetic peptide digestions with recombinant ERAP1 variants and estimated peptide-binding affinity with standard algorithms. In the BD-associated ERAP1 context of B*51:08, longer peptides were generated; of the two major HLA-B*51 subpeptidomes with Pro-2 and Ala-2, the former one was significantly reduced, and the latter was increased and showed more ERAP1-susceptible N-terminal residues. These effects were readily explained by the low activity of Hap10 and the differential susceptibility of X–Pro and X–Ala bonds to ERAP1 trimming and together resulted in a significantly altered peptidome with lower affinity. The differences due to ERAP1 were clearly distinguished from those due to HLA-B*51 subtype polymorphism, which affected residue frequencies at internal positions of the peptide ligands. The alterations in the nature and affinity of HLA-B*51·peptide complexes probably affect T-cell and natural killer cell recognition, providing a sound basis for the joint association of ERAP1 and HLA-B*51 with BD.
Keywords: aminopeptidase, antigen presentation, antigen processing, mass spectrometry (MS), peptides, proteomics
Introduction
Behçet's disease (BD)3 is an autoinflammatory disorder characterized by oral aphthae, genital ulcerations, skin lesions, joint inflammation, and uveitis. It is rare in Western countries and has a higher incidence in Turkey (1). The pathogenetic mechanism remains elusive, and both genetic and environmental factors seem to be involved. Of the many genes known to increase the risk for BD (2), HLA-B*51 is the main one, with an odds ratio of ∼5.78 (3). Besides the main B*51:01 subtype, the less frequent B*51:08, which differs from B*51:01 by the E152V and L156D changes (4), is also associated with BD in multiple studies (5–11) (supplemental Table S1).
Like other MHC class I (MHC-I) molecules, HLA-B*51 presents peptides derived from cellular protein degradation at the cell surface for recognition by cytotoxic T lymphocytes and natural killer (NK) cells. Endogenous proteins are degraded in the cytosol by the proteasome and other proteases, and the resulting peptides are translocated into the endoplasmic reticulum by the transporter associated with antigen processing (12). Once in the ER, the endoplasmic reticulum amino peptidase 1 (ERAP1) and ERAP2 trim many of the peptides to the correct length for MHC-I binding (13, 14).
The two enzymes share about 50% amino acid identity but differ in their specificity, being distinctly affected by the length and N-terminal residues of peptide substrates (reviewed in Ref. 15). ERAP1 is encoded by a highly polymorphic gene. The most common variants, designated as haplotypes (Hap), or combinations of non-synonymous SNPs, are named Hap1 to Hap10 (16). Several amino acid changes alter the enzymatic activity of ERAP1 (17–22), and the multiple polymorphisms co-occurring in each natural variant result in its unique specificity (23). The D575N and R725Q mutations have been associated with BD in epistasis with HLA-B*51 (2). This combination is uniquely present in Hap10 (16), a variant with low enzymatic activity that is the single risk haplotype for BD, again in epistasis with HLA-B*51 (24).
The peptidome of HLA-B*51:01 consists mainly of two subpeptidomes, one with Ala at peptide position 2 (P2) (Ala-2), which confers relatively lower affinity for HLA-B*51:01, and one with Pro-2 and higher affinity (25). In the present study, we analyzed the peptidome presented by HLA-B*51:08 in a B lymphoblastoid cell line (LCL) expressing the BD-associated Hap10 variant of ERAP1. To assess the effect of this ERAP1 context, as well as the effect of polymorphism between two BD-associated subtypes, on the HLA-B*51 peptidome, the B*51:08 ligands were compared with those of HLA-B*51:01 generated in a more active ERAP1 background and identified in our previous study (25). Using this approach, we addressed two issues concerning the relationship between antigen processing and presentation by HLA-B*51 and BD: 1) the effects of the BD-associated ERAP1 variant on the HLA-B*51 peptidome and 2) how this peptidome is affected by polymorphism between two BD-associated B*51 subtypes. The differences observed allowed us to establish the effects of ERAP1 on the HLA-B*51 peptidome and to distinguish them from those due to subtype polymorphism.
Results
ERAP1 and ERAP2 genotype and expression
BCH-30 was homozygous for the BD-associated Hap10 variant of ERAP1. It was heterozygous for ERAP2, although only the Lys-392 allotype of ERAP2 is expressed due to linkage disequilibrium of the Asn-392 variant with a polymorphism (rs2248374) that impairs the expression of the protein (26). Full genomic sequencing of ERAP1 from the 721.221-B*51:01 cell line (27) revealed that it was heterozygous for Hap1 and Hap8 (Table 1). The protein expression levels of ERAP1 were similar (0.9 ± 0.1 versus 0.8 ± 0.1), and those of ERAP2 were somewhat higher (1.4 ± 0.1 versus 1.0 ± 0.3) in BCH-30 compared with 721.221-B*51:01 cells (Fig. 1).
Table 1.
ERAP1 and ERAP2 variants in BCH-30 and 721.221-B*51:01 cells
Changes in the coding strands are shown. Nucleotide and amino acid residue numbering and consensus sequence are from human ERAP1 isoform 2 (UniProt/Swiss-Prot database accession no. Q9NZ08-2) and ERAP2 isoform 1 (UniProt/Swiss-Prot database accession no. Q6P179-1). The ERAP1 and ERAP2 variants in BCH-30 were determined by sequencing of the corresponding exons. Those in 721.221-B*51:01 cells were reported previously (25, 27). Single-letter codes for amino acids are used. ERAP1 haplotypes are as described previously (16). AA, amino acid.
| Single-nucleotide polymorphism | Exon | Nucleotide/AA position | Nucleotide/AA: consensus | Nucleotide/AA: BCH-30 | Nucleotide/AA: 721.221-B*51:01 |
|---|---|---|---|---|---|
| ERAP1 | |||||
| rs26653 | 2 | 380/127 | g/R | c/P | c/P |
| rs26618 | 5 | 828/276 | a/I | a/I | g,a/M,I |
| rs27895 | 6 | 1037/346 | g/G | g/G | g/G |
| rs2287987 | 6 | 1045/349 | a/M | g/V | a/M |
| rs30187 | 11 | 1583/528 | a/K | g/R | g,a/R,K |
| rs10050860 | 12 | 1723/575 | g/D | a/N | g/D |
| rs17482078 | 15 | 2174/725 | g/R | a/Q | g/R |
| rs27044 | 15 | 2188/730 | c/Q | g/E | g,c/E,Q |
| ERAP1 haplotype | Hap10 | Hap1 + Hap8 | |||
| ERAP2 | |||||
| rs2549782 | 7 | 1176/392 | g/K | g,t/K,N | g/K |
| ERAP2 expression | + | + | |||
Figure 1.
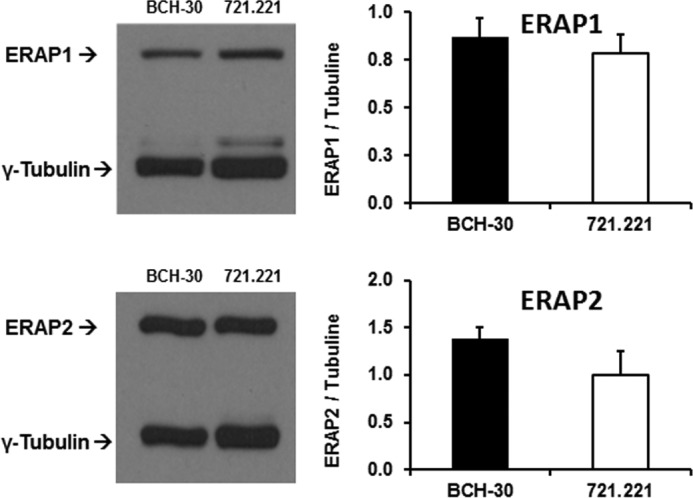
Expression of ERAP1 and ERAP2 proteins in 721.221-B*51:01 and BCH-30 cells. Representative Western blots and quantification of enzyme expression from three (for ERAP1) or four (for ERAP2) independent experiments (mean ± S.D. (error bars)) are shown. ERAP1 and ERAP2 expression in 721.221-B*51:01 was reported previously (25), but the blots are repeated here in parallel with BCH-30 for comparison.
Identification of HLA-B*51:08 ligands
A total of 5072 peptides were identified from BCH-30 cells. These peptides included the ligands of all of the MHC-I molecules in this cell line, due to the lack of allotypic and locus specificity of W6/32. Peptides with 8–12 residues (n = 4502) were selected. B*51:08 ligands were assigned on the basis of the major B*51 motif defined in our previous study (25), consisting of Pro or Ala at P2 and Val or Ile at the C-terminal position (PΩ). After restricting for the combined P2+PΩ motif, 624 peptides were reliably assigned as B*51:08 ligands (supplemental Table S2). When the same filter was applied to the 1620 B*51:01 ligands from 721.221-B*51:01 cells (25), 1271 peptides (78.5%) remained, indicating that this stringent filtering selected the majority of the HLA-B*51 peptidome. Peptides lacking this motif could not be reliably assigned as B*51:08 ligands and were excluded. Among these excluded peptides, 1188 and 1479 could be assigned as A*02:01 or B*40:01 ligands on the basis of the major motifs of these allotypes: Leu/Met at P2 + Leu/Val/Ile at PΩ for A*02:01 and Glu at P2 + Leu at PΩ (Immunoepitope database). The lower number of B*51:08 ligands may be partially a consequence of the stringent filtering applied but also due to the tendency of this allotype to be expressed at the cell surface at lower levels, compared with other MHC-I molecules (25). The peptides assigned to either A*02:01 or B*40:01 showed much higher affinity for their cognate molecule than for the other HLA-A,B molecules expressed in BCH-30 cells (supplemental Fig. S1), confirming the reliability of the assignments. In subsequent analyses, B*51:08 ligands with Pro/Ala at P2 and Val/Ile at PΩ were compared with the B*51:01 ligands from 721.221-B*51:01 cells showing the same motif.
Length distribution of B*51:08 ligands
The B*51:08 ligands from BCH-30 consisted of 13.6% 8-mers, 77.9% 9-mers, 4.2% 10-mers, and 4.3% longer peptides. When compared with those of B*51:01 from 721.221-B*51:01 cells, a significantly lower percentage of 8-mers and a corresponding increase of 9-mers was observed both in the whole B*51:08 peptide set and in the Pro-2 and Ala-2 subpeptidomes (Fig. 2, A and B).
Figure 2.
Length and subpeptidome distribution of HLA-B*51 ligands. A, the B*51:08 ligands with Pro-2/Ala-2 and Ile/Val at PΩ from the LCL BCH-30 (black) were classified by length and compared with the equivalent set of previously reported B*51:01 ligands from 721.221 transfectant cells (25) with the same motif (white). A total of 624 and 1271 peptides from B*51:08 and B*51:01, respectively, were included. B, comparison of the Pro-2 and Ala-2 subpeptidomes. A total of 285 and 833 peptides with Pro-2 and 339 and 438 peptides with Ala-2 from B*51:08 and B*51:01, respectively, were included. C, percent residue frequency at P2 of B*51:08 ligands (8–12 residues) from BCH-30 cells (black) with Pro-2/Ala-2 + Val/Ile at PΩ (n = 624), compared with the equivalent set of previously identified B*51:01 ligands (n = 1271) from 721.221-B*51:01 transfectant cells (white). Statistically significant differences, assessed by the χ2 test, and their p values are shown.
ERAP1 polymorphism affects the relative size of HLA-B*51 subpeptidomes
The B*51:08 ligands from BCH-30 showed 45.7% Pro and 54.3% Ala at P2 (Pro-2/Ala-2 ratio of 0.84). This was in strong contrast to the B*51:01 ligands from 721.221 cells: 65.5% Pro-2 and 34.5% Ala-2 (Pro-2/Ala-2 ratio of 1.9). This large difference (Fig. 2C) can hardly be explained by the amino acid changes between B*51:01 and B*51:08 because these are located away from the B pocket. However, the distinct activity of the ERAP1 variants in BCH-30 and 721.221 cells may account for the observed differences. In particular, peptides with Pro-2 are not degraded by ERAP1, regardless of the activity of the enzymatic variant, whereas peptides with Ala-2 could be more extensively overtrimmed by the more active variants in 721.221 cells, compared with BCH-30.
To further support that the distinct residue usage at P2 is due to differences in ERAP1 activity, 12 synthetic precursors of HLA-B*51 ligands with two N-terminal extensions were digested in vitro with three recombinant ERAP1 variants: Hap2 (high activity), Hap8 (intermediate activity), and Hap10 (low activity). Hap8 and Hap10 are expressed in 721.221 and BCH-30 cells, respectively. Hap2 is functionally undistinguishable from Hap1 (28), which is also expressed in 721.221 cells.
Ligands with Pro-2 were generated, but not destroyed, by Hap2, Hap8, and Hap10 in all six cases analyzed. For five of the six substrates, generation of the ligand was faster with Hap2 and Hap8 than with Hap10. It reached 100% in all cases with the two former variants and only in three of the six ligands with Hap10 (Fig. 3A). Ligands with Ala-2 were generated, but also destroyed to various extents, by all three ERAP1 variants. The time course of their generation/destruction balance was variant and peptide-dependent, but in five of six peptides, the yield of the ligand at the largest time analyzed was higher with Hap10, due to more overtrimming with the Hap2 and Hap8 enzymes. Only in one case (SA.IAKQEDVSV) (where the dot separates the two N-terminal extensions, as present in the parental proteins, from the sequence of the natural ligand) did the generation of the ligand prevail over its destruction and result in higher yields with Hap2 and Hap8 than with Hap10 at all times (Fig. 3B).
Figure 3.
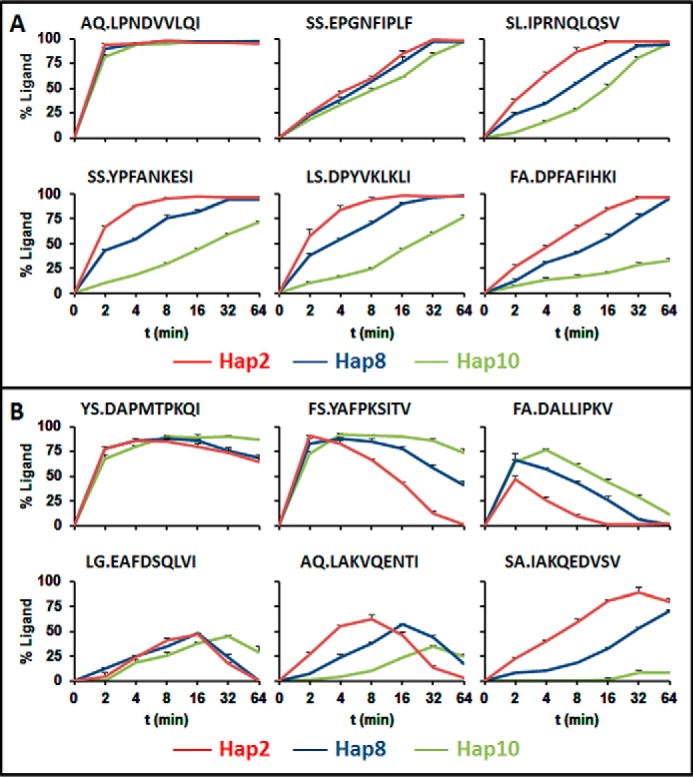
Digestion of synthetic precursors of HLA-B*51 ligands by recombinant ERAP1 variants. A, six synthetic peptides consisting of N-terminally extended precursors of natural HLA-B*51 ligands with Pro-2 were digested with recombinant Hap2 (red), Hap8 (blue), or Hap10 (green) ERAP1 variants. The percentage yield of each ligand is represented as a function of the digestion time. The amino acid sequence of each synthetic substrate is indicated. Dots separate the two N-terminal extensions, as present in the parental proteins, from the sequence of the natural ligand. B, the same experiments, in identical conditions as in A, were performed with six precursors of HLA-B*51 ligands with Ala-2. Error bars, S.E.
These results indicate that peptides with Pro-2 are less efficiently generated by the low activity Hap10 variant than by the more active ones. In contrast, at least at long digestion times, most peptides with Ala-2 are produced with higher yield by the less active variant. This pattern reproduces the one found in live cells, where the Ala-2 prevails over the Pro-2 subpeptidome in the Hap10 context of BCH-30, whereas the Pro-2 subpeptidome prevails over the Ala-2 one in the more active ERAP1 context of 721.221 cells (Fig. 2C). Thus, in vitro digestions strongly support that the large differences in the relative amounts of the Pro-2 and Ala-2 subpeptidomes found between B*51:08 and B*51:01 ligands are essentially determined by ERAP1.
ERAP1 polymorphism shapes the Ala-2 subpeptidome of HLA-B*51 through differential P1 trimming
In our previous study (25), we found that whereas B*51:01 ligands with Pro-2 showed a predominance of ERAP1-susceptible residues at P1, Ala-2 ligands showed a high frequency of ERAP1-resistant residues, mainly Asp, at this position. It was inferred that ERAP1 accounted for these differences due to its inefficient overtrimming of Ala-2 ligands with ERAP1-resistant P1 residues and to its inability to trim X–Pro bonds. Thus, P1 usage among peptides with Pro-2 should be determined by the binding preferences of HLA-B*51 and the relative amounts of the corresponding residues in the proteome, but not by their susceptibility to ERAP1 trimming.
Experimental demonstration of these assumptions was provided by the analysis of B*51:08 ligands from BCH-30. Their P1 residue frequencies were similar to those found among B*51:01 ligands from 721.221 cells, except for a higher frequency of Ala-1, an ERAP1-susceptible residue, in B*51:08 (Fig. 4A). The Pro-2 subpeptidomes showed no differences, indicating a lack of effect of either ERAP1 activity or subtype polymorphism on this position. The Ala-2 subpeptidome of B*51:08 and B*51:01 also showed similar patterns of residue usage, including a high frequency of Asp-1, albeit lower among B*51:08 ligands, and an increased frequency of Ala-1 in B*51:08, which accounted for that in the total peptidome. When grouped according to their ERAP1 susceptibility (Fig. 4B), a statistically significant increase of susceptible P1 residues and a corresponding decrease of resistant ones were found among B*51:08 ligands with Ala-2, relative to B*51:01. Thus, the low-activity ERAP1 context of BCH-30 cells resulted in less destruction of HLA-B*51 ligands with Ala-2 and ERAP1-susceptible P1 residues, relative to the more active context of 721.221 cells. The results reveal a significant effect of ERAP1 polymorphism on shaping the Ala-2, but not the Pro2, subpeptidomes of HLA-B*51 through differential processing of N-terminal residues.
Figure 4.
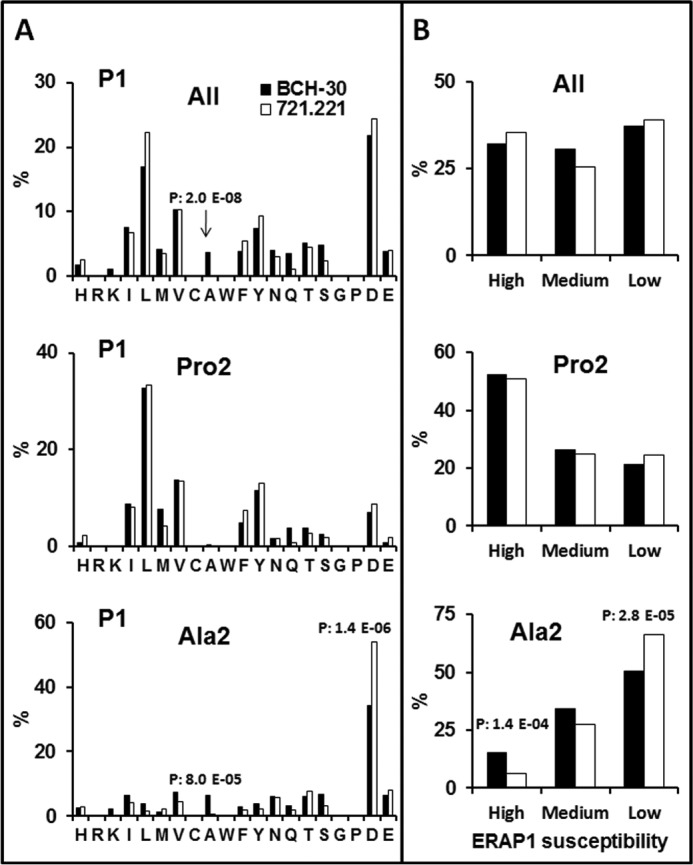
N-terminal residue usage among B*51:08 ligands and its comparison with B*51:01. A, the percent frequency of P1 residues among 624 B*51:08 ligands (8–12 residues) from BCH-30 cells with the Pro-2/Ala-2 + Val-9/Ile-9 motif and among the Pro-2 (n = 285) and Ala-2 (n = 339) subpeptidomes (black) were compared with the equivalent sets of 1271, 833, and 438 B51:01 ligands, respectively, from 721.221 transfectant cells (white) identified in Ref. 25. Amino acid residues are ordered according to their chemical features: basic (H to K), aliphatic (I to A), aromatic (W to Y), polar (N to S), G, P, and acidic (D and E). B, the P1 residues of the same peptide sets as in A were grouped according to their susceptibility to ERAP1 trimming (35) in high (Y, M, A, and L), medium (C, I, F, S, T, H, Q, G, and N), and low (E, W, D, K, V, R, and P) susceptibility. Statistically significant differences were assessed by the χ2 test with Bonferroni correction, and their p values are specified.
BD-associated ERAP1 polymorphism lowers the binding affinity of the HLA-B*51 peptidome
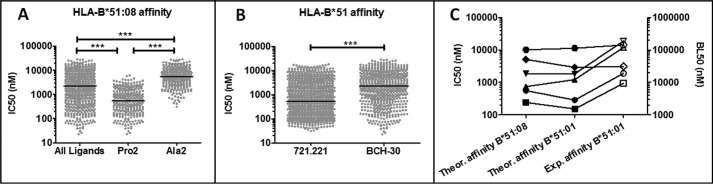
The Pro-2 subpeptidome of B*51:08 showed higher affinity than the Ala-2 counterpart (Fig. 5A), as also observed in HLA-B*51:01, both theoretically (25) and experimentally (29). The overall affinity of B*51:08 ligands from BCH-30 was significantly lower than that of B*51:01 ligands from 721.221 cells for its corresponding subtype (Fig. 5B). This difference was accounted for by the higher percentage of Ala-2 peptides in B*51:08, compared with B*51:01, and was therefore determined by the low activity of the Hap10 variant of ERAP1 in BCH-30 cells.
Figure 5.
Affinity of HLA-B*51 ligands.
A, theoretical binding affinity of the B*51:08 ligands (n = 624) and of the respective Pro-2 (n = 285) and Ala-2 (n = 339) subsets. B, comparison of the theoretical affinity of B*51:01 ligands from 721.221 cells (n = 1271) and the B*51:08 ligands (n = 624) from BCH-30 for their respective HLA-B*51 subtypes. The data concerning B*51:01 were previously reported (25) and are included here for comparison. Symbols represent individual peptides. The median of each data set is indicated by horizontal lines. Statistical differences were analyzed by the Mann-Whitney U test. ***, p < 0.0001. C, the theoretical affinity of various HIV-derived HLA-B*51 ligands for B*51:01 and B*51:08 (black symbols) is compared with their experimental binding affinities for B*51:01 (white symbols), reported in independent studies (29, 30). Experimental binding values were determined in those studies by an epitope stabilization assay and are expressed as their half-maximal binding level (BL50), defined as the peptide concentration yielding the half-maximal mean fluorescence intensity. The peptides shown are the following: FPISPIETV (■), LPPVVAKEI (●), LPCRIKQII (▴), NPPIPVGEI (▾), YAPPIGGQI (♦), and DARAYDTEV ( ).
).
The correspondence between theoretical and experimentally determined affinity values was assessed with a number of HLA-B*51 ligands whose experimental affinity for B*51:01 had been determined in earlier studies (29, 30). A comparison of these experimental values with their theoretical affinity for both B*51:01 and B*51:08 (Fig. 5C) showed a good correlation. Both the theoretical and experimental affinities showed a similar distribution, although the relative affinity between two given individual peptides was not always concordant. Thus, the theoretical affinity of the peptidome, as determined here, is a valid method to assess global affinity variations among peptide sets.
Subtype polymorphism affects the HLA-B*51 peptidome at internal sequence positions
The following analysis was performed only with 9-mers to ensure uniform sequence alignments. A total of 486 B*51:08 and 821 B*51:01 ligands from BCH-30 and 721.221 transfectants, respectively, were compared. Residue usage at each peptide position was determined for the whole peptide sets (Fig. 6) and the Pro-2 (supplemental Fig. S2) and the Ala-2 subpeptidomes (supplemental Fig. S3). Besides P1 and P2, statistically significant differences were found at P3 and P5–P8. At P3, an increased frequency of Lys, Met, and Gln and decreased frequency of Phe and Tyr in HLA-B*51:08, relative to B*51:01, was observed both with all of the 9-mers (Fig. 6) and in the Pro-2 subset (supplemental Fig. S2). Only the differences in Lys, Phe, and Tyr reached statistical significance in the Ala-2 subpeptidome (supplemental Fig. S3). Because the P3 position can be properly aligned among peptides of different lengths, this analysis was also performed using all B*51 ligands, with very similar results. Differential P3 frequencies were related to both the size and chemical nature of the side chains (supplemental Fig. S4); smaller and hydrophilic residues were more frequent in B*51:08.
Figure 6.
Residue usage among B*51:08 ligands and its comparison with B*51:01. The percent residue frequency at each peptide position among 486 nonameric ligands of B*51:08 from BCH-30 cells with the Pro-2/Ala-2 + Val-9/Ile-9 motif (black) was compared with the equivalent set of 821 ligands of B51:01 from 721.221 transfectant cells (white) identified in Ref. 25. Statistically significant differences were assessed by the χ2 test with Bonferroni correction and are labeled with an asterisk.
Ile-5, Val-5, Val-6, His-7, and Ala-8 were increased, whereas Pro-7 and Leu-8 were decreased, in B*51:08, relative to B*51:01 (Fig. 6). Most of these differences also reached statistical significance in the Pro-2 or Ala-2 subpeptidomes. In addition, Val-7 and Lys-7 were statistically increased only in the Pro-2 and Ala-2 subpeptidomes of B*51:08, respectively (supplemental Figs. S2 and S3).
To discern whether the observed changes at P5–P8 resulted directly from the polymorphism between B*51:01 and B*51:08 or were due to ERAP1-dependent alterations in the Pro-2/Ala-2 subpeptidomes, we compared the residue frequencies of the nonameric Pro-2 with Ala-2 subpeptidomes within each subtype. As observed previously (25), Pro-7 was increased in the Pro-2 relative to the Ala-2 subpeptidome of B*51:01. In contrast, in B*51:08, Lys-7 was increased in the Ala-2 relative to the Pro-2 subpeptidome (supplemental Fig. S5). No other statistically significant differences at internal peptide positions were observed in this subtype. Therefore, the changes between B*51:08 and B*51:01 at P5–P8, except those of Pro-7 and Lys-7, are a direct effect of subtype polymorphism.
Discussion
In this study, we analyzed the effects of the BD-associated Hap10 variant of ERAP1 and those of polymorphism between two BD-associated HLA-B*51 subtypes on their constitutive peptidomes. The strategy used was based on our previous description of the B*51:01 peptidome in a relatively active ERAP1 context (25) and the availability of a cell line from a patient expressing the BD-associated, but less frequent, B*51:08 subtype and the low-activity Hap10 variant.
Due to the lack of specific antibodies for HLA-B*51, the assignment of B*51:08 ligands from the pool of MHC-I sequences obtained from BCH-30 cells followed stringent criteria. These were based on selecting for the most frequent P2 (Pro/Ala) plus PΩ (Ile/Val) anchor residues of HLA-B*51. In addition to B*51:08, BCH-30 expresses HLA-A*02:01, B*40:01, C*16:02, and C*03. The main P2 motifs of A*02:01 and B*40:01 consist in Leu/Met (31) and Glu (32, 33), respectively. The peptidomes of C*16:02 and C*03 are not well characterized. However, based on their binding motifs defined by in vitro assays (34), selecting for the major P2+PΩ motif of B*51 should exclude most HLA-C ligands. The filter applied may exclude some bona fide B*51:08 ligands, but restricted our analyses to highly reliable assignments.
The comparison between the peptidomes of B*51:08 in the Hap10 context and of B*51:01 in a more active ERAP1 background provided information at two levels: 1) it allowed us to define the changes due to the BD-associated ERAP1 variant, which are presumably relevant to the pathogenesis of this disease, and 2) it revealed the effect of polymorphism between two BD-associated HLA-B*51 subtypes, therefore defining peptidome changes not affecting disease susceptibility. We observed differences in five features: 1) peptide length, 2) relative amounts of the Ala-2 and Pro-2 subpeptidomes, 3) N-terminal residue frequencies, 4) binding affinity, and 5) residue usage at internal peptide positions.
The smaller percentage of octamers among B*51:08 ligands is best explained by the lower activity of Hap10 in BCH-30, relative to the Hap1/Hap8 background of 721.221 cells, because the trimming of 9-mers is strongly dependent on ERAP1 activity.
A major effect of the low activity ERAP1 background on the B*51:08 peptidome was reversing the Ala-2/Pro-2 ratio, compared with B*51:01 expressed in a more active context. This is explained by the ability of ERAP1 to trim X–Ala, but not X–Pro bonds (35). Thus, peptides with Pro-2 are generated with higher efficiency by the more active ERAP1 variants but are not destroyed by them. In contrast, peptides with Ala-2 are generated more efficiently, but also more extensively destroyed, by the more active enzymes. As a result, Pro-2 peptides prevail over those with Ala-2 in the more active context of B*51:01, whereas peptides with Ala-2 prevailed in the less active ERAP1 context of B*51:08. That differences at P2 are not due to HLA-B*51 subtype polymorphism is also supported by the identical P2 preferences of HLA-B*44:02 and HLA-B*44:03, two subtypes that only differ by the D156L change, also present in HLA-B*51:08 and B*51:01 (36). Similarly, the V152E change did not alter P2 preferences among HLA-B*27 subtypes (37).
The higher affinity of the Pro-2 compared with the Ala-2 subpeptidome and the larger size of the latter in B*51:08 explain the low affinity of its peptidome. Thus, Hap10 confers a particularly low affinity to HLA-B*51·peptide complexes, by increasing the Ala-2/Pro-2 subpeptidome ratio, relative to more active ERAP1 variants. The larger contribution of Pro-2, relative to Ala-2, to HLA-B*51 binding was experimentally established in previous studies (29). Likewise, the good correlation between the theoretical affinity distribution of several HLA-B*51 ligands and the experimental binding data available from these peptides from earlier studies (29, 30) strongly supports that the collective affinity of large peptide sets is reliably established by theoretical predictive algorithms.
A fourth effect of ERAP1 on the B*51:08 peptidome was at the level of P1 residues. We previously reported that the Ala-2 and Pro-2 subpeptidomes of B*51:01 differed drastically at P1 in a way that correlated with the distinct cleavage of X–Pro and X–Ala bonds (25). Thus, Ala-2 peptides with ERAP1-resistant P1 residues, predominantly Asp-1, prevailed over those with ERAP1-susceptible residues, whereas among Pro-2 peptides, P1 residue usage was determined by the binding preferences of HLA-B*51:01 and by the abundance of the various amino acids in the proteome, and not by ERAP1. The similar pattern found in B*51:08 fully supports this interpretation. Moreover, the lower activity of Hap10, although not affecting P1 residue usage among Pro-2 ligands, diminished overtrimming among Ala-2 peptides, as confirmed by in vitro digestions, leading to higher prevalence of B*51:08 ligands with ERAP1-susceptible P1 residues, relative to B*51:01. Thus, N-terminal shaping of the HLA-B*51 peptidome is essentially due to ERAP1 polymorphism.
Whereas the differences in peptide length and P1/P2 residue usage can be reliably assigned to ERAP1, those at positions downstream are mainly determined by the amino acid differences between B*51:08 and B*51:01. Their polymorphic residues 152 and 156 are located in or close to the D and E pockets (38). In particular, the L156D change in B*51:08 affects the size and polarity of the D pocket, which is the binding site of the peptidic P3 residues, shifting the preference of B*51:01 for bulky aromatic residues at this position to smaller and polar ones, including Lys, in B*51:08.
Likewise, the decreased polarity due to the E152V change may explain the increased frequency of nonpolar residues at P5 and P6 among B*51:08 ligands. In contrast, the differences in Pro-7 and Lys-7 between both subtypes are explained by the distinct P7 frequencies between the Pro-2 and Ala-2 subpeptidomes of B*51:01 and B*51:08, respectively, and may reflect an interacting effect of the P2 residue on P7 usage that is subtype-dependent. Thus, without ruling out some reciprocal influence between changes at P1/P2 and those at positions downstream, the major influence of ERAP1 and subtype polymorphism, respectively, on both peptidic regions can be clearly distinguished.
The mechanism underlying the association of HLA-B*51 and ERAP1 with BD is unknown. It could be due to altered presentation of specific epitopes or to changes in other properties of HLA-B*51, such as its peptide-binding affinity. An involvement of specific epitopes would be compatible with the profound effects of ERAP1 on the Pro-2 and Ala-2 subpeptidomes, which implies the differential generation and destruction of many HLA-B*51 ligands, depending on the particular ERAP1 context. For instance, a protective epitope might not be generated by the risk Hap10 variant of ERAP1. Alternatively, a pathogenic epitope may only be efficiently presented in this low activity context due to overtrimming by more active enzyme variants. There is some evidence for an involvement of specific T-cell epitopes in BD. For instance, HLA-B*51-restricted autoreactive T-cell responses specific for the MICA-derived AAAAAIFVI peptide were selectively detected in BD patients with active disease (39). The sequence of this peptide includes five Ala residues at the N-terminal positions, which could be digested by high-activity ERAP1 variants more extensively than by the BD-associated Hap10 variant. However, a possible argument against a pathogenetic role of specific T-cell antigens is that both the E152V and L156D changes between B*51:01 and B*51:08 are critical for T-cell recognition (36, 37, 40). Thus, the T-cell repertoires recognizing B*51:01 and B*51:08 should presumably be quite different, despite the association of both subtypes with BD.
Alternatively, the pathogenic role of ERAP1 in BD might be related to its modulation of NK recognition of HLA-B*51. The role of NK cells in this disease is widely acknowledged, although their precise mechanism is incompletely understood (41–44). In one study, the down-regulation of ERAP1 expression in human tumor cell lines resulted in an increase of NK cell-mediated killing, especially in target cells expressing HLA allotypes that bind peptides with Pro-2, and this effect was reverted by the addition of high-affinity HLA ligands (45). Based on these observations, it might be suggested that the lower affinity of HLA-B*51·peptide complexes expressed in the risk Hap10 context might enhance NK activity in B*51/Hap10-positive individuals.
In conclusion, our study reveals that the BD-associated Hap10 haplotype of ERAP1 induces extensive changes in the peptide repertoires available for loading onto HLA-B*51, which alter its antigen-presenting specificity and can destabilize the molecule through generating a peptidome of lower affinity. These alterations can upset the recognition of HLA-B*51 by both cytotoxic T lymphocytes and NK cells, which may directly influence disease.
Experimental procedures
Cell lines and MHC-specific antibodies
The BCH-30 LCL (HLA-A*02:01; HLA-B*51:08, B*40:01; HLA-C*16:02, C*03) was generated by Epstein-Barr virus transformation from a Spanish BD patient. The study was approved by the ethical committee of the Hospital Universitario Virgen del Rocío (Sevilla, Spain), and the patient signed a written informed consent. The 721.221 cell line lacks expression of its endogenous MHC-I molecules. The 721.221-B*51:01 transfectants were kindly provided by Prof. Masafumi Takiguchi. Both BCH-30 and 721.221-B*51:01 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Sigma-Aldrich), 25 mm HEPES buffer, and 2 mm l-glutamine. The mAb W6/32 (IgG2a; specific for a monomorphic HLA class I determinant) (46) was used for the immunopurification of MHC-I molecules.
ERAP1 and ERAP2 genotyping and Western blotting
The individual from whom the BCH-30 LCL was derived was typed for five non-synonymous SNPs of ERAP1: rs27044 (Q730E), rs17482078 (R725Q), rs10050860 (D575N), rs30187 (R528K), and rs2287987 (V349M), as well as for the ERAP2 SNPs rs2549782 (K392N), and rs2248374, rs2287988, and rs2548538, located in non-coding regions, using TaqMan® SNP genotyping assays (Applied Biosystems, Barcelona, Spain) in a LightCycler 480 (Roche, Barcelona, Spain). In addition, the relevant exons of ERAP1 and ERAP2 were sequenced from the BCH-30 cell line as described previously (47). The ERAP1/ERAP2 genotype from 721.221-B*51:01 transfectant cells was described elsewhere (25, 27). Expression of the ERAP1, ERAP2, and γ-tubulin proteins was measured by Western blotting using the mAb 6H9 (a generous gift from Dr. Peter Van Endert), 3F5 (R&D Systems), and GTU88 (Sigma-Aldrich), respectively, as described previously (47). The scanned autoradiograms were quantified using ImageJ version 1.48 software (National Institutes of Health).
Isolation of HLA-bound peptides
The immunopurification of MHC-I-bound peptides from 109 BCH-30 cells was performed at 4 °C as described previously (28) with minor modifications. Cells were lysed in the presence of protease inhibitors, and the supernatant was passed through a column containing 1 mg of W6/32 bound to CNBr-activated Sepharose 4B (GE Healthcare, Buckinghamshire, UK) and washed with 40 column volumes each of 20 mm Tris-HCl, pH 8.0, containing 1) 150 mm NaCl, 2) 400 mm NaCl, 3) 150 mm NaCl, and 4) 80 column volumes of buffer without NaCl. Peptides were eluted with 1% trifluoroacetic acid (Sigma-Aldrich). The unfractionated peptide pool was filtered through a Vivaspin 2 concentrator, cutoff 5000 Da (Sartorius Stedim Biotech, Gottingen, Germany), and dried in a SpeedVac. Three preparations were independently obtained from the same cell amounts to provide for biological replicates.
Mass spectrometry
Each sample was analyzed in a Q-Exactive-Plus mass spectrometer (Thermo Fisher Scientific) as described previously (25). The peptides were resolved using 7–40% acetonitrile gradients with 0.1% formic acid for 180 min and 0.15 μl/min on a capillary column pressure-packed with Reprosil C18-Aqua (Dr. Maisch, GmbH, Ammerbuch-Entringen, Germany) as described previously (48). The dynamic exclusion was set to 20 s; the selected masses were fragmented from the survey scan of mass to charge ratio (m/z) 300–1,800 atomic mass units at resolution of 70,000. MS/MS spectra were acquired starting at m/z 200 with a resolution of 17,500. The target value was set to 1 × 105, and the isolation window was set to 1.8 m/z. Peptide sequences were assigned from the MS/MS spectra using MaxQuant software (version 1.5.0.25) (49) with the Andromeda search engine (50) and the human UniProt/Swiss-Prot database (release 2015_07: 69,693 entries) under the following parameters: precursor ion mass and fragment mass tolerance, 20 ppm; false discovery rate, 0.01; peptide-spectrum matching FDR, 0.05; and oxidation (Met), acetyl (protein N terminus), and Gln to Pyro-Glu as variable modifications. Identifications derived from the reverse database and known contaminants were eliminated. The raw MS data are available via ProteomeXchange with identifier PXD006215.
Recombinant ERAP1 variants and peptide-trimming assays
Recombinant proteins of 3 ERAP1 variants, Hap2, Hap8, and Hap10 (16) were obtained, and in vitro peptide digestions were performed, as described previously (25).
MHC-binding affinity and hydropathy analyses
Theoretical peptide-binding affinities were calculated using the NetMHCcons 1.1 server, which combines three predictive algorithms: NetMHC, NetMHCpan, and PickPocket (51). The first algorithm predicts binding only to MHC molecules on which it has been trained, whereas NetMHCpan and PickPocket are pan-specific and can be used to calculate binding to MHC alleles with little or no experimental binding data. NetMHCcons integrates these three algorithms and gives the most accurate binding prediction to any given MHC-I allele. The hydropathy of amino acid residues was estimated using the index of Kyte and Doolittle (52). A higher hydropathy index indicates higher hydrophobicity.
Statistical analyses
Differences in residue frequencies were analyzed by the χ2 test with Bonferroni correction. Differences in MHC binding affinities were analyzed by the Mann-Whitney test. p values < 0.05 were considered as statistically significant.
Author contributions
P. G. was responsible for study conception; data acquisition, analysis, and interpretation; and drafting of the article. E. B., M. F. G.-E., A. J.-R., J. R. R., and A. A. were responsible for data acquisition and analysis. J. A. L. C. was responsible for study conception and coordination, data analysis, and writing of the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Masafumi Takiguchi (Center for AIDS Research, Kumamoto University, Japan) and Peter van Endert (Hôpital Necker, INSERM U1151, Paris) for providing 221.221-B*51:01 cells and the 6H9 mAb, respectively. We also thank Sergio Ciordia and María C. Mena (Proteomics Unit, Centro Nacional de Biotecnología, Madrid) for technical assistance in MS.
This work was supported by Plan Nacional de I+D+i Research Grant SAF2014/51931-R (to J. A. L. C.); Binational Science Foundation Grant 2009393 (to A. A.); Complutense University Grants SAF2014–54708R, SAF2016-81876-REDT (Plan Nacional de I+D+i), and 920631 (to J. R. R.); Ministry of Health, Social Services and Equality Grant FIS PI13/01118 (to M. F. G. E.); and an institutional grant of the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2 and Figs. S1–S5.
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD006215.
- BD
- Behçet's disease
- NK
- natural killer
- ERAP
- endoplasmic reticulum aminopeptidase
- Hap
- haplotype
- P
- peptide position
- LCL
- lymphoblastoid cell line.
References
- 1. Sakane T., Takeno M., Suzuki N., and Inaba G. (1999) Behçet's disease. N. Engl. J. Med. 341, 1284–1291 [DOI] [PubMed] [Google Scholar]
- 2. Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F. S., Erer B., Inoko H., Emrence Z., Cakar A., Abaci N., Ustek D., Satorius C., et al. (2013) Genome-wide association analysis identifies new susceptibility loci for Behçet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 45, 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Menthon M., Lavalley M. P., Maldini C., Guillevin L., and Mahr A. (2009) HLA-B51/B5 and the risk of Behçet's disease: a systematic review and meta-analysis of case-control genetic association studies. Arthritis Rheum. 61, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vilches C., Bunce M., de Pablo R., Murray A. K., McIntyre C. A., and Kreisler M. (1997) Complete coding regions of two novel HLA-B alleles detected by phototyping (PCR-SSP) in the British caucasoid population: B*5108 and B*5002. Tissue Antigens 50, 38–41 [DOI] [PubMed] [Google Scholar]
- 5. González-Escribano M. F., Rodríguez M. R., Walter K., Sanchez-Roman J., García-Lozano J. R., and Núñez-Roldán A. (1998) Association of HLA-B51 subtypes and Behçet's disease in Spain. Tissue Antigens 52, 78–80 [DOI] [PubMed] [Google Scholar]
- 6. Kera J., Mizuki N., Ota M., Katsuyama Y., Pivetti-Pezzi P., Ohno S., and Inoko H. (1999) Significant associations of HLA-B*5101 and B*5108, and lack of association of class II alleles with Behçet's disease in Italian patients. Tissue Antigens 54, 565–571 [DOI] [PubMed] [Google Scholar]
- 7. Yabuki K., Ohno S., Mizuki N., Ando H., Tabbara K. F., Goto K., Nomura E., Nakamura S., Ito N., Ota M., Katsuyama Y., and Inoko H. (1999) HLA class I and II typing of the patients with Behçet's disease in Saudi Arabia. Tissue Antigens 54, 273–277 [DOI] [PubMed] [Google Scholar]
- 8. Mizuki N., Ota M., Katsuyama Y., Yabuki K., Ando H., Yoshida M., Onari K., Nikbin B., Davatchi F., Chams H., Ghaderi A. A., Ohno S., and Inoko H. (2001) HLA class I genotyping including HLA-B*51 allele typing in the Iranian patients with Behçet's disease. Tissue Antigens 57, 457–462 [DOI] [PubMed] [Google Scholar]
- 9. Kötter I., Günaydin I., Stübiger N., Yazici H., Fresko I., Zouboulis C. C., Adler Y., Steiert I., Kurz B., Wernet D., Braun B., and Müller C. A. (2001) Comparative analysis of the association of HLA-B*51 suballeles with Behçet's disease in patients of German and Turkish origin. Tissue Antigens 58, 166–170 [DOI] [PubMed] [Google Scholar]
- 10. Paul M., Klein T., Krause I., Molad Y., Narinsky R., and Weinberger A. (2001) Allelic distribution of HLA-B*5 in HLA-B5-positive Israeli patients with Behçet's disease. Tissue Antigens 58, 185–186 [DOI] [PubMed] [Google Scholar]
- 11. Mizuki N., Ota M., Katsuyama Y., Yabuki K., Ando H., Shiina T., Palimeris G. D., Kaklamani E., Ito D., Ohno S., and Inoko H. (2002) Sequencing-based typing of HLA-B*51 alleles and the significant association of HLA-B*5101 and -B*5108 with Behçet's disease in Greek patients. Tissue Antigens 59, 118–121 [DOI] [PubMed] [Google Scholar]
- 12. Rock K. L., York I. A., Saric T., and Goldberg A. L. (2002) Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 80, 1–70 [DOI] [PubMed] [Google Scholar]
- 13. Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., and Goldberg A. L. (2002) An IFN-γ-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 14. Saveanu L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G., and van Endert P. M. (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6, 689–697 [DOI] [PubMed] [Google Scholar]
- 15. López de Castro J. A., Alvarez-Navarro C., Brito A., Guasp P., Martín-Esteban A., and Sanz-Bravo A. (2016) Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: towards a unifying view. Mol. Immunol. 77, 193–204 [DOI] [PubMed] [Google Scholar]
- 16. Ombrello M. J., Kastner D. L., and Remmers E. F. (2015) Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: genetics. Curr. Opin. Rheumatol. 27, 349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goto Y., Hattori A., Ishii Y., and Tsujimoto M. (2006) Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 580, 1833–1838 [DOI] [PubMed] [Google Scholar]
- 18. Evans D. M., Spencer C. C., Pointon J. J., Su Z., Harvey D., Kochan G., Oppermann U., Opperman U., Dilthey A., Pirinen M., Stone M. A., Appleton L., Moutsianas L., Moutsianis L., Leslie S., et al. (2011) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evnouchidou I., Kamal R. P., Seregin S. S., Goto Y., Tsujimoto M., Hattori A., Voulgari P. V., Drosos A. A., Amalfitano A., York I. A., and Stratikos E. (2011) Cutting edge: coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J. Immunol. 186, 1909–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kochan G., Krojer T., Harvey D., Fischer R., Chen L., Vollmar M., von Delft F., Kavanagh K. L., Brown M. A., Bowness P., Wordsworth P., Kessler B. M., and Oppermann U. (2011) Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U.S.A. 108, 7745–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martín-Esteban A., Gómez-Molina P., Sanz-Bravo A., and López de Castro J. A. (2014) Combined effects of ankylosing spondylitis-associated ERAP1 polymorphisms outside the catalytic and peptide-binding sites on the processing of natural HLA-B27 ligands. J. Biol. Chem. 289, 3978–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stamogiannos A., Koumantou D., Papakyriakou A., and Stratikos E. (2015) Effects of polymorphic variation on the mechanism of endoplasmic reticulum aminopeptidase 1. Mol. Immunol. 67, 426–435 [DOI] [PubMed] [Google Scholar]
- 23. Reeves E., Edwards C. J., Elliott T., and James E. (2013) Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J. Immunol. 191, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeuchi M., Ombrello M. J., Kirino Y., Erer B., Tugal-Tutkun I., Seyahi E., Özyazgan Y., Watts N. R., Gül A., Kastner D. L., and Remmers E. F. (2016) A single endoplasmic reticulum aminopeptidase-1 protein allotype is a strong risk factor for Behçet's disease in HLA-B*51 carriers. Ann. Rheum. Dis. 75, 2208–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guasp P., Alvarez-Navarro C., Gomez-Molina P., Martín-Esteban A., Marcilla M., Barnea E., Admon A., and López de Castro J. A. (2016) The peptidome of the Behçet's disease-associated HLA-B*51:01 includes two sub-peptidomes differentially shaped by ERAP1. Arthritis Rheumatol. 68, 505–515 [DOI] [PubMed] [Google Scholar]
- 26. Andrés A. M., Dennis M. Y., Kretzschmar W. W., Cannons J. L., Lee-Lin S. Q., Hurle B., NISC Comparative Sequencing Program, Schwartzberg P. L., Williamson S. H., Bustamante C. D., Nielsen R., Clark A. G., and Green E. D. (2010) Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6, e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guasp P., Alvarez-Navarro C., Gomez-Molina P., Martin-Esteban A., Marcilla M., Barnea E., Admon A., and López de Castro J. A. (2017) Heterozygosity of the 721.221-B*51:01 cell line used in the study by Guasp et (Arthritis Rheumatol, February 2016). Arthritis Rheumatol. 10.1002/art.40073 [DOI] [PubMed] [Google Scholar]
- 28. Martín-Esteban A., Guasp P., Barnea E., Admon A., and López de Castro J. A. (2016) Functional interaction of the ankylosing spondylitis associated endoplasmic reticulum aminopeptidase 2 with the HLA-B*27 peptidome in human cells. Arthritis Rheumatol. 68, 2466–2475 [DOI] [PubMed] [Google Scholar]
- 29. Sakaguchi T., Ibe M., Miwa K., Kaneko Y., Yokota S., Tanaka K., and Takiguchi M. (1997) Binding of 8-mer to 11-mer peptides carrying the anchor residues to slow assembling HLA class I molecules (HLA-B*5101). Immunogenetics 45, 259–265 [DOI] [PubMed] [Google Scholar]
- 30. Sakaguchi T., Ibe M., Miwa K., Yokota S., Tanaka K., Schönbach C., and Takiguchi M. (1997) Predominant role of N-terminal residue of nonamer peptides in their binding to HLA-B* 5101 molecules. Immunogenetics 46, 245–248 [DOI] [PubMed] [Google Scholar]
- 31. Falk K., Rötzschke O., Stevanović S., Jung G., and Rammensee H. G. (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 [DOI] [PubMed] [Google Scholar]
- 32. Falk K., Rötzschke O., Takiguchi M., Gnau V., Stevanović S., Jung G., and Rammensee H. G. (1995) Peptide motifs of HLA-B58, B60, B61, and B62 molecules. Immunogenetics 41, 165–168 [DOI] [PubMed] [Google Scholar]
- 33. Hillen N., Mester G., Lemmel C., Weinzierl A. O., Müller M., Wernet D., Hennenlotter J., Stenzl A., Rammensee H. G., and Stevanović S. (2008) Essential differences in ligand presentation and T cell epitope recognition among HLA molecules of the HLA-B44 supertype. Eur. J. Immunol. 38, 2993–3003 [DOI] [PubMed] [Google Scholar]
- 34. Rasmussen M., Harndahl M., Stryhn A., Boucherma R., Nielsen L. L., Lemonnier F. A., Nielsen M., and Buus S. (2014) Uncovering the peptide-binding specificities of HLA-C: a general strategy to determine the specificity of any MHC class I molecule. J. Immunol. 193, 4790–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hearn A., York I. A., and Rock K. L. (2009) The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J. Immunol. 183, 5526–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fleischhauer K., Avila D., Vilbois F., Traversari C., Bordignon C., and Wallny H. J. (1994) Characterization of natural peptide ligands for HLA-B*4402 and -B*4403: implications for peptide involvement in allorecognition of a single amino acid change in the HLA-B44 heavy chain. Tissue Antigens 44, 311–317 [DOI] [PubMed] [Google Scholar]
- 37. García F., Rognan D., Lamas J. R., Marina A., and López de Castro J. A. (1998) An HLA-B27 polymorphism (B*2710) that is critical for T-cell recognition has limited effects on peptide specificity. Tissue Antigens 51, 1–9 [DOI] [PubMed] [Google Scholar]
- 38. Maenaka K., Maenaka T., Tomiyama H., Takiguchi M., Stuart D. I., and Jones E. Y. (2000) Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes. J. Immunol. 165, 3260–3267 [DOI] [PubMed] [Google Scholar]
- 39. Yasuoka H., Okazaki Y., Kawakami Y., Hirakata M., Inoko H., Ikeda Y., and Kuwana M. (2004) Autoreactive CD8+ cytotoxic T lymphocytes to major histocompatibility complex class I chain-related gene A in patients with Behçet's disease. Arthritis Rheum. 50, 3658–3662 [DOI] [PubMed] [Google Scholar]
- 40. Cowan E. P., Jelachich M. L., Coligan J. E., and Biddison W. E. (1987) Site-directed mutagenesis of an HLA-A3 gene identifies amino acid 152 as crucial for major-histocompatibility-complex- restricted and alloreactive cytotoxic-T-lymphocyte recognition. Proc. Natl. Acad. Sci. U.S.A. 84, 5014–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamzaoui K., Ayed K., Hamza M., and Touraine J. L. (1988) Natural killer cells in Behçet's disease. Clin. Exp. Immunol. 71, 126–131 [PMC free article] [PubMed] [Google Scholar]
- 42. Suzuki Y., Hoshi K., Matsuda T., and Mizushima Y. (1992) Increased peripheral blood γδ+ T cells and natural killer cells in Behçet's disease. J. Rheumatol. 19, 588–592 [PubMed] [Google Scholar]
- 43. Yamaguchi Y., Takahashi H., Satoh T., Okazaki Y., Mizuki N., Takahashi K., Ikezawa Z., and Kuwana M. (2010) Natural killer cells control a T-helper 1 response in patients with Behçet's disease. Arthritis Res. Ther. 12, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hasan M. S., Ryan P. L., Bergmeier L. A., and Fortune F. (2017) Circulating NK cells and their subsets in Behçet's disease. Clin. Exp. Immunol. 188, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cifaldi L., Romania P., Falco M., Lorenzi S., Meazza R., Petrini S., Andreani M., Pende D., Locatelli F., and Fruci D. (2015) ERAP1 regulates natural killer cell function by controlling the engagement of inhibitory receptors. Cancer Res. 75, 824–834 [DOI] [PubMed] [Google Scholar]
- 46. Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., and Ziegler A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens: new tools for genetic analysis. Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 47. García-Medel N., Sanz-Bravo A., Van Nguyen D., Galocha B., Gómez-Molina P., Martín-Esteban A., Alvarez-Navarro C., and López de Castro J. A. (2012) Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol. Cell Proteomics 11, 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishihama Y., Rappsilber J., Andersen J. S., and Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 49. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 50. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome. Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 51. Karosiene E., Lundegaard C., Lund O., and Nielsen M. (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 [DOI] [PubMed] [Google Scholar]
- 52. Kyte J., and Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.