Abstract
The vaccinia virus F10 protein is one of two virally encoded protein kinases. A phenotypic analysis of infections involving a tetracycline-inducible recombinant (vΔiF10) indicated that F10 is involved in the early stages of virion morphogenesis, as previously reported for the mutants ts28 and ts15. The proteins encoded by ts28 and ts15 have primary defects in enzymatic activity and thermostability, respectively. Using a transient complementation assay, we demonstrated that the enzymatic activity of F10 is essential for its biological function and that both its enzymatic and biological functions depend upon N-terminal sequences that precede the catalytic domain. An execution point analysis indicated that in addition to its role at the onset of morphogenesis, F10 is also required at later stages, when membrane crescents surround virosomal contents and develop into immature virions. The F10 protein is phosphorylated in vivo, appears to be tightly associated with intracellular membranes, and can bind to specific phosphoinositides in vitro. When F10 is repressed or impaired, the phosphorylation of several cellular and viral proteins appears to increase in intensity, suggesting that F10 may normally intersect with cellular signaling cascades via the activation of a phosphatase or the inhibition of another kinase. These cascades may drive the F10-induced remodeling of membranes that accompanies virion biogenesis. Upon the release of ts28-infected cultures from a 40°C-induced block, a synchronous resumption of morphogenesis that culminates in the production of infectious virus can be observed. The pharmacological agents H89 and cerulenin, which are inhibitors of endoplasmic reticulum exit site formation and de novo lipid synthesis, respectively, block this recovery.
Protein phosphorylation plays a vital role in the regulation of key cellular processes. From signal transduction to cell cycle control, the phosphorylation state of a protein has been known to influence its intracellular localization, enzymatic activity, and interactions with proteins, nucleic acids, ligands, or downstream effectors. Protein kinases and phosphatases can act as molecular switches, regulating the activation or inhibition of cellular pathways in response to intrinsic or extrinsic stimuli.
The analogous regulation of the vaccinia virus life cycle by reversible protein phosphorylation is illustrated by the fact that the viral genome encodes one protein phosphatase (H1) and two protein kinases (B1 and F10), all of which are essential for virus viability. The 20-kDa phosphatase encoded by the H1 gene was the first member of the dual-specificity class of phosphatases to be identified (19). The protein is expressed at late times during infection, and ∼200 to 300 copies of H1 are encapsidated in wild-type (wt) virions. An analysis of a recombinant virus in which the expression of H1 is regulated by the lac operator-repressor system (vindH1) (33) revealed that the repression of H1 does not compromise the number of viral particles produced but instead affects the fraction of viral particles that are infectious. Phosphatase-deficient virions appear to contain a more fragile internal structure and are impaired in the ability to direct early gene expression from the encapsidated viral genome. The first virally encoded Ser/Thr kinase identified was the 34-kDa product of the B1 early gene (3, 31, 54). Temperature-sensitive (ts) mutants with lesions in B1 are defective in DNA replication (39). Although some reports have indicated that B1 is encapsidated, the major protein kinase activity packaged in the viral core (∼200 to 300 copies per virion) is the product of the F10 gene (30).
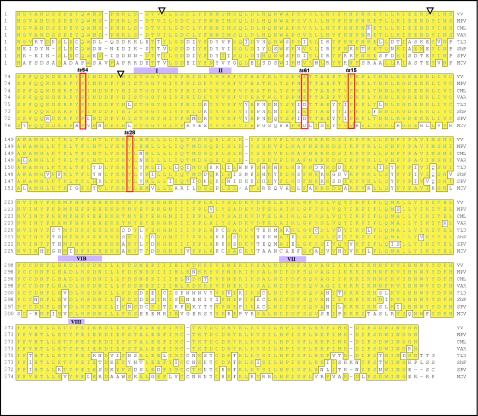
The 439-amino-acid F10 protein has a calculated molecular weight of 52,130 and a predicted pI of 7.47. The kinase activity of F10 was determined empirically rather than predicted by bioinformatic analysis, and indeed F10 shows little similarity to protein kinases other than poxviral orthologs (Fig. 1). The vaccinia virus protein shows identities of 99, 70, and 60% to the orthologs encoded by variola virus, swinepox virus, and molluscum contagiosum virus, respectively. Retention of an F10 ortholog extends beyond the chordopoxviruses to the entomopoxviruses, as distantly related proteins (∼15% identity) have been identified in the genomes of the Amsacta moorei and Melanoplus sanguinipes entomopoxviruses (14) (www.poxvirus.org). A comparison of the F10 sequence with those of cellular Ser/Thr protein kinases revealed that F10 has an atypical structure (20). The F10 sequence has recognizable motifs corresponding to subdomain II (ATP binding pocket) and subdomain VI (catalytic loop). Other subdomains are either highly divergent from consensus motifs or difficult to identify at all. The characteristic consensus sequence in subdomain I, GxxGxG, has been modified in the F10 sequence such that the first glycine (G) is replaced with a serine (S). The usually invariant sequence in subdomain VII (DFG) cannot be readily identified in F10, although D343FD or D345FS is the most likely substitute based on the predicted positions between preceding and subsequent subdomains. We have previously shown that F10 is a dual-specificity protein kinase (DSK) capable of phosphorylating Ser, Thr, and Tyr residues (13). Because no signature motifs have been identified that are diagnostic of DSK activity, a sequence analysis of the F10 protein was also not informative in this regard.
FIG. 1.
Alignment of F10 protein sequence with those of homologs encoded by other poxviruses. Using the Clustal method, we aligned the vaccinia virus (WR strain) (VV) F10 ORF with those of F10 homologs in monkeypox virus (MPV), camelpox virus (CML), variola virus (VAR), Yaba-like disease virus (YLD), sheeppox virus (ShP), swinepox virus (SPV), and molluscum contagiosum virus (MCV). Residues that are boxed and shaded in yellow are identical to those in vaccinia virus F10. Several conserved kinase subdomains are marked with purple boxes and Roman numerals, and residues that are changed in various ts F10 mutants (ts28, ts15, ts54, and ts61) are also indicated (red outlines). Positions at which N-terminal deletion mutants (discussed in Results) initiate are marked by inverted triangles.
There are four temperature-sensitive mutants from the Condit collection that map to a single complementation group affecting the F10 kinase: they are ts15, ts28, ts54, and ts61 (53, 55, 61). ts15 and ts28 form no macroscopic plaques at the nonpermissive temperature and show a 100- to 1,000-fold reduction in virus production at the nonpermissive temperature relative to infections performed at the permissive temperature. ts54 and ts61 have a leakier phenotype, forming small plaques at the nonpermissive temperature and showing a more modest reduction in virus production. In all cases, cells infected with the ts mutants at the nonpermissive temperature were shown to have completed genome replication and all phases of gene expression but demonstrated a profound defect in virion morphogenesis.
Virion morphogenesis proceeds through a succession of visually distinct stages. Proteins destined for encapsidation into virions coalesce into electron-dense foci known as virosomes. Membrane crescents appear at the periphery of these virosomes. The origin of these crescents, either de novo formation or the restructuring of intracellular membranes, as well as their structure, comprising one or two lipid bilayers, remains unresolved (11, 22, 40, 46, 47). Crescents enlarge, encircling a portion of the virosome, to form oval immature virions (IV). The viral DNA is then packaged into the IV, forming immature virions with nucleoids (IVn). The maturation of IVn into intracellular mature virions (IMV), the most abundant infectious form of the virus, is a complex and poorly understood process which involves proteolytic cleavage events as well as numerous structural changes.
Electron microscopic analyses of nonpermissive ts28 and ts15 infections indicated that an impairment of F10 causes an arrest of the infectious cycle at the earliest point in viral morphogenesis (55, 61). In the majority of cells, none of the visible hallmarks of morphogenesis, ranging from virosome formation to membrane biogenesis, were apparent. Instead, the cells contained only a cleared-out area of cytoplasm devoid of any cellular organelles (55). In more extensive analyses, we noted that some cells contained atypical “curdled” virosomes (A. Punjabi and P. Traktman, unpublished data, and this study).
Since the initial characterization of these ts F10 mutants, several viral proteins have been shown to be substrates for F10 in vivo and/or in vitro, and several of these proteins have been shown to play essential roles in virion morphogenesis. For example, the A14 and A17 proteins are essential components of the virion membrane; when either is absent, membranous vesicles and/or tubules accumulate and morphogenesis arrests prior to the development of functional crescents (42, 44, 56). A14 is phosphorylated on Ser residues in an F10-dependent manner, and A17 is phosphorylated on Ser, Thr, and Tyr residues in an F10-dependent manner (5, 13, 35). The A30, G7, and H5 proteins have been implicated in virosomal function and are all destined for encapsidation in the virion core (12, 48, 49, 52). Evidence for the phosphorylation of these proteins by F10, in some cases in vivo and in other cases in vitro, has also been obtained (51; J. Mercer, A. Punjabi, and P. Traktman, unpublished data). One ts H5 mutant has been generated and characterized; during nonpermissive infections, all biochemical events of the life cycle are completed normally, but morphogenesis arrests at an early stage, with the appearance of curdled virosomes and the absence of any evidence of membrane biogenesis (12). The phenotype of this mutant, therefore, is quite similar to that observed for the ts F10 mutants. The repression of either A30 or G7 also causes the appearance of atypical virosomes as well as the accumulation of aberrant membrane precursors (48, 52).
Despite the extensive progress made in cataloging and characterizing the proteins involved in virion biogenesis, the question of how F10 drives the initiation of morphogenesis remains unresolved. Moreover, the role played by protein phosphorylation itself has not been clarified. The studies described herein were designed to answer these two questions. We present a characterization of F10's intracellular localization, a demonstration that the proteins encoded by the ts F10 mutants are either thermolabile or catalytically inactive, an analysis of the phenotype seen upon repression of F10 via the use of a tetracycline (TET)-inducible recombinant virus, the development of a transient complementation system to enable further structure-function analysis of F10, and evidence that F10 is also involved in later stages of viral morphogenesis.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, Taq DNA polymerase, T4 DNA ligase, calf intestinal phosphatase, pancreatic RNase, and DNA molecular weight standards were purchased from either Roche Molecular Biochemicals (Indianapolis, Ind.) or New England Biolabs, Inc. (Beverly, Mass.) and were used as specified by the manufacturer. Cytosine β-d-arabinofuranoside (araC) and rifampin (RIF) were obtained from Sigma Chemical Co. (St. Louis, Mo.). 35S-labeled methionine, 32P-labeled nucleoside triphosphates, and orthophosphoric acid (32PPi) were purchased from Perkin-Elmer Life Sciences, Inc. (Boston, Mass). Lipofectamine Plus and protein molecular weight markers were acquired from GIBCO BRL Life Technologies (Gaithersburg, Md.) or Invitrogen Life Technologies (Carlsbad, Calif.) and used as specified. 14C-labeled markers were purchased from Amersham Pharmacia Biotech (Piscataway, N.J.).
Cells and viruses.
BSC40 African green monkey kidney cells and human thymidine kinase-negative (TK−) cells were maintained as monolayer cultures in Dulbecco modified Eagle medium (GIBCO BRL Life Technologies) containing 5% fetal calf serum. Viral stocks of wt vaccinia virus (strain WR), ts15, ts28, ts54, and ts61 (kind gifts of R. Condit) were prepared by ultracentrifugation of infected cytoplasmic extracts through a 36% sucrose cushion. The titers of all viral stocks were determined by plaque assays performed on BSC40 cells. For analyses of ts mutants, 31.5 and 39.7°C (referred to as 40°C in the figures) served as the permissive and nonpermissive temperatures, respectively.
Primers.
Construction of the plasmids and viruses described below involved the primers listed in Table 1.
TABLE 1.
Primers used for this study
| Primer | Sequencea |
|---|---|
| pATH constructs | |
| pATH U | 5′-GTGGATCCGAGAGAAAGGACAGTAT-3′ |
| pATH D | 5′-GCTCTAGATTAGTTTCCGCCATTTA-3′ |
| pET constructs | |
| pET U | 5′-CGGGATCCATATGGGTGTTGCCAATGA-3′ |
| pET/pUC D | 5′-CGGGATCCTTAGTTTCCGCCATTTAT-3′ |
| ts28 U | 5′-CGGATCCCATATGGGTGTTGCCAA-3′ |
| ts28 D | 5′-CGGATCCCATATGAAGAATCCGTTTATACAGAGT-3′ |
| ts15 ups | 5′-GAAATTTGGATACTGTGAACTCCG-3′ |
| ts15 dns | 5′-TCACAGTATCCAAATTTCTATAC-3′ |
| pTM constructs | |
| pTM U | 5′-ATGGATCCATGGGTGTTGCCAATGA-3′ |
| pTM D | 5′-CCATATCCTCCTGTAGA-3′ |
| FLAG U | 5′-GAAGATCTCCATGGACTACAAAGAC-3′ |
| FLAG D | 5′-ATGGATCCATATGCTTGTCATCGTCATCCT-3′ |
| pUC constructs | |
| pUC U | 5′-CCATCGATGGGTGTTGCCAATGA-3′ |
| pET/pUC D | 5′-CGGGATCCTTAGTTTCCGCCATTTAT-3′ |
| K→A ups | 5′-ATACAAATGCTACTACATAGTTATCTA-3′ |
| K→A dns | 5′-TGTAGTAGCATTTGTATTCGAGGCCAC-3′ |
| D→G ups | 5′-GTTTAAGACCTGCGTGTAAAAAGTTGT-3′ |
| D→G dns | 5′-ACACGCAGGTCTTAAACCCGATAA-3′ |
| ΔN91 U | 5′-CCATCGATGGATTTTTATCACATATC-3′ |
| ΔN69 U | 5′-CCATCGATCATATGACACTACTAAAGATAGA-3′ |
| ΔN23 U | 5′-CCATCGATCATATGTTAGGAGACTGTTTGTA-3′ |
| vΔiF10 constructs | |
| iF10-1 | 5′-CCATCGATTAGTTTCCGCCATTTA-3′ |
| iF10-2 | 5′-GCGGATCCAAGCTTATCAAACCTGGACTTT-3′ |
| iF10-B | 5′-ATCCCTATCAGTGATAGAGAATGGGTGTTGCCAATGAT-3′ |
| iF10-C | 5′-CTATCACTGATAGGGATATTTATTGTACAAAAAGC-3′ |
| F9-1 | 5′-ATGGTACCGCACTAGGTACATCG-3′ |
| F9-2 | 5′-GCGGATCCCAATTAGTTTCTTGG-3′ |
| F11-1 | 5′-GCGGATCCAAGCTTACAAAAAGCCCCAATT-3′ |
| F11-2 | 5′-GCTCTAGACACAGGCAACAATTGG-3′ |
Restriction sites are underlined. Nucleotide changes are indicated in bold. For the construction of the pTM3X-FLAG vector, sequences from the termini of the 3X-FLAG epitope were included in the FLAG U and D primers (bold), and the complement of the ATG (CAT) introduced by the NdeI site is italicized in FLAG D. For the vΔiF10 construct, the 19-bp tet operator sequence introduced by iF10-B and -C is shown in bold and italicized.
Preparation of recombinant His-tagged F10 kinase.
To incorporate the ts28 mutation (T504A) into the F10 open reading frame (ORF), we took advantage of an internal NdeI site at position 508 of the F10 ORF. A 513-bp 5′ fragment of F10 was amplified with the ts28 U and ts28 D primers (to incorporate the ts28 mutation and introduce NdeI sites at both ends). The fragment was digested with NdeI and cloned into the pET16b-F10 plasmid, which had been similarly digested. To facilitate the cloning of other F10 variants with the restriction enzyme NdeI, we introduced a silent mutation to remove the internal NdeI site (C508ATATG→CAAATG) from the F10 ORF. The resulting F10ΔNde ORF fragment was digested with NdeI and BamHI and cloned into pET16bF10 to generate pET16bF10ΔNde. Variants encoding ts15-F10, D307G-F10, and K110A-F10 were generated either by overlap PCRs (using ts15 ups and ts15 dns as internal primers [Table 1]) or PCRs using the corresponding pUC1246-F10 constructs (see below) as templates. As described previously (13), these constructs were maintained in Escherichia coli BL21(DE3), and mid-log-phase cultures were induced to express N′His-F10 proteins, which were then purified by chromatography on Ni-nitrilotriacetic acid agarose (Qiagen, Valencia, Calif.).
Construction of recombinant viruses vTM-F10 and vTM-3XFLAG-F10. (i) Plasmid construction.
The F10 ORF was subcloned into the expression vector pTM1 (15). A 5′ 280-bp F10 fragment was amplified by the use of primers pTM U and pTM D, which introduced an upstream NcoI and a downstream SfcI site, respectively. A 3′ 1,050-bp SfcI/BamHI F10 fragment was released from the pUC1246-F10 plasmid (see below). After digestion of the 5′ 280-bp fragment with NcoI and SfcI, the two fragments were mixed and ligated to pTM1 vector DNA that had been digested with NcoI and BamHI, generating pTM-F10.
For the pTM-3XFLAG-F10 construct, we first generated the pTM-3XFLAG vector by amplifying the FLAG sequence from the p3XFLAG vector (Sigma Chemical Co.) with the FLAG U and FLAG D primers (Table 1). These primers inserted an NcoI site (upstream) and BamHI and NdeI sites (downstream). The NdeI site was positioned such that the ATG codon was in frame with the 3XFLAG epitope sequences. The fragment was digested with NcoI and BamHI and ligated to pTM1 vector DNA that had been previously digested with the same enzymes. An NdeI/BamHI fragment containing the entire F10 ORF was subcloned from pET16bΔNde-F10 into the resulting pTM-3XFLAG vector. In some cases, constructs containing N′ deletions of F10 (ΔN′F10; see below) were also subcloned into pTM-3XFLAG and used for the subsequent construction of vTM-F10 viruses.
(ii) Recombinant virus construction.
The TK sequences found in the pTM1 vectors enable the insertion of T7-regulated F10 genes into the TK locus of the viral genome by homologous recombination. pTM-F10 and pTM-3XFLAG-F10 were linearized with ScaI and introduced into wt-infected cells as previously described (34), but with the Lipofectamine Plus reagent used for transfection. TK− viruses were plaque purified on human TK− cells in the presence of bromodeoxyuridine (BrdU) (25 μg/ml). Viral isolates that direct the expression of a 47-kDa (F10) or 52-kDa (3XFLAG-F10) protein upon coinfection with vTF7.5 were used for overexpression studies.
Purification of FLAG-F10 and FLAG-ΔN′F10 proteins that were overexpressed by use of the vaccinia virus-T7 system.
BSC40 cells were coinfected with vTF7.5 and either vTM-F10, vTM-3XFLAG-F10, or vTM-3XFLAG-ΔN′F10 (multiplicity of infection [MOI] of 2 to 5 for each). Infections were allowed to proceed for 8 h. The cells were harvested, washed with phosphate-buffered saline (PBS), and lysed with FLAG lysis buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). The lysates were mixed in an end-over-end rotator at 4°C for 15 min and clarified by centrifugation at 8000 × g at 4°C for 15 min. FLAG-agarose beads (Sigma Chemical Co.) were added, and incubation was allowed to proceed overnight at 4°C with end-over-end rotation. The beads were retrieved, washed with FLAG-TBS (50 mM Tris-Cl [pH 7.4], 150 mM NaCl), and eluted with the 3XFLAG elution peptide (150 μg/ml). Eluates were analyzed for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were tested for kinase activity.
Construction of kinase-null and N′ deletion mutants of F10.
The wt F10 ORF and five mutant alleles designed to introduce mutations at key catalytic residues or to remove portions of the N terminus of F10 were amplified by PCR from the vaccinia virus HindIII F fragment or the pET16b-F10ΔNde construct (see above). For the K110A and D307G mutations, overlap PCR was used to incorporate the internal substitutions. The following two sets of primer pairs were used to amplify the F10 ORF: (i) the upstream pUC U primer and a primer which introduces the complement of the mutation (K→A or D→G ups) and (ii) a primer which introduces the mutation (K→A or D→G dns) and the downstream pUC D primer. PCRs performed with these primer pairs yielded products that overlapped by 17 nucleotides (nt). A mixture of these products then served as the template for a second round of PCR performed with the pUC U and D primers. The resulting PCR products were digested with ClaI and BamHI (sites introduced by pUC U and D, respectively) and cloned into a similarly digested pUC1246 vector, which placed the F10 allele downstream of the cowpox ATI promoter (35, 37). For the wt and N′ deletion constructs, the relevant upstream primers were used with the pUC D primer in single PCRs to generate products which were similarly introduced into pUC1246. These constructs, pUC1246 D307G-F10, K110A-F10, ΔN23-F10, ΔN69-F10, and ΔN91-F10, were subjected to DNA sequencing prior to being used in transient complementation studies as described below.
Transient complementation analysis of catalytic and structural F10 mutants.
Confluent 35-mm-diameter dishes of BSC40 cells were infected with ts28 at an MOI of 5 and maintained at 39.7°C. At 6 h postinfection (hpi), constructs (5 μg) expressing wt F10 or different catalytic (D307G and K110A) or structural (ΔN′) mutants of F10 were introduced by transfection with the Lipofectamine Plus reagent. The cells were harvested at 28 hpi and then titrated at 31.5°C in order to monitor the restoration of virus production during the nonpermissive ts28 infection. Expression of the F10 protein from the transfected plasmids was confirmed by immunoblot analysis performed with an anti-F10 antiserum.
Construction of vTKiF10.
Overlap PCR was used to generate a 1.4-kb F10 fragment which included the F10 ORF and 48 bp of upstream regulatory sequence (towards F11) and to insert the 19-bp tet operator sequence between the transcriptional and translational start sites of the F10 gene. All primers used are listed in Table 1. Primers iF10-1 and iF10-B were used to amplify a 1.3-kb fragment (F10 ORF plus 19-bp tet operator sequence), while primers iF10-C and iF10-2 amplified a 67-bp fragment (upstream 48 bp plus 19-bp tet operator sequence). The resulting PCR products were used as templates in a second round of PCR amplification with the outside primers (iF10-1 and -2). The final PCR product was digested with HindIII and ClaI (sites introduced by the outside primers) and ligated to similarly digested pJS4/tetR vector DNA (58). The final plasmid carried the tet repressor under the control of a vaccinia virus constitutive promoter and the tet-inducible F10 allele under the regulation of its own promoter. The pJS4tetR↔iF10 construct (5 μg) was linearized with ScaI and introduced at 3 hpi into BSC40 cells infected with the wt virus (MOI = 0.03) by use of the Lipofectamine Plus reagent. The cells were harvested at 48 hpi, and the TK− virus was purified by two rounds of plaque purification on TK− cells in the presence of BrdU (25 μg/ml). The identification of plaques in which the tetR↔iF10 cassette had become inserted into the genomic TK locus by homologous recombination was performed by PCRs with tet repressor-specific primers (58).
Construction of vΔiF10.
For construction of a cassette which would enable the insertion of the Neor gene in place of the endogenous F10 locus, regions of the flanking F9 and F11 genes were amplified with the primers listed in Table 1. F9-1 and -2 were used to amplify a 352-bp fragment of F9 from the HindIII F fragment of the vaccinia virus genome and to introduce unique BamHI and Asp718 sites at the 5′ and 3′ termini of the F9 fragment, respectively. F11-1 and -2 were used to amplify a 335-bp fragment of F11 and to introduce unique XbaI and BamHI-HindIII sites at the 5′ and 3′ termini of the F11 fragment, respectively. These PCR products were digested with Asp718 and BamHI and with BamHI and XbaI, respectively, and then ligated simultaneously to pBSIIKS DNA that had been previously digested with Asp718 and XbaI. This construct, which contained abutted F9 and F11 sequences in the same orientation as that found in the genome, was then digested with BamHI and HindIII to allow the insertion of a 1.3-kb HindIII/BamHI fragment containing the neo gene under the regulation of the viral p7.5 promoter (26, 33). The insert in the final plasmid contained 352 bp of F9, 1.3 kb of neo, and 335 bp of F11. Cells were infected with vTKiF10 at an MOI of 0.03 and transfected at 3 hpi with the F9-neo-F11 construct (5 μg), which had been linearized with ScaI. From this point on, TET (1 μg/ml) was included in the culture medium. At 20 hpi, the medium was replaced with fresh medium that also contained G418 (2.5 mg/ml), and the infection was allowed to proceed for another 48 h. The cells were then harvested, and G418r viruses were isolated by two rounds of plaque purification in the presence of G418. Plaques chosen for expansion were tested to confirm the replacement of the endogenous F10 allele with the Neor cassette by two PCR strategies. These included the ability to amplify a Neo-F11 fragment as well as the inability to amplify an F10-F11 fragment. Plaques containing the correct genomic structure were expanded, and the virus was designated vΔiF10.
Preparation of polyclonal anti-F10 antiserum.
The 3′ half of the F10 ORF, nt 595 to 1320, was amplified by use of the primers pATH U and pATH D (Table 1), with the HindIII F fragment of the vaccinia virus genome as a template. The primers introduced an upstream BamHI site and a downstream XbaI site (underlined in Table 1). The 725-bp product was digested with BamHI and XbaI and ligated to pATH11 (27) vector DNA which had been similarly digested. E. coli HB101 transformants containing the pATH11-F10 construct directed the expression of a 64-kDa TrpE fusion protein containing 37 kDa of TrpE and 27 kDa of F10. The band corresponding to the F10-TrpE fusion protein was excised from an SDS-PAGE gel and used to immunize rabbits. Antibody specificity was determined by immunoblot analyses of lysates of vaccinia virus-infected BSC40 cells, purified virions, and E. coli cells expressing recombinant F10 kinase.
Immunoblot analysis.
Cell extracts were fractionated by SDS-PAGE, and the proteins were transferred electrophoretically to nitrocellulose (Schleicher & Schuell, Keene, N.H.) in Tris-glycine buffer (25 mM Tris, 192 mM glycine, 20% methanol). Blots were incubated with an anti-F10 serum (1:500 dilution) or an anti-D8 serum (a kind gift of Ed Niles, SUNY at Buffalo) and subsequently incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (1:20,000) (Bio-Rad Laboratories, Hercules, Calif.). In some cases, blots were developed with the His-probe reagent (Pierce, Rockford, Ill.). Immunoreactive proteins were visualized by chemiluminescence with Super Signal reagents (Pierce).
Lipid binding analysis.
The ability of purified 3XFLAG-F10 and the N′ deletion mutants to interact with various lipids was determined by “fat Western” analysis. PIP strips (Echelon BioSciences, Salt Lake City, Utah) were blocked for 1 h in PIP-TTBS (10 mM Tris [pH 8.0], 150 mM NaCl, 0.1% Tween 20) supplemented with 3% fatty acid-free bovine serum albumin (BSA) (Sigma). Filters were then incubated overnight with the protein of interest (∼80 ng/ml in PIP-TTBS plus BSA) at 4°C with gentle agitation. The strips were washed well with PIP-TTBS and then developed with an anti-FLAG antibody (except where noted) followed by a horseradish peroxidase-conjugated secondary antibody; the bound protein was visualized by chemiluminescence as described above. Quantitation of the chemiluminescent signal was performed on an Alpha Innotech FluorChem 8800 documentation system.
Metabolic labeling and immunoprecipitation analysis.
At designated times postinfection, infected cells were rinsed with methionine- or phosphate-free Dulbecco modified Eagle medium and then incubated with the same medium containing 100 μCi of [35S]methionine or [32P]orthophosphoric acid/ml, respectively. The cells were harvested and prepared for immunoprecipitation analysis as previously described (26). In some cases, lysates were precleared with protein A-Sepharose beads (Sigma) prior to immunoprecipitation. After incubation with either preimmune, anti-F10, or anti-FLAG antiserum, complexes were captured with protein A-Sepharose beads and washed extensively. Immunoprecipitates were resolved by SDS-PAGE and visualized by fluorography (35S) or autoradiography (32P).
Immunofluorescence.
BSC40 cells grown on Lab-Tek four-well chamber slides (Nalge Nunc International, Naperville, Ill.) or in 12-well plates (Corning Incorporated, Corning, N.Y.) were coinfected with vTF7.5 and vTM-F10 or vTM-3XFLAG-F10 at an MOI of 2 to 5 (each) for 8 h. The cells were fixed with 4% (vol/vol) paraformaldehyde in PBS and then permeabilized with 0.1% (vol/vol) Triton X-100 prior to being processed for immunofluorescence. The cells were blocked in 5% (wt/vol) BSA-PBS and then incubated with the primary antiserum at a dilution of 1:75 (anti-F10) or 1:1,000 (anti-p63 or anti-ERGIC53) in 1% (wt/vol) BSA-PBS for 1 h. After several washes with PBS, the cells were incubated with a fluor-conjugated secondary antibody (goat anti-rabbit Alexa fluor 488 or goat anti-mouse Alexa fluor 594 [Molecular Probes, Eugene, Oreg.]) in 1% (wt/vol) BSA-PBS. The cells were washed extensively before viewing under a Diaphot 200 inverted fluorescence microscope (Nikon, Inc., Melville, N.Y.). Images were captured with a SPOT-2 camera system (Diagnostic Instruments, Inc.) and were analyzed with Adobe Photoshop (Adobe Systems, Inc., San Jose, Calif.).
Kinase assays.
The activity of purified F10 (4 to 20 ng for 3XFLAG-F10 preparations and 35 to 100 ng for N′His-F10 preparations) was assayed in reaction mixes (25 μl) containing 50 mM Tris (pH 7.4), 10 mM MgCl2, 5 μM [γ-32P]ATP, 1 mM dithiothreitol, and 2.5 μg of myelin basic protein (MBP). The reactions were incubated for 30 min at room temperature, stopped by the addition of 5× protein sample buffer (1× PSB is 50 mM Tris [pH 6.8], 1% SDS, 1% β-mercaptoethanol, 10% glycerol), and resolved electrophoretically. Phosphorylated MBP was visualized by autoradiography.
Electron microscopy.
Confluent 60-mm-diameter dishes of BSC40 cells were infected with the vtetR, ts28, or vΔiF10 virus at an MOI of 5 and incubated at 31.5, 37, or 39.7°C for the indicated times. Cells were fixed in situ with 2% glutaraldehyde in PBS for 30 min on ice, scraped gently, and collected by centrifugation. Fresh fixative was added, and the samples were then processed for conventional transmission electron microscopy as previously described (12). Thin sections were examined with a Hitachi H-600 transmission electron microscope operating at 75 kV.
TX-114 and sodium carbonate extraction of PNS from infected cells.
BSC40 cells were infected with wt virus at an MOI of 10 and harvested at 8 hpi. The cells were washed in PBS, resuspended in 600 μl of 10 mM Tris, pH 9, and then disrupted with a Dounce homogenizer. Lysates were clarified by low-speed centrifugation, and the postnuclear supernatant (PNS) was subjected to either TX-114 extraction or sodium carbonate extraction. For TX-114 extraction, the PNS was solubilized with an equal volume of buffer containing 10 mM Tris (pH 10), 300 mM NaCl, and 2% TX-114 and then incubated at 37°C for 10 min before being subjected to sedimentation at 1,000 × g for 10 min at room temperature to separate the phases (10). The extraction was repeated with the aqueous phase, while the detergent phase was re-extracted with 10 volumes of buffer containing 10 mM Tris (pH 10), 150 mM NaCl, and 1 mM EDTA. For alkaline carbonate extraction (17), the PNS was adjusted to pH 11 by the addition of an equal volume of 200 mM Na2CO3 and was incubated on ice for 30 min before being subjected to centrifugation in an Optima TL ultracentrifuge (Beckman Instruments Inc., Palo Alto, Calif.) at 150,000 × g for 30 min to sediment the membranes. Samples were concentrated by acetone precipitation when required, and equivalent amounts were fractionated by SDS-PAGE and subjected to immunoblot analysis with the indicated antisera.
Computer analysis.
Autoradiographic films were scanned with a SAPHIR scanner (Linotype Hell Co., Hauppauge, N.Y.) and adjusted with Adobe Photoshop (Adobe Systems, Inc.) or Canvas 8.0 (Deneba Systems, Miami, Fla.) software. Plaque assays were photographed with an AlphaImager (Alpha Innotech) digital camera. Figures were labeled with Canvas 8.0. Alignments were prepared with Lasergene (DNASTAR Inc., Madison, Wis.), using sequences obtained from www.poxvirus.org.
RESULTS
Synthesis, modification, and intracellular localization of F10 protein. (i) Temporal expression and posttranslational modification of F10 protein.
To facilitate analyses of the expression, processing, and localization of wt and mutant forms of F10 in vivo, we developed a polyclonal anti-F10 antibody. The most effective antigen was found to be a fusion protein containing the E. coli TrpE protein fused to the C-terminal half (residues 199 to 439) of the F10 protein. Reactivity against F10 was confirmed by immunoblot analyses of bacterial lysates expressing the recombinant His-tagged F10 protein, purified virions, and extracts of infected cell lysates (not shown). The apparent molecular mass of the F10 protein found within infected cells and within purified virions was determined to be 47.4 kDa, while recombinant His-F10 migrated with an apparent molecular mass of 53.9 kDa.
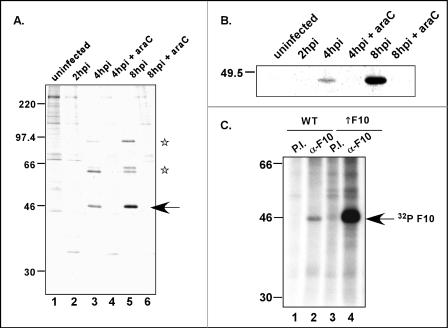
The initiating ATG codon of the F10 ORF lies within the consensus sequence TAAATG, which is the hallmark of late promoters. To monitor the temporal profile of F10 synthesis, we pulse labeled cells with [35S]methionine at different times postinfection, retrieved the F10 protein by immunoprecipitation, and analyzed the protein by SDS-PAGE and fluorography. An ∼46-kDa band was recognized by the F10 antiserum in infected but not uninfected cell extracts (Fig. 2A). The protein was first detected at 4 hpi, but the signal was more intense at 8 hpi. No synthesis was seen when infections were performed in the presence of the DNA replication inhibitor araC, confirming that the expression of F10 is indeed part of the late, postreplicative phase of infection. Two other proteins, of ∼60 and ∼90 kDa, were also immunoprecipitated by the F10 antiserum (Fig. 2A, lanes 3 and 5). An analysis of total 35S-labeled lysates (not shown) revealed that these proteins comigrate with abundant late proteins and presumably represent the major core proteins p4a (95 kDa when uncleaved) and p4b (65 kDa when uncleaved) (59, 60). Whether the coimmunoprecipitation of these proteins with F10 reflects bona fide protein-protein interactions or is merely an artifact of their high abundance remains to be determined.
FIG. 2.
F10 is expressed as a late protein and is phosphorylated in infected cells. (A) Cells were infected with wt virus at an MOI of 5 in either the presence or absence of araC (50 μM) and were maintained at 37°C. At the indicated times postinfection, the cells were labeled with [35S]methionine for 1 h and then harvested as described in Materials and Methods, and extracts were immunoprecipitated with the anti-F10 antiserum. An ∼46-kDa band (arrow) was retrieved from wt-infected cells but not from uninfected cells. Coimmunoprecipitating bands at ∼60 and ∼90 kDa (stars) are believed to represent highly abundant core proteins. (B) Thirty-five-millimiter-diameter dishes of BSC40 cells were infected as described above (MOI = 10) but were instead processed for immunoblot analysis. The same pattern of expression was observed. (C) The F10 protein is phosphorylated in vivo. Cells infected with wt virus (MOI = 5) (lanes 1 and 2) or coinfected with vTF7.5 and vTM-F10 (lanes 3 and 4) (MOI = 2.5 [each]) were labeled with 32PPi from 3 to 12 hpi; lysates were subjected to immunoprecipitation with either preimmune (lanes 1 and 3) or anti-F10 (lanes 2 and 4) antiserum. The anti-F10 serum specifically retrieved [32P]F10 (arrow) from both lysates. The electrophoretic profile of protein standards is shown at the left in panels A and C; molecular masses are indicated in kilodaltons.
When cultures from infected cells were harvested at various times postinfection and subjected to immunoblot analysis with the anti-F10 antiserum, similar profiles of expression were observed. F10 accumulation began at approximately 4 hpi in wt-infected cells, with more protein having accumulated by 8 hpi (Fig. 2B). F10 did not accumulate in infections performed in the presence of araC, again supporting the observation that F10 is a late protein.
The F10 protein has been shown to undergo autophosphorylation in vitro (30, 61). To determine if F10 is phosphorylated in vivo, we metabolically labeled cells that were infected with wt virus or coinfected with vTF7.5 and vTM-F10 with 32PPi. The latter infection led to the T7-induced ∼10-fold overexpression of F10. Extracts prepared from these infections were subjected to immunoprecipitation analyses with both preimmune and anti-F10 antisera. As shown in Fig. 2C, a specific 32P-labeled protein of the expected size was retrieved from both lysates by the anti-F10, but not the preimmune, serum; as expected, the signal was greatly amplified when F10 was overexpressed. From these data, we conclude that the F10 protein is indeed phosphorylated in vivo.
(ii) The F10 protein colocalizes with cellular markers of the ER and ERGIC.
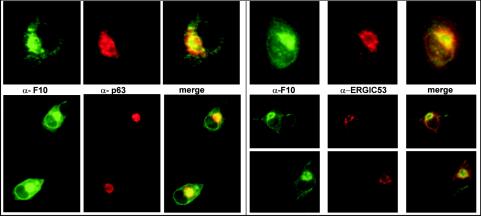
The early stages of virion morphogenesis are thought to intersect with the cellular membrane machinery (40, 46). Moreover, the A14 and A17 proteins, which are F10 substrates and play essential roles in the biogenesis of the virion membrane, appear to insert cotranslationally into the endoplasmic reticulum (ER) (43, 45). Therefore, it was intuitively appealing to propose that F10 localizes to this region of the cell. Attempts to visualize F10 by immunofluorescence analysis of wt-infected cells were not successful, most probably due to an inadequate sensitivity of our anti-F10 antiserum. To boost the levels of intracellular F10 protein, we coinfected cells with vTM-F10 and vTF7.5 for 8 h prior to permeabilizing and processing them for immunofluorescence microscopy using the anti-F10 serum. F10 appeared to be concentrated in the perinuclear region and showed a reticular pattern of cytoplasmic localization. Samples were simultaneously labeled with antibodies directed against markers of the ER and the ER-Golgi intermediate compartment (ERGIC). The perinuclear staining of F10 (Fig. 3, green) was found to overlap with that seen for the ER marker (α-p63, red) and the ERGIC marker (α-ERGIC53, red), as evidenced by the merged images (Fig. 3, rightmost panels). No colocalization was observed with markers of the Golgi apparatus (not shown). These data indicated that at 8 hpi, F10 is concentrated at or near the intracellular membrane systems of the ER and ERGIC.
FIG. 3.
F10 localizes to the perinuclear region of cells, colocalizing with ER- and ERGIC-specific markers. BSC40 cells infected under conditions which direct the overexpression of the F10 protein were fixed and permeabilized at 8 hpi and subjected to immunofluorescence analysis with the anti-F10 antiserum (green staining). The infected cells were also labeled with an anti-p63 antibody (left panels, red staining), specific for the ER, or an anti-ERGIC53 antibody (right panels, red staining), specific for the ERGIC. The merged images on the right illustrate that F10 shows significant colocalization with these ER- and ERGIC-specific markers.
(iii) The F10 protein is tightly associated with membranes in wt-infected cells.
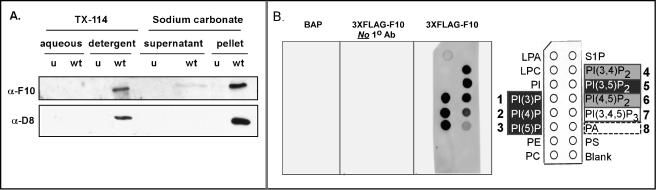
As described above, the pattern of intracellular localization of the F10 protein appeared to coincide with that of the ER and ERGIC markers. To determine whether the F10 protein was indeed membrane associated, we subjected wt-infected cells to two fractionation protocols and monitored the partitioning of the F10 protein by immunoblot analysis. Extraction of the PNS of infected cells with TX-114 leads to the partitioning of proteins between the detergent (membrane-associated, hydrophobic, and amphipathic proteins) and the aqueous (soluble, hydrophilic proteins) phases (6, 10). Alkaline carbonate extraction of the PNS followed by high-speed sedimentation separates integral membrane proteins from peripheral membrane proteins and soluble proteins (17). Both methods were utilized to test whether F10 is indeed associated with intracellular membranes. As shown in Fig. 4A, F10 was present in the detergent fraction after TX-114 extraction of the PNS. After alkaline carbonate extraction of the PNS, the vast majority of F10 was present in the pellet (membrane fraction), although a low level of F10 reactivity was seen in the supernatant (soluble fraction). In both cases, F10 fractionation mimicked that of a known component of the virion membrane, the D8 protein (36). Taken together, these observations strongly support the conclusion that F10 is primarily associated with intracellular membranes at 8 hpi.
FIG. 4.
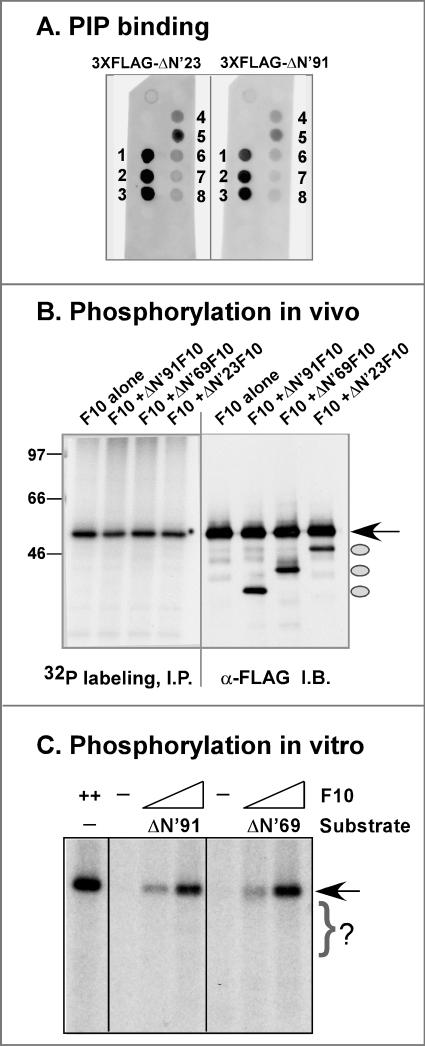
(A) F10 is tightly associated with membranes during wt infections. Cells were left uninfected (u) or infected with wt virus at an MOI of 5 and were harvested at 8 hpi. A PNS was prepared and subjected to either TX-114 or sodium carbonate (Na2CO3) extraction as described in Materials and Methods. Equivalent amounts of the aqueous and detergent phases (TX-114 extraction) and the supernatant and pellet fractions (Na2CO3 extraction) were resolved by SDS-12% PAGE and transferred to a nitrocellulose filter. Immunoblot analysis was performed with the anti-F10 antiserum. The lower half of the blot was probed with an antiserum specific for a known viral membrane protein (D8) as a control for extraction conditions. The F10 protein partitioned into the detergent phase upon TX-114 extraction of the PNS and into the pellet after Na2CO3 extraction of the PNS. (B) F10 binds phosphoinositides in vitro. PIP strip membranes containing immobilized phosphoinositides (Echelon Biosciences) were incubated overnight with 80 ng of purified FLAG-F10 protein/ml (middle and right blots) or with the control protein 3XFLAG-BAP (left blot). For visualization of the bound FLAG-F10, an immunoblot analysis was then performed with the anti-FLAG antibody (left and right blots), an appropriate secondary antibody, and chemiluminescent development. The immobilized species were as follows: LPA, lysophosphatidic acid; LPC, lysophosphocholine; PI, phosphoinositide; PI(3)P, phosphoinositide-3-phosphate; PI(4)P; PI(5)P; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PI(3,4)P2; PI(3,4)P2; PI(4,5)P2; PI(3,4,5)P3; PA, phosphatidic acid; and PS, phosphatidylserine. The chemiluminescent signals were quantitated, and the hierarchy of binding is shown schematically as follows: highest binding, spots 1, 2, 3, and 5 (white text in black boxes); strong binding, spots 4 and 6 (gray boxes); modest binding, spot 7 (white box); and minimal binding, spot 8 (dashed box).
(iv) The F10 protein binds phosphoinositides in vitro.
An analysis of the predicted amino acid sequence of F10 (Kyte-Doolittle hydropathy plot analysis [not shown]) indicated that the protein is mainly hydrophilic. Neither transmembrane domains (PROSITE database analysis) nor membrane targeting sequences (PSORT database analysis) have been identified by computer algorithms. Several cellular proteins are known to associate with membranes by virtue of binding to lipids such as phosphoinositides. There are numerous phosphoinositide binding domains, such as the PH domain, the FYVE domain, and the more recently identified PX domain (29). To determine whether the observed association of F10 with membranes could be explained by its ability to bind to phosphoinositides, we performed a lipid binding assay in which PIP strips (nitrocellulose membranes containing immobilized phosphoinositides and phospholipids) were incubated first with the purified FLAG-F10 protein and then with an anti-FLAG antibody. Using this fat Western approach, we found that the recombinant FLAG-F10 protein bound to various lipid species, with the hierarchy of signal strength being PI(4)P = PI(5)P > PI(3)P = PI(3,5)P2 ≫ PI(3,4)P2 = PI(4,5)P2 ≫ PI(3,4,5)P3 ≫ PA (Fig. 4B). No binding was seen for the other species immobilized on the filter. Moreover, no signal was obtained when an unrelated FLAG-tagged protein (FLAG-bovine alkaline phosphatase) served as the primary probe or when the primary antiserum was omitted during development. These data indicate that F10 binds specifically to a subset of phosphoinositides, raising the possibility that F10 may associate with membranes via its interactions with lipids.
What features are required for F10's biological function, and what stages of viral morphogenesis are dependent upon F10? (i) Characterization of the F10 proteins encoded by ts mutants defective in F10.
A complementation group comprising four temperature-sensitive mutants with defects in the F10 kinase has been described previously (55, 61). Phenotypic characterizations of nonpermissive infections performed with these ts mutants have shown that F10 plays an essential role at the earliest stages of viral morphogenesis. Sequencing of the lesions within the F10 ORFs of these ts mutants revealed that the mutations cause amino acid substitutions at the following highly conserved residues: for ts28, Val168→Ile; for ts15, Pro131→Ser; for ts54, Pro85→Ser; and for ts61, Glu123→Lys (61) (Fig. 1). As a means of elucidating the important properties of wt F10, we wanted to characterize the catalytic and/or structural deficiencies of the proteins encoded by the ts F10 alleles.
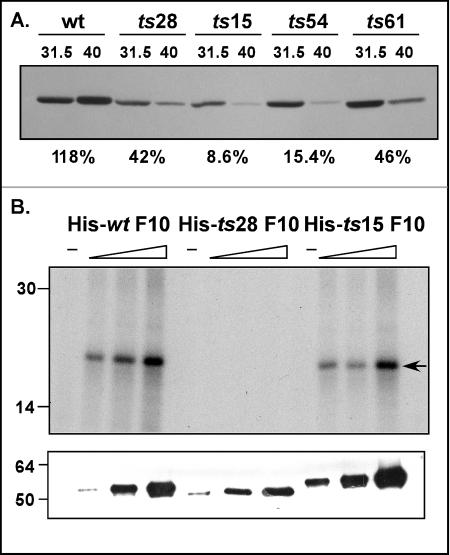
To establish whether the phenotypic defect in the ts F10 mutants was due to thermolability of the mutant proteins, we prepared extracts at 12 hpi from cells infected with ts28, ts15, ts54, and ts61 under permissive and nonpermissive conditions. These extracts were subjected to immunoblot analysis with the anti-F10 antibody. The amino acid substitutions in the ts F10 alleles affect the N-terminal half of the protein (61) (Fig. 1) and are therefore unlikely to affect recognition by the anti-F10 antibody, which was raised against the C-terminal half of the protein. As shown in Fig. 5A, at 39.7°C, the wt F10 protein accumulated to levels even higher than those seen at 31.5°C (118%). However, accumulation of the ts15 and ts54 F10 proteins was appreciably compromised at the nonpermissive temperature, with the levels of F10 at 39.7°C being reduced to 8.6% (ts15) and 15.4% (ts54) of those seen at the permissive temperature. The accumulation of the F10 proteins expressed by ts28 and ts61 was modestly affected at the nonpermissive temperature, with the levels at 39.7°C being reduced to 42% (ts28) and 46% (ts61) of those seen at 31.5°C. It is interesting that the proteins expressed by ts15 and ts54 contain amino acid substitutions at Pro residues, which suggests that these substitutions induce structural changes that contribute to the thermolability of the proteins (61). Because ts15 has a tight ts phenotype in vivo, we examined the thermolability of this protein in more detail (55, 61). Cultures were subjected to pulse-chase analysis as described previously (26); the radiolabeled F10 protein present within each sample was retrieved by immunoprecipitation and quantitated by SDS-PAGE and spot densitometry. The half-life (t1/2) of the ts15 F10 protein at 39.7°C was calculated to be ∼30 min, while its t1/2 at the permissive temperature was ∼75 min (not shown). The t1/2 of the wt F10 protein was ∼75 min at either temperature, comparable to that seen for the ts15 F10 protein at the permissive temperature.
FIG. 5.
(A) F10 proteins encoded by ts15 and ts54 are thermolabile in vivo. Cells were infected with wt virus or the tsF10 mutant viruses at an MOI of 5 and maintained at either 31.5 or 40°C for 12 h. Extracts were subjected to immunoblot analysis with the anti-F10 antiserum; the relative amount of F10 protein seen in each lane was determined by spot densitometry. For each virus, the ratio of the amount of F10 accumulated at 40°C to that accumulated at 31.5°C is shown as a percentage below the relevant lanes of the immunoblot. (B) Recombinant His-ts28 F10 lacks detectable kinase activity in vitro. Purified N′ His-tagged versions of wt F10, ts28 F10 (Val167→Ile), and ts15 F10 (Pro131→Ser) were assayed in vitro for protein kinase activity, using the generic substrate MBP. For each enzyme preparation, three assays containing increasing amounts of enzyme (∼35 to 140 ng) were performed. Reactions were fractionated in an SDS-15% PAGE gel and analyzed by autoradiography (top); phosphorylated MBP is indicated by the arrow. The specific activities of the wt F10 and ts15 F10 preparations were comparable, whereas the ts28 F10 preparation had no detectable enzymatic activity (<2% of the wt activity). An immunoblot developed with a His probe (bottom) confirmed that similar amounts of the three preparations were used in the assays. The migration of protein standards is shown at the left, with molecular masses indicated in kilodaltons.
Of the four ts mutants in the F10 complementation group, ts28 and ts15 display the tightest ts phenotypes (55, 61). It is notable that whereas the protein expressed by ts15 was markedly thermolabile in vivo, the protein expressed by ts28 was far less so: at 39.7°C, the amount of F10 which accumulated was only 2.4-fold less than that seen under permissive conditions. We have previously shown that an approximately fourfold reduction in the level of wt F10 protein has a minimal impact on virus production (B. Unger and P. Traktman, unpublished data). Wang and Shuman (61) reported that recombinant N′ His-tagged versions of the F10 proteins encoded by ts54 and ts61 exhibited 30 and 50% of the specific kinase activity of wt N′His-F10, respectively, but they did not address the proteins encoded by ts28 and ts15. We therefore generated pET16b plasmids encoding N′ His-tagged versions of the F10 proteins encoded by ts28 (Val168→Ile) and ts15 (Pro131→Ser). E. coli transformants bearing these plasmids or a comparable plasmid encoding N′His-wt F10 were induced at 18°C to enable the expression of F10 proteins, which were purified by Ni2+ affinity chromatography. The three proteins were tested for kinase activity in vitro, using the generic substrate MBP. As shown in Fig. 5B, increasing levels of phosphorylated MBP were seen in reactions containing increasing amounts of N′His-wt F10 or N′His-ts15 F10, but essentially no MBP phosphorylation was seen when equivalent amounts of N′His-ts28 F10 were used. In summary, the lesion in ts28 eliminates catalytic activity, while the protein encoded by the ts15 allele is markedly thermolabile in vivo. Therefore, a significant reduction in either the abundance (∼12-fold) or the catalytic activity (∼50-fold) of the F10 protein is sufficient to block virion morphogenesis.
(ii) Does repression of F10 recapitulate the ts F10 phenotype?
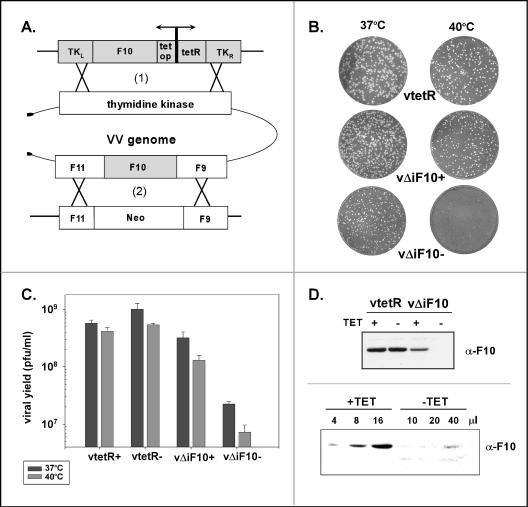
The data described above for ts15 support the hypothesis that a marked reduction in F10 levels arrests the progression of virion morphogenesis at a very early stage. To enable the controlled depletion of wt F10, we chose to construct a TET-inducible recombinant virus in which the expression of F10 is dependent upon the inclusion of tetracycline in the medium. A copy of the F10 gene under the regulation of its own promoter and the tet operator was inserted into the TK locus along with a cassette directing the constitutive expression of the tet repressor. This construct was transfected into wt-infected BSC40 cells, and the intermediate virus was then plaque purified on TK− cells via selection with BrdU. Once this first recombinant was isolated, the endogenous F10 allele was replaced by the gene conferring G418 resistance (neo). The resulting F10-inducible virus is referred to as vΔiF10. In this genomic configuration, read-through transcripts from upstream genes did not compromise the Tetr-mediated repression of F10 expression; we have used the same approach to analyze the viral A13 protein (58).
To characterize the plaque-forming ability of vΔiF10, we infected cells with an equal amount of vtetR or vΔiF10 in the presence or absence of TET; infected cells were incubated at either 37 or 39.7°C for 48 h prior to staining with crystal violet. The stimulus for investigating the phenotype at both temperatures was the previously published observation that repression of the A30 gene resulted in temperature-dependent ultrastructural changes (52). As shown in Fig. 6B, vtetR produced large plaques at both temperatures in the presence or absence of TET. In the presence of TET, vΔiF10 produced large plaques at 37°C and slightly smaller plaques at 39.7°C. In the absence of TET, vΔiF10 produced small plaques at 37°C and fewer, pinpoint plaques at 39.7°C. To extend these observations, we infected cells with vtetR or vΔiF10 in the presence or absence of TET for 24 h at either 37 or 39.7°C (MOI = 5). The viral yield was then determined by titration in the presence of TET at 37°C. The viral yield from cells infected with vΔiF10 in the absence of TET at 37°C was 45-fold lower than that obtained from parallel infections performed with vtetR (Fig. 6C); at 39.7°C, a 74-fold difference in viral yields was observed.
FIG. 6.
Construction and phenotypic analysis of TET-inducible vΔiF10 recombinant virus. (A) Schematic illustration of virus construction. An inducible copy of the F10 gene, regulated by its own promoter and by the tet operator (tet op), was inserted into the nonessential TK locus of the viral genome along with a copy of the tet repressor (tetR) under the regulation of a constitutive promoter (1). Once this intermediate virus was isolated, the endogenous F10 allele was replaced with the gene conferring G418 resistance (Neo) (2). The resulting virus is referred to as vΔiF10. Details on the construction of this virus are provided in Materials and Methods. (B) Repression of F10 expression inhibits plaque formation in a temperature-dependent manner. BSC40 cells were infected with an equivalent number of PFU of vtetR (expresses the tet repressor) or the inducible recombinant vΔiF10 in the presence (+) or absence (−) of TET and were maintained at either 37 or 40°C for 48 h prior to staining with crystal violet. Relative to the plaques seen in the other infections, the plaques formed by vΔiF10 in the absence of TET showed a modest reduction in size at 37°C and a dramatic reduction in size at 40°C. (C) Repression of F10 expression causes a decrease in viral yield at 24 h. Cells were infected with the indicated viruses (with or without TET) at an MOI of 5 and were maintained at either 37 or 40°C for 24 h; viral yields were determined by titration at 37°C in the presence of TET. The data represent the averages of two experiments. The viral yields from vΔiF10 infections in the absence of TET performed at 37 and 40°C were <3% and <2%, respectively, of the yields obtained from the comparable vtetR infections. (D) F10 protein accumulation is repressed in cells infected with vΔiF10 in the absence of TET. (Top) BSC40 cells were infected with vtetR or vΔiF10 (with or without TET) and were incubated at 40°C. The cells were harvested after 24 h, and lysates were fractionated in an SDS-12% PAGE gel and subjected to immunoblot analysis with the anti-F10 antiserum. Only the relevant portion of the blot is shown. F10 accumulation was diminished in samples prepared from vΔiF10 infections in the presence of TET and was undetectable in samples prepared from vΔiF10 infections in the absence of TET. (Bottom) Serial dilutions of extracts prepared from vΔiF10 (with or without TET) infections were fractionated and analyzed by immunoblot analysis with the anti-F10 antiserum. The amount of extract loaded (in microliters) is shown above each lane. Comparable levels of F10 protein were seen with 40 μl of the vΔiF10 −TET sample and with 4 μl of the vΔiF10 +TET sample.
To monitor the levels of F10 protein that accumulated during infections performed under induced and repressed conditions, we infected cells as described above and then subjected lysates to immunoblot analysis with the polyclonal anti-F10 antiserum (Fig. 6D). F10 accumulated to similar levels during vtetR infections whether or not TET was included in the culture medium. When TET was present during vΔiF10 infections, the level of F10 accumulation was approximately fivefold lower than that seen for vtetR infections. In contrast, no F10 protein could be detected when a sample from the vΔiF10 infection in the absence of TET was analyzed.
To further quantitate the TET dependence of F10 expression during vΔiF10 infections at 37°C, we examined serial dilutions of these extracts by immunoblot analysis. As shown in Fig. 6D, 4 μl of the +TET extract appeared to contain an equivalent amount of F10 as 40 μl of the −TET extract, indicating that the omission of TET decreased the steady-state levels of F10 10-fold. Since 5-fold less F10 protein accumulated in vΔiF10 infections in the presence of TET than in vtetR infections (see above), we concluded that ∼50-fold less F10 protein is found in cells infected with vΔiF10 in the absence of TET than in those infected with vtetR.
Our expectation was that a controlled repression of F10 would cause an arrest in the viral life cycle similar to that seen during nonpermissive infections with ts15 and ts28. An electron microscopic analysis of cells infected with vΔiF10 in the absence of TET indicated that this was largely the case (not shown). At 40°C, only the earliest stages of morphogenesis were seen; ∼70% of the cells exhibited no signs of viral membrane biogenesis, although many did contain curdled virosomes. However, ∼30% of the cells examined also contained membrane crescents and a few immature virions (IV). In summary, these data suggest that the repression of F10 leads to a somewhat leakier phenotype than that observed for ts28: the expression of ∼2% of the wt F10 level may be less deleterious than the expression of a stable but catalytically impaired protein. Similar observations were reported by the Moss laboratory for a recombinant virus encoding an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible F10 gene (50).
(iii) ts F10 mutants are also impaired in later stages of morphogenesis.
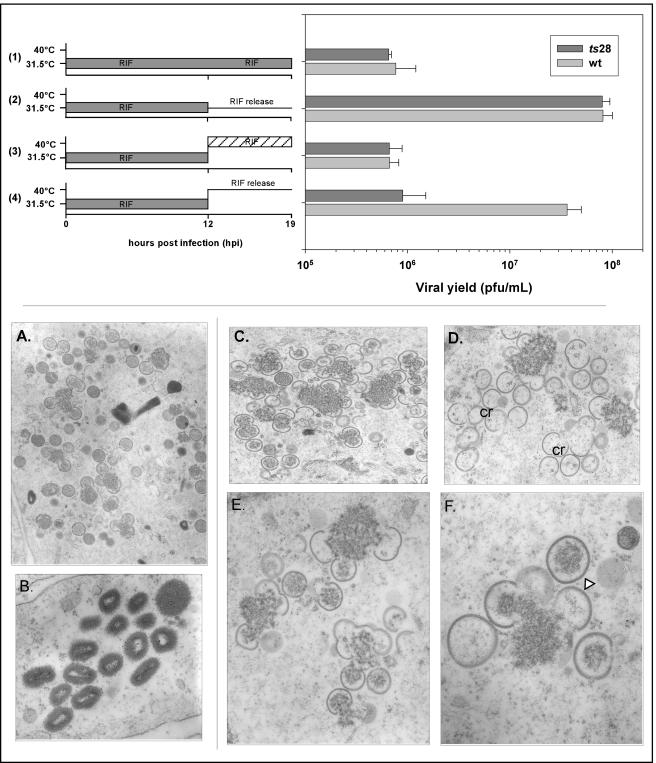
One of the intrinsic caveats for the use of ts mutants to elucidate the function of a protein is that only the first lethal phenotype is uncovered. Hence, we know that in the absence of active F10, morphogenesis arrests prior to the appearance of any discernible viral membranes. Because of this early arrest, any role of F10 in later facets of virion assembly or infectivity has remained unexplored. To approach this question, we performed the following experiment: cultures were infected with wt virus or ts F10 mutants and maintained at the permissive temperature for 12 h in the presence of RIF. This protocol leads to a synchronous and characteristic arrest in morphogenesis at the RIF-sensitive step. Cells accumulate virosomes that are surrounded by precursor membranes which lack the D13 protein and hence fail to acquire the rigidity and curvature of normal crescents (62). At 12 hpi, these cultures were either harvested immediately or fed with fresh medium ± RIF and then were maintained at the permissive temperature or shifted to the nonpermissive temperature. Cultures were harvested 3 h after RIF release for examination by electron microscopy and 7 h after RIF release for determination of the viral yield. For wt- and ts28-infected cultures maintained at the permissive temperature throughout the experiment, incubation in the absence of RIF from 12 to 19 hpi led to an ∼100-fold increase in the viral yield (Fig. 7, row 2). In contrast, for cultures that were shifted to the nonpermissive temperature at 12 hpi, this recovery of virus production was restricted to wt-infected cultures (Fig. 7, row 4). These data strongly suggest that active F10 is also required for events in morphogenesis that occur subsequent to the RIF block. Electron microscopic analyses of ts28-infected cells that were shifted to the nonpermissive temperature upon release from the RIF block provided further support for this hypothesis (Fig. 7C to F). At 3 h postshift, virosomes had a curdled appearance and were surrounded both by empty crescents and by crescents that were only loosely associated with virosomal contents. Few, if any, immature or mature virions were observed. In contrast, identically treated wt-infected cultures contained the full spectrum of normal morphogenetic intermediates, including virosomes, crescents engulfing virosomal contents, immature virions with or without nucleoids, and mature virions (Fig. 7A and B). These data support an essential role for the F10 kinase in the incorporation of virosomal material into nascent virions.
FIG. 7.
Rifampin release experiments reveal that the ts28 mutation also impairs later stages of virion morphogenesis. (Top) Production of infectious virus does not resume in ts28-infected cells shifted to 40°C after release from a rifampin block. BSC40 cells were infected with wt virus or ts28 (MOI = 5) at 31.5°C in the presence of 100 μg of RIF/ml. At 12 hpi, the cells were either maintained at 31.5°C (rows 1 and 2) or shifted to 40°C (rows 3 and 4). In half of the cultures, RIF was removed and cultures were fed with fresh, prewarmed medium (rows 2 and 4, RIF release). The other half of the cultures (rows 1 and 3) were maintained in the presence of RIF. Cells were then harvested at 19 hpi (7 h after RIF washout and/or temperature shift), and lysates were titrated at 31.5°C in the absence of RIF. A graphic representation of the viral yields is shown. Whereas wt-infected cultures recovered from the RIF block when they were shifted to 40°C in the absence of drug, ts28-infected cultures did not (row 4). (Bottom) Empty crescents and abnormal immature virions accumulate in ts28-infected cells shifted to 40°C after release from a rifampin block. BSC40 cells were infected with wt virus or ts28 (MOI = 5) at 31.5°C in the presence of RIF; at 12 hpi, the cultures were shifted to 40°C in the absence of RIF. At 3 h postshift, the cultures were fixed and processed for transmission electron microscopy. wt cultures recovered fully from the RIF treatment, with morphogenesis proceeding through the formation of immature (A) and mature (B) virions. In contrast, only a partial recovery was seen in ts28-infected cells, as curdled virosomes, empty crescents (cr), and empty or partially filled immature virions (triangle) were seen (C to F). Normal immature virions and mature virions were not formed. Magnification: ×12,000 (A), ×30,800 (B), ×15,800 (C), ×16,900 (D), ×25,000 (E), or ×40,000 (F).
(iv) Transient complementation analysis shows that the catalytic activity of F10 is essential for its biological function.
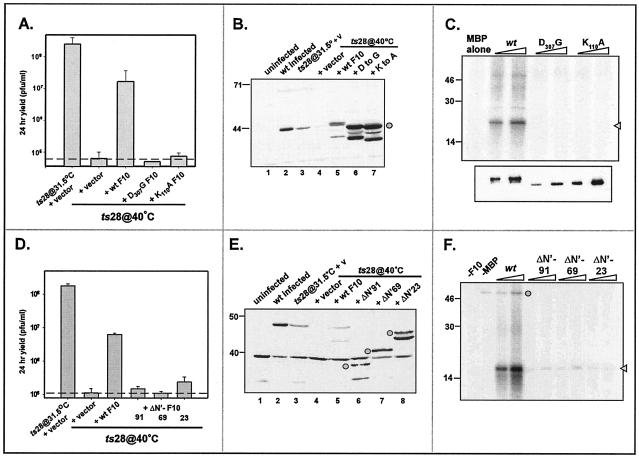
The F10 protein encoded by ts28 is stable in vivo but lacks kinase activity in vitro. These data suggest that the catalytic activity of F10 is essential for its biological function. To test this hypothesis more directly and to determine which regions or residues within the F10 protein are important for its function, we performed a transient complementation assay. Briefly, plasmids directing the expression of wt or mutant copies of the F10 kinase were transfected into cells infected with ts28 at the nonpermissive temperature. The ability of these constructs to complement the ts28 defect was determined by measuring the viral yield at 24 h. Initially, we compared the complementation activities of an empty vector, a plasmid encoding wt F10, and plasmids encoding mutant F10 alleles containing substitutions in residues that play essential roles in catalysis, i.e., K110 (Walker B box) and D307 (catalytic loop). As shown in Fig. 8A, the expression of wt F10 was able to restore virus production during the nonpermissive ts28 infection, with the viral yield being almost 2 orders of magnitude higher than that seen with the negative control. In contrast, the K110A and D307G alleles were unable to complement nonpermissive ts28 infections. To confirm that all of the proteins were expressed equally in these experiments, we performed immunoblot analyses of lysates prepared from the infected or transfected cells (Fig. 8B). The mutant F10 proteins had indeed accumulated to levels that matched or exceeded that of the wt protein and had the predicted molecular weights. We also examined the impact of transient expression of the wt and mutant F10 proteins during infections at 37 or 39.7°C with the wt virus; the viral yield was not affected by the transient expression of any of these proteins (not shown), ruling out the possibility that they exerted a dominant-negative phenotype. To verify that the mutations in the critical kinase domain residues did in fact abolish the kinase activity, we produced recombinant N′ His-tagged F10 proteins containing the K110A and D307G mutations and tested them in kinase assays by use of the generic substrate MBP. In contrast to N′His-wt F10, neither N′His-K110A F10 nor N′His-D307G F10 showed any detectable kinase activity in this assay (Fig. 8C). These data indicate that the catalytic activity of the F10 kinase is critical for its biological function.
FIG. 8.
Transient complementation of nonpermissive ts28 infections. (A to C) The kinase activity of F10 is required for its biological function in vivo. BSC40 cells were infected with ts28 (MOI = 5) at 40°C and then transfected at 6 hpi with an empty vector or plasmids directing the expression of either wt F10 or kinase-null alleles of F10 (K110A F10 or D307G F10). As a positive control, an additional culture was infected at 31.5°C and transfected with an empty vector. At 28 hpi, the cells were harvested and viral yields were titrated at 31.5°C to evaluate the restoration of virus production (A). The data represent the average of two experiments, and the dashed line indicates the baseline titer obtained from cells transfected with the empty vector. Only the wt F10 construct restored virus production in the context of a nonpermissive ts28 infection. Extracts were also subjected to immunoblot analysis with the anti-F10 antiserum to verify the stable expression of the plasmid-borne proteins (B, lanes 5 to 7, circle). Endogenous F10 protein was also detected in wt-infected cells (lane 2) or cells infected with ts28 at the permissive temperature (lane 3). To verify that the K110A and D307G substitutions did in fact disrupt enzymatic activity, we tested equivalent amounts of purified recombinant N′His-wt F10, -K110A F10, and -D307G F10 proteins for their ability to phosphorylate MBP in vitro (C); an autoradiograph of the kinase assays (top) and an immunoblot developed with a His probe (bottom) are shown. No phosphorylation of MBP was seen in the reactions performed with N′His-K110A or -D307G F10. (D to F) The N terminus of F10 is required for its biological function in vivo and its enzymatic activity in vitro. Cells were infected as described above and then transfected with an empty vector or plasmids directing the expression of either wt F10 or alleles containing N-terminal deletions (ΔN′-F10) of 91, 69, and 23 aa. The 28-h viral yield was determined by titration at 31.5°C (D), and the stable expression of the various proteins was confirmed by immunoblot analysis with the anti-F10 serum (E) (the ΔN′-F10 proteins are indicated by circles). The ∼38-kDa protein seen in all samples in panel E represents a cross-reactive cellular protein. Only the wt F10 construct restored virus production in the context of a nonpermissive ts28 infection. To test the impact of the N-terminal deletions on enzymatic activity, we tested increasing amounts of purified 3XFLAG-tagged wt F10 and ΔN-F10 proteins (4 to 20 ng) for their ability to phosphorylate MBP in vitro (F). Phosphorylated MBP is indicated by the arrowhead; autophosphorylated 3XFLAG-F10 is indicated by the circle. The ΔN-F10 proteins showed significant reductions in specific activity compared to wt F10. For panels B, C, E, and F, the electrophoretic migration of protein standards is shown at the left, with molecular masses indicated in kilodaltons.
(v) The extracatalytic N-terminal 91 amino acids of the F10 protein are crucial for its biological function in vivo and its kinase activity in vitro.
F10 contains 91 amino acids (aa) upstream of the minimal kinase domain. To test whether this region of the protein is essential in vivo, we prepared three constructs in which the N-terminal 23 amino acids (ΔN23-F10), the N-terminal 69 aa (ΔN69-F10), or the entire extracatalytic N-terminal region of F10 (ΔN91-F10) was deleted. None of these constructs were able to rescue virus production in the context of a nonpermissive ts28 infection (Fig. 8D), indicating that the entire N-terminal region plays a critical role in the protein's function. Immunoblot analyses of lysates prepared for these transient rescue experiments indicated that all of these truncated forms of F10 were stably expressed (Fig. 8E). As described above, we also determined that the ΔN-F10 proteins did not exert a dominant-negative effect during wt infections (not shown). Because we encountered difficulty in producing recombinant N′ His-tagged ΔN-F10 proteins in E. coli, we utilized a different method for producing the ΔN-F10 recombinant proteins. We generated vaccinia virus recombinants encoding 3XFLAG-tagged versions of wt F10 and the ΔN-F10 ORFs under the regulation of the T7 promoter (15). Upon coinfection of cells with these recombinants and vTF7.5, a virus which expresses the T7 RNA polymerase, a robust expression of these 3XFLAG-ΔN F10 proteins was induced. The 3XFLAG-tagged F10 proteins were purified by affinity chromatography, and their catalytic activities were tested in kinase assays performed with MBP as a substrate (Fig. 8F). Compared to the wt protein, the 3XFLAG-ΔN F10 proteins showed severely impaired kinase activities (specific activities were reduced ≥10-fold). This result was unanticipated because these proteins retained all of the domains that are known to be required for kinase activity. Nevertheless, it appears that the N terminus of F10 is required, either directly or indirectly, for its enzymatic activity.
To further investigate how the N terminus of F10 might contribute to its function, we investigated the ability of the truncated forms to interact with phospholipids and to undergo phosphorylation in vivo and in vitro. Purified preparations of 3XFLAG-ΔN23 F10 and 3XFLAG-ΔN91 F10 were analyzed by the fat Western assay described above. As shown in Fig. 9A, both preparations retained the ability to interact with the same spectrum of lipid species as that bound by the wt protein (compare Fig. 9A and Fig. 4B, which were probed and developed simultaneously). Quantitation of the signals seen for the wt, ΔN23, and ΔN91 proteins indicated that the hierarchy of binding was largely conserved; although higher levels of overall binding were observed for the wt protein in this particular experiment, this difference was not seen consistently.
FIG. 9.
Deletion of the N terminus of F10 does not block protein-lipid interactions but compromises phosphorylation. (A) Protein-lipid overlay. 3XFLAG-ΔN′23 F10 and 3XFLAG-ΔN′91 F10 were analyzed in a fat Western assay (see Fig. 4B for the wt 3XFLAG-F10 panel from the same experiment). The lipids to which the proteins were found to bind, numbered 1 to 8, are further described in Fig. 4B and its legend. (B) Phosphorylation in vivo. BSC40 cells were coinfected with vTF7.5 and vTM-3XFLAG-F10 or were triply infected with these two viruses and either vTM-3XFLAG-ΔN′23 F10, 3XFLAG-ΔN′69 F10, or 3XFLAG-ΔN′91 F10 (MOI = 2 for each virus). The cells were metabolically labeled with 32PPi from 3.5 to 9 hpi, and cell lysates were then subjected to immunoprecipitation with the anti-FLAG serum. The immunoprecipitates were resolved electrophoretically and either visualized by autoradiography (left) or further analyzed by immunoblot analysis with the anti-FLAG serum (right). Full-length 3XFLAG-F10 is indicated by the arrow; the ovals indicate the ΔN′23, ΔN′69, andΔN′91 forms of the 3XFLAG-F10 protein. The numbers at the left indicate the molecular masses (in kilodaltons) of electrophoretically resolved protein standards. Only the full-length form was phosphorylated in vivo. (C) Phosphorylation in vitro. Kinase reactions were performed with or without wt 3XFLAG-F10 (5 or 20 ng) and with or without purified 3XFLAG-ΔN′91 F10 or 3XFLAG-ΔN′69 F10 (65 to 100 ng) as a substrate in the presence of [γ-32P]ATP. Samples were resolved electrophoretically and visualized by autoradiography. Autophosphorylated 3XFLAG-F10 is indicated by the arrow; although the ΔN′ proteins were present in molar excess and would migrate within the region indicated by the bracket, no phosphorylated bands corresponding to these species were seen.
As discussed above (Fig. 8F), the N-terminally truncated proteins lacked significant kinase activity even though they retained the motifs associated with catalysis. We considered the possibility that deletion of the N terminus might remove the site of an activating, autophosphorylation event. To assess the ability of these proteins to undergo trans-phosphorylation in vivo, we overexpressed them along with 3XFLAG-wt F10 in the presence of 32PPi. Immunoprecipitation was then performed with the anti-FLAG serum, and the retrieved proteins were visualized by autoradiography; the accumulation of all of the F10 species was confirmed by immunoblot analysis. As shown in Fig. 9B, only the full-length F10 protein was radiolabeled in vivo, indicating that wt 3XFLAG-F10 could not trans-phosphorylate 3XFLAG-ΔN′23F10, -ΔN′69F10, or -ΔN′91 F10. A similar result was obtained when purified preparations of the proteins were analyzed in vitro, as shown in Fig. 9C; the ΔN′69 and ΔN′91 proteins were not phosphorylated in trans by full-length F10, even when they were present in a >30-fold molar excess.
What trafficking and signaling pathways are involved in the F10-dependent process of IMV assembly? (i) Development of a temperature release assay for ts28, with synchronous resumption of morphogenesis upon shift from 39.7 to 31.5°C at 12 hpi.
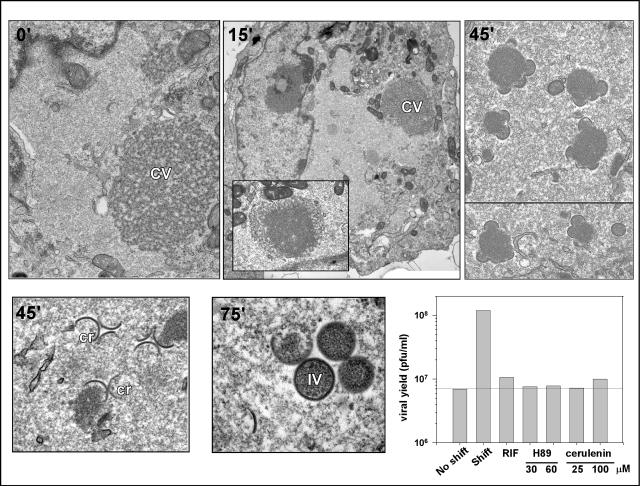
As described above and in earlier published reports (55, 61), morphogenesis arrests at a very early stage during ts28 infections performed at 39.7°C: virosomes are either absent or appear curdled, and no intermediates in viral membrane biogenesis are seen. We hypothesized that we might be able to observe a synchronous resumption of morphogenesis upon release of the temperature block. We therefore performed a series of shift-down experiments with ts28-infected cells and analyzed the progression of morphogenesis by electron microscopy. After 12 h of infection at the nonpermissive temperature, the cultures were either fixed immediately or shifted to 31.5°C for 15, 45, or 75 min. In the 39.7°C culture, no morphogenesis intermediates were seen. A large, cleared area of cytoplasm was observed, while in some cells, curdled virosomes were also seen, often in the proximity of numerous mitochondria (Fig. 10). After 15 min of incubation at 31.5°C, the curdled virosomes appeared to be regaining a uniform electron density, beginning within the interior of the virosome and then extending towards the periphery. After 45 min at 31.5°C, the first signs of membrane biogenesis were observed, with crescents appearing at or near the virosomes. Morphogenesis appeared to be well under way by 75 min after the temperature shift, as evidenced by the presence of numerous virosomes with adjacent crescents and the appearance of some immature virions. To confirm that this morphological recovery was biologically relevant, we monitored the production of infectious virus after the temperature release. We found that by 6 to 7 h postrelease, the yield of virus was at least 10-fold higher than that obtained from cultures maintained at the nonpermissive temperature (not shown). This assay therefore provided an opportunity to evaluate the pathways involved in moving forward from the point of ts28 arrest to the formation of mature, infectious, intracellular virions.
FIG. 10.
Synchronous recovery of viral morphogenesis upon shift of ts28-infected cultures from 40 to 31.5°C at 12 hpi: dissection of pharmacological sensitivities by electron microscopic visualization of recovery. Cells were infected with ts28 at 40°C (MOI = 5) for 12 h and then shifted to 31.5°C; at 0, 15, 45, or 75 min postshift, cells were fixed and processed for transmission electron microscopy. Prior to the temperature shift (0′), cells displayed the characteristic phenotype of tsF10 infections at 40°C, containing cleared-out areas of cytoplasm devoid of any cellular organelles. In some cells, curdled virosomes (CV) were also apparent. At 15 min postshift (15′), no clear signs of morphogenesis were seen, although smoothing out of the curdled virosomes was observed in most cells (inset). Virosomes surrounded by crescents (cr) were apparent at 45 and 75 min postshift (45′ and 75′), with immature virions (IV) first appearing at 75 min postshift. Magnification: ×13,300 (0′), ×5,500 (15′), ×10,200 (inset, 15′), ×13,800 (45′, upper right panel, top image), ×12,700 (45′, upper right panel, bottom image), 28,500 (45′, lower left panel), or 37,400 (75′). The titration of viral yields following release in the absence or presence of pharmacological inhibitors was also performed. Cells were infected as described above for 12 h and then either maintained at 40°C or shifted to 31.5°C in the absence of inhibitors or the presence of 100 μg of RIF/ml, 30 or 60 μM H89 (blocks the formation of ER exit sites), or 25 or 100 μM cerulenin (inhibitor of de novo lipid synthesis). Cells were harvested 7 h after the temperature shift, and the viral yield was determined by titration at 31.5°C. H89, cerulenin, and rifampin blocked the restoration of virus production after the temperature shift.
(ii) Release from ts F10 block requires de novo lipid synthesis and is sensitive to the kinase inhibitor H89.
The mechanism by which F10 diverts vesicles and/or tubules from the cellular membrane machinery for use in virion assembly is currently unknown. Neither treatment with brefeldin A or nordihydroguiaretic acid nor the use of a cell line encoding a ts allele of ɛ-COP had any impact on IMV formation (not shown). This process was also unaffected by the expression of dominant-negative mutants of Sar1p (23; data not shown). Taken together, these data provide strong evidence that vaccinia virus membrane biogenesis does not require the COPI or COPII vesicle systems. To obtain new insights into the mechanisms that might be involved in virion assembly, we used our synchronous temperature release assay to test the impact of various pharmacological agents on the resumption of virion morphogenesis. Cells were infected nonpermissively with ts28 for 12 h and then shifted to the permissive temperature for 7 h in the presence of H89, a protein kinase inhibitor which has been shown to interfere with the formation of ER exit sites (2, 28), or cerulenin, an inhibitor of de novo lipid synthesis (41). As controls, parallel cultures were shifted to the permissive temperature in the presence of RIF, which arrests morphogenesis at a stage later than the ts28 arrest but prior to IV formation, or in the absence of any inhibitors. As shown in Fig. 10, release in the absence of a drug led to an ∼10-fold increase in the viral yield. The addition of H89 or cerulenin was as effective at blocking virus production as the addition of RIF. These data indicate that virion morphogenesis involves de novo lipid synthesis and relies on an H89-dependent signaling pathway.
(iii) Repression or impairment of F10 kinase alters the phosphoprotein profile of infected cells.
To date, the search for F10 substrates has focused on viral proteins known to play roles in morphogenesis (13, 56). It is likely, however, that the reorganization of intracellular membranes involves the F10-mediated phosphorylation of cellular proteins and perhaps of viral proteins that may not themselves be major components of nascent virions. We therefore compared the global phosphoprotein profiles of infected cells expressing sufficient wt F10 (vtetR), trace levels of wt F10 (vΔiF10 in the absence of TET), or catalytically deficient F10 (ts28 at 39.7°C). Cells infected under these three conditions were metabolically labeled with 32PPi from 4 to 8 hpi; cytoplasmic lysates were then fractionated by SDS-PAGE and analyzed by autoradiography. As shown in Fig. 11 and as would have been predicted, several phosphorylated species found in the vtetR-infected samples were absent from the lysates prepared from ts28 and vΔiF10 (−TET) infections (↓). However, the more striking observation was the increased intensity of several phosphoproteins in these F10-deficient lysates (↑) (compare lane 1 to lanes 2 and 3). The same pattern was seen when F10 protein levels were reduced (vΔiF10 infections in the absence of TET) or when F10's kinase activity was compromised (ts28 infections). This unanticipated result implies that F10 may exert its function by intersecting with other regulatory cascades, either through the activation of cellular phosphatases or through the inhibition of cellular kinases.
FIG. 11.
Repression or inactivation of F10 alters the phosphoprotein profile of infected cells. Cells were infected (MOI = 5) with either vtetR (lane 1), ts28 (lane 2), or vΔiF10 in the absence of TET (lane 3) at 40°C and were metabolically labeled with 32PPi from 4 to 8 hpi. Cytoplasmic extracts were fractionated in an SDS-15% PAGE gel and analyzed by autoradiography. While several phosphorylated proteins were found to decrease in intensity during nonpermissive vΔiF10 and ts28 infections (↓), the more striking result was the appearance of prominent phosphorylated proteins (↑) under these conditions (compare lanes 2 and 3 to lane 1). The migration of protein standards is shown on the left, with molecular masses indicated in kilodaltons.
DISCUSSION
Genetic analyses of the F10 kinase have shown that it plays an essential role at the onset of vaccinia virus assembly. Moreover, several of the key components of the virion membrane and core are indeed substrates of F10. Although it was presumed that the kinase activity of F10 was instrumental in orchestrating virion morphogenesis, this presumption had not been tested. Since ∼300 copies of the protein are encapsidated in mature virions, it was also possible that F10 plays a structural role in virion assembly. This present study was initiated in part to ascertain which properties of the F10 protein were important for its biological function. We developed several additional reagents for these analyses, including an anti-F10 antiserum and three viruses that facilitated the experimental manipulation of F10 levels, namely, a TET-inducible recombinant that enabled the repression of F10 and two T7-regulated recombinants that allowed the overexpression of F10 and 3XFLAG-F10.
We confirmed that, as expected, the F10 protein is expressed at late times after infection. Although F10 contains an internal A350GI motif, which conforms to the consensus site for cleavage by the viral I7 protease (7), it is not subjected to proteolytic processing. We and others have observed that purified and recombinant forms of F10 undergo autophosphorylation in vitro (30, 61). Here we showed, by using the anti-F10 antiserum that we were able to immunoprecipitate radiolabeled F10 from 32P-labeled lysates of wt-infected cells or cells induced to overexpress F10. Although we have confirmed that phosphorylation in vivo and in vitro occurs on Ser residues (not shown), the site(s) on which F10 undergoes phosphorylation and the impact of this modification on F10's activity remain to be determined. Interestingly, alleles of F10 containing N-terminal deletions of 23, 69, and 91 aa did not undergo phosphorylation in vivo or in vitro when coexpressed or assayed with wt F10. Although these truncations may alter the accessibility of an internal site of phosphorylation, the simplest interpretation is that the site(s) of phosphorylation is found within the N-terminal region preceding the catalytic domain. We have previously shown that F10 is a dual-specificity kinase, and the catalytic activities of other members of this enzyme class have been shown to be regulated by autophosphorylation (21, 25).
Immunolocalization of overexpressed F10 indicated that the protein was distributed in perinuclear foci that overlapped with the distribution of the cellular ER and ERGIC. This localization was certainly consistent with the central role of F10 in virion membrane biogenesis. Even more significant was the observation that, upon subcellular fractionation of wt-infected cells, F10 partitioned with the membrane fraction after TX-114 or Na2CO3 extraction of the PNS. These data indicate that F10 is in fact strongly associated with membranes. Although the F10 protein contains a relatively high percentage (38%) of hydrophobic residues, the sequence contains no discernible transmembrane domains. It seemed possible that the membrane association of F10 might be mediated by direct binding to lipids such as phosphoinositides, although the F10 sequence does not appear to contain any of the known PI-binding domains (24). We demonstrated empirically that recombinant FLAG-F10 binds PIPs in vitro. F10 shows selectivity, binding most strongly to the singly phosphorylated species PI(3)P, PI(4)P, and PI(5)P and the doubly phosphorylated species PI(3,5)P. This binding profile is not predictive of an association with any particular intracellular membrane compartment or of involvement with any distinct signaling pathway. Determining whether PIPs modulate the kinase activity of F10 or whether F10 has lipid kinase activity and alters the PIP profile of key intracellular membrane compartments will be important areas of future study. Furthermore, it will be important to test the role that PIP binding plays in the association of F10 with intracellular membranes. Finally, since F10 has been shown to interact with the core proteins A30 and G7 (51) and is itself found solely within the core of the mature virus particle (30), it will be important to determine how the seeming shift in F10's localization is regulated.
Placement of F10 at the top of the morphogenetic hierarchy was a consequence of analysis of the phenotype of nonpermissive infections performed with ts F10 mutants. In this study, we determined that the protein encoded by ts28 is relatively thermostable in vivo but lacks detectable kinase activity in vitro, whereas that encoded by ts15 is quite thermolabile in vivo but retains full enzymatic activity in vitro. These data indicated that compromising the level or enzymatic activity of the F10 protein can eliminate morphogenesis. To test this conclusion more directly, we developed a transient complementation assay for ts28 that enabled us to test the biological function of catalytically inert alleles of F10 and an inducible recombinant (vΔiF10) that enabled us to repress F10 in a controlled manner. Constructs expressing catalytically inactive F10 mutants were unable to complement a nonpermissive ts28 infection, whereas the transient expression of wt F10 did restore significant virus production. The catalytic activity of F10 is therefore essential to its biological function. Characterizations of infections performed with vΔiF10 in the absence of TET made it clear that the repression of F10 did not reproduce the precise phenotype observed with the tightest ts F10 mutants. ts15 and ts28 formed no plaques at 39.7°C, and the relative viral yields from 24-h infections performed at 39.7°C compared to those from infections performed at 31.5°C decreased by ∼3 orders of magnitude (55). In contrast, vΔiF10 infections performed in the absence of TET formed pinpoint plaques, and the relative viral yield from cells infected with vΔiF10 in the absence of TET compared to that from infections performed with the parental vtetR virus decreased 74-fold under the most stringent conditions (39.7°C). Apparently, a reduction in the level of wt protein does not have as significant an impact on the viral life cycle as does an impairment of the protein's catalytic activity. Comparable results and similar conclusions were recently reported by the Moss laboratory (50). This finding supports the model that the primary role of F10 is catalytic rather than structural.
The extracatalytic regions of kinases have been known to play a role in regulating the enzymatic activity or intracellular localization. The F10 protein contains a 91-aa N-terminal region outside its minimal kinase domain. Using our transient complementation system, we determined that the expression of several N-terminally deleted versions of F10 (ΔN′-F10) was unable to restore virus production in the context of a nonpermissive ts28 infection. The deletion of 23, 69, or 91 aa from the N terminus of the F10 protein abolished its biological function and, unexpectedly, its enzymatic activity. As stated above, the N terminus of F10 may contain a site(s) of regulatory autophosphorylation. Additionally, the N terminus may be required for the proper intracellular localization of F10. For example, the subnuclear localization of DYRK1, a DSK involved in brain development, depends on N-terminal elements outside the minimal kinase domain (4). In the case of Myt1Hu, a DSK involved in cdc2 regulation, a stretch of hydrophobic and uncharged residues at the C terminus is responsible for targeting to the ER and the Golgi apparatus (32). A possible role for the N terminus in F10 localization is supported by preliminary observations that a fusion protein comprising the N-terminal 91 aa of F10 and enhanced green fluorescent protein appears to localize to perinuclear regions of transfected cells (not shown).
F10's central role in the early events of membrane biogenesis is consistent with the observation that the A14 and A17 membrane proteins, which are required for membrane formation, are both F10 substrates. The observation that proteins involved in subsequent steps of morphogenesis are also phosphorylated in an F10-dependent manner (e.g., A30 and G7) (51; Mercer et al., unpublished data) and the fact that F10 is encapsidated in virion cores (30) stimulated the hypothesis that F10 might be involved in multiple facets of virion assembly. We therefore induced a synchronous arrest in viral morphogenesis at a point beyond the early ts28 arrest by infecting cells with the wt virus or ts28 at a low temperature in the presence of RIF. When both wt and ts28 infections were released from RIF arrest at a low temperature, morphogenesis resumed and culminated in the production of mature infectious virus within 7 h. However, when cultures were shifted to a high temperature at the time of RIF release, only the wt infections proceeded to form mature virions. In the ts28-infected cultures, the flaccid membranes that accumulated in the presence of RIF did recover their rigidity and curvature, but they appeared unable to associate with and engulf the virosomal contents. Moreover, the virosomes assumed the curdled appearance which has been seen previously during nonpermissive infections with ts F10, H5, and A30 mutants (12, 52; this report). No immature or mature virions developed. These data suggest that F10 mediates virosome structure and function, perhaps via known and predicted interactions with the H5, A30, and/or G7 proteins (51; Mercer and Traktman, unpublished data). Moreover, F10 appears to modulate the interaction of crescents with virosomal contents and to enable crescents to mature into the ovoid membrane that surrounds immature virions.
The mechanism through which F10 drives the assembly of the virion membrane remains to be discovered (47). Early investigators suggested that the membrane was assembled de novo in the cytoplasm, an idea which was then supplanted by the suggestion that the viral membrane was derived from the ER and ERGIC. However, neither brefeldin A nor dominant-negative forms of Sar1p inhibit viral morphogenesis. These data refute any role for the COPI or COPII vesicle systems, which comprise the ERGIC, in virion membrane biogenesis (23, 57). In order to address additional mechanisms by which F10 might mediate membrane formation, we took advantage of the reversibility of the ts28 phenotype (55) and performed shift-down experiments. Normal intermediates in membrane biogenesis could be observed by electron microscopy within 45 to 75 min after the shift to the permissive temperature, and an ∼10-fold increase in infectious virus was seen by 6 to 7 h after the shift. Shift-down experiments were also performed in the presence of a variety of pharmacological inhibitors, and H89 and cerulenin completely blocked recovery. Cerulenin inhibits de novo lipid biosynthesis by binding covalently to the active site of β-ketoacyl acyl carrier protein synthase, a key enzyme in the fatty acid synthesis pathway (38). Cerulenin has also been shown to inhibit viral RNA replication in several RNA viruses, including poliovirus, grapevine fanleaf virus, and cowpea mosaic virus, by hindering their ability to utilize ER-derived membranes for replication (8, 16, 41). Our data strongly suggest that viral membrane formation requires de novo lipid synthesis.
H89, originally identified as a protein kinase C (PKC) inhibitor, also inhibits PKA and PKD, albeit at different concentrations (nanomolar versus micromolar range) (9, 28). H89 does not inhibit the in vitro kinase activity of purified F10 at concentrations ranging from 10 to 500 nM (not shown); presumably, the inhibition of membrane biogenesis by H89 occurs by the inhibition of a target other than F10. Inhibitors of PKD membrane association, such as fumonisin B1 and propanolol, had no impact on our ts28 release assay (not shown), suggesting that PKD may also not be a relevant target. It has been reported that there is an H89-sensitive step early in the process of ER export that involves a protein kinase distinct from PKA, PKC, and PKD (2). The absence of cellular markers and glycosylated proteins from virions argues that perhaps vaccinia virus “hijacks” sites of vesicle formation in the ER, generating specialized exit sites that exclude cellular membrane proteins and enrich for viral membrane proteins. The generation and diversion of these sites would therefore involve both the cellular H89-sensitive protein kinase and the viral F10 kinase.
It is noteworthy that although the membrane-associated F10 substrates A14 and A17 and the F10 kinase itself are essential for virus viability, removing the site of F10 phosphorylation in A14 (35) or the major site of Tyr phosphorylation in A17 (Unger and Traktman, unpublished data) did not appear to disrupt virus production. This led us to hypothesize that F10's regulation of vesicle diversion may involve unidentified cellular substrates and/or additional viral substrates. It was therefore intriguing that we observed an increased intensity of some phosphorylated proteins as well as a diminished intensity of other phosphorylated proteins in an autoradiographic profile of cell extracts prepared under conditions in which F10 was repressed or catalytically defective. This observation lends additional credence to the hypothesis that F10 may intersect with a cellular signaling pathway(s) in order to drive membrane biogenesis and/or virosome formation. wt F10 might negatively regulate a cellular kinase or positively regulate a cellular phosphatase. The regulation of kinase activity by other kinases is best exemplified by the cascade of phosphorylation events in the mitogen-activated protein (MAP) kinase family (18). A comparable regulation of phosphatases is illustrated by the case of VHR, the vaccinia virus H1-related dual-specificity phosphatase; this phosphatase, which negatively regulates the MAP kinases Erk and Jnk, undergoes activating phosphorylation by the tyrosine kinase ZAP70 (1).
In summary, these studies further our understanding of the structure and function of the F10 protein kinase. F10 is a membrane-associated phosphoprotein that functions at multiple steps in morphogenesis in a manner that is dependent upon its enzymatic activity. Moreover, morphogenesis appears to involve de novo lipid synthesis and the involvement of additional kinases and/or phosphatases. Future studies that build on these new observations should provide a deeper understanding of vaccinia virus biology and should elucidate the plasticity of the intracellular membrane machinery.
Acknowledgments
This work was supported by NIH grant 5 R01 GM53601 (P.T.).
We thank Clive Wells for expert assistance with electron microscopy, Bethany Unger and Jason Mercer for sharing unpublished data, Jon Skarie for assistance with the lipid overlay assays, and T. Irma Naytor for technical assistance.
REFERENCES
- 1.Alonso, A., S. Rahmouni, S. Williams, M. van Stipdonk, L. Jaroszewski, A. Godzik, R. T. Abraham, S. P. Schoenberger, and T. Mustelin. 2003. Tyrosine phosphorylation of VHR phosphatase by ZAP-70. Nat. Immunol. 4:44-48. [DOI] [PubMed] [Google Scholar]
- 2.Aridor, M., and W. E. Balch. 2000. Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J. Biol. Chem. 275:35673-35676. [DOI] [PubMed] [Google Scholar]
- 3.Banham, A. H., and G. L. Smith. 1992. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology 191:803-812. [DOI] [PubMed] [Google Scholar]
- 4.Becker, W., Y. Weber, K. Wetzel, K. Eirmbter, F. J. Tejedor, and H. G. Joost. 1998. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J. Biol. Chem. 273:25893-25902. [DOI] [PubMed] [Google Scholar]
- 5.Betakova, T., E. J. Wolffe, and B. Moss. 1999. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J. Virol. 73:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 7.Byrd, C. M., T. C. Bolken, and D. E. Hruby. 2002. The vaccinia virus I7L gene product is the core protein proteinase. J. Virol. 76:8973-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carette, J. E., M. Stuiver, J. Van Lent, J. Wellink, and A. Van Kammen. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chijiwa, T., A. Mishima, M. Hagiwara, M. Sano, K. Hayashi, T. Inoue, K. Naito, T. Toshioka, and H. Hidaka. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265:5267-5272. [PubMed] [Google Scholar]
- 10.Cudmore, S., R. Blasco, R. Vincentelli, M. Esteban, B. Sodeik, G. Griffiths, and L. J. Krijnse. 1996. A vaccinia virus core protein, p39, is membrane associated. J. Virol. 70:6909-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dales, S., and E. H. Mosbach. 1968. Vaccinia as a model for membrane biogenesis. Virology 35:564-583. [DOI] [PubMed] [Google Scholar]
- 12.DeMasi, J., and P. Traktman. 2000. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J. Virol. 74:2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien, M., A. Punjabi, M. Khanna, O. Grubisha, and P. Traktman. 1999. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J. Virol. 73:7287-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehlers, A., J. Osborne, S. Slack, R. L. Roper, and C. Upton. 2002. Poxvirus orthologous clusters (POCs). Bioinformatics 18:1544-1545. [DOI] [PubMed] [Google Scholar]
- 15.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogg, M. H., N. L. Teterina, and E. Ehrenfeld. 2003. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J. Virol. 77:11408-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrington, T. P., and G. L. Johnson. 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11:211-218. [DOI] [PubMed] [Google Scholar]
- 19.Guan, K. L., S. S. Broyles, and J. E. Dixon. 1991. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature 350:359-362. [DOI] [PubMed] [Google Scholar]
- 20.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 21.Himpel, S., P. Panzer, K. Eirmbter, H. Czajkowska, M. Sayed, L. C. Packman, T. Blundell, H. Kentrup, J. Grotzinger, H. G. Joost, and W. Becker. 2001. Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem. J. 359:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husain, M., and B. Moss. 2003. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J. Virol. 77:11754-11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh, T., and T. Takenawa. 2002. Phosphoinositide-binding domains: functional units for temporal and spatial regulation of intracellular signalling. Cell Signal. 14:733-743. [DOI] [PubMed] [Google Scholar]
- 25.Kentrup, H., W. Becker, J. Heukelbach, A. Wilmes, A. Schurmann, C. Huppertz, H. Kainulainen, and H. G. Joost. 1996. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J. Biol. Chem. 271:3488-3495. [DOI] [PubMed] [Google Scholar]
- 26.Klemperer, N., J. Ward, E. Evans, and P. Traktman. 1997. The vaccinia virus I1 protein is essential for the assembly of mature virions. J. Virol. 71:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koerner, T. J., J. E. Hill, A. M. Myers, and A. Tzagoloff. 1991. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 194:477-490. [DOI] [PubMed] [Google Scholar]
- 28.Lee, T. H., and A. D. Linstedt. 2000. Potential role for protein kinases in regulation of bidirectional endoplasmic reticulum-to-Golgi transport revealed by protein kinase inhibitor H89. Mol. Biol. Cell 11:2577-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemmon, M. A. 2003. Phosphoinositide recognition domains. Traffic 4:201-213. [DOI] [PubMed] [Google Scholar]
- 30.Lin, S., and S. S. Broyles. 1994. Vaccinia protein kinase 2: a second essential serine/threonine protein kinase encoded by vaccinia virus. Proc. Natl. Acad. Sci. USA 91:7653-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, S., W. Chen, and S. S. Broyles. 1992. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J. Virol. 66:2717-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, F., J. J. Stanton, Z. Wu, and H. Piwnica-Worms. 1997. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol. Cell. Biol. 17:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, K., B. Lemon, and P. Traktman. 1995. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 69:7823-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald, W. F., and P. Traktman. 1994. Overexpression and purification of the vaccinia virus DNA polymerase. Protein Expr. Purif. 5:409-421. [DOI] [PubMed] [Google Scholar]
- 35.Mercer, J., and P. Traktman. 2003. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J. Virol. 77:8857-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niles, E. G., and J. Seto. 1988. Vaccinia virus gene D8 encodes a virion transmembrane protein. J. Virol. 62:3772-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, D. D., C. A. Ray, R. P. Drucker, and D. J. Pickup. 1988. A poxvirus-derived vector that directs high levels of expression of cloned genes in mammalian cells. Proc. Natl. Acad. Sci. USA 85:9431-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price, A. C., K. H. Choi, R. J. Heath, Z. Li, S. W. White, and C. O. Rock. 2001. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J. Biol. Chem. 276:6551-6559. [DOI] [PubMed] [Google Scholar]
- 39.Rempel, R. E., and P. Traktman. 1992. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J. Virol. 66:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritzenthaler, C., C. Laporte, F. Gaire, P. Dunoyer, C. Schmitt, S. Duval, A. Piequet, A. M. Loudes, O. Rohfritsch, C. Stussi-Garaud, and P. Pfeiffer. 2002. Grapevine fanleaf virus replication occurs on endoplasmic reticulum-derived membranes. J. Virol. 76:8808-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez, D., M. Esteban, and J. R. Rodriguez. 1995. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J. Virol. 69:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. K. Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 48.Szajner, P., H. Jaffe, A. S. Weisberg, and B. Moss. 2003. Vaccinia virus G7L protein interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 77:3418-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szajner, P., A. S. Weisberg, and B. Moss. 2001. Unique temperature-sensitive defect in vaccinia virus morphogenesis maps to a single nucleotide substitution in the A30L gene. J. Virol. 75:11222-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szajner, P., A. S. Weisberg, and B. Moss. 2004. Evidence for an essential catalytic role of the F10 protein kinase in vaccinia virus morphogenesis. J. Virol. 78:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szajner, P., A. S. Weisberg, and B. Moss. 2004. Physical and functional interactions between vaccinia virus F10 protein kinase and virion assembly proteins A30 and G7. J. Virol. 78:266-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szajner, P., A. S. Weisberg, E. J. Wolffe, and B. Moss. 2001. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 75:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, C. L., and R. C. Condit. 1986. Marker rescue mapping of vaccinia virus temperature-sensitive mutants using overlapping cosmid clones representing the entire virus genome. Virology 150:10-20. [DOI] [PubMed] [Google Scholar]
- 54.Traktman, P., M. K. Anderson, and R. E. Rempel. 1989. Vaccinia virus encodes an essential gene with strong homology to protein kinases. J. Biol. Chem. 264:21458-21461. [PubMed] [Google Scholar]
- 55.Traktman, P., A. Caligiuri, S. A. Jesty, K. Liu, and U. Sankar. 1995. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J. Virol. 69:6581-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulaeto, D., D. Grosenbach, and D. E. Hruby. 1995. Brefeldin A inhibits vaccinia virus envelopment but does not prevent normal processing and localization of the putative envelopment receptor P37. J. Gen. Virol. 76:103-111. [DOI] [PubMed] [Google Scholar]
- 58.Unger, B., and P. Traktman. 2004. Vaccinia virus morphogenesis: A13 phosphoprotein is required for assembly of mature virions. J. Virol. 78:8885-8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanslyke, J. K., C. A. Franke, and D. E. Hruby. 1991. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J. Gen. Virol. 72:411-416. [DOI] [PubMed] [Google Scholar]
- 60.Vanslyke, J. K., S. S. Whitehead, E. M. Wilson, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]
- 61.Wang, S., and S. Shuman. 1995. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J. Virol. 69:6376-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., and B. Moss. 1992. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology 187:643-653. [DOI] [PubMed] [Google Scholar]