Abstract
The complete DNA sequence of grouper iridovirus (GIV) was determined using a whole-genome shotgun approach on virion DNA. The circular form genome was 139,793 bp in length with a 49% G+C content. It contained 120 predicted open reading frames (ORFs) with coding capacities ranging from 62 to 1,268 amino acids. A total of 21% (25 of 120) of GIV ORFs are conserved in the other five sequenced iridovirus genomes, including DNA replication, transcription, nucleotide metabolism, protein modification, viral structure, and virus-host interaction genes. The whole-genome nucleotide pairwise comparison showed that GIV virus was partially colinear with counterparts of previously sequenced ranaviruses (ATV and TFV). Besides, sequence analysis revealed that GIV possesses several unique features which are different from those of other complete sequenced iridovirus genomes: (i) GIV is the first ranavirus-like virus which has been sequenced completely and which infects fish other than amphibians, (ii) GIV is the only vertebrate iridovirus without CpG sequence methylation and lacking DNA methyltransferase, (iii) GIV contains a purine nucleoside phosphorylase gene which is not found in other iridoviruses or in any other viruses, (iv) GIV contains 17 sets of repeat sequence, with basic unit sizes ranging from 9 to 63 bp, dispersed throughout the whole genome. These distinctive features of GIV further extend our understanding of molecular events taking place between ranavirus and its hosts and the iridovirus evolution.
Iridoviruses are large icosahedral cytoplasmic deoxyriboviruses with viral particle sizes ranging from 120 to 350 nm in diameter. The family Iridoviridae can be subdivided into at least four genera, including Iridovirus, Chloriridovirus, Ranavirus, and Lymphocystivirus (59). Members of the family are notable for their variability in infecting a diverse range of vertebrate (Lymphocystivirus, Ranavirus) and invertebrate (Chloriridovirus, Iridovirus) hosts, causing diseases that range in severity from subclinical to lethal (58). The iridovirus genome is both circularly permutated and terminally redundant and is a double-stranded DNA genome ranging from 103 to 212 kbp in length (30). Moreover, the genome of the iridoviruses that infect vertebrates is highly methylated at the cytosine residues in CpG sequences by a virus-encoded methyltransferase (53, 60).
Vertebrate iridoviruses are found in fish, amphibians, and reptiles. Many of them are confirmed to be pathogens that cause systemic diseases in aquaculture. Iridovirus infection of fish and frogs is a serious problem in modern aquaculture, fish farming, and wildlife conservation because of its epidemic morbidity and ability to cause mortality. Systemic iridoviral infections have been previously described for a variety of freshwater and marine food fish species (18, 22, 32, 37), tropical freshwater ornamental fish (40), frogs (20), and salamanders (30). Since the discovery of the first fish iridovirus, LCDV (lymphocystis disease virus), in 1962 (56), many new iridovirus-like pathogens have been reported from over 140 different species of fish worldwide (15). Intriguingly, most of these fish iridoviruses have shown to be more closely related to frog virus 3, the typical species of the genus Ranavirus, than to Lymphocystivirus. Although the genus Ranavirus was originally thought to infect only amphibians, the ranavirus-like pathogens can infect fish and reptiles as well (35). This has prompted speculations that viruses belonging to genus Ranavirus are capable of infecting cross-species organisms and that one class of poikilothermic vertebrates might serve as a reservoir, alternate host, or amplifying host for viruses infecting another class of lower vertebrates (35). These observations raise several questions and research interests.
(i) The taxonomic position of newly isolated iridoviruses.
In previous studies, identification of a newly isolated virus as an iridovirus was performed on the basis of characteristic virion morphology or of the use of a phylogenic algorithm employing a few common genes or of a single gene such as the major capsid protein gene (37, 47, 54). However, there has been considerable debate on whether a phylogenetic tree constructed on the basis of any single gene could accurately represent the evolution of a species, due to the possibility of horizontal gene transfer and degradation of the phylogenetic signal because of saturation for amino acid substitutions (10). Besides, these methods cannot provide enough information for clear identification and determination of isolated viruses. There is no common or universal phylogenetic tree to support the taxonomic position of a virus because of the differences in the rates of evolution of different genes from the same virus species (38). Developing tools and criteria for the identification and classification of virus taxonomy would be a big step toward understanding the evolution process of iridoviruses.
(ii) The essence of iridovirus.
The recent advances in genome sequencing technology have made it possible to scan the whole genome to define the genes that are homologous between iridoviruses. Comparison of different iridovirus genomes has revealed that many genes have been conserved during evolution, and this comparison would enable understanding of relationship between the phenotype and genotype. Besides, the genomic comparison would also allow the identification of genes which define iridoviruses and their host range.
Recently, five iridovirus genomes have been completely sequenced, including those of Lymphocystis disease virus 1 (LCDV-1; genus Lymphocystivirus) (51), Chilo iridescent virus (CIV; genus Iridovirus) (29), Tiger frog virus (TFV; genus Ranavirus) (24), Infectious spleen and kidney necrosis virus (ISKNV; unclassified genus) (23) and Ambystoma tigrinum virus (ATV; genus Ranavirus) (30). Considering the disease problems caused by viruses in fish aquaculture and their impact on fish farming economy and to facilitate the understanding of the molecular mechanism of infection, we describe here another complete genome sequencing and analyses of an iridovirus, the grouper iridovirus (GIV), which was isolated from diseased yellow grouper, Epinephelus awoara (34, 37).
MATERIALS AND METHODS
Isolation of virus and its DNA.
The GIV used in this study was isolated from spleen tissue of a diseased yellow grouper (Epinephelus awoara). The tissue was ground with a mortar and pestle. Leibovitz's L15 medium containing no fetal bovine serum was added to homogenates and centrifuged at 6,000 × g for 10 min. The supernatant fluid was filtered through a 0.2-μm-pore-size membrane. A total of 10 μl of the filtrate was inoculated into a 25-cm2 Nunc culture flask containing a 50% confluent monolayer of grouper kidney (GK) cells prepared 24 h earlier, and the mixture was incubated at 28°C until a cytopathic effect was observed. The supernatant fluid from virus-infected cells was filtered through a 0.2-μm-pore-size membrane and used for virus replication and purification. The virus was propagated in GK cells and purified by ultracentrifugation as described previously (34). For extraction of viral genomic DNA, purified GIV virions were soaked and incubated with genomic DNA extraction buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl) containing 0.2% (wt/vol) sodium dodecyl sulfate and 200 μg of proteinase K/ml at 55°C for 2 h. Subsequently, viral DNA was extracted with phenol-chloroform and precipitated with alcohol. Viral DNA pellets were dissolved in TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
GIV DNA cloning and sequence determination.
The GIV genome was sequenced to 3,273 sequences for 11-fold of coverage by a shotgun approach, and the gaps were filled by a chromosome walking method. The viral DNA was sheared (HydroShear; GeneMachines, San Carlos, Calif.) by nebulization into fragments with average sizes of 2,000 and 5,000 bp. DNA fragments were size fractionated by gel electrophoresis and cloned into the EcoRV site of pBluescript SK(+/-) (Stratagene). After transformation into Escherichia coli DH10B competent cells (Gibco-BRL), 1,446 recombinant colonies with a fragment size of 5 kbp and 576 recombinant colonies with a fragment size of 2 kbp were picked randomly. DNA templates for sequencing were isolated using Multiscreen kits (Millipore, Bedford, Mass.). Sequencing was performed using an ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit with FS AmpliTaq DNA polymerase (Perkin-Elmer, Palo Alto, Calif.) and analyzed on an ABI 3730 DNA analyzer. Shotgun sequences were base called by using the PHRED basecaller and assembled with the PHRAP assembler. PHRAP-assembled data were stored in a Sun workstation assembly database by using a Sun workstation interface. The Sun workstation interface and its features were then used for editing and completing the sequence. The Sun workstation was used with a probabilistic consensus algorithm based on expected error rates for output by PHRED to perform the consensus calculation with a quality cutoff value of 40. Primers were designed for chromosome walking through the gaps of the existing contiguous fragments.
DNA sequence analysis.
Open reading frames (ORFs) encoding more than 30 amino acids (90 bp), starting with the ATG codon and ending with the termination codon, were considered protein encoding and hence were designated putative genes. The maximal 570 ORFs were predicted and analyzed using Artemis 4.0 software (42). The amino acid sequence comparisons of all 570 ORFs were carried out using the PSI-BLAST program of the National Center for Biotechnology Information, and comparisons with the Swiss-Prot sequence databases and the Conserved Domain Database (CDD) (1, 36) were also performed. The predicted ORF homologues to any previously identified ORFs in the sequence database were incorporated in our analysis list. The percentages of identity were determined on the basis of the percentages of identical residues between two complete sequences. In addition, ORFs with significant homologies to ORFs in the protein sequence database and ORFs larger than 19 kDa without any significant homology to ORFs in the sequence database were also included. The molecular weight selection criterion was selected to coincide with our previously determined viral particle sodium dodecyl sulfate-polyacrylamide gel electrophoresis pattern (37). DNA repeats were identified by using a DNA tandem-repeat finder (6). The transmembrane domains of the predicted ORFs were predicted using the TMHMM server, v. 2.0 (33, 45). Dot plotting comparisons were performed by using the pairwise program of prFLAG (http://bioinformatics.itri.org.tw/prflag/prflag.php) from the Biomedical Engineering Center, Industrial Technology Research Center, Hsinchu, Taiwan.
Nucleotide sequence accession number.
The complete GIV sequence can be obtained from GenBank (accession no. AY666015).
RESULTS AND DISCUSSION
The features of GIV genome.
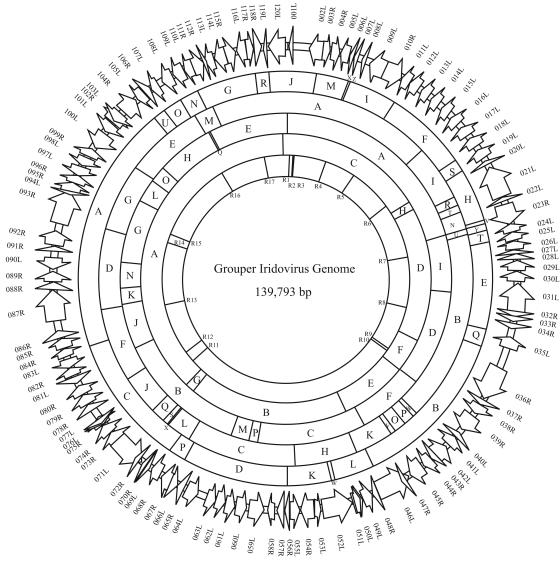
The assembled GIV genome was 139,793 bp in length, which was similar to our earlier prediction based on the restriction enzyme fragment analysis. Four restriction enzyme cutting maps are presented in Fig. 1. The genome of GIV was shown to be circularly permutated, possibly assembled into a circular form, which is a major characteristic feature of iridovirus genomes (24). The genome size of GIV is larger than those of ATV (106,332 bp) (30), TFV (105,057 bp) (24), LCDV (102,653 bp) (51), and ISKNV (111,362 bp) (23) but much smaller than that of CIV (212,482 bp) (29). It seems that the invertebrate iridovirus needs a larger genome than the vertebrate iridovirus. The percentage of G+C content of GIV (49%) is comparable to those of the genomes of ATV (54%), TFV (55%), and ISKNV (55%) but much higher than those of CIV (29%) and LCDV (29%) (Table 1). Artemis analysis detected a total of 570 ORFs as methionine-initiated ORFs comprising more than 30 amino acids. Of these ORFs, 120 present in GIV genome were homologous to ORFs in the Swiss-Prot database or were larger than 19 kDa without having homology to ORFs in the sequence database (Fig. 1). The ORFs are distributed evenly along the genome in clockwise (right, 47%) and anticlockwise (left, 53%) directions. The first ORF, 001L, is referred to as the first ORF present in the A fragment of XhoI-digested GIV genome (Fig. 1). The locations, orientations, sizes, and PSI-BLAST results for the putative ORFs are listed in Table 2. Predicted ORFs, with an average length of 987 nucleotides (329 amino acids), represented 83% coding density.
FIG. 1.
Circular representation of the GIV genome. The putative 120 ORFs are shown as blank arrows in the first circle (outer ring). The second to fifth rings are the sites of EcoRI, HindIII, XhoI, and NotI restriction enzymes, respectively (the fragments are shown by alphabetic letters in descending order). The repeat sequences (R1 to R17) are indicated in the sixth ring (the innermost ring).
TABLE 1.
Summary of genomic sequence data for six viral species representing four genera within the family Iridoviridae
| Characteristic | Data for indicated genus and species
|
|||||
|---|---|---|---|---|---|---|
| Iridovirus CIV | Ranavirus
|
Lymphocystivirus LCDV | Unassigned (SKNV) | |||
| ATV | TFV | GIV | ||||
| Genome size (bp) | 212,482 | 106,332 | 105,057 | 139,793 | 102,653 | 111,362 |
| GC (%) | 29 | 54 | 55 | 49 | 29 | 55 |
| No. of putative ORFs | 468 | 96 | 105 | 120 | 195 | 124 |
| ORF size (no. of amino acids) | 40-2,432 | 32-1,294 | 40-1,294 | 62-1,268 | 40-1,199 | 40-1,208 |
| Accession no. | AF303741 | AY150217 | NC003407 | AY666015 | NC001824 | NC001824 |
TABLE 2.
Characterization of predicted open reading frames of Grouper Indovirus (GIV)a
| ORFb | Nucleotide position | Lengthc
|
Conserved domain or signature | CDD accession no.d | Best match homolog
|
||||
|---|---|---|---|---|---|---|---|---|---|
| nt | a.a. | MW | Predicted function and/or similarity | % Identity | Accession no.e | ||||
| 001L | 1:513 | 513 | 170 | 18,976 | 47 | AAP33240 | |||
| 002L | 2295:3536 | 1,242 | 413 | 46,218 | 40 | AAP33180 | |||
| 003R | 3564:4421 | 858 | 285 | 32,309 | 37 | AAP33179 | |||
| 004R | 4443:5399 | 957 | 318 | 34,343 | DUF230, Poxvirus proteins of unknown function | pfam03003 | Myristylated membrane protein | 70 | AAP33178 |
| 005L | 5453:6421 | 969 | 322 | 35,362 | 40 | AAP33250 | |||
| 006L | 6475:6900 | 426 | 141 | 16,629 | 59 | AAP33251 | |||
| 007L | 6950:7486 | 537 | 178 | 19,419 | Inositol-1,4,5-triphosphate 5-phosphatase | KOG0566 | Neurofilament triplet Hl-like protein | 41 | AAP33252 |
| 008L | 7528:7983 | 456 | 151 | 17,269 | 27 | AAP33254 | |||
| 009L | 8165:9682 | 1,518 | 505 | 55,796 | SAP, Putative DNA-binding motif | smart00513 | 43 | AAP33256 | |
| 010R | 9761:11461 | 1,701 | 566 | 63,213 | 39 | AAP33257 | |||
| 011Lf | 11546:12559 | 1,014 | 337 | 36,667 | |||||
| 012L | 12643:13659 | 1,017 | 338 | 37,611 | IG, Immunoglobulin | smart00409 | MGC52801 protein | 30 | AAH41253 |
| 013Lf | 13816:14850 | 1,035 | 344 | 38,125 | |||||
| 014Lf | 15362:16384 | 1,023 | 340 | 37,766 | |||||
| 015Lf | 16846:17904 | 1,059 | 352 | 39,680 | |||||
| 016Lf | 17964:19010 | 1,047 | 348 | 38,155 | |||||
| 017L | 19511:20638 | 1,128 | 375 | 42,281 | Neural cell adhesion molecule L1 | KOG3513 | 25 | AAH62581 | |
| 018L | 20771:21835 | 1,065 | 354 | 39,733 | Drosophila melanogaster CG15570-PA | 55 | NP_572154 | ||
| 019L | 21988:23016 | 1,029 | 342 | 37,096 | Neural cell adhesion molecule L1 | KOG3513 | |||
| 020L | 23044:23556 | 513 | 170 | 19,029 | 46 | AAP33260 | |||
| 021L | 23583:26738 | 3,156 | 1,051 | 118,626 | 37 | AAP33261 | |||
| 022L | 26865:27296 | 432 | 143 | 15,072 | Drosophila melanogaster CG15570-PA | 42 | NP_572154 | ||
| 023R | 27608:29605 | 1,998 | 665 | 73,550 | 40 | AAP33262 | |||
| 024Lf | 29679:30407 | 729 | 242 | 22,813 | |||||
| 025L | 30467:31204 | 738 | 245 | 23,473 | COG5295: Autotransporter adhesin | 39 | ZP_00122019 | ||
| 026L | 31247:32401 | 1,155 | 384 | 43,712 | NrdF, Ribonucleotide reductase, β subunit | COG0208 | 73 | AAP33216 | |
| 027L | 32549:32824 | 276 | 91 | 10,505 | CARD, caspase recruitment domain | pfam00619 | CARD-like caspase | 42 | AAP33218 |
| TFIIF-interacting CTD phosphatase | KOG0323 | ||||||||
| 028L | 32857:33336 | 480 | 159 | 17,483 | dUTPase | KOG3370 | dUTPase | 66 | AAH19979 |
| 029L | 33367:34029 | 663 | 220 | 23,582 | TNFR domain | cd00185 | Tumor necrosis factor receptor superfamily, member a | 29 | NP_571915 |
| 030L | 34201:34896 | 696 | 231 | 25,959 | TNFR domain | cd00185 | Tumor necrosis factor receptor superfamily member 14 precursor | 33 | XP_345617 |
| 031L | 35097:37997 | 2,901 | 966 | 109,744 | Predicted ATPase | COG3378 | D5 family NTPase | 71 | AAP33258 |
| Poxvirus D5 protein-like | pfam03288 | ||||||||
| 032R | 38100:38747 | 648 | 215 | 25,128 | 70 | AAP33259 | |||
| 033R | 38770:39492 | 723 | 240 | 22,715 | ATV nucleotide position 39,711-40,067 | 50 | |||
| 034R | 39521:40261 | 741 | 246 | 23,009 | Collagen-like protein | 51 | NP_078660 | ||
| 035L | 40327:43833 | 1,168 | 389 | 131,512 | LCDV ORF 2-like protein | 54 | AAK37740 | ||
| 036R | 44348:47200 | 2,853 | 950 | 107,211 | DNA repair protein, SNF2 family | KOG0390 | Helicase | 61 | AAP33184 |
| helicase_C, helicase conserved C-terminal domain | pfam00271 | ||||||||
| 037R | 47235:47849 | 615 | 204 | 23,319 | CPDc, catalytic domain of Ctd-like phosphatase | smart00577 | 50 | AAP33244 | |
| TFIIF-interacting CTD phosphatase | KOG1605 | ||||||||
| 038R | 47914:48708 | 795 | 264 | 27,641 | IGF-like growth factor | smart00078 | 38 | NP_705854 | |
| Amphiphysin | KOG3771 | ||||||||
| 039R | 48760:50478 | 1,719 | 572 | 63,753 | NrdA, ribonucleotide reductase, α subunit | COG0209 | Ribonucleoside-diphosphate reductase, α subunit-like protein | 71 | NP_571996 |
| 040L | 51254:51829 | 576 | 191 | 21,640 | Mitochondrial thymidine kinase 2-deoxyguanosine kinase | KOG4235 | Deoxyribonucleoside kinase-thymidine kinase | 47 | AAP33196 |
| 041L | 51863:52681 | 819 | 272 | 29,652 | ISKNV ORF112R | 27 | NP_612334 | ||
| 042L | 52668:54314 | 1,647 | 548 | 61,930 | 37 | AAP33194 | |||
| 043R | 54341:54799 | 459 | 152 | 17,035 | Ervl/Alr family | pfam04777 | Thiol oxidoreductase | 53 | AAP33193 |
| 044R | 54830:55654 | 825 | 274 | 31,589 | 32 | AAP33192 | |||
| 045R | 55751:57142 | 1,392 | 463 | 50,520 | Capsid iridovirus | pfam04451 | Major capsid protein | 72 | AAO32315 |
| 046L | 57221:60532 | 3,312 | 1,103 | 123,289 | RNA polymerase II, second largest subunit | KOG0214 | DNA-directed RNA polymerase II β subunit | 66 | NP_572001 |
| 047R | 57819:58085 | 267 | 88 | 9,173 | 73 | AAP33222 | |||
| 048R | 60586:61122 | 537 | 178 | 19,947 | Sensory box histidine kinase-response regulator | 26 | NP_793671 | ||
| 049L | 61207:62064 | 858 | 285 | 30,332 | Purine nucleoside phosphorylase | KOG3984 | Purine nucleoside phosphorylase | 50 | NP_000261 |
| 050Lf | 62188:63096 | 909 | 302 | 34,096 | |||||
| 051Lf | 62282:62944 | 663 | 220 | 25,860 | |||||
| 052L | 63202:66156 | 2,955 | 984 | 110,839 | CAP10, putative lipopolysaccharide-modifying enzyme | smart00672 | Putative tyrosine protein kinase | 44 | NP_571995 |
| 053Lf | 66271:66939 | 669 | 222 | 24,364 | |||||
| 054R | 67019:68356 | 1,338 | 445 | 50,545 | 41 | AAP33203 | |||
| 055L | 68413:69540 | 1,128 | 375 | 41,572 | Rnc, double-stranded RNA-specific ribonuclease | COG0571 | RNase III | 47 | NP_572005 |
| 056R | 69598:69876 | 279 | 92 | 10,570 | RPB9, DNA-directed RNA polymerase, subunit M-Transcription elongation factor TFIIS | COG1594 | Putative transcription elongation factor SII | 55 | NP_572006 |
| C2C2 zinc finger | smart00440 | ||||||||
| 057R | 69938:70402 | 465 | 154 | 17,127 | Immediate-early protein ICP-18 | 46 | P03298 | ||
| 058Rf | 70728:71375 | 648 | 215 | 25,146 | |||||
| 059L | 71835:73355 | 1,521 | 506 | 53,963 | Myristylated membrane protein A | 57 | AAK54492 | ||
| 060L | 73416:74588 | 1,173 | 390 | 45,521 | 23 | AAP33232 | |||
| 061L | 74673:75794 | 1,122 | 373 | 43,483 | 21 | AAP33232 | |||
| 062Lf | 75915:77051 | 1,137 | 378 | 44,664 | |||||
| 063L | 77240:78373 | 1,134 | 377 | 44,411 | 20 | AAP33232 | |||
| 064L | 78437:79654 | 1,218 | 405 | 47,514 | 22 | AAP33232 | |||
| 065R | 80301:80771 | 471 | 156 | 16,872 | TNFR domain | cd00185 | Tumor necrosis factor receptor superfamily, member 11b | 35 | NP_037002 |
| 066L | 80778:81926 | 1,149 | 382 | 43,338 | XPG, Xeroderma pigmentosum G | cd00128 | Putative DNA repair protein RAD2 | 54 | NP_572012 |
| 067R | 81580:82383 | 804 | 267 | 30,517 | RRV 8.141-kb PstI fragment | 57 | AAK53746 | ||
| 068Rf | 82906:83847 | 942 | 313 | 35,127 | |||||
| 069L | 83928:84161 | 234 | 77 | 8,547 | Ubiquitin-60s ribosomal protein L40 fusion protein | KOG0003 | Ubiquitin GP37 fusion protein | 81 | NP_258300 |
| 070R | 84246:84539 | 294 | 97 | 11,059 | 38 | AAP33269 | |||
| 071L | 84600:88406 | 3,807 | 1,268 | 139,162 | RNA polymerase III, large subunit | KOG0261 | DNA-dependent RNA polymerase α subunit | 61 | AAP33183 |
| RNA polymerase II, large subunit | KOG0260 | ||||||||
| 072R | 84652:84885 | 234 | 77 | 8,106 | CIV 344R | 45 | AAK82205 | ||
| 073R | 88462:89088 | 327 | 208 | 21,540 | Ati or CPXV158 protein | 29 | NP_619942 | ||
| 074R | 89401:90168 | 768 | 255 | 29,136 | 36 | AAP33270 | |||
| 075R | 90223:91326 | 1,104 | 367 | 35,962 | Collagens (type XV) | KOG3546 | Procollagen, type VI, α 3 | 38 | NP_034065 |
| 076L | 90579:90971 | 393 | 130 | 12,179 | B antigen | 38 | CAA42442 | ||
| 077L | 90975:91304 | 330 | 109 | 10,453 | Levansucrase | 38 | CAD48195 | ||
| 078R | 91793:92251 | 459 | 152 | 17,210 | BCL | smart00337 | Bak protein | 33 | AAF89533 |
| 079R | 92371:93147 | 777 | 258 | 30,130 | Putative replication factor | 58 | AAP33272 | ||
| 080R | 93463:94422 | 960 | 319 | 35,749 | 61 | AAP33268 | |||
| 081L | 94779:95162 | 384 | 127 | 14,315 | 47 | AAP33204 | |||
| 082Lf | 95643:96275 | 633 | 210 | 24,255 | |||||
| 083L | 96328:97416 | 1,089 | 362 | 41,459 | 31 | AAL13097 | |||
| 084Rf | 98151:98705 | 555 | 184 | 21,078 | |||||
| 085Rf | 98692:99330 | 639 | 212 | 23,960 | |||||
| 086Rf | 99403:99924 | 522 | 173 | 19,877 | |||||
| 087R | 100788:103817 | 3,030 | 1,009 | 114,730 | DNA polymerase family B | pfam00136 | DNA polymerase-like protein | 69 | NP_572000 |
| 088Rf | 104499:105053 | 555 | 184 | 19,822 | |||||
| 089R | 105071:105898 | 828 | 275 | 31,303 | 46 | AAP33263 | |||
| 090L | 106273:107244 | 972 | 323 | 36,485 | AAA, ATPases associated with a variety of cellular activities | smart00382 | ATPase-like protein | 68 | NP_571992 |
| 091R | 107692:108006 | 315 | 104 | 11,634 | LITAF, LPS-induced tumor necrosis factor α factor | smart00714 | 60 | AAP33206 | |
| 092R | 108028:109413 | 1,386 | 461 | 49,680 | 36 | AAP33207 | |||
| 093R | 110141:113554 | 3,414 | 1,137 | 119,286 | Trypanosoma cruzi R27-2 protein | 37 | T30296 | ||
| 094L | 113639:113878 | 240 | 79 | 8,967 | 49 | AAP33212 | |||
| 095R | 113959:114417 | 459 | 152 | 16,748 | Fibroblast growth factor | KOG3885 | Fibroblast growth factor 22 | 35 | NP_570107 |
| 096R | 114476:114973 | 498 | 165 | 18,060 | FGF, acidic and basic fibroblast growth factor family | cd00058 | Fibroblast growth factor 10 | 31 | NP_032028 |
| 097L | 115076:116050 | 975 | 324 | 36,684 | Predicted E3 ubiquitin ligase | KOG1814 | NTPase-helicase | 43 | AAP33208 |
| 098L | 116186:117220 | 1,035 | 344 | 39,379 | 25 | AAP33224 | |||
| 099R | 117323:117802 | 480 | 159 | 17,613 | 45 | AAP33210 | |||
| 100L | 118300:119826 | 1,527 | 508 | 57,187 | Phosphotransferase | 36 | AAP33226 | ||
| 101L | 119847:120434 | 588 | 195 | 22,222 | 35 | AAP33227 | |||
| 102R | 120533:121771 | 1,239 | 412 | 46,549 | SSL2, DNA or RNA helicases of superfamily II | COG1061 | Putative helicase | 44 | CAB37349 |
| 103L | 121088:121660 | 573 | 190 | 21,910 | FV3 40K protein | 30 | S49999 | ||
| 104R | 122172:123896 | 1,725 | 574 | 64,611 | Semaphorins | KOG3611 | Sema domain, immunoglobulin domain (1g), and GPI membrane anchor | 27 | NP_003603 |
| 105Lf | 124043:124852 | 810 | 269 | 31,085 | |||||
| 106R | 124940:125464 | 525 | 174 | 19,604 | 59 | AAP33225 | |||
| 107L | 126500:126988 | 489 | 162 | 18,861 | RNA polymerase, 25-kDa subunit | KOG3218 | 41 | AAP33238 | |
| 108L | 127649:128797 | 1,149 | 382 | 44,105 | 49 | AAP33190 | |||
| 109L | 128963:130138 | 1,176 | 391 | 46,396 | 21 | AAP33232 | |||
| 110Lf | 130166:131080 | 915 | 304 | 34,638 | |||||
| 111R | 131127:132272 | 1,146 | 381 | 43,194 | 3-β hydroxysteroid dehydrogenase-isomerase family | pfam01073 | 3-β-hydroxy-δ5-C27-steroid oxidoreductase-like protein | 53 | NP_571998 |
| 112R | 132248:132442 | 195 | 64 | 7,326 | Neuroblastoma-amplified protein | KOG1797 | |||
| 113L | 132345:133442 | 1,098 | 365 | 41,746 | 20 | AAP33232 | |||
| 114L | 133510:134652 | 1,143 | 380 | 40,361 | Transmembrane amino acid transporter protein | pfam01490 | N system amino acid transporter NAT-1 | 26 | AF61849 |
| 115R | 134680:135453 | 774 | 257 | 28,943 | 67 | AAP33234 | |||
| 116L | 135520:136425 | 906 | 301 | 29,638 | COG5295: autotransporter adhesin | 25 | ZP_00122019 | ||
| 117L | 136280:136972 | 693 | 230 | 22,247 | Collagen-like protein | 37 | NP_078660.1 | ||
| 118L | 137066:137530 | 465 | 154 | 16,433 | ATV nucleotide position 71,894-72,235 | 28 | |||
| 119L | 138030:138218 | 189 | 62 | 6,633 | 61 | AAP33242 | |||
| 120L | 138307:139776 | 1,470 | 489 | 56,353 | 45 | AAP33240 | |||
Nucleotides in the GIV virus genome were numbered sequentially, beginning with the nucleotide in the XhoI-A fragment.
The direction of the transcripts are indicated by R (right) or L (left).
nt, nucleotides; a.a., amino acids; MW, molecular weight.
Accession numbers obtained from the Conserved Domain Database by use of PSI-BLAST software.
GenBank accession numbers of homologous proteins.
ORFs unique to GIV.
Repeat sequences of GIV.
Repeat sequences have been observed in ATV, ISKNV, LCDV, and CIV genomes (23, 29, 30, 51). Although no repeat sequences are reported for the TFV genome (24), such extensive repeat regions were identified in TFV as well. Besides iridoviruses, the poxviruses, herpesviruses, baculoviruses, adenoviruses, and retroviruses also possess repeat sequences (30). These repeat sequences could be associated with important regulatory functions during viral replication. Seventeen sets of repeat sequences (R1 to R17) were found throughout the GIV genome (Fig. 1). The repeat sequences were of a basic unit length ranging from 9 to 63 bp, with copy numbers of 1.9 to 36.5. When compared to individual consensus repeat units, the repeat sequences matched from 82 to 100%. Repeat sequence R14 was 1,533 bp long and is the longest repeat sequence among the complete sequenced iridovirus genomes reported so far. R14 was composed of 25 sets of 42-bp-long complete direct-repeat units and 23 sets of 21-bp half-direct-repeat units. The function of this repeat sequence needs to be further characterized. Besides, R2, R3, R13, and R16 of GIV were located in the noncoding regions and 13 were within the putative ORFs (001L, 007L, 012L, 018L, 025L, 034R, 038R, 073R, 075R, 093R, and 116L). Two putative Goldberg-Hogness boxes, each comprising an AT-rich sequence (TATTTTA), were observed in the repeat sequences R1 and R16. A similar TATTTTA box was also found in upstream of the FV3 immediate-early gene ICR 169 and FV3 late gene ICR 534 and was demonstrated to be transcription dependent (61). Besides the TATTTTA box, repeat sequence R1 also contained a CCAAT box, which is similar to the upstream sequence result observed for the FV3 immediate-early gene ICR 489 (62). The CCAAT box was shown to bind a cellular transcriptional factor (62). In the coding region, repeat sequences R9 and R10 were found within ORF 038R; repeat sequence R9 was located in a putative amphiphysin domain (CDD accession no. KOG3771). Apart from repeat sequence R9, none of the repeat sequences was located within or formed any possible functional domain. In addition, we did not find any homology between the repeat sequences in GIV genome and those of the other five complete sequenced iridovirus genomes. However, repeat sequence homology was observed between ATV and TFV, suggesting that ATV may be closer to TFV than to GIV in taxonomic position.
Protein coding content.
Among the 570 ORFs identified by Artemis analysis, 97 of them had significant homology to ORFs in the protein database and 2 (019L and 112R) had homology to ORFs encoding a significant protein domain; however, no identifiable functional protein was found. Two ORFs (033R and 118L) had no corresponding homolog in the database, but they were found in the ATV genome (ATV nucleotide positions 39,711 to 40,067 and positions 71,894 to 72,235). A total of 19 ORFs considered to be unique to the GIV genes had no corresponding homologues in protein database. In total, 120 GIV ORFs ranging from 62 to 1,268 amino acids in length are listed in the Table 2. Fifty-six ORFs showed 40 to 81% identity to ORFs encoding known proteins from other viruses or organisms or encoding an identifiable functional domain, and 41 showed partial homology (20 to 39% identity) to ORFs encoding a known protein or an identifiable functional domain.
GIV ORFs were, for the most part, nonoverlapping, which is consistent with the results seen with other iridoviruses (30). However, 12 overlapping ORFs were observed in the GIV genome (Fig. 1 and Table 2). ORF 047R had homology with ORFs in ATV, CIV, and TFV, and it was located within the β subunit of RNA polymerase II (046L). ORF 067R had 57% sequence identity with RRV (Regina ranavirus) and overlapped with ORF 066L, which was similar to the results seen with DNA repair protein RAD2. ORF 072R, which shared 45% sequence identity with CIV 344R, was located within 071L (in similarity to the results seen with the DNA-dependent RNA polymerase α subunit). ORF 076L (similar to an ORF encoding B antigen) and ORF 077L (similar to an ORF encoding levansucrase) were located within the 075R, which shared 38% sequence identity with an ORF encoding procollagen, type VI. ORF 103L (similar to an ORF encoding FV3 40K protein) was located within 102R (similar to an ORF encoding helicase).
GIV ORF analysis. (i) Replication.
The GIV genome contains four homologues of the iridovirus genes associated with DNA replication machinery. Like those of the other five iridoviruses, the GIV genome encoded D5 family NTPase (031L), helicase (036R), DNA repair protein RAD2 (066L), and DNA polymerase (087R) (Table 3). The function prediction analysis of ORF 031L revealed that it encoded not only an ATPase domain but also a poxvirus D5 protein-like domain in its N terminus. This D5 protein is necessary for viral DNA replication and is a nucleic acid-independent nucleoside triphosphatase. ORF 036R encoded a protein containing not only a DNA repair protein, the SNF2 family domain, but also a helicase superfamily C-terminal domain. This helicase domain is found in a wide variety of helicases and helicase-related proteins. All helicases share the ability to unwind nucleic acid duplexes; however, the exact function of ORF 036R needs further investigation. ORF 066L encoded a DNA repair-like protein, RAD2, and the corresponding ORFs can also be found in the other five iridoviruses. In the ATV, LCDV, ISKNV, and CIV genomes, however, the start site of the RAD2 ORFs is different from that of TFV RAD2 gene. The TFV RAD2 (TFV 101R) had a point mutation which shifted the RAD2 start site upstream (30). Interestingly, alignment of the GIV RAD2 gene (066L) with the TFV RAD2 gene revealed that amino acid position 24 of ORF 066L matches amino acid position 28 of TFV 101R. When the start sites of the RAD2 genes of GIV and TFV are aligned, it is observed that GIV and TFV are in the same frame except for ORF 066L, which is short four amino acids. The product of ORF 087R shows significant homology to a DNA polymerase of cellular and viral origin, belonging to the family B DNA polymerase. These conserved DNA replication machinery-dependent genes may represent an advantage for viral genome replication that might ultimately lead to persistence of infection and a broad host range for viral infection. These conserved genes might have been acquired from a host or from infection of a common host by ancient iridoviruses (41). The acquired cellular genes could have remained as essential genes for virus replication or could represent nonessential genes which were lost or replaced by other genes in the virus during evolution (51).
TABLE 3.
ORF sequence homologies between GIV and other iridovirusesaa
| ORF group and putative function | GIV ORFs | ATV corresponding ORFs | TFV corresponding ORFs | LCDV corresponding ORFs | ISKNV corresponding ORFs | CIV corresponding ORFs |
|---|---|---|---|---|---|---|
| ORFs conserved in all six sequenced iridovirus genomes | ||||||
| Replication | 031L | 77L | 022R | 6 | 109L | 184R |
| 036R | 7L | 009L | 4 | 063L | 022L | |
| 066L | 10L | 101R | 34 | 027L | 369L | |
| 087R | 44L | 063R | 5 | 019R | 037L | |
| Transcription | 037R | 64R | 040R | 64 | 005L | 355R |
| 046L | 43R | 065L | 3 | 034R | 428L | |
| 055L | 25R | 085L | 44 | 087R | 142R | |
| 056R | 24L | 086R | 105 | 029L | 349L | |
| 071L | 6R | 008R | 1 | 028L | 176R | |
| Nucleotide metabolism | 026L | 38R | 071L | 26 | 024R | 376L |
| 040L | 19L | Position 90,715-91,302 | 60 | 032R | 143R | |
| Protetin modification | 043R | 16L | 094R | 79 | 043L | 347L |
| Structure proteins | 004R | 1L | 002L | 29 | Position 83,701-84,663 | 337L |
| 045R | 14L | 096R | 91 | 006L | 274L | |
| 059L | 51L | 055R | 20 | 007L | 118L | |
| Virus-host interaction | 052L | 58R | 029R | 8 | 114L | 179R |
| Other | 021L | 80L | 019R | 14 | 055L | 380R |
| 032R | 78R | 021L | 70 | 056L | 067R | |
| 035L | 69R | 045R | 2 | 076L | 295L | |
| 067R | 11R | 100L | 71 | 086L | 307L | |
| 079R | 91R | 105R | 43 | 061L | 282R | |
| 080R | 87R | 012L | 39 | 096L | 287R | |
| 090L | 83L | 016R | 46 | 122R | 075L | |
| 100L | 47L | 059R | 17 | 013R | 380R | |
| 108L | 13L | 097R | 27 | 115R | 393L | |
| ORFs conserved in the ranavirus genomes | ||||||
| Transcription | 091R | 29R | 080L | |||
| Virus-host interaction | 007L | 72L | 048L | |||
| 027L | 40L | Position 73,946-74,209 | ||||
| Other | 005L | 70L | 046L | |||
| 008L | 73L | 049L | ||||
| 023R | 81R | 018L | ||||
| 044R | 15L | 095R | ||||
| 054R | 26L | 084R | ||||
| 057R | 23L | 087L | ||||
| 060L | 53R | 023R | ||||
| 061L | 53R | 023R | ||||
| 064L | 53R | 023R | ||||
| 070R | 88L | 011R | ||||
| 083L | 45R | 062L | ||||
| 092R | 30R | 079L | ||||
| 094L | 35L | 074R | ||||
| 098L | 45R | 062L | ||||
| 106R | 46L | 061R | ||||
| 109L | 53R | 023R | ||||
| 113L | 53R | 023R | ||||
| 119L | 62R | 034R | ||||
| ORFs showing significant homologies to other iridoviruse genomes | ||||||
| Transcription | 009L | 75L | 051L | 58 | ||
| 102R | 50R | 056L | 161L | |||
| 107L | 59R | 030R | 75 | |||
| Nucleotide metabolism | 028L | 42L | 068R | 438L | ||
| 039R | 65R | 041R | 12 | 085L | ||
| Protein modification | 097L | 31R | 078L | 36 | ||
| Structure proteins | 033R | Position 39,711-40,067 | 37 | |||
| 034R | 37 | |||||
| 075R | 37 | |||||
| 117L | 37 | |||||
| ORF group and putative function | GIV ORFs | ATV corresponding ORFs | TFV corresponding ORFs | LCDV corresponding ORFs | ISKNV corresponding ORFs | CIV corresponding ORFs |
| Virus-host interaction | 038R | 7 | 261R | |||
| 096R | 37R | |||||
| 111R | 52R | 054L | 31 | |||
| Other | 001L | 61R | ||||
| 002L | 3R | 004R | 229L | |||
| 003R | 2L | 25 | ||||
| 006L | 71L | 047L | 88 | |||
| 010R | 76R | 053R | 9 | |||
| 020L | 79L | 020R | 84 | 117L | ||
| 041L | 45 | 112R | ||||
| 042L | 17R | 093L | 11 | |||
| 047R | 43L | Position 69,879-70,073 | 430R | |||
| 048R | 55 | |||||
| 063L | 53R | |||||
| 072R | 344R | |||||
| 074R | 89R | 103R | 48 | |||
| 081L | 27R | 082L | 102 | |||
| 089R | 82R | |||||
| 099R | 33L | 74 | ||||
| 101L | 48L | 058R | 83 | |||
| 114L | 001L | |||||
| 115R | 55R | 025R | 49 | 118L | ||
| 118L | Position 71,894-72,235 | |||||
| 120L | 61R |
The comparison analysis was performed in a bidirectional manner, meaning that every ORF in each individual genome was used to query all the other genomes.
(ii) Transcription.
The genes involved in virus transcription processes included two genes encoding subunits of a DNA-dependent RNA polymerase (046L and 071L) and ORFs encoding a SAF-A/B, Acinus, and PIAS (SAP) domain (009L), a putative Ctd-like phosphatase (037R), RNase III (055L), transcription elongation factor SII (056R), lipopolysaccharide-induced tumor necrosis factor alpha (TNF-α) factor (LITAF) (091R), a putative helicase (102R), and a 25-kDa subunit RNA polymerase (107L). ORF 46L and ORF 071L showed significant homologies to a RNA polymerase II β and α subunit, respectively. Both of them are conserved in six iridoviruses as well as in DNA virus genomes. The C terminus of ORF 009L contained a SAP domain with 91.4% sequence similarity to the SAP domain consensus sequence (smart00513). This motif can be observed in ATV 75L, TFV 051L, and LCDV 58, with 43, 45, and 43% sequence identity, respectively, to the GIV 009L motif. The SAP motif contained several positions enriched with positively charged amino acids that might make contact with the backbone of the DNA and may play a role in sequence- or structure-specific DNA binding involving DNA transcription or repair (2). ORF 037R not only matched 93.3% of the amino acid sequence of catalytic domain of Ctd-like (CPDc) but also matched a TFIIF-interacting CTD phosphatase domain. However, the function of ORF 037R remains to be studied.
ORF 055L encoded an RNase III, which is an important enzyme in the processing of cellular and viral precursor RNAs to produce mature rRNAs, mRNAs, and tRNAs (65). ORF 056R contained a DNA-directed RNA polymerase subunit M and a C2C2 zinc finger domain in its C terminus. This ORF was conserved in all six iridovirus genomes (Table 3) and may be a subunit of DNA-dependent RNA polymerase. ORF 091R encoded a protein with a LITAF domain (smart00714) in its C terminus, and it also can be found both in ATV 29R and TFV 080L, with 60 and 58% sequence identity, respectively. This domain is a LITAF and had been suggested to be a transcription factor which regulates the TNF-α gene expression (50). The product of ORF 102R showed 88.8% sequence similarity to sequences of SSL2, DNA, or RNA helicases of superfamily II, and this ORF is also observed in ATV 50R, TFV 056L, and CIV 161L, with 44, 44, and 29% of sequence identity to GIV 102R, respectively. The amino acid sequence of the C terminus of ORF 107L was homologous to that of an RNA polymerase 25-kDa subunit (KOG3218), and this domain can also be found in ATV 59R and TFV 30R. Both ATV 59R and TFV 30R showed 41% amino acid sequence identity to GIV 107L. Although LCDV 75 shares 37% sequence identity over 115 amino acids with GIV 107L, we could not find this conserved domain in LCDV 75.
(iii) Nucleotide metabolism.
GIV-encoded enzymes involved in nucleic acid metabolism include the two subunits of the ribonucleotide reductase (026L and 039R), dUTPase (028L), deoxyribonucleoside kinase-thymidine kinase (040L), and purine nucleoside phosphorylase (PNP) (049L). The product of ORF 026L showed 97.1% sequence similarity to a ribonucleotide reductase β subunit conserved domain (COG0208) in the CDD database. ORF 039R showed significant homology to a ribonucleotide reductase α subunit. Both ribonucleotide reductase α and β subunits are conserved in GIV, ATV, TFV, LCDV, and CIV genomes (Table 3). However, the ribonucleotide reductase α subunit was not observed in the ISKNV genome (Table 3) (23). Ribonucleotide reductase reduced ribonucelotides into deoxyribonucleotides as immediate precursors of DNA (31). The lack of the ribonucleotide reductase α subunit in ISKNV means that this enzyme may not be functional or that its function may be replaced by other genes. Similarly, not only ISKNV but also some large DNA viruses, such as betaherpesviruses and poxviruses, do not possess a ribonucleotide reductase (52) or show a lack of homology to one of the ribonucleotide reductase subunits, such as that of Rhesus cytomegalovirus (21). Whether the ribonucleotide reductase in GIV or in other iridoviruses is virus replication dependent or not is worth exploring. ORF 028L showed 66% sequence identity to a mouse dUTPase over 159 amino acids. This enzyme cleaves the alpha-beta phosphodiester bonds of dUTP to form pyrophosphate and dUMP, preventing the incorporation of uracil into DNA and providing the substrate for thymine synthesis. dUTP has been shown to be essential for the replication of DNA viruses (4). However, it was not observed in the ISKNV and LCDV genomes (Table 3). ORF 040L showed 85.7% sequence similarity to a mitochondrial thymidine kinase 2-deoxyguanosine kinase domain (KOG4235). Deoxyribonucleoside kinase-thymidine kinase is an enzyme that catalyzes the transfer of the gamma phosphoryl group of ATP to thymidine to generate dMTP in the deoxyribonucleoside salvage pathway. It is a key enzyme in viral DNA replication and is conserved in all six sequenced iridovirus genomes (7) (Table 3). The product of ORF 049L had over 50% sequence identity to a human PNP enzyme. PNP enzyme is a key enzyme of purine salvage pathway, which can catalyze the reversible phosphorylysis of (2′-deoxy) purine ribonucleotides to free bases and (2′-deoxy) ribose-1-phosphates (14). Despite its distribution in nature, a unique viral PNP gene has just been reported for the GIV genome (55). PNP is not found in any iridoviruses or any existing virus, according to comparisons of ORF 049L by use of the protein database. This suggests that GIV might acquire this gene from its host during evolution. However, such an assumption requires more data to support it.
(iv) Protein modification.
ORF 043R encoded a modification enzyme, namely, thiol oxidoreductase. ORF 43R contained a C-X-X-C motif with a homology to an Erv1/Alr family domain. Erv1/Alr family proteins are encoded by all eukaryotes and many cytoplasmic DNA viruses. The conserved domain of the Erv1/Alr family consisted of 100 amino acids and contained a C-X-X-C motif, suggesting that Erv1/Alr family proteins might function as thiol oxidoreductases and facilitate the disulfide bond formation of viral proteins in the host cytoplasm (44). Two ORFs (069L and 097L) are involved in the ubiquitin protein degradation process. ORF 069L encoded a protein with 77 amino acids and has 81% sequence identity with a Spodoptera litura nucleopolyhedrovirus (SpltMNPV) ubiquitin-60s ribosomal protein L40 fusion protein. The function of ubiquitin in cells is to signal the degradation of protein by the 26S proteosome (25). ORF 097L was homologous to a predicted E3 ubiquitin ligase domain (KOG1814) and showed 50.5% amino acid sequence similarity to this domain. The ubiquitin-mediated proteolysis is part of the regulated turnover of proteins required for controlling cell cycle progression. The role of E3 ubiquitin involved a three-step proteolysis mechanism. The E3 ubiquitin ligase-coupled E2 ubiquitin enzyme binds to substrates and assembles a multiubiquitin chain on the substrates. The E3 ubiquitin ligase transfers the activated ubiquitin from the E2 to the substrate lysine residue (28). It was found to be a host immunity modulator in bovine herpesvirus 4, Kaposi's sarcoma-associated herpesvirus, and swine-pox virus (11, 19). This ligase domain is also observed in ATV 31R, TFV 078L, and LCDV 36, and they share 43, 44, and 28% sequence identity, respectively, to GIV 097L. The E3 ubiquitin ligase domain can also be found in ISKNV 120R and CIV 095L. However, the sequence alignment similarities of ISKNV 120R and CIV 095L with this E3 ubiquitin ligase were as low as 10 and 12%, respectively, and no significant similarity were found in comparisons to GIV 097L.
(v) Structure proteins.
GIV encoded three putative viral structural proteins, ORFs 004R, 045R, and 059L, and these three viral structural proteins are conserved through all six sequenced iridovirus genomes. ORF 004R encoded a putative myristylated membrane protein with a poxvirus conserved region (pfam03003) and a transmembrane domain in its C terminus. The TMHMM transmembrane prediction software showed that the poxvirus conserved region might be located outside the cell membrane. The corresponding ORF of GIV 004R was not described for ISKNV, but it could be found within nucleotides 83,701 to 84,663 of the ISKNV genome. ORF 059L also showed sequence homology to a putative myristylated membrane protein, but we could not find any conserved domain or identifiable functional motif in this ORF. Besides, ORF 059L contained two transmembrane domains at amino acids 189 to 211 and 216 to 238, in the middle of this ORF. The functions of both putative membrane proteins GIV 004R and 059L are yet to be investigated. ORF 045R encodes a major capsid protein gene which is 463 amino acids long and is of the same peptide size as those of ATV and TFV. Moreover, ORF 045R shares 72% amino acid sequence identity with ORFs of both ATV and TFV. ORFs 033R, 034R, 075R, and 117L all contained glycine-rich fragments and showed homology to a collagen-like protein, LCDV 37. Besides the glycine-rich domain, ORF 075R contained a collagen type XV domain. The glycine-rich domain in the collagen-like protein was also found in the structural proteins of Ectocarpus siliculosus virus 1 (EsV-1) (12), herpesvirus saimiri (HSV) (17), and bacteriophage PRD1 (5). The glycine-rich domains in these structural proteins may play only a supplementary role in protein function (64).
(vi) Virus-host interaction.
The product of ORF 007L showed just 9.4% weakly significant sequence homology to a inositol-1,4,5-triphosphate 5-phosphatase, INP51/INP52/INP53 family domain (KOG0566). This ORF can be observed in the ATV genome (ATV 72L, with 41% sequence identity to GIV 007L) and TFV genome (TFV 048L, with 43% sequence identity to GIV 007L) (Table 3), but its function is yet to be defined. Both ORF 017L and 019L showed weak sequence homologies to a neural cell adhesion molecule L1 domain, but neither of them had sequence homology to any functional protein in database. Both ORF 017L and 019L contained two transmembrane domains in their N and C termini, respectively. The neural cell adhesion molecule L1 domain of ORFs 017L and 019L were located between these two transmembrane domains and were predicted to be located outside the cell membrane. However, the functions of these two ORFs are unknown. The N terminus of ORF 038R was homologous to an insulin-like growth factor (IGF) domain and an amphiphysin domain which follows the IGF domain. ORF 038R shows 38% sequence identity to LCDV 7. Viral proteins containing an IGF domain might be involved in virus-host interactions (51), and the amphiphysin has been implicated in synaptic vesicle endocytosis (57).
ORFs 095R and 096R show 35 and 31% sequence identity, respectively, to rat and mouse fibroblast growth factors (FGF). ORF 096R has a weak homology with ATV 37R, but 095R has no sequence similarity to ATV 37R. Besides the GIV genome, an FGF-like gene is also encoded in the nuclear polyhedrosis virus genome (3). However, the function of FGF superfamily-like proteins in the virus genome is still unknown. ORF 111R encoded a protein with homology to 3-beta-hydroxy-delta 5-C27-steroid oxidoreductase, and the corresponding ORF can also be found in TFV 054L, with 53% sequence identity to GIV 111R. This enzyme can be found in the ATV 52R, but it is truncated to 53 amino acids and shares no sequence homology with GIV 111R. This protein has been suggested to be involved in modulating the host response to poxvirus infection (43). However, the exact function of this gene in virus has yet to be defined. The identifiable domain or predicted function of these ORFs suggested that they might play a role in the signal transduction process of virus and cell interaction.
ORF 012L encoded a protein with an immunoglobulin domain in its N terminus and a transmembrane domain in its C terminus. The TMHMM prediction revealed that the immunoglobulin domain of this ORF was located outside the cell membrane. ORF 104R has 27% sequence identity to a human semaphorin K1. Semaphorin-like sequences have been identified in the genomes of poxviruses and alcelaphine herpesvirus-1 (13). Viral semaphorins have been proposed to be a part of a defense mechanism used by certain viruses to suppress the immune response of the host (63). Many viruses encode proteins that can function as immune modulators, for example, by interfering with antigen presentation, acting as cytokines or cytokine antagonists, inhibiting apoptosis, or interrupting the complement cascade (46). Many of these immunity-modulating genes have been described as nonconserved genes, revealing the possibility of coevolution of viruses and hosts (46).
ORF 027L encoded a caspase recruitment domain (CARD) (pfam00619) possessing protein. CARD can be observed in the ATV 40L and within nucleotides 73946 to 74209 of the TFV genome, with 42 and 39% sequence identities to GIV 027L, respectively. CARD-containing proteins are suggested to be involved in apoptotic signaling. Although the function of ORF 027L remains to be investigated, it might regulate apoptosis in virus-infected cells (30). The products of ORF 029L, 030L, and 065R were homologous to a TNF receptor (TNFR) domain, and these three predicted ORFs shared 26 to 30% amino acid sequence identity with each other. ORF 030L contained a transmembrane domain in its C terminus, and its TNFR domain was predicted to be located outside the cell membrane. Although ORF 029 also contained a transmembrane domain in its C terminus, the TMHMM prediction result could not reveal whether the TNFR domain is inside or outside of the membrane. ORF 065R did not contain any transmembrane domain in its amino acid sequence. A TNFR domain-containing ORF was also described for the LCDV genome (LCDV 40), but no sequence similarity could be found between LCDV 40 and GIV 029L, 030L, and 065R. The TNFR-like viral factor of LCDV has been considered to be a “viroceptor.” It could bind to the fish cytokine TNF and neutralize its effects (16).
ORF 078R encoded a protein with a BCL domain containing BH1 and BH2 regions and contained a transmembrane domain in its N terminus. Bcl-2 homologues were found in gamma herpesviruses, avipoxvirus (fowlpox), asf virus (African swine fever), alpha herpesvirus and the most-studied E1B of adenoviruses. Although advances in understanding the Bcl-2 family protein have been made in recent years, the precise biochemical mechanisms by which any Bcl-2 family protein inhibits cell death are still unknown and are the subject of a lively debate (39). ORFs 029L, 030L, 065R, 078R, and 104R all possessed putative apoptosis modulator domains and might be involved in the regulation of cell apoptosis; such observations might be coincident with our previous observations of a high virus infection titer for GIV, 108 50% tissue culture infective doses/ml (34), and the phenomenon of a lack of infected GK cell apoptosis.
ORF 52L contained a putative lipopolysaccharide modifying enzyme domain (CAP10) in its C terminus, and this ORF was conserved in all six iridovirus genomes. The protein CAP-10 was demonstrated to be required for capsule formation, and its deletion abolished fungus virulence (8). However, whether ORF 52L plays a role in virus virulence or not needs further investigation.
(vii) Other.
ORF 047R encoded a putative protein with 88 amino acids and shared sequence identities of 73 and 46% with ATV 43L and CIV 430R, respectively. ORF 47R is located within the RNA polymerase II β subunit (046L), as ATV 43L is located within the RNA polymerase II β subunit of the ATV genome (43R). Although this ORF was not mentioned with respect to the TFV genome (24), it can be seen in within nucleotides 69,879 to 70,073 bp of the TFV genome and it is also located within the RNA polymerase II β subunit (65L) of the TFV genome. This TFV ORF has 75% sequence identity with GIV 047R. The reason for the continuing presence of this ORF in these iridoviruses and its function in different iridoviruses remain to be investigated further. ORF 057R encoded a protein with 46% sequence identity to FV 3 immediate-early protein ICP-18, and it can be found in the ATV genome (23L) and TFV genome (087L), with 44 and 45% sequence identities to GIV 057R, respectively. ORF 090L was homologous to an AAA ATPase domain in its N terminus, with 97.4% sequence similarity. The AAA ATPases are enzymes containing a P-loop NTPase domain and function as molecular chaperones and ATPase subunits of protease-, helicase-, or nucleic-acid-stimulated ATPase (27). However, the exact function of ORF 090L needs to be further investigated.
ORF 100L encodes a phosphotransferase; however, the predicted function of this ORF was determined only on the basis of sequence homology with the sequence database; no identifiable kinase domains could be found. The product of ORF 114L had a homology to a mouse N-system amino acids transporter, NAT-1, and it shares 21% of sequence identity with ISKNV 001L. The amino acid sequence of ORF 114L could be aligned to 87.7% of the putative transmembrane amino acid transporter sequence in the CDD. This ORF encoded an 11-transmembrane domain protein predicted by TMHMM, and the transcription product was observed in GIV-infected GK cells by Northern blotting (data not shown). However, its function is due for elucidation. In addition to these ORFs with identifiable functions, there are another seven ORFs of the GIV genome which are conserved in all six sequenced iridoviruses, including ORFs 021L, 032R, 035L, 067R, 079R, 080R, and 108L (Table 3). The sequence identity between these seven GIV ORFs and those of the other five iridoviruses ranged from a highest identity of 70% over 215 amino acids (GIV 032R compared to ATV 78R) to a lowest identity of 20% over 282 amino acids (GIV 108L compared to ISKNV 115R). All of the ORFs of these seven iridoviruses were without significant identifiable functional domains. Their functions remain to be characterized. The conservation of these unknown ORFs may reveal that these conserved iridovirus common genes might be the essential genes of the iridovirus. ORFs 060L, 061L, 063L, 064L, 109L, and 113L all show about 20% of sequence identity with ATV 53R and TFV 023R. Although their functions are yet to be investigated, these ORFs seem to be redundant gene copies in the GIV genome and share a level of sequence identity from 36 to 21% to each other.
The genes conserved in other five iridoviruses but not observed in the GIV genome.
ATV 22L showed homology to thymidylate synthase, and this enzyme is conserved in other two genomes (TFV 088R and CIV 225R) but has not yet been found in a GIV genome. Thymidylate synthase catalyzes the methylation of dUTP to yield the nucleotide precursor dTMP. This is an important step in the de novo pathway of biosynthesis of pyrimidine. Most viruses do not contain it, and they depend mostly on the host enzyme machinery for the replication of their genome to enable a reduction of genome size in these viruses (64). However, lack of such an important enzyme in DNA viruses (GIV and ISKNV) larger than ATV and TFV is surprising. ATV 57R encodes a homology of eIF-2α, and the corresponding ORF can also be found in the TFV genome (TFV 027R). Previous studies suggested that this gene is conserved throughout the genus Ranavirus (30), but the corresponding ORF was not found in the GIV genome. Vertebrate iridoviruses were found to be highly methylated at the cytosine residues in CpG sequences, and so a conserved DNA methyltransferase enzyme can be found in four vertebrate iridovirus genomes (ATV 21L, TFV 089R, ISKNV 046L, and LCDV 51) and the sequence identities ranged from 95 to 49%. But this enzyme is not found in the GIV genome, and restriction enzyme analysis revealed that the GIV genome was not methylated at all (data not shown). The DNA methylation was previously suggested to play a role in the viral restriction modification system (51). In eukaryote cells, the DNA methylation is involved in the regulation of specific gene expression. More sequence data would be required to answer questions such as these: is there such a viral restriction modification system that exists in other DNA methyltransferase containing vertebrate iridoviruses, and why it is absent in the GIV? Does GIV regulate its gene expression with a different regulatory mechanism other than the host gene expression regulatory mechanisms? Did GIV lose this enzyme during evolution, or was this virus a mutant virus? ATV 20 had homology to a proliferating cell nuclear antigen, and this ORF was conserved in other vertebrate iridoviruses (TFV 090R, ISKNV 112R, and LCDV 45). However, the function of this ORF is yet to be investigated.
The relationship of GIV to other five iridoviruses.
Among the 120 GIV ORFs, 25 ORFs (21%) are conserved in the other five iridoviruses (Table 3). The GIV genome shared 68.4, 54.6, 37.5, 27.5, and 23.3% of BLAST significant ORFs with the ATV, TFV, LCDV, CIV, and ISKNV genomes, respectively (Table 4). In addition to 25 iridovirus-conserved ORFs, 21 GIV ORFs showed significant sequence homologies with ORFs in the ATV and TFV genomes, the genomes of the Ranavirus genus (Table 3). The sequence analysis revealed that these 21 corresponding ORFs in ATV showed very high sequence identities, generally more than 90%, to the corresponding ORFs in TFV. These genes occupied 17.5% of the whole putative gene encoding content of GIV and are conserved in all complete sequenced ranavirus genomes. They could be classified as Ranavirus-specific genes. However, such an inference might be changed as new complete sequenced ranavirus genomes become available. Classification of the ranavirus- or iridovirus-specific genes requires more iridovirus genome sequences or individual viral ORF studies.
TABLE 4.
Common gene content in iridovirus genomes
| Virus | No. or % of genesa
|
|||||
|---|---|---|---|---|---|---|
| GIV | ATV | TFV | LCDV | ISKNV | CIV | |
| GIV | 120 | 68.4 | 54.6 | 37.5 | 23.3 | 27.5 |
| ATV | 67 | 98b | 85.7 | 38.8 | 23.5 | 34.7 |
| TFV | 59 | 84 | 108b | 38.9 | 20.4 | 24.1 |
| LCDV | 45 | 38 | 42 | 195 | 16.8 | 15.4 |
| ISKNV | 28 | 23 | 22 | 21 | 125b | 16 |
| CIV | 33 | 34 | 26 | 30 | 20 | 468 |
The number of genes shared between genomes (lower left triangle) or the percentage of genes shared between genomes (the total number divided by the number of genes in the smallest genome; upper right triangle) and the numbers of genes per genome (bold).
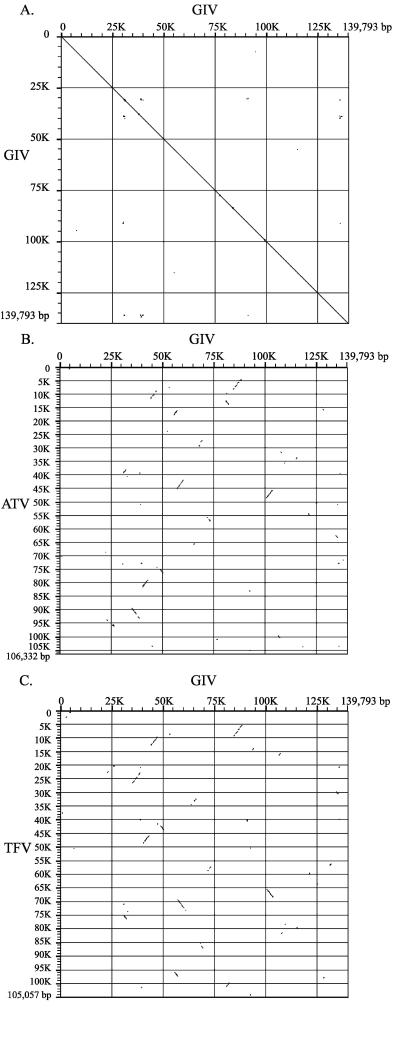
Besides the high percentage of ORFs shared between GIV, ATV, and TFV, we also can find the partial colinearity by whole-genome nucleotide dot plotting (Fig. 2). In comparisons made using GIV itself, the dot plot graph revealed the −45° diagonal line with intragenic repeat sequence “dots” on it (Fig. 2A). The intragenic repeat sequence dots indicate short palindrome repeat sequences. Most of these palindrome sequences were located in the noncoding regions except those palindrome sequences located within the ORFs 002L, 006L, 024R, 025L, 026L, 033R, 034R, 053L, 063L, 065L, 090L, and 116L. Sequence analysis reveals that most of these sequences contain short imperfect palindromes and form hairpins with a variety of sequence sizes. The functions of these coding region-hairpin palindromes need further investigations. Furthermore, no consensus promoter regions were found within the noncoding palindrome sequences. The lacking of consensus promoter regions of these palindrome regions might suggest that GIV genes are regulated differentially within viral replication life cycle and that the GIV genes might be regulated by unique transcription factors or promoter regions. Besides the intragenic repeat sequence dots on the −45° line, the dot plot also revealed the repeat sequence dots out of the line (Fig. 2A). Most of these repeat sequences were located within GIV ORFs and contributed to the formation of redundant genes in the GIV genome. Although we could not find perfect sequence colinearity when comparing GIV to ATV and GIV to TFV, the dot plot revealed short parallel 45° or −45° lines (Fig. 2B and C). These lines indicated most of the iridovirus replication, transcription, nucleotide metabolism, and structural conserved ORFs.
FIG. 2.
prFLAG analysis of the GIV genome (horizontal axis) versus the ATV and TFV genomes (vertical axis). (A) The GIV genome; (B) the ATV genome; (C) the TFV genome.
Moreover, the short parallel lines also indicated the conserved ranavirus gene clusters, including gene clusters of ORF 020L-021R, 031L-032R, 037R-039R, and 066L-067R (Table 5). However, we could not find any sequence colinearity when comparing GIV to LCDV, ISKNV, and CIV (data not shown). In addition to the gene cluster revealing by dot plotting, comparing GIV genomic structure to those of ATV and TFV also revealed conserved gene clusters between these three ranaviruses (Table 5). Although the ORFs of gene cluster VIII are found in all six sequenced iridoviruses, they are not found to be clustered in LCDV, ISKNV, and CIV. Besides, gene cluster I and X were composed of ranavirus conserved ORFs, and so it can be considered that these gene clusters may be specific for the Ranavirus genus. However, this assumption needs more sequenced iridovirus genomes to support it.
TABLE 5.
Gene clusters of ranaviruses
| Gene cluster | ORFs of ranavirus genome
|
||
|---|---|---|---|
| GIV | ATV | TFV | |
| I | 005L, 006L, 007L, 008L | 70L, 71L, 72L, 73L | 046L, 047L, 048L, 049L |
| II | 020L, 021L | 79L, 80L | 020R, 019R |
| III | 031L, 032R | 77L, 78R | 022R, 021L |
| IV | 037R, 039R | 64R, 65R | 040R, 041R |
| V | 042L, 043R, 044R, 045R | 17R, 16L, 15L, 14L | 093L, 094R, 095R, 096R |
| VI | 046L, 047R | 43R, 43L | 065L, position 69, 879-70, 073 |
| VII | 054R, 055L, 056R, 057R | 26L, 25R, 24L, 23L | 084R, 085L, 086R, 087L |
| VIII | 066L, 067R | 10L, 11R | 101R, 100L |
| IX | 083L, 087R | 45R, 44L | 062L, 063R |
| X | 091R, 092R | 29R, 30R | 080L, 079L |
| XI | 100L, 101L, 102R | 47L, 48L, 50R | 059R, 058R, 056L |
The ATV gene order of gene clusters V, VII, and IX and gene clusters II, III, VIII, X, and XI of TFV is inverse to the corresponding gene order of GIV. GIV ORFs of these gene clusters shared 20 to 70% of amino acid sequence identity with those of ATV and TFV. Although these three viruses are categorized into the same genus, Ranavirus, the comparison result suggested that ATV is closer to TFV than to GIV (30). The results of a similar genomic comparison of GIV to ISKNV, LCDV, and CIV genomes did not reveal such gene order conservation. It suggests that a greater extent of gene order is conserved in GIV, ATV, and TFV than in ISKNV, LCDV, and CIV.
The advances in high-throughput genomic sequencing technologies make the whole-genome comparison possible. Sequencing genes, which are homologous between different organisms, makes it obvious that many genes have been conserved over hundreds of millions of years of independent evolution. Such understanding makes the gene order of whole genomes a tool to distinguish the relations between different organisms (48). Generally the gene order is well preserved at close phylogenetic distance (49). When the species are not closely related, the degree of gene order conservation is usually low, and consequently it can be proposed that conservation of gene order is easily lost during evolution (26). The pairwise nucleotide alignment and genomic comparison suggest that the phylogenetic distance between GIV and ATV or between GIV and TFV could be greater than the distance between ATV and TFV (30). Repeat sequences and gene cluster inversions of GIV, ATV, and TFV genomes may suggest the higher recombination and genomic rearrangements of ranaviruses. High recombination rates have been described for the FV3 virus (9). The higher recombination rate may reflect the following facts about GIV. (i) GIV has more redundant genes. ORFs 060L, 061L, 063L, 064L, 109L, and 113L are similar to ATV 53R and TFV 23R. ORFs 017L and 019L are similar to neural cell adhesion molecule L1. ORFs 029L, 030L, and 065R are similar to TNFR. (ii) Unique genes exist in the GIV genome. Recombination may result in the insertion of new genes from host cells or other coinfected viruses such as PNP gene (ORF 049L), Bcl-2 like gene (ORF 078R), and host immunity interfering genes (ORF 104R). (iii) Conserved genus Ranavirus genes have been deleted. GIV genome lacks several conserved genes in Ranavirus, including DNA methyltransferase, thymidylate synthase, proliferating cell nuclear antigen, and eIF-2α. These facts may reflect the host range of GIV and the adaptation of GIV during host infection.
The similarity of G+C content and protein-coding content, the high percentage of ORF sharing, the high-level homology of ORF sequences, and the similar gene orders suggest that the GIV genome we describe here belongs to the genus Ranavirus along with the ATV and TFV. Syntenic conservation between GIV, ATV, and TFV provides us not only a more macroscopic analysis of the GIV genome's taxonomic position than analysis on the basis of one to a few individual genes but also facilitates our prediction of the approximate location of the relevant genes in different iridoviruses. However, to achieve a more precise prediction or understanding of iridovirus genome organization, more iridovirus genome information would be required.
Acknowledgments
We thank Teh-Yuan Chow of the Institute of Botany, Academia Sinica, for technical help and preliminary analysis of the genomic sequence. We thank J. A. Christopher John for advising and proofreading the manuscript.
This study was supported by thematic projects AS92IZ3 and AS92-AB-IZ-01 from Academia Sinica.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 3.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 4.Baldo, A. M., and M. A. McClure. 1999. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J. Virol. 73:7710-7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford, J. K. H., and D. H. Bamford. 1990. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology 177:445-451. [DOI] [PubMed] [Google Scholar]
- 6.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347-384. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinchar, V. G., and A. Granoff. 1986. Temperature-sensitive mutants of frog virus 3: biochemical and genetic characterization. J. Virol. 58:192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coenye, T., and P. Vandamme. 2003. Extracting phylogenetic information from whole-genome sequencing projects: the lactic acid bacteria as a test case. Microbiology 149:3507-3517. [DOI] [PubMed] [Google Scholar]
- 11.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaroque, N., S. Wolf, D. G. Müller, and R. Knippers. 2000. Characterization and immunolocalization of major structural proteins in the brown algal virus EsV-1. Virology 269:148-155. [DOI] [PubMed] [Google Scholar]
- 13.Ensser, A., and B. Fleckenstein. 1995. Alcelaphine herpesvirus type 1 has a semaphorin-like gene. J. Gen. Virol. 76:1063-1067. [DOI] [PubMed] [Google Scholar]
- 14.Erion, M. D., Takabayashi, K. Smith, H. B. Kessi, J. Wagner, S. Hönger, S. Shames, S. L., and S. E. Ealick. 1997. Purine nucleoside phosphorylase. 1. Structure-function studies. Biochemistry 36:11725-11734. [DOI] [PubMed] [Google Scholar]
- 15.Essbauer, S., and W. Ahne. 2001. Viruses of lower vertebrates. J. Vet. Med. Ser. B 48:403-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Essbauer, S., U. Fischer, S. Bergmann, and W. Ahne. 2004. Investigations on the ORF 167L of lymphocystis disease virus (Iridoviridae). Virus Genes 28:19-39. [DOI] [PubMed] [Google Scholar]
- 17.Geck, P., S. A. Whitaker, M. M. Medveczky, and P. G. Medveczky. 1990. Expression of collagen-like sequences by a tumor virus, herpesvirus saimiri. J. Virol. 64:3509-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson-Kueh, S., P. Netto, G. H. Ngoh-Lim, S. F. Chang, L. L. Ho, Q. W. Qin, F. H. C. Chua, M. L. Ng, and H. W. Ferguson. 2003. The pathology of systemic iridoviral disease in fish. J. Comp. Pathol. 129:111-119. [DOI] [PubMed] [Google Scholar]
- 19.Goto, E., S. Ishido, Y. Sato, S. Ohgimoto, K. Ohgimoto, M. Nagano-Fujii, and H. Hotta. 2003. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J. Biol. Chem. 278:14657-14668. [DOI] [PubMed] [Google Scholar]
- 20.Granoff, A., P. E. Came, and K. A. Rafferly. 1965. The isolation and properties of viruses from Rana pipiens: their possible relationship to the renal adenocarcinoma of the leopard frog. Ann. N. Y. Acad. Sci. 126:237-255. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, S. G., L. I. Strelow, D. C. Franchi, D. G. Anders, and S. W. Wong. 2003. Complete sequence and genomic analysis of Rhesus cytomegalovirus. J. Virol. 77:6620-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, J. G., S. P. Wang, K. Zeng, Z. J. Huang, and S.-M. Chan. 2000. Systemic disease caused by an iridovirus-like agent in cultured mandarinfish, Siniperca chuatsi (Basilewsky), in China. J. Fish Dis. 23:219-222. [Google Scholar]
- 23.He, J. G., M. Deng, S. P. Weng, Z. Li, S. Y. Zhou, Q. X. Long, X. Z. Wang, and S.-M. Chan. 2001. Complete genome analysis of the mandarin fish infectious spleen and kidney necrosis iridovirus. Virology 291:126-139. [DOI] [PubMed] [Google Scholar]
- 24.He, J. G., L. Lü, M. Deng, H. H. He, S. P. Weng, X. H. Wang, S. Y. Zhou, Q. X. Long, X. Z. Wang, and S. M. Chan. 2002. Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology 292:185-197. [DOI] [PubMed] [Google Scholar]
- 25.Hochstrasser, K. 1996. Protein degradation or regulation: Ub the judge. Cell 84:813-815. [DOI] [PubMed] [Google Scholar]
- 26.Huynen, M. A., and P. Bork. 1998. Measuring genome evolution. Proc. Natl. Acad. Sci. USA 95:5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer, L. M., D. D. Leipe, E., V. Koonin, and L. Aravind. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146:11-31. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, P. K. A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. R. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 29.Jakob, N. J., K. Müller, U. Bahr, and G. Darai. 2001. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286:182-196. [DOI] [PubMed] [Google Scholar]
- 30.Jancovich, J. K., J. Mao, V. G. Chinchar, C. Wyatt, S. T. Case, S. Kumar, G. Valente, S. Subramanian, E. W. Davidson, J. P. Collins, and B. L. Jacobs. 2003. Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America. Virology 316:90-103. [DOI] [PubMed] [Google Scholar]
- 31.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 32.Jung, S. J., and M. J. Oh. 2000. Iridovirus-like infection associated with high mortalities of striped beakperch, Oplegnathus fasciatus (Temminck et Schlegel), in southern coastal areas of the Korean peninsula. J. Fish Dis. 23:223-226. [Google Scholar]
- 33.Krogh, A., B. Larsson, G. V. Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 34.Lai, Y. S., S. Murali, H. Y. Ju, M. F. Wu, I. C. Guo, S. C. Chen, K. Fang, and C. Y. Chang. 2000. Two iridovirus-susceptible cell lines established from kidney and liver of grouper, Epinephelus awoara (Temminck & Schlegel), and partial characterization of grouper iridovirus. J. Fish Dis. 23:379-388. [Google Scholar]
- 35.Mao, J., R. P. Hedrick, and V. G. Chinchar. 1997. Molecular characterization, sequence analysis, and taxonomic position of newly isolated fish iridoviruses. Virology 229:212-220. [DOI] [PubMed] [Google Scholar]
- 36.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murali, S., M. F. Wu, I. C. Guo, S. C. Chen, H. W. Yang, and C. Y. Chang. 2002. Molecular characterization and pathogenicity of a grouper iridovirus (GIV) isolated from yellow grouper, Epinephelus awoara (Temminck & Schlegel). J. Fish Dis. 25:91-100. [Google Scholar]
- 38.Pollock, D. D., J. A. Eisen, N. A. Doggett, and M. P. Cummings. 2000. A case for evolutionary genomics and the comprehensive examination of sequence biodiversity. Mol. Biol. Evol. 17:1776-1788. [DOI] [PubMed] [Google Scholar]
- 39.Polster, B. M., J. Pevsner, and J. M. Hardwick. 2004. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim. Biophys. Acta 1644:211-227. [DOI] [PubMed] [Google Scholar]
- 40.Rodger, H. D., M. Kobs, A. Macartney, and G. N. Frerichs. 1997. Systemic iridovirus infection in freshwater angelfish, Pterophyllum scalare (Lichtenstein). J. Fish Dis. 20:69-72. [Google Scholar]
- 41.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 43.Senkevich, T. G., J. J. Bugert, J. R. Sisler, E. V. Koonin, G. Darai, and B. Moss. 1996. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science 273:813-816. [DOI] [PubMed] [Google Scholar]
- 44.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. USA 97:12068-12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnhammer, E. L. L., G. von Heiine, and A. Krogh. 1998. A hidden markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 46.Spriggs, M. K. 1996. One step ahead of the game: viral immunomodulatory molecules. Annu. Rev. Immunol. 14:101-130. [DOI] [PubMed] [Google Scholar]
- 47.Sudthongkong, C., M. Miyata, and T. Miyazaki. 2002. Viral DNA sequences of genes encoding the ATPase and the major capsid protein of tropical iridovirus isolates which are pathogenic to fishes in Japan, South China Sea and Southeast Asian countries. Arch. Virol. 147:2089-2109. [DOI] [PubMed] [Google Scholar]
- 48.Tamames, J. 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2:Research 0020.1-0020.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamames, J., G. Casari, C. Ouzounis, and A. Valencia. 1997. Conserved clusters of functionally related genes in two bacterial genomes. J. Mol. Evol. 44:66-73. [DOI] [PubMed] [Google Scholar]
- 50.Tang, X., M. J. Fenton, and S. Amar. 2003. Identification and functional characterization of a novel binding site on TNF-α promoter. Proc. Natl. Acad. Sci. USA 100:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tidona, C. A., and G. Darai. 1997. The complete DNA sequence of lymphocystis disease virus. Virology 230:207-216. [DOI] [PubMed] [Google Scholar]
- 52.Tidona, C. A., and G. Darai. 2000. Iridovirus homologues of cellular genes—implications for the molecular evolution of large DNA viruses. Virus Genes 21:77-81. [PubMed] [Google Scholar]
- 53.Tidona, C. A., D. Schnitzler, R. Kehm, and G. Darai. 1996. Identification of the gene encoding the DNA (cytosine-5) methyltransferase of lymphocystic disease virus. Virus Genes 12:219-229. [DOI] [PubMed] [Google Scholar]
- 54.Tidona, C. A., P. Schnitzler, R. Kehm, and G. Darai. 1998. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 16:59-66. [DOI] [PubMed] [Google Scholar]
- 55.Ting, J. W., M. F. Wu, C. T. Tsai, C. C. Lin, I. C. Guo, and C. Y. Chang. 2004. Identification and characterization of a novel gene of grouper iridovirus (GIV) encoding a purine nucleoside phosphorylase. J. Gen. Virol. 85:2883-2892. [DOI] [PubMed] [Google Scholar]
- 56.Walker, R. 1962. Fine structure of lymphocystis virus of fish. Virology 18:503-505. [DOI] [PubMed] [Google Scholar]
- 57.Wigge, P., K. Köhler, Y. Vallis, C. A. Doyle, D. Owen, S. P. Hunt, and H. T. McMahon. 1997. Amphiphysin heterodimers: Potential role in clathrin-mediated endocytosis. Mol. Biol. Cell 8:2003-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams, T. 1996. The iridoviruses. Adv. Virus Res. 46:345-412. [DOI] [PubMed] [Google Scholar]
- 59.Williams, T., V. G. Chinchar, G. Darai, A. Hyatt, J. Kalmakoff, and V. Seligy. 2000. Iridoviridae, p. 167-182. In M. H. V. van Regenmortel, D. H. L. Bishop. E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Seventh Report of the International Committee on the Taxonomy of Viruses. International Committee on Taxonomy of Viruses (ICTV), Orlando, Fla.
- 60.Willis, D. B., and A. Granoff. 1980. Frog virus 3 DNA is heavily methylated at CpG sequences. Virology 107:250-257. [DOI] [PubMed] [Google Scholar]
- 61.Willis, D. B., and A. Granoff. 1985. trans activation of an immediate-early frog virus 3 promoter by a virion protein. J. Virol. 56:495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis, D. B., J. P. Thompson, and W. Beckman. 1990. Transcription of frog virus 3, p. 173-186. In G. Darai (ed.), Molecular biology of iridoviruses, Kluwer Academic Publishers, Boston, Mass.
- 63.Xu, X., S. Ng, Z.-L. Wu, D. Nguyen, S. Homburger, C. Seidel-Dugan, A. Ebens, and Y. Luo. 1998. Human semaphorin K1 is glycosylphosphatidylinositol-linked and defines a new subfamily of viral-related semaphorins. J. Biol. Chem. 273:22428-22434. [DOI] [PubMed] [Google Scholar]
- 64.Yang, F., J. He, X. Lin, Q. Li, D. Pan, X. Zhang, and X. Xu. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., I. Calin-Jageman, J. R. Gurnon, T.-J. Choi, B. Adams, A. W. Nicholson, and J. L. V. Etten. 2003. Characterization of a chlorella virus PBCV-1 encoded ribonuclease III. Virology 317:73-83. [DOI] [PubMed] [Google Scholar]