Abstract
Background
In the past few years, eleven new human viruses have joined the two previously known members JCPyV and BKPyV of the Polyomaviridae family, by virtue of molecular methods. Serology data suggest that infections with human polyomaviruses (HPyVs) occur since childhood and the viruses are widespread in the general population. However, the viral persistence sites and transmission routes are by and large unknown. Our previous studies demonstrated that the four new HPyVs – KIPyV, WUPyV, MCPyV and TSPyV – were present in the tonsils, and suggested lymphoid tissue as a persistent site of these emerging human viruses.
We developed a Luminex-based multiplex assay for simultaneous detection of all 13 HPyVs known, and explored their occurrence in tonsillar tissues of children and adults mostly with tonsillitis or tonsillar hypertrophy.
Methods
We set up and validated a new Luminex-based multiplex assay by using primer pairs and probes targeting the respective HPyV viral protein 1 (VP1) genes. With this assay we tested 78 tonsillar tissues for DNAs of 13 HPyVs.
Results
The multiplex assay allowed for simultaneous detection of 13 HPyVs with high analytical sensitivity and specificity, with detection limits of 100–102 copies per microliter, and identified correctly all 13 target sequences with no cross reactions. HPyV DNA altogether was found in 14 (17.9%) of 78 tonsils. The most prevalent HPyVs were HPyV6 (7.7%), TSPyV (3.8%) and WUPyV (3.8%). Mixed infection of two HPyVs occurred in one sample.
Conclusions
The Luminex-based HPyV multiplex assay appears highly suitable for clinical diagnostic purposes and large-scale epidemiological studies. Additional evidence was acquired that the lymphoid system plays a role in HPyV infection and persistence. Thereby, shedding from this site during reactivation might take part in transmission of the newly found HPyVs.
Keywords: HPyV, PCR, Luminex, Tonsil
Background
Thirteen human polyomaviruses (HPyVs) have been identified to date. Up to 2007, only two HPyVs had been introduced by cell culture: BK polyomavirus (BKPyV) and JC polyomavirus (JCPyV), coincidentally in 1971 [1, 2]. Since then, eleven HPyVs have been discovered by modern molecular techniques [3–14]. All HPyVs have high seroprevalences in human populations ranging from 40 to 90% in adults [15–26]. Typically, the primary infections occur asymptomatically during childhood and are followed by lifelong DNA persistence. In immunocompromised hosts some of these viruses give rise to severe disorders such as nephropathy, progressive multifocal leukoencephalopathy, Merkel cell carcinoma or trichodysplasia spinulosa [5, 7, 27, 28]. The other HPyVs, i.e. KIPyV, WUPyV, HPyV6, HPyV7, HPyV9, HPyV10 (MWPyV), Saint Louis polyomavirus (STLPyV), HPyV12 and New Jersey polyomavirus (NJPyV)-2013, have up to date not been definitively associated with specific diseases.
On the other hand, evidence suggests that tonsils are permissive for JCPyV and BKPyV, indicating that this tissue might play a role in the viral persistence [29–34]. KIPyV and WUPyV, the first two novel polyomaviruses discovered in the respiratory secretions of children with acute respiratory symptoms, [3, 4] might also remain in lymphoid tissue as suggested by detection of the corresponding sequences in tonsils [29, 35, 36]. We have reported the occurrence of DNA of MCPyV and TSPyV in tonsillar tissue, suggesting persistence in lymphoid tissue or mucosa [36, 37]. MCPyV is a common skin commensal and causes about 80% of cases of Merkel cell carcinoma (MCC) [5, 38–40]. TSPyV induces the rare skin disease trichodysplasia spinulosa (TS) in immunocompromised patients [7]. HPyV6 and HPyV7 identified in skin and eyebrow hairs, have in one study been isolated from malignant and non-malignant tonsils [6, 38, 40–42]. HPyV9 was identified in 2010 in the serum of a kidney transplant patient under immunosuppressive treatment [8]. Its DNA was found in serum, PBMCs and skin [38, 43, 44]. HPyV10 was originally detected in stool, while the isolate MWPyV was first encountered in the skin of a patient with the “warts, hypogammaglobulinaemia, infections and myelokathexis” (WHIM) syndrome. MXPyV, another HPyV10 isolate, was found in stool and in respiratory samples, [9, 10, 45] while STLPyV DNA was amplified in stool and urine [13]. A STLPyV variant sharing 92% genome identity with the originally described MA138 and WD972 strains, was amplified from skin warts of a patient suffering from WHIM syndrome [46]. HPyV12 was found in organs of the digestive tract, particularly in the liver but also in colon, rectum and stool [47]. NJPyV DNA sequences and virions were originally detected in a muscle biopsy of a pancreatic transplant recipient with viral sequences also found in endothelial cells in muscle and skin [48].
To demonstrate clinical correlates and disease associations for HPyVs, as with most other viruses, the diagnostic cornerstones are nucleic acid detection and serodiagnosis. To this end, we have developed and validated several PCR protocols [37, 49–51]. The Luminex technology offers a novel platform for sensitive and specific, high-throughput multiplex DNA detection. An assay has earlier been set up for the detection of 10 HPyVs [52, 53]. We here describe the further development of this multiplex nucleic acid assay for the detection of all 13 HPyVs currently known, in a clinically applicable format. By combining multiplex PCR amplification with bead based hybridization and flow cytometric analysis, the resulting Luminex-based multiplex assay can simultaneously identify all the 13 HPyVs in a single reaction.
Herein, the multiplex assay is evaluated for specificity, sensitivity and reproducibility. Furthermore, we aimed to determine to what extent the lymphoid system plays a role in HPyV infection and persistence by exploring the frequencies of occurrence of these viral genomes in tonsillar biopsies from children and adults with tonsillar disease.
Methods
Clinical specimens
The clinical material comprised tonsillar tissue from 78 subjects: 31 children and 47 adults (Fig. 1). The pediatric donors ranged in age from 2 to 15 years (average, 6.6), and the adults from 16 to 69 (average, 30.3). Of the specimens, 31 were from males and 47 from females (Fig. 1). The tonsillectomies and tonsillotomies were performed in most cases due to chronic tonsillitis or tonsillar hypertrophy (Fig. 1). All tissues were collected and used in accordance with the ethical rules of the Ethics Committee of the Hospital District of Helsinki and Uusimaa. All tonsil tissues were freshly obtained directly after surgical resection at the operation theatre. The tonsils were cut with disposable scalpels and cell suspensions were prepared by mechanical homogenisation with a syringe plunge, followed by a wash with PBS and filtration through a 70 μm mesh (Corning Life Sciences). The cells were resuspended into final volume of 100 μL PBS.
Fig. 1.
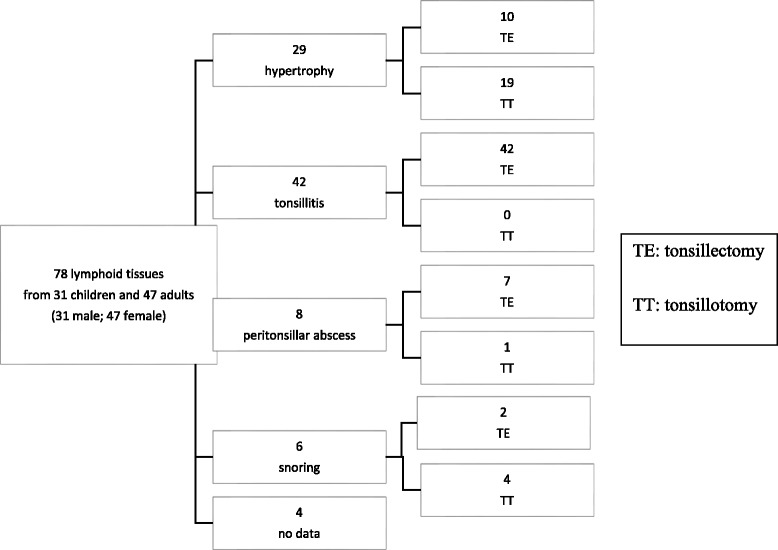
Tonsillar samples from children and adults; clinical indications
Nucleic acid extraction
Whole DNA was extracted from cell suspension by the KingFisher Duo DNA Extraction Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Standard precautions to avoid contamination were taken. The extracted DNA was stored at −20 °C.
Primer pairs and probes
Primer pairs and probes for 13 HPyVs (BKPyV, JCPyV, KIPyV, WUPyV, MCPyV, HPyV6, HPyV7, TSPyV, HPyV9, HPyV10 (MWPyV), STLPyV, HPyV12 and NJPyV), were based on GenBank sequences (Table 1). The primers and probes for the first 10 except MCPyV were the same as published by Gustafsson et al. [53] while the primers and probes for STLPyV, HPyV12 and NJPyV were designed for the present study. Primers for MCPyV were reselected due to dimerization of the published primers with primers for STLPyV. The primers were designed in current study by using the Amplify 3X software, version 3.1.4 (Bill Engels, University of Wisconsin) and assessed using an online NCBI Blast analysis. The 5′ ends of the reverse primers of the viral protein 1 (VP1) region were labeled with biotin (5′ biotinylation). The probes were 5′ amine-C12-modified (5′ Aminolink C12).
Table 1.
Sequences of primers and probes, amplicon sizes, amplicon positions (in VP1 region) in target genome and reference strain used in the 13-plex Luminex assay [59]
| Virus | Forward/Reverse (5′ biotinylated) Primers sequence (5′-3′) Probe (5′ amine-C12) sequence (5′-3′) |
Amplicon length (bp) | Amplicon position (bp) in target genome | Reference strain (GenBank accession no.) |
|---|---|---|---|---|
| JCPyV | AATGAGGATCTAACCTGTGGAA/CTGCACCATTGTCATGAGTTGCTTG ATGAATGTGCACTCTAATGG |
127 | 1742–1868 | J02226 |
| BKPyV | ACAGAGGTTATTGGAATAACTAG/ACTCCCTGCATTTCCAAGGG CTTAACCTTCATGCAGGGTC |
143 | 1952–2094 | DQ305492 |
| KIPyV | TTGGATGAAAATGGCATTGG/TAACCCTTCTTTGTCTAAAATGTAGCC CTTGGAACAGCTAATAGTAGAATC |
142 | 2263–2404 | EF127906 |
| WUPyV | TTGGATGAAAATGGCATTGG/TAACCCTTCTTTGTCTAAAATGTAGCC GAGTACATACAGGGCTTTCCAG |
142 | 2411–2552 | EF444554 |
| MCPyV | TTCCATCTTTATCTAATTTTGCTT/AGGCCTAGTTTTAGATTACCAGAC GTAATAGGCCCACCATTTGT |
144 | 3757–3900 | EU375803 |
| HPyV6 | TTGCTTCTGGATCCAATACTGC/GGCCTCAGGAATTTCAGGCAA TGGATGCTGGTTCATCTCTG |
131 | 1426–1556 | HM011558 |
| HPyV7 | AAGCAGCTACAACTGGGAACTT/GGCCTCAGGAATTTCAGGCAA GCCTACCTTATCCTATGAGTG |
125 | 1450–1574 | HM011566 |
| TSPyV | AGAATGTATGATGACAAAGGTAT/TCTGTAGTTTCCAGTTAGAAAC TGAGGGAATGAATTTCCATATGTT |
111 | 1722–1832 | GU989205 |
| HPyV9 | ATCTATGGCTCATCCTCAGG/GTAGAGCTAGCAACTAGGCCT AGTGCAGGGTACCACTCTC |
107 | 1862–1968 | KC831440 |
| HPyV10 | GTCCAGTTCCTACTAAAGTTCCT/TACATCATTGCCCATCCTTGGTT GCCGGACACCACAATGACA |
106 | 1501–1628 | JQ898292 |
| STPyV | TGAATATGATCCGTGCCAAA/ACTGCATCAGGGCCTACTTG CCTCCTCCAACATGTGTTCC |
129 | 1318–1446 | JX463184 |
| HPyV12 | GTAATGGCACCCAAGAGGAA/GGGGATTTAGAAAGGCCTCA CCCAGCAGTGTCCCTAAATT |
157 | 1402–1558 | JX308829 |
| NJPyV | TGTGTGCCAAAGAAGTGTCCT/TCTGTCACCTGTTGGAGCATT CTGATGCTACTACTGAAATTGAA |
159 | 1113–1271 | KF954417 |
Sequences for primers and probes for all except MCPyV, STPyV, HPyV12 and NJPyV as in Gustafsson et al [53].
Plasmid clones
For use as positive controls and to determine assay sensitivities by limiting dilution analysis, plasmids containing each HPyV genome were used. Plasmid DNA was purified using the High Pure Plasmid Isolation Kit (Roche Diagnostics). The concentration of the purified plasmid DNA was determined with the NanoDrop 2000 (Invitrogen, Carlsbad, CA, USA) and the corresponding genome copies were calculated based on the concentration and molecular weight of the plasmid. A 10-fold dilution series of 108 to 100 copies/μL of HPyV DNA was prepared for each of the 13 HPyVs in PCR-grade H2O, aliquoted, and stored at −20 °C.
Multiplex PCR for HPyV detection using the Luminex platform
All the HPyV assays were at first performed in singleplex format and then multiplexed. After confirming that each individual primer pair amplified specifically its plasmid sequence, the primer pairs were combined. The multiplex nucleic acid amplification was conducted as below. In brief, 5 μL DNA templates were mixed in a 20 μL multiplex reaction consisting of 12.5 μL of 2× multiplex PCR mastermix (Qiagen), 0.2 μM of each forward primer and 1 μM of each biotinylated reverse primer. The amplification conditions were 95 °C for 15 min, 40 cycles at 94 °C for 20 s, 50 °C for 90 s, 71 °C for 1 min and 20 s, and a final extension at 71 °C for 10 min.
HPyV singleplex nested PCR
Each positive specimen was amplified and confirmed with the corresponding HPyV singleplex nested PCR (JCPyV, WUPyV, MCPyV, HPyV, and TSPyV). PCR-grade H2O was included in each experiment. In brief, 3 μL of amplified DNA template was mixed in a 22 μL multiplex reaction consisting of 12.5 μL of 2× multiplex PCR mastermix (Qiagen), 0.2 μM of the corresponding forward primer and 0.2 μM of the corresponding biotinylated reverse primer. The amplification conditions were the same as with the multiplex PCR.
Luminex-based suspension array procedure (oligonucleotide coupling, hybridization, and measurement)
Oligonucleotide coupling; thirteen capture probes were included in the multiplex assay (Table 1). Each probe sequence represented the reverse complement to the target region of the biotinylated PCR product. Different sets of carboxylated fluorescent microbeads were obtained from Luminex Corp. (‘s-Hertogenbosch, The Netherlands), and oligonucleotide probes for target viruses were assigned to individual bead sets. The oligonucleotide coupling was done according to the manufacturer’s instructions (xMAP cookbook, Luminex). The probe-coupled beads were counted using a hemocytometer and were stored in the dark at +4 °C. Hybridization; probe-beads and PCR products were hybridized as published [53] except that the streptavidin-phycoerythrin (SAPE, Invitrogen) incubation temperature was 48 °C. After three washes the amplicons were labeled with 4 μg/mL SAPE conjugate in 2 M tetramethylammonium chloride (Sigma), 75 mM Tris, 6 mM EDTA and 1.5 g7 L sarkosyl (Sigma), pH 8.0; for 20 min in the dark. Measurement; after three washes, the beads and the SAPE signal were analyzed in a Bio-Plex 200 (Bio-Rad).
Data analysis and cutoff definition
The results were measured and calculated by the software Xponent 2.1. The data were expressed as Median Fluorescence Intensity (MFI) of 100 microspheres of each bead type. The cut-off value for a positive result was defined as net MFI two times background mean plus 15 MFI.
Specificity of HPyV multiplex assay
To evaluate the specificity of the multiplex assay, plasmids of each HPyV alone (at 105 copies/μL) and combined were used as template to hybridize with a mixture of 13 type-specific probe-bead sets. Specificity of the multiplex platform was also investigated with virus-free SF9 and HEK 293 cells.
Detection limits of HPyV multiplex assay
To determine the sensitivity of the single and multiplex assays, we tested each virus with 10-fold serial dilutions of the DNA standards. The detection limit was defined as the dilution containing the fewest copies of viral genome (in duplicate testing) that still gave a positive result.
Reproducibility of HPyV multiplex assay
To test the multiplex assay reproducibility, a mixture of all 13 HPyV plasmids (at 103 copies/μL/each HPyV) was analyzed with a mixture of 13 type-specific probe-bead sets. Intra-assay variation was calculated with triplicates in the same run, and inter-assay variation in 3 distinct runs.
Results
We designed a multiplex assay for the detection and identification of 13 HPyVs, by extension of a previously designed multiplex assay for 10 HPyVs [53]. The assay conditions in this study were optimized for the following parameters: amounts of HPyV-specific probes, each coupled to a distinct Luminex bead, amounts of forward primer and biotinylated reverse primer; hybridization conditions; SAPE concentration; incubation time for SAPE staining; and numbers of washing cycles (data not shown).
Specificity of HPyV multiplex assay
PCR-grade H2O was included in each experiment to determine background. Specificity of the assay was assessed with plasmids containing the respective HPyV VP1 inserts. Plasmids of each of the 13 HPyVs alone and combined (105 copies/μl/each HPyV) were used as template for singleplex/multiplex amplification and hybridization. The multiplex assay did identify correctly all the 13 target sequences, with no cross reactions (Table 2). Furthermore, we tested the specificity of the multiplex assay with virus-free SF9 and HEK 293 cells, with no positive signals obtained (data not shown).
Table 2.
Specificities of 13 type-specific probes employed in multiplex HPyV genotyping
| Target sequence | HPyV type-specific probe (MFI) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BKPyV | JCPyV | KIPyV | WUPyV | MCPyV | HPyV6 | HPyV7 | TSPyV | HPyV9 | HPyV10 | HPyV11 | HPyV12 | HPyV13 | |
| BKPyV | 2105 | 43 | 35 | 36 | 33 | 54 | 42 | 27 | 34 | 34 | 44 | 35 | 19 |
| JCPyV | 39 | 1145 | 32 | 32 | 32 | 34 | 34 | 24 | 39 | 28 | 47 | 32 | 20 |
| KIPyV | 49 | 48 | 2522 | 45 | 42 | 43 | 43 | 38 | 48 | 44 | 59 | 47 | 17 |
| WUPyV | 33 | 36 | 29 | 5704 | 31 | 31 | 65 | 18 | 35 | 26 | 43 | 29 | 19 |
| MCPyV | 46 | 46 | 37 | 38 | 2736 | 45 | 41 | 31 | 40 | 37 | 49 | 39 | 21 |
| HPyV6 | 38 | 38 | 34 | 32 | 33 | 3196 | 35 | 30 | 34 | 31 | 44 | 35 | 23 |
| HPyV7 | 43 | 46 | 38 | 52 | 35 | 42 | 2574 | 29 | 41 | 41 | 51 | 37 | 18 |
| TSPyV | 52 | 52 | 45 | 74 | 42 | 46 | 64 | 2894 | 47 | 42 | 58 | 47 | 19 |
| HPyV9 | 50 | 48 | 44 | 40 | 47 | 47 | 39 | 29 | 1388 | 39 | 54 | 49 | 18 |
| HPyV10 | 38 | 36 | 33 | 32 | 34 | 36 | 32 | 21 | 33 | 5116 | 47 | 31 | 24 |
| HPyV11 | 47 | 56 | 42 | 51 | 45 | 66 | 61 | 57 | 59 | 58 | 1782 | 66 | 17 |
| HPyV12 | 55 | 50 | 42 | 48 | 45 | 50 | 46 | 38 | 46 | 39 | 61 | 681 | 20 |
| HPyV13 | 39 | 42 | 36 | 36 | 37 | 39 | 40 | 27 | 40 | 35 | 48 | 37 | 2327 |
| Mix HPyVs | 745 | 1367 | 1518 | 2884 | 1511 | 1586 | 1125 | 733 | 603 | 2639 | 612 | 326 | 1631 |
| cutoff | 85 | 81 | 77 | 81 | 77 | 83 | 77 | 75 | 81 | 72 | 107 | 73 | 127 |
Each line represents a single well with each HPyV plasmid (105 copies/μL) or plasmid mix hybridized to a mixture of 13 distinct beads
Limit of detection of HPyV multiplex assay
To determine the sensitivities of the single- and multiplex assays, each HPyV plasmid was diluted serially from 108 to 100 in PCR-grade H2O. The limit of detection was the dilution containing the fewest copies in duplicate of viral genome that still gave a positive result. The analytical sensitivities in singleplex and multiplex format ranged from 100 to 102 copies per microliter with all 13 HPyVs (Table 3).
Table 3.
Limits of detection of 13 HPyVs in singleplex and multiplex format
| HPyV | Limits of detection per μL of DNA extract | |
|---|---|---|
| Singleplex | Multiplex | |
| BKPyV | 101 | 101 |
| JCPyV | 101 | 101 |
| KIPyV | 101 | 101 |
| WUPyV | 101 | 101 |
| MCPyV | 101 | 102 |
| HPyV6 | 101 | 101 |
| HPyV7 | 102 | 102 |
| TSPyV | 101 | 101 |
| HPyV9 | 101 | 102 |
| HPyV10 | 100 | 101 |
| HPyV11 | 101 | 102 |
| HPyV12 | 100 | 101 |
| HPyV13 | 101 | 101 |
Intra-assay and inter-assay reproducibility of HPyV multiplex assay
The reproducibility of HPyV multiplex assay was determined with a mixture of all 13 HPyV plasmids (103 copies/μL/each HPyV) and a mixture of 13 type-specific probe-bead sets. The coefficient of variation obtained from triplicates in a single run ranged from 2.9% to 9.2% and from three independent runs ranged from 5.7% to 18.8%.
Application of HPyV multiplex assay on tissues
The multiplex assay was applied to tonsillar samples obtained from children and adults. Among the 78 tissue donors the assay tested positive as follows: BKPyV (n = 0), JCPyV (n = 1), KIPyV (n = 0), WUPyV (n = 3), MCPyV (n = 1), HPyV6 (n = 6), HPyV7 (n = 0), TSPyV (n = 3), HPyV9 (n = 0), and HPyV10 (n = 0), STLPyV (n = 0), HPyV12 (n = 0), and NJPyV (n = 0). Co-infection of WUPyV and TSPyV was observed in one tissue. Altogether 13 specimens tested positive: JCPyV DNA was found in a female (22 year old) with tonsillitis and peritonsillar abscess; MCPyV DNA in a female (46 y) with tonsillitis; and HPyV6 DNA in 4 males and 2 females (33 y median age [range, 14 to 69]) with diverse tonsillar conditions. WUPyV DNA occurred in two children (male, aged <5 years; one with snoring and the other with hypertrophy). TSPyV DNA was detected in a 2-y child (male) with hypertrophy and an 11-y child (male) with no clinical data available. Co-infection with WUPyV and TSPyV was seen in a 6-y child (female) with hypertrophy. Each positive specimen was confirmed with the corresponding PyV singleplex nested PCR(s). No product was generated from the negative control.
Discussion
The number of HPyVs known has multiplied explosively during the past decade. Infections by these viruses appear ubiquitous since birth. Their DNAs have been found widely in the human body, including lymphoid organs, albeit in low copy numbers [54–60]. Whereas JCPyV, BKPyV, MCPyV and TSPyV are known to be associated with diseases, the others are orphan as regards clinical manifestations.
The currently existing PCR assays are of limited value in multi-HPyV assessment. To this end, sensitive detection methods covering all the HPyVs are needed. By extension and optimization of an earlier one [53], we here describe the development and validation of a Luminex-based multiplex assay that allows for simultaneous detection of 13 HPyVs. The new assay exhibited a high analytical sensitivity, i.e., ability to detect all HPyVs with detection limits of 100–102 copies per microliter, and suitability for high-throughput analysis. We consider the method advantageous also for analyzing multiple infections. This was demonstrated by the detection of up to 13 types in mixes of HPyV plasmids, and of 2 HPyVs in a single tonsillar sample.
In a previous study [61] addressing the detection of members of Polyomaviride in tonsillar tissues from Chinese children with chronic tonsillar disease, WUPyV, MCPyV, TSPyV, KIPyV, MWPyV, and STLPyV were detected in eleven (11%), four (4.0%), three (3.0%), two (2.0%), two (2.0%), and two (2.0%) of 99 samples, respectively. A single HPyV infection was found in 24 (24%) of the samples, and dual HPyV infections (WUPyV and MCPyV) in 2 (2%) samples. In our previous study, we investigated 229 matched pairs of tonsillar tissue biopsies and corresponding serum samples for the presence of TSPyV and found this emerging HPyV in 8 (3.5%) tonsils, and in none of the sera [37]. Each of the 8 PCR-positive subjects had antiviral IgG of high avidity but not IgM, disclosing persistence. In the present study, we detected JCPyV, WUPyV, MCPyV, HPyV6, and TSPyV DNA altogether in 14 (18%) of 78 tonsils, and co-infection of two HPyVs (WUPyV and TSPyV) in one tissue. All of these data provide evidence of tonsils serving as shedding site in HPyV reactivation and thus contributing to HPyV transmission.
Note added in submission
After completion of our work, Herberhold et al. (Med Microbiol Immunol, DOI 10.1007/s00430%E2%80%93016%E2%80%930486-6) published tonsillar HPyV DNA prevalence data highly similar to ours by using real-time quantitative PCRs in singleplex format.
Conclusion
A Luminex-based multiplex assay was developed for epidemiological and diagnostic studies to address whether any of the emerging HPyVs, or infection with HPyV thereof, is associated with disease development. Based on the observed frequent occurrence of HPyVs in human tonsils, we suggest that lymphoid tissue may be a general persistence site for these viruses. Thereby, shedding from this site during reactivation might play a role in HPyV transmission.
Acknowledgments
We wish to thank Eeva Auvinen for providing the JC and BK plasmids; Mariet Feltkamp the TSV; John A. Lednicky the HPyV9; James A. DeCaprio the HPyV10; Bernhard Ehlers the HPyV12; and W. Ian Lipkin the NJPyV plasmids, We are grateful to Sari Tillander at Finnish Institute of Occupational Health and Maija Lappalainen at HUSLAB for permission of use of the Bioplex devices.
Funding
This study was supported by the Sigrid Jusélius Foundation, The Jane and Aatos Erkko Foundation, The Finnish Academy, The Medical Foundation Liv och Hälsa, the Helsinki University Research Fund, The Ida Montin Foundation, and The Finnish-Norwegian Medical Foundation.
Availability of data and materials
All the data supporting our findings are contained within the manuscript. The raw datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
MS and YW carried out the experiments, acquired and analyzed the data, and drafted the manuscript. TR contributed to conception, study design and provision of study materials. L-MA provided tonsillar samples and the clinical data. LP and MT participated in the DNA work. MS-V and KH designed, coordinated the project, and participated in manuscript writing. All authors read, revised, and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (Dnro 450/13/03/04/08 HUS). The patients provided written consent before entry into the study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BBMA
Bead-based multiplex assay
- BKPyV
BK polyomavirus
- HPyVs
Human polyomaviruses
- JCPyV
JC polyomavirus
- MCC
Merkel cell carcinoma
- MCPyV
Merkel cell polyomavirus
- MFI
Median fluorescence intensity
- NJPyV
New Jersey polyomavirus
- SAPE
Streptavidin-phycoerythrin
- STLPyV
Saint Louis polyomavirus
- TS
Trichodysplasia spinulosa
- TSPyV
Trichodysplasia spinulosa
- VP1
Viral protein 1
- WHIM
Warts, hypogammaglobulinaemia, infections and myelokathexis
Contributor Information
Mohammadreza Sadeghi, Email: rsadeghi@bloodsystems.org.
Yilin Wang, Email: yilin.wang@helsinki.fi.
Torbjörn Ramqvist, Email: torbjorn.ramqvist@ki.se.
Leena-Maija Aaltonen, Email: leena-maija.aaltonen@hus.fi.
Lari Pyöriä, Email: lari.pyoria@helsinki.fi.
Mari Toppinen, Email: mari.toppinen@helsinki.fi.
Maria Söderlund-Venermo, Email: maria.soderlund-venermo@helsinki.fi.
Klaus Hedman, Phone: +358-5052-49086, Email: klaus.hedman@helsinki.fi.
References
- 1.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1(7712):1253–1257. doi: 10.1016/S0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 2.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1(7712):1257–1260. doi: 10.1016/S0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 3.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, et al. Identification of a third human polyomavirus. J Virol. 2007;81(8):4130–6. [DOI] [PMC free article] [PubMed]
- 4.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3(5):e64. [DOI] [PMC free article] [PubMed]
- 5.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7(6):509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6(7) doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, et al. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85(9):4586–90. [DOI] [PMC free article] [PubMed]
- 9.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. Complete genome sequence of a tenth human polyomavirus. J Virol. 2012;86(19):10887. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, et al. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol. 2012;86(19):10321–6. [DOI] [PMC free article] [PubMed]
- 11.Yu G, Greninger AL, Isa P, Phan TG, Martinez MA, de la Luz SM, et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One. 2012;7(11):e49449. [DOI] [PMC free article] [PubMed]
- 12.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, et al. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One. 2013;8(3):e58021. [DOI] [PMC free article] [PubMed]
- 13.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology. 2013;436(2):295–303. [DOI] [PMC free article] [PubMed]
- 14.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, et al. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis. 2014;210(10):1595–9. [DOI] [PMC free article] [PubMed]
- 15.Trusch F, Klein M, Finsterbusch T, Kuhn J, Hofmann J, Ehlers B. Seroprevalence of human polyomavirus 9 and cross-reactivity to African green monkey-derived lymphotropic polyomavirus. J Gen Virol. 2012;93:698–705. doi: 10.1099/vir.0.039156-0. [DOI] [PubMed] [Google Scholar]
- 16.Nicol JTJ, Touze A, Robinot R, Arnold F, Mazzoni E, Tognon M, et al. Seroprevalence and cross-reactivity of human polyomavirus 9. Emerg Infect Dis. 2012;18(8):1329–32. [DOI] [PMC free article] [PubMed]
- 17.Sadeghi M, Riipinen A, Vaisanen E, Kantola K, Karikoski R, Taskinen H, Soderlund-Venermo M, Hedman K: Are the newly discovered human polyomaviruses associated with fetal death or miscarriage? J Clin Virol 2009, 46:S60.
- 18.Sadeghi M, Riipinen A, Vaisanen E, Chen TT, Kantola K, Surcel HM, et al. Newly discovered KI, WU, and Merkel cell polyomaviruses: no evidence of mother-to-fetus transmission. Virol J. 2010;7:251. [DOI] [PMC free article] [PubMed]
- 19.Sadeghi M, Aronen M, Chen TT, Jartti L, Jartti T, Ruuskanen O, et al. Merkel cell polyomavirus and trichodysplasia spinulosa-associated polyomavirus DNAs and antibodies in blood among the elderly. BMC Infect Dis. 2012;12:383. [DOI] [PMC free article] [PubMed]
- 20.Kardas P, Sadeghi M, Weissbach FH, Chen TT, Hedman L, Auvinen E, et al. Inter- and Intralaboratory comparison of JC polyomavirus antibody testing using two different virus-like particle-based assays. Clin Vaccine Immunol. 2014;21(11):1581–8. [DOI] [PMC free article] [PubMed]
- 21.Kantola K, Sadeghi M, Ewald MJ, Weissbrich B, Allander T, Lindau C, et al. Expression and serological characterization of polyomavirus WUPyV and KIPyV structural proteins. Viral Immunol. 2010;23(4):385–93. [DOI] [PubMed]
- 22.Chen TT, Mattila PS, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. Seroepidemiology of the newly found Trichodysplasia Spinulosa-associated polyomavirus. J Infect Dis. 2011;204(10):1523–1526. doi: 10.1093/infdis/jir614. [DOI] [PubMed] [Google Scholar]
- 23.Stolt A, Sasnauskas K, Koskela P, Lehtinen M, Dillner J. Seroepidemiology of the human polyomaviruses. J Gen Virol. 2003;84(Pt 6):1499–1504. doi: 10.1099/vir.0.18842-0. [DOI] [PubMed] [Google Scholar]
- 24.Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5(3) doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen NL, Le BM, Wang D. Serologic evidence of frequent human infection with WU and KI polyomaviruses. Emerg Infect Dis. 2009;15(8):1199–1205. doi: 10.3201/eid1508.090270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis. 2009;199(6):837–46. [DOI] [PubMed]
- 27.Hirsch HH, Steiger J. Polyomavirus BK. Lancet Infect Dis. 2003;3(10):611–623. doi: 10.1016/S1473-3099(03)00770-9. [DOI] [PubMed] [Google Scholar]
- 28.Tan CS, Koralnik IJ. Progressive multifocal leukoencephalopathy and other disorders caused by JC virus: clinical features and pathogenesis. Lancet Neurol. 2010;9(4):425–437. doi: 10.1016/S1474-4422(10)70040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Astegiano S, Terlizzi ME, Elia M, Cavallo GP, Costa C, Cavallo R, et al. Prevalence of polyomaviruses BK, JC, SV40, KI, and WU in non-malignant tonsil specimens. Minerva Med. 2010;101(6):385–9. [PubMed]
- 30.Comar M, Zanotta N, Bovenzi M, Campello C. JCV/BKV and SV40 viral load in lymphoid tissues of young immunocompetent children from an area of north-east Italy. J Med Virol. 2010;82(7):1236–1240. doi: 10.1002/jmv.21786. [DOI] [PubMed] [Google Scholar]
- 31.Meneses A, Lopez-Terrada D, Zanwar P, Killen DE, Monterroso V, Butel JS, et al. Lymphoproliferative disorders in Costa Rica and simian virus 40. Haematologica. 2005;90(12):1635–42. [PubMed]
- 32.Monaco MC, Jensen PN, Hou J, Durham LC, Major EO. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J Virol. 1998;72(12):9918–9923. doi: 10.1128/jvi.72.12.9918-9923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monaco MC, Atwood WJ, Gravell M, Tornatore CS, Major EO. JC virus infection of hematopoietic progenitor cells, primary B lymphocytes, and tonsillar stromal cells: implications for viral latency. J Virol. 1996;70(10):7004–7012. doi: 10.1128/jvi.70.10.7004-7012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato A, Kitamura T, Takasaka T, Tominaga T, Ishikawa A, Zheng HY, et al. Detection of the archetypal regulatory region of JC virus from the tonsil tissue of patients with tonsillitis and tonsilar hypertrophy. J Neuro-Oncol. 2004;10(4):244–9. [DOI] [PubMed]
- 35.Babakir-Mina M, Ciccozzi M, Bonifacio D, Bergallo M, Costa C, Cavallo R, et al. Identification of the novel KI and WU polyomaviruses in human tonsils. J Clin Virol. 2009;46(1):75–9. [DOI] [PubMed]
- 36.Kantola K, Sadeghi M, Lahtinen A, Koskenvuo M, Aaltonen LM, Mottonen M, et al. Merkel cell polyomavirus DNA in tumor-free tonsillar tissues and upper respiratory tract samples: implications for respiratory transmission and latency. J Clin Virol. 2009;45(4):292–5. [DOI] [PMC free article] [PubMed]
- 37.Sadeghi M, Aaltonen LM, Hedman L, Chen T, Soderlund-Venermo M, Hedman K. Detection of TS polyomavirus DNA in tonsillar tissues of children and adults: evidence for site of viral latency. J Clin Virol. 2014;59(1):55–8. [DOI] [PubMed]
- 38.Hampras SS, Giuliano AR, Lin HY, Fisher KJ, Abrahamsen ME, McKay-Chopin S, et al. Natural history of polyomaviruses in men: the HPV infection in men (HIM) study. J Infect Dis. 2015;211(9):1437–46. [DOI] [PMC free article] [PubMed]
- 39.Gunel C, Kirdar S, Omurlu IK, Agdas F. Detection of the Epstein-Barr virus, human Bocavirus and novel KI and KU polyomaviruses in adenotonsillar tissues. Int J Pediatr Otorhinolaryngol. 2015;79(3):423–427. doi: 10.1016/j.ijporl.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Bellaud G, Gheit T, Pugin A, Pretet JL, Tommasino M, Mougin C, et al. Prevalence of human polyomavirus DNA in eyebrow hairs plucked from patients with psoriasis treated with TNF inhibitors. J Eur Acad Dermatol Venereol. 2015;29(5):1019–21. [DOI] [PubMed]
- 41.Salakova M, Koslabova E, Vojtechova Z, Tachezy R, Sroller V. The detection of human polyomaviruses MCPyV, HPyV6, and HPyV7 in malignant and non-malignant tonsillar tissues. J Med Virol. 2016;88(4):695–702. [DOI] [PubMed]
- 42.Wieland U, Silling S, Hellmich M, Potthoff A, Pfister H, Kreuter A. Human polyomaviruses 6, 7, 9, 10 and Trichodysplasia spinulosa-associated polyomavirus in HIV-infected men. J Gen Virol. 2014;95:928–32. [DOI] [PubMed]
- 43.van der Meijden E, Wunderink HF, van der Blij-de Brouwer CS, Zaaijer HL, Rotmans JI, Bavinck JN, et al. Human polyomavirus 9 infection in kidney transplant patients. Emerg Infect Dis. 2014:20(6):991–9. [DOI] [PMC free article] [PubMed]
- 44.Lednicky JA, Butel JS, Luetke MC, Loeb JC. Complete genomic sequence of a new human polyomavirus 9 strain with an altered noncoding control region. Virus Genes. 2014;49(3):490–492. doi: 10.1007/s11262-014-1119-z. [DOI] [PubMed] [Google Scholar]
- 45.Yu GX, Greninger AL, Isa P, Phan TG, Martinez MA, Sanchez MD, Contreras JF, Santos-Preciado JI, Parsonnet J, Miller S et al. Discovery of a Novel Polyomavirus in Acute Diarrheal Samples from Children. PLoS One 2012;7(11):e49449. [DOI] [PMC free article] [PubMed]
- 46.Pastrana DV, Fitzgerald PC, Phan GQ, Raiji MT, Murphy PM, McDermott DH, et al. A divergent variant of the eleventh human polyomavirus species, saint louis polyomavirus. Genome Announcements. 2013:1(5). [DOI] [PMC free article] [PubMed]
- 47.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, et al. Identification of a Novel Human Polyomavirus in Organs of the Gastrointestinal Tract. PLoS One. 2013;8(3):e58021. [DOI] [PMC free article] [PubMed]
- 48.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, et al. Identification of a Novel Polyomavirus in a Pancreatic Transplant Recipient With Retinal Blindness and Vasculitic Myopathy. J Infect Dis. 2014. [DOI] [PMC free article] [PubMed]
- 49.Kantola K, Sadeghi M, Lahtinen A, Koskenvuo M, Aaltonen LM, Mottonen M, et al. Merkel cell polyomavirus DNA in tumor-free tonsillar tissues and upper respiratory tract samples: implications for respiratory transmission and latency. J Clin Virol. 2009;45(4):292–5. [DOI] [PMC free article] [PubMed]
- 50.Sadeghi M, Aronen M, Chen T, Jartti L, Jartti T, Ruuskanen O, et al. Merkel cell polyomavirus and trichodysplasia spinulosa-associated polyomavirus DNAs and antibodies in blood among the elderly. BMC Infect Dis. 2012;12:383. [DOI] [PMC free article] [PubMed]
- 51.Sadeghi M, Riipinen A, Vaisanen E, Chen T, Kantola K, Surcel HM, et al. Newly discovered KI, WU, and Merkel cell polyomaviruses: no evidence of mother-to-fetus transmission. Virol J. 2010;7:251. [DOI] [PMC free article] [PubMed]
- 52.Ramqvist T, Nordfors C, Dalianis T, Ragnarsson-Olding B. DNA from human polyomaviruses, TSPyV, MWPyV, HPyV6, 7 and 9 was not detected in primary mucosal melanomas. Anticancer Res. 2014;34(2):639–643. [PubMed] [Google Scholar]
- 53.Gustafsson B, Priftakis P, Rubin J, Giraud G, Ramqvist T, Dalianis T. Human polyomaviruses were not detected in cerebrospinal fluid of patients with neurological complications after hematopoietic stem cell transplantation. Future Virol. 2013;8(8):809–814. doi: 10.2217/fvl.13.55. [DOI] [PubMed] [Google Scholar]
- 54.Rinaldo CH, Tylden GD, Sharma BN. The human polyomavirus BK (BKPyV): virological background and clinical implications. APMIS. 2013;121(8):728–745. doi: 10.1111/apm.12134. [DOI] [PubMed] [Google Scholar]
- 55.Rinaldo CH, Hirsch HH. The human polyomaviruses: from orphans and mutants to patchwork family. APMIS. 2013;121(8):681–684. doi: 10.1111/apm.12125. [DOI] [PubMed] [Google Scholar]
- 56.Kazem S, van der Meijden E, Feltkamp MC. The trichodysplasia spinulosa-associated polyomavirus: virological background and clinical implications. APMIS. 2013;121(8):770–782. doi: 10.1111/apm.12092. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch HH, Kardas P, Kranz D, Leboeuf C. The human JC polyomavirus (JCPyV): virological background and clinical implications. APMIS. 2013;121(8):685–727. doi: 10.1111/apm.12128. [DOI] [PubMed] [Google Scholar]
- 58.Ehlers B, Wieland U. The novel human polyomaviruses HPyV6, 7, 9 and beyond. APMIS. 2013;121(8):783–795. doi: 10.1111/apm.12104. [DOI] [PubMed] [Google Scholar]
- 59.Coursaget P, Samimi M, Nicol JT, Gardair C, Touze A. Human Merkel cell polyomavirus: virological background and clinical implications. APMIS. 2013;121(8):755–769. doi: 10.1111/apm.12122. [DOI] [PubMed] [Google Scholar]
- 60.Babakir-Mina M, Ciccozzi M, Perno CF, Ciotti M. The human polyomaviruses KI and WU: virological background and clinical implications. APMIS. 2013;121(8):746–754. doi: 10.1111/apm.12091. [DOI] [PubMed] [Google Scholar]
- 61.Peng J, Li K, Zhang C, Jin Q. MW polyomavirus and STL polyomavirus present in tonsillar tissues from children with chronic tonsillar disease. Clin Microbiol Infect. 2016;22(1):97.e1-3. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data supporting our findings are contained within the manuscript. The raw datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


