Abstract
Severe acute respiratory syndrome (SARS) was caused by a novel virus now known as SARS coronavirus (SARS-CoV). The discovery of SARS-CoV-like viruses in masked palm civets (Paguma larvata) raises the possibility that civets play a role in SARS-CoV transmission. To test the susceptibility of civets to experimental infection by different SARS-CoV isolates, 10 civets were inoculated with two human isolates of SARS-CoV, BJ01 (with a 29-nucleotide deletion) and GZ01 (without the 29-nucleotide deletion). All inoculated animals displayed clinical symptoms, such as fever, lethargy, and loss of aggressiveness, and the infection was confirmed by virus isolation, detection of viral genomic RNA, and serum-neutralizing antibodies. Our data show that civets were equally susceptible to SARS-CoV isolates GZ01 and BJ01.
Severe acute respiratory syndrome (SARS) first appeared in Guangdong, China, in November 2002, and it subsequently spread to many parts of the world, making it the first major infectious disease outbreak of the 21st century (8, 13, 19). The etiological agent was a newly emerged and previously unrecognized coronavirus, now known as SARS coronavirus (SARS-CoV) (2, 3, 5-7, 12), which is classified within the order Nidovirales, family Coronaviridae, genus Coronavirus (9, 14, 15). Epidemiological data obtained from the early stage of the SARS outbreak suggest an animal origin for SARS-CoV, although the reservoir host has yet to be identified (11, 20-22). The isolation of SARS-CoV-like viruses in masked palm civets and the relationship of their genomic sequences with those of viruses isolated from humans (1, 4) raise the possibility that civets play a role in SARS-CoV transmission to the human population. A striking difference between the vast majority of SARS-CoV genomes from humans and those from civets is the presence in the latter of an additional 29-nucleotide (nt) sequence 246 nt upstream of the start codon of the N gene. Only human SARS-CoV isolated from the earliest stage of the outbreak contains this same 29-nt additional sequence (1). In other words, most human SARS-CoV isolates had a 29-nt deletion in this region of the genome. Thus, while it is clear that SARS-CoV with and without the 29-nt deletion can replicate in humans, the influence of the 29-nt deletion on the capacity of the virus to replicate in civets has not been determined. Here we show that civets are equally susceptible to experimental infection with two different human SARS-CoV isolates, one containing and the other lacking the 29-nt sequence, and that all animals display clinical signs during the early stage of infection.
SARS-CoV isolates GZ01 and BJ01 used in this study were originally isolated in Vero E6 cells at the Institute of Microbiology and Epidemiology, Academy of Military Medical Sciences, Beijing, China, and were propagated in Vero E6 cells for two additional passages at our institute in Harbin to generate virus stocks with titers of 106 50% tissue culture infective doses (TCID50)/ml. BJ01 has the 29-nt deletion found in most human SARS-CoV isolates, whereas GZ01 resembles the viruses isolated from civets in that it does contain the 29-nt sequence (1, 4).
To test the susceptibility of civets to SARS-CoV, 11 masked palm civets (Paguma larvata) were purchased from a farm in Hebei Province. All animals were approximately 1 year old, and none contained anti-SARS-CoV antibodies when tested by virus neutralization prior to the infection experiment. The animals were observed in the laboratory for approximately 1 month. No clinical signs were detected during the observation period, and no SARS-CoV-related RNA was detected in throat or anal swabs analyzed by reverse transcription-PCR (RT-PCR). Ten animals were each housed in separate biosafety isolators and were divided into two groups (n = 5 per group). Animals in groups A and B were inoculated with 3 ml of virus solution containing 3 × 106 TCID50 of GZ01 and BJ01 isolates, respectively, with 2 ml given intratracheally and 1 ml given intranasally. A control civet was mock infected in an identical fashion with 3 ml of Vero E6 cell culture supernatant. Animal experiments were conducted in accordance with animal ethics guidelines and approved protocols by the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, and were carried out in an approved animal biosafety level 3 facility.
The clinical signs of animals were checked daily. Throat swabs, anal swabs, and blood samples were taken on 0, 3, 8, 13, 18, 23, 28, and 33 days postinfection (dpi) and were subjected to virus isolation and RT-PCR analysis. Blood samples were also subjected to leukocyte counting. One animal from each group was sacrificed on 3, 13, 23, 34, and 35 dpi. On the day of euthanasia, lung, heart, spleen, lymph node, kidney, and liver samples were taken from each animal and homogenized in phosphate-buffered saline (PBS) for virus isolation and RT-PCR analysis. Serum samples were also taken for virus neutralization analysis.
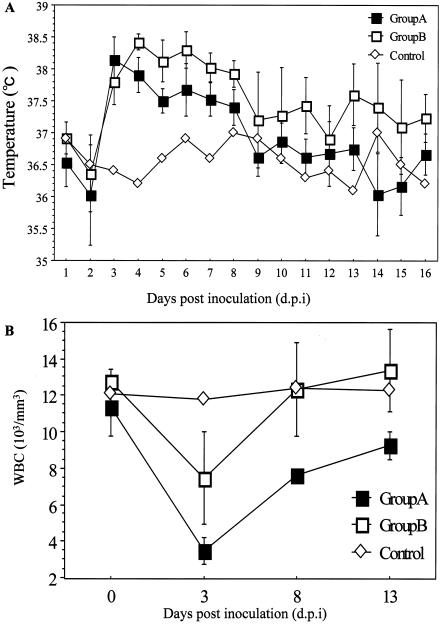
From 3 dpi, all animals became lethargic and less aggressive. Febrile episodes commenced around 3 dpi, and temperatures remained elevated for up to 7 days in infected civets (Fig. 1A). Leucopenia was also observed, with white blood cell counts reaching a minimum at approximately 3 dpi and returning to normal from 13 dpi onwards. This trend was similar for animals in groups A and B (Fig. 1B). Two civets (nos. 14 and 8) in group A and one in group B developed diarrhea between 3 and 14 dpi. Conjunctivitis was observed for one animal (no. 14) in group A and two (nos. 7 and 11) in group B.
FIG. 1.
Clinical changes in civets after inoculation with SARS coronavirus (SARS-CoV). (A) Daily average temperature of animals in groups A and B plotted together with the daily temperature of the control animal. Febrile episodes commenced around 3 dpi, and temperatures remained elevated for up to 7 days in infected civets. (B) White blood cell (WBC) counts measured on day 0 and at 3, 8, and 13 dpi for the control animal and animals in groups A and B. For animals in groups A and B, the average counts are used in the plot. Leucopenia was observed, with white blood cell counts reaching a minimum at approximately 3 dpi and recovering to about normal levels from 13 dpi.
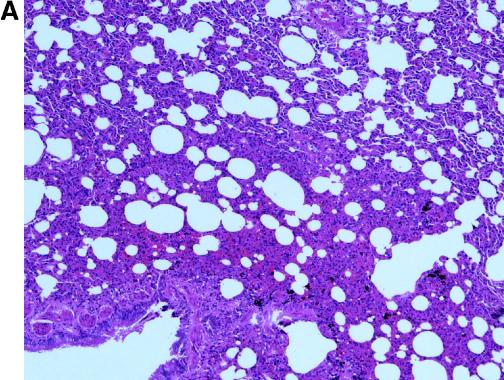
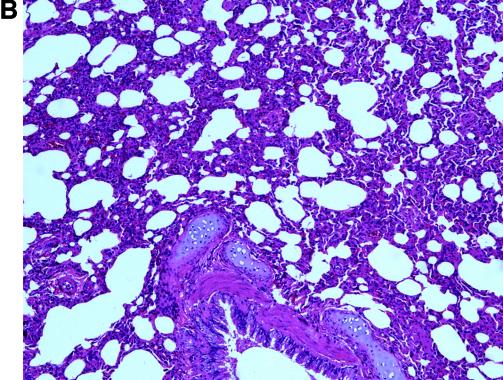
For histological examination, lung tissues were fixed in 10% neutral-buffered formalin, embedded in paraffin, and processed for routine histology. No gross pathological changes were found in necropsied animals. Histologically, interstitial pneumonia lesions were observed in both groups of animals on days 13 to 35 postinfection. The lesions were similar to those described for the SARS-CoV-infected macaques (3), but the absence of syncytia (Fig. 2A and B) resembled the observation made with experimentally infected ferrets (10).
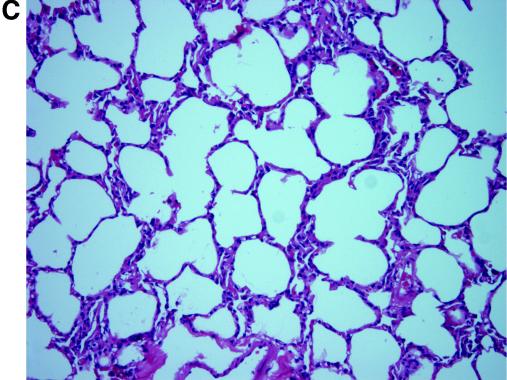
FIG. 2.
Pathological changes in civets after inoculation with SARS-CoV. Lung tissues were taken at 13 dpi from animal no. 5 of group A (A) and no. 7 of group B (B). Alveolar septa enlargement with macrophage and lymphocyte infiltration was evident in both animals. (C) Hematoxylin and eosin stain (magnification, ×20) of lung tissue of the control animal showed no abnormal changes.
For virus isolation, collected samples were inoculated on Vero E6 cell monolayers in 96-well plates and passaged up to three times. For samples showing cytopathic effect (CPE), the presence of SARS-CoV was confirmed by electron microscopy and RT-PCR analysis. For RT-PCR analysis, viral RNA was isolated from swabs, serum, supernatants of homogenates, and tissue culture by using the QIAamp viral RNA Mini kit (QIAGEN). First-strand cDNA was made using random hexamer primer and the RNA LA PCR (AMV) kit (TaKaRa), and it was subjected to amplification using a nested PCR. The first PCR was performed with primers VNUP (5′GATAA TGGAC CCCAA TCAAA CCAA3′) and VNLOW (5′CTGAG TTGAA TCAGG AGAAG CTCC3′), and the second PCR was performed with primers N355UP (5′GAACT GGCCC AGAAG CTTCA CT3′) and N355LOW (5′TTGGC CTTTA CCAGA AACTT TG3′). The size of the nested PCR product was 355 bp. All PCR products were confirmed by nucleotide sequencing.
Results presented in Table 1 indicate that viral genome was detected by RT-PCR in throat and anal swabs from 3 to 18 dpi, live virus was isolated at 3 dpi from animals in both groups, and live virus was isolated at 8 dpi from animals in group A only (Table 1). Morphologically, the virus recovered was identical to that used for inoculation (data not shown). The detection frequency of virus in blood samples was very low, with only one animal (no. 4) in group A giving positive results for both virus isolation and RT-PCR at 3 dpi, and a second animal (no. 14) in group A showing positive RT-PCR for samples taken at 3, 8, and 13 dpi. In contrast, virus was readily detected up to 13 dpi in a variety of organs, including lung, liver, kidney, and heart (Table 2). Virus was still detected by RT-PCR at the end of the experiment (34 to 35 dpi) in lymph nodes and spleen.
TABLE 1.
Detection of SARS-CoV in throat swab (T), anal swab (A), and blood (B) by virus isolation and RT-PCR
| Group (virus isolate) | Animal no. | Detection at dpia:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0
|
3
|
8
|
13
|
18
|
23
|
28
|
33
|
||||||||||||||||||
| T | A | B | T | A | B | T | A | B | T | A | B | T | A | B | T | A | B | T | A | B | T | A | B | ||
| A (GZ01) | 6 | −/− | −/− | −/− | −/+ | −/+ | −/− | ||||||||||||||||||
| 5 | −/− | −/− | −/− | −/+ | −/+ | −/− | −/− | +/+ | −/− | −/− | −/− | −/− | |||||||||||||
| 14 | −/− | −/− | −/− | −/+ | −/+ | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/− | −/− | −/− | −/− | −/− | −/− | |||||||
| 4 | −/− | −/− | −/− | +/+ | +/+ | +/+ | −/+ | +/+ | −/− | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
| 3 | −/− | −/− | −/− | +/+ | +/+ | −/− | +/+ | +/+ | −/− | −/+ | −/− | −/− | −/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
| B (BJ01) | 13 | −/− | −/− | −/− | −/+ | +/+ | −/− | ||||||||||||||||||
| 7 | −/− | −/− | −/− | NDb | ND | ND | −/− | −/− | −/− | −/− | −/− | −/− | |||||||||||||
| 11 | −/− | −/− | −/− | +/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/− | −/− | −/− | −/− | −/− | |||||||
| 10 | −/− | −/− | −/− | +/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
| 8 | −/− | −/− | −/− | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/+ | −/− | −/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | |
Virus isolation/RT-PCR results are depicted. +/+ indicates positive for both virus isolation and PCR, whereas −/+ means that particular specimen gave positive PCR signals but virus isolation was negative.
ND, not determined.
TABLE 2.
Detection of neutralizing antibodies in serum and SARS-CoV in postmortem tissues
| Group (virus isolate) | Animal no. | Time of euthanasia (dpi) | Neutralizing antibody titera | Detection in postmortem tissuesc
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Lung | Heart | Spleen | Lymph node | Kidney | Liver | ||||
| A (GZ01) | 6 | 3 | NDb | +/+ | −/− | −/+ | +/+ | +/+ | +/+ |
| 5 | 13 | 20 | −/− | −/+ | −/+ | −/ND | −/− | −/− | |
| 14 | 23 | 80 | −/− | −/− | −/+ | −/+ | −/− | −/− | |
| 4 | 34 | 40 | −/− | −/− | −/+ | −/+ | −/− | −/− | |
| 3 | 35 | 40 | −/− | −/− | −/− | −/+ | −/− | −/− | |
| B (BJ01) | 13 | 3 | <10 | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| 7 | 13 | 20 | −/− | −/− | −/+ | −/+ | −/+ | −/+ | |
| 11 | 23 | 80 | −/− | −/− | −/+ | −/+ | −/− | −/− | |
| 10 | 34 | 80 | −/− | −/− | −/+ | −/+ | −/− | −/− | |
| 8 | 35 | 80 | −/− | −/− | −/− | −/+ | −/− | −/− | |
All animals were tested prior to inoculation, and none of the animal had neutralizing antibodies to SARS-CoV in their prebleed sera.
ND, not determined.
Virus isolation/RT-PCR results are depicted. +/+ indicates positive for both virus isolation and PCR, whereas −/+ means that particular tissue gave positive PCR signals but virus isolation was negative.
For antibody detection, twofold dilutions of serum were tested in a microneutralization assay for the presence of antibodies that neutralized the infectivity of 200 TCID50 of SARS-CoV in Vero E6 cell monolayers, with 4 wells per dilution on a 96-well plate. The presence of CPE was read on days 3 and 4, and neutralizing titers were determined from the dilution factor of serum that completely prevented CPE in 50% of the wells. Serum samples taken on the day of euthanasia were analyzed, and neutralizing antibodies were detected in samples taken from 13 dpi onwards, with the antibody titers varying from 20 at 13 dpi to 80 at 34 or 35 dpi (Table 2).
Due to the limited number of civets that we could house in our biosafety isolators, we were not able to sample animals more frequently during the first few days of the infection, so we were unable to determine the peak of virus shedding during the experiment. However, the main focus of the present study was to test and compare the susceptibility of farmed civets to that of two different human SARS-CoV isolates. In that regard, our data conclusively show that civets are readily susceptible to experimental infection by SARS-CoV, and all infected animals developed clinical symptoms and produced both virological and serological evidence of infection with the two different virus isolates used in this study. The susceptibility and clinical symptoms observed also suggest that civets could be another useful animal model for development of vaccines and antiviral drugs against SARS.
One interesting finding is that infection by isolate BJ01 seemed to result in higher average body temperature (Fig. 1A) and induced a slightly stronger antibody response (Table 2). However, because of the relatively small number of animals used in each group, future experiments will be required to confirm these differences.
Another observation which warrants further study is the detection of viral genomic RNA in spleen and lymph nodes up to 34 and 35 dpi (Table 2). Whether these tissues are able to support persistent infection of SARS-CoV in civets remains to be investigated. Once we have access to a larger PC3 or PC4 facility which allows cohousing of civets, further experiments need to be carried out to assess other important issues, including virus shedding and interanimal transmission either by contact or aerosol.
In addition to civets, experimental infection of SARS-CoV has also been demonstrated in monkeys, ferrets, cats, mice, and pigs (3, 10, 16, 17, 18). It should be noted that different SARS-CoV isolates were used by each of the groups. It is therefore not clear at this stage whether one particular animal would be better than others as a model for SARS-CoV. These studies, however, do demonstrate the diverse range of mammalian species that are susceptible to experimental infection by SARS-CoV. In a previous study of wildlife animals in a market, it was shown that Chinese ferret badgers (Melogale moschata) and raccoon dogs (Nyctereutes procyonoides) could also be infected with SARS-CoV (4). Altogether, there are now at least eight different animal species known to be susceptible to SARS-CoV infection. This undoubtedly poses a formidable challenge for any future investigation into the origin of SARS-CoV.
In summary, our present study demonstrated equal susceptibility of farmed civets to infection by two different human SARS-CoV isolates under experimental conditions, and all animals displayed clinical signs of infection. Our results support the notion that civets may play an important role for transmission of SARS-CoV from animals to humans. However, further field study of wild civets is required to assess whether civets are a natural reservoir host of SARS-CoV.
.
Acknowledgments
We thank Qingyu Zhu of the Institute of Microbiology and Epidemiology, Academy of Military Medical Sciences, Beijing, China, for providing SARS-CoV isolates BJ01 and GZ01 and the Vero E6 cell line.
This research was supported by the national high-technology research and development program (863; grant no. 2003AA208407) and the Guangdong Provincial Project on SARS Prevention and Treatment (2003FD02-06).
REFERENCES
- 1. Chinese SARS Molecular Epidemiology Consdortium. 2004. Molecular evolution of SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. [DOI] [PubMed] [Google Scholar]
- 2.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterization of viruses related to the SARS corona-virus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 5.Holmes, K. V. 2003. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Investig. 111:1605-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and the SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 7.Kuiken, T., R. A. M. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. S. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. M. Peiris, and A. D. M. E. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 9.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffin, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. E. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 10.Martina, B. E. E., B. L. Haagmans, T. Kuiken, R. A. M. Fouchier, G. F. Rimmelzwaan, G. V. Amerongen, J. S. M. Peiris, W. Lim, and A. D. M. E. Osterhaus. 2003. SARS virus infection of cats and ferrets. Nature 425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Normile, D., and M. Enserink. 2003. SARS in China—tracking the roots of a killer. Science 301:297-299. [DOI] [PubMed] [Google Scholar]
- 12.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, and Members of the SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, A. J. McGeer, Canadian National Microbiology Laboratory, and the Canadian Severe Acute Respiratory Syndrome Study Team. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 14.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. X. Tong, A. Tamin, L. Lowe, M. Frace, J. L. Derisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 15.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W.-J. Shih, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weingartl, H. M., J. Copps, M. A. Drebot, P. Marszal, G. Smith, J. Gren, M. Andonova, J. Pasick, P. Kitching, and M. Czub. 2004. Susceptibility of pigs and chickens to SARS coronavirus. Emerg. Infect. Dis. 10:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wentworth, D. E., L. Gillim-Ross, N., Espina, and K. A. Bernard. 2004. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 10:1293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. 2004. Report to the W.H.O. executive board on severe acute respiratory syndrome (SARS). [Online] http://www.who.int/gb/EB_WHA/PDF/EB113/eeb11333.pdf.
- 20.Xu, R.-H., J.-F. He, M. R. Evans, G.-P. Peng, H. E. Field, D.-W. Yu, C.-K. Li, H.-M. Luo, W.-S. Lin, P. Lin, L.-H. Li, W.-J. Liang, J.-Y. Lin, and A. Schnur. 2004. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 10:1030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, D., H. Li, R. Xu, J. He, J. Lin, L. Li, W. Li, H. Xu, S. Huang, and J. Huang. 2003. Prevalence of IgG antibody to SARS-associated coronavirus in animal traders—Guangdong Province, China, 2003. Morb. Mortal. Wkly. Rep. 52:986-987. [PubMed] [Google Scholar]
- 22.Zhong, N. S., B. J. Zheng, Y. M. Li, L. L. M. Poon, Z. H. Xie, K. H. Chan, P. H. Li, S.Y. Tan, Q. Chang, J. P. Xie, X. Q. Liu, J. Xu, D. X. Li, K. Y. Yuen, J. S. M. Peiris, and Y. Guan. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February 2003. Lancet 362:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]