Abstract
Local translation of specific mRNAs is regulated by dynamic changes in their subcellular localization, and these changes are due to complex mechanisms controlling cytoplasmic mRNA transport. The budding yeast Saccharomyces cerevisiae is well suited to studying these mechanisms because many of its transcripts are transported from the mother cell to the budding daughter cell. Here, we investigated the translational control of ASH1 mRNA after transport and localization. We show that although ASH1 transcripts were translated after they reached the bud tip, some mRNAs were bound by the RNA-binding protein Puf6 and were non-polysomal. We also found that the DEAD-box helicase Dhh1 complexed with the untranslated ASH1 mRNA and Puf6. Loss of Dhh1 affected local translation of ASH1 mRNA and resulted in delocalization of ASH1 transcript in the bud. Forcibly shifting the non-polysomal ASH1 mRNA into polysomes was associated with Dhh1 dissociation. We further demonstrated that Dhh1 is not recruited to ASH1 mRNA co-transcriptionally, suggesting that it could bind to ASH1 mRNA within the cytoplasm. Of note, Dhh1 bound to the 5′-UTR of ASH1 mRNA and inhibited its translation in vitro. These results suggest that after localization to the bud tip, a portion of the localized ASH1 mRNA becomes translationally inactive because of binding of Dhh1 and Puf6 to the 5′- and 3′-UTRs of ASH1 mRNA.
Keywords: protein translocation, RNA-binding protein, RNA transport, translation, translation control
Introduction
Local translation of particular mRNAs involves dynamic changes in subcellular localization. To this end, cells employ various and complex mechanisms to control specific cytoplasmic localization of mRNAs and their site of translation (1). The budding yeast Saccharomyces cerevisiae has emerged as a good system to study these mechanisms because a substantial number of transcripts have been identified as transporting from the mother cell to the budding daughter cell for such a purpose (2). One of these transcripts, ASH1, is transcribed in the mother cell and then transported to the bud tip, where the protein is translated and translocated into the daughter cell nuclei during late anaphase (3). The asymmetric localization of Ash1 within the daughter cell nuclei represses the transcription of HO endonuclease, which is crucial for the mating-type switch (3–5). The localization of ASH1 mRNA relies on She2, an RNA-binding protein that is co-transcriptionally bound to the cis-acting elements (or “zipcodes”) of the mRNA (6, 7). After transcription, the ASH1 mRNP2 complex is exported to the cytoplasm, where it interacts with She3, an adaptor protein for the mRNA localization machinery (8). She3 connects the ASH1 mRNP complex to the motor protein She1 (Myo4), which utilizes the actin cytoskeleton to transport the complex to the bud tip (9).
In addition to She2, other RNA-binding proteins, such as Puf6 and Khd1, have been identified as essential for complete localization of ASH1 mRNP complex and subsequent local translation of Ash1. Khd1p was identified in a systematic survey of potential candidate RNA-binding proteins for ASH1 mRNA localization (10). Binding of Khd1 decreased translational initiation of ASH1 mRNA and reduced leakage of Ash1 into the mother cell nucleus (11). Puf6 is a member of the Pumilio/FBF (fem-3 mRNA-binding factor) family of RNA-binding proteins that was purified from She2-associated mRNPs (12). Puf6 directly binds to the 3′-UTR of ASH1 mRNA and represses its translation during the transport process. At the bud tip, phosphorylation of Puf6 releases translational repression of ASH1 mRNA and leads to Ash1 synthesis (13). These studies indicate that translation of ASH1 mRNA is precisely regulated by RNA-binding proteins, and Puf6 and Khd1 could function in the linkage between ASH1 mRNA localization and its translation. Although translational repression of ASH1 mRNA during transport has been well studied, the regulatory mechanism for ASH1 mRNA translation after its proper localization remains to be studied.
In this study, we show that although translation of ASH1 mRNA occurs after localization, a portion of the transcripts remain non-polysomal and therefore could still bind to Puf6. We identified Dhh1, which interacts with untranslated ASH1 mRNA and co-precipitates with Puf6. Deletion of the DHH1 gene not only results in diffusion of ASH1 mRNA in the bud but also affects translation of the mRNA. Forcibly shifting non-polysomal ASH1 mRNA into polysomes causes dissociation of Dhh1 with the transcript. Dhh1 is not recruited to ASH1 mRNA co-transcriptionally but is possibly associated with the messenger RNA within the cytoplasm. Interestingly, Dhh1 did not directly interact with Puf6 but bound to the 5′-UTR of ASH1 mRNA, and this binding repressed translation of the mRNA in vitro. These results suggest a novel function of Dhh1 in regulating localized ASH1 mRNA translation after its bud-tip localization.
Results
Translation occurs after ASH1 mRNA localization to the bud tip
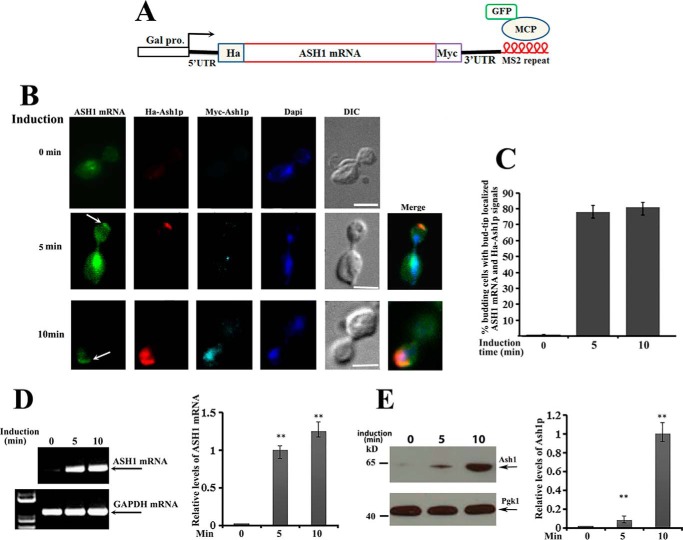
It has been reported previously that the average time to transcribe a yeast gene is about 25–50 s/kb (14–17), and the transport time for an ASH1 mRNA reporter (from mother to the bud tip) is about 128 s (18). Based on these results, the process from transcription to localization of ASH1 mRNA would be around 4–5 min. Due to difficulties in investigating the local translation of endogenous ASH1 mRNA, we transformed two centromeric plasmids, one expressing HA-ASH1-Myc-MS26 mRNA (a 2.5-kb transcript) under Gal promoter control and another expressing a GFP-MCP (MS2-binding coat protein) fusion protein into an ASH1 gene deletion strain. The fusion protein contains a nuclear localization signal, such that the protein unbound to ASH1 mRNA is sequestered within the nuclei (Fig. 1A). This system allowed us to study the coordinated localization and translation of ASH1 mRNA in the anaphase cells because translational initiation of the protein could be imaged by immunostaining with HA antibodies, and fully translated Ash1 would be translocated into the bud nucleus and be detected by Myc antibodies. Fig. 1B shows representative images in a time course of 0, 5, and 10 min after galactose induction. GFP-labeled ASH1 mRNA was visualized at the bud tip after 5 min of galactose induction (Fig. 1B, middle panels, arrow), where translational initiation could be seen by Cy3-labeled anti-HA antibody (red). After a 10-min induction, ASH1 mRNA and HA-Ash1 signals were still obviously distinguished at the bud tip, whereas the fully translated Ash1, which was detected by both anti-HA and Cy5-labeled anti-Myc antibodies, was translocated into the nucleus (cyan) (Fig. 1B, bottom panels). Localized translation of ASH1 mRNA after 5 and 10 min of galactose induction could be observed in about 80% of anaphase cells (Fig. 1C). Consistent with the imaging results, ASH1 mRNA was identified within 5 and 10 min of induction by RT-PCR (Fig. 1D), and the full-length Ash1 was detected by the Myc antibody after 10 min of induction (Fig. 1E). Statistical analyses of the relative levels of ASH1 mRNA and Ash1 are shown in the right panels of Fig. 1, D and E. These data suggest that translational initiation of ASH1 mRNA could occur directly after localization.
Figure 1.
Translation of ASH1 mRNA occurs after bud-tip localization. A, schematic presentation of the centromeric construct used to visualize ASH1 mRNA localization and translation in ash1 cells. The ASH1 gene was placed under the control of a galactose-inducible promoter (Gal1). HA and Myc tags were separately fused onto the N and C termini of Ash1. Six repeats of MS2 stem loops were fused to the 3′-UTR of ASH1 mRNA, which were recognized by GFP-MCP (MS2 RNA coat protein). B, yeast cells were cultured in 2% raffinose medium at 30 °C and induced with galactose (2%). After 0, 5, and 10 min of induction, cells were harvested and subjected to immunostaining assays. Localization of ASH1 mRNA was visualized by GFP-MCP, and translation of Ash1 was detected by Cy3-labeled HA (red) or Cy5-labeled Myc (cyan) antibodies. The arrows indicate the bud-tip-localized ASH1 mRNA. Bar, 3 μm. C, percentage of anaphase cells with localized ASH1 mRNA and translated HA-Ash1 signals after galactose induction from three independent experiments. An average of 50–60 cells were counted each time. D, RT-PCR was used to detect the expression of ASH1 mRNA after 0, 5, and 10 min of galactose induction. GAPDH mRNA was used as a positive control. Right, error bars indicate S.E. from three independent experiments. **, p < 0.01. E, full-length Ash1 after 0, 5, and 10 min of galactose induction was detected using anti-Myc antibodies by Western blotting, in which Pgk1 was used as a loading control. Right, mean values ± S.D. (error bars) were from three independent experiments. **, p < 0.01.
A portion of ASH1 mRNAs could be translationally repressed after localization
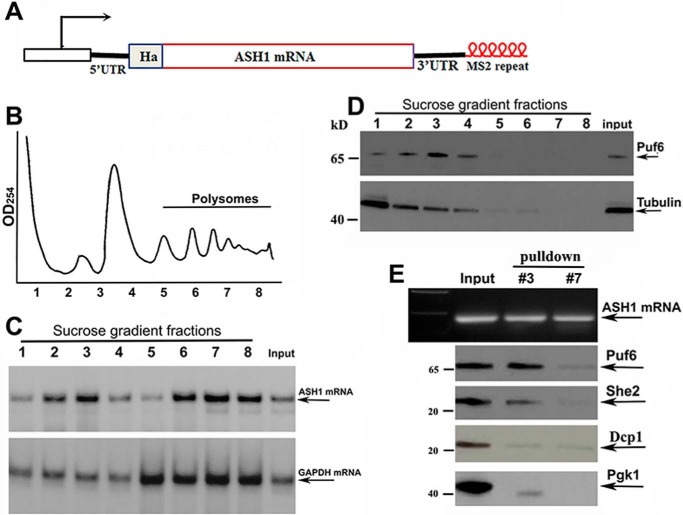
To examine whether all ASH1 mRNA was translated after localization, we performed sucrose-gradient fractionation of cells expressing an endogenous HA-tagged ASH1-MS26 transcript and a His-tagged Puf6 (Fig. 2A). An A254 plot corresponding to the polysomal fractions is shown in Fig. 2B. Total RNAs were isolated from the sucrose fractions and were used to analyze the distribution of ASH1 mRNA by Northern blotting (Fig. 2C). In comparison with GAPDH mRNA that was mostly polysomal, a substantial amount of ASH1 mRNA was detected in non-polysomal fractions (fractions 2 and 3) as well as in the polysomal fractions (fractions 6–8). Disrupting the polysomes with EDTA shifted ASH1 mRNA into non-polysomal fractions (supplemental Fig. S1). Because binding of Puf6 repressed ASH1 mRNA translation during transport (12) and release of the protein by phosphorylation resulted in translational activation of the mRNA (13), we analyzed the distribution of Puf6 in the sucrose fractions. Western blotting demonstrated that Puf6 was present in the non-polysomal fractions (Fig. 2D, fractions 2 and 3 of the top panel). We then used recombinant chimeric MBP-MCP (supplemental Fig. S2A; MBP binds to amylose resin) to precipitate ASH1-MS26 mRNA in selected sucrose fractions (Fig. 2E, top). Puf6 co-precipitated with ASH1 mRNA in the non-polysomal fraction 3 but not in the polysomal fraction 7. She2, an essential protein of the ASH1 mRNA localization machinery, was also detected in the precipitates of the non-polysomal fraction. An mRNA-binding and -decapping protein, Dcp1 and an abundant Pgk1 (phosphoglycerate kinase 1) were not present in the precipitates of either fraction 3 or fraction 7 (Fig. 2E, bottom). These data suggest that the non-polysomal fractions contained localized and translationally repressed ASH1 mRNA.
Figure 2.
Puf6 associates with non-polysomal ASH1 mRNA. A, schematic representation of the modified endogenous ASH1 gene. HA tag was fused to the N terminus of Ash1. Six repeats of MS2 stem loops were fused to the 3′-UTR of ASH1 mRNA. B, extracts of yeast cells were fractionated in 10–50% linear sucrose gradients. An A254 plot corresponding to the polysomal fractions is shown. C, total RNAs were isolated from the input and sucrose fractions. An equal amount of RNA from each fraction was subjected to Northern blotting to detect the distribution of ASH1 mRNA in the sucrose gradient (arrows) with GAPDH mRNA as an internal control. D, equal amounts of protein from each fraction were subjected to Western blotting for detecting the expression of Puf6. Tubulin was used as an internal control. E, ASH1-MS2 mRNA was precipitated using recombinant MBP-MCP (MBP binds to amylose resins), as shown in the top panel. Fractions 3 and 7 are non-polysomal and polysomal fractions. Puf6 and She2 were detected in the input and the precipitates of the non-polysomal fraction. Dcp1 and Pgk1 were used as negative controls.
Dhh1 binds to bud-tip-localized and translationally inactive ASH1 mRNA
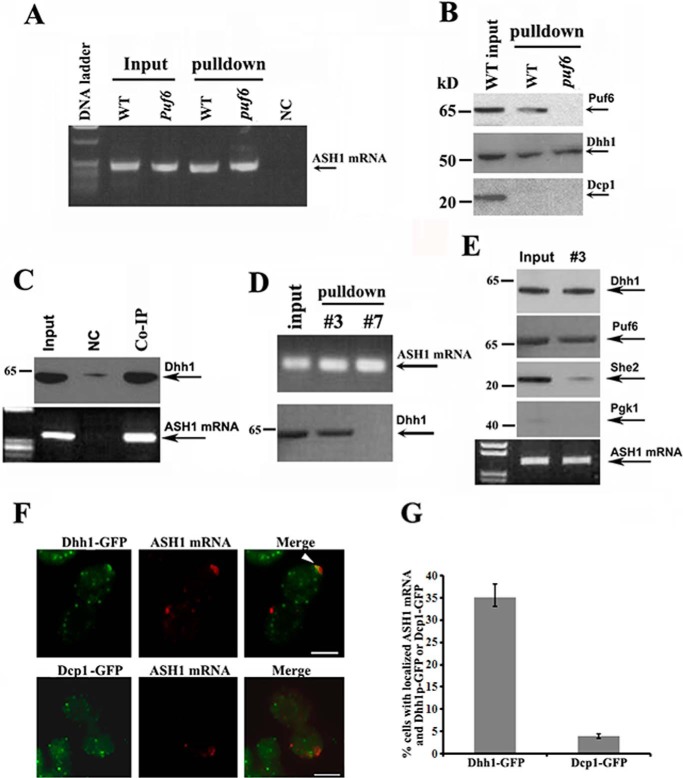
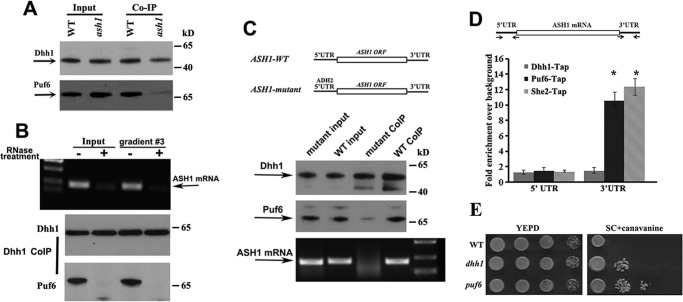
In yeast, Dhh1 is an abundant DEAD-box helicase, implicated in translational repression and enhancement of mRNA decay (19–21). The role of Dhh1 in storing and maintaining untranslated mRNAs has been well reported (22). To test the possibility that Dhh1 or other P-body proteins could be involved in the regulation of ASH1 mRNA translation, we prepared extracts from WT and puf6 strains expressing an endogenous HA-tagged ASH1-MS26 transcript. We then used the recombinant MBP-MCP to pull down the ASH1-MS26 mRNA. RT-PCR analysis indicated that ASH1 mRNA was successfully precipitated in both wild-type and puf6 cells (Fig. 3A). The specificity of the experiment was tested by a control strain in which the MS2 stem loops were not fused to ASH1 mRNA (Fig. 3A, NC). Analyzing the proteins co-precipitated with the ASH1 mRNA, we found that in addition to Puf6, Dhh1 also existed in the precipitates of both WT and puf6 cells (Fig. 3B), suggesting that the in vivo binding of the Dhh1 with ASH1 mRNA could be Puf6-independent. Another P-bodies component, a decapping enzyme, Dcp1, was not detected in the precipitates. The interaction of Dhh1 with ASH1 mRNA was confirmed by a reciprocal approach, in which untagged endogenous ASH1 mRNA was detected in the precipitates of Dhh1 antibodies (Fig. 3C). We next precipitated ASH1 mRNA from sucrose fractions 3 and 7 (Fig. 2A) and found that Dhh1 was associated with ASH1 mRNA specifically in the non-polysomal fraction (Fig. 3D). Dcp1 was not detected in the precipitates (not shown). Interestingly, further experiments revealed that although Dhh1 and She2 were both identified to associate with ASH1 mRNA, precipitation of Dhh1-bound ASH1 mRNA in the non-polysomal fraction detected little She2 (Fig. 3E). This indicates that Dhh1 could bind to localized and translationally inactive ASH1 mRNA independent of She2. Using fluorescent in situ hybridization (FISH) assays for ASH1 mRNA in a yeast strain expressing Dhh1-GFP or Dcp1-GFP fusions, we found that ASH1 mRNA was co-localized with Dhh1 in >30% of the budding cells (Fig. 3, F and G).
Figure 3.
Dhh1 binds to localized and translationally inactive ASH1 mRNA. A, ASH1 mRNA was precipitated from extracts of WT and puf6 cells by MBP-MCP. The precipitates were analyzed by RT-PCR. Arrow, amplified ASH1 cDNA. NC, negative control in which the MS26 motif was not fused into ASH1 mRNA. B, the potential interaction of ASH1 mRNA with Puf6, Dhh1, or Dcp1 was analyzed by immunoblotting with the precipitates from A. C, co-IP experiments were used to analyze the association of endogenous ASH1 mRNA with Dhh1 using Protein A beads conjugated with Dhh1 antibodies in wild-type cells. The precipitates were subjected to Western blotting and PT-PCR analyses. NC, negative control using Protein A beads unconjugated with the antibodies. All Western blotting experiments were carried out at least two times. D, ASH1 mRNA was precipitated with MBP-MCP from total cell extracts (input) and the sucrose fractions 3 and 7 (Fig. 2B). Dhh1 was associated with ASH1 mRNA in the input and non-polysomal fraction. E, co-IP experiments were performed to analyze the binding status of ASH1 mRNA with Dhh1 and She2 in cell extracts (Input) and non-polysomal fraction (#3) using Protein A beads conjugated with Dhh1 antibodies. The precipitates were subjected to Western blotting and PT-PCR analyses. F, FISH was performed to detect the localization of ASH1 mRNA in yeast strains expressing Dhh1-GFP or Dcp1-GFP. The arrowhead in the representative images indicates colocalization of ASH1 mRNA with Dhh1-GFP. Bar, 3 μm. G, the percentage of ASH1 mRNA colocalizing with Dhh1 or Dcp1 was shown from three independent experiments. On average, 40–50 cells were counted in each experiment. Error bars, S.D.
The roles of Dhh1 in bud-tip localization, translation, and stability of ASH1 mRNA
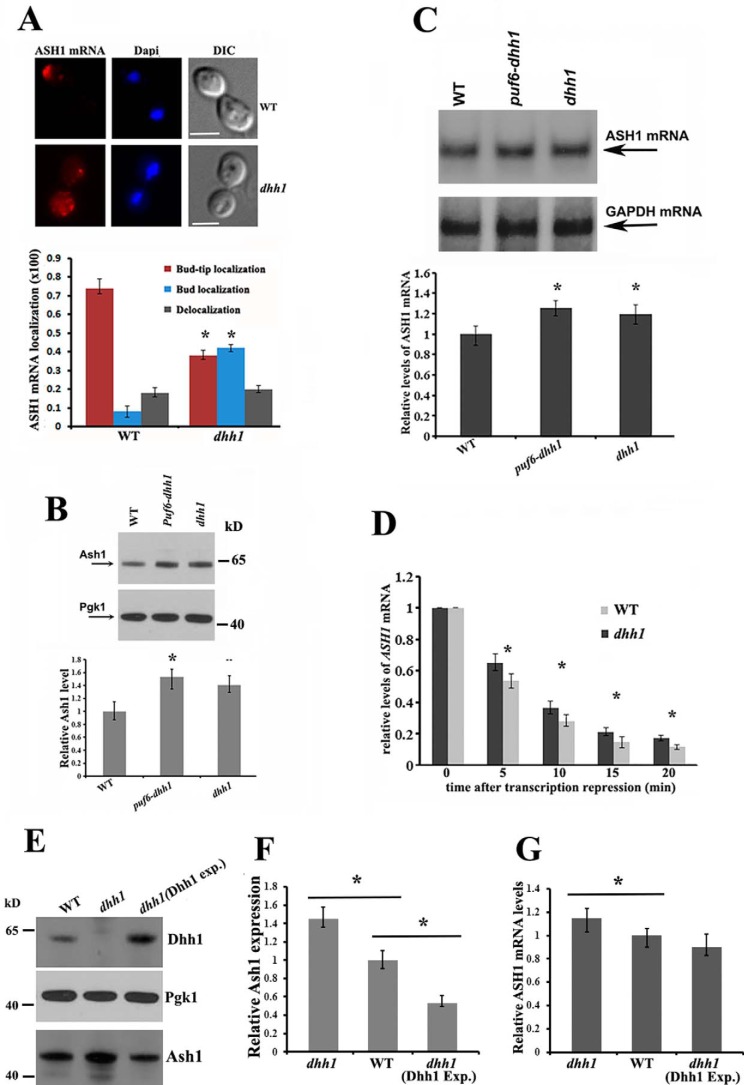
We have shown previously that binding of Puf6 to the PUF consensus sequences in the 3′-UTR of ASH1 mRNA regulated localization and translation of the mRNA (12). Here, we analyzed how the Dhh1-ASH1 mRNA interactions affect the localization and translation of ASH1 mRNA. Whereas >70% of the wild-type budding cells showed localized ASH1 mRNA at the bud tip (red), a diminution of bud-tip localization of ASH1 mRNA (∼40%) was observed in dhh1 cells, where about 42% of ASH1 mRNA was diffuse in the bud (Fig. 4A, bottom left). Compared with the phenotype observed in she2 or puf6 cells, where the asymmetric distribution of ASH1 mRNA was completely disrupted (12, 23), deletion of Dhh1 only resulted in delocalizing ASH1 mRNA in the bud.
Figure 4.
Effects of Dhh1 in ASH1 mRNA localization, translation, and stability. A, FISH to determine ASH1 mRNA localization in wild-type and dhh1 cells. Representative images show ASH1 mRNA localization in a wild-type budding cell and in the bud of a dhh1 cell (top panels). Bar, 3 μm. Bottom, cells showing different patterns of ASH1 mRNA localization were classified as follows: anchored (tightly localized ASH1 mRNA at the distal bud tip); delocalized in the bud (delocalized ASH1 mRNA confined to the bud); or delocalized in mother and bud (ASH1 mRNA in both mother cell and bud). The percentage of localized ASH1 mRNA in wild-type and dhh1 cells was shown from three independent experiments. An average of 50–80 cells was counted each time. *, p < 0.05. B, Western blotting of Ash1 and Pgk1 in WT, dhh1, and dhh1-puf6 cells. The arrows indicate the detected proteins. Relative levels of Ash1 expression were normalized to Pgk1. Mean values ± S.D. (error bars) from three independent experiments are shown. *, p < 0.05. C, Northern blotting analysis of ASH1 mRNA levels in WT, dhh1, and dhh1-puf6 cells. ASH1 mRNA levels were normalized to GAPDH mRNA. Mean values ± S.D. were from three independent experiments. *, p < 0.05. D, cells expressing ASH1 mRNA under the Gal promoter control were grown in synthetic medium with 2% raffinose to A600 ∼0.8, followed by the addition of galactose to 2% to induce ASH1 mRNA expression. 30 ml of culture was collected as t0. Glucose was added to a final concentration of 3% to stop ASH1 transcription. Cells were collected at 5, 10, 15, and 20 min. ASH1 mRNA levels were measured by qPCR and normalized to GAPDH mRNA for each sample. Bars, S.E. from three independent experiments. *, p < 0.05. E, a centromeric plasmid expressing Dhh1 was transformed into dhh1 cells. Expression of Dhh1, Pgk1, and Ash1 proteins in WT, dhh1, and dhh1(Dhh1 exp.) strains were detected by Western blotting. F, Ash1 expression in WT, dhh1, and dhh1(Dhh1 exp.) cells. The relative levels of Ash1 were normalized to Pgk1. Mean values ± S.D. from three independent experiments were shown. *, p < 0.05. C, RT-qPCR to analyze the levels of ASH1 mRNA in WT, dhh1, and dhh1(Dhh1 exp.) cells. ASH1 mRNA levels were normalized to GAPDH mRNA. Mean values ± S.D. were from three independent experiments. *, p < 0.05.
To examine the potential effect of Dhh1 in ASH1 mRNA translation and stability, we examined the intracellular levels of Ash1 and ASH1 mRNA in the wild-type, dhh1, and dhh1-puf6 strains. The levels of Ash1 were clearly increased in the dhh1 (∼1.38-fold) and dhh1-puf6 cells (∼1.5-fold), in contrast to the wild-type strain (Fig. 4B). The ASH1 mRNA levels were also relatively higher in the dhh1 (∼1.2-fold) and dhh1-puf6 strains (∼1.3-fold) when normalized to GAPDH mRNA (Fig. 4C). To analyze whether the increased levels of ASH1 mRNA resulted from DHH1 deletion, we transformed a plasmid expressing ASH1 mRNA under the Gal promoter control into the ash1 and ash1-dhh1 strains. Cells were grown in medium with 2% raffinose to ∼0.6 at A600 and were induced with 2% galactose for 10 min. The Gal promoter was repressed by the addition of glucose, and the ASH1 mRNA levels were quantitated using RT-qPCR. The steady-state levels of ASH1 mRNA were not obviously different between the two cell strains. In the dhh1 cells, levels of ASH1 mRNA were decreased by 35% at 5 min and by 62% at 10 min compared with reduction of 46% at 5 min and 70% at 10 min in the control cells (Fig. 4D). To further determine the role of Dhh1 in ASH1 mRNA translation, we transformed a centromeric plasmid expressing Dhh1 protein into dhh1 cells. We found that when Dhh1 was overexpressed, expression of Ash1 was considerably decreased, whereas the levels of ASH1 mRNA were still mildly reduced (Fig. 4, E–G). These results indicate that Dhh1 affects both the levels of ASH1 mRNA and protein. It seems that the impact of Dhh1 on protein levels can at least partially be explained by effects that are distinct from the effects on mRNA levels. One possibility, consistent with previous studies on Dhh1 as a translation repressor (19), is that it can act to repress translation of the ASH1 mRNA.
Translational initiation of ASH1 mRNA is accompanied with the dissociation of Dhh1 from the transcript
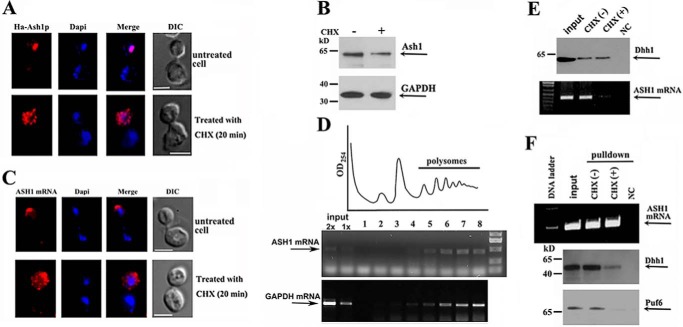
We hypothesized that binding of Dhh1 to the non-polysomal ASH1 mRNA could lead to regulation of its localized translation. To address this, we treated yeast cells with low levels of cycloheximide (CHX), which inhibited translational elongation but not initiation (24), to force the shift of ribosome equilibrium toward the polysomes (25). Using the HA-ASH1-Myc-MS26 reporter, we showed that in CHX-untreated growing cells, matured Ash1 (red) was synthesized and translocated into the bud nucleus, and ASH1 mRNA was tightly localized at the bud tip (top panels of Fig. 5, A and C, respectively). However, in CHX-treated cells, strong HA signal was mostly seen in the bud cytoplasm, whereas very little matured Ash1 was seen in the nucleus (Fig. 5A, bottom panels). Repression of Ash1 elongation was confirmed by Western blotting using Myc antibodies, which indicated that cells treated with CHX expressed less full-length Ash1 (Fig. 5B). Previous studies have reported that translation is required for proper localization of ASH1 mRNA. Either prevention or inhibition of ASH1 mRNA translation delocalized the mRNA in the bud (10, 26). As a result of CHX treatment, we also found that ASH1 mRNA (red) was diffusely localized in the cytoplasm of the bud (Fig. 5C, bottom), probably resulting from the mRNA that was trapped in polysomes with the nascent Ash1 peptide. Sucrose gradient assays showed that ASH1 mRNA, like GAPDH mRNA, was mostly distributed in the polysomal fractions in CHX-treated cells (Fig. 5D). In addition, Dhh1 and Puf6 GAPDH levels were modestly decreased in CHX-treated cells (supplemental Fig. S3, A and B). To determine the effect of the cellular association of Dhh1 with ASH1 mRNA after CHX treatment, we immunoprecipitated Dhh1 (Fig. 5E, top) and tested for the presence of ASH1 mRNA by RT-PCR. ASH1 mRNA was shown in the precipitates of the untreated growing cells but was greatly decreased in that of CHX-treated cells (Fig. 5E, bottom). We alternatively pulled down ASH1 mRNA by MBP-MCP (Fig. 5F, top) and analyzed the presence of Dhh1 and Puf6 in the precipitates. Western blotting (Fig. 5F, bottom panels) and quantitative analysis (supplemental Fig. S3C) indicated significantly decreased amounts of Puf6 and Dhh1 associated with ASH1 transcript in cells treated with CHX, suggesting that the shift of non-polysomal ASH1 mRNA into polysomes was accompanied by the dissociation of Dhh1 and Puf6.
Figure 5.
Activation of translational initiation results in disassociation of Dhh1 with ASH1 mRNA. A, fluorescent experiments were performed in the cells without or with CHX treatment. The top panels show a representative normal cell, in which Ash1 was fully translated and translocated into the cell nucleus, whereas in a CHX-treated cell, detected Ash1 was mostly cytoplasmic, due to the inhibition of protein elongation (bottom panels). Bar, 3 μm. B, Western blots showing the expression of Ash1 and GAPDH in normal and CHX-treated cells. C, FISH analyses showed ASH1 mRNA that was localized at the bud tip in a normal growing cell (top panels) but was diffuse in the bud in a CHX-treated cell (bottom panels). Bar, 3 μm. D, extracts of yeast cells were prepared after treatment with cycloheximide for 20 min and were fractionated in 10–50% linear sucrose gradients. An A254 plot corresponding to the polysomal fractions is shown. Total RNAs were isolated from the input and sucrose fractions. An equal amount of RNA from each fraction was subjected to RT-PCR to detect the distribution of ASH1 mRNA and GAPDH mRNA in the sucrose gradient (arrows). E, immunoprecipitation assays to analyze the association of Dhh1 with ASH1 mRNA in the cells treated with CHX. ASH1 mRNA was co-precipitated with Dhh1 in normal growing cells but little in CHX-treated cells (right). NC, a negative control in which no antibody was used. F, ASH1 mRNA was pulled down by MBP-MCP (top). Western blotting indicated that Dhh1 and Puf6 were co-precipitated in normal growing cells but not in CHX-treated cells (bottom panels). NC, a negative control using empty beads. The results are representative of three independent experiments.
Involvement of Dhh1 in translational regulation of other bud-localized mRNAs
Reports have shown that a number of bud-localized transcripts, including MID2 (YLR332w) and SRL1 (YOR247w) mRNAs, were associated with Puf6 post-transcriptionally (1). Interestingly, these two mRNAs were also shown to complex with Dhh1 in vivo (27). To determine whether blocking of translational elongation of mRNAs would also result in the disassociation of Puf6 and Dhh1 with the two transcripts, we treated a TAP-tagged Puf6 strain (12). After treatment with CHX, we immunoprecipitated Puf6 and analyzed the precipitates by Western blotting and RT-PCR. The results indicated that the two transcripts (MID2 and SLR1) as well as Dhh1 co-precipitated with Puf6 in normal growing cells. However, in CHX-treated cells, the binding capability of Puf6 to the transcripts and Dhh1 was dramatically reduced (supplemental Fig. S4A). This suggests that in addition to ASH1 mRNA, Dhh1 could also be involved in the translation of other bud-localized mRNAs.
Dhh1 does not directly interact with Puf6 but binds to the 5′-UTR of ASH1 mRNA
Because Puf6 binds to the 3′-UTR of the ASH1 mRNA (12) and Dhh1 co-precipitated with Puf6 and ASH1 mRNA (Fig. 3), we tested whether the two proteins could interact with each other. The results show that although Dhh1 antibodies could pull down Puf6 in WT strain, it co-precipitated much less Puf6 in ash1 cells (Fig. 6A). The small amount of coprecipitated Puf6 could indicate the interaction of Dhh1 and Puf6 with other mRNAs, such as MID2 and SRL1. Furthermore, when yeast extracts and the non-polysomal fraction 3 (Fig. 2) were treated with RNase A and RNase One, Dhh1 was not able to co-precipitate with Puf6 (Fig. 6B, bottom panels), indicating that association of the two proteins was mediated by ASH1 or perhaps the other mRNAs. We then prepared yeast strains expressing ASH1 WT mRNA (5′UTR-ASH1ORF-3′UTR) and ASH1 mutant (ADH2-5′UTR-ASH1ORF-3′UTR), in which the 5′-UTR of ASH1 mRNA was replaced by the 5′-UTR of ADH2 mRNA (Fig. 6C, top panels), and performed co-IP assays using Dhh1 antibodies. ASH1 mRNA was co-precipitated with Dhh1 in the extracts of WT cells but not in the mutant cells when the 5′-UTR of the ASH1 mRNA was absent (Fig. 6C, bottom panels). Precipitation of Puf6 was also greatly decreased in ASH1 mRNA mutant cells (Fig. 6C, middle panels). Because Puf6 binds to the 3′-UTR of ASH1 mRNA (12), the result suggested that Dhh1 could bind to the 5′-UTR of ASH1 mRNA. To further address this, we fused MS26 repeats into the 3′-ends of 5′UTR-ASH1ORF-3′UTR (WT) and ADH2-5′UTR-ASH1ORF-3′UTR (5′mut) constructs and used recombinant chimeric MBP-MCP to precipitate ASH1-MS26 mRNA in the cell extracts. Puf6 was co-precipitated with both WT and mutant ASH1 mRNA, whereas Dhh1 was not present in the precipitates of the mutant ASH1 mRNA (supplemental Fig. S4B). These results confirmed that Dhh1 bound to the transcript through the 5′-UTR and that the association of Dhh1 with Puf6 required both 5′-UTR and 3′-UTR of ASH1 mRNA.
Figure 6.
Dhh1 does not directly interact with Puf6; it could bind to the 5′-UTR of ASH1 mRNA. A, co-IP experiments using Protein A beads conjugated with Dhh1 antibodies were performed in the extracts of WT and ash1 cells. Western blotting indicated that co-precipitation of Dhh1 with Puf6 required ASH1 mRNA. B, co-IP experiments were performed in the yeast extracts and the non-polysomal fraction 3 treated with or without RNase A and RNase One. Dhh1 did not co-precipitate Puf6 when total mRNA in the extracts was degraded. C, top panels, schematic presentation of yeast strains expressing ASH1-WT (5′UTR-ASH1–3′UTR) and ASH1-mutant (ADH2–5′ UTR-ASH1-3′UTR) mRNAs. The 5′-UTR of ASH1 is a 150-nucleotide sequence upstream of the translational initiation site of the mRNA. The 3′-UTR of ASH1 is a 160-nucleotide sequence containing binding sites for Puf6. The 5′-UTR of ADH2 is a 150-nucleotide sequence upstream of the translational initiation site of the mRNA. Bottom panels, Dhh1 was immunoprecipitated in the extracts of ASH1-WT and ASH1-mutant cells. Puf6 and ASH1 mRNA were detected in the precipitates of WT extracts. A small amount of Puf6 and no ASH1 mRNA were detected in the precipitates of ASH1-mutant extracts. Arrows, positions of Dhh1, Puf6, and ASH1 mRNA. The results are representative of three independent experiments. D, top, schematic presentation of the ASH1 gene. The 3′-UTR contains the E3 localization element for Puf6 and She2 binding. Arrows, primers used for qPCR analysis. Amplicon lengths for 5′-UTR and 3′-UTR are 150 and 160 bp, respectively. Bottom, TAP-tagged Puf6, She2, and Dhh1 were used for CHIP assays. After immunoprecipitation, amplicons corresponding to 5′-UTR and 3′-UTR were amplified and analyzed. E, effect of DHH1 deletion on repression of the HO promoter. 10-Fold serial dilutions of exponentially growing wild-type, dhh1, or puf6 cells were spotted on YPD or SC medium containing 0.03% canavanine and incubated for 2 and 5 days at 30 °C, respectively. Error bars, S.E.
Dhh1 is not recruited on ASH1 mRNA co-transcriptionally
Puf6 is co-transcriptionally recruited onto nascent ASH1 mRNA (1). To test whether Dhh1 could also be recruited onto ASH1 mRNA during transcription, ChIP assays were performed using strains expressing endogenous C-terminal TAP-tagged Dhh1, She2, or Puf6 (supplemental Fig. S5A). In these assays, chromatin that was associated with She2, Puf6, or Dhh1 was immunopurified by using IgG-Sepharose, and the enriched specific regions of the ASH1 gene were measured by qPCR. Two specific amplicons from the ASH1 gene were analyzed; one was 5′-UTR, and the other was 3′-UTR, which contained the She2- and Puf6-binding motifs and was used as an internal control (Fig. 6D). As shown in the figure, control amplicons of the 3′-UTR were specifically enriched after Puf6 and She2 CHIPs, whereas no enrichment of the 3′-UTR was identified in Dhh1 CHIP. There was also no enrichment for the 5′-UTR amplicon in all ChIP assays. In contrast, ASH1 mRNA was successfully co-precipitated with Dhh1-TAP from the cell extracts (supplemental Fig. S5B). These results suggest that Dhh1 was not co-transcriptionally recruited on the 5′-UTR of the ASH1 gene.
Loss of Dhh1 function affects the HO promoter activity
To examine the physiological relevance of the DHH1 gene to the cell's ability to control mating-type switch, we tested the influence of Dhh1 on the regulation of the HO promoter. DHH1 was deleted in a strain in which the endogenous CAN1 gene was under the control of the HO promoter (12, 28). The cells were sensitive to canavanine when CAN1 was expressed, whereas inhibition of the CAN1 gene led to tolerance to the drug. We observed that deletion of DHH1 decreased the sensitivity to canavanine (Fig. 6E), although to a lesser extent than the puf6 cells that have been known to more intensely interfere with the localization of ASH1 mRNA. This indicates a role for Dhh1 in regulation of HO expression and, therefore, mating-type switch.
Binding of Dhh1 to the 5′-UTR of ASH1 mRNA represses its translation in vitro
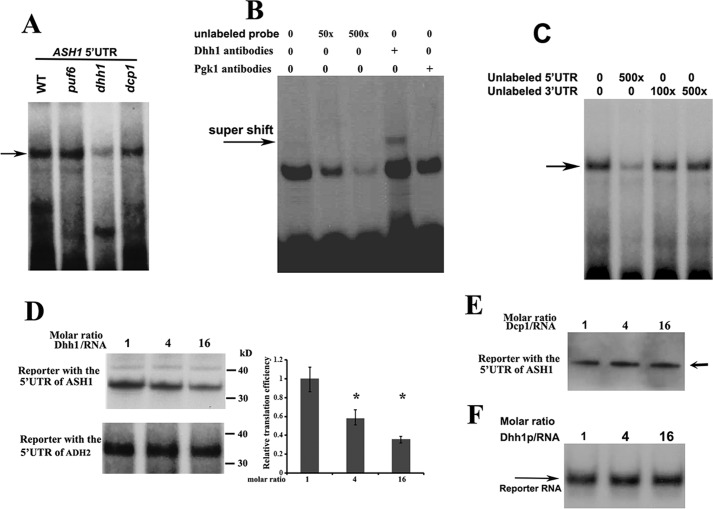
We used in vitro RNA mobility-shift experiments to further identify the binding ability of Dhh1 to the 32P-labeled 5′-UTR of ASH1 mRNA. An RNA-protein complex was formed when the RNA probe was incubated with the extracts of WT, puf6, and dcp1 cells. However, the complex formation was significantly attenuated upon incubation with the extracts of dhh1 cells (Fig. 7A; complex is indicated by the arrow), indicating that the formation of the complex relied on Dhh1. We next expressed His-tagged Dhh1 in Escherichia coli and used the purified Dhh1 (supplemental Fig. S2B) to perform binding assays. Incubation of 32P-labeled 5′-UTR of ASH1 mRNA with recombinant Dhh1 formed a distinct RNA-protein complex, which was effectively competed by excess amounts of the unlabeled probe (Fig. 7B). The complex formation was specific because a supershift band was observed when Dhh1 antibody was added into the reaction (arrow). Dhh1 did not interact with the 3′-UTR of ASH1 mRNA because the 3′-UTR of ASH1 mRNA did not show any competition for the complex formation (Fig. 7C). Thus, the in vitro experiments supported the in vivo binding of Dhh1 to the 5′-UTR of untranslated ASH1 mRNA.
Figure 7.
Dhh1 binds to the 5′-UTR of ASH1 mRNA and represses its translation in vitro. A, aliquots of 32P-labeled 5′-UTR of ASH1 mRNA were incubated with extracts prepared from WT, puf6, dhh1, and dcp1 strains. RNA-protein complexes (arrow) were formed in the extracts when Dhh1 was expressed. B, 32P-labeled 5′-UTR of ASH1 mRNA was incubated with recombinant Dhh1 and an increased amount of unlabeled RNAs or Dhh1 antibodies. Arrow, a supershift of the 5′-UTR-Dhh1 antibody. C, binding of Dhh1 to the 5′-UTR of ASH1 mRNA was not competed by the unlabeled 3′-UTR of ASH1 mRNA. D, left, autoradiography of in vitro translated [35S]methionine-labeled luciferase protein using the RNA templates containing the 5′-UTR of ASH1 mRNA or the 5′-UTR of ADH2 mRNA in the presence of a 1-, 4-, and 16-fold excess molar ratio of recombinant Dhh1. Right, relative translational efficiency of RNA template containing the 5′-UTRs of ASH1 mRNA was shown after normalizing to the RNA templates containing the 5′-UTR of ADH2 mRNA. Mean values ± S.D. (error bars) were from three independent experiments. *, p < 0.05. E, in vitro translation of [35S]methionine-labeled luciferase protein using the RNA templates containing the 5′-UTR of ASH1 mRNA and 1-, 4-, and 16-fold excess molar ratio of recombinant Dcp1. F, a urea gel showing that the 32P-labeled reporter RNA used for in vitro translation did not degrade over the course of the experiments in D. The experiments were carried out three times.
To evaluate whether binding of Dhh1 to the 5′-UTR of ASH1 mRNA could affect translation, we performed in vitro cell-free translation assays in a rabbit reticulocyte lysate system using two R-luciferase reporters; one contained the 5′-UTR of ASH1 mRNA (160 bp), and the other contained the 5′-UTR of ADH2 mRNA (150 bp). A progressive reduction of translation was observed when the 5′-UTR of ASH1 reporter was incubated with increasing amounts of recombinant Dhh1. The efficiency of in vitro translation was reduced by > 50% when the molar ratio of the reporter RNA/Dhh1 reached 1:16, whereas no obvious reduction was observed when Dhh1 was incubated with the 5′-UTR of ADH2 reporter (Fig. 7D). In addition, the translational efficiency was not changed when the 5′-UTR of ASH1 reporter was incubated with increased amounts of recombinant Dcp1 (Fig. 7E). Recombinant Dhh1-repressed in vitro translation of the ASH1 5′-UTR reporter was also observed in a yeast extract prepared from dhh1 cells (supplemental Fig. S6). The translational inhibition did not result from selective RNA degradation, because the amount of the reporter RNA in the reactions did not change with increasing Dhh1 levels (Fig. 7F). Thus, binding of Dhh1 to the 5′-UTR of ASH1 mRNA conferred repression of translation.
Discussion
We have reported previously that binding of Puf6 to the 3′-UTR of ASH1 mRNA repressed its translation during transport, and this repression could be released by CK2 phosphorylation in the N-terminal region of Puf6 when the mRNA was localized to the bud tip (12, 13). Our new observations showed that although translation occurred after localization, a fraction of ASH1 mRNA was still associated with Puf6 and was translationally inactive. The 5′-UTR of the untranslated ASH1 mRNA was simultaneously bound to Dhh1, which served to maintain the non-polysomal status of the mRNA and cause the mRNA to be tightly localized at the bud cortex.
Post-transcriptional control of mRNA localization and translation was achieved through the concerted action of RNA-binding factors and the translation machinery (10, 29–31). The fact that Dhh1 coordinates with Puf6 to regulate the translation of ASH1 mRNA provides additional information for the importance of localized translation in relation to cell phenotype. The fact that activation of the non-polysomal ASH1 mRNA was accompanied with Dhh1 dissociation strongly suggested that Dhh1 was indeed associated with translationally repressed ASH1 mRNA, and this interaction requires the 5′-UTR of the transcript. The suppressive role of Dhh1 for Ash1 translation was also determined by the in vivo overexpression analysis and the in vitro translation assays. Binding of Dhh1 to the 5′-UTR of ASH1 mRNA was most likely due to the involvement of the protein in the regulation of translational initiation. This could be partly supported by the finding that Ash1 expression was decreased when Dhh1 was overexpressed and that increasing translational initiation by CHX treatment resulted in the dissociation of the protein from ASH1 mRNA. Because phosphorylation of Puf6 at the bud tip by CK2 decreased its binding ability to ASH1 mRNA and eliminated the translational repression (13), it is possible that CK2 could also act on Dhh1 to release its binding.
ASH1 mRNA contains four localization elements for She2 binding, which make the localization of the mRNA more precise and efficient (7). Localized translation of ASH1 mRNA could also be finely controlled. This explains why Dhh1 is involved in the regulation of localized translation of ASH1 mRNA. However, compared with puf6 cells, in which ASH1 mRNA was widely delocalized in both the mother and daughter cells (12), the mRNA in the dhh1 cells was mostly diffusely localized in the buds. As a result, we observed a milder effect of Dhh1 on the regulation of the HO promoter when compared with the mutation in puf6. In addition, unlike Puf6 that was recruited into ASH1 mRNA during transcription (1), Dhh1 was shown to associate with the transcript within the cytoplasm, suggesting that Dhh1 might be more involved in the regulation of ASH1 mRNA translation after localization. We hypothesized that after bud-tip localization, some ASH1 mRNA could be programmed for immediate translation, whereas other ASH1 mRNA could be stored. In this case, binding of Dhh1 to translationally inactive ASH1 mRNA could primarily result in temporal repression, storage, and then decay if no more protein synthesis is required.
In S. cerevisiae, Dhh1 is a DEAD-box RNA helicase and is also a major P-body component involved in translational repression and mRNA decapping (20). Recent studies indicated that Dhh1 represses mRNA translation in a way that does not rely on the translation initiation factors and that Dhh1 connects translation to mRNA decay by monitoring codon optimality (32, 33). Because Dhh1 binds to the 5′-UTR of ASH1 mRNA, we feel that Dhh1 could be more implicated in the control of translational initiation of the mRNA. Deletion of the DHH1 gene not only resulted in a defect in mating through the down-regulation of Ste12 expression (34) but also affected the mating-type switch due to the dysregulation of HO expression. In addition, the roles of the Dhh1 family in translational repression and enhancement of mRNA degradation have also been well studied in higher eukaryotes (19, 21). Dhh1 homologs in Drosophila (Me31B), Caenorhabditis elegans (CGH-1), and Xenopus (Xp54) have been implicated in translational repression and storage of maternal mRNAs (35–37).
A recent global analysis of yeast mRNPs failed to identify consensus sequences for Dhh1 binding (27); however, an in vitro analysis has indicated that recombinant Dhh1 showed a strong preference for binding to adenine-rich nucleotides (38). We found that Dhh1 interacted with the 5′-UTR of ASH1 mRNA that is not adenine-rich. Although the exact sequence element for Dhh1 binding has yet to be identified, our results indicated that interaction of Dhh1 with the 5′-UTR of ASH1 mRNA is required for regulation of Ash1 translation. Binding of the Dhh1 to ASH1 mRNA did not result from using the system of MS2-tagged RNA and MCP-GFP (39), because the binding was also confirmed when the mRNA was not MS2-tagged and when the MCP-GFP was not used. Our future work will be to identify the signal that determines the fate of untranslated ASH1 mRNA: to be translated or to be stored for degradation.
In conclusion, we identified a potential regulatory mechanism for localized ASH1 mRNA translation in which not all of the mRNA was translated after localization. Untranslated mRNA was repressed and temporally stored through the interaction with Dhh1 and Puf6. Thus, translation of localized ASH1 mRNA could be precisely regulated by Dhh1, which bound to the 5′-UTR, and Puf6, which bound to the 3′-UTR of the mRNA. Interference of the coordination due to the lack of Dhh1 could affect ASH1 mRNA localization and translation.
Experimental procedures
Growth medium, yeast strains, and plasmids
Strains used in this study are listed in Table 1. Yeast cells were grown in either synthetic medium lacking the nutrients indicated or rich medium. Transformation was performed according to the protocol of Gietz et al. (40). Unless otherwise stated, standard genetic techniques were used. Tagging, insertion, and deletion were performed by PCR-based homologous recombination using cassettes described previously (12, 41). The deletion was verified by colony-PCR analysis to confirm that the insertion and replacement were in the expected locus.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source/Reference |
|---|---|---|
| K4452 | Mata, leu2-3, 112, ura3, his3, ade2-1, ho can1-100 | Ref. 28 |
| K4452-ash1 | K4452, ash1::KAN | Ref. 28 |
| K4452-ash1, Ash1-Myc9 | K4452-ash1, ASH1-myc9::LEU | Ref. 3 |
| K4452-ash1, puf6 | K4452-ash1, puf6::TRP | Ref. 12 |
| K699 | Mata, ura3, leu2-3, 112, his3-11, trp1-1, ade2-1, ho can1-100 | Ref. 28 |
| K699-puf6 | K699, puf6::TRP | Ref. 12 |
| K699-puf6, Puf6-His6 | K699-puf6, Puf6-HIS6::LEU | This study |
| K4452-ash1, HA-ASH1-MS26 | K4452-ash1, HA-ASH-MS26::URA | This study |
| K4452-ash1, HA-ASH1-Myc-MS26, MCP-GFP | K4452-ash1, HA-ASH1-myc-MS26::LEU, MCP-GFP::TRP | This study |
| K4452-ash1, HA-ASH1-MS26, Puf6-His6 | K4452-ash1, HA-ASH1-myc-MS26::LEU, Puf6-His6::TRP | This study |
| K4452-ash1, 5′UTR-ASH1–3′UTR | K4452-ash1, 5′UTR-ASH1–3′UTR::LEU | This study |
| K4452-ash1, ADH2 5′-UTR ASH1–3′-UTR | K4452-ash1, ADH2 5′UTR ASH1–3′UTR::LEU | This study |
| K4452-dhh1 | K4452, dhh1::URA | This study |
| K699-dhh1 | K699, dhh1::URA | This study |
| K4452, Dhh1-GFP | K4452, DHH1-GFP::TRP | This study |
| K4452-puf6, dhh1 | K4452-puf6, dhh1::URA | This study |
| K699-puf6, dhh1 | K699-puf6, dhh1::URA | This study |
| K4452, Dhh1-His6 | K4452, Dhh1-HIS6::LEU | This study |
| K699, Dhh1-TAP | K699, DHH1-TAP::TRP | This study |
| K699, Puf6-TAP | K699 PUF6-tap:TRP | Ref. 12 |
| K699, She2-TAP | K699 SHE2-tap:TRP | Ref. 12 |
Plasmid construction
Plasmids used in this study were constructed using standard techniques. The plasmids YCP111-Gal-HA-ASH1-Myc-MS2 and YCP111-HA-ASH1-MS2 were derived from plasmid YCP111-Gal-MS2-LacZ-E3. These plasmids contain full-length 5′-UTR as well as the coding region of ASH1 mRNA and six MS2 sites after the 3′-UTR of ASH1 mRNA for MCP binding. Plasmid YCP33-GFP-MCP was constructed previously (18). Plasmids YCP111-GFP-Dhh1 and YCP111-GFP-MCP were derived from plasmid YCP33-GFP-MCP. Plasmids YIP211-HAASH1-MS2, YIP211-5′UTR-ASH1-3′UTR, and YIP211-ADH2-5′UTR-ASH1-3′UTR were derived from the plasmid pXR193 (42). Plasmid pSP64-ASH-5′UTR was derived from plasmid pSP64-ASH1-3′UTR (12).
In situ hybridization and immunofluorescence
Yeast cells were grown in the appropriate culture medium to early or mid-log phase and were processed for FISH as described previously (12). To perform FISH, yeast spheroplasts were hybridized with a pool of Cy3-conjugated ASH1 DNA oligonucleotide probes or Cy3-conjugated MS2 probes (3, 43). Immunofluorescence was performed using a protocol described previously (42). For detecting HA-tagged or Myc-tagged Ash1 proteins, the mouse Cy3-conjugated anti-HA or Cy5-conjugated anti-Myc antibodies (Roche Applied Science) were used in a 1:200 dilution in 1× PBS and 0.1% BSA.
Preparation of yeast extracts
Yeast cells were cultured to A495 1.0 and were rapidly harvested by centrifugation at 3,200 × g for 5 min at 4 °C. Where indicated, cycloheximide (50 μg/ml) was added to the cultures for 20 min. All subsequent steps were carried out in a cold room. After washing with 10 ml of cold buffer (50 mm Tris-Cl (pH 6.8), 100 mm NaCl, 30 mm MgCl2, or 50 μg/ml), the cells were pelleted again and resuspended in 800 μl of the buffer containing 20 units/ml RNasin and 2× protease inhibitors (2 mm phenylmethylsulfonyl fluoride (PMSF), 100 μg/ml aprotinin, 100 μg/ml pepstatin, 100 μg/ml leupeptin) in 2-ml tubes containing 800 μl of glass beads. Cells were lysed in a FastPrep (MP Biomedicals) 4 times for 60 s each time. Cell debris and beads were removed from the extract by centrifugation for 15 min at 8,000 × g. To prepare the extracts for in vitro translation assays, the obtained supernatant was subjected to centrifugation (4 °C, 15,000 × g, 30 min) again.
Sucrose-gradient fractionation
500 μl of yeast extracts were loaded onto 10-ml, 10–50% linear sucrose gradients and fractionated at 35,000 rpm for 2 h in an SW41 rotor (Beckman). Fractions (1.25 ml each) were collected from top to bottom, and the A254 profile was monitored.
Northern and Western blotting
For Northern blotting, total RNA was extracted, and equal amounts of total RNA were separated by agarose gel electrophoresis. RNA was transferred to a Hybond membrane and incubated with dCTP-32P-labeled cDNA probes for the ASH1 gene and the GAPDH gene. Hybridization and autoradiography were performed as described previously (44). For immunoblotting, equal amounts of cell extracts were resolved into a 4–12% gradient SDS-PAGE (Invitrogen), followed by electroblotting onto a nitrocellulose membrane. Western blotting was performed using one of the following primary antibodies: anti-His, anti-HA, anti-Myc, anti-Dhh1, anti-Dcp1, anti-She2, anti-GAPDH, or anti-Pgk1. After incubation with HRP-conjugated secondary antibodies, proteins were detected with an enhanced chemiluminescence detection kit (Millipore). The intensity of the protein bands was quantified using ImageJ software (NIH). Relative protein levels were determined after normalization to an internal protein control.
Expression and purification of recombinant MBP-MCP and Dhh1
Recombinant MBP-MCP protein was expressed and purified as described previously (45). A His6-tagged cDNA encoding Dhh1 or Dcp1 protein was PCR-amplified from yeast genomic DNA and cloned into a donor plasmid pET23a plasmid (Novagen) into the NdeI and BamHI sites. The plasmid, after DNA sequence analysis, was transformed into E. coli (DE21) cells and expressed. The method for purifying recombinant His6-tagged Dhh1 or Dcp1 protein was as mentioned previously (45).
In vitro pulldown experiments, RT-PCR, and Western blotting
Yeast extracts from exponentially growing cell cultures were prepared as described above. For precipitation of Dhh1-associated proteins, a total of 100 μl of extracts were incubated with 2 μg of anti-Dhh1 IgG (Santa Cruz Biotechnology, Inc.) or control IgG and 20 μl of Protein A beads (Sigma) for 4–6 h at 4 °C with gentle shaking. Where indicated, RNase A (10 μg/100 μl) and RNase One (100 units; Promega) were added. After washing extensively with DEPC-treated PBS, the beads were suspended in 40 μl of water, and the proteins were eluted by boiling for 5 min. Alternatively, His-tagged recombinant Dhh1 was attached to nickel-nitrilotriacetic acid beads for pulldown experiments. The methods for precipitation of the ASH1 mRNA complex using MBP-MCP recombinant protein were performed as mentioned previously (45). For reverse transcription, TRIzol-extracted RNA was used as template. All PCRs were performed for 25–30 cycles using the primers specific for ASH1 mRNA and an internal control. The intensity of the DNA bands was quantified using ImageJ software (National Institutes of Health). Relative levels of ASH1 mRNA were normalized to the levels of the control. Primers used for RT-PCR and PCR experiments are shown in Table 2.
Table 2.
List of primers used for qRT-PCR and qPCR
| Gene | Forward primers | Reverse primers |
|---|---|---|
| ASH1 5′-UTR | GGCTCCTGCTCAAAAAAAGAGG | CCTCTAGAGTCGACTTTTTTTGC |
| ASH1 3′-UTR | TACATGGATAACTGAATCTCTTTC | GCGGCGTGTCGAATGAAAATG |
| ASH1 mRNA | GATCATCCTCACCTGTGCGTC | CTCTACTGTCTCACCGTTCAAG |
| GAPDH | GAGTCAACGGATTTGGTCGT | TGGGATTTCCATTGATGACA |
| DHH1 | TCCTAATAGAAAGATAGACGCAG | CCGTTTCGTCATTTTCAGCCAC |
| DCP1 | GTACATCTCTGCGCATTTTTCTC | GTAGCACGCCCTCTTCTACTAC |
| ADH2 5′-UTR | TCGCTACTGGCACTCTATTTATAG | CTATCAACTATTAACTATATCGT |
ChIP
Puf6-TAP and She2-TAP strains were established previously, and the construction of the C-terminal insertion cassette for tap-tagging of the DHH1 gene was described previously (12). Expression of Dhh1-Tap, Puf6-Tap, and She2-Tap was from their corresponding endogenous loci. For each ChIP, three independent 50-ml early log phase cultures were used. To increase the number of ASH1 transcripts, cells were synchronized for 2 h using Nocodazole at the final concentration of 15 μg/ml. IgG-Sepharose 6 Fast Flow (GE Healthcare) was used for the experiments, and the procedure for CHIP assays was as mentioned previously (1). Quantification of the immunoprecipitated DNA was performed by RT-qPCR (S1000TM thermal cycler Bio-Rad) using the SYBR Green PCR core reagent system. Each 20-μl PCR contained 10 μl of qPCR mix, 2 μl of DNA, and a 300 nm concentration of each of the primers. PCR was performed in the following conditions: activation of the reaction for 30 s at 95 ºC, followed by 35 cycles of 10 s at 95 °C and then 30 s at 57 °C for the annealing and extension steps. Cycle thresholds (Ct) for each triplicate of samples were averaged, and ChIP enrichment was calculated by dividing the amount of ChIP DNA over control DNA using the formula, 2−ΔΔCt.
HO promoter activity assay
The effect of DHH1 on HO expression was determined as described by Jansen et al. (28). Briefly, 10-fold serial dilutions of exponentially growing wild-type (K4535), puf6, or dhh1 cells were spotted on YPD or SD medium contain 0.03% canavanine and incubated for 2 and 5 days at 30 °C, respectively.
Gel mobility-shift assay
A 150-bp cDNA fragment encoding 5′-UTR of ASH1 mRNA was PCR-amplified and subcloned into pSP64 plasmid (Promega). The plasmid encoding 160 bp of the 3′-UTR of ASH1 mRNA was constructed previously (12). 32P-Labeled RNA probes were in vitro generated by SP6 RNA polymerase from pSP64-ASH1 5′-UTR and pSP64-ASH1 3′-UTR constructs. Transcribed RNA probes were purified after resolving in a 6% denaturing gel. RNA-protein gel-shift assays were performed at room temperature as described previously (12). The RNA-protein complexes formed were separated by electrophoresis in a 4% native gel and visualized by autoradiography. To establish the specificity of RNA-protein interactions, competition assays were performed by preincubating the cell extracts or the recombinant Dhh1 with unlabeled specific or nonspecific RNA competitors.
In vitro translation assays
In vitro translation assays were performed as described previously (12). Briefly, capped mRNA reporters with the 5′ UTR of ASH1 mRNA or the 5′ UTR of ADH2 mRNA were transcribed from the plasmid, pC3.1-5′UTR-Luciferase, using a T7Message Machine kit (Ambion). In vitro translation was performed in 25 μl of rabbit reticulocyte lysate (Promega) containing 10 nm of the mRNA templates and [35S]methionine as a tracer to monitor the translated proteins in the presence of increasing amounts of recombinant Dhh1 and Dcp1. Where indicated, a yeast extract prepared from dhh1 cells was used for the in vitro translation assays. Translated proteins were detected by autoradiography after separation by a 12% SDS-polyacrylamide gel. Signal intensities were determined by a PhosphorImager screen (Molecular Dynamics) and quantified using ImageQuant software.
Analysis of ASH1 mRNA and Ash1 localization
The method of quantitative measurement on the localization of ASH1 mRNA has been described previously (42). By microscopy, yeast cells in late anaphase were scored for localized or delocalized ASH1 mRNA in each experiment. ASH1 mRNA was considered as bud-tip-localized when it was predominantly in the bud tip (crescent localization). ASH1 mRNA was considered as diffuse in the bud when it was not tightly localization at the bud tip. For each experiment, ∼100 budding cells were scored.
Author contributions
Q. Z. carried out the molecular and biochemical studies. X. M. carried out the gene expression analysis and genetic studies. S. C., D. L., J. L., and L. Z. contributed to the biochemical studies and immunofluorescent experiments. R. H. S. participated in supervision and manuscript editing. W. G. designed the experiments, carried out some molecular and biochemical studies, coordinated studies, and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank members of the Gu and Singer laboratories for technical support.
This work was supported by National Natural Science Foundation of China Grant 31171209 (to W. G.) and National Institutes of Health Grant GM57071 (to R. H. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S6.
- mRNP
- messenger ribonucleoprotein
- MBP
- maltose-binding protein
- MCP
- MS2-binding coat protein
- qPCR
- quantitative PCR
- CHX
- cycloheximide
- IP
- immunoprecipitation.
References
- 1. Shahbabian K., Jeronimo C., Forget A., Robert F., and Chartrand P. (2014) Co-transcriptional recruitment of Puf6 by She2 couples translational repression to mRNA localization. Nucleic Acids Res. 42, 8692–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shepard K. A., Gerber A. P., Jambhekar A., Takizawa P. A., Brown P. O., Herschlag D., DeRisi J. L., and Vale R. D. (2003) Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 100, 11429–11434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., and Jansen R. P. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383–387 [DOI] [PubMed] [Google Scholar]
- 4. Bobola N., Jansen R. P., Shin T. H., and Nasmyth K. (1996) Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84, 699–709 [DOI] [PubMed] [Google Scholar]
- 5. Sil A., and Herskowitz I. (1996) Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84, 711–722 [DOI] [PubMed] [Google Scholar]
- 6. Shen Z., St-Denis A., and Chartrand P. (2010) Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 24, 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chartrand P., Meng X. H., Singer R. H., and Long R. M. (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9, 333–336 [DOI] [PubMed] [Google Scholar]
- 8. Müller M., Heym R. G., Mayer A., Kramer K., Schmid M., Cramer P., Urlaub H., Jansen R. P., and Niessing D. (2011) A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 9, e1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takizawa P. A., and Vale R. D. (2000) The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. U.S.A. 97, 5273–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Irie K., Tadauchi T., Takizawa P. A., Vale R. D., Matsumoto K., and Herskowitz I. (2002) The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paquin N., Ménade M., Poirier G., Donato D., Drouet E., and Chartrand P. (2007) Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26, 795–809 [DOI] [PubMed] [Google Scholar]
- 12. Gu W., Deng Y., Zenklusen D., and Singer R. H. (2004) A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18, 1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng Y., Singer R. H., and Gu W. (2008) Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 22, 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pérez-Ortin J. E., Alepuz P. M., and Moreno J. (2007) Genomics and gene transcription kinetics in yeast. Trends Genet. 23, 250–257 [DOI] [PubMed] [Google Scholar]
- 15. Zenklusen D., Larson D. R., and Singer R. H. (2008) Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larson D. R., Zenklusen D., Wu B., Chao J. A., and Singer R. H. (2011) Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hocine S., Raymond P., Zenklusen D., Chao J. A., and Singer R. H. (2013) Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods 10, 119–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., and Long R. M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 [DOI] [PubMed] [Google Scholar]
- 19. Presnyak V., and Coller J. (2013) The DHH1/RCKp54 family of helicases: an ancient family of proteins that promote translational silencing. Biochim. Biophys. Acta 1829, 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coller J., and Parker R. (2005) General translational repression by activators of mRNA decapping. Cell 122, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coller J. M., Tucker M., Sheth U., Valencia-Sanchez M. A., and Parker R. (2001) The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7, 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., and Parker R. (2005) Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chartrand P., Singer R. H., and Long R. M. (2001) RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 17, 297–310 [DOI] [PubMed] [Google Scholar]
- 24. Schneider-Poetsch T., Ju J., Eyler D. E., Dang Y., Bhat S., Merrick W. C., Green R., Shen B., and Liu J. O. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zobel-Thropp P., Yang M. C., Machado L., and Clarke S. (2000) A novel post-translational modification of yeast elongation factor 1A: methylesterification at the C terminus. J. Biol. Chem. 275, 37150–37158 [DOI] [PubMed] [Google Scholar]
- 26. Gonzalez I., Buonomo S. B., Nasmyth K., and von Ahsen U. (1999) ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 9, 337–340 [DOI] [PubMed] [Google Scholar]
- 27. Mitchell S. F., Jain S., She M., and Parker R. (2013) Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 20, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jansen R. P., Dowzer C., Michaelis C., Galova M., and Nasmyth K. (1996) Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell 84, 687–697 [DOI] [PubMed] [Google Scholar]
- 29. Shen Z., Paquin N., Forget A., and Chartrand P. (2009) Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol. Biol. Cell 20, 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hinnebusch A. G. (2009) Active destruction of defective ribosomes by a ubiquitin ligase involved in DNA repair. Genes Dev. 23, 891–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sonenberg N., and Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Radhakrishnan A., Chen Y. H., Martin S., Alhusaini N., Green R., and Coller J. (2016) The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122–132.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sweet T., Kovalak C., and Coller J. (2012) The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 10, e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ka M., Park Y. U., and Kim J. (2008) The DEAD-box RNA helicase, Dhh1, functions in mating by regulating Ste12 translation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 367, 680–686 [DOI] [PubMed] [Google Scholar]
- 35. Minshall N., Thom G., and Standart N. (2001) A conserved role of a DEAD box helicase in mRNA masking. RNA 7, 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakamura A., Amikura R., Hanyu K., and Kobayashi S. (2001) Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233–3242 [DOI] [PubMed] [Google Scholar]
- 37. Minshall N., Kress M., Weil D., and Standart N. (2009) Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell 20, 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dutta A., Zheng S., Jain D., Cameron C. E., and Reese J. C. (2011) Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J. Biol. Chem. 286, 27454–27470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia J. F., and Parker R. (2015) MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA 21, 1393–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gietz R. D., Schiestl R. H., Willems A. R., and Woods R. A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 [DOI] [PubMed] [Google Scholar]
- 41. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., and Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 42. Chartrand P., Meng X. H., Huttelmaier S., Donato D., and Singer R. H. (2002) Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell 10, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 43. Lionnet T., Czaplinski K., Darzacq X., Shav-Tal Y., Wells A. L., Chao J. A., Park H. Y., de Turris V., Lopez-Jones M., and Singer R. H. (2011) A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gu W., Wells A. L., Pan F., and Singer R. H. (2008) Feedback regulation between zipcode binding protein 1 and β-catenin mRNAs in breast cancer cells. Mol. Cell Biol. 28, 4963–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Song T., Zheng Y., Wang Y., Katz Z., Liu X., Chen S., Singer R. H., and Gu W. (2015) Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J. Cell Sci. 128, 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.