Abstract
As coordinators of energy demands and nutritional supplies, the PGC-1 family of transcriptional coactivators regulates mitochondrial biogenesis to control the cellular bioenergetic state. Aside from maintaining normal and adapted cell physiology, recent studies indicate that PGC-1 coactivators also serve important functions in cancer cells. In fact, by balancing mitochondrial energy production with demands for cell proliferation, these factors are involved in almost every step of tumorigenesis. In this review, we discuss the interplay between PGC-1 coactivators and cancer pathogenesis, including tumor initiation, metastatic spread, and response to treatment. Given their involvement in the functional biology of cancers, identification of regulatory targets that influence PGC-1 expression and activity may reveal novel strategies suitable for mono- or combinatorial cancer therapies.
Keywords: Transcriptional coactivators, mitochondrial biogenesis, cancer metabolism
An Emerging Role of PGC-1 coactivators in Cancer Metabolism
Metabolic reprogramming represents one of the hallmarks of human cancers [1, 2] – a concept first recognized by Otto Warburg and based on observations that glycolytic fermentation in cancer cells persists even in the presence of sufficient oxygen. Although “defective mitochondria” were initially thought to be the cause of aerobic glycolysis [3], we know today that mitochondrial components are relatively rarely defective in tumors [4]. In fact, there is a broad requirement for this organelle to sustain cancer cell survival and growth [5]. In addition to serving as energy producing organelles, which allow efficient ATP generation through oxidative phosphorylation (OxPhos), mitochondria act as biosynthetic macromolecular hubs that permit biomass accumulation [5], and also regulate cellular calcium homeostasis and apoptosis. It is therefore not surprising that tumor cells coerce mitochondrial regulatory pathways to support their degenerate growth.
Mitochondrial mass is regulated by the dynamic balance between biogenesis and degradation. The biogenesis of mitochondria is a highly orchestrated process by which cells replenish this energy-producing organelle with newly synthesized enzymes, respiratory chain components and membrane lipids. Because mitochondria have their own genome (mtDNA), mitochondrial biogenesis requires replication of the mtDNA as well as assembly of both nuclear and mitochondrial encoded proteins. Although two genomes are involved, the nuclear encoded factors play a dominant role [6]. Specifically, mitochondrial biogenesis is critically regulated by the peroxisome proliferator-activated receptor γ (PPARγ) coactivator (PGC) family, which consists of PGC-1α (PPARGC1A), PGC-1β (PPARGC1B) and PRC (PPRC1) (Box 1: Molecular Biology of PGC-1s). The PGC-1 family members interact with and potentiate the activity of a diverse set of transcription factors, exemplified by PPARs, nuclear respiratory factor-1/2 (NRF1/2), yin yang 1 (YY1), and estrogen-related receptors (ERRs), which collectively control expression of a large number of proteins involved in mitochondrial biology. By integrating environmental signaling cues and cellular energy demand, the PGC-1 factors have a main role in the regulation of cellular metabolism (Figure 1). It is therefore intuitive that variances in PGC-1 expression and activity directly affect the metabolic status of tumors, while also shaping responses to metabolic perturbations and modulating functional aspects of tumorigenesis. While the contributions of individual PGC-1-partnering transcription factors such as PPARs and ERRs to oncogenic processes have been elegantly summarized recently [7, 8], in this review we discuss the multifaceted biological roles of the PGC-1 coactivators in cancer biology.
Box 1. Molecular Biology of PGC-1s.
The PGC-1 coactivator family consists of three members, PGC-1α, PGC-1β and PRC that are able to facilitate gene transactivation through of a diverse set of nuclear receptors and transcription factors. The first member, PGC-1α, was initially discovered as an interacting partner for the nuclear receptor PPARγ in brown adipose tissue, driving mitochondrial biogenesis and thermogenesis [65]. Human PGC-1α is a 798-amino acid (797 in mice) protein that is encoded by the PPARGC1A gene located on chromosome 4 (chromosome 5 in mice). Differential promoter usage and alternative splicing of the single PPARGC1A locus gives rise to multiple PGC-1α isoforms. Although these isoforms shared a high level of similarity, their distinct protein structures result in modulation of function across tissue types, at least in mice [66]. Whether these alternative PGC-1α isoforms are differentially involved in tumor pathogenesis remains to be determined.
PGC-1β and PRC were identified by sequence homology with PGC-1α and share functional similarities in the regulation of mitochondrial biogenesis [67-69]. PGC-1β protein, which contains 1.023 amino acids and is encoded by the PPARGC1B locus on chromosome 5 (chromosome 18 in mice), is highly homologous to PGC-1α (about 50% amino acid identity in the N- and C-terminal regions). The third member, PRC, is a significantly larger protein comprising of 1,664 amino acids, and is encoded by the PPRC1 gene on chromosome 10 (chromosome 19 in mice). All the PGC-1 family members promote mitochondrial biogenesis, improving oxidative metabolism and antioxidant responses. However, their expression across tissues reflects inherent oxidative requirements, wherein uniformly high levels are detected in the heart, skeletal muscle, brown fat, liver, brain, and kidney.
All three PGC-1 coactivator family members have an activation domain at the N-terminal containing the nuclear receptor coactivator motif LXXLL [70]. This activation domain recruits multiple protein complexes that facilitate transcription, such as histone acetyltransferases (CBP/p300 and SRC1) [71]. In addition, a proline-rich domain separates the two distinct recognition motifs for the nuclear respiratory factor-1 (NRF1)[67]. Preceding the C-terminus is an interaction domain for the Mediator complex—also known as the thyroid receptor-associated protein/vitamin D receptor-interacting protein (TRAP/DRIP) complex, which mediates transcription initiation [72]. This region is also thought to recruit the SWI/SNF chromatin remodeling complex [73], thus providing another layer of transcription activation. The C-terminal contains an Arginine/Serine-rich domain (RS) and a RNA recognition motif (RRM), both of which can couple pre-mRNA splicing with transcription [74]. Collectively, the PGC-1 family members are docking platforms for the assembly of the transcription machinery, orchestrating the functions of transcription factors, chromatin modifying complexes, and transcription initiators that act in concert to drive target gene expression.
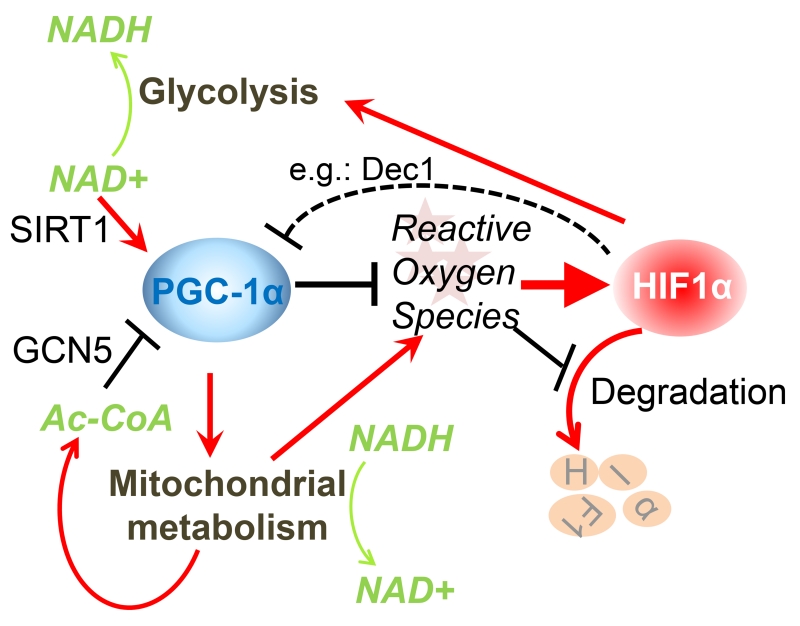
Figure 1. Interplay between PGC-1α and HIFs in metabolic regulation.
Both PGC-1α and HIF-1α serve as sensors and effectors for mitochondrial oxidative metabolism and cytoplasmic glycolysis. PGC-1α promotes oxidative metabolism, in a manner that is directly regulated through reversible acetylation. High acetyl-CoA levels increase the acetyl transferase activity of GCN5 that suppresses PGC-1α function. Conversely, an increase in NAD+ levels indicates energy demand and activates SIRT1 to increase PGC-1α function by deacetylation. To meet the harmful effects of increased mitochondrial energy production, PGC-1α orchestrates a cellular antioxidant program that prevents ROS accumulation, which in turn destabilizes HIF-1α. On the other hand, HIF-1α impedes mitochondrial activity, either by suppressing PGC-1α transcription levels through promoting expression of the repressor Dec1, or by promoting glycolysis at the expense of mitochondrial glucose flux and reduction of NAD+ levels. In the figure, red arrows indicate activation while black bars represent suppression.
PGC-1s and Cancer Initiation
Senescence is an important barrier to tumorigenesis following oncogene activation or genotoxic stresses and is characterized by permanent withdrawal from the cell cycle. Excessive reactive oxygen species (ROS) can induce senescence [9], and because mitochondria are the major cellular source of ROS, reducing mitochondrial oxidative processes may drive escape from oncogene-induced senescence (OIS) [10]. Considering that PGC-1α is able to orchestrate both mitochondrial bioenergetics and a ROS detoxification program [11], a link between PGC-1s and senescence is therefore intuitive. A recent study indicated that mitochondrial biogenesis is indispensable for the maintenance of the replicative senescent phenotype[12]. Upon genotoxic insults, the activated DNA damage response triggers mTORC1-mediated induction of PGC-1α and PGC-1β, leading to increased mitochondrial biogenesis and induction of senescence. Depletion of PGC-1β in mouse embryonic fibroblasts suppresses senescence, concomitant with attenuated induction of p16INK4a and the pro-inflammatory secretome. While demonstrating how PGC-1s impact OIS needs to be firmly demonstrated, current studies certainly suggest a role for PGC-1 factors in ROS attenuation and senescence execution.
Given the association of PGC-1s with control of senescence, it is plausible that these coactivators may possess tumor-suppressive functions in certain contexts. Evidence in support of this notion exists: for example, clinical correlative analyses indicated that PGC-1α expression is lower in colon and ovarian tumors than their respective adjacent normal tissues [13, 14]. Furthermore, and in line with these data, nullizygousity for PGC-1α within engineered tumor-prone mouse backgrounds enhances colorectal and prostate carcinoma development [15, 16]. On the other hand, the oxidative and lipogenic functions of PGC-1s, which will be discussed in detail in the following sections, also suggest a role in tumor promotion. For instance, genetic ablation of PGC-1α or PGC-1β protects mice from chemical-induced liver, colon and small intestine carcinogenesis [17, 18], and mice with specific PGC-1β overexpression in the intestine have enhanced incidence of tumors [18]. These studies taken together clearly indicate that PGC-1s impose safeguards against oxidative stress and enforce senescence, but they also endow cells with certain survival functions required for tumor development. Thus, PGC-1 coactivators might promote or suppress tumor initiation in a manner that is dependent on the tissue context and tumor type (Figure 2).
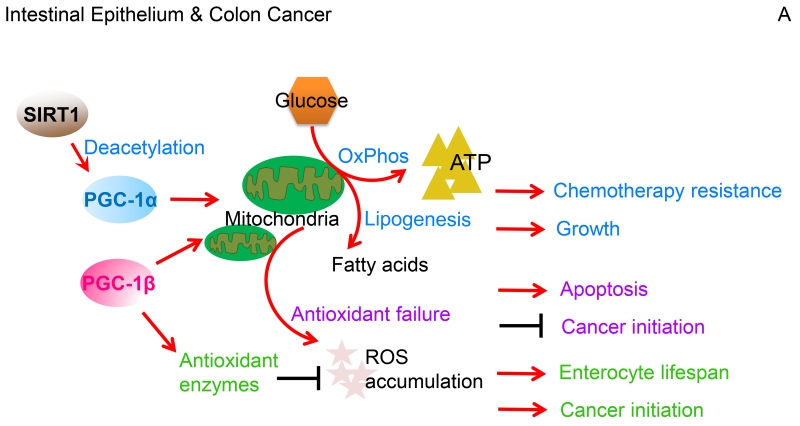
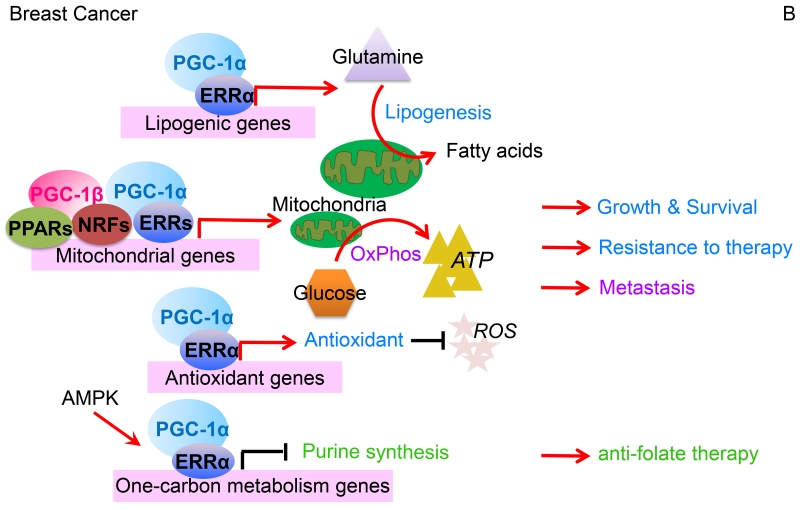
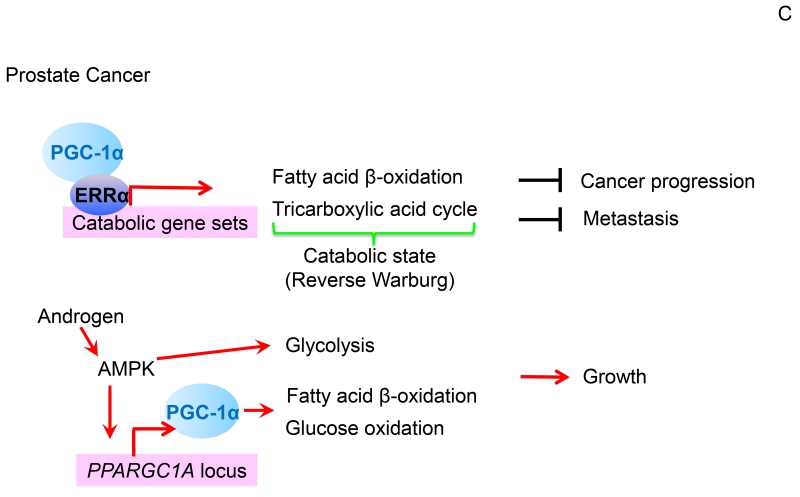
Figure 2. Different functions of PGC-1s across cancer types.
(A). In colon cancer, PGC-1α-mediated mitochondrial biogenesis promotes glucose oxidation into energy and fatty acid synthesis, which in response to chemotherapy facilitates tumor cell growth and survival. SIRT1 accentuates these effects by deacetylating PGC-1α to potentiate transcriptional co-activation. Intestinal epithelial enterocytes have an elevated expression of the PGC-1 factors and thereby enhance oxidative metabolic pathways. Furthermore, PGC-1β coordinates ROS levels through the cellular antioxidant scavenging, which increases enterocyte survival and increases the likelihood for cancer development. However, when ROS exceeds the detoxification capacity, induction of apoptosis clears redox-compromised cells, and hence, blocks tumorigenic transformation. (B). In breast cancer, PGC-1 coactivators stimulate certain nuclear receptors and transcription factors, including PPARs, ERRs, and NRFs to drive mitochondrial biogenesis, which enhances OxPhos to elevate ATP generation. Through ERRα, PGC-1α co-activates expression of lipogenic genes that drive an increased conversion of glutamine into fatty acids. Additionally, the PGC-1α/ERRα axis coordinates cellular redox balance by regulating antioxidant scavenging. Through allowing adaptation to metabolic stress, these PGC-1-mediated cellular processes afford breast cancer growth, survival, metastatic spread, and resistance to therapy. In response to AMPK activation, however, PGC-1α/ERRα downregulates expression of one-carbon metabolism (folate cycle) enzymes, thus increasing the susceptibility of breast cancer cells to anti-folate therapy. (C). In highly glycolytic prostate cancers, the PGC-1α/ERRα complex executes catabolic pathways that include fatty acid β-oxidation and tricarboxylic acid cycle metabolism, thus reversing the Warburg effect towards a less aggressive cancerous state. In a subset of androgen-dependent prostate cancers, however, AMPK-signaling equally promotes glycolysis and PGC-1α-dependent catabolic pathways that overall culminates in accelerated tumor growth. In this Figure, red arrows indicate activation while black bars represent suppression.
Balancing Nutrient Usage to Sustain Cancer Cell Proliferation and Survival
Tumor cells must balance their metabolic demands and provide anabolic biomolecular synthesis to sustain proliferation, while maintaining sufficient catabolic pathways to produce energy. An elevated glycolytic flux provides cancer cells with ATP; however, the mitochondrial tricarboxylic acid (TCA) cycle and coupled OxPhos efficiently supply cancer cells with large amounts of ATP, macromolecular intermediates for biosynthesis and acetyl/methyl sources for epigenetic regulation of gene expression [19]. Consequently, well-tuned mitochondria allow efficient production of energy from nutrients and leave energy surplus for other metabolic uses, such as anabolic pathways. Evidence has highlighted de novo lipogenesis as a distinctive anabolic feature of malignant cells [20]. Lipogenic fluxes consist of carbons from glucose and glutamine that enter the TCA cycle as acetyl coenzyme A or as α-ketoglutarate, respectively, and depending on metabolic demands, supply cytoplasmic citrate for fatty acid synthesis with the help of lipogenic enzymes [20]. PGC-1β is a positive regulator of lipogenesis in liver [21, 22] and this anabolic function is adopted by PGC-1s in tumors. In liver and colon cancer cells, PGC-1α drives the expression of key lipogenic enzymes and of mitochondrial citrate exporter, enabling TCA carbon flux towards anabolic processes that fuel tumor growth (Figure 2A)[17].
Glutamine, which is the most abundant amino acid in plasma, can serve as an anaplerotic mitochondrial fuel and seems to be critical for tumor survival [23]. In ERBB2-positive breast cancer cells, the PGC-1α/ERRα complex directly modulates the expression of glutamine metabolism enzymes, which enhance glutamine uptake and influx into the TCA cycle, leading to the donation of glutamine carbons to de novo fatty acid biosynthesis [24]. PGC-1α overexpression, or ERRα activation, confers breast cancer cells growth advantages even under limited nutrients, echoed by correlative clinical data that high expression of PGC-1α, and its downstream glutamine pathway target genes, are associated with poor patient prognosis [24]. By orchestrating different transcriptional programs governing anabolic and catabolic metabolisms, PGC-1α balances the bioenergetic supply with macromolecular biosynthesis to optimize cancer cell growth and survival (Figure 2B).
Transcriptional Circuits that Regulate PGC-1 Expression in Tumors
The expression of PGC-1 coactivators in tumors is influenced by various transcriptional pathways. One example occurs in melanoma, where elevated expression of PGC-1α controls growth and survival downstream of the melanocyte-lineage transcription master regulator and oncogene MITF [25, 26]. High PGC-1α expression drives dependence on mitochondrial respiration and resistance to oxidative stress in melanoma cells, and conversely, melanomas with low PGC-1α levels are more reliant on glycolysis and display increased sensitivity to oxidative stress (Figure 3A). While melanoma cells with high PGC-1α expression are addicted to OxPhos, acute PGC-1α deletion produces some cells that survive by reprogramming their metabolism towards enhanced glycolysis. Furthermore, when glycolysis is inhibited in these PGC-1α-depleted melanoma cells, a metabolic shift to increased glutamine utilization protects the cells from complete elimination [27]. These findings not only suggest that high metabolic and bioenergetic flexibility exists in cancer cells, but also imply that targeting cancer metabolic components will require combinatorial approaches.
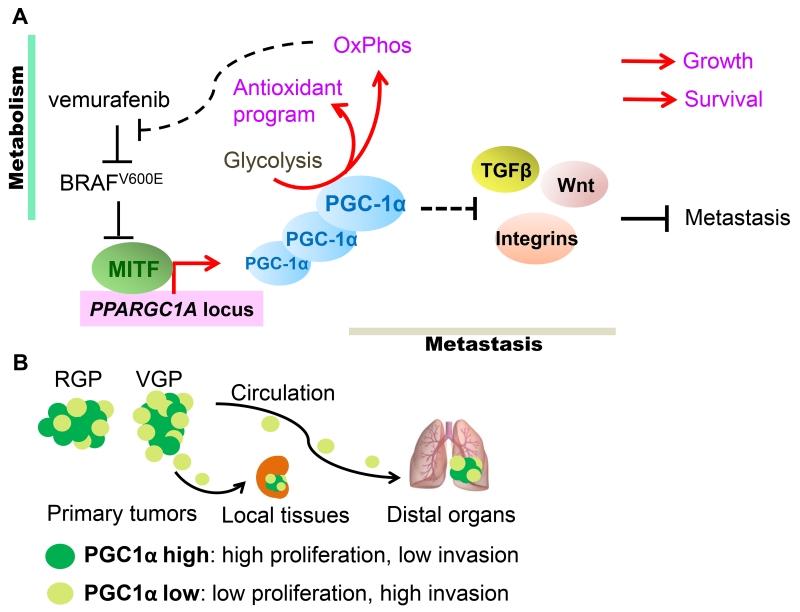
Figure 3. The dualistic nature of PGC-1α in melanomas.
(A). In melanomas, expression of PGC-1α is regulated by the melanocyte master-transcription factor MITF, whose expression in turn is suppressed by signaling downstream of oncogenic NRAS and BRAFV600E. In addition to promoting OxPhos, heightened PGC-1α expression also induces ROS detoxification that ensures cellular survival. Furthermore, in response to BRAF-targeted therapies, such as vemurafenib, mitochondrial biogenesis affords metabolic survival benefits to melanoma cells. However, PGC-1α also attenuates the expression of integrins, TGFβ and Wnt components involved in metastatic dissemination. Consequently, PGC-1α promotes melanoma growth and survival, but through regulation of parallel acting transcriptional programs, also suppresses metastatic spread. (B). Melanoma tumors contain subpopulations of PGC-1α-high and -low cells. The proliferative PGC-1α-high cells are supported by oxidative metabolism, while the PGC-1α-low cells display increased metastatic ability. Specifically, radial growth melanomas (RGP) tend to express more PGC-1α while the invading vertically growth melanomas (VGP) have lessα. These PGC-1α-low cells are endowed with the capacity to locally invade and/or disseminate to circulation and seed distal organs, however once seeded, increased PGC-1α expression facilitates their growth.
Akin to the role of PGC-1α in melanomas, the androgen receptor - AMP-activated protein kinase (AMPK) signaling axis controls expression of PGC-1α in prostate tumors and drives growth advantages (Figure 2C) [28]. Similarly, in breast tumors overexpressing ERBB2, increased PGC-1β expression is associated with elevated mitochondrial respiration, enhanced ROS detoxification and a more aggressive clinical phenotype (Figure 2B) [29]. However, using an ERBB2-negative mammary carcinoma model, PGC-1α overexpression, despite enhancing bioenergetic efficiency, fails to accelerate tumor growth, which may relate to precocious activation of the mesenchymal-to-epithelial transition [30].
Advantages conferred by PGC-1α can also be illustrated in the context of tumor TP53 status. PGC-1α expression is significantly higher in lung adenocarcinomas with wild-type p53 than in tumors with mutant p53 [31]. Following metabolic stress, PGC-1α promotes the expression of genes involved in mitochondrial biogenesis, and in complex with p53 also coactivates transcription of cell cycle inhibitors, which are cooperative functions that enhance cell survival. Interestingly, oxidative stress in the context of PGC-1α genetic suppression results in p53-induced apoptosis [32]. Increased levels of PGC-1α may also prevent p53-induced cell death by maintaining an adequate balance between OxPhos and glycolysis [33].
Although PGC-1α is beneficial to some cancers, it is detrimental to others. Clear cell renal cell carcinomas (ccRCC) are characterized by frequent inactivation of the von Hippel-Lindau (VHL) E3 ubiquitin ligase, leading to the accumulation of HIFs and accelerated glycolysis [34]. HIF-1α regulates expression of the transcriptional repressor Dec1, which in turn suppresses PGC-1α expression and reduces the efficiency of mitochondrial respiration (Figure 1) [35]. Although PGC-1α re-expression in VHL-deficient cells restores mitochondrial function, the increased oxidative stress potently inhibits ccRCC growth and enhances sensitivity to cytotoxic therapies [35]. This is consistent with clinical ccRCC data indicating that higher mitochondrial mass correlates with reduced tumor aggressiveness [36], and conversely, that lower PGC-1α levels associate with worse patient outcome [35]. Similarly, in colorectal cancer cells with high glycolysis, PGC-1α overexpression suppresses tumor growth by elevating intrinsic oxidative stress [15]. Although more in depth mechanistic insights are required, it is conceptually interesting to note that, by interacting with heat-shock factor 1 (HSF1), PGC-1α attenuates stress responses necessary for cancer survival [37], providing an additional mechanism for PGC-1α’s tumor suppressive effects.
PGC-1s, Tumor Microenvironment and Metastasis
Apart from exerting tumor intrinsic effects, PGC-1 coactivators also contribute to shaping the tumor microenvironment, largely through regulation of angiogenesis. PGC-1α promotes vascularization in multiple myeloma and some breast cancers [38, 39]. In breast tumors, increased nutrient availability following tumor angiogenesis alleviates the unfolded protein response (UPR) stress and allows ERBB2 translation and tumor growth [39]. The mechanism by which PGC-1α enhances cancer angiogenesis is still elusive, although in the normal physiology of tissues such as the skeletal muscle, PGC-1α drives angiogenesis through direct up-regulation of vascular endothelial growth factor (VEGF) [40], or by indirectly stabilizing HIFs [41, 42].
Metastasis, which involves matrix invasion, intra-/extravasation of vessels, and capacity to seed and grow in a distal microenvironment, represents the ultimate and fatal trait of tumors. In order to metastasize while maintaining cellular integrity and survival, malignant cells reorganize their functional behaviors which involves switching from a fast proliferating state to a slow growing, but highly invasive state [43]. The metabolic requirements that endow metastatic dissemination and allow cells to switch between proliferation and migration/invasion are by large unknown. Interestingly, recent evidence suggests that PGC-1α is involved in this metastatic switch. For instance, circulating mammary epithelium cancer cells exhibit increased expression of PGC-1α and enhanced mitochondrial biogenesis, which associates with distant metastasis and poor patient outcome [44]. PGC-1α knockdown compromises ATP production, reduces actin cytoskeleton remodeling, lowers anchorage-independent survival, and decreases intra-/extravasation, all checkpoints that prevent metastasis (Figure 2B) [44]. In contrast, PGC-1α suppresses metastatic spread of prostate carcinomas, which appears to be due to an ERRα-dependent transcriptional program (Figure 2C). Not only the expression of PGC-1α itself but also of the ERRα-regulated transcriptional program is progressively decreased from human primary prostate tumor to metastasis, highlighting the prognostic value of these gene sets [16].
Further evidence underscoring the role of PGC-1α in the regulation of metastasis comes partly from our recent study of melanoma. Melanoma tumors are thought to comprise functionally heterogeneous cells with variable proliferative and invasive predisposition, but each endowed with some ability to switch between these two functional phenotypes [43]. Interestingly, highly metastatic melanoma cells contain fewer mitochondria and express lower levels of PGC-1α [45, 46]. This PGC-1α-low subpopulation also displays a pro-metastatic expression program that includes integrin, TGFβ, and Wnt signaling pathway components. In poorly invasive melanoma cells, genetic depletion of PGC-1α turns on the pro-metastatic program and enables metastatic spread in immunocompromised mice. Conversely, boosting PGC-1α expression by either ectopic expression or exposure to BRAFV600E inhibitor vemurafenib suppresses metastasis, which involves direct regulation of ID2 and consequent inhibition of TCF4-mediated gene transcription (Figure 3A) [46]. Given its role in promoting the growth of melanoma tumors, PGC-1α acts at a critical juncture between metabolic survival and metastatic dissemination. Heterogeneous PGC-1α expression within a tumor marks different subpopulations with proliferative or invasive properties, while its dynamic regulation likely allows phenotype switching and ensures metabolic adaption to local environment (Figure 3B).
In summary, through regulation of different transcriptional programs, PGC-1α serves as a positive and negative regulator of metastasis. Increasing vascularization in the tumor niche and mitochondrial biogenesis within tumor cells paves the road for cancer dissemination, while boosting a catabolic state in glycolytic tumors and inhibiting invasive signature genes result in suppression of metastasis. The fact that PGC-1α expression displays heterogeneity within tumors and that PGC-1α levels are dynamically regulated during different stages of cancer progression reflects the dualistic nature of PGC-1α in adjusting metabolic states to optimize metastatic processes.
PGC-1s and Therapeutic Responses
In normal physiological conditions, the expression of PGC-1s in metabolic tissues, including fat, liver and muscle is low; however, it is robustly induced upon stresses such as cold, fasting, and exercise. Induction of PGC-1α counteracts various stresses by stimulating the production of heat (mitochondrial uncoupling), glucose (gluconeogenesis), and activating the UPR to constrain inflammation, and hence, maintain cellular and organismal homeostasis. Considering these many cytoprotective functions, it is not surprising that the PGC-1 proteins are involved in responses to anti-cancer drugs. For example, targeted inhibition of BRAFV600E using vemurafenib leads to potent induction of PGC-1α and mitochondrial respiration that contributes to melanoma survival [26]. Increased mitochondrial biogenesis underlies the cytoprotective effects of PGC-1α, because inhibition of either the respiratory chain or mitochondrial chaperones increases the treatment efficacy of vemurafenib (Figure 3A) [26, 47, 48]. Analogous to what was shown for PGC-1-mediated adaptive chemoresistance in colon and non-small cell lung cancers [49, 50], enhancing oxidative metabolism also drives resistance to cytotoxic agents, such as cisplatin, in melanoma cells [51]. Correspondingly, during both normal and hyperglycemic conditions, depletion of PGC-1α elevates ROS levels and increases apoptosis following treatment with chemotherapeutic drugs in multiple myelomas [52, 53]. There also seems to be a consistent upregulation of the PGC-1 coactivators by treatment with chemotherapeutic drugs that is linked to cell survival cues [54]. However, the question remains whether the multidrug-resistance phenotype in tumor cells is empowered by efficient mitochondrial biogenesis and improved ATP production, or driven mainly by increased expression of multidrug efflux transporters [55].
While induction of PGC-1α following drug treatment promotes therapy evasion, the basal expression of PGC-1α in cancer cells may enable specific therapeutic targeting. Pancreatic cancer stem cells (CSCs) exhibit dependency on OxPhos, unlike the differentiated progeny that critically rely on glycolysis. The increased expression of PGC-1α in the CSC fraction of pancreatic cancers restricts their metabolic plasticity, rendering them particularly susceptible to mitochondrial inhibitors such as the anti-diabetic drug metformin, which is thought to target the respiratory chain Complex I [56]. Increased glutamine metabolism and ROS detoxification by the PGC-1α/ERRα axis in breast cancers not only afford growth advantages, as discussed above, but also promote metabolic adaptation to therapeutic stresses such as receptor tyrosine kinase inhibition by lapatinib. Using an ERRα antagonist (Compound 29) to dampen PGC-1α-mediated metabolic reprogramming, lapatinib resistance can be successfully reversed in an experimental mammary tumor model (Figure 2B) [57]. Furthermore, a reduction in one-carbon metabolism rates represents another functional consequence of PGC-1α expression in breast cancer [58]. Folate intermediates generated in the one-carbon metabolic pathways provide basic building units for the production of purines and thymidylate necessary for nucleotide synthesis and cell proliferation. In response to AMPK stimulation, the PGC-1α/ERRα complex directly impedes the transcription of several one-carbon biosynthetic enzymes, resulting in substantial reductions in purine levels. Accordingly, breast cancers with elevated PGC-1α activity are more reliant on a resilient folate cycle and thereby more vulnerable to anti-folate drugs such as methotrexate (Figure 2B) [58].
Concluding Remarks
Although recent work underscored new aspects of the PGC-1 coactivators in cancer initiation, progression and response to treatment, a coherent picture of PGC-1s in tumor pathogenesis has yet to emerge. Depending on variations in tissue origin, disease stage, specific microenvironment and probably even the experimental setting, different cancer cells might manifest differential preference for the expression and activity of PGC-1 coactivators. Unlike for genuine tumor suppressors or oncogenes, genetic missense mutations, deletions or amplifications have rarely been detected for PGC-1 coactivators; however, cancer cells instead exploit the use of PGC-1α function through dynamic modulation of its expression, which allows rapid optimization of their metabolic status. Adding to the complexity, as cancer is increasingly recognized to be a heterogeneous disease with different subpopulations adapting to distinct metabolic states to fulfill their unique functional demands, the consequences of overexpression or suppression of PGC-1s in each subpopulation may vary. Considering the adaptive roles of PGC-1s in shaping cellular metabolism, it is tempting to speculate that these factors play an important role in tumor heterogeneity, and may help to explain the distinct expression patterns observed across cancer types (Table 1).
Table 1. Summary of the expression and functions of PGC-1 coactivators in various cancers.
| Tissue/Tumor | Experimental Model |
Expression Level | Cellular Process regulated | Reference |
|---|---|---|---|---|
| Breast Cancer | Human specimens and cell lines |
Heterogeneous PGC-1α
levels: high expression correlates with reduced survival & metastases |
Promotes glutamine metabolism in
ERBB2+ breast cancers to enhance growth & chemoresistance; induces mitochondrial respiration & ATP production to drive metastasis; promotes vascularization; dictates sensitivity to anti-folate drugs |
[24, 39, 44,57, 58] |
| Colon Cancer | Human specimens |
Decreased PGC-1α in
tumors compared with normal mucosa |
Not determined | [13] |
| Human specimens and cell line |
Not specified | PGC-1α promotes chemoresistance | [50] | |
| Mouse model & human cell line |
Not specified | PGC-1α promotes tumor growth by
regulating mitochondrial & fatty acid metabolism |
[17] | |
|
Intestinal
Epithelium |
Mouse model & human cell line |
PGC-1α is highly expressed | PGC-1α regulates enterocyte cell fate
& protects against tumorigenesis |
[15] |
| Mouse model | PGC-1β is highly expressed | PGC-1β stimulates mitochondrial
biogenesis & induces antioxidant enzymes |
[18] | |
|
Kidney
Cancer |
Human specimens and cell line |
Decreased PGC-1α correlates with poor outcome |
PGC-1α induces mitochondrial function,
oxidative stress & sensitivity to cytotoxic therapies |
[35] |
| Liver Cancer | Mouse model & human cell line |
Not specified | PGC-1α promotes tumor growth by
regulating mitochondrial & fatty acid metabolism; enhances cell survival with p53 |
[17, 32] |
| Lung Cancer | Human cell lines |
Heterogeneous PGC-1α levels | PGC-1α correlates with an oxidative
metabolism signature; enhances cell survival with wild-type p53. |
[25, 31, 33] |
| Human cell line |
Not specified | PGC-1β promotes chemoresistance | [49] | |
| Melanoma | Human specimens and cell lines |
Increased PGC-1α in
~20% melanomas; highly heterogeneous among individual cells; PGC- 1α level associates with reduced primary melanoma invasion and improved outcome, but with increased aggressiveness of metastatic disease |
PGC-1α promotes mitochondrial
biogenesis, protects against oxidative stress to influence drug sensitivity & survival; suppresses migration/invasion gene sets to inhibit metastasis |
[25, 26, 46] |
|
Multiple
Myeloma |
Human cell line |
Not specified | PGC-1α promotes vascularization
& chemoresistance |
[38, 53] |
|
Ovarian
Cancer |
Human specimens and cell line |
Decreased PGC-1α in
tumors compared with normal ovaries |
PGC-1α induces apoptosis | [14] |
|
Pancreatic
Cancer |
Human cells | Not specified | PGC-1α promotes oxidative metabolism
to maintain cancer stem cell function |
[56] |
|
Prostate
Cancer |
Human specimens, cell line and mouse model |
Progressively decreased
PGC-1α from primary tumors to metastasis |
PGC-1α promotes catabolic metabolism
to suppress cancer progression & metastasis |
[16] |
| Human specimens, cell line and mouse model |
Increased PGC-1α in ~5%
of patients |
PGC-1α promotes mitochondrial
biogenesis, ATP production & cell growth downstream of androgen-AMPK axis. |
[28] |
Furthermore, it remains unclear whether the PGC-1 proteins can represent tangible and effective targets for anti-cancer therapy (see Outstanding Questions). These factors are ubiquitous and their expression varies across tissues, and hence, broad approaches to inhibit or boost their function may prove ineffective – and even counterproductive. Because the function of PGC-1s is regulated by phosphorylation, acetylation, and methylation (Box 2: The Regulation of PGC-1s Expression and Activity), therapeutic modulation of enzymes that alter post-translational modifications of PGC-1s is likely the most straightforward approach for anti-cancer strategies. As transcriptional coactivators, all PGC-1 family members interact with DNA-bound factors. Therefore, molecular approaches that intervene in their specific recruitment can also be exploited. For example, interfering with the interaction between PGC-1α and ERRα by antagonists, such as Compound 29 as mentioned above, is effective in multiple experimental models of human cancers [57, 59]. Almost without exception, all studies so far have focused on PGC-1 transcriptional coactivator functions, neglecting the fact that each of the PGC-1 family members harbors a conserved RNA-binding motif. Functional characterization of the RNA binding motif could help developing aptamer-based methods to interfere with this yet uncharted domain.
OUTSTANDING QUESTIONS.
In the functional biology of different tumor types, which transcription factors recruit PGC-1 family members?
Are there non-transcriptional PGC-1 functions that contribute to tumor growth?
Which intrinsic pathways with functional relevance affect PGC-1 levels in tumor cells?
To which extent can tumor heterogeneity be explained by nutrient-dependent cues acting on PGC-1 function?
How can the PGC-1 coactivators be used as rational targets for anti-cancer therapy?
Box 2. Regulation of PGC-1s Expression and Activity.
The expression and activity of PGC-1 coactivators are regulated at multiple levels. Expression of PGC-1 family members is collectively regulated by transcriptional cues involving: myocyte, enhancer factor 2 (MEF2s), cAMP response element-binding protein (CREB), activating transcription factor 2 (ATF2), and microphthalmia-associated transcription factor (MITF) [26, 75-77]. The abundance of PGC-1 coactivators is also regulated through changes in protein stability. For example, phosphorylation by glycogen synthase kinase 3β (GSK3β) is able to enhance PGC-1α proteasomal degradation [78], largely mediated by N-terminal ubiquitination by Skp1/Cullin/F-box-cell division control 4 (SCFCdc4) or Ring Finger Protein 34 (RNF34)[79, 80]. Additionally, phosphorylation by various kinases differentially affects the activity of PGC-1α, for example, AMP-activated protein kinase (AMPK) and p38 MAPK stimulate[81, 82], while Akt and CDC-like kinase 2 (Clk2) suppress PGC-1α activity [83-85]. Other post-translational modifications such as lysine acetylation (as maintained by the dynamic balance between acetyltransferase GCN5 and deacetylase SIRT1) also control the transcriptional activity of PGC-1s [86-88]. As both GCN5 and SIRT1 sense energy availability and demand, respectively, acetylation coordinates PGC-1 activity with the cellular energy status. PGC-1s can be furthermore activated by methylation via protein arginine methyltransferase 1 (PRMT1) [89]. In depth analysis of converging signals that regulate the function of PGC-1 have been previously reviewed [90].
The stromal microenvironment constitutes an important contributor to cancer growth. Whether PGC-1 members serve functions in fibroblasts or endothelial cells involved in the creation of stromal niches is entirely unknown at this time. However, there is evidence for roles in tumor-immune cell interactions; for example, PGC-1β has an important function in M2 macrophages, the macrophage population that promotes tumor escape and progression [60]. Furthermore, activation of cytolytic immune cells, such as natural killer (NK) cells or CD8+ T-cell subsets seems to rely on glycolysis [61, 62]; however, recent studies indicate a pivotal role for PGC-1α and oxidative metabolism in proper functions of NK cells and tumor-infiltrating T cells [63, 64], suggesting the multiple facets of PGC-1 functions within tumor niches. Understanding the involvement of PGC-1 coactivators between tumor and immune cells will be instrumental to help improve cancer immunotherapies.
In conclusion, recent progress in our understanding of the function of the PGC-1 coactivators have shifted the focus from being mere facilitators of mitochondrial processes to critical regulators of cancer progression. Characterization of pathways that regulate PGC-1s expression and identification of interacting proteins though which PGC-1s exert functions may provide future strategies to curb cancer growth, metastatic spread, and drug resistance.
Trends Box.
The PGC-1 family of transcriptional co-activators exerts central control of cellular bioenergetics through adjustable regulation of mitochondrial biogenesis.
Identifying and targeting metabolic dependencies unique to specific tumor types hold promise for effective cancer treatment.
Although increased anabolic metabolism is a characteristic of growing tumors, the dynamic ability of PGC-1 coactivators to restore mitochondrial catabolism ensures cancer cell survival.
To promote oxidative metabolic pathways, PGC-1 coactivators simultaneously orchestrate cellular antioxidant programs that maintain redox homeostasis.
PGC-1 factors integrate catabolic energy production with anabolic biomass needs, thus leading to an optimized metabolic state that promotes cancer progression as well as resistance to drug-induced stress conditions.
There is a marked heterogeneity in PGC-1α expression among tumor subpopulations that affects the proliferative and invasive functional phenotype; high PGC-1α expression promotes mitochondrial bioenergetics to support proliferation, while low PGC-1α enables transcription of metastasis-promoting genes.
The identification of various pathways regulated by PGC-1s and different components that affect PGC-1 functionality will provide future strategies to target cancer.
Acknowledgements
The authors are grateful to Dr. Ana Batista for editorial supervision, Ms. Katrina Zaralides, Drs. Amy Rines and Meghan Soustek for manuscript assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors have no financial conflicts of interests with regard to the contents of this review.
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O, et al. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8(6):519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zong WX, et al. Mitochondria and Cancer. Mol Cell. 2016;61(5):667–76. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–98. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominy JE, Puigserver P. Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb Perspect Biol. 2013;5(7) doi: 10.1101/cshperspect.a015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deblois G, et al. The PGC-1/ERR signaling axis in cancer. Oncogene. 2013;32(30):3483–90. doi: 10.1038/onc.2012.529. [DOI] [PubMed] [Google Scholar]
- 8.Peters JM, et al. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12(3):181–95. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–67. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498(7452):109–12. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 11.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 12.Correia-Melo C, et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016;35(7):724–42. doi: 10.15252/embj.201592862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feilchenfeldt J, et al. Peroxisome proliferator-activated receptors (PPARs) and associated transcription factors in colon cancer: reduced expression of PPARgamma-coactivator 1 (PGC-1) Cancer Lett. 2004;203(1):25–33. doi: 10.1016/j.canlet.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. PGC-1alpha induces apoptosis in human epithelial ovarian cancer cells through a PPARgamma-dependent pathway. Cell Res. 2007;17(4):363–73. doi: 10.1038/cr.2007.11. [DOI] [PubMed] [Google Scholar]
- 15.D’Errico I, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc Natl Acad Sci U S A. 2011;108(16):6603–8. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrano V, et al. The metabolic co-regulator PGC1alpha suppresses prostate cancer metastasis. Nat Cell Biol. 2016;18(6):645–56. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhalla K, et al. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 2011;71(21):6888–98. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellafante E, et al. PGC-1beta promotes enterocyte lifespan and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. 2014;111(42):E4523–31. doi: 10.1073/pnas.1415279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Deberardinis RJ, et al. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120(2):261–73. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 22.Chambers KT, et al. PGC-1beta and ChREBP partner to cooperatively regulate hepatic lipogenesis in a glucose concentration-dependent manner. Mol Metab. 2013;2(3):194–204. doi: 10.1016/j.molmet.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hensley CT, et al. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuirk S, et al. PGC-1alpha supports glutamine metabolism in breast cancer. Cancer Metab. 2013;1(1):22. doi: 10.1186/2049-3002-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez F, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23(3):302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim JH, et al. Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization. Cancer Res. 2014;74(13):3535–45. doi: 10.1158/0008-5472.CAN-13-2893-T. [DOI] [PubMed] [Google Scholar]
- 28.Tennakoon JB, et al. Androgens regulate prostate cancer cell growth via an AMPK-PGC-1 alpha-mediated metabolic switch. Oncogene. 2014;33(45):5251–61. doi: 10.1038/onc.2013.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Victorino VJ, et al. PGC-1beta regulates HER2-overexpressing breast cancer cells proliferation by metabolic and redox pathways. Tumour Biol. 2016;37(5):6035–44. doi: 10.1007/s13277-015-4449-0. [DOI] [PubMed] [Google Scholar]
- 30.Tiraby C, et al. Estrogen-related receptor gamma promotes mesenchymal-to-epithelial transition and suppresses breast tumor growth. Cancer Res. 2011;71(7):2518–28. doi: 10.1158/0008-5472.CAN-10-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taguchi A, et al. Proteomic signatures associated with p53 mutational status in lung adenocarcinoma. Proteomics. 2014;14(23-24):2750–9. doi: 10.1002/pmic.201400378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sen N, et al. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Mol Cell. 2011;44(4):621–34. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 33.Chen W, et al. RIP1 maintains DNA integrity and cell proliferation by regulating PGC-1alpha-mediated mitochondrial oxidative phosphorylation and glycolysis. Cell Death Differ. 2014;21(7):1061–70. doi: 10.1038/cdd.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 35.LaGory EL, et al. Suppression of PGC-1alpha Is Critical for Reprogramming Oxidative Metabolism in Renal Cell Carcinoma. Cell Rep. 2015;12(1):116–27. doi: 10.1016/j.celrep.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonnet H, et al. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23(5):759–68. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 37.Minsky N, Roeder RG. Direct link between metabolic regulation and the heat-shock response through the transcriptional regulator PGC-1alpha. Proc Natl Acad Sci U S A. 2015;112(42):E5669–78. doi: 10.1073/pnas.1516219112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao D, et al. PGC-1alpha integrates glucose metabolism and angiogenesis in multiple myeloma cells by regulating VEGF and GLUT-4. Oncol Rep. 2014;31(3):1205–10. doi: 10.3892/or.2014.2974. [DOI] [PubMed] [Google Scholar]
- 39.Klimcakova E, et al. PGC-1alpha promotes the growth of ErbB2/Neu-induced mammary tumors by regulating nutrient supply. Cancer Res. 2012;72(6):1538–46. doi: 10.1158/0008-5472.CAN-11-2967. [DOI] [PubMed] [Google Scholar]
- 40.Arany Z, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451(7181):1008–12. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 41.Rasbach KA, et al. PGC-1alpha regulates a HIF2alpha-dependent switch in skeletal muscle fiber types. Proc Natl Acad Sci U S A. 2010;107(50):21866–71. doi: 10.1073/pnas.1016089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Hagan KA, et al. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A. 2009;106(7):2188–93. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoek KS, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68(3):650–6. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 44.LeBleu VS, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16(10):992–1003. 1–15. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–91. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo C, et al. A PGC1alpha-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016 doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016;126(5):1834–56. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo C, et al. Breaking BRAF(V600E)-drug resistance by stressing mitochondria. Pigment Cell Melanoma Res. 2016;29(4):401–3. doi: 10.1111/pcmr.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao Z, et al. PGC-1beta mediates adaptive chemoresistance associated with mitochondrial DNA mutations. Oncogene. 2013;32(20):2592–600. doi: 10.1038/onc.2012.259. [DOI] [PubMed] [Google Scholar]
- 50.Vellinga TT, et al. SIRT1/PGC1alpha-Dependent Increase in Oxidative Phosphorylation Supports Chemotherapy Resistance of Colon Cancer. Clin Cancer Res. 2015;21(12):2870–9. doi: 10.1158/1078-0432.CCR-14-2290. [DOI] [PubMed] [Google Scholar]
- 51.Roesch A, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23(6):811–25. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu W, et al. PGC-1alpha is responsible for survival of multiple myeloma cells under hyperglycemia and chemotherapy. Oncol Rep. 2015;33(4):2086–92. doi: 10.3892/or.2015.3809. [DOI] [PubMed] [Google Scholar]
- 53.Cao D, et al. Inhibition of PGC-1alpha after chemotherapy-mediated insult confines multiple myeloma cell survival by affecting ROS accumulation. Oncol Rep. 2015;33(2):899–904. doi: 10.3892/or.2014.3635. [DOI] [PubMed] [Google Scholar]
- 54.Mouler Rechtman M, et al. The metabolic regulator PGC-1alpha links anti-cancer cytotoxic chemotherapy to reactivation of hepatitis B virus. J Viral Hepat. 2013;20(1):34–41. doi: 10.1111/j.1365-2893.2012.01622.x. [DOI] [PubMed] [Google Scholar]
- 55.Wong K, et al. Towards understanding promiscuity in multidrug efflux pumps. Trends Biochem Sci. 2014;39(1):8–16. doi: 10.1016/j.tibs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Sancho P, et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22(4):590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 57.Deblois G, et al. ERRalpha mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat Commun. 2016;7:12156. doi: 10.1038/ncomms12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Audet-Walsh E, et al. The PGC-1alpha/ERRalpha Axis Represses One-Carbon Metabolism and Promotes Sensitivity to Anti-folate Therapy in Breast Cancer. Cell Rep. 2016;14(4):920–31. doi: 10.1016/j.celrep.2015.12.086. [DOI] [PubMed] [Google Scholar]
- 59.Park S, et al. ERRalpha-Regulated Lactate Metabolism Contributes to Resistance to Targeted Therapies in Breast Cancer. Cell Rep. 2016;15(2):323–35. doi: 10.1016/j.celrep.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vats D, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4(1):13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Sullivan D, Pearce EL. Targeting T cell metabolism for therapy. Trends Immunol. 2015;36(2):71–80. doi: 10.1016/j.it.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finlay DK. Metabolic regulation of natural killer cells. Biochem Soc Trans. 2015;43(4):758–62. doi: 10.1042/BST20150116. [DOI] [PubMed] [Google Scholar]
- 63.Miranda D, et al. PGC-1alpha-Dependent Mitochondrial Adaptation Is Necessary to Sustain IL-2-Induced Activities in Human NK Cells. Mediators Inflamm. 2016;2016:9605253. doi: 10.1155/2016/9605253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scharping NE, et al. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity. 2016;45(2):374–88. doi: 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 66.Martinez-Redondo V, et al. The hitchhiker’s guide to PGC-1alpha isoform structure and biological functions. Diabetologia. 2015;58(9):1969–77. doi: 10.1007/s00125-015-3671-z. [DOI] [PubMed] [Google Scholar]
- 67.Andersson U, Scarpulla RC. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21(11):3738–49. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin J, et al. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277(3):1645–8. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 69.Kressler D, et al. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277(16):13918–25. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 70.Heery DM, et al. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 71.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–71. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 72.Wallberg AE, et al. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12(5):1137–49. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 73.Li S, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8(2):105–17. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monsalve M, et al. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6(2):307–16. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 75.Akimoto T, et al. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280(20):19587–93. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 76.Handschin C, et al. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100(12):7111–6. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–83. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 78.Anderson RM, et al. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7(1):101–11. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Olson BL, et al. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22(2):252–64. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei P, et al. RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1alpha and modulates brown fat cell metabolism. Mol Cell Biol. 2012;32(2):266–75. doi: 10.1128/MCB.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jager S, et al. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104(29):12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Puigserver P, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8(5):971–82. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 83.Li X, et al. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447(7147):1012–6. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 84.Rodgers JT, et al. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 2010;11(1):23–34. doi: 10.1016/j.cmet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tabata M, et al. Cdc2-like kinase 2 suppresses hepatic fatty acid oxidation and ketogenesis through disruption of the PGC-1alpha and MED1 complex. Diabetes. 2014;63(5):1519–32. doi: 10.2337/db13-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 87.Lerin C, et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3(6):429–38. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 88.Kelly TJ, et al. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1beta through lysine acetylation. J Biol Chem. 2009;284(30):19945–52. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teyssier C, et al. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19(12):1466–73. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93(4):884S–90. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]