Abstract
The proteasome-associated deubiquitinating enzyme Usp14/Ubp6 inhibits protein degradation by catalyzing substrate deubiquitination and by poorly understood allosteric actions. However, upon binding a ubiquitin chain, Usp14 enhances proteasomal degradation by stimulating ATP and peptide degradation. These studies were undertaken to clarify these seemingly opposite regulatory roles of Usp14 and their importance. To learn how the presence of Usp14 on 26S proteasomes influences its different activities, we compared enzymatic and regulatory properties of 26S proteasomes purified from wild-type mouse embryonic fibroblast cells and those lacking Usp14. The proteasomes lacking Usp14 had higher basal peptidase activity than WT 26S, and this activity was stimulated to a greater extent by adenosine 5′-O-(thiotriphosphate) (ATPγS) than with WT particles. These differences were clear even though Usp14 is present on only a minor fraction (30–40%) of the 26S in WT mouse embryonic fibroblast cells. Addition of purified Usp14 to the WT and Usp14-defficient proteasomes reduced both their basal peptidase activity and the stimulation by ATPγS. Usp14 inhibits these processes allosterically because a catalytically inactive Usp14 mutant also inhibited them. Proteasomes lacking Usp14 also exhibited greater deubiquitinating activity by Rpn11 and greater basal ATPase activity than WT particles. ATP hydrolysis by WT proteasomes is activated if they bind a ubiquitinated protein, which is loosely folded. Surprisingly, proteasomes lacking Usp14 could be activated by such proteins even without a ubiquitin chain present. Furthermore, proteasomes lacking Usp14 are much more active in degrading non-ubiquitinated proteins (e.g. Sic1) than WT particles. Thus, without a ubiquitinated substrate present, Usp14 suppresses multiple proteasomal activities, especially basal ATP consumption and degradation of non-ubiquitinated proteins. These allosteric effects thus reduce ATP hydrolysis by inactive proteasomes and nonspecific proteolysis and enhance proteasomal specificity for ubiquitinated proteins.
Keywords: allosteric regulation, ATPase, deubiquitylation (deubiquitination), peptidase, proteasome, protein degradation, gate opening, Usp14
Introduction
In eukaryotic cells, 26S proteasomes are the major site for protein degradation. Most proteins digested by proteasomes are first tagged with ubiquitin (Ub)2 chains (1). The 26S proteasome consists of the 20S proteolytic particle and one or two 19S regulatory particles (1, 2). The cylindrical 20S proteasome is a hollow four-ring particle that contains in each of its central β-rings three types of active sites, which are chymotrypsin-like, trypsin-like, and caspase-like in specificity (3). Its outer α-rings contain a gated channel for substrate entry (4, 5). The 19S regulatory particle performs several enzymatic and non-enzymatic functions that are required for the degradation of ubiquitinated proteins, including binding and disassembly of the Ub chain, ATP hydrolysis, and unfolding and translocating the substrate protein (1, 2). The 19S base consists of six homologous AAA ATPases, which form a ring, plus three associated subunits, Rpn1, Rpn2, and Rpn13 (6, 7). The ATPases bind, unfold, and translocate the polypeptide through their central channel and then through the gated entry channel into the inner chamber of the 20S particle where proteolysis occurs (1, 8). The gate of the 20S is opened upon nucleotide binding when the ATPases' C-terminal HbYX (where Hb is a hydrophobic residue) residues bind to intersubunit pockets in the outer α-ring of 20S (9). Most steps in the degradation process are linked to ordered cycles of ATP binding and hydrolysis (10). Initially, the Ub chain binds reversibly to one of the 19S receptor subunits, Rpn10/S5a, Rpn13, or Rpn1 (11–13), but commitment to degradation occurs through an ATP-driven step in which a loosely folded region of the polypeptide becomes tightly bound to the ATPase ring (14).
During degradation of a Ub conjugate, the Ub chain is disassembled so that the Ub molecules can be reutilized. Three different deubiquitinating enzymes (DUBs) are associated with the 19S regulatory particle in higher eukaryotes. Two are cysteine proteases, Usp14/Ubp6 (15) and Uch37/UchL5 (16, 17), and the third, Rpn11, is a metalloprotease (18, 19). Rpn11 is an integral 19S subunit, but Usp14 and Uch37 associate reversibly with the proteasome and are usually present at substoichiometric amounts. Like its homolog, Ubp6, in budding yeast, Usp14 contains an N-terminal UBL domain, which binds to Rpn1 on the proteasome (13), and a C-terminal USP domain, which is essential for deubiquitination (20). Deubiquitination of substrates by Rpn11 (unlike Usp14 and Uch37) is an ATP-stimulated process that is directly linked to the degradation process (18, 19). Rpn11 appears to remove the Ub chain en bloc by cleaving the isopeptide bond adjacent (or near) to the substrate and facilitates the translocation of the polypeptide through the ATPase ring into the 20S particle (18).
Surprisingly, the binding of a Ub conjugate to Usp14 or Uch37 also allosterically activates the proteasome's degradative mechanism (21, 22). This stimulatory role of Usp14 requires occupancy of its active site by either a Ub chain or the inhibitor ubiquitin aldehyde (Ub-al). This step, and not the initial binding of the ubiquitinated substrate to the particle, somehow enhances substrate entry into the 20S (“gate opening”) (21). If in addition to interacting with Usp14 the ubiquitinated protein contains a loosely folded domain, ATP hydrolysis is also activated (22). Unlike gate opening, this stimulation of ATP hydrolysis, which drives conjugate degradation, thus requires two signaling events, a Ub chain (or Ub-al) binding to Usp14 and a polypeptide interacting probably with the ATPase (22).
These activating functions of Usp14 were surprising because Usp14/Ubp6 was previously shown to inhibit the degradation of some substrates (23, 24). Studies of Ubp6-deficient yeast mutants had shown that Ubp6 reduces proteolysis both by acting as a DUB and an allosteric mechanism, which remains unexplained (25, 26). This deubiquitinating activity of Usp14 is dramatically stimulated upon its association with the 19S particle, which requires the UBL domain (24, 27). Although Usp14 was believed to cleave sequentially Ub moieties from the distal end of the Ub chain (25, 28, 29), recently Usp14/Ubp6 was shown to release these chains en bloc from the protein, especially from proteins bearing multiple Ub chains (23). The DUB activity of Ubp6/Usp14 thus helps prevent the prolonged residence of a ubiquitinated protein on the 26S, and it also serves as a timing device that limits the time for hydrolysis of a potential substrate and that can promote the release of the substrate without its degradation (23, 26). Based on this insight, Finley, King, and co-workers (24) developed inhibitors of the DUB activity of Usp14 (e.g. IU1) that by prolonging the substrate's residence time on the proteasome can enhance its degradation.
The role of Usp14 as a central regulator of 26S function has been further supported by the recent cryoelectron microscopy (cryo-EM) studies by Baumeister and co-workers (30), Martin and co-workers (31), and Shi and co-workers (32) that revealed that the association of Ub-al with Usp14/Ubp6 induces marked changes in the structure of the 19S complex. After binding of this substrate analog, the central channel in the ATPase ring is enlarged, and the catalytic domain of Ubp6 becomes located closer to the ATPase ring and Rpn11.
These findings indicate that Usp14 has seemingly opposite actions and that it functions both as an inhibitor and an activator of proteolysis. The simplest hypothesis to resolve these apparently conflicting roles would be that Usp14/Ubp6 normally inhibits proteolysis, but when a Ub chain on a potential substrate is bound, Usp14 allosterically activates the proteasome's degradative functions (21, 22). This activated state is then maintained until the Ub chain is no longer bound to the DUB as occurs when proteolysis is complete or the substrate is released. The present studies were undertaken to examine further this hypothesis and to determine how Usp14 functions as an allosteric inhibitor of proteolysis and how it influences key steps in conjugate degradation. By comparing the enzymatic properties of WT and Usp14-deficient proteasomes, we show here that 26S particles lacking Usp14 exhibit a greater capacity to degrade non-ubiquitinated proteins, greater ATPase activity, and even greater DUB activity than WT proteasomes. Together, these findings imply that, in the absence of a substrate, Usp14 normally maintains the 26S proteasome in a quiescent state, but upon binding a Ub conjugate, it allosterically activates several enzymatic processes essential for efficient destruction of Ub conjugates.
Results
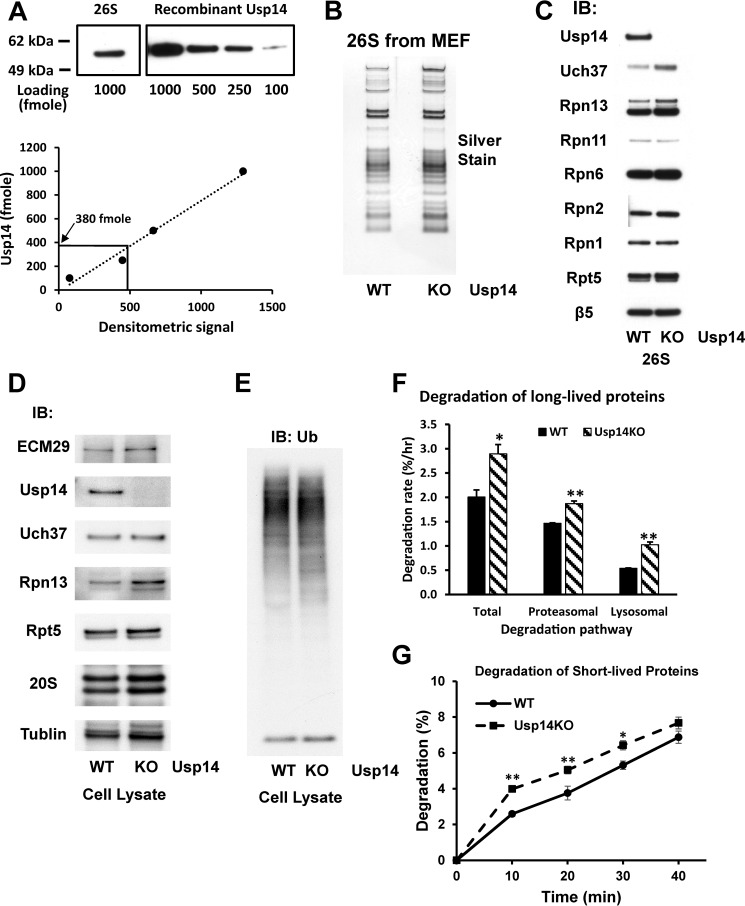
Usp14 deficiency increases 26S content of Uch37 and Rpn13 and overall proteolysis
To understand the regulatory role of Usp14, we compared subunit composition and various enzymatic activities of 26S proteasomes purified from WT mouse embryonic fibroblast (MEF) cells and MEFs lacking Usp14 (Usp14KO). In yeast, Ubp6 is present on a small fraction of the proteasomes (30). In the WT MEF cells, we determined that Usp14 was present on ∼40% of the 26S proteasomes based on Western blotting using increasing amounts of purified Usp14 as a standard and a proteasome with molecular mass of 2.5 MDa (Fig. 1A). The proteasomes purified from the WT and Usp14KO MEFs appeared very similar in subunit composition (Fig. 1, B and C) with two clear exceptions. The other cysteine protease DUB, Uch37, and the Ub receptor, Rpn13, to which Uch37 binds were both increased. Interestingly, the total quantity of Uch37 in the two types of cell lysates did not differ, although Rpn13 expression did increase in the Usp14KO cells (Fig. 1D). This increased association of Uch37 and Rpn13 with the proteasome presumably occurred to compensate for the loss of Usp14. In the Δubp6 yeast, degradation of a number of model proteins was increased (25) above control levels. We therefore tested whether overall rates of protein degradation differed in WT and KO MEF cells by pulse-chase methods using [3H]phenylalanine. To follow the breakdown of long-lived proteins, which comprise the bulk of cell proteins (33, 34), cell proteins were radiolabeled for 24 h and regrown in chase medium (DMEM containing non-radioactive phenylalanine and cycloheximide to repress protein synthesis) for 3 h to allow breakdown of short-lived cell components. Then the cells were regrown in the fresh chase medium, and degradation rates were assayed. The total protein degradation rate was about 45% greater in Usp14KO cells (p < 0.05) than in controls (Fig. 1F).
Figure 1.
MEF cells lacking Usp14 have increased content of Rpn13 and Uch37 in the 26S proteasomes. Proteasomes were purified from WT and Usp14KO MEFs using the UBL method (see “Experimental procedures”). Concentrations of each proteasome preparation were determined using the BCA method, and molarities were calculated using 2.5 MDa as the molecular mass of 26S proteasomes. A, Usp14 is a substoichiometric component of WT MEF 26S. The content of Usp14 in 1 pmol of WT 26S proteasomes was determined by Western blotting using recombinant Usp14 as the standard. The content of Usp14 in 1 pmol of WT MEF 26S was calculated as 380 fmol. Upper panel, Western blot. Lower panel, quantitation from upper panel using ImageJ software. B, SDS-PAGE of purified 26S proteasomes from WT and Usp14KO MEFs. Proteins were silver-stained after electrophoresis. C, the contents of Uch37 and Rpn13 were increased in the purified 26S proteasomes from Usp14KO MEFs even though Usp14KO 26S contains the same levels of other subunits as WT 26S. Levels of proteins in 26S proteasomes purified from WT and Usp14KO MEF were determined by Western blotting after SDS-PAGE. D, the expression of Rpn13 was increased in Usp14KO MEFs. Levels of proteasome subunits were determined by Western blotting of cell lysates of WT and Usp14KO MEFs after SDS-PAGE. Unlike the purified proteasomes, only the content of Rpn13, but not that of Uch37, was increased in Usp14KO MEF cells. Therefore, this increase in Uch37 content must be due to increased binding of Rpn13. The content of tubulin was monitored as an input control. E, the cellular content of Ub conjugates did not change significantly in Usp14KO MEFs. The levels of Ub conjugates were determined in lysates of WT and Usp14KO MEFs by Western blotting after SDS-PAGE. F, MEF cells lacking Usp14 degrade long-lived cell proteins faster than WT MEFs. WT and Usp14KO MEF cells were labeled with [3H]phenylalanine (5 μCi/ml) for 24 h. After washing the cells with PBS and then chase medium (DMEM containing 2 mg/ml nonradioactive phenylalanine and 100 μm cycloheximide), cells were grown for a further 2 h to let short-lived labeled proteins be degraded. Then cells were grown in the chase medium with either DMSO (control) or 10 μm bortezomib/Velcade dissolved in DMSO to block proteasome activity. After 1 h of inhibitor treatment, degradation of cellular proteins was measured in the chase medium for up to 4 h. Proteasomal degradation rates were calculated by subtracting the degradation rate with bortezomib treatment from the total degradation rate. Error bars represent S.D. The remaining proteasome-independent proteolysis represents lysosomal proteolysis. *, p < 0.05; **, p < 0.01 compared with WT MEF by Student's t test. G, MEF cells lacking Usp14 degrade short-lived cell proteins faster than WT MEFs. WT and Usp14KO MEF cells were labeled with [3H]phenylalanine (10 μCi/ml) for 20 min. After washing the cells as in F, cells were incubated for 10 min in the chase medium. Degradation of cellular proteins was then measured in the chase medium for up to 40 min. Error bars represent S.D. *, p < 0.05; **, p < 0.01 compared with WT MEF by Student's t test. n = 6. IB, immunoblotting.
We then determined the contribution of the ubiquitin-proteasome pathway using bortezomib to selectively inhibit the proteasome as described elsewhere (33). Total substrate flux through the proteasome was increased (Fig. 1F) by about 27% in the Usp14KO MEFs (p < 0.01). However, in addition to the enhanced proteasomal proteolysis, there was also greater protein degradation by lysosomes (calculated by subtracting the proteasomal degradation from the total proteolysis) in the Usp14KO MEFs, which presumably reflects the greater rate of autophagy recently reported in other Usp14-deficient cells (35). However, as shown previously (33), the great majority of the intracellular proteolysis in the MEF cells was by proteasomes.
To also evaluate the degradation rate of short-lived proteins, the MEF cells were initially labeled for 20 min with [3H]phenylalanine. During the subsequent chase period, the most short-lived cell proteins were hydrolyzed about 53% faster in Usp14KO cells (p < 0.05) than in controls (Fig. 1G). Despite the greater rates of proteolysis by the ubiquitin-proteasome and autophagic pathways and despite the loss of a major proteasomal DUB in the Usp14KO MEF cells, the content of ubiquitinated proteins did not differ significantly in the Usp14KO and WT cells (Fig. 1E).
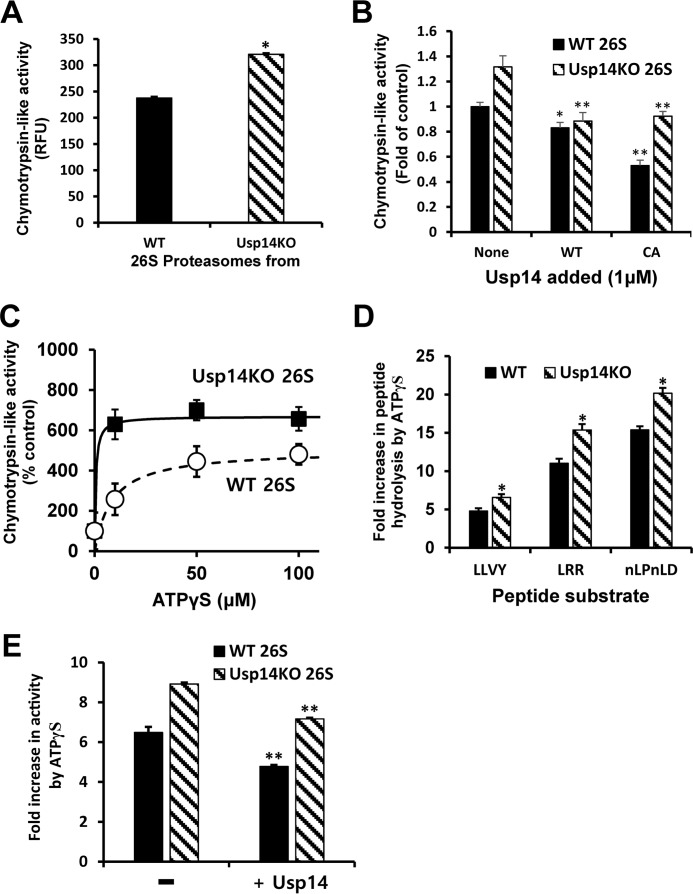
Binding of Usp14 to the 26S proteasomes inhibits basal peptide hydrolysis
The chymotrypsin-like activity of the proteasomes from Usp14KO cells was slightly (35%) higher than that of the particles from WT MEFs (Fig. 2A). Because this difference in peptidase activities was seen even though Usp14 is present on less than half the WT particles (Fig. 1A), this difference must underestimate the actual increase in peptide hydrolysis in the Usp14KO cells. We therefore studied the effects of increasing Usp14 content by adding recombinant Usp14 to the purified 26S proteasomes. With WT Usp14 added, the absolute chymotrypsin-like activity of the 26S decreased to a similar extent in WT and KO particles (17–33%) (Fig. 2B). In general, the present inhibition appeared larger in Usp14KO 26S where the basal activity was greater. Addition of the enzymatically inactive Cys → Ala Usp14 mutant also decreased the chymotrypsin-like activity in both WT and Usp14KO proteasomes. Thus, this inhibition is due to allosteric action of Usp14 (Fig. 2B).
Figure 2.
Usp14 reduces peptidase activities in the proteasome and the activation of peptide entry by ATPγS. Peptidase activities of purified WT or Usp14KO 26S were assayed by measuring hydrolysis of fluorogenic peptide-amc substrates (10 μm; Suc-LLVY-amc for chymotrypsin-like activity, Boc-LRR-amc for trypsin-like activity, and Ac-nLPnLD-amc for caspase-like activity) in the presence of 100 μm ATP or ATPγS. The basal peptidase activity (control) of the proteasomes was measured in the presence of ATP. Error bars represent S.D. of at least three independent measurements. A, chymotrypsin-like activity of 26S purified from Usp14KO cells (4 nm) is consistently greater (35%) than that of WT 26S in the presence of ATP (100 μm) even though only a small fraction (∼40%) of the proteasomes from WT cells contained Usp14 (Fig. 1A). Cleavage of the fluorogenic substrate Suc-LLVY-amc (10 μm) by each type of proteasomes was monitored. *, p < 0.05 compared with WT 26S by Student's t test. B, binding of full-length recombinant Usp14 protein to the 26S particles inhibits the chymotrypsin-like activity of WT and Usp14KO 26S. Purified proteasomes (100 nm) were preincubated for 10 min at room temperature with bacterially expressed full-length WT or an inactive Cys → Ala (CA) mutant Usp14 (1 μm) before the start of the assays. *, p < 0.05; **, p < 0.01 compared with control proteasomes (without Usp14 added) by Student's t test. C, increasing amounts of ATPγS stimulate the chymotrypsin-like activity of both WT and Usp14KO proteasomes in a saturable manner. Both the maximal stimulation by ATPγS and the apparent affinity for ATPγS were greater for Usp14KO 26S than for the WT particles. D, ATPγS (100 μm) markedly stimulated all three peptidase activities of both WT and Usp14KO proteasomes (4 nm) but consistently caused a 30–40% larger stimulation of these activities in the Usp14KO 26S. *, p < 0.05 compared with WT 26S by Student's t test. E, addition of Usp14 reduced the stimulation of the chymotrypsin-like activity by ATPγS. After incubation of purified proteasomes (100 nm) with a molar excess of Usp14 (1 μm) at room temperature for 10 min, the chymotrypsin-like activities of WT and Usp14KO 26S with ATPγS (100 μm) were assayed as in D. **, p < 0.01 compared with control samples without Usp14 added by Student's t test. RFU, relative fluorescence units.
Unlike ATP, the non-hydrolyzable ATP analog ATPγS causes a marked stimulation of proteasomal peptide hydrolysis because it induces major conformational changes in the 19S particle, leading to an alignment of the ATPases' central channel with the 20S gate (36) and gate opening by the ATPases' C termini (10). Adding increasing amounts of ATPγS stimulated the chymotrypsin-like activities of both the WT and Usp14KO particles (Fig. 2C). However, the maximum stimulation of Usp14KO 26S proteasomes was consistently larger than that of WT particles (Fig. 2C). With ATPγS (100 μm), the chymotrypsin-like activity was stimulated maximally 5–6-fold, the trypsin-like activity was stimulated 11–15-fold, and the caspase-like activity was stimulated 15–20-fold over those activities with ATP (Fig. 2D), and in each case, the maximal activity and the degree of stimulation were greater in the Usp14KO proteasomes than in WT particles (Fig. 1D).
We also tested the effects of increasing Usp14 content on this stimulation of proteasomal peptide hydrolysis by ATPγS. Incubation with the recombinant Usp14 in the presence of ATPγS decreased by 20–26% the chymotrypsin-like activity of both types of proteasomes (Fig. 2E). Thus, although Usp14 increases peptide hydrolysis (21) when it binds a Ub chain, the presence of Usp14 by itself inhibits 20S gate opening and the activation by nucleotides.
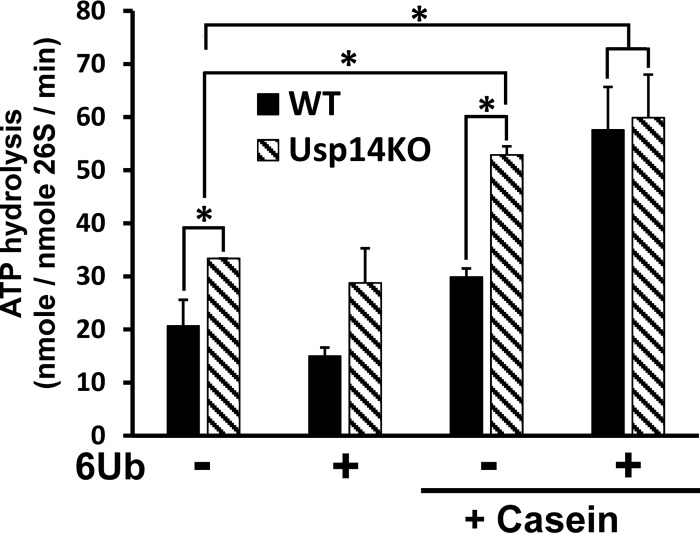
Usp14 inhibits the basal activity of proteasomal ATPases
A general feature of bacterial ATP-dependent proteases and the archaeal PAN ATPase, which are all hexameric complexes homologous to the 26S proteasomal ATPases Rpt1–6, is that their ATPase activity is stimulated upon binding a loosely folded polypeptide (37–39). All these ATPases also bind and translocate polypeptides into an associated proteolytic compartment. Unlike these enzymes, the stimulation of the 26S ATPases requires the simultaneous binding of a Ub chain to Usp14 (or to Uch37) in addition to the unstructured region of the polypeptide, although the Ub chain and polypeptide need not be covalently linked (22). To evaluate the role of Usp14 in regulating the ATPases, we compared the activation of ATP hydrolysis by WT and Usp14KO particles (Fig. 3). Surprisingly, the Usp14KO 26S had a 60% higher basal ATPase activity than WT 26S (33.4 versus 20.7 nm ATP/nm 26S/min; p < 0.05) (Fig. 3). When either type of the proteasomes was incubated with linear hexa-Ub chains, there was no significant stimulation of ATPase (unlike some previous reports (22, 40)). However, when the Ub chains were added with the structureless protein β-casein, there was a 2.7-fold stimulation of ATP hydrolysis with the WT and a 1.8-fold stimulation with Usp14KO 26S (Fig. 3). As a result, the stimulated rates of ATP hydrolysis by the 26S from the two cell types were both about 60 μm ATP/μm 26S/min.
Figure 3.
ATPases of Usp14KO proteasomes, unlike those of WT 26S, are stimulated by unfolded proteins in the absence of a Ub chain. ATP hydrolysis by 26S proteasomes (20 nm) was measured using the malachite green assay (54). The basal ATPase activity of Usp14KO 26S was consistently (60%) higher than that of WT 26S. Upon addition of casein (1 μm) and a hexa-Ub chain (1 μm), which together mimic the binding of a Ub conjugate (22), ATP hydrolysis of WT 26S increased 2–3-fold and resembled the ATPase activity of Usp14KO 26S under the same conditions. However, hexa-Ub alone did not increase ATP hydrolysis of either type of proteasomes. ATP hydrolysis by Usp14KO 26S, but not that by WT 26S, was enhanced by addition of casein alone and reached the same level as seen with 26S upon binding both a Ub chain and casein. 6Ub, linear hexa-Ub chain. *, p < 0.05 compared with control by Student's t test. Error bars represent S.D.
Although β-casein by itself had no effect on ATP hydrolysis by the WT proteasomes, surprisingly, ATP hydrolysis by the Usp14KO 26S was consistently stimulated by β-casein alone (1.7-fold), which is similar to the stimulation seen with a Ub conjugate or when a Ub chain and casein were present together. In this respect, the Usp14KO particles resemble the AAA ATPases that regulate proteolysis in Escherichia coli and the archaeal PAN proteasome-regulatory ATPases, which hydrolyze ATP faster in the presence of casein (37–39). Thus, Usp14 must normally inhibit the activation of ATP hydrolysis by protein alone, and this inhibition is eliminated when Usp14 interacts with a Ub chain (22). This inhibition by Usp14 of both the basal ATP hydrolysis and its activation by proteins in the absence of a Ub chain must help prevent ATP consumption that is not linked to degradation of ubiquitinated proteins.
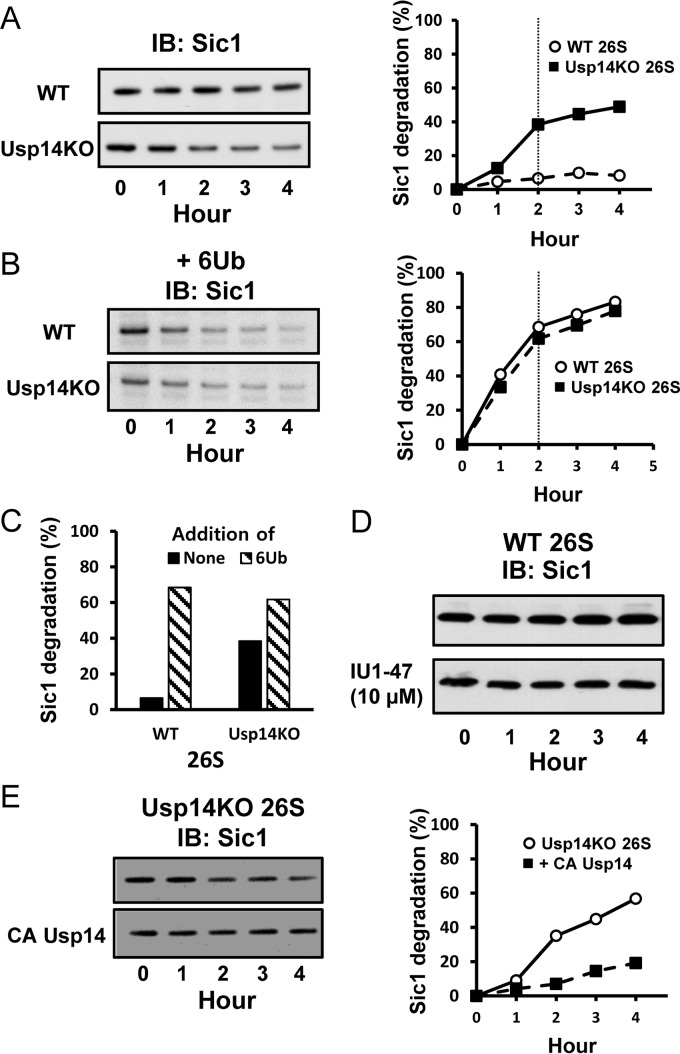
Usp14 reduces the degradation of non-ubiquitinated unfolded proteins
These findings predict that proteasomes lacking Usp14 should degrade some non-ubiquitinated proteins faster than the WT 26S. In fact, in the absence of Ub, the intrinsically disordered protein PY-Sic1 (41) is degraded very slowly by WT proteasomes; however, as predicted, Usp14KO 26S degraded Sic1 at least 6-fold faster than WT 26S did (Fig. 4, A and C). By contrast, the addition of linear hexa-Ub chains to mimic substrate ubiquitination stimulated the breakdown of Sic1 by WT 26S about 10-fold and by Usp14KO 26S at most 2-fold (Fig. 4, B and C). Consequently, the maximal rates of degradation achieved by the two types of particles with the Ub chains present were similar. These findings provide further evidence that Usp14 inhibits proteasomal proteolysis until a Ub chain binds and reverses this inhibition. These inhibitory effects are allosteric because a specific inhibitor of Usp14's catalytic activity, IU1, did not enhance the degradation of non-ubiquitinated Sic1 (Fig. 4D). Furthermore, the addition of the inactive Cys → Ala mutant Usp14 to Usp14KO 26S decreased the degradation of Sic1 (Fig. 4E).
Figure 4.
Usp14 reduces proteasomal degradation of unstructured proteins lacking ubiquitination. Degradation of the intrinsically unstructured protein PY-Sic1 (100 nm) by 26S proteasomes (2 nm) was assayed by Western blotting. A and B, left panels, after reaction for the indicated times, the remaining Sic1 was analyzed by Western blotting. Right panels, degradation of Sic1 was plotted by measuring Sic1 density using ImageJ software. Each point is representative of at least two independent experiments. A, unlike 26S proteasomes from WT cells, Usp14KO 26S proteasomes degrade non-ubiquitinated PY-Sic1. These data are representative of three independent experiments. B, although the basal degradation of PY-Sic1 by 26S from Usp14KO MEFs was much higher than that of WT proteasomes, the degradation rates by both types of 26S particles were quite similar in the presence of linear hexa-Ub chain (1 μm). Thus, the Ub chains stimulate degradation by the WT proteasomes much more than by the Usp14KO 26S. Similar data were obtained in three independent experiments. 26S proteasomes were incubated with the Ub chain at room temperature for 15 min before the reaction started. C, the dependence of proteasomal degradation of PY-Sic1 on the presence of a Ub chain was much greater in WT than Usp14KO 26S. Addition of Ub chain stimulated degradation 10-fold in WT 26S and 60% in Usp14KO particles. Degradation rates were compared after incubation for 2 h when reaction rates slowed below initial rates (at the vertical line in A and B). D, although knock-out of Usp14 stimulates the degradation of PY-Sic1, a Usp14-specific inhibitor IU1 (10 μm) does not stimulate the degradation of PY-Sic1 by the purified WT 26S particles nor does it affect peptide hydrolysis (data not shown) even though knock-out of Usp14 stimulates their degradation. E, addition of inactive Cys → Ala (CA) Usp14 mutant inhibits degradation of Sic1 by Usp14KO 26S. These proteasomes were preincubated with Usp14 as in Fig. 2B before the degradation reactions. These data are representative of two independent experiments. IB, immunoblotting.
Usp14 inhibits deubiquitination by Rpn11
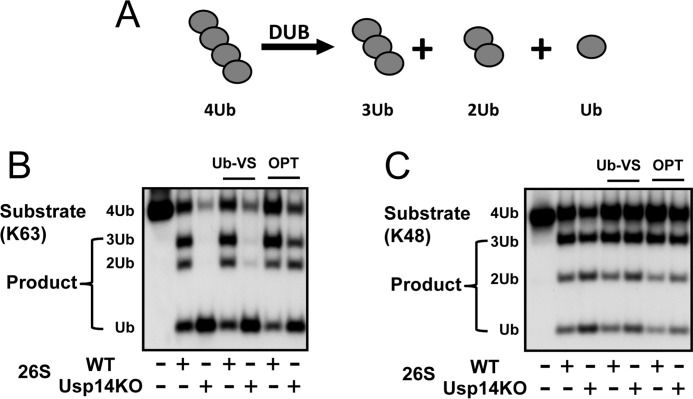
Rapid degradation of Ub conjugates by the proteasome requires the removal of the Ub chain from the substrate (1). Of the three DUBs associated with mammalian 26S proteasomes, only Rpn11 seems essential for the degradation of ubiquitinated proteins (18). Because Usp14 upon binding a Ub chain stimulates proteasomal peptide and ATP hydrolysis, we tested whether it may also enhance the proteasome's other DUB activities by comparing the rates of breakdown of free tetra-Ub chains (chains not conjugated to a protein) by WT and Usp14KO 26S (Fig. 5A). Surprisingly, even though Usp14KO proteasomes lack the important DUB Usp14, they showed a much greater capacity to disassemble Lys-63 Ub chains (Fig. 5B) than WT particles as shown by their more rapid generation of tri-, di-, and mono-Ubs. The loss of Usp14 also mildly increased the rates of disassembly of Lys-48 tetra-Ub (Fig. 5C). Because the Zn2+ chelator o-phenanthroline markedly suppressed this accelerated breakdown of tetra-Ub to the level seen with WT 26S, the faster chain hydrolysis in the Usp14KO must be due to activation of the Zn2+-containing metalloprotease Rpn11. In fact, Rpn11 is the most active proteasomal DUB against Lys-63 chains (42). Furthermore, although Uch37 and Rpn13 levels were increased in the Usp14KO proteasomes (Fig. 1C), Ub-VS, which inhibits Uch37, did not decrease the rapid breakdown of tetra-Ub chains by Usp14KO 26S (Fig. 5B). Thus, the activity of Rpn11 (but not Uch37) is stimulated by the absence of Usp14 in the 26S. These findings are also consistent with the recent structure of the 26S by cryo-EM, which shows that Ubp6 can normally block substrate access to Rpn11 (31).
Figure 5.
Usp14 inhibits deubiquitination by Rpn11. A, disassembly of tetra-Ub chain (368 nm) by WT or Usp14KO 26S (5 nm) was assayed after incubation at 37 °C for 20 min. The generation of tri-, di-, and mono-Ubs was analyzed by Western blotting after electrophoresis. B and C, Usp14KO MEF 26S proteasomes disassembled tetra-Ub chains much faster than WT 26S particles. Although Ub-VS (1.5 μm) did not inhibit Ub chain hydrolysis, o-phenanthroline (OPT) (1 mm) markedly inhibits Ub chain hydrolysis and reduces deubiquitination by Usp14KO 26S to the level of WT 26S. Thus, this process involves the Zn2+ metalloprotease Rpn11. Similar results were also obtained with Lys-63 tetra-Ub chains (B) and Lys-48 tetra-Ub chains (C) as substrates and were obtained in at least three independent experiments.
Discussion
By helping release Ub from the substrate, especially from proteins bearing multiple Ub chains (23), Usp14/Ubp6 promotes Ub recycling and prevents the degradation of Ub by the proteasome (43). However, in addition to this catalytic role, as emphasized by the present results, Usp14 is a major allosteric regulator of proteasome function that has the unusual capacity to both activate and inhibit multiple steps in Ub conjugate degradation. The first evidence of this regulatory role was the finding in yeast that Ubp6 inhibits the degradation of several proteins (25), leading to the proposal that Usp14/Ubp6 DUB activity serves as a timing device for proteasomal degradation (26). Thus, by removing Ub moieties from the substrate, Usp14 functions in a kinetic competition between substrate release after deubiquitination and substrate destruction (26). Accordingly, low-molecular-weight selective inhibitors of Usp14's catalytic activity, e.g. IU1, were shown to enhance the degradation of certain proteins (24), presumably by slowing their deubiquitination and providing more time for proteasomal proteolysis. However, unlike a Usp14 substrate or substrate analog (e.g. Ub-al), IU1 does not cause allosteric activation of ATP hydrolysis (22), peptide entry, or degradation of non-ubiquitinated PY-Sic1 (Fig. 4D). Nevertheless, by slowing substrate deubiquitination by Usp14, IU1 may prolong substrate association with Usp14 and thus prolong proteasomal activation.
Because of Usp14's inhibitory actions, it was surprising to find that binding of a Ub chain to Usp14/Ubp6 allosterically stimulates peptide entry into the 20S (21) as well as 26S ATPase activity provided the substrate contains an unfolded domain (22). The present observations bridge these seemingly contradictory roles and show that Usp14 by itself inhibits at least three key functions in substrate degradation, but upon binding a Ub chain, Usp14 releases this inhibition and activates these processes further.
Interestingly, in Usp14-deficient cells, the proteasomes contain significantly greater amounts of Rpn13 and Uch37 (Fig. 1B) even though the cells contained only slightly increased cellular levels of Rpn13 (Fig. 1C). Although this Ub receptor is often classified as a stoichiometric subunit, several investigators have reported that the Ub receptor Rpn13 is present in proteasomes in substoichiometric levels (44–46). Presumably, the increased Uch37 and Rpn13 levels on the proteasome help compensate for the loss of Usp14's catalytically activity, but how this loss of Usp14 is recognized and leads to increased Uch37 and Rpn13 binding are interesting questions for future study.
In the absence of a substrate and thus independently of its role as a DUB, Usp14 was found to inhibit multiple proteasomal processes. For example, the 26S lacking Usp14 has a 20–30% greater peptidase activity than WT 26S (Fig. 2A). Because at most 30–40% of the WT proteasomes contain Usp14 (Fig. 1E), this greater activity must underestimate the actual effect of Usp14 on peptide entry into the 20S particle and ATP hydrolysis. In addition, all three peptidase activities of the Usp14KO 26S were consistently stimulated by ATPγS more than the WT (Fig. 2D). Accordingly, addition of recombinant WT or Cys → Ala Usp14 further represses allosterically both the basal and ATPγS-stimulated peptidase activities (Fig. 2, B and E). Nucleotides, like ATPγS, open the gated entry of the 20S particle by stimulating interaction of ATPases' C-terminal HbYX motifs with intersubunit pockets in the 20S α-ring (47), but recent cryo-EM studies showed that ATPγS also induces structural reorganization of the 19S complex that aligns the central pore in the ATPase ring with the 20S entry channel (36). Because Usp14 suppresses all three peptidase activities in the presence of ATPγS (Fig. 2D), Usp14 must inhibit substrate entry rather than altering the catalytic activity of the three sites. Although ATPγS was previously assumed to cause maximal increases in substrate entry, Usp14KO can be stimulated further than WT 26S in the presence of ATPγS. Thus, Usp14 must influence substrate entry by additional mechanisms distinct from those activated by ATPγS.
One important surprising finding was that after loss of Usp14 the 26S exhibited a greater ability to disassemble tetra-Ub chains due to increased activity of Rpn11 (Fig. 5). Thus, under basal conditions, Usp14 allosterically inhibits Rpn11 and reduces its capacity to digest Ub conjugates. Recently, the catalytic domain of Usp14 (the “USP domain”) upon association with Ub-VS was shown by cryo-EM to block sterically substrate access to Rpn11 (31). Therefore, the loss of Usp14 probably exposes the active site of Rpn11 and enables it to digest Ub chains more readily. However, it seems likely that in addition to this steric effect Usp14 may allosterically inhibit Rpn11's catalytic activity just as it inhibits the particle's other proteasomal activities.
In the Usp14KO proteasomes, ATPase activities are also regulated differently than in WT (Fig. 3). Unlike the WT, which requires the binding of both a Ub chain and an unfolded protein to stimulate ATP hydrolysis (22), in the Usp14-deficient 26S, such a protein can stimulate this process even without a Ub chain present (Fig. 3). In this respect, the ATPases in the Usp14KO proteasomes resemble the homologous, ATP-dependent proteases from bacteria (Lon, ClpAP, ClpXP, and HslUV) and the direct 26S ancestor, PAN, which regulates the activity of archaeal 20S proteasomes. These enzymes are all protein substrate-activated AAA ATPases (38, 39). During the evolution of eukaryotes, ubiquitin conjugation became linked to proteolysis, and through Usp14, the 26S proteasome evolved the requirement for Ub chain binding to activate peptide entry and ATP hydrolysis. This enhancement of ATPase activity is a critical event because it drives substrate translocation, and the amount of ATP consumed is directly proportional to the amount of Ub conjugates degraded (48). Although the structural basis for these different allosteric effects are not yet clear, it is noteworthy that Ubp6 binds to Rpn1, but when Ub aldehyde occupies the active site on Usp14, these proteins also interact with the ATPases Rpt1 and -2 (30–32).
The inhibition of multiple proteasomal activities by Usp14 can account for the finding that MEF cells lacking Usp14 degrade cellular proteins, both short-lived and long-lived components (Fig. 1, F and G), faster than WT MEFs. This observation of enhanced degradation of cell proteins generally by proteasomes extends the findings on breakdown of several model proteins in Δubp6 yeast (25). Surprisingly, protein degradation in lysosomes was also enhanced in these Usp14KO cells (Fig. 1F), and the lack of this DUB has been reported recently to stimulate autophagy in other cell types (35). These observations are also intriguing because they are further evidence that under physiological conditions the ubiquitin-proteasome pathway and autophagy are coordinately regulated even though the proteasomes remain the primary site of protein hydrolysis (33, 49).
It is also noteworthy that, unlike the WT, proteasomes lacking Usp14 can efficiently degrade certain unstructured proteins without ubiquitination (Fig. 4A). This degradation of non-ubiquitinated proteins must result from enhanced translocation of such unstructured proteins into the 20S particle and presumably from the ability of such non-ubiquitinated proteins to activate ATP hydrolysis. The inhibition of Ub-independent proteolysis by Usp14 must also be an allosteric effect because there is no Ub chain on these substrates to hydrolyze and because the inactive Cys → Ala mutant Usp14 also inhibited degradation (Fig. 4E).
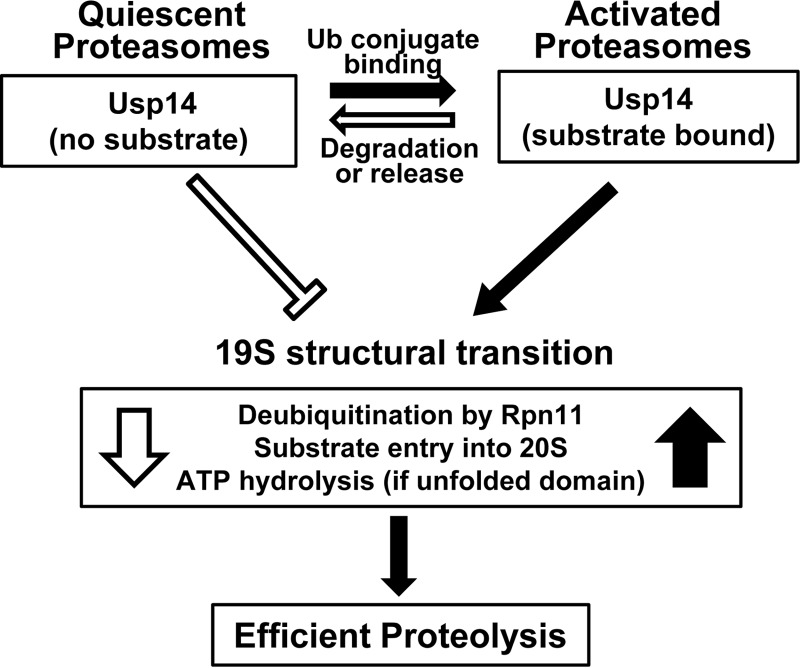
Although these several inhibitory actions of Usp14 probably contribute to its ability to suppress overall protein degradation in vivo (25), its inhibition of ATPase activation by non-ubiquitinated proteins appears to be critical in preventing Ub-independent proteolysis in vivo. These findings together also suggest a clear rationale for these bidirectional effects of Usp14 on proteasome function (Fig. 6). In the absence of a substrate, Usp14 helps prevent wasteful ATP consumption and non-selective proteolysis by the proteasome, especially degradation of non-ubiquitinated proteins. The activation of the proteasome upon binding a Ub conjugate thus enhances the selectivity for ubiquitinated substrates, especially those with unfolded domains.
Figure 6.
Summary of Usp14's allosteric regulation of proteasomal degradation. Without a Ub conjugate bound, proteasomes are relatively inactive, and Usp14 inhibits ATP hydrolysis, substrate entry into the 20S, and deubiquitination by Rpn11. These actions reduce futile ATP consumption and the degradation of non-ubiquitinated proteins, which occur in the Usp14KO 26S but not in WT 26S. However, upon Ub chain binding, Usp14 (and the 19S particle) undergoes a major structural transition and thus activates Rpn11, peptide entry into 20S, and ATP hydrolysis (if an unfolded protein also is bound). These actions promote efficient degradation of ubiquitinated substrates. This activated state should be maintained as long as Usp14 binds a Ub chain. These processes return to the quiescent basal state when the substrate is degraded or is deubiquitinated and dissociates. These allosteric actions are not dependent on Usp14's catalytic activity, which functions to degrade the Ub chain and thus limits the duration of the active state and the substrate's “dwell time” on the 19S complex.
As noted above, the greater activity of the 26S from cells lacking Usp14 is striking because Usp14 is present on only a fraction of the WT particles. It seems likely, therefore, that additional factors can suppress the basal activities of the particles lacking Usp14. One possible inhibitory factor might be Uch37 because its content on the 26S is increased in the Usp14KO MEFs and because the interaction of Ub chains with Uch37 can activate gate opening and ATP hydrolysis (22). Recent cryo-EM studies suggest that only a small fraction of 26S proteasomes in cells is active in proteolysis, and it is probably the fraction containing Usp14 (50). Because our recent studies show that the binding of Ub conjugates to the proteasome stimulates the association of cytosolic Usp14 with the particles,3 the subsequent interaction between the Ub chain and Usp14 can lead to more efficient and selective breakdown of the ubiquitinated substrates.
Experimental procedures
Purification of proteins
GST-UBL derived from hHR23B was expressed in E. coli and purified with GSH-Sepharose as described previously (51). GST-fused Usp14 was also expressed in E. coli and purified using GSH-Sepharose (24). Resin-bound Usp14 was treated with thrombin to cut off GST from Usp14. Residual thrombin was cleared from Usp14 using benzamidine-Sepharose. His-UIM derived from UIM2 of S5a (51) and His-tagged PY-Sic1 (52) were expressed in E. coli and purified with a Ni-NTA resin.
Purification of 26S proteasomes
WT and Usp14KO MEFs (24) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin. Affinity purification of 26S proteasomes was carried out as described previously (53). Briefly, cell lysates prepared with sonication (15 s six times at 18 watts) were spun for 1 h at 100,000 × g. The soluble supernatants were incubated at 4 °C with GST-UBL derived from hHR23B and a corresponding amount of GSH-Sepharose. The slurry containing 26S proteasomes bound to GST-UBL was poured into an empty column, washed, and incubated with His-UIM. The eluate was collected and incubated with Ni-NTA-agarose for 20 min at 4 °C. The Ni-NTA-bound His-UIM was spun out, and the remaining supernatant contained purified 26S proteasomes. Protein concentration was determined using BCA reagent. The molarity of 26S proteasome particles was calculated based on an average molecular mass of 2.5 MDa.
Antibodies
Polyclonal rabbit anti-Ub (A-100) was purchased from Boston Biochem. Anti-Rpn1 (sc-68352), anti-Rpn2 (sc-166038), and anti-Sic1 (sc-50441) were from Santa Cruz Biotechnology. Anti-Rpn6 (14303), anti-Rpn11 (4197), and anti-Rpt5 (13923) were from Cell Signaling Technology. Anti-β5 (A303-847A), anti-Usp14 (A300-920A), and anti-Rpn13 (A302-554A) were purchased from Bethyl Laboratories. Anti-Uch37 (3904-1) was from Epitomics. HRP-conjugated secondary antibodies were from Promega.
Immunoblot analysis
Samples for immunoblotting were run on 4–12% Bis-Tris gels (Life Technologies) with MES buffer (NP0002). Proteins were analyzed following SDS-PAGE and transferred onto 0.45-μm PVDF membranes (Whatman). Immunoblots were blocked and incubated with appropriate primary and secondary antibodies. Membranes were developed with enhanced chemiluminescence reagent (Immobilon Western HRP substrate and Luminol reagent WBKLS0500, Millipore) onto X-ray film. The ImageJ program (W. S. Rasband, National Institutes of Health) was utilized to quantify the signal from the film.
Proteasome activity assays
As indicated, proteasomes were generally incubated with or without casein, linear hexa-Ub chains, Usp14, or a DUB inhibitor at room temperature for 15 min before the start of reactions. Peptide hydrolysis by MEF 26S proteasomes was measured with 10 μm Boc-Leu-Arg-Arg-amc (Boc-LRR-amc), Suc-Leu-Leu-Val-Tyr-amc (Suc-LLVY-amc), or Ac-Nle-Pro-Nle-Asp-amc (Ac-nLPnLD-amc) (Bachem) (λex, 380 nm; λem, 460 nm) and the indicated concentration of 26S proteasomes at 37 °C. Proteasomal activities were calculated from 30 to 60 min after the start of the reaction. The reaction mixture was composed of 50 mm Tris (pH 7.6), 100 mm KCl, 0.1 mm ATP or ATPγS, 0.5 mm MgCl2, 1 mm DTT, and 25 ng/μl BSA (Sigma). Degradation of PY-Sic1 (100 nm) by 26S proteasomes (2 nm) was carried out in the presence of 50 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 1 mm ATP, 1 mm DTT, and 0.01 mg/ml BSA (Sigma) for the indicated time (0–4 h) at 37 °C and measured by Western blotting with an anti-Sic1 antibody. ATP hydrolysis by proteasomes was measured using the malachite green assay (54). Deubiquitination by 26S particles (5 nm) was assayed with tetra-Ub chains (368 nm) in HEPES buffer (25 mm) containing 100 mm KCl, 5 mm MgCl2, 1 mm DTT, and 1 mm ATP at 37 °C for 20 min. After the reaction, the products were analyzed by Western blotting with anti-Ub antibodies.
In vivo proteolysis
The overall rates of degradation of long-lived proteins in WT and Usp14KO MEF cells were determined as described previously (33, 49). MEF cells were grown with [3H]phenylalanine (5 μCi/ml) for 24 h to label cell proteins and then washed twice with PBS and once with chase medium (i.e. fresh DMEM containing 2 mm nonradioactive phenylalanine and 100 μm cycloheximide). Cells were grown in chase medium for 2 h to let short-lived proteins be degraded. Then fresh chase medium containing DMSO or 10 μm bortezomib/Velcade was added. One hour after addition of the inhibitors, samples of the medium were collected for up to 4 h. Then each sample was mixed with 10% trichloroacetic acid (final concentration) to precipitate proteins. The acid-soluble radioactivity reflects the amount of prelabeled, long-lived cell proteins degraded at each time and was expressed relative to the total amount of radioactivity initially incorporated into protein. The absolute rate of proteasomal proteolysis was determined by subtracting the rates of proteolysis in cells treated with bortezomib from that in untreated cells. Lysosomal proteolysis was determined by subtracting the rates of proteasomal degradation from that of overall proteolysis.
The overall rates of degradation of short-lived proteins were determined by a pulse-chase protocol by incubating MEF cells with [3H]phenylalanine (10 μCi/ml) for 20 min to label cell proteins. The cells were then washed twice with PBS and once with the chase medium containing large amounts of nonradioactive phenylalanine to block incorporation of labeled amino acids. After incubation in chase medium for 20 min more, samples were collected every 10 min for up to 40 min. The degradation rate was calculated using the same method as used for evaluating the degradation rate of long-lived proteins.
Author contributions
H. T. K. and A. L. G. designed the studies and wrote the manuscript. H. T. K. performed the experiments.
Acknowledgments
We are grateful to Megan LaChance for valuable assistance in preparing the manuscript and to Dan Finley and Byung Hoon Lee for providing cells and reagents.
This work was supported by NIGMS, National Institutes of Health Grant R01 GM51923, by the Cure Alzheimer's Fund, and by the Project ALS Foundation (2015-06). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
C. -L. Kuo and A. L. Goldberg. (2017) Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc. Natl. Acad. Sci. U.S.A. 114, E3404–E3413.
- Ub
- ubiquitin
- MEF
- mouse embryonic fibroblast
- UBL
- ubiquitin-like
- DUB
- deubiquitinating enzyme
- UIM
- ubiquitin-interacting motif
- Ub-al
- ubiquitin aldehyde
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- USP
- ubiquitin-specific protease
- Ni-NTA
- nickel-nitrilotriacetic acid
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- amc
- 7-amino-4-methylcoumarin
- Boc
- t-butoxycarbonyl
- Suc
- succinyl
- Nle/nL
- norleucine
- Ub-VS
- ubiquitin vinyl sulfone
- PAN
- proteasome-activating nucleotidase.
References
- 1. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith D. M., Benaroudj N., and Goldberg A. (2006) Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 156, 72–83 [DOI] [PubMed] [Google Scholar]
- 3. Kisselev A. F., Garcia-Calvo M., Overkleeft H. S., Peterson E., Pennington M. W., Ploegh H. L., Thornberry N. A., and Goldberg A. L. (2003) The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J. Biol. Chem. 278, 35869–35877 [DOI] [PubMed] [Google Scholar]
- 4. Groll M., and Huber R. (2003) Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 35, 606–616 [DOI] [PubMed] [Google Scholar]
- 5. Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., and Finley D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 6. Lander G. C., Martin A., and Nogales E. (2013) The proteasome under the microscope: the regulatory particle in focus. Curr. Opin. Struct. Biol. 23, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Förster F., Unverdorben P., Sledź P., and Baumeister W. (2013) Unveiling the long-held secrets of the 26S proteasome. Structure 21, 1551–1562 [DOI] [PubMed] [Google Scholar]
- 8. Matyskiela M. E., Lander G. C., and Martin A. (2013) Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., and Goldberg A. L. (2007) Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's α ring opens the gate for substrate entry. Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith D. M., Fraga H., Reis C., Kafri G., and Goldberg A. L. (2011) ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., and Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verma R., Oania R., Graumann J., and Deshaies R. J. (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 13. Shi Y., Chen X., Elsasser S., Stocks B. B., Tian G., Lee B. H., Shi Y., Zhang N., de Poot S. A., Tuebing F., Sun S., Vannoy J., Tarasov S. G., Engen J. R., Finley D., et al. (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351, aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peth A., Uchiki T., and Goldberg A. L. (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 40, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., and Ploegh H. L. (2001) A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li T., Naqvi N. I., Yang H., and Teo T. S. (2000) Identification of a 26S proteasome-associated UCH in fission yeast. Biochem. Biophys. Res. Commun. 272, 270–275 [DOI] [PubMed] [Google Scholar]
- 17. Stone M., Hartmann-Petersen R., Seeger M., Bech-Otschir D., Wallace M., and Gordon C. (2004) Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J. Mol. Biol. 344, 697–706 [DOI] [PubMed] [Google Scholar]
- 18. Yao T., and Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 19. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R. 3rd, Koonin E. V., and Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 20. Wyndham A. M., Baker R. T., and Chelvanayagam G. (1999) The Ubp6 family of deubiquitinating enzymes contains a ubiquitin-like domain: SUb. Protein Sci. 8, 1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peth A., Besche H. C., and Goldberg A. L. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peth A., Kukushkin N., Bossé M., and Goldberg A. L. (2013) Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J. Biol. Chem. 288, 7781–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee B. H., Lu Y., Prado M. A., Shi Y., Tian G., Sun S., Elsasser S., Gygi S. P., King R. W., and Finley D. (2016) USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532, 398–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., and Finley D. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., and Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 26. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., and Finley D. (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 27. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., and Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 28. Lam Y. A., Xu W., DeMartino G. N., and Cohen R. E. (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385, 737–740 [DOI] [PubMed] [Google Scholar]
- 29. Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., Cohen R. E., and Shi Y. (2005) Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aufderheide A., Beck F., Stengel F., Hartwig M., Schweitzer A., Pfeifer G., Goldberg A. L., Sakata E., Baumeister W., and Förster F. (2015) Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 112, 8626–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bashore C., Dambacher C. M., Goodall E. A., Matyskiela M. E., Lander G. C., and Martin A. (2015) Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol. 22, 712–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang X., Luan B., Wu J., and Shi Y. (2016) An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 23, 778–785 [DOI] [PubMed] [Google Scholar]
- 33. Zhao J., Zhai B., Gygi S. P., and Goldberg A. L. (2015) mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. U.S.A. 112, 15790–15797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao J., Garcia G. A., and Goldberg A. L. (2016) Control of proteasomal proteolysis by mTOR. Nature 529, E1–E2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu D., Shan B., Sun H., Xiao J., Zhu K., Xie X., Li X., Liang W., Lu X., Qian L., and Yuan J. (2016) USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 30, 1718–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Śledź P., Unverdorben P., Beck F., Pfeifer G., Schweitzer A., Förster F., and Baumeister W. (2013) Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 7264–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benaroudj N., Zwickl P., Seemüller E., Baumeister W., and Goldberg A. L. (2003) ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 11, 69–78 [DOI] [PubMed] [Google Scholar]
- 38. Goldberg A. L. (1992) The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur. J. Biochem. 203, 9–23 [DOI] [PubMed] [Google Scholar]
- 39. Menon A. S., and Goldberg A. L. (1987) Protein substrates activate the ATP-dependent protease La by promoting nucleotide binding and release of bound ADP. J. Biol. Chem. 262, 14929–14934 [PubMed] [Google Scholar]
- 40. Li X., and Demartino G. N. (2009) Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem. J. 421, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brocca S., Samalíková M., Uversky V. N., Lotti M., Vanoni M., Alberghina L., and Grandori R. (2009) Order propensity of an intrinsically disordered protein, the cyclin-dependent-kinase inhibitor Sic1. Proteins 76, 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., and Cohen R. E. (2009) K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanna J., Meides A., Zhang D. P., and Finley D. (2007) A ubiquitin stress response induces altered proteasome composition. Cell 129, 747–759 [DOI] [PubMed] [Google Scholar]
- 44. Fabre B., Lambour T., Garrigues L., Ducoux-Petit M., Amalric F., Monsarrat B., Burlet-Schiltz O., and Bousquet-Dubouch M. P. (2014) Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 13, 3027–3037 [DOI] [PubMed] [Google Scholar]
- 45. Bohn S., Beck F., Sakata E., Walzthoeni T., Beck M., Aebersold R., Förster F., Baumeister W., and Nickell S. (2010) Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 20992–20997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berko D., Herkon O., Braunstein I., Isakov E., David Y., Ziv T., Navon A., and Stanhill A. (2014) Inherent asymmetry in the 26S proteasome is defined by the ubiquitin receptor RPN13. J. Biol. Chem. 289, 5609–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., and Cheng Y. (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peth A., Nathan J. A., and Goldberg A. L. (2013) The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J. Biol. Chem. 288, 29215–29222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao J., Brault J. J., Schild A., and Goldberg A. L. (2008) Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy 4, 378–380 [DOI] [PubMed] [Google Scholar]
- 50. Asano S., Fukuda Y., Beck F., Aufderheide A., Förster F., Danev R., and Baumeister W. (2015) Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442 [DOI] [PubMed] [Google Scholar]
- 51. Besche H. C., Haas W., Gygi S. P., and Goldberg A. L. (2009) Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saeki Y., Isono E., and Toh-E A. (2005) Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399, 215–227 [DOI] [PubMed] [Google Scholar]
- 53. Besche H. C., and Goldberg A. L. (2012) Affinity purification of mammalian 26S proteasomes using an ubiquitin-like domain. Methods Mol. Biol. 832, 423–432 [DOI] [PubMed] [Google Scholar]
- 54. Lanzetta P. A., Alvarez L. J., Reinach P. S., and Candia O. A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]