Abstract
Despite the immune-reconstitution with antiretroviral therapy (ART), HIV-infected individuals remain highly susceptible to tuberculosis (TB) and have an enrichment of oral anaerobes in the lung. Products of bacterial anaerobic metabolism, like butyrate and other short chain fatty acids (SCFAs), induce regulatory T cells (Tregs). We tested if SCFAs contribute to poor TB control in a longitudinal cohort of ART treated HIV-infected South Africans. Increase in serum SCFAs was associated with increased TB susceptibility. SCFAs inhibited IFN-γ and IL-17A production in peripheral blood mononuclear cells from HIV-infected ART-treated individuals in response to M. tuberculosis antigen stimulation. Pulmonary SCFAs correlated with increased oral anaerobes such as Prevotella in the lung and with M tuberculosis antigen-induced Tregs. Metabolites from anaerobic bacterial fermentation may therefore increase TB susceptibility by suppressing IFN-γ and IL-17A production during the cellular immune response to M. tuberculosis.
Keywords: HIV, tuberculosis, lung, dysbiosis, short chain fatty acids, FoxP3, FoxP1
eTOC
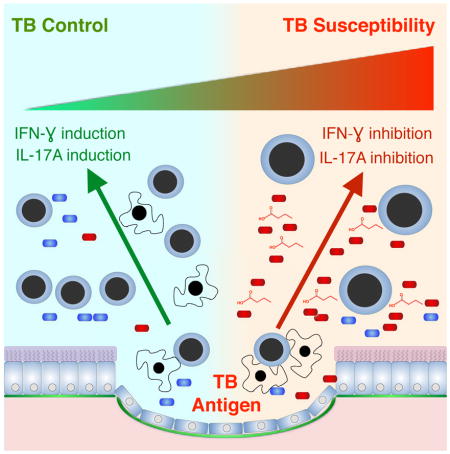
HIV patients on anti-retroviral therapy (ART) are vulnerable to tuberculosis. Segal et al. show that short chain fatty acids (SCFAs) produced by the increased abundance of lung anaerobic bacteria in these patients inhibit the immune response to M. tuberculosis likely enhancing tuberculosis susceptibility.
Introduction
HIV-infected individuals on ART have residual immune compromise that has contributed to the disastrous tuberculosis (TB) epidemic in South Africa (Churchyard et al., 2014; Group et al., 2015). Immunologically, post-ART lung lymphocytes have a deficiency of IFN-γ production in response to TB antigens (Jambo et al., 2014). The nature of the persistent immune compromise despite ART is poorly understood.
The lower airway contains complex bacterial communities that correlate with immune competence and levels of inflammation (Lozupone et al., 2013; Morris et al., 2013; Segal et al., 2013; Twigg et al., 2016). Similar to the gut, specific lung microbiomes are associated with Th17 immunity (Segal et al., 2016). In addition to generating immune responses, bacterial metabolites occur in the lower airway consistent with active bacterial metabolism (Segal et al., 2017). Investigating the lung microbiome, however, has been hampered by two sets of confounders. First, the lower airways have low bacterial burden. Approximately half of HIV uninfected individuals have bacterial communities that cluster with background observed in laboratory methods controls (Segal et al., 2016). Second, aspiration from the upper airway could contribute to individuals’ lower airway microbiome, challenging our ability to discern lower airway viability of microbes. More functional data is required to establish the degree to which oral microbes live and grow in the lung.
Compared with HIV uninfected individuals, post-ART HIV infected individuals with median CD4+ lymphocytes of 375 cells/μL have increased relative abundance of lower airway anaerobic bacteria oral commensals like Prevotella and Veillonella (Twigg et al., 2016). Anaerobes like Prevotella use fermentation for energy production during hypoxia. The major products of bacterial fermentation are short chain fatty acids (SCFAs) such as acetate (C2), propionate (C3), and butyrate (C4)(Bourriaud et al., 2005). In the gut, SCFA drive CD4+ lymphocytes to the forkhead transcription factor FoxP3 expressing Treg phenotype with beneficial anti-inflammatory effects (Trompette et al., 2014). Alternately, Treg can be immune compromising, inhibiting TB control (Semple et al., 2013).
IFN-γ and IL-17A producing CD4+ and CD8+ lymphocytes provide mucosal immunity to TB (Gopal et al., 2013; Khader et al., 2007). Post-ART, both of these lymphocyte populations remain dysfunctional (Cosgrove et al., 2013; Leeansyah et al., 2013). We have completed a longitudinal study of ART-treated HIV-infected individuals in Cape Town, South Africa with high post-enrollment TB incidence. Since the lungs of post-ART individuals are enriched with anaerobes capable of SCFA synthesis and SCFA increase Treg that inhibit TB control, we used cells and metabolites from this cohort to test the hypothesis that baseline SCFA is a risk factor for active TB. We also assessed the lung microbiome and metagenome to understand the characteristics of the bacterial communities associated with high pulmonary SCFA concentration defined by detectable BAL propionate. We observed baseline serum butyrate and propionate were associated with the subsequent increasing hazard of TB. Further, high pulmonary SCFA correlated with enrichment of anaerobes in the lung and Treg induction by TB antigens. Finally, butyrate inhibited TB antigen induced IFN-γ and IL-17A. These data suggest metabolic products of anaerobic fermentation increase TB susceptibility.
Results
Serum IFN-γ, IL-17A, butyrate and propionate predict subsequent TB hazard
We prospectively followed 193 HIV infected individuals on ART in Cape Town South Africa over three years with a TB rate of 2.6 cases per 100 person years. The cohort CD4+ lymphocyte count was 383±203 cells/μl and 46% had cellular immunity to M. tuberculosis defined by a commercial interferon-gamma release assay (Quantiferon) (Table S1). Serum SCFAs were measured at enrollment. Compared with HIV uninfected blood donors, HIV infected individuals have higher blood SCFA concentration (Figure S1). Those who proceeded to TB had a trend toward higher butyrate concentration (16.4 [15.2–19.8] μM vs. 14.7 [12.1–17.4] μM p=0.063 Table S1). IFN-γ and IL-17 were also assayed in enrolment serum. TB cases had significantly lower levels of IFN-γ than those who did not develop TB (Median [IQR]=6.1[5.4–10.0] pg/ml vs. 10[7–12.1] pg/ml, p=0.031) and a trend to lower IL-17A (1.6 [0.65–4.7] pg/ml vs. 3.6 [2.2–6.1] pg/ml, p=0.056 Table S1).
To assess the contribution of SCFA and cytokines to subsequent TB hazard, we used four independent Cox multivariable models, two with IFN-γ combined with SCFAs and two with IL-17A combined with SCFAs. Doubling butyrate or halving IFN-γ increased subsequent TB hazard 2.0 fold (p=0.008) and 5.8 fold (p=0.024) respectively (Table 1). Doubling propionate or halving IFN-γ increased subsequent TB hazard 2.1 fold (p=0.006) and produced a trend for increased TB hazard 3.5 fold (p=0.079). Similarly, doubling butyrate or halving IL-17A increased TB hazard 1.6 fold (p=0.011) and 5.3 fold (p=0.017) respectively; doubling propionate or halving IL-17A increased TB hazard 1.7 fold (p=0.005) and 3.7 fold (p=0.047) respectively.
Table 1.
Cox Multi-variable models of Incident TB*
| TB predictors in enrollment serum | HR | 95% Cl | p-value | |
|---|---|---|---|---|
| Models with IFN-γ | ||||
| 1) | Halving IFN-γ | 2.0 | 1.2–3.5 | 0.008 |
| Doubling Butyrate | 5.8 | 1.3–26 | 0.024 | |
| 2) | Halving IFN-γ | 2.1 | 1.2–3.7 | 0.006 |
| Doubling Propionate | 3.5 | 0.86–14 | 0.079 | |
|
| ||||
| Models with IL-17A | ||||
| 1) | Halving IL-17A | 1.6 | 1.1–2.2 | 0.011 |
| Doubling Butyrate | 5.3 | 1.4–21 | 0.017 | |
| 2) | Halving IL-17A | 1.7 | 1.2–2.4 | 0.005 |
| Doubling Propionate | 3.7 | 1.0–13 | 0.047 | |
Adjusted for age, gender, vital capacity, body mass index and serum acetate
Propionate and butyrate are present in the lungs of HIV-infected individuals on ART and butyrate impairs cellular immunity to tuberculosis
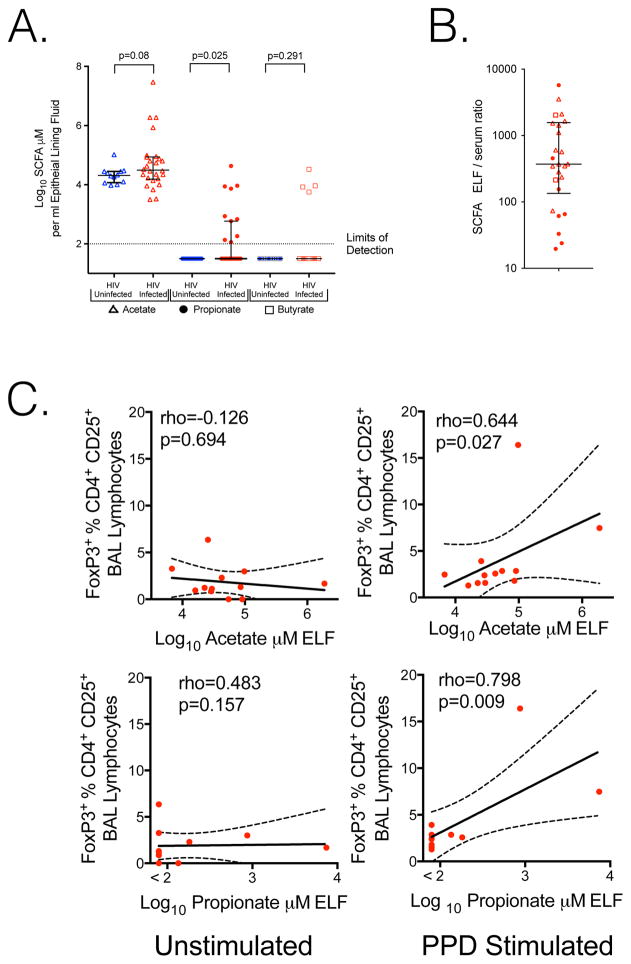
Propionate and butyrate are anti-inflammatory via induction of Treg (Trompette et al., 2014). The concentration and effects of SCFA in the lung, however, have not been explored. To better understand the lower airway environment and measured levels of lung SCFA we performed bronchoscopy with broncho-alveolar lavage (BAL) on 27 HIV-infected (19 from Cape Town and 8 from New York) and 12 HIV-uninfected individuals from New York (Table S2). Butyrate or propionate was detectable in epithelial lining fluid (ELF, Rennard et al., 1986) of 10/27 HIV-infected individuals (8 from Cape Town and 2 from New York), but not in any of 12 HIV uninfected individuals from New York (Figure 1A p=0.025 Mann–Whitney U test). HIV infected individuals also had a trend to increased acetate (p=0.08) but butyrate was not significantly different in BAL (p=0.291) due to the small number of individuals (4/27) with butyrate above the limits of detection. ELF propionate correlated with butyrate and acetate (rho=0.686, p<0.0001 and rho=0.722, p<0.0001 respectively) making detectable propionate a suitable measure of elevated pulmonary SCFA. SCFA in ELF were 370 fold higher than serum in HIV infected individuals with paired samples (Figure 1B). Since Tregs restrict control of M. tuberculosis in the lung (Semple et al., 2013), we evaluated the effects of lower airway SCFA concentration on FoxP3 induction by M tuberculosis antigen derived from purified protein derivative (PPD). Elevated acetate or propionate correlated with greater FoxP3 expressing Tregs in BAL cells after PPD stimulation (Figure 1C rho=0.644, p=0.027 and rho=0.798, p=0.009 respectively).
Figure 1. SCFA are present in the lungs of HIV-infected individuals on ART; inhibiting IFN-γ and IL-17A production after M. tuberculosis antigen stimulation.
A. Acetate (open triangles), Propionate (closed circles) and butyrate (open boxes) are present in BAL. Concentrations are corrected for BAL induced dilution of epithelial lining fluid (ELF). Propionate is significantly elevated in HIV-infected subjects, Mann–Whitney U test. B. In HIV infected individuals with paired samples, ELF SCFA is higher than serum in all subjects, with a median ratio of 370. C. Addition of PPD to BAL cells demonstrates induction of FoxP3 CD4+ cells significantly correlates with in vivo ELF acetate or propionate at the time of bronchoscopy, Spearman correlation.
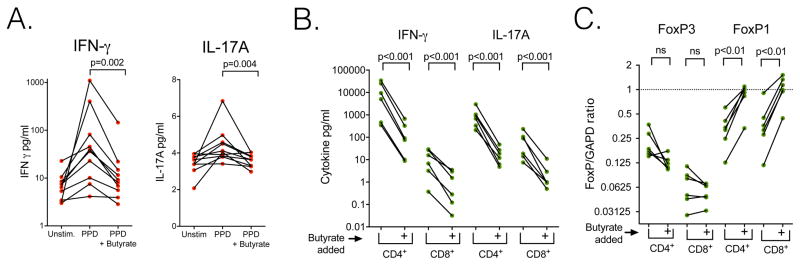
To better understand the impact of SCFA on the immune response to tuberculosis we measured IFN-γ or IL-17A production in peripheral blood mononuclear cells (PBMC) from 10 HIV infected ART treated individuals who had latent tuberculosis infection. PPD stimulation induced both IFN-γ and IL-17A (Figure 2A). Two mM of butyrate, the dose that inhibits inflammatory cytokine production in macrophages and lymphocytes (Dillon et al., 2017), inhibited PPD-stimulated IFN-γ 87% and IL-17A 21% (Figure 2A p=0.002 and p=0.004 respectively). The IFN-γ and IL-17A concentrations released by PBMC cultured ex vivo without PPD were similar to those in PPD stimulated PBMC cultures with exogenous butyrate added (Figure 2A Unstim. vs. PPD+butyrate, ns for both comparisons).
Figure 2. Butyrate inhibits IFN-γ and IL-17A production after M. tuberculosis antigen stimulation and induces FoxP1 mRNA in CD4+ and CD8+ lymphocytes.
A. Compared with PPD, butyrate inhibits IFN-γ and IL-17A production by PPD stimulation. Peripheral blood mononuclear cells of HIV-infected individuals on ART with latent tuberculosis were cultured in vitro without stimulation for three days (Unstim.), with PPD for three days (PPD) or with 2mM butyrate added with PPD three days (PPD+ Butyrate). Paired t test of log transformed data. B. Addition of 2mM butyrate inhibits IFN-γ and IL-17A expression in CD4+ and CD8+ lymphocytes stimulated with anti-CD3/CD28. Lymphocytes from 6 different individuals are shown. Paired t test of log transformed data. C. Addition of 2mM butyrate increases FoxP1 mRNA expression in CD4+ and CD8+ lymphocytes. Paired t test of log transformed FoxP/GAPDH.
To assess which cell types were butyrate targets, we isolated human CD4+ and CD8+ lymphocytes and stimulated cytokine production with CD3/CD28 antibodies. Butyrate reduced IFN-γ secretion 98% in CD4+ and 90% in CD8+ lymphocytes. Butyrate also reduced IL-17A secretion 98% in CD4+ and 96% in CD8+ lymphocytes (Figure 2B p<0.001 for all comparisons). Butyrate did not alter relative FoxP3 mRNA expression in either CD4+ or CD8+ lymphocytes but did increase FoxP1/GAPDH mRNA 7 fold in CD4+ and 9 fold in CD8+ lymphocytes (Figure 2C p<0.01 for both comparisons).
Lung microbiome of propionate-detectable individuals is enriched with anaerobes
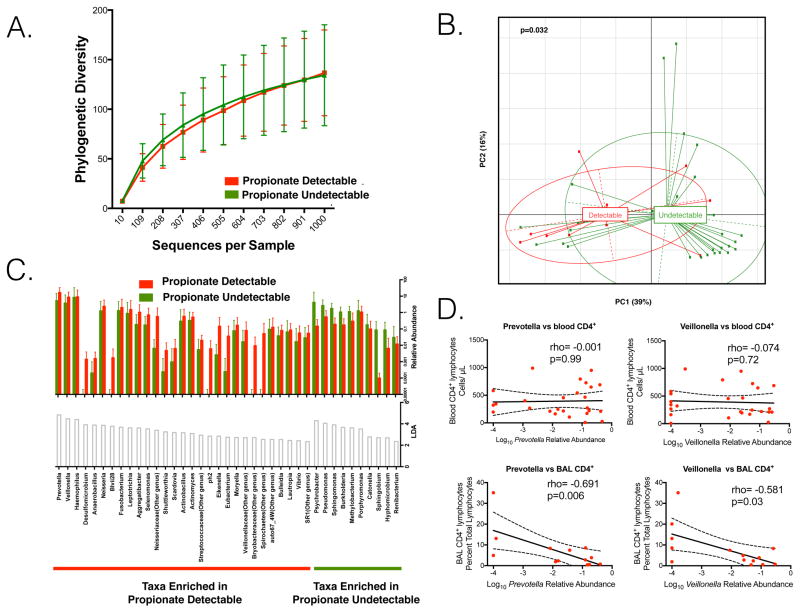
Since SCFA are end-products of bacterial anaerobic metabolism, we utilized 16S rRNA gene sequencing to examine the lower airway microbiota in the bronchoscopy cohort. We compared the microbiome from the 10 BAL samples with detectable propionate levels to the microbiome from the 29 BAL samples with undetectable propionate (Table S2). The BAL microbiome from propionate-detectable individuals did not significantly differ from supraglottic samples based on β diversity analysis (PERMANOVA p=0.17). The propionate-undetectable BAL samples more closely resembled background samples rather than supraglottic samples, although it was significantly different from background samples (PERMANOVA p<0.01, Figure S2 and Table S3). The bacterial rRNA gene concentration (Table S2) and α diversity was similar in propionate undetectable and detectable samples (Figure 3A). However, β diversity based on weighted UniFrac distances demonstrated distinct clustering of subjects with BAL propionate-detectable when compared to propionate-undetectable individuals (Figure 3B, p=0.032 PERMANOVA). There is a trend to difference in β diversity when HIV infected individuals with BAL propionate-detectable are compared to HIV infected individuals with propionate-undetectable (p=0.07 PERMANOVA). To further explore for taxonomic differences between propionate-detectable and undetectable individuals, we utilized Linear-discriminant analysis (LDA) of Effect Size (LEfSe) where significant differences are defined as an LDA>2 (Segata et al., 2011). The lung microbiome of those with propionate-detectable in BAL was enriched with anaerobes such as Prevotella, Veillonella and Haemophilus (Figure 3C red bars LDA>2). The lung microbiome of those with propionate-undetectable in BAL was enriched with Psychrobacter, Pseudomonas and Sphingomonas (Figure 3C green bars LDA>2). As many different taxa can produce SCFA, we characterized bacterial metabolic potential by estimating the entire complement of bacterial genes, i.e. the metagenome. The inferred potential of the lower airway microbiome using taxonomic data (Langille et al., 2013) demonstrated reduction of genes in the Propionate and Butyrate pathways in propionate detectable samples (Figure S3), a finding confirmed with shotgun sequencing of bacterial DNA from BAL (Figure S4)
Figure 3. Propionate-detectable individuals have increased relative abundance of oral anaerobic bacteria in BAL.
A. Rarefaction curve on taxonomic 16S rRNA gene annotation demonstrates no difference in α-diversity between the taxa in the microbiomes of 10 propionate-detectable and 29 propionate-undetectable individuals. B. A principle component analysis based on UniFrac distance demonstrates a significant difference in the β-diversity between propionate-detectable and propionate-undetectable individuals (p value based on PERMANOVA). C. LEfSe demonstrates enrichment of anaerobes such as Prevotella or Veillonella in the lung microbiome of propionate-detectable individuals. The histograms on top of the panel represent relative abundance of each taxa while the histograms below represent the LDA value. The name of the enriched taxa is at the bottom of the panel. D. There is a significant inverse correlation between BAL CD4+ lymphocytes and log10 transformed relative abundance of Prevotella or Veillonella in BAL. Spearman correlation.
We then measured blood and BAL CD4+ lymphocytes to assess if the effectiveness of immune reconstitution correlated with enrichment of anaerobic taxa in the lung. There was no association between blood CD4+ lymphocytes and lung Prevotella or Veillonella relative abundance in post-ART individuals (rho= −0.001, p=0.99 and rho= −0.074, p= 0.72 respectively). However, as BAL CD4+ lymphocytes decreased in HIV-infected individuals, the relative abundances of Prevotella or Veillonella significantly increased (Figure 3D rho= −0.691, p=0.006 and rho= −0.581, p=0.03 respectively).
Discussion
In this report, we investigated a South African HIV-infected ART treated cohort with high incidence of TB to evaluate the characteristics of post-ART TB susceptibility. Similar to a recent lung microbiome study that demonstrated enrichment of oral anaerobes, such as Prevotella, in the lower airways of HIV subjects on ART (Twigg et al., 2016), the subjects evaluated in this study also had incomplete immune reconstitution. In the current study, blood butyrate and propionate, products of anaerobic bacterial fermentation, are increased in HIV infected individuals and are associated with increased TB hazard. Using samples from the lower respiratory tract, we found that increased anaerobes, such as Prevotella, correlated with pulmonary SCFA. In vitro experiments confirm that butyrate inhibits IFN-γ and IL-17A induced by TB antigens at concentrations similar to those found in the lung. Our observation that immunologically active anaerobic metabolites are associated with enrichment in the lung of oral anaerobes, such as Prevotella, illustrates potential consequences of post-ART lung dysbiosis with increased Prevotella (Twigg et al., 2016). These data suggest pulmonary SCFAs, from either lower airway commensal or aspirated microbes, likely contribute to deficiency of IFN-γ induction by TB antigens (Jambo et al., 2014) and increase TB risk in HIV infected individuals on ART (Churchyard et al., 2014; Group et al., 2015; Samandari et al., 2011).
Post-ART, relative abundance of Prevotella and Veillonella is increased in the lung (Twigg et al., 2016). We extend this observation demonstrating Prevotella and Veillonella correlate with CD4+ lymphocytes in the lung but not blood. The inverse correlation between lung oral anaerobes and pulmonary but not systemic CD4+ lymphocytes could be due to reduced lymphocyte recruitment to the lung or due to increased destruction at mucosal sites. SCFA produced by oral anaerobes activates latent HIV in lymphocytes (Imai et al., 2009) including primary human Th-17 cells (Das et al., 2015). Butyrate stimulated HIV replication of latent virus in the lung (Twigg Iii et al., 2008) could produce pulmonary CD4+ lymphocyte destruction. SCFA induced viral replication is a possible mechanism for persistent Th-17 dysfunction observed in HIV infected individuals on ART (Brenchley et al., 2008; El Hed et al., 2010).
In this cohort, reduced serum IFN-γ and IL-17A also increased TB hazard, consistent with the known protective roles of CD4 expressing TH1 and Th17 responses in TB (Gopal et al., 2013; Jambo et al., 2014; Khader et al., 2007). Increased serum butyrate also increased subsequent TB hazard in this cohort. Our data show that butyrate directly inhibits IFN-γ and IL-17 release after TB antigen stimulation of peripheral blood mononuclear cells from HIV infected individuals with latent tuberculosis. Similar to data from lamina propria derived T cells(Dillon et al., 2017), butyrate also inhibits IFN-γ and IL-17A elaboration by both CD4+ and CD8+ lymphocytes. Like CD4+ Th17 cells, CD8+ mucosal associated invariant T cells (MAIT) produce IFN-γ and IL-17A and are important for TB control (Gold et al., 2013). Both CD4+ Th-17 cells and CD8+ MAIT are dysfunctional or depleted in ART-treated HIV-infected individuals (Cosgrove et al., 2013; Leeansyah et al., 2013). The inhibitory effect of butyrate on Th-1 and Th-17 cytokine induction by both CD4+ and CD8+ lymphocytes could contribute to increased TB susceptibility in HIV infected individuals with pulmonary enrichment of oral anaerobes.
Similar to SCFA mediated Treg induction in the gut (Trompette et al., 2014), high pulmonary SCFA was associated with induction of FoxP3-expressing Treg after TB antigen stimulation. Since Treg inhibit control of M. tuberculosis replication in BAL cells (Semple et al., 2013), this could increased TB risk. In isolated lymphocytes, however, butyrate did not induce FoxP3 mRNA. We therefore evaluated the effects of SCFA on other related forkhead transcription factors. Butyrate strongly induces FoxP1 mRNA in both CD4+ and CD8+ lymphocytes. FoxP1 is a repressor that inhibits lymphocyte development as well as CD4+, CD8+, and B cell activation (Durek et al., 2016; Feng et al., 2011; Stephen et al., 2014; Wang et al., 2014; Wei et al., 2016). FoxP1 is a candidate to mediate some of the inhibitory effects of SCFA on CD4+ and CD8+ lymphocyte mediated immune response, but the role of FoxP1 induction by SCFA in the lung host microbial interface needs confirmation in animal models with cell specific conditional deletion.
Since lung SCFA are on average 370 fold higher than blood, diffusion of SCFA from blood cannot explain SCFA observed in the lung. Pulmonary production of SCFA would require hypoxic niches since SCFA are an end product of fermentation used as an energy source by anaerobes under hypoxic conditions (Bourriaud et al., 2005). Obligate anaerobes could survive the oxygen stress of the lower airway by forming multicellular complexes within biofilm that enables hypoxic microenvironments (Lone et al., 2015; Williamson et al., 2012). Failure of the immune response to inhibit biofilm formation could predispose to dysbiosis with anaerobes (Singh et al., 2002; Wakabayashi et al., 2009). A distinct feature of the lower airway environment is a high concentration of surfactant, 90% of which is phospholipids; one of these, arachidonic acid induces biofilm formation (Rao et al., 2011) and is associated with increased relative abundance of the oral anaerobes such as Prevotella and Veillonella in the lung (Segal et al., 2016). The supraglottic space has high anaerobic bacterial burden (Segal et al., 2013) and so could be a source of SCFAs that are aspirated into the lower airways. Further investigations are required to discern the role of aspirated metabolites from in situ lower airway production of SCFA.
Lower airway microbial metagenomic functional capacity correlates with circulating metabolites and mortality in HIV-infected patients with pneumonia (Shenoy et al., 2016), providing evidence that active microbial metabolism affects the outcome of immune-compromised host. Paradoxically, genes encoding metabolic pathways for both propionate and butyrate were decreased in subjects with detectable propionate and butyrate. It is possible that the reduced genomic potential observed in propionate-detectable individuals preferentially affects SCFA catabolic genes limiting SCFA degradation. Alternately, biofilm formation potential and the ability to generate a hypoxic niche in an oxygen rich pulmonary environment may supersede the genomic potential for SCFA metabolism (Humphries et al., 2017; Lone et al., 2015; Williamson et al., 2012). Transcriptional profiling is required to define pathway activity and identify the bacterial species that contribute most to SCFA production.
This study has several limitations. The source of butyrate in the blood of the longitudinal cohort is currently unclear. Since gut and vaginal microbiomes also have HIV associated dysbiosis with epithelial dysfunction (Borgdorff et al., 2016; Serrano-Villar et al., 2016) non-pulmonary sources may contribute to the HIV associated SCFA elevation in the blood. Additionally, the BAL studies are cross sectional. The results are consistent with the microbiome contributing to immune suppression in the lungs of HIV-infected subjects, but this study is unable to assess causality. It is likely that the differences in microbiome that enables production of SCFA are consequences of residual immune suppression. An assessment of the contribution of bacterial SCFA production to local immune suppression will require placebo-controlled randomized clinical trials using SCFA as a measurable outcome. If anaerobically-produced SCFA are immune-compromising, then antibiotic or prebiotic treatments that reduce SCFA (Serrano-Villar et al., 2016) might improve immune response.
In summary, we have expanded a prior observation that HIV infected individuals on ART have pulmonary dysbiosis with enrichment with oral anaerobes (Twigg et al., 2016). Anaerobic overgrowth could lead to elevated pulmonary SCFA that was associated with induction of Treg after M. tuberculosis antigen stimulation of lower airway cells. The pulmonary targeting of SCFA induced Treg (Trompette et al., 2014) in combination with the ability of Treg to inhibit M. tuberculosis control in human alveolar macrophages (Semple et al., 2013) may impose an immune compromising burden in the lung that blunts IFN-γ production (Jambo et al., 2014). Further, the inhibitory effect of butyrate on IFN-γ and IL-17A production by CD4+ and CD8+ lymphocytes is likely to increase TB susceptibility. A better understanding of the immune defect in HIV-infected individuals on ART may inspire future therapeutic strategies to improve pulmonary immunity to TB.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for reagents may be directed to and will be fulfilled by the lead contact, Dr. Michael D. Weiden (Michael.Weiden@nyumc.org). Sequences are available from the NCBI Sequence Read Archive (accession number PRJNA357622).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Lung HIV Longitudinal Cohort
The UCT Lung-HIV cohort enrolled 193 HIV infected individuals on ART. The UCT HIV positive participants were drawn from the NIH-sponsored multicenter Lung HIV study (RO1HL090316). They were enrolled from November 2009 to December 2010. At study enrollment, all subjects had serum stored and complete pulmonary function tests (PFTs). Inclusion criteria: Subjects were HIV-infected on stable anti-retroviral therapy. Exclusion criteria: Active infection at the time of enrolment. The age (38 years old), gender (56%female) and immune status (all HIV positive) of 193 UCT study subjects with were followed for incident tuberculosis are presented in Table S1. Individuals received three yearly assessments for incident TB. Broncho-alveolar lavage Cohort We performed bronchoscopy in 58 HIV infected subjects, and in 27 HIV negative subjects. Procedures were approved by the Institutional Review Boards of New York University (8 HIV infected, 12 HIV uninfected), the University of Cape Town (19 HIV infected), University of Pittsburgh (7 HIV infected, 4 HIV uninfected) and Ohio State University Columbus (28 HIV infected, 4 HIV uninfected). Demographics are shown in Table S2 (range of average age for the groups= 42 to 55 years old; gender range in the groups= 17 to 47% female). Nasal route was used for all bronchoscopies avoiding suctioning until wedged to avoid contamination with upper airway secretions. A single bronchoscope was used for Lung HIV patients. Our prior published data showed that there is no systematic carry over of oral microbiome into the lower airway (Segal et al., 2013; Segal et al., 2016). None of these subjects enrolled had respiratory symptoms or had received any inhaled treatment.
Human primary cell culture
For the ex vivo PPD stimulation of human cells we utilized Ficoll gradient PBMCs and BAL cells. Cells were resuspended in RPMI with 10% FCS and stimulated with 12 micrograms/mL PPD. PBMC supernatants for cytokine measurement were collected in 72 hours. BAL cells for flow cytometry were collected in 12 hours. and PBMCs BAL cells were stained with the following antibodies from BD bioscience: anti-CD3 cat# 557872, anti-CD4 cat# 560650, anti-CD8 cat#555366, anti-CD25 cat# 560987, and anti FoxP3 cat# 17-4776-42. For the ex vivo CD4/CD8 cell experiments, Ficoll gradient isolated PBMCs were stained with anti-CD3 cat# 557872, anti-CD4 cat# 560650, anti-CD8 cat#555366 and FACS-sorted into 24 well plates coated with 5 μg/mL mouse anti-CD3 cat#555337. Then, 1 μg/ml anti-CD28 cat#556620 was added with or without butyrate. We utilized 2mM butyrate (Dillon et al., 2017), a concentration slightly lower than the range of 5.6–33 mM butyrate observed in ELF on those BAL samples with butyrate-detectable levels. Supernatants for cytokine measurement and cell pellets for RT PCR were collected in 72 hours.
METHOD DETAILS
Blood and BAL SCFA measurement
Targeted SCFA assays in BAL fluid (19 from Cape Town, 20 from NYU) and serum (193 from Cape Town) was performed at the University of Michigan Metabolomics core using gas chromatography-mass spectrometry (GCMS). Fifty de-identified blood donor samples provide by the blood bank to the University of Michigan Metabolomics core were run as HIV negative controls. Pooled blood donor samples were included in each of the four runs required to assay the serum samples as an internal control. TB cases were included in each run to avoid batch bias. Samples were kept on ice to prevent SCFA evaporation. Deuterated acetic, butyric and hexanoic acids were added as internal standards. The BAL was acidified and extracted with cold diethyl ether, then injected directly onto a GCMS (ZB-WAXplus column, Agilent 6890- 5975C or equivalent) instrument for measurement of C2 to C9 SCFA species. Internal standards yielded measures of absolute SCFA concentrations with a detection limit of 0.5–7 μM for C3 and C4. Measurements were adjusted by the ratio of BAL/blood urea to account for dilution of ELF during BAL (Rennard et al., 1986).
Cytokine, urea and mRNA measurement
All Serum and supernatants INF-γ and IL-17A were assayed with EMD Milipore Human High Sensitivity T Cell panel HSTCMAG28SPMX21. In serum cytokine assays, TB cases were included in each plate to avoid batch bias. Urea was measured by Elisa (cat# Z5030016). Quantitative RT PCR for FoxP mRNA assay: lymphocyte cDNA was conducted with Biorad PrimePCR™ FAM probes and primers: FoxP1 probe qHsaCIP0027768, FoxP3 probe qHsaCIP0026547 and GAPDH probe qHsaCEP0041396.
Bacterial rRNA gene qPCR and sequencing
All samples were processed at NYU. For DNA isolation, lysis was ensured by a freeze-thaw cycle, use of lysozyme and a heat step (56°C) at the beginning of the DNA isolation process. DNA was extracted with an ion exchange column (Qiagen). Total bacterial levels were determined by quantitative PCR (qPCR) using the LightCycler FastStart TaqMan Probe Master (Roche, Germany) in a Roche Lightcycler 480 Real-Time PCR system (Roche, Germany). Universal primers and probes were used to target conserved regions of the 16S rRNA gene (sequence 5′-3′ primers: 8F=AGAGTTTGATYMTGGCTCAG; EUB361R= CGYCCATTGBGBAADATTCC; and probe = 6-FAM-TACGGGAGGCAGCAGT-BHQ1)(Gao et al., 2007). Duplicate 20 μl reactions were performed under the following reaction conditions: 10 μl Probe Master Mix, 6 μl PCR-grade H2O, 1 μl of forward and reverse primer (10 pM/μl each), 1 μl TaqMan Probe (5 pM/μl) and 1 μl template DNA. PCR cycling conditions were as follow: initial denaturing at 95°C for 10 min, foll owed by 45 cycles of denaturation at 95°C for 10 seconds, annealing at 54°C for 30 seconds, and extension at 72 C for 20 seconds.
High-throughput sequencing of bacterial 16S rRNA gene amplicons encoding the V4 region (150 bp read length, paired-end protocol) was performed using a MiSeq Illumina Sequencer. For each sample, the V4 region of the bacterial 16S rRNA gene was amplified in duplicate reactions, using primer set 515F/806R, which nearly universally amplifies bacterial and archaeal 16S rRNA genes.(Caporaso et al., 2012) Each unique barcoded amplicon was generated in pairs of 25μl reactions with the following reaction conditions: 11μl PCR-grade H2O, 10μl Hot MasterMix (5 Prime Cat# 2200410), 2μl of forward and reversed barcoded primer (5μM) and 2μl template DNA. Reactions were run on a C1000 Touch Thermal Cycler (Bio-Rad) with the following cycling conditions: initial denaturing at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 45 seconds, a nnealing at 58°C for 1 minute, and extension at 72 C for 90 seconds, with a final extension of 10 min at 72°C. Amplicons were quantified using Agilent 2200 TapeStation system and pooled. Purification was then performed using Ampure XT (Beckman Coulter Cat# A63882) as per the manufacturer instructions. Sequencing was then performed in MiSeq (Illumina) to produce 150 base-paired end reads.
Bacterial whole genome shotgun sequencing
Shotgun sequencing of isolated DNA was performed using a HiSeq Illumina Sequencer. Approximately, 20 million reads were obtained per sample (median[IQR]= 20,720,613 [19,553,497-21,154,173]). Adapter sequences were trimmed with Cutadapt(Martin, 2011), and overlapping Read1 and Read2 sequences were stitched into a single read using fastq-join (ea-utils package, http://code.google.com/p/ea-utils). Sequence quality control was performed with Prinseq (Schmieder and Edwards, 2011) with average PHRED cut-off of 25 and minimum quality cut-off score of 10.
QUANTIFICATION AND STATISTICAL ANALYSIS
Cox proportion hazard models
Multivariable models of TB rate were calculated in SPSS with incident TB as the outcome and serum cytokines and SCFA levels as predictors. Models were adjusted for age, gender, vital capacity, body mass index and serum acetate.
16S rRNA gene sequencing analysis
The 16S rRNA gene sequences were analyzed using the Quantitative Insights into Microbial Ecology (QIIME) pipeline for analysis of microbiome data(Caporaso et al., 2010). Reads were demultiplexed and quality filtered with default parameters. Sequences were then clustered (closed reference OTU picking) into operational taxonomic units (OTUs) using a 97% similarity threshold with UCLUST and the Greengenes 16S reference dataset and taxonomy (McDonald et al., 2012). After curating and removal of sequences potentially derived from reagent controls, the absolute OTU sequence counts were normalized to obtain the relative abundances of the taxa within each sample. The proportion of reads at the OTU or genus levels was used as a measure of the relative abundance of each type of bacteria. A median of 11,984 reads were obtained per sample (IQR= 6,793–27,387]. Further analysis was performed on rarefied data (depth of 5,000 reads in each sample). Rarefaction curves of OTUs assignment at variable sequence depth were constructed to evaluate α diversity. Weighted UniFrac was used to measure β diversity of bacterial communities and to perform principal coordinate analysis (PCoA) (Lozupone and Knight, 2005). We used the ade4 package in R to PCoA on weighted UniFrac distances. To avoid negative eigenvalues in the analysis, we used the Cailliez method to convert the weighted UniFrac distance matrix into a closest corresponding matrix with Euclidean properties, which was further used for PCoA (Cailliez, 1983). Microbiome analysis of BAL samples was compared with 16S data from background and upper airways.
To determine the genomic potential of these two pneumotypes, we computationally predicted the metagenome using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)(Langille et al., 2013). This software tool uses the obtained 16S rRNA gene sequence data to predict the functional profile of a bacterial community based on an existing reference genome database. In a first step, 16S rRNA gene annotation is normalized by copy number. Then, the gene content is inferred based on a reference phylogenetic gene tree. Metagenomic pathway analysis was performed using STAMP with default parameters (Parks et al., 2014). Shannon Diversity index was utilized to evaluate α diversity of inferred metagenome. Bray Curtis dissimilarity index was used to measure β diversity of gene composition and to perform principal coordinate analysis (PCoA).
Bacterial whole genome shotgun sequencing analysis
NCBI’s Best Match Tagger (BMTagger) was utilized to remove human reads. Quality control removed approximately 8.9 million reads per sample. Of the remaining reads, approximately 3.5 million were human and were removed. Of the remaining genetic information, an average of 16.6% were of bacterial origin (range:4.0 to 30.1%), yielding on average 1.1 million bacterial reads/sample. High-quality sequences were then sequentially queried against a bacterial amino acid database constructed from IMG genomes as previously described(Fierer et al., 2013). This approach yield a median of 648,766 reads annotated to a KO pathway (IQR= 618,285–709,552]. Shannon Diversity index of rarified data (depth 500,000 reads/sample) was utilized to evaluate α diversity of shotgun metagenome. Differential taxa were identified using Linear Discriminant Analysis (LDA) Effect Size (Segata et al., 2011) with alpha value 0.01 and an “all-against-all” multi-class analysis strategy.
Supplementary Material
Highlights.
Increased short chain fatty acids (SCFAs) predict TB risk in HIV patients on ART
SCFAs block IFN-γ and IL-17A induced by M. tuberculosis antigens
SCFAs induce FoxP1 and correlate with TB antigen induced pulmonary Tregs
SCFAs in the lower airways are associated with increased lung anaerobic bacteria
Acknowledgments
Sources of support: This work was supported by K23 AI102970, RO1HL090316, 5U01CA086137-13, K24 AI080298A, UL1 TR000038, U24 DK097153
The authors thank the contributions of Peter Meyn and Adriana Heguy at the NYUMC Genome Technology Center, that is supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center, and a generous gift from Helga R. Dawn.
Footnotes
Conflict of interest. No conflicts of interest are reported by any authors.
Author Contributions: LNS, and MDW conceived the study, designed experiments and wrote the manuscript. AM, PD, GC, RD, RNZS, KD, and WNR collected clinical data and managed clinical samples. CR, and JC performed the SCFAs assays. LNS, ST, and KD performed flow cytometry. GC, RD, and RNZS performed PPD stimulation assays. YL, and MDD performed cytokine measurements and data analyses. LNS, JCC, BGW, MDW performed 16S sequencing and analysis. All authors discussed the results, edited and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Borgdorff H, Gautam R, Armstrong SD, Xia D, Ndayisaba GF, van Teijlingen NH, Geijtenbeek TB, Wastling JM, van de Wijgert JH. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal immunology. 2016;9:621–633. doi: 10.1038/mi.2015.86. [DOI] [PubMed] [Google Scholar]
- Bourriaud C, Robins RJ, Martin L, Kozlowski F, Tenailleau E, Cherbut C, Michel C. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J Appl Microbiol. 2005;99:201–212. doi: 10.1111/j.1365-2672.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailliez F. The analytical solution of the additive constant problem. Psychometrika. 1983;48:305–310. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchyard GJ, Fielding KL, Lewis JJ, Coetzee L, Corbett EL, Godfrey-Faussett P, Hayes RJ, Chaisson RE, Grant AD, Thibela TBST. A trial of mass isoniazid preventive therapy for tuberculosis control. N Engl J Med. 2014;370:301–310. doi: 10.1056/NEJMoa1214289. [DOI] [PubMed] [Google Scholar]
- Cosgrove C, Ussher JE, Rauch A, Gartner K, Kurioka A, Huhn MH, Adelmann K, Kang YH, Fergusson JR, Simmonds P, et al. Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood. 2013;121:951–961. doi: 10.1182/blood-2012-06-436436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Dobrowolski C, Shahir AM, Feng Z, Yu X, Sha J, Bissada NF, Weinberg A, Karn J, Ye F. Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology. 2015;474:65–81. doi: 10.1016/j.virol.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, Kibbie J, Lee EJ, Guo K, Santiago ML, Austin GL, Gianella S, Landay AL, Donovan AM, Frank DN, et al. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. Aids. 2017;31:511–521. doi: 10.1097/QAD.0000000000001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Nordstrom K, Gasparoni G, Salhab A, Kressler C, de Almeida M, Bassler K, Ulas T, Schmidt F, Xiong J, et al. Epigenomic Profiling of Human CD4+ T Cells Supports a Linear Differentiation Model and Highlights Molecular Regulators of Memory Development. Immunity. 2016;45:1148–1161. doi: 10.1016/j.immuni.2016.10.022. [DOI] [PubMed] [Google Scholar]
- El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, Borkowsky W, Valentine F, Littman DR, Unutmaz D. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis. 2010;201:843–854. doi: 10.1086/651021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol. 2011;12:544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL. Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science. 2013;342:621–624. doi: 10.1126/science.1243768. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MC, Eid T, Smyk-Pearson S, Eberling Y, Swarbrick GM, Langley SM, Streeter PR, Lewinsohn DA, Lewinsohn DM. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal immunology. 2013;6:35–44. doi: 10.1038/mi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal R, Rangel-Moreno J, Slight S, Lin Y, Nawar HF, Fallert Junecko BA, Reinhart TA, Kolls J, Randall TD, Connell TD, et al. Interleukin-17-dependent CXCL13 mediates mucosal vaccine-induced immunity against tuberculosis. Mucosal immunology. 2013;6:972–984. doi: 10.1038/mi.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group TAS, Danel C, Moh R, Gabillard D, Badje A, Le Carrou J, Ouassa T, Ouattara E, Anzian A, Ntakpe JB, et al. A Trial of Early Antiretrovirals and Isoniazid Preventive Therapy in Africa. N Engl J Med. 2015;373:808–822. doi: 10.1056/NEJMoa1507198. [DOI] [PubMed] [Google Scholar]
- Humphries J, Xiong L, Liu J, Prindle A, Yuan F, Arjes HA, Tsimring L, Suel GM. Species-Independent Attraction to Biofilms through Electrical Signaling. Cell. 2017;168:200–209. e212. doi: 10.1016/j.cell.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Ochiai K, Okamoto T. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J Immunol. 2009;182:3688–3695. doi: 10.4049/jimmunol.0802906. [DOI] [PubMed] [Google Scholar]
- Jambo KC, Banda DH, Afran L, Kankwatira AM, Malamba RD, Allain TJ, Gordon SB, Heyderman RS, Russell DG, Mwandumba HC. Asymptomatic HIV-infected individuals on antiretroviral therapy exhibit impaired lung CD4(+) T-cell responses to mycobacteria. Am J Respir Crit Care Med. 2014;190:938–947. doi: 10.1164/rccm.201405-0864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, Somsouk M, Deeks SG, Martin JN, Moll M, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121:1124–1135. doi: 10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone AG, Atci E, Renslow R, Beyenal H, Noh S, Fransson B, Abu-Lail N, Park JJ, Gang DR, Call DR. Staphylococcus aureus induces hypoxia and cellular damage in porcine dermal explants. Infect Immun. 2015;83:2531–2541. doi: 10.1128/IAI.03075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, et al. Widespread Colonization of the Lung by Tropheryma whipplei in HIV Infection. Am J Respir Crit Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011;17 [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, et al. Comparison of the Respiratory Microbiome in Healthy Non-Smokers and Smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J, DiGiandomenico A, Artamonov M, Leitinger N, Amin AR, Goldberg JB. Host derived inflammatory phospholipids regulate rahU (PA0122) gene, protein, and biofilm formation in Pseudomonas aeruginosa. Cell Immunol. 2011;270:95–102. doi: 10.1016/j.cellimm.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennard SI, Basset G, Lecossier D, O’Donnell KM, Pinkston P, Martin PG, Crystal RG. Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of dilution. Journal of applied physiology. 1986;60:532–538. doi: 10.1152/jappl.1986.60.2.532. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, Li Y, Shen N, Ghedin E, Morris A, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal LN, Clemente JC, Wu BG, Wikoff WR, Gao Z, Li Y, Ko JP, Rom WN, Blaser MJ, Weiden MD. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax. 2017;72:13–22. doi: 10.1136/thoraxjnl-2016-208599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple PL, Binder AB, Davids M, Maredza A, van Zyl-Smit RN, Dheda K. Regulatory T cells attenuate mycobacterial stasis in alveolar and blood-derived macrophages from patients with tuberculosis. Am J Respir Crit Care Med. 2013;187:1249–1258. doi: 10.1164/rccm.201210-1934OC. [DOI] [PubMed] [Google Scholar]
- Serrano-Villar S, Vazquez-Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando-Martinez S, Rojo D, Martinez-Botas J, Del Romero J, Madrid N, et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal immunology. 2016 doi: 10.1038/mi.2016.122. [DOI] [PubMed] [Google Scholar]
- Shenoy MK, Iwai S, Lin DL, Worodria W, Ayakaka I, Byanyima P, Kaswabuli S, Fong S, Stone S, Chang E, et al. Immune Response and Mortality Risk Relate to Distinct Lung Microbiomes in HIV-Pneumonia Patients. Am J Respir Crit Care Med. 2016 doi: 10.1164/rccm.201603-0523OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Stephen TL, Rutkowski MR, Allegrezza MJ, Perales-Puchalt A, Tesone AJ, Svoronos N, Nguyen JM, Sarmin F, Borowsky ME, Tchou J, et al. Transforming growth factor beta-mediated suppression of antitumor T cells requires FoxP1 transcription factor expression. Immunity. 2014;41:427–439. doi: 10.1016/j.immuni.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Twigg HL, 3rd, Knox KS, Zhou J, Crothers KA, Nelson DE, Toh E, Day RB, Lin H, Gao X, Dong Q, et al. Effect of Advanced HIV Infection on the Respiratory Microbiome. Am J Respir Crit Care Med. 2016;194:226–235. doi: 10.1164/rccm.201509-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg HL, Iii, Weiden M, Valentine F, Schnizlein-Bick CT, Bassett R, Zheng L, Wheat J, Day RB, Rominger H, Collman RG, et al. Effect of highly active antiretroviral therapy on viral burden in the lungs of HIV-infected subjects. J Infect Dis. 2008;197:109–116. doi: 10.1086/523766. [DOI] [PubMed] [Google Scholar]
- Wakabayashi H, Yamauchi K, Kobayashi T, Yaeshima T, Iwatsuki K, Yoshie H. Inhibitory effects of lactoferrin on growth and biofilm formation of Porphyromonas gingivalis and Prevotella intermedia. Antimicrob Agents Chemother. 2009;53:3308–3316. doi: 10.1128/AAC.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, Tas J, Choi YS, Takata H, Day TJ, et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol. 2014;15:667–675. doi: 10.1038/ni.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Geng J, Shi B, Liu Z, Wang YH, Stevens AC, Sprout SL, Yao M, Wang H, Hu H. Cutting Edge: Foxp1 Controls Naive CD8+ T Cell Quiescence by Simultaneously Repressing Key Pathways in Cellular Metabolism and Cell Cycle Progression. J Immunol. 2016;196:3537–3541. doi: 10.4049/jimmunol.1501896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. Journal of bacteriology. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.