Abstract
Split-hand and foot malformation (SHFM; MIM 183600) is a rare human genetic limb malformation. It is characterized by missing digital rays in the hands and feet. SHFMs vary in severity from mild abnormalities affecting a single limb to acute malformations involving all 4 limbs. It is inherited, as part of both a syndromic and nonsyndromic disorder, in an autosomal recessive, autosomal dominant, and X-linked patterns. So far, 9 loci of hand and foot malformation have been mapped on human chromosomes. The present study describes a family with 2 affected individuals segregating SHFM in an autosomal dominant fashion. Sanger sequencing of the genes involved in SHFM was performed to identify the disease-causing variant. Sequence analysis revealed the first heterozygous missense variant (c.632T>A, p.Val211Glu) in the distal-less homeobox 6 (DLX6) gene, located in chromosome 7q21, causing SHFM in the present family. This study supports the evidence of DLX6 as an SHFM-causing gene.
Key Words: Autosomal dominant SHFM1, DLX6, Missense variant, Sanger sequencing
Split-hand/foot malformation (SHFM) is a rare human inherited limb developmental disorder characterized by missing medial digital rays, hypoplasia of phalanges, and syndactyly of fingers and/or toes. Clinical features of SHFM vary from mild abnormalities of fused digits in a single limb to more severe malformations involving central digital rays leading to lobster-claw-shaped hands and/or feet. Inter- and intrafamilial variations in features associated with SHFM have been reported [Klopocki et al., 2012]. In a few cases, variations were observed in different limbs in the same individual as well [Khan et al., 2012; Aziz et al., 2014; Ullah et al., 2016].
To date, 9 loci including SHFM1-6, SHFLD3, chromosome 8q21.11q22.3, and chromosome 19p13.11 have been reported being involved in causing SHFM. For these 9 loci, intragenic mutations have been identified in 4 genes (DLX5, ZAK, TP63, and WNT10B) [Ianakiev et al., 2000; Ugur et al., 2008; Khan et al., 2012; Shamseldin et al., 2012; Simonazzi et al., 2012; Aziz et al., 2014; Lango et al., 2014; Wang et al., 2014; Spielmann et al., 2016; Ullah et al., 2016]. Four of the SHFM forms (SHFM1, SHFM3, SHFM4, and SHFM5) transmit in autosomal dominant, SHFM2 in X-linked, and SHFM6 in an autosomal recessive pattern of inheritance. The malformation occurs either as an isolated limb defect or in association with some other phenotypes, including hearing loss, intellectual disability, nail anomaly, and a cleft palate/lip [Aten et al., 2009; Spielmann et al., 2016].
Split-hand/foot malformation-1 (SHFM1) is caused by chromosomal aberrations such as deletions, inversions, translocations, and duplications in a region in chromosome 7q21 encompassing the genes DLX5, DLX6 and exonic enhancers of DYNC1I1 [Wieland et al., 2004; van Silfhout et al., 2009; Velinov et al., 2012; Delgado and Velinov, 2015]. Intragenic mutations in the DLX5 gene have been reported in families segregating SHFM in autosomal dominant fashion. A homozygous mutation in the DLX5 gene was found causing SHFM1 associated with hearing loss and segregating in an autosomal recessive pattern in a family of Turkish origin [Shamseldin et al., 2012]. Recently, we reported a 4-bp duplication in the DLX5 gene causing SHFM1, segregating in an autosomal dominant manner in a family of Pakistani origin [Ullah et al., 2016]. Both DLX5 and DLX6 genes are members of the DLX gene family showing expression in the head and limbs of the developing embryo. Robledo et al. [2002] described the role of the DLX genes in craniofacial, limb and bone development.
Materials and Methods
Blood Sampling and DNA Extraction
A family segregating SHFM1 in autosomal dominant manner was examined from the Punjab province of Pakistan. Information provided by the father of the proband (III-3) validated that his wife was not among his blood relatives. In the family pedigree, the affected mother (II-2) has an affected son, suggesting an autosomal dominant pattern of inheritance of the disease (Fig. 1a). Peripheral blood samples were collected from the proband (III-3) and 6 unaffected members of the family. DNA was extracted from blood samples using the standard protocol of Sigma-Aldrich GenElute Blood Genomic DNA Kit (St. Louis, MO, USA).
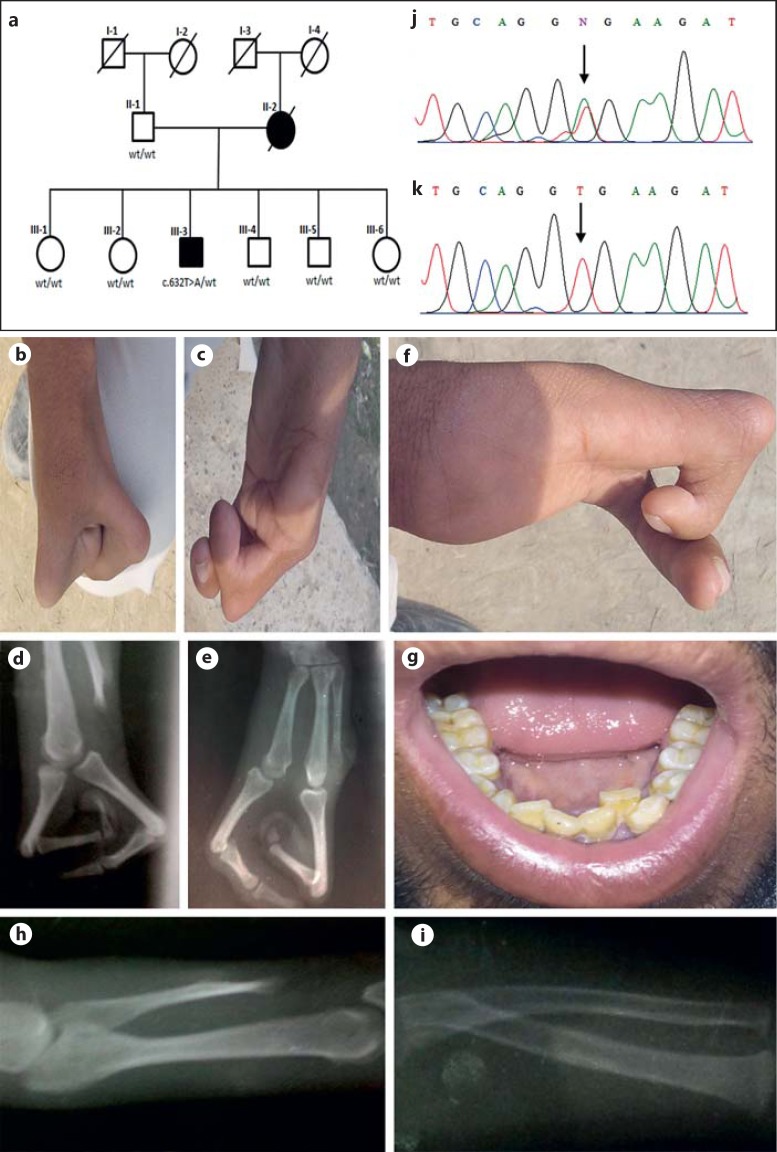
Fig. 1.
a Pedigree of a Pakistani family segregating SHFM1 in an autosomal dominant pattern. Members affected with SHFM1 are represented by black symbols. White symbols indicate unaffected individuals. Affected members screened for the heterozygous missense variant (c.632T>A) are shown. Deceased individuals are marked with a diagonal line. b-i Clinical features of the proband (III-3). b Right hand showing agenesis of the central ray with lobster claw deformity. c Left hand with aplasia of the thumb and fingers 2–3, and anonychia of the 4th finger. d, e Radiographs of the hands showing aplasia of phalanges and turning of phalanges towards the palms at metatarsophalangeal joints. f Right hand with nail clubbing. g Dental crowding. h, i Short radio-ulna. j, k Sequencing results of exon 3 in DLX6 (5′-3′). A heterozygous mutation (c.632T>A) in the proband is indicated by an arrow (j). Sequencing results of exon 3 in normal family members. The arrow indicates site of mutation (k). w/t, wild type.
Karyotype Analysis
Karyotype analysis was performed on peripheral blood lymphocytes collected from the proband (III-3) using G-banding technique. Phytohemagglutinin-stimulated culture was established and harvested after 72 h according to international standard protocols. Twenty metaphases were analyzed after Giemsa trypsin banding.
PCR and Sanger Sequencing
All coding exons and flanking intronic sequences of DLX5, DLX6, TP63, WNT10B as well as exon 15 and 17 of DYNC1I1 genes were PCR amplified using gene-specific primers, (PRIMER 3 software; http://frodo.wi.mit.edu/ primer3). Primer sequences for PCR-amplification of the WNT10B gene were the same as reported previously [Khan et al., 2012]. These sequences are available upon request. The primers were checked for specificity using the basic local alignment search tool (BLAST; http://www.ncbi.nlm. nih.gov/blast). In silico PCR for these primers was performed using In-silico PCR tool (http://genome.ucsc.edu/cgi-bin/hgPcr). PCR was carried out in 25 μL reaction volumes containing 40 ng genomic DNA, 20 pmol of each primer, 200 mM of each deoxynucleoside triphosphate, 2.5 μL reaction buffer (MBI Fermentas, Life Sciences, York, UK), and 1 unit Taq DNA polymerase (MBI Fermentas). The standard thermal cycle conditions, as described earlier [Ullah et al., 2015], were used to perform PCR reactions.
PCR products were detected by agarose gel electrophoresis and purified with a commercially available kit (AXYGEN, Union City, CA, USA). DNA sequencing was performed on ABI Prism 310 Genetic Analyzer (Applera, Foster City, CA, USA) using the Big Dye Terminator v3.1 Cycle Sequencing Kit. Sequence variants were identified using BIOEDIT sequence alignment editor version 6.0.7 (Ibis Biosciences, Carlsbad, CA, USA; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The pathogenicity score for the identified variant was calculated using mutation Taster (http://www.mutationtaster.org/), Polymorphism Phenotyping V2 (PolyPhen-2; http://genetics.bwh.harvard.edu/pph2/) and Sorting Intolerant from Tolerant (SIFT; http://sift.bii.a-star.edu.sg/).
Results
In the present family, the mother with SHFM has an affected son, genuinely validating transmission of the disorder in an autosomal dominant pattern. The proband (III-3) was 21 years old showing classical features of SHFM. He has aplasia of phalanges in his hands and has only 2 digits on the right hand which are turned towards the palms at the metacarpophalangeal joints with aplasia of carpals and metacarpals of the remaining digits showing a classical central ray defect. His left hand has agenesis of preaxial components. The first 3 digits and 2 corresponding carpals in the left hand are absent. Additional clinical features observed included short radius/ulna, nail clubbing of the right hand, anonychia of the second finger on the left hand, synophrys, and dental crowding (Fig. 1b,1c,1d,1e,1f,1g,1h1i).
Radiographic analysis showed aplasia of 3 metacarpals and phalanges in the right hand, aplasia of preaxial 3 digits and 2 metacarpals in the left hand, and short radius/ulna (Fig. 1d, 1e, 1h, 1i). The proband's feet were normal.
Anomalies such as dysmorphic facial features, hearing loss, and intellectual disability were not observed in the affected son. The proband's deceased mother had similar phenotypes as observed in our case. The unaffected members of the family were healthy.
Cytogenetic analysis of the patient (III-3) revealed an apparently normal male karyotype of 46,XY after G-banding. Therefore, no gross structural/numerical chromosomal abnormality was detected.
Mutation Screening
Five genes (DLX5, DLX6, and exons 15 and 17 of DYNC1I1, TP63, WNT10B), known for causing SHFM, were sequenced in the DNA of the proband (III-3). The analysis revealed a heterozygous sequence variant (c.632T>A) in exon 3 of the DLX6 gene mapped on chromosome 7q21.2q21.3 (Fig. 1j, 1k). The variant (c.632T>A) was not present in the other healthy family members, in 200 ethnically matched control individuals, 1000 Genomes (0.0%), Exome Aggregation Consortium (0.0%), and in the Human Gene Mutation Database. Mutation Taster predicted the variant (c.632T>A) in the DLX6 gene as probably damaging (score = 121). Protein prediction tool Polyphen2 revealed that the substitution of valine by glutamic acid at amino acid position 211 (p.Val211Glu) could potentially have a damaging effect (score = 1.000) on the DLX6 structure. The nonpathogenic nature of this variant was excluded by sequencing 200 ethnically matched individuals.
Discussion
SHFM is a rare limb developmental disorder involving central rays of autopods and is inherited in an autosomal dominant, recessive, and X-linked pattern. The proband, segregating SHFM1 in an autosomal dominant fashion, presented phenotypes restricted to the upper limbs with a bilateral cleft of the hands. His feet were apparently normal. Hypoplasia of radius and ulna, nail clubbing of the right hand, anonychia of the second finger on the left hand, crowding teeth, and synophrys were observed. Klopocki et al. [2012] reported phenotypes with aplasia/hypoplasia of radius and ulna, and microdontia in individuals affected with SHFM due to the duplication of 17p13.3. Shamseldin et al. [2012] reported a Yemeni family segregating SHFM1 due to a missense mutation in DLX5. In addition to the SHFM1 phenotype, the 6-year-old girl showed other features including hearing impairment, synophrys, low anterior hair line, circumferential nails, deformed legs and feet, restriction of joint flexion at all the metacarpophalangeal and interphalangeal joints, mild scoliosis, and malposition of left tibia and fibula. The second affected individual in the same family was a 4-year-old girl who revealed associated features including hearing loss, frontal bossing, and a high frontal hair line. Certain features including hearing loss, frontal bossing, scoliosis, and intellectual disability were not observed in our family.
Sequence analysis of the DLX6 gene in our proband (III-3) revealed a novel heterozygous mutation (p.Val211Glu). Previously, chromosomal aberrations including deletions, duplications, translocations, and inversions in 7q21.2q21.3 encompassing DLX5, DLX6, and DYNC1I1 have been reported in families segregating SHFM1 in an autosomal dominant pattern. Three intragenic heterozygous mutations (p.Gln186His, p.Glu39*, and p.Ala163-Profs*55) in the DLX5 gene causing the autosomal dominant form of SHFM1 [Sowińska-Seidler et al., 2014; Wang et al., 2014; Ullah et al., 2016] and a homozygous missense variant (p.Gln178Pro) in the same gene causing the autosomal recessive form of SHFM1 have been reported [Shamseldin et al., 2012] (Table 1, 2). Lango et al. [2014] reported that disruption of exon 15 and 17 of the DYNC1I1 gene, enhancers of DLX5/6, caused the autosomal dominant form of SHFM1 in humans.
Table 1.
List of mutations reported in DLX5 and DLX6 genes to date
| Mutation | Codon change | Amino acid change | Nucleotide change | Protein | Reported phenotype | Reference |
|---|---|---|---|---|---|---|
| Nonsense (DLX5) | GAG-TAG | Glu39Term | c.115G>T | p.E39* | SHFM | Sowińska-Seidler et al., 2014 |
| Missense (DLX5) | CAA-CCA | Gln178Pro | c.533A>C | p.Q178P | SHFM |
Shamseldin et al., 2012 Wang et al., 2014 |
| Missense (DLX5) | CAG-CAT | Gln186His | c.558G>T | p.Q186H | SHFM | Wang et al., 2014 |
| Missense (DLX5) | ATC-ATG | Ile192Met | c.576C>G | p.I192M | RHS, SHFM | Wolf et al., 2014 |
| Small insertion (DLX5) | CAG∧161TACCTacctCGC | c.482_485dupACCT | p.Ala163Profs*55 | SHFM | Ullah et al., 2016 | |
| Missense (DLX6) | GTG-GAG | Val211Glu | c.632T>A | p.V211E | SHFM | Present study |
RHS, Rapp-Hodgkin syndrome; SHFM, split-hand/foot malformation.
Table 2.
Chromosomal rearrangements encompassing DLX5 and DLX6 to date
| Mutation | Rearrangements | Reported phenotype | Reference |
|---|---|---|---|
| Gross deletion (DLX5 and DLX6) | ~8.478-Mb deletion including entire DLX5, DLX6, and >50 others genes | SHFM | Vera-Carbonell et al., 2012 |
| Gross deletion (DLX5 and DLX6) | 0.9–1.8-Mb deletion including entire DLX5, DLX6, and DSS1 genes | SHFM with Mondini dysplasia | Wieland et al., 2004 |
| Gross insertion (DLX5 and DLX6) | 719-kb duplication including entire DLX5 and DLX6 genes | SHFM | Velinov et al., 2012 |
The DLX6 gene encodes a homeobox protein DLX-6 and maps next to its paralog DLX5 in chromosome 7q21. As a transcription factor, it functions in osteoblast differentiation, induction of bone formation, and in the development of neural crest cells and branchial arch.
During limb patterning along the 3 axes (proximodistal, anterior-posterior, and dorsoventral), proximal/distal growth and digit extension are controlled by the apical ectodermal ridge by producing proteins of the fibroblast growth factor family [Lu and Werb, 2008; Mariani et al., 2008]. Anterior-posterior patterning is controlled by the zone of polarizing activity that secretes the sonic hedgehog protein. Dorsoventral patterning is performed by the activation of the Lmx1b transcription factor by WNT7A signaling protein, when the activity of EN1 transcription factor restricts the expression of Wnt7a to the dorsal ectoderm [Parr and McMahon, 1995; Vogel et al., 1995; Loomis et al., 1996; Niswander, 2002].
Dlx and Msx homeobox transcription factors are expressed in the apical ectodermal ridge and play an important morphogenetic role in proximal/distal limb development. Single knockout of Dlx5, Dlx6, Msx1, and Msx2 did not show any limb abnormality [Merlo et al., 2002; Satokata et al., 2000]. Double knock out of Dlx5/6 in mice resulted in phenotypes similar to human SHFM1 [Merlo et al., 2002; Robledo et al., 2002]. Double inactivation of Msx1 and Msx2 in mice caused polydactyly in the forelimbs and oligodactyly in the hindlimbs [Lallemand et al., 2005; Bensoussan-Trigano et al., 2011]. Triple knockout mice involving Msx1, Dlx5, and Dlx6 developed phenotypes similar to those of Msx1 and Msx2 double knockout mice, while phenotypes of Msx2, Dlx5, and Dlx6 triple knockout mice were similar to Dlx5/6 double knockout mice. These experiments showed that Dlx5/6 control the expression of Msx2 [Lallemand et al., 2005; Vieux-Rochas et al., 2013]. Loss of DLX5 and DLX6 causes degeneration of the apical ectodermal ridge, resulting in the phenotypes of SHFM1 [Robledo et al., 2002].
In conclusion, we have reported the first intragenic mutation in the DLX6 gene in a family segregating SHFM1 in an autosomal dominant pattern. In silico analysis predicts the heterozygous missense mutation (p.Val211Glu) in DLX6 is most likely the cause for SHFM phenotypes in the present family. However, due to the nonavailability of DNA of the proband's mother, further characterization of the missense mutation to validate its pathogenicity was not possible.
Statement of Ethics
Approval of the study was obtained from the Institutional Review Board (IRB), Quaid-i-Azam University Islamabad, Pakistan. Informed written consent was obtained from all those who participated in the study.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgment
We gratefully acknowledge the family members for participating in the present research study.
References
- Aten E, den Hollander N, Ruivenkamp C, Knijnenburg J, van Bokhoven H, et al. Split hand-foot malformation, tetralogy of Fallot, mental retardation and a 1 Mb 19p deletion – evidence for further heterogeneity? Am J Med Genet A. 2009;149A:975–981. doi: 10.1002/ajmg.a.32748. [DOI] [PubMed] [Google Scholar]
- Aziz A, Khan S, Zimri FK, Muhammad N, et al. Novel homozygous mutations in the WNT10B gene underlying autosomal recessive split hand/foot malformation in three consanguineous families. Gene. 2014;534:265–271. doi: 10.1016/j.gene.2013.10.047. [DOI] [PubMed] [Google Scholar]
- Bensoussan-Trigano V, Lallemand Y, Saint Cloment C, Robert B. Msx1 and Msx2 in limb mesenchyme modulate digit number and identity. Dev Dyn. 2011;240:1190–1202. doi: 10.1002/dvdy.22619. [DOI] [PubMed] [Google Scholar]
- Delgado S, Velinov M. 7q21.3 Deletion involving enhancer sequences within the gene DYNC1I1 presents with intellectual disability and split hand-split foot malformation with decreased penetrance. Mol Cytogenet. 2015;8:37. doi: 10.1186/s13039-015-0139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Basit S, Zimri FK, Ali N, Ali G, et al. A novel homozygous missense mutation in WNT10B in familial split-hand/foot malformation. Clin Genet. 2012;82:48–55. doi: 10.1111/j.1399-0004.2011.01698.x. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Lohan S, Doelken SC, Stricker S, Ockeloen CW, et al. Duplications of BHLHA9 are associated with ectrodactyly and tibia hemimelia inherited in non-Mendelian fashion. J Med Genet. 2012;49:119–125. doi: 10.1136/jmedgenet-2011-100409. [DOI] [PubMed] [Google Scholar]
- Lallemand Y, Nicola MA, Ramos C, Bach A, Cloment CS, Robert B. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005;132:3003–3014. doi: 10.1242/dev.01877. [DOI] [PubMed] [Google Scholar]
- Lango AH, Caswell R, Xie W, Xu X, Wragg C, et al. Next generation sequencing of chromosomal rearrangements in patients with split-hand/split-foot malformation provides evidence for DYNC1I1 exonic enhancers of DLX5/6 expression in humans. J Med Genet. 2014;51:264–267. doi: 10.1136/jmedgenet-2013-102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Lu P, Werb Z. Patterning mechanisms of branched organs. Science. 2008;322:1506–1509. doi: 10.1126/science.1162783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo GR, Paleari L, Mantero S, Genova F, Beverdam A, et al. Mouse model of split hand/foot malformation type I. Genesis. 2002;33:97–101. doi: 10.1002/gene.10098. [DOI] [PubMed] [Google Scholar]
- Niswander L. Interplay between the molecular signals that control vertebrate limb development. Int J Dev Biol. 2002;46:877–881. [PubMed] [Google Scholar]
- Parr BA, McMahon AP. Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature. 1995;374:350–353. doi: 10.1038/374350a0. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16:1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- Shamseldin HE, Faden MA, Alashram W, Alkuraya FS. Identification of a novel DLX5 mutation in a family with autosomal recessive split hand and foot malformation. J Med Genet. 2012;49:16–20. doi: 10.1136/jmedgenet-2011-100556. [DOI] [PubMed] [Google Scholar]
- Simonazzi G, Miccoli S, Salfi N, Bonasoni MP, Bocciardi R, et al. A novel p63 mutation in a fetus with ultrasound detection of split hand/foot malformation. Prenat Diagn. 2012;32:296–298. doi: 10.1002/pd.2932. [DOI] [PubMed] [Google Scholar]
- Sowińska-Seidler A, Badura-Stronka M, Latos-Bieleńska A, Stronka M, Jamsheer A. Heterozygous DLX5 nonsense mutation associated with isolated split hand/foot malformation with reduced penetrance and variable expressivity in two unrelated families. Birth Def Res A Clin Mol Teratol. 2014;100:764–771. doi: 10.1002/bdra.23298. [DOI] [PubMed] [Google Scholar]
- Spielmann M, Kakar N, Tayebi N, Leettola C, Nürnberg G, et al. Exome sequencing and CRISPR/Cas genome editing identify mutations of ZAK as a cause of limb defects in humans and mice. Genome Res. 2016;26:183–191. doi: 10.1101/gr.199430.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur SA, Tolun A. Homozygous WNT10B mutation and complex inheritance in split-hand/foot malformation. Hum Mol Genet. 2008;17:2644–2653. doi: 10.1093/hmg/ddn164. [DOI] [PubMed] [Google Scholar]
- Ullah A, Raza SI, Ali RH, Naveed AK, Jan A, et al. A novel deletion mutation in the DSG4 gene underlies autosomal recessive hypotrichosis with variable phenotype in two unrelated consanguineous families. Clin Exp Dermatol. 2015;40:78–84. doi: 10.1111/ced.12457. [DOI] [PubMed] [Google Scholar]
- Ullah A, Ullah MF, Khalid ZM, Ahmad W. A novel heterozygous frameshift mutation in DLX5 gene underlies isolated split hand foot malformation type 1. Pediatr Int. 2016 doi: 10.1111/ped.13023. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- van Silfhout AT, van den Akker PC, Dijkhuizen T, Verheij JB, Olderode-Berends MJ, et al. Split hand/foot malformation due to chromosome 7q aberrations (SHFM1): additional support for functional haploinsufficiency as the causative mechanism. Eur J Hum Genet. 2009;17:1432–1438. doi: 10.1038/ejhg.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velinov M, Ahmad A, Brown-Kipphut B, Shafiq M, Blau J, et al. A 0.7 Mb De Novo duplication at 7q21.3 including the genes DLX5 and DLX6 in a patient with split-hand/split-foot malformation. Am J Med Genet Part A. 2012;158A:3201–3206. doi: 10.1002/ajmg.a.35644. [DOI] [PubMed] [Google Scholar]
- Vera-Carbonell A, Moya-Quiles MR, Ballesta-Martinez M, López-González V, Bafallíu JA, et al. Rapp-Hodgkin syndrome and SHFM1 patients: delineating the p63-Dlx5/Dlx6 pathway. Gene. 2012;497:292–297. doi: 10.1016/j.gene.2012.01.088. [DOI] [PubMed] [Google Scholar]
- Vieux-Rochas M, Bouhali K, Mantero S, Garaffo G, Provero P, et al. BMP-mediated functional cooperation between Dlx5;Dlx6 and Msx1;Msx2 during mammalian limb development. PLoS One. 2013;8:e51700. doi: 10.1371/journal.pone.0051700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Rodriguez C, Warnken W, Izpisúa Belmonte JC. Dorsal cell fate specified by chick Lmx1 during vertebrate limb development. Nature. 1995;378:716–720. doi: 10.1038/378716a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Xin Q, Li L, Li J, Zhang C, et al. Exome sequencing reveals a heterozygous DLX5 mutation in a Chinese family with autosomal dominant split-hand/foot malformation. Eur J Hum Genet. 2014;22:1105–1110. doi: 10.1038/ejhg.2014.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland I, Muschke P, Jakubiczka S, Volleth M, Freigang B, Wieacker PF. Refinement of the deletion in 7q21.3 associated with split hand/foot malformation type 1 and Mondini dysplasia. J Med Genet. 2004;41:e54. doi: 10.1136/jmg.2003.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ZT, Leslie EJ, Arzi B, Jayashankar K1, Karmi N, et al. A LINE-1 insertion in DLX6 is responsible for cleft palate and mandibular abnormalities in a canine model of Pierre Robin sequence. PLoS Genet. 2014;10:e1004257. doi: 10.1371/journal.pgen.1004257. [DOI] [PMC free article] [PubMed] [Google Scholar]