Abstract
The genomes of herpesviruses contain a number of genes which are conserved throughout the family of Herpesviridae, indicating that the proteins may serve important functions in the replication of these viruses. Among these are several envelope glycoproteins, including glycoprotein M (gM) and gN, which form a complex that is covalently linked via disulfide bonds in some herpesviruses. However, deletion of gM and/or gN from most alphaherpesviruses has limited effects on replication of the respective viruses in vitro. In contrast, insertional inactivation of the gM gene of the betaherpesvirus human cytomegalovirus (HCMV) results in a replication-incompetent virus. We have started to analyze the structural and functional aspects of the interaction between gM and gN of HCMV. We show that large parts of gM are dispensable for the formation of a gM/gN complex that is transported to distal parts of the cellular secretory pathway. In addition, we demonstrate that the disulfide bond is between the cysteine at position 44 in gM and cysteine 90 in gN. However, disulfide linkage is not a prerequisite for modification and transport of the gM/gN complex. Moreover, mutant viruses that lack a disulfide bridge between gM and gN replicate with efficiencies similar to that of wild-type viruses.
The genomes of herpesviruses contain a number of genes which are conserved throughout the family Herpesviridae. Among these are several envelope glycoproteins, including glycoprotein B (gB), gH, gL, gM, and gN. Although these genes are conserved in different herpesviruses, the corresponding proteins appear to have different functional roles in the replication of herpesviruses from different subgroups. An obvious example is the central role of gB in virus attachment and fusion in both alpha- and betaherpesviruses whereas the gB homolog in the gammaherpesvirus Epstein-Barr virus is either not present in the virion or present in only limited amounts (13). Perhaps most striking is the finding that deletion of gM and gN from the alphaherpesviruses pseudorabies virus and herpes simplex virus (HSV) have only a minimal effect on in vitro virus assembly and replication, whereas a deletion of gM in the alphaherpesvirus Marek's disease virus and the betaherpesvirus human cytomegalovirus (HCMV) results in a null phenotype (3, 14, 15, 27, 38). Deletion of gN from Epstein-Barr virus results in an intermediate phenotype with impaired assembly and infection (21). Thus, it has been difficult to assign a role for the gM/gN complex of proteins to a common function in the replication of herpesviruses.
Two observations from studies of Pseudorabies virus suggest that gM may have a role in virus-induced fusion/penetration and also in cytoplasmic envelopment (12, 19). The latter hypothesis was based on the observation that a Pseudorabies virus mutant that was deficient in gK, gI, and gM failed to undergo cytoplasmic envelopment (6). More recent studies in HSV have suggested that although gM is not essential for the production of infectious virions, viral mutants lacking gM are less efficiently enveloped in the cytoplasm of infected cells (7). Together, these data argue that gM almost certainly has an important role in the infectious cycle of herpesviruses and that the function of this protein may only be observable under more stringent conditions of replication such as occur in vivo. In fact, attenuation has been noted after infection of animals with viruses lacking gM and/or gN (9, 26, 30). The contribution of gN to the function(s) of gM is unknown.
The gM and gN proteins from different herpesviruses share a number of structural properties. The most characteristic structural feature of gM is that it is a highly hydrophobic protein with seven predicted membrane-spanning regions and is either modified minimally by glycosylation (4, 20, 23, 31, 39) or not glycosylated (10). It also aggregates readily when heated, a biophysical property that delayed identification of complexes containing gM. In contrast, the gNs are small typical type I glycoproteins with no or low levels of sugar modification (1, 16, 20, 22, 24, 33). The gN of HCMV contains an undetermined number of side chains that contribute over 40 kDa of mass to the 15-kDa polypeptide backbone (25). Interestingly, these carbohydrate modifications are primarily O-linked.
Together, gM and gN form a complex, which in some viruses, including pseudorabies virus and HCMV, is covalently linked through disulfide bonds (15, 25). Because these proteins were felt to be nonessential for virus replication in alphaherpesviruses, little information is available on the nature of the gM/gN complex in these viruses or on the intracellular trafficking of the components of the complex. More extensive studies have been done in betaherpesviruses, primarily HCMV (25). Thus, although these two glycoproteins are absolutely conserved in herpesviruses, their structural characteristics and role in virus replication suggest that they have assumed different functions in the replicative cycle of these viruses during their evolution within their respective hosts.
The gM/gN complex of betaherpesviruses, more specifically that of HCMV, represents the disulfide complex previously designated gCII (18). Early studies identified this disulfide-linked complex, but definitive evidence that it was composed of gM and gN has been presented only recently (25). These studies documented that gM and gN were present in a higher-molecular-weight complex that was at least partially dependent on intact disulfide bonds. The analysis of this complex was accelerated by the availability of a virus-neutralizing murine monoclonal antibody (MAb) that specifically reacted with gN but only when gN was glycosylated and complexed with gM. Immature forms of gN were not recognized by this reagent. With this reagent, it was shown that gM and gN must interact in the endoplasmic reticulum (ER) to form a transport-competent complex that eventually enters the cytoplasmic assembly compartment in virus-infected cells. When either gM or gN was transiently expressed in the absence of the other component of the gM/gN complex or other viral proteins; both viral glycoproteins were retained in the ER, whereas coexpression of the genes encoding these glycoproteins resulted in the formation of a complex that was transported to more distal sites in the secretory pathway (25). Thus, these data indicated that gM/gN complex formation, likely including disulfide bond formation, was required for transport of this virion envelope glycoprotein to the assembly compartment and that deletion of either viral gene could be expected to result in loss of the complex and also loss of virion infectivity.
The function of the gM/gN complex in the assembly of the virions of betaherpesviruses is unknown. Studies with mass spectrometry to analyze the HCMV virion proteome have suggested that the gM/gN complex appears to be the most abundant protein component of the viral envelope, perhaps in >5-fold excess of gB (D. Streblow, Oregon Health Sciences University, Portland, personal communication). These recent data would argue that gM/gN could have a major structural role in either the formation or maintenance of the virion envelope. Thus, it is not surprising that insertional inactivation of the gene encoding gM results in a replication-incompetent virus, and because both gM and gN are required for complex formation and intracellular transport, it is likely that gN is also essential for virus replication (14).
Antibodies derived from convalescent human serum react with the gM/gN complex and neutralize virus infectivity, suggesting that this complex is expressed during in vivo infection. An additional and unexpected characteristic of gN has been the finding of significant variation in predicted amino acid sequence between the gN of unrelated clinical HCMV isolates (28, 32). It is uncertain if this strain diversity has biological significance, but analysis of sequence variability from over 200 isolates has raised the possibility of immune selective pressure in the generation of these genotypes (32). This finding, together with the extensive carbohydrate modification of gN, suggested that gN could have a role in the complex and therefore in the virion envelope other than providing a chaperone or transport function for gM.
To begin a comprehensive analysis of this glycoprotein complex in the biology of betaherpesviruses, we have determined the structural requirements for the interaction(s) between gM and gN. Our results have demonstrated that the gM/gN complex consists of both covalently linked and non-convalently associated gM and gN. The covalently linked complexes arise from disulfide bonding between cysteine 44 of gM and cysteine 90 in gN. Covalent gM/gN interaction via disulfide bonds, however, is not necessary for the generation of infectious virions. Transport-competent and MAb-reactive gM/gN complexes could be formed between carboxyl truncation mutants of gM containing as few as 120 amino acids of the gM sequence and full-length gN. This result suggested that only minimal domains of gM were required for interaction with gN. Finally, we have formally demonstrated that gN is essential for the production of infectious virions. These studies will serve as the foundation for more specific and targeted mutations that will allow assignment of functions to gM and gN and the gM/gN complex in the replicative cycle of HCMV.
MATERIALS AND METHODS
Cells and viruses
HCMV strain AD169 and recombinant viruses were propagated in primary human foreskin fibroblasts (HFF) grown in minimal essential medium (Invitrogen, Karlsruhe, Germany) supplemented with 5% fetal calf serum (FCS), glutamine (100 mg/liter), and gentamicin (350 mg/liter). Virions were isolated by glycerol-tartrate gradient centrifigation as described (37). 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, glutamine, and gentamicin. Cos7 cells were passaged in DMEM supplemented with 10% FCS, glutamine, and gentamicin. For immunofluorescence, cells were grown on 13-mm glass coverslips in 24-well plates. For growth curves, HFF plated in six-well dishes were infected at a multiplicity of infection (MOI) of approximately 0.1. After adsorption of virus (2 h), the inoculum was removed and replaced by fresh medium. Supernatants were harvested at the indicated time points and stored at −70°C until use. Virus titers were determined by an indirect immunfluorescence assay with a monoclonal antibody (MAb) against the IE1 protein of HCMV as described (2).
Plasmids.
Construction of expression plasmids for gM and gN has been described (25). Plasmids coding for mutated forms of gN or gM were constructed by inserting the appropriate DNA fragment into the vector pcDNA3, pcDNA3.1myc/his, or pEF-1/myc/his (Invitrogen, Karlsruhe, Germany). Mutagenesis of the coding sequence was achieved in a two-step PCR with degenerate primers (35). The cysteine residues at positions 90, 125, and 126 of gN were individually changed to serine. The DNA fragments were introduced into the vector pcDNA3.1myc/his, and the plasmids were designated gN, gNc90, gNc125, and gNc126, respectively. The correct DNA sequence of the gN orf of all constructs was confirmed by nucleotide sequencing. The gM constructs utilized in this study were construct by carboxyl-terminal truncation and insertion of the remaining nucelotides of the UL100 reading frame into the pEF-1/myc/his vector. The gM mutants are designated by the sequence location of the final carboxyl-terminal amino acid.
Construction of pHB5-ΔUL73 and BAC mutagenesis.
Mutagenesis of the HCMV-BAC HB5 (5) was performed with linear DNA fragments for homologous recombination. To create pHB5-ΔUL73, plasmid pCPo-Delta73 was constructed, which contained the entire UL72 open reading frame (nucleotides [nt] 104560 to 105730) at the 5′ end and the entire UL74 ORF (nt 106095 to 107587) at the 3′ end of the kanamycin resistance gene in plasmid pCP-o-15-Link2. pCPo15-LINK2 is a derivative of pCP15 (8) containing multiple cloning sites at the 5′ and 3′ ends of the kanamycin resistance gene. Primers that were used included 72-5 (nt 104560 to 104576), 72-3 (nt 105730 to 105713), 74-5 (nt 106094 to 106111), and 74-3 (nt 107567 to 107551) (nomenclature according to GenBank accession number X17403). From this plasmid, a PCR fragment containing ORF UL72, the kanamycin resistance gene, and UL74 was generated and electroporated into Escherichia coli DH10B carrying the bacterial artificial chromosome (BAC) pHB5 and the plasmid pBAD for RecE/T-mediated recombination (29). Bacterial colonies were selected on agar plates containing kanamycin (30 μg/ml) and chloramphenicol (30 μg/ml).
To confirm the integrity of the recombined BAC, digestion of DNA with the appropriate restriction enzymes was carried out and analyzed via agarose gel electrophoresis in comparison to the parental pHB5. To confirm recombination at the predicted site, Southern blot analysis, PCR analysis, and DNA sequence analysis of the UL71-UL75 region was performed.
To create BAC pHB5-UL73c90mf, a PCR fragment containing UL73c90 was generated from plasmid gNc90 and the amplimer was inserted into the 5′-lanking region of the kanamycin resistance gene in pCPo15-Link2. The 3′-lanking region consisted of the entire ORF UL74. A PCR fragment encompassing UL73c90-kan-UL74 was generated and used for recombination in pHB5 as described above. To remove the kanamycin resistance gene after successful recombination, plasmid pT322 encoding the flp recombinase was used as described (8). Nucleotide sequence was established with primers 73-up5 (nt 105682 to 105700) and 73-rec3 (nt 106271 to 106251).
Reconstitution of BAC-derived virus.
MRC-5 cells (2.5 × 105 cells per well) were seeded into six-well dishes. Two days later 2 μg of BAC DNA together with 1 μg of pcDNApp71tag plasmid DNA (kindly provided by B. Plachter, University of Mainz, Mainz, Germany) was cotransfected with the Superfect reagent (Qiagen, Hilden, Germany) according to the manufacturer's instruction. At 24 h after transfection, the culture medium was replaced by fresh medium, and the cells were cultivated for 7 days. All cells from the six-well culture was transferred to a 24-cm2 flask and cultured until a cytopathic effect was observed. Propagation of the infectious virus was either through cocultivation of infected and uninfected cells or infection of fibroblasts with the supernatant from infected cells. To test for reversion of the recombinant viruses, primers 72up5 (nt 105682 to 105699) and 73rec3 (nt 1062721 to 106251) were used.
Immunoprecipitation.
Immunoprecipitation of Myc-containing proteins was carried out with the μMACS c-Myc-tagged protein isolation kit (Miltenyi Biotec, Bergisch-Gladbach, Germany) according to the manufacturer's instructions. To avoid aggregation of the gM protein, samples were eluted from the columns in urea-containing sodium dodecyl sulfate (SDS) sample buffer as described below.
SDS-PAGE and immunoblotting.
To avoid formation of high-molecular-weight aggregates after boiling, transfected cells or extracellular HCMV particles were incubated in sample buffer containing 15 mM Tris-Cl (pH 6.8) 8 M urea 4% (wt/vol) SDS, 2% (vol/vol) β-mercaptoethanol, 10% (vol/vol) glycerol, and 0.01% bromophenol blue for 2 to 3 h at room temperature. Separation of proteins was carried out by conventional 8% to 11% polyacrylamide gel electrophoresis (PAGE) except that solutions for stacking and separating gels contained 3 M urea and 0.5% (vol/vol) Triton X-100 (final concentrations). All solutions containing urea were prepared fresh. Gel electrophoresis and transfer of samples to nitrocellulose membranes were carried out by standard procedures. For detection of antigen, antibodies were diluted in phosphate-buffered saline (PBS) containing 0.1% Tween 20. Antibody binding was detected with horseradish peroxidase-coupled anti-murine immunoglobulin and the ECL detection system (Pharmacia Biotech, Freiburg, Germany).
Image analysis.
Cos7 cells grown on glass coverslips in 24-well plates were transfected with 1 to 2 μg of plasmid DNA with Lipfectamine (Invitrogen, Karlsruhe, Germany). Fibroblasts also grown on glass coverslips in 24-well plates, were infected with the viruses at a multiplicity at infection of 0.4. Two days later, the coverslips were washed and fixed in 3.0% paraformaldehyde in PBS. The fixed cells were permeabilized with Triton X-100-containing buffer and then blocked in PBS-1% bovine serum albumin (25). Primary antibodies, including a Myc-specific MAb, rabbit anticalreticulin (ER marker; Dianova, Hamburg, Germany), and rabbit anti-gm130 (Golgi marker; Dianova) were then added. Following washing, antibody binding was detected with the appropriate secondary antibody conjugated with either fluorescein or tetramethylrhodamine isothiocyanate (Dianova). Images were collected with a Zeiss Axioplan 2 fluorescence microscope fitted with a Visitron System (Puchheim, Germany) CCD camera. Images were processed with MetaView software and Adobe Photoshop. In some cases, images were collected with a Leica Diavert fluorescence microscope fitted with a Photometrics CCD and processed with Image Pro software (MediaCybernetics, Gaithersburg, Md.).
RESULTS
Sequences involved in gM/gN interaction.
Although gN represents a typical type I glycoprotein which is predicted to be modified cotranslationally by high-mannose N-linked sugars in the ER, the finding that transient expression of gN in the absence of gM results in an unmodified protein of 18 kDa suggested that it required either the direct chaperone function of gM or an associated cellular protein for localization and posttranslational modification (25). Furthermore, when expressed in the absence of gM, gN was not distributed throughout the ER, as would have been expected for a misfolded protein retained in the ER, but was found in discrete perinuclear dots (see Fig. 1). Thus, both carbohydrate modification and transport of HCMV gN to distal sites in the secretory pathway were absolutely dependent on the coexpression of gM.
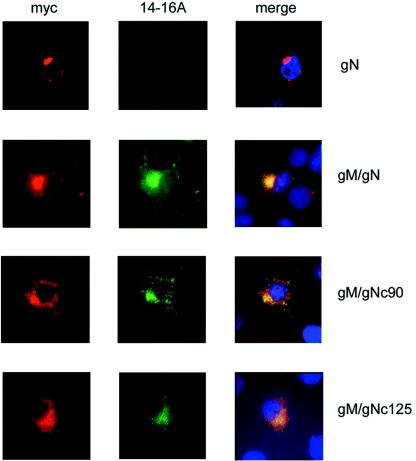
FIG. 1.
Mutations in cysteine residues of gN do not prevent recognition by MAb 14-16A. Cos7 cells were transfected with the indicated plasmids, and protein expression was assayed by reactivity with MAb 14-16A (anti-gM/gN) and MAb 9E10 (anti-Myc). Reactivity was detected with an anti-mouse • • chain-specific antibody. MAb 9E10 was directly coupled with Cy3. The appearance of yellow in the merged pictures indicates colocalization of signals.
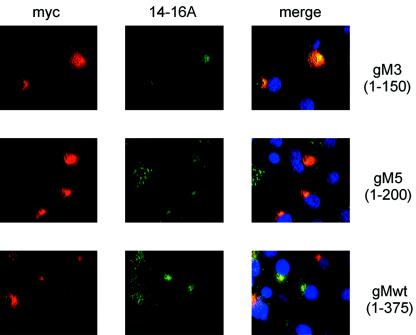
To develop a greater understanding of the structural requirements for the formation of a transport-competent gM/gN complex, we designed experiments to define the sequences and domains of gM that were necessary for complex formation. Initially, we created a panel of gM truncation mutants by carboxyl-terminal deletion of gM amino acid sequences at positions that were predicted to lie outside of transmembrane domains. These sites are listed in Table 1. Truncations were generated by PCR amplification, and amplimers were cloned into an expression plasmid that encoded a Myc tag at the carboxyl terminus of each mutant. Proteins were transiently expressed in Cos7 cells with and without coexpression of gN, and expression was monitored by reactivity with anti-Myc antibodies and with MAb 14-16A in an immunofluorescence assay. We utilized MAb 14-16A as a marker for complex formation because this antibody has previously been shown to be reactive with gN only after the protein is modified by glycosylation and transported distal to the ER-Golgi intermediate compartment in the secretory pathway. Within cells, gN complex formation with gM is also prerequisite for the recognition of gN by MAb 14-16A (25). In agreement with this previous observation, we did not detect reactivity of gN with MAb 14-16A in the absence of gM or mutant forms of gM (Fig. 1 and data not shown).
TABLE 1.
gM carboxyl-terminal truncation mutants and complex formation
| gM | Amino acida changed | Predicted TM domainb | Reactive with MAb 14-16Ac |
|---|---|---|---|
| gM-2 | 118 | 119-141 | Yes |
| gM-3 | 150 | 151-173 | Yes |
| gM-5 | 200 | 201-223 | Yes |
| gM-6 | 224 | 201-223 | Yes |
| gM-7 | 236 | 238-260 | Yes |
| gM-8 | 262 | 238-260 | Yes |
| gM-11 | 298 | 299-321 | Yes |
| gM-12 | 322 | 299-321 | Yes |
| gM-wt | 358 | Yes |
Amino acids numbered according to HCMV strain AD169.
The predicted transmembrane domains were generated with the TMHMM server http://www.cbs.dtu.dk/services/TMHMM. The predicted topology of gM is that the amino terminus of the molecule lies within the lumen (extracytoplasmic) and the carboxyl terminus of the molecule (321 to 358) is cytoplasmic.
Reactivity was determined in indirect immunofluorescence following cotransfection of the indicated gM plasmids together with a wild-type gN-expressing plasmid.
We observed that gM formed complexes with gN even after extensive deletions of the gM amino acid sequence. In fact, we could delete nearly the entire sequence of gM, leaving only the first 118 amino acids that contained the first two predicted membrane-spanning domains, and still maintain complex formation with gN (Table 1; Fig. 2). Biochemical characterization of these mutant forms of gM were consistent with their predicted sizes, and complex formation could be demonstrated by immunoblot (data not shown). Thus, the structural requirements for gM and gN interactions are minimal and located in the amino terminus of the molecule. These findings argue that gN may complex with gM through interactions utilizing sequences between the first two membrane-spanning domains or possibly even through interactions within the membrane-spanning domains. Interestingly, the gM of strain AD169 contains seven cysteine residues (positions 44, 79, 98, 163, 222, 278, and 316), of which six are localized in membrane-spanning segments by computer predictions. The cysteine at position 44 is in a predicted hydrophilic domain and thus could be involved in the formation of the disulfide bridge between gM and gN.
FIG. 2.
Regions of gM are sufficient to enable gN modification and transport. Cos7 cells were transfected with the indicated plasmids, and protein expression was assayed by reactivity with MAb 14-16A (anti-gM/gN) and MAb 9E10 (anti-Myc). Reactivity was detected with a fluorescein isothiocyanate-conjugated anti-mouse • •chain-specific antibody. MAb 9E10 was detected with a Texas Red-conjugated anti-mouse IgG1 second antibody. The appearance of yellow in the merged pictures indicates colocalization of signals.
Role of disulfide linkage between gM and gN for complex formation and intracellular transport.
A major difference between the structure of the complex between gM and gN in different herpesviruses is the presence of a covalent linkage between the proteins in the form of disulfide bonds. We have previously demonstrated the presence of both disulfide-linked and non-convalently linked gM/gN complexes in HCMV-infected cells (25). In these experiments, we investigated if disulfide bond formation was required for generation of the gM/gN complex and ultimately for the assembly of an infectious virion. The results from the studies above suggested that Cys44 of gM likely participated in disulfide bond formation between gM and gN. To extend these studies, we determined which Cys residues in both gN and gM were responsible for disulfide linkages between the two molecules.
The gN of strain AD169 contains four cysteine residues at amino acid positions 90, 119, 125, and 126. Cysteine 90 is predicted to be localized in the lumenal part of the protein, Cys119 within the membrane anchor, and Cys125/Cys126 in the cytoplasmic tail of gN. All cysteine residues are located in a region of the molecule that is absolutely conserved between all HCMV strains that have been sequenced thus far. In order to investigate the contribution of each of these cysteine residues to gM/gN complex formation, plasmids which expressed gN molecules with the individual cysteine residues mutated to serines were constructed. The coding sequences were inserted into a vector that allowed expression of Myc-tagged gN. Expression of the protein was monitored by immunofluorescence analysis following transient transfection into Cos7 cells with a Myc-specific antibody and/or MAb 14-16A. In the absence of gM, the intracellular distribution of gN or the mutant proteins was confined to perinuclear dots (wild-type gN shown in Fig. 1). No reactivity was detected with antibody 14-16A.
When the different gN mutants were expressed in the presence of gM, a significantly different intracellular distribution of gN was observed, and we could detect MAb 14-16A reactivity with all gN mutants when coexpressed with gM. Examples for mutants gNc90 and gNc125 are shown in Fig. 1. Colocalization of the signals derived from the Myc-specific antibody and MAb 14-16A indicated that most of the gN protein was converted to the modified form and transported through the secretory pathway. This was also confirmed by colocalization of the MAb 14-16A signal with signals derived from marker specific for the Golgi and the trans-Golgi network, and colocalization was not seen with markers for the ER (data not shown). Thus, mutation of cysteine residues that were potentially involved in the formation of disulfide bridging to gM had no detectable influence on the reactivity with MAb 14-16A or transport of the protein through the secretory pathway and presumably did not alter the posttranslational modifications of gN based on the maintenance of reactivity with MAb 14-16A.
gN Cys90 and gM Cys44 form the disulfide linkage between gM and gN.
To define which cysteine residues were responsible for the disulfide bridging of gN to gM, we carried out immunoprecipitation analysis of complexes and analyzed the immune complexes in the presence and absence of a reducing agent. After transient expression of the gNs together with gM in 293T cells, gN was precipitated with an antibody directed at the Myc epitope present on all gN molecules. Precipitates were subjected to Western blot analysis, and proteins were detected with antibody 14-16A. In the presence of β-mercaptoethanol, we could detect modified gN as the characteristic smear between 50 and 60 kDa (Fig. 3A). The signal intensity was more intense in the lane containing wild-type gM/gN compared to lanes containing gN mutants, presumably secondary to more efficient expression of the complex in this transfection; however, roughly comparable amounts of gNs were present in the lysates of cells transfected with the mutant forms of gN. An additional signal between 80 and 100 kDa, representing nonreduced gM/gN complex, was obtained in samples containing gM/gN, gM/gNc125, and gM/gN126 but not in gM/gNc90 complexes. Since all incubation steps must be carried out at room temperature in order to prevent aggregation of gM, it is likely that the reduction of the disulfide bonds was less than complete.
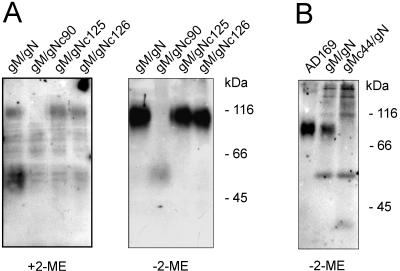
FIG. 3.
Cysteine 90 of gN and cysteine 44 of gM are involved in disulfide linkage in the gM/gN complex. 293T cells were transfected with gM and the gN mutant plasmids (A) or gN and the gM mutant plasmid (B). Cell lysates were subjected to immunoprecipitation with Myc-specific MAb 9E10. SDS-PAGE was performed in the presence (+) or absence (−) of the reducing agent 2-mercaptoethanol (2-ME). Proteins were detected in Western blots with MAb 14-16A (anti-gN) (A) or MAb 14-16A and the anti-gM MAb IMP 91-3/1 (22) (B). The sizes are indicated on the right of each panel.
When the Western blot analysis was carried out under nonreducing conditions, we could detect the gM/gN complex in all samples except gM/gNc90 (Fig. 3A). In this sample; modified gN was detected at 50 to 60 kDa, indicating that modification of gN occurred but not complex formation. The weaker signal obtained with lysates from gM/gNc90 was not the result of loading less protein in these lanes but secondary to the binding characteristics of MAb 14-16A, which exhibits a higher affinity to the complex than to gN alone (unpublished observations). This difference in binding affinity is also illustrated by the intensity of the signal from the nonreduced complexes compared to the signal from noncomplexed gN (Fig. 3A).
As mentioned above, only the cysteine at position 44 of gM is in a hydrophilic area of the molecule and thus a potential candidate for disulfide linkage to gN. In order to test this hypothesis, a plasmid was constructed which encoded a gM protein with a Cys→Ser exchange at position 44. Following transient expression of the protein together with gN, reactivity with MAb 14-16A was considered evidence for the presence of modified gN. Similar to the experiments with mutants of gN, we observed reactivity with MAb 14-16A indicating that gM/gN complex formation had taken place and that gN was transported from the ER (data not shown). Using immunoprecipitations, we found that a mutation at cysteine 44 resulted in loss of the signal at 80 to 100 kDa characteristic of the gM complex when the mutant forms of gM was coexpressed with gN and analyzed in the absence of 2-mercaptoethanol. Instead, a higher-molecular-weight smear was observed, which is indicative of gM aggregation in the absence of the complex partner (Fig. 3B). This result suggested that the cysteine in gM at position 44 participated with the cysteine at position 90 in gN in the disulfide binding of gM and gN. Interestingly, our results indicated that in the absence of disulfide bonding, noncovalent interactions occur leading to modification of gN. This finding was consistent with our previous studies in virus-infected cells and infectious virions and suggested that complex formation was required for gM/gN transport and acquistion of the MAb 14-16A binding site but that neither of these characteristics of the gM/gN complex were absolutely dependent on native disulfide bond formation.
Influence of gM on transport of additional HCMV glycoproteins.
Since covalent linkage to gM is not necessary for modification and transport of gN and only the extreme amino terminus of the gM protein was required for complex formation with gN, a possible role for gM could be to function as a membrane-associated chaperone for HCMV glycoproteins or to recruit cellular chaperones for the folding of viral glycoproteins. We examined this possibility with two different approaches. In the first experiments, we compared complex formation between gM derived from strain AD169 HCMV and HCMV gNs representing prototype sequences of genotypes 1, 2, 3a, and 4a (32). These gNs differed considerably in the predicted amino acid sequence of their ectodomain from the protoypic gN from strain AD169, and homologous sequence longer than 9 residues could not be identified. All gN molecules formed complexes with gM that were recognized by MAb 14-16A and that were transported through the secretory pathway (data not shown).
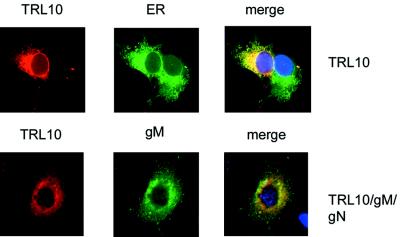
Next we examined the possibility of nonspecific interaction between gM and unrelated viral glycoproteins by coexpression of gM and gpTRL10, an HCMV virion glycoprotein whose expression is restricted to the ER in the absence of additional viral functions (36). When coexpressed with gM, gpTRL10 remained localized to the ER, indicating that gM did not provide a chaperone function for this HCMV glycoprotein (Fig. 4). From these experiments, it appears that gM does not act as a nonspecific chaperone for HCMV glycoproteins and that sequence variability within the gN molecule did not limit gM/gN complex formation.
FIG. 4.
Interaction of gN with gM is specific. Cos7 cells were transfected with the indicated plasmids, and protein expression was assayed by reactivity with MAb 9E10 (anti-Myc/gpTRL10), anticalreticulin (ER), and a rabitt serum specific for gM (gM). Reactivity was detected with anti-mouse or anti-rabbit antibodies. MAb 9E10 was directly coupled with Cy3. The appearance of yellow in the merged pictures indicates colocalization of signals.
Loss of disulfide bonding between gM and gN does not affect virus replication.
In order to determine the role of disulfide bonding between gM and gN in the replication of HCMV, we generated a series of recombinant viruses by a strategy that included the use of an infectious clone of HCMV maintained as a bacterial artificial chromosome (BAC) in E. coli, pHB5 (5). Our strategy required that we initially determine if gN was an essential envelope protein for virus assembly and infectivity. To generate a gN null mutant, we used a kanamycin resistance cassette that was flanked by ORFs 72 and 74. This cassette was recombined into the HCMV BAC pHB5, and kanamycin-resistant colonies were selected and characterized. DNA restriction analysis, Southern blotting, and DNA sequence analysis confirmed correct targeting of the antibiotic resistance gene and deletion of the UL73 ORF between nt 105738 and 106095, corresponding to residues 1 to 118 of gN (data not shown). When DNA purified from this UL73 deletion mutant, pHB5-ΔUL73, was transfected into MRC-5 cells, we failed to obtain infectious virus on numerous attempts.
Identical results were obtained with a different approach, in which an ampicillin resistance gene-containing cassette was inserted into the UL73 ORF (data not shown). However, cotransfection of the UL73 deletion mutant BAC DNA together with a PCR fragment encompassing UL72-UL74 (nt 104556 to 107567) of strain AD169 resulted in replication-competent virus that was indistinguishable with respect to replication kinetics to the parent virus RVHB5 (RV-UL73resc in Fig. 5C). PCR analysis of the resulting virus showed that recombination of the PCR fragment with pHB5-ΔUL73 had taken place and not rescue by providing gN in trans (data not shown). Thus, ORF UL73 of HCMV AD169 appears to be essential for replication of the virus in fibroblasts, a finding consistent with previous studies in which global mutagensis of the HCMV genome was accomplished (11, 40). From these findings, we could then experimentally determine if disulfide bonding between gM and gN was necessary for the replication phenotype of wild-type virus.
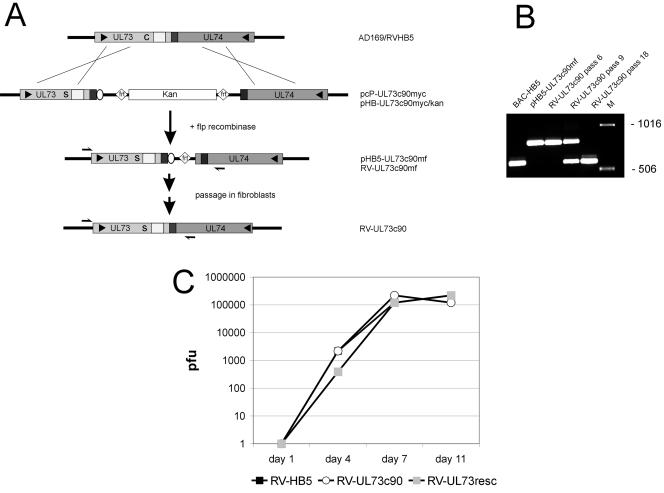
FIG. 5.
Construction of a recombinant virus expressing gNc90. (A) Cartoon of the genomic region encompassing UL73 and UL74 of HCMV AD169 and RVHB5, the linear recombination fragment from plasmid pcp-UL73c90myc, the resulting BAC after homologous recombination (pHB-UL73c90myc/kan), the BAC after removal of the kanamycin resistance gene (pHB5-UL73c90mf), and the recombinant virus (RV-UL73c90mf). RV-UL73c90 represents the recombinant virus after several passages in fibroblasts. The putative transmembrane regions of gN are indicated by light grey boxes, overlap between the 3′ ends of UL73 and UL74 by the dark grey box, and the Myc epitope by the open oval. The positions of cysteine 90 and serine 90 are indicated in the UL73 ORF. The binding site for primers 72-up5 and 73-rec3 are indicated by arrows. (B) PCR fragments were generated from the indicated templates and analyzed via agarose gel electrophoresis and ethidium bromide staining. The primers were 73-up5 and 73-rec3. (C) Growth curves of the mutant virus RV-UL73c90, the parental virus RVHB5 and the rescued virus RV-UL73resc. Fibroblasts were seeded in six-well dishes and infected at a multiplicity of infection of 0.1. At the indicated days postinfection, supernatants from the infected cultures were harvested, and infectious virus was titered by indirect immunofluorescence with an antibody directed against immediate-early antigen 1. Each data point represents the mean of three independent wells.
The results shown above demonstrated that in transient transfection assays, the gNc90 mutation had no detectable influence on transport and modification of gN but precluded covalent complex formation of gM and gN. To test the importance of disulfide bonding on the function of the gM/gN complex in virus replication, we generated a recombinant virus containing a gNc90 mutation. The recombinant virus carrying the gNc90 mutation was also produced in the BAC system, and the strategy is outlined in Fig. 5A. Initially, plasmid pcP-UL73c90myc was constructed. On the 5′ flank of the kanamycin resistance gene, the plasmid encoded the UL73 reading frame with the Cys→Ser exchange on position 90 as well as the coding sequence for a Myc epitope. The 3′ homology region consisted of the UL74 ORF. Note that in HCMV AD169 the 3′ ends of ORFs UL73 and UL74 overlap, and consequently these DNA sequences are present in both homology regions. A DNA fragment encompassing UL73c90, the kanamycin resistance gene, and UL74 was created by PCR and electroporated into E. coli harboring the pHB5 BAC. Following homologous recombination of the cassette with pHB5, the kanamycin coding sequences were removed with flp recombinase. DNA from the resulting BAC, pHB5-UL73c90mf, was transfected into MRC-5 cells to reconstitute the recombinant virus RV-UL73c90mf. A cytopathic effect was observed 3 weeks after transfection.
Nucleotide sequencing of the UL73/UL74 region at that stage indicated that the recombination cassette had been inserted into RVHB5, as predicted. Unexpectedly, upon passaging RV-UL73c90mf through a fibroblast culture, a recombinant virus emerged which had restored the prototypical UL73/UL74 gene arrangement with the exception of the Cys→Ser exchange at position 90 of gN. This virus was designated RV-UL73c90. The PCR analysis shown in Fig. 5B demonstrated the transition of RV-UL73c90mf to RV-UL73c90. Whereas in the first few passages in fibroblasts the recombinant virus produced the same fragments as the parental BAC, a smaller fragment having the size of the prototypic arrangement of UL73/74 in pHB5 appeared after approximately five passages. Following passage 15, only the smaller fragment was amplified from infected cultures (Fig. 5B).
The nucleotide sequence of the entire UL72-UL74 region confirmed that RV-UL73c90 contained the prototypical UL73/UL74 arrangement with the exception of the Cys→Ser mutation in position 90 of UL73 (data not shown). Note that during this process the Myc epitope from gN was also removed. The reversion to the UL73/UL74 phenotype was not an isolated occurrence, because it was observed with different DNA preparations in multiple recombination experiments (data not shown).
To investigate possible effects of the gN cysteine mutation on the kinetics of virus replication, a multistep growth curve analysis was performed. Fibroblasts were infected with identical amounts of infectious virus from either the parental virus RVHB5 or the mutant RV-UL73c90, and the virus titers in the tissue culture supernatant were determined. No significant difference was observed between the growth kinetics of the two viruses (Fig. 5C).
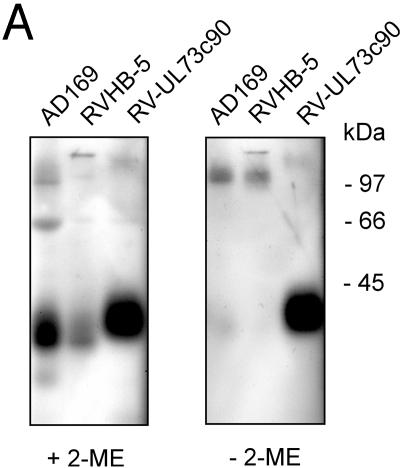
Complex formation by gN was investigated in immunoblot analysis with lysates from extracellular virus particles. In contrast to the parent virus RVHB5 or AD169, no disulfide-linked complex was detected in particles from RV-UL73c90 (Fig. 6A). Interestingly, gM in RV-UL73c90 appeared to migrate more slowly than the gM from the parent virus or AD169, indicating that modification of gM was altered in the recombinant virus. The intracellular distribution of gM and gN, as determined by indirect immunofluorescence analysis with gM- and gN-specific monoclonal antibodies, indicated complete colocalization of gM and gNc90 as well as colocalization of both proteins with additional viral structural glycoproteins such as gB (Fig. 6B and data not shown). Taken together, these results suggested that the absence of a disulfide bridge between gM and gN had no apparent influence on viral replication in human fibroblasts.
FIG. 6.
gM/gN complex formation in RV-UL73c90-infected cells and virions. (A) Extracellular virus was purified from cultures infected with the viruses and subjected to immunoblot analysis with the gM-specific MAb IMP91/3-1. SDS-PAGE was performed in the presence (+) or absence (−) of 2-mercaptoethanol (2-ME). (B) Human fibroblasts were infected with RV-UL73c90, and protein expression was assayed 72 h later in indirect immunofluorescence analysis by reactivity with monoclonal antibodies specific for gN (MAb 14-16A), gM (MAb IMP91-3/1), and gB (MAb C23).
DISCUSSION
The envelope of HCMV is exceedingly complex, and the functions of many of the more abundant protein components have been assigned based on either sequence conservation with glycoprotein homologs from other herpesviruses or positional homologies in the genomes of different herpesviruses. The gM and gN glycoproteins are found in alpha-, beta-, and gammaherpesviruses, although the function of these glycoproteins in the replicative cycle in each of these families of viruses is unclear. This observation is particularly apparent in alphaherpesviruses, in which the genes encoding these envelope proteins can be deleted with only minimal alteration in the phenotype of the resulting mutant for some viruses but not for others (3, 15, 38). Only recently have investigators suggested a possible role for gM in envelopment; however, this phenotype is clearly seen only in conjunction with the deletion of other envelope glycoproteins, raising the possibility that gM has some type of regulatory and/or modulatory function (6). In contrast, gM and gN are both essential for the assembly of infectious HCMV, and recent studies indicate that these two proteins may be the most abundant proteins in the envelope of the virion.
Our findings described in this report provide the initial characterization of the gM/gN complex of HCMV and will serve as the basis for more extensive studies that will define the function of these proteins and the complex in the replicative cycle of this virus. Furthermore, the results of these studies also provided some unexpected findings, including only minimal structural requirements for gM/gN complex formation and an apparent lack of an absolute requirement for disulfide bonding between gM and gN for the production of infectious virus.
In an earlier study of the gM/gN complex, we noted that the complex contained two forms of gM/gN, one linked through disulfide bonds and one associated through noncovalent interactions (25). In the present study, we have presented evidence that the cysteines at position 44 in gM and 90 in gN were involved in this intermolecular bonding. Several other findings were also consistent with disulfide bonding between these residues in gM and gN. These included the finding that the predicted primary sequence of gNs from wild-type HCMVs exhibited significant amino acid variability but contained a conserved cysteine residue at position 90. Two of the gNs that were derived from clinical isolates of HCMV complexed with gM from the laboratory strain of HCMV and exhibited a wild-type phenotype when transiently expressed. Both of the gNs exhibited significant predicted primary sequence divergence in the ectodomain of this glycoprotein compared to the sequence of the AD169 laboratory strain gN (data not shown). Sequence analysis of other UL100 genes (gM) from unrelated HCMVs also revealed a conserved cysteine at position 44 (W. Britt and M. Mach, unpublished results).
Interestingly, a significant proportion of gM and gN in the gM/gN complex was not covalently linked through disulfide bonding, and thus it was not unexpected that even after mutation of these cysteines and elimination of disulfide bonding, the gM/gN complex still formed and was transported through the secretory pathway. Evidence of complex formation and terminal modification was obtained with an MAb that was specific for gN after it has been incorporated into the complex and transported into the distal secretory pathway. Thus, our results suggest that the disulfide linkages between gM and gN contribute only minimally to the stability of this complex and perhaps form in the oxidizing environment of the ER secondary to the conformation of gM and gN within the complex.
The importance of nonconvalent interactions between gM and gN initially led us to investigate the sequences in these molecules that accounted for these interactions. We examined the importance of amino acid sequences in gM in this study because of the unique multiple-membrane-spanning structure of this protein predicted from its primary sequence. We attempted to define the domains of gM involved in complex formation by generating carboxyl-terminal truncations at sites selected to be outside the predicted seven transmembrane domains in this protein. Our results were surprising in that we could delete nearly the entire gM molecule yet continue to see an interaction with gN, as assayed both by MAb 14-16A reactivity and by transport of the complex to a distal secretory compartment. The smallest deletion mutant of gM that was produced was 118 amino acids in length and still contained Cys44, and thus it is possible that even this mutant form of gM was complexed with gN through this disulfide bond. We are currently exploring this possibility. More recently we generated an even smaller gM truncation mutant that contains only the first 90 residues. This protein has been shown to complex with gN, and this complex is transported to distant sites in the secretory pathway (data not shown). Together, our results indicated that most of gM could be deleted and only the extreme amino terminus was necessary for complex formation with gN.
The gM molecule exhibits the predicted structure of a multiple-membrane-spanning protein and contains several motifs in its predicted cytoplasmic domain that could facilitate interaction with the cellular machinery of the secretory and endocytic pathway, including acidic domains in the cytoplasmic tail and motifs associated with endocytosis. With these exceptions, it is difficult to assign significant structure or identify a characteristic motif in sequences of the molecule that are predicted to be outside the membrane because of their limited size. Thus, sequence analysis has not identified a region of the amino terminus of this molecule other than Cys44 that is likely to interact with gN. Interestingly, HCMV gN can form a complex with HSV gM (data not shown), yet these two different molecules share little primary sequence but structurally resemble one another in that both are multiple-membrane-spanning proteins (4). Furthermore, HSV gM also contains a Cys residue at position 42. Whether HCMV gN and HSV gM can form a disulfide-linked complex remains unstudied.
From these results, it was therefore not unexpected that gM derived from HCMV AD169 will form complexes with gN derived from unrelated clinical HCMV isolates, yet this interaction is not nonspecific, as evidenced by the lack of complex formation with an unrelated glycoprotein, gpTRL10. Another possible mechanism leading to gM/gN complex formation that we have considered based on the studies of other membrane-spanning proteins with multiple transmembrane domains is that the interaction between gM and gN could take place within the membrane itself.
To confirm the result from our studies with transient expression assays, we constructed a recombinant HCMV in which the cysteine at position 90 of gN was mutated to a serine. This recombinant virus replicated with wild-type kinetics, indicating that disulfide bonding between Cys90 of gN and Cys44 of gM was not essential for the assembly and infectivity of HCMV. This finding also confirmed results from studies with transient expression of gM and gN that indicated that disulfide bond formation was not required for either complex formation or intracellular transport. The gM/gN complex did form in cells infected with this mutant virus, and this result suggested that complex formation between these proteins even in the absence of the interchain disulfide bonding was essential for intracellular transport of gM and gN and, by inference, assembly of an infectious virion. Infectious virus cannot be recovered from HCMV BACs containing insertional mutations in either the UL100 or UL73 gene, suggesting that the function of either of these glycoproteins in the replicative cycle is dependent on the expression of the other partner.
We would argue that at least one aspect of the codependence is complex formation and normal intracellular trafficking. Alternative explanations for this finding were excluded. These included the possibility that gN is a nonessential glycoprotein of HCMV. Several published findings argue against this possibility. These include the recently published finding that gN appears to be under immune pressure in the population, a finding that would argue that if this glycoprotein was nonessential for replication, it would be underrepresented in the virus population secondary to this immune pressure (32). Second, previous studies utilizing genomewide transposon-mediated insertional mutagenesis have indicated that gN is essential for the production of infectious virus (11, 40).
In the current series of studies, we have formally demonstrated that the UL73 gene product is essential for the production of infectious virions by deletion of the UL73 (gN) gene by two different approaches. In neither case could virus be recovered from the deletion mutant (data not shown). Second, revertant viruses have been generated from these deletion mutants, and both revertants replicated like the wild-type parent virus, indicating that the deletion mutation in UL73 was specific for the observed null phenotype. Thus, the capacity of the gNc90 mutant virus to replicate like the wild-type virus suggested that covalent linkage between gM and gN is not necessary for the incorporation or function of this glycoprotein complex in the virion envelope, at least during short-term passage in vitro.
The finding that the carboxyl-terminal truncation mutant gM2 could still form a complex with gN and that this complex could be transported into distal secretory pathway compartments indicated that a considerable amount of gM was redundant, at least for this specific function of gM. It is unlikely that the predicted seven membrane-spanning domains of gM would be maintained and conserved amongst HCMVs and other herpesviruses if these regions of gM were redundant. Clearly these domains are likely required for other functions of gM. Although far from understood, in alphaherpesviruses some reports suggest that these functions are intrinsic to the process of virion envelopment in the cytoplasm of infected cells, perhaps by modulating intracellular fusion during virion envelopment (6). Yet it appears that these functions of gM that facilitate envelopment can only be readily observed when the gene encoding gM is deleted together with other envelope glycoproteins. These findings have been provided as arguments that the function of gM in virion assembly of alphaherpesviruses is in the final cytoplasmic envelopment of the particle.
The unique structure of herpesvirus gMs would argue for similar functions in the replicative cycle of these viruses, and therefore it is likely that HCMV gM and gN also play some role in the envelopment of the particle. Consistent with this proposed function of gM and gN is the finding that these proteins concentrate in the juxtanuclear asembly compartment of HCMV-infected cells, a presumed site of virus envelopment (34). Other functions of gM and gN can also be envisioned, such as virion attachment or penetration of cells during the initial stages of infection. Early studies of HCMV gCII complexes now known to be gM/gN demonstrated the heparin binding activity of these glycoproteins (17). Together, with recent findings demonstrating the abundance of the gM/gN complex in the virion envelope, it is also possible that gM/gN plays at least some role in the early steps of infection, likely attachment. Finally, the abundant O-linked carbohydrate modifications of gN are unique to HCMV gN and unique among known HCMV glycoproteins. The requirement for this type of carbohydrate and the extent of glycosylation (>200% of mass) of gN suggests that this envelope glycoprotein likely functions in interactions with molecules on the cell surface during the early steps of infection.
The finding that gN is essential for the production of infectious virus could result from either an assembly defect, i.e., failure of gM transport and function, or secondary to loss of a key virion surface protein. Our studies with transient expression of gM and gN favor the former mechanism for the loss of infectious virus production by cells transfected with the gN null virus, based on our observations that gM cannot traffic in the secretory pathway unless complexed with gN and that the gM/gN complex represents a major component of the virion envelope. However, it is also possible that loss of either of these glycoproteins could result in a lethal defect(s) in assembly as well as loss of an essential virion function required for early steps in infection. Studies are currently in progress to address these various possibilities.
Preliminary studies have suggested that truncations of gN led to a more significant effect on the phenotype of this protein and gM/gN complex formation. The deletion of the cytoplasmic domain of gN had little effect on the intracellular localization of this type I glycoprotein or its subsequent complex formation with gM. However, deletions of the carboxyl terminus at position 90, 61, or 41 all resulted in loss of MAb 14-16A reactivity even in the presence of gM, suggesting that sequences within the ectodomain of gN were essential for recognition by this complex-specific anti-gN MAb (data not shown). Furthermore, the intracellular localization of truncated forms of gNs appeared to be similar to that of proteins associated with peripheral membranes and very distinct from that of wild-type gN (data not shown). We are currently confirming these findings and mapping the domains in gN that are essential for its interactions with gM. As noted previously, our preliminary findings suggest that these two proteins may interact through sequences within their membrane-spanning domains or perhaps through specialized host cell membranes modified by the multiple-membrane-spanning domains of gM.
In summary, we have reported on the sequence requirements for formation of the gM/gN complex. We have determined that conserved cysteines at position 44 in gM and position 90 in gN are required for covalent interactions between these molecules; however, these interactions are not required for the production of infectious virions. In addition, we have determined that a large amount of the gM/gN complex is noncovalently associated and that the domains required for these noncovalent interactions are located in the extreme amino terminus of gM and the ectodomain of gN. The nature of these interactions is unknown, but they could take place within the membrane. Finally, the gN of HCMV was formally shown to be essential for replication of infectious virus.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant MA929/6-1) and by grants from the HHS, NIH (AI35602 and AI50189).
REFERENCES
- 1.Adams, R., C. Cunningham, M. D. Davison, C. A. MacLean, and A. J. Davison. 1998. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J. Gen. Virol. 79:813-823. [DOI] [PubMed] [Google Scholar]
- 2.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. [DOI] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D. and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein m in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Dijkstra, J. M., V. Gerdts, B. G. Klupp, and T. C. Mettenleiter. 1997. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J. Gen. Virol. 78:2147-2151. [DOI] [PubMed] [Google Scholar]
- 10.Dijkstra, J. M., T. C. Mettenleiter, and B. G. Klupp. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology 237:113-122. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stole, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong, M., and E. Kieff. 1990. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J. Virol. 64:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: Mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jons, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jons, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kari, B., and R. Gehrz. 1992. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J. Virol. 66:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kari, B., R. Goertz, and R. Gehrz. 1990. Characterization of cytomegalovirus glycoproteins in a family of complexes designated gC-II with murine monoclonal antibodies. Arch. Virol. 112:55-65. [DOI] [PubMed] [Google Scholar]
- 19.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 21.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehner, R., H. Meyer, and M. Mach. 1989. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J. Virol. 63:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, X., B. Chow, C. Raggo, and L. A. Babiuk. 1996. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J. Virol. 70:1448-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mach, M., B. Kropff, P. Dal-Monte, and W. J. Britt. 2000. Complex formation of human cytomegalovirus gylcoprotein M (gpUL100) and glycoprotein N (gpU173). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLean, C. A., L. M. Robertson, and F. E. Jamieson. 1993. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J. Gen. Virol. 74:975-983. [DOI] [PubMed] [Google Scholar]
- 27.Masse, M. J., A. Jons, J. M. Dijkstra, T. C. Mettenleiter, and A. Flamand. 1999. Glycoproteins gM and gN of pseudorabies virus are dispensable for viral penetration and propagation in the nervous systems of adult mice. J. Virol. 73:10503-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monte, P. D., S. Pignatelli, M. Mach, and M. P. Landini. 2001. The product of human cytomegalovirus UL73 is a new polymorphic structural glycoprotein (gpUL73). J. Hum. Virol. 4:26-34. [PubMed] [Google Scholar]
- 29.Muyrers, J. P., Y. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neubauer, A., M. Beer, C. Brandmuller, O. R. Kaaden, and N. Osterrieder. 1997. Equine herpesvirus 1 mutants devoid of glycoprotein B or M are apathogenic for mice but induce protection against challenge infection. Virology 239:36-45. [DOI] [PubMed] [Google Scholar]
- 31.Osterrieder, N., A. Neubauer, B. Fakler, C. Brandmuller, C. Seyboldt, O. R. Kaaden, and J. D. Baines. 1997. Synthesis and processing of the equine herpesvirus 1 glycoprotein M. Virology 232:230-239. [DOI] [PubMed] [Google Scholar]
- 32.Pignatelli, S., P. Dal Monte, G. Rossini, S. Chou, T. Gojobori, K. Hanada, J. J. Guo, W. Rawlinson, W. Britt, M. Mach, and M. P. Landini. 2003. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 84:647-655. [DOI] [PubMed] [Google Scholar]
- 33.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234:186-195. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoppel, K., E. Hassfurther, W. J. Britt, M. Ohlin, C. A. Borrebaeck, and M. Mach. 1996. Antibodies specific for the antigenic domain 1 (AD-1) of glycoprotein B (gpUL55) of human cytomegalovirus bind to different substructures. Virology 216:133-145. [DOI] [PubMed] [Google Scholar]
- 36.Spaderna, S., H. Blessing, E. Bogner, W. Britt, and M. Mach. 2002. Identification of glycoprotein gpTRL10 as a structural component of human cytomegalovirus. J. Virol. 76:1450-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talbot, P., and J. D. Almeida. 1977. Human cytomegalovirus: purification of enveloped virions and dense bodies. J. Gen. Virol. 36:345-349. [DOI] [PubMed] [Google Scholar]
- 38.Tischer, B. K., D. Schumacher, M. Messerle, M. Wagner, and N. Osterrieder. 2002. The products of the UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 83:997-1003. [DOI] [PubMed] [Google Scholar]
- 39.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the UL49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]