Abstract
It was previously shown that the second open reading frame of the avian reovirus S1 gene encodes a 146-amino-acid nonstructural protein, designated p17, which has no known function and no sequence similarity to other known proteins. The results presented in this report demonstrate that p17 accumulates in the nucleoplasm of infected and transfected cells. An examination of the deduced amino acid sequence of p17 revealed the presence of a putative monopartite nuclear localization signal (NLS) between residues 119 and 128. Mutagenesis analysis revealed both that this sequence is indeed a functional NLS and that two of its basic residues are critical for the normal nuclear distribution of p17. An interspecies heterokaryon assay further showed that p17 shuttles continuously between the nucleus and the cytoplasm and that this activity is restricted to its NLS-containing C-terminal tail. Finally, an analysis of the intracellular distribution of p17 in the presence of inhibitors of both RNA polymerase II and CRM1 further revealed that the nucleocytoplasmic distribution of p17 is coupled to transcriptional activity and that the viral protein exits the nucleus via a CRM1-independent pathway.
Avian reoviruses and mammalian reoviruses are members of the Orthoreovirus genus, 1 of the 11 genera of the Reoviridae family (22, 37). These agents, which replicate in the cytoplasm of infected cells, lack a lipid envelope and contain a fragmented double-stranded RNA genome enclosed within a double protein capsid shell with a 70- to 80-nm external diameter. Their genome segments have been grouped into three classes according to size, namely, large (L1, L2, and L3), medium (M1, M2, and M3), and small (S1, S2, S3, and S4). Each segment expresses an mRNA that is identical to the positive strand of its encoding double-stranded RNA; reoviral mRNAs lack a 3′ poly(A) tail and contain a 5′ type 1 cap (20, 21).
Most reoviral mRNAs are monocistronic, with each encoding a single primary translation product. However, there are some exceptions to this rule. For example, the S1 genome segment of most mammalian reoviruses is bicistronic and encodes one structural and one nonstructural polypeptide from overlapping open reading frames (ORFs) (10, 46). The S1 genes of the fusogenic avian reoviruses and Nelson Bay mammalian reovirus express one structural and two nonstructural proteins from partially overlapping ORFs (2, 49). The S4 segment of the fusogenic baboon reovirus specifies two nonstructural proteins from partially overlapping ORFs (6). The bicistronic S1 gene of the fusogenic reptilian reovirus encodes a structural and a nonstructural protein (8). Finally, the M3 genes of avian and mammalian reoviruses express two isoforms of the nonstructural protein μNS, apparently by initiation of translation at two different AUG codons (31, 55).
We have recently demonstrated that the three S1 ORFs of various avian reovirus strains are expressed in infected cells (2). The first ORF specifies p10, a nonstructural fusion-associated small transmembrane protein that displays membrane destabilization activity (3, 48). The second ORF encodes p17, a nonstructural protein of unknown function. Finally, the third ORF expresses σC, an elongated trimeric structural protein responsible for the initial attachment of the virus to cell receptors (30, 47).
When we initiated this study, nothing was known about the activity or properties of the avian reovirus nonstructural p17 protein. Furthermore, this polypeptide has no significant sequence similarity to other known proteins, so its amino acid sequence offers no clues about its function. On the other hand, the fact that the p17 ORF is conserved in every avian reovirus S1 gene sequence reported so far suggests that p17 plays an important function in virus-host interactions. All of these facts and also the broadly accepted assumption that nonstructural viral proteins play key roles in virus-host interactions prompted us to perform an initial characterization of this viral protein. The results of this study demonstrate that p17 is a nuclear targeting protein that shuttles between the nucleus and the cytoplasm in a transcription-dependent manner and that exits the nucleus via a CRM1-independent pathway. We have also identified a monopartite-type functional nuclear localization signal (NLS) near the C terminus of p17 which is necessary and sufficient for nuclear import.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Primary cultures of chicken embryo fibroblasts (CEF) were prepared from 9- to 10-day-old chicken embryos and grown in medium 199 supplemented with 10% tryptose-phosphate broth and 5% calf serum. Monkey Vero and human HeLa cells were grown in monolayers in medium 199 supplemented with 10% fetal bovine serum. Strain S1133 of avian reovirus was grown on semiconfluent monolayers of primary CEF as previously described (16).
A polyclonal antiserum against the N terminus of p17 was raised by Sigma Genosys Ltd. (Cambridge, United Kingdom). Specifically, the peptide CDFEREYKSWRFDADL, comprising residues 132 to 146 of the deduced amino acid sequence of p17 and containing an extra cysteine residue at its N terminus for coupling, was chemically synthesized, cross-linked to carrier keyhole limpet hemocyanin, and then used as an immunogen for antibody production in rabbits. The production of an anti-σNS monoclonal antibody was described previously (55). The following antibodies were purchased from Sigma-Aldrich (Madrid, Spain): an anti-tubulin mouse monoclonal antibody, an anti-nucleolin rabbit polyclonal antibody, and alkaline phosphatase-, fluorescein isothiocyanate (FITC)-, and tetramethyl rhodamine isocyanate (TRITC)-conjugated goat antibodies against mouse and rabbit immunoglobulin G (IgG). An anti-fibrillarin monoclonal antibody was purchased from Cytoskeleton, Inc. (Denver, Colo.).
Cloning and transfections.
All manipulations of DNA were performed according to standard cloning protocols (45). Cloning of the S1 ORF2 into the eukaryotic pCINeo expression vector to generate pCINeo-p17 was described previously (2). For the expression of fusions with green fluorescent protein (GFP), ORF2 sequences were amplified by PCR, digested with appropriate restriction enzymes, and inserted into the pGFP-C1 vector (Clontech). To generate inserts for the production of pGFP-C1 recombinant plasmids, we used the following primers: for the production of GFP-p17, the sense primer was 5′-GGAATTCTATGCAATGGCTCCGCCATACG-3′ (EcoRI site is underlined) and the antisense primer was 5′-CGGGATCCTCATAGATCGGCGTCAAATCGC-3′ (BamHI site is underlined); for the production of GFP-p17(1-103), the sense primer was 5′-GGAATTCTATGCAATGGCTCCGCCATACG-3′ (EcoRI) and the antisense primer was 5′-CGGGATCCTCAATCGGATGCAGGCAATGG-3′ (BamHI); and for the production of GFP-p17(104-146), the sense primer was 5′-CGAATTCTCGGCGGTCTTGTCTTATAGTTC-3′ (EcoRI) and the antisense primer was 5′-CGGGATCCTCATAGATCGGCGTCAAATCGC-3′ (BamHI). The correct orientation of the inserts was confirmed by restriction analysis and nucleotide sequencing.
The pCINeo-p17 plasmid and a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) were used according to the manufacturer's specifications to generate recombinant plasmids that expressed K122A and/or R123A mutant versions of p17. The following mutagenic oligonucleotide primers were used: for the production of pCINeo-p17(K122A,R123A), the sense primer was 5′-CAATCCATCGCAGCGGCGGCAGGTCGTCAGCTTGATAC-3′ and the antisense primer was 5′-GTATCAAGCTGACGACCTGCCGCCGCTGCGATGGATTG-3′; for the production of pCINeo-p17(K122A), the sense primer was 5′-CAATCCATCGCAGCGGCGAGAGGTCGTCAGCTTG-3′ and the antisense primer was 5′-CAAGCTGACGACCTCTCGCCGCTGCGATGGATTG-3′; and for the production of pCINeo-p17(R123A), the sense primer was 5′-CAATCCATCGCAGCGAAGGCAGGTCGTCAGCTTG-3′ and the antisense primer was 5′-CAAGCTGACGACCTGCCTTCGCTGCGATGGATTG-3′.
Transfections of preconfluent cell monolayers were done by use of the Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells were incubated at 37°C for 24 h, unless otherwise stated.
Viral infections, metabolic radiolabeling, and electrophoretic analysis of proteins.
The infection of CEF by avian reovirus S1133 and metabolic radiolabeling have been described previously (29). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed in 15% polyacrylamide gels as described previously (26). After electrophoresis, the gels were stained in a solution containing 0.25% Coomassie blue in 33% methanol and 10% acetic acid and then were destained in the same solution without the dye. For visualization of the radioactive bands, the gels were dried and exposed to X-ray films (Agfa-Curix AFW).
Subcellular fractionation and immunoblot analysis.
The fractionation of avian reovirus-infected cells into cytosolic and nuclear fractions was performed essentially as described previously (39). Briefly, a cell monolayer (30 × 106 cells) was washed with PBS, and the cells were subsequently collected in 0.5 ml of TBN buffer (10 mM Tris-HCl [pH 6.5], 140 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 100 μM tosylsulfonyl phenylalanyl chloromethyl ketone, and 50 μg of aprotinin/ml) and incubated for 10 min at 4°C. The resulting extract was centrifuged (5,000 × g for 5 min at 4°C), and the supernatant was collected and stored as the cytosolic fraction. The pellet was washed twice with TBN buffer, resuspended in 0.4 ml of a buffer containing 62.5 mM Tris-HCl (pH 6.8), 40% glycerol, 4% sodium dodecyl sulfate, and 3% dithiothreitol, and incubated for 5 min at 4°C. The sample was then passed 20 times through a 25-gauge needle and centrifuged at 12,000 × g for 5 min at 4°C, and the supernatant was collected as the nuclear fraction. Western blotting analysis was done as described previously (2).
Heterokaryon formation and immunofluorescence microscopy.
Heterokaryons of transfected monkey Vero cells and untransfected mouse L929 cells were formed as previously described (11). For indirect immunofluorescence microscopy, cell monolayers grown on coverslips were infected or transfected as indicated in the figure legends. At the indicated times, the monolayers were washed twice with PBS and fixed either in methanol at −20°C for 30 min or in 4% paraformaldehyde at 37°C for 10 min. Paraformaldehyde-fixed cells were washed twice with PBS, incubated for 10 min in permeabilizing buffer (0.5% Triton X-100 in PBS), and then incubated for 1 h at room temperature with primary antibodies diluted in blocking buffer (2% bovine serum albumin in PBS). The cells were washed three more times with PBS and then incubated with secondary antibodies and with either 1 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml, 10 μg of propidium iodide/ml, or 0.05 μg of Hoechst dye/ml for 1 h at room temperature. The coverslips were then washed six times with PBS and mounted on glass slides. Images were obtained by sequential scanning with a Leica TCS SP2 confocal microscope using a 100× 1.3 oil immersion objective. FITC was detected by using the 488-nm excitation line and analyzing the emission between 500 and 535 nm. TRITC was detected by using the 543-nm excitation line and analyzing the emission between 575 and 650 nm. Images were processed with Adobe Photoshop (Adobe Systems).
RESULTS
Avian reovirus nonstructural protein p17 localizes to the nucleoplasm of infected and transfected cells.
To study the subcellular localization of p17, we first generated a polyclonal antiserum against a hydrophilic peptide comprising the 15 most C-terminal amino acid residues of the viral protein. This antiserum recognized p17 in extracts of avian reovirus-infected cells, but not in extracts of mock-infected cells, by both immunoblotting and immunoprecipitation (data not shown).
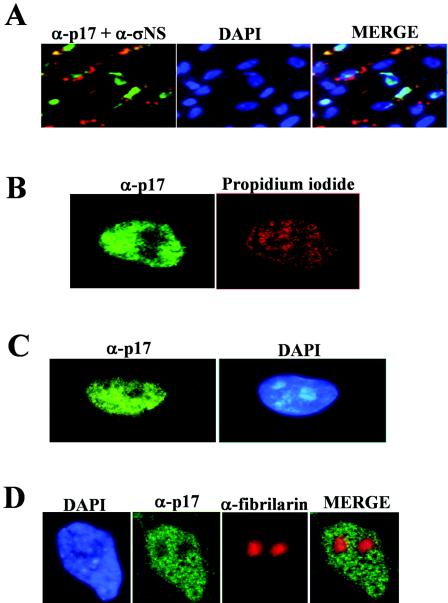
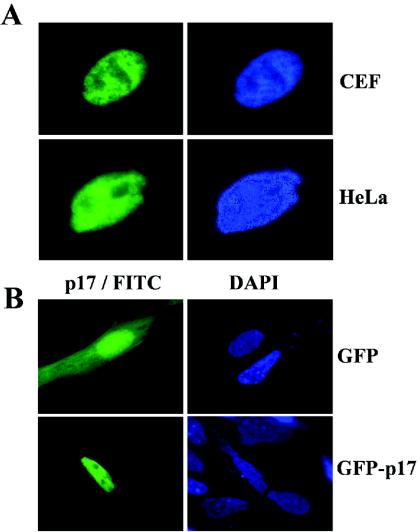
The anti-p17 antiserum was next used to evaluate the subcellular distribution of p17 in avian reovirus-infected cells by indirect immunofluorescence. To this end, avian reovirus S1133-infected cells were stained at 9 h postinfection (hpi) with DAPI and then immunostained with antibodies against both p17 and the avian reovirus nonstructural protein σNS. An examination of the triply stained cells by fluorescence microscopy (Fig. 1A) revealed that while σNS was found in cytoplasmic globular phase-dense perinuclear inclusions, as previously described (55), p17 was concentrated within the nucleus. The nuclear accumulation of the nonstructural protein was confirmed by subcellular fractionation (see Fig. 7A) as well as by confocal microscopy using optical sectioning parameters that eliminated the out-of-focus cytosolic fluorescence above or below the nucleus (Fig. 1B). On the other hand, p17 nuclear targeting was not dependent on the virus strain or the host cell type, since this protein also exhibited a robust nuclear signal in CEF infected with different avian reovirus strains (data not shown) and in a monkey cell line infected with avian reovirus S1133 (Fig. 1C).
FIG. 1.
Intracellular distribution of σNS and p17 in infected cells. Mock-infected or avian reovirus-infected cells (10 PFU/cell) were incubated for 9 h. (A) Infected CEF were fixed with methanol and incubated with DAPI, anti-p17 serum, and a monoclonal antibody against the avian reovirus nonstructural protein σNS. The cells were subsequently immunostained with FITC-conjugated goat anti-rabbit serum and a TRITC-conjugated anti-mouse monoclonal antibody. The p17 protein is shown in green, and σNS is shown in red. Stained cells were visualized by fluorescence microscopy. (B) Infected CEF were stained with propidium iodide and immunostained for p17. The cells were subsequently visualized by confocal microscopy. (C) Infected monkey Vero cells were stained with DAPI and immunostained for p17. Stained cells were visualized by fluorescence microscopy. (D) Infected CEF were stained with DAPI and immunostained for both p17 and fibrillarin. Stained cells were visualized by fluorescence microscopy. The p17 protein is shown in green, and fibrillarin is shown in red.
FIG. 7.
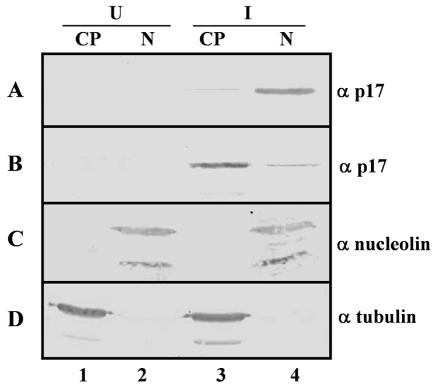
Subcellular fractionation. At 9 hpi, mock-infected (U) (lanes 1 and 2) and infected (I) (lanes 3 and 4) CEF cells were mock treated (A, C, and D) or treated for 5 h at 37°C with 20 μg of cycloheximide/ml and 100 μM DRB (B). The cells were then lysed, and the lysates were fractionated into cytoplasmic (lanes 1 and 3) and nuclear (lanes 2 and 4) fractions as described in Materials and Methods. The fractions were analyzed by Western blotting with specific antibodies against p17, nucleolin, and tubulin, as indicated on the right.
Visualization of the nuclei of infected cells by confocal microscopy suggested that p17 was not uniformly distributed within the nucleus and that its signal was absent from nuclear structures resembling nucleoli (Fig. 1B). To assess whether p17 was indeed absent from the nucleolus, we stained avian reovirus-infected cells with DAPI and with antibodies against both p17 and the nucleolar protein fibrillarin. The results revealed that p17 was present in the nucleoplasm but was excluded from fibrillarin-stained nucleoli, as clearly shown in the superimposed image (Fig. 1D). Similar results were obtained when nucleoli were stained with anti-nucleolin antibodies (data not shown).
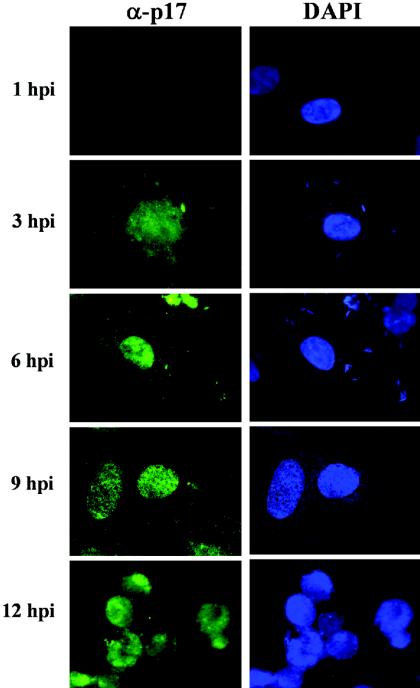
Examinations of p17-immunostained cells at different infection times showed that p17 was first detectable at 3 hpi in both the nucleus and the perinuclear region (Fig. 2); as infection progressed, p17-associated staining was restricted to the nucleus, and at 12 hpi, the protein was observed inside the nuclei of the typical multinucleated cells that are formed at late infection times following infection with fusogenic avian reoviruses.
FIG. 2.
Intracellular distribution of p17 during infection. At the infection times indicated on the left, avian reovirus-infected cells (20 PFU of S1133/cell) were fixed, stained with DAPI, and immunostained for p17. Stained cells were visualized by fluorescence microscopy.
To determine whether the intranuclear location of p17 is dependent on viral factors and/or viral infection, we next examined its intracellular distribution in transfected cells. To this end, the second open reading frame of the avian reovirus S1 gene was cloned into the mammalian expression vector pCINeo, and the recombinant pCINeo-ORF2 plasmid was transfected into avian CEF and human HeLa cells by the use of Lipofectamine. Cells were subsequently stained with DAPI and immunostained for p17. Visualization of the cells by fluorescence microscopy (Fig. 3A) revealed that p17 accumulated in the nuclei of the transfected cells. Furthermore, when p17 was fused to the C terminus of GFP, the resulting chimera, GFP-p17, also accumulated in the nuclei of the transfected cells, while GFP alone was located in both the nuclear and cytoplasmic compartments (Fig. 3B). This result demonstrated that p17 is able to target appended GFP to the nucleus, indicating that GFP fusions may be used for further studies.
FIG. 3.
Distribution of p17 within transfected cells. (A) Semiconfluent monolayers of CEF and HeLa cells were transfected with the pCINeo-p17 plasmid by the use of Lipofectamine, and 24 h later the cells were fixed, stained with DAPI, and immunostained for p17. Stained cells were visualized by fluorescence microscopy. (B) Fluorescence microscopy analysis of CEF that were transfected with the pGFP (top row) or pGFP-p17 (bottom row) plasmid by the use of Lipofectamine.
Identification of a functional NLS in p17.
Most nuclear proteins enter the nucleus through specific interactions between the nuclear import machinery and specific protein sequences known as nuclear localization signals (NLSs). The following two types of canonical NLSs have been described: (i) monopartite NLSs, which contain one continuous array of basic amino acid residues; and (ii) bipartite NLSs, which possess two basic regions separated by an unconserved linker region of approximately 10 to 12 amino acid residues (reviewed in reference 14).
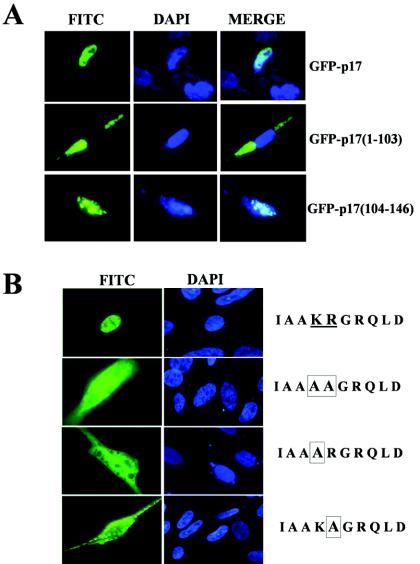
A close examination of the deduced amino acid sequence of p17 revealed the presence of a basic region, 119IAAKRGRQLD128 (underlined in Fig. 4A), which shares a high degree of similarity with the functional monopartite NLS of the c-Myc protein (5) and which is highly conserved among the p17 proteins from different avian reovirus isolates and from the fusogenic mammalian Nelson Bay reovirus (Fig. 4B). As a first approach to assess whether this basic region represents a functional NLS, we evaluated the nuclear targeting capacity of two p17 fragments comprising the first 103 or the last 43 residues of the viral protein. For this experiment, constructs producing GFP fused to the N terminus of p17 or to the p17 regions spanning residues 1 to 103 and 104 to 146 were transfected into CEF, and 24 h after transfection, the subcellular localization of the fused proteins was monitored by fluorescence microscopy. The results shown in Fig. 5A revealed that while GFP-p17 and GFP-p17(104-146) were efficiently imported into the nuclei of the transfected cells, GFP-p17(1-103), which lacks the putative NLS sequence, was excluded from the nucleus and distributed evenly in the cytoplasmic compartment. These results demonstrate both that a functional NLS is present in the p17 region spanning residues 104 to 146 and that residues 1 to 103 are dispensable for the normal nuclear localization of p17.
FIG. 4.
Amino acid sequences. (A) Deduced primary sequence of avian reovirus p17 nonstructural protein. The basic putative monopartite NLS is underlined. (B) Alignment of putative NLSs of several avian reovirus strains and the mammalian Nelson Bay reovirus and of the reported functional NLS of the human c-Myc protein.
FIG. 5.
Intracellular distribution of deleted and mutated p17 versions. (A) Fluorescence microscopy analysis of the GFP fusion proteins indicated on the right. (B) Immunofluorescent localization of wild-type p17 (top row) and of three mutated versions in which residues K-122 and R-123 (underlined) were mutated to alanines (boxed), either together (second row) or individually (bottom two rows).
To assess whether the region 119IAAKRGRQLD128 has NLS activity as well as to identify specific residues that may be essential for nuclear targeting, we generated three recombinant plasmids that expressed p17 versions in which the two conserved basic residues K-122 and R-123 were mutated to alanines, either together or individually. The intracellular distribution of the transiently expressed p17 versions in CEF was subsequently analyzed by fluorescence microscopy 24 h after transfection. As shown in Fig. 5B, in contrast to the strong nuclear signal of wild-type p17, all three mutants were distributed evenly over the cell, indicating that K-122 and R-123 are necessary for the normal nuclear localization of p17, which in turn suggests that these two basic residues belong to a functional NLS motif that mediates p17 nuclear import.
Inhibition of transcription redistributes p17 to the cytoplasm.
An inhibition of RNA polymerase II activity causes a redistribution of some nuclear proteins to the cytoplasm, suggesting that active transcription is required for the normal nuclear localization of these proteins (56). To assess whether this is also true for p17, we analyzed the intracellular distribution of p17 in the presence and absence of the RNA polymerase inhibitors actinomycin D (AMD) and 5,6-dichlorobenzimidazole riboside (DRB). To this end, avian reovirus-infected CEF and pCINeo-p17-transfected Vero cells were treated with or without 5 μg of AMD/ml or 100 μM DRB for 5 h in the presence of 20 μg of cycloheximide/ml, and the intracellular distribution of p17 was subsequently assessed by immunofluorescence microscopy. Visualization of the cells by indirect immunofluorescence revealed a shift in the distribution of p17 from the nucleus to the cytoplasm when infected and transfected cells were incubated in the presence of cycloheximide plus either DRB (Fig. 6A and B, compare panels 1 and 2) or AMD (see Fig. 9), but not when cells were incubated with cycloheximide alone (Fig. 6A and B, panels 1). Cytoplasmic accumulation in the presence of RNA polymerase inhibitors occurred in ∼75% of the antigen-positive cells. In contrast, p17 remained within the nucleus when the cells were incubated with AMD (data not shown) or DRB (Fig. 6A and B, panels 3) at 4°C, suggesting that p17 is exported from the nucleus through an active, energy-dependent process. Furthermore, the DRB-induced cytoplasmic accumulation of p17 could be reversed in ∼80% of the antigen-positive cells by subsequent incubation of the cells in DRB-free medium for 5 h at 37°C (Fig. 6A and B, panels 4), but not at 4°C (results not shown), indicating both that the normal nuclear localization of p17 can be restored by the activation of transcription and that p17 is imported into the nucleus through an energy-mediated process. A DRB-induced redistribution of p17 from the nucleus to the cytoplasm in avian reovirus-infected cells was further demonstrated by subcellular fractionation and immunoblot analysis (Fig. 7A and B).
FIG. 6.
Effect of RNA polymerase II inhibitors. At 9 hpi of CEF with avian reovirus (A) and 24 h after the transfection of Vero cells with the pCINeo plasmid (B), the cells were incubated as follows: (1), 5 h at 37°C with 20 μg of cycloheximide/ml; (2), 5 h at 37°C with 20 μg of cycloheximide/ml and 100 μM DRB; (3), 5 h at 4°C with 20 μg of cycloheximide/ml and 100 μM DRB; and (4), 5 h at 37°C with 20 μg of cycloheximide/ml and 100 μM DRB, followed by another 5-h incubation at 37°C with 20 μg of cycloheximide/ml. At the end of the incubation period, the cells were fixed, stained with DAPI, and immunostained for p17 (FITC). The stained cells were visualized by fluorescence microscopy.
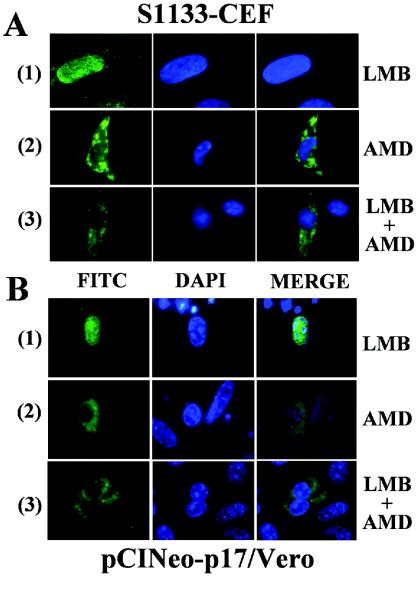
FIG. 9.
Effect of leptomycin B. At 9 hpi of CEF with avian reovirus (A) and 24 h after the transfection of Vero cells with the pCINeo-p17 plasmid (B), the cells were incubated for 5 h at 37°C in the presence of 20 μg of cycloheximide/ml plus the following: (1), 30 ng of leptomycin B (LMB)/ml; (2), 5 μg of actinomycin D (AMD)/ml; or (3), both inhibitors. The cells were then stained with DAPI and immunostained for p17 (FITC). The stained cells were subsequently visualized by fluorescence microscopy.
p17 is a nucleocytoplasmic shuttling protein.
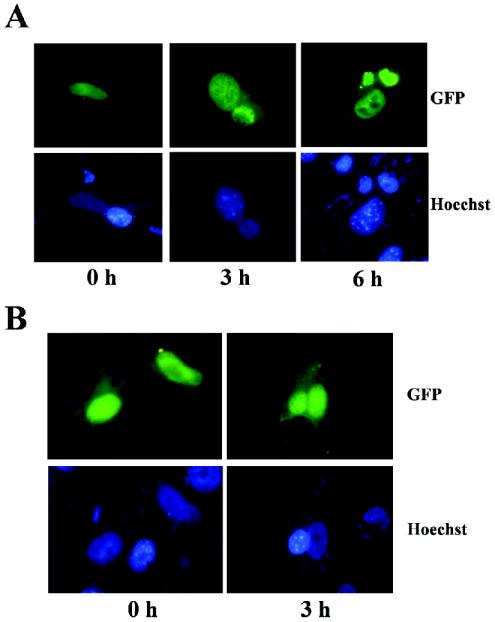
There are proteins that are always confined to the nucleus, while others shuttle continuously between the nucleus and the cytoplasm (38). Our finding that p17 redistributes to the cytoplasmic compartment upon inhibition of transcription raised the possibility that p17 shuttles between the nucleus and the cytoplasm. To test this hypothesis, we performed a very sensitive in vivo interspecies heterokaryon assay that has been previously used to characterize the shuttling properties of various proteins (11). Monkey Vero cells were transfected with the recombinant plasmid pGFP-p17, and 24 h later these cells were fused with untransfected mouse L cells in the presence of 50% polyethylene glycol. The fused cells were then incubated for 3 h in the presence of 100 μg of cycloheximide/ml. The appearance of p17 in mouse L nuclei of interspecies heterokaryons indicated that p17 was exported out of the transfected monkey cells into untransfected mouse nuclei. Monkey and mouse nuclei in heterokaryons can be distinguished by Hoechst staining, since monkey nuclei stain relatively diffusely, whereas mouse nuclei exhibit a characteristic punctate pattern due to the staining of intranuclear bodies (11). As shown in Fig. 8A, GFP-associated fluorescence was detected in both monkey and mouse nuclei of heterokaryons, indicating that p17 was able to travel from donor-transfected nuclei into recipient mouse nuclei. However, a similar assay performed with pcDNA-σA-transfected Vero cells failed to reveal translocation of the avian reovirus nuclear protein σA into the mouse nucleus (results not shown), suggesting both that σA is not a nucleocytoplasmic shuttling protein and that our heterokaryon assay is able to discriminate between shuttling and nonshuttling proteins. The nucleocytoplasmic shuttling of p17 was also evident in an avian-murine heterokaryon assay performed with pGFP-p17-transfected avian CEF (data not shown). Taken together, our findings demonstrate that p17 is a nucleocytoplasmic shuttling protein.
FIG. 8.
Heterokaryon assays. Semiconfluent monkey Vero cell monolayers were transfected with the pGFP-p17 (A) or pGFP-p17(104-146) (B) plasmid by the use of Lipofectamine, and 24 h later the cells were scraped off the plate. Aliquots of 5 × 105 transfected cells were then fused with 5 × 105 untransfected mouse L cells in the presence of 50% polyethylene glycol 4000 and 100 μg of cycloheximide/ml. At the indicated postfusion times, the cells were fixed and stained with Hoechst 33258.
To assess whether the shuttling property of p17 is conserved in its NLS-containing C-terminal fragment, we performed a monkey-mouse heterokaryon assay with GFP-p17(104-146)-transfected Vero cells (Fig. 8B). The data demonstrated that p17(104-146) is also able to translocate from nuclei of donor monkey cells into nuclei of recipient mouse cells, indicating that the shuttling properties of p17 are retained in the region spanning amino acid residues 104 to 146.
p17 exits the nucleus through a CRM1-independent pathway.
Many shuttling proteins are exported from the nucleus to the cytoplasm via a pathway that involves the chromosome region maintenance 1 (CRM1) protein. CRM1 is a cellular transport receptor that specifically binds nuclear export signals (NESs) of such proteins for their transfer through the nuclear pore complex. This receptor mediates the nuclear export of numerous cellular proteins with leucine-rich NESs (13, 40, 53). The antifungal cytotoxin leptomycin B (LMB) is a highly specific and potent inhibitor of CRM1 function through a covalent modification blocking the CRM1-dependent nuclear export pathway (25). To investigate the role of CRM1 in p17 nuclear export, we assessed whether LMB can inhibit the AMD-induced cytoplasmic accumulation of p17 in both transfected cells and avian reovirus-infected cells. CEF cells that had been infected with 20 PFU of avian reovirus S1133/cell for 9 h (Fig. 9A) or transfected with pCIneo-p17 for 24 h (Fig. 9B) were incubated for 5 h in the presence of 20 μg of cycloheximide/ml with either 30 ng of LMB/ml (panels 1), 5 μg of AMD/ml (panels 2), or 30 ng of LMB/ml plus 5 μg of AMD/ml (panels 3). The cells were then fixed, stained with both DAPI and anti-p17 serum, and visualized by immunofluorescence microscopy. The results revealed that LMB did not alter the normal nuclear localization of p17 in AMD-untreated cells (Fig. 9A and B, panels 1) or the cytoplasmic accumulation of the viral protein in AMD-treated cells (Fig. 9A and B, compare panels 2 and 3). These results strongly suggest that p17 does not exit the nucleus through a CRM1-dependent pathway.
DISCUSSION
The second cistron of the avian reovirus S1 gene expresses a nuclear protein.
The results presented in the first part of this study demonstrate that the protein encoded by the second open reading frame of the avian reovirus S1 gene accumulates in the nucleoplasm of the infected host cell, with nucleolar exclusion. Furthermore, our finding that p17 also targets the nuclei of transfected cells indicates that it does not require other viral factors or viral infection for nuclear import, thus suggesting that the viral protein itself has the ability to translocate across the nuclear pore complex and to accumulate in the nucleus.
Eukaryotic cell nuclei possess an exclusionary double membrane containing nuclear pores that regulates the transport of macromolecules into and out of the nucleus during interphase. Transport proceeds through the nuclear pore complex (NPC), a macromolecular assembly of nucleoporins which allows the passive diffusion of globular proteins of up to 40 to 60 kDa. Molecules and particles that exceed the diffusion limit of NPCs are actively imported and exported via specific pathways that are mainly governed by a class of soluble protein receptors of the karyopherin-β family. Regarding proteins that are smaller than the diffusion limit of NPCs, some are able to diffuse into the nucleus passively, whereas others are dependent on signal-mediated import (reviewed in reference 15). Because p17 has a mass of ∼17 kDa, it was formally possible that it might enter the nucleus by passive diffusion. However, the data presented in this study argue against this and suggest that p17 is actively imported into the nucleus, as (i) p17 and the C-terminal fragment p17(104-146) are both able to transport the GFP protein, which lacks an NLS signal, into the nucleus, but this property is not displayed by the p17(1-103) N-terminal fragment; (ii) mutation of either K-122 or R-123 to alanine completely eliminates p17 accumulation in the nucleus; and (iii) the viral protein redistributes to the cytoplasm upon inhibition of transcription, but after release from inhibition, the return of the viral protein to the nucleus is temperature dependent. Collectively, our data suggest that p17 is imported into the nucleus through the NPC by an energy- and signal-dependent mechanism.
Our finding that avian reovirus, which replicates exclusively in the cytoplasm of the infected host cell, expresses a nuclear protein was at first surprising. However, this is not a unique situation, since several other cytoplasmic viruses have also been shown to express nuclear proteins. As with avian reovirus p17, most of these viral proteins are sufficiently small to cross the NPC by passive diffusion, yet they are imported into the nucleus actively and contain NLSs that resemble cellular motifs (see reference 17 and references therein). The mammalian reovirus S1 gene also expresses a 14-kDa nonstructural protein, designated σ1s, which, like avian reovirus p17, accumulates in the nuclei of transfected and infected cells (1, 4, 43). However, a comparative analysis of the deduced amino acid sequences of p17 and σ1s revealed no significant similarities in their primary sequences and hydropathy profiles and showed an absence of conserved functional motifs (49). This and the fact that, unlike p17, σ1s was detected in the nucleoli of transfected cells (1), suggest that these two proteins are unrelated in evolutionary and functional terms.
A functional monopartite NLS is present in the C-terminal tail of p17.
Deletion and mutagenesis analyses allowed us to identify a functional NLS near the C terminus of p17. The deletion of the 43 most C-terminal residues completely eliminated the ability of p17 to transport appended GFP into the nucleus, suggesting that the first 103 amino acid residues of p17 do not have nuclear import activity. This observation and our finding that the p17 fragment spanning residues 104 to 146 redirects GFP to the nucleus suggest that the p17 nuclear import signal activity resides in its 43-residue C-terminal tail. An examination of the deduced amino acid sequence of this fragment revealed the presence of the sequence 119IAAKRGRQLD128, which is a good candidate for a monopartite-type NLS and is highly conserved among the p17 proteins of all known avian reovirus strains and of the fusogenic mammalian Nelson Bay reovirus. Our finding that the replacement of K-122 and/or R-123 with alanines causes an even distribution of p17 throughout the cell suggests that these residues form part of a functional NLS motif that is sufficient for the translocation of p17 into the nucleus. This motif is reminiscent of the previously reported functional monopartite NLSs of c-Myc proteins from different species (Fig. 5B) (5), of the C-terminal nuclear targeting signal of the polyomavirus large T antigen (PPKKARED) (42), and of the solitary NLS present in the simian virus 40 large T antigen (PKKKRKV) (24).
The p17 protein contains a CRM1-independent shuttling sequence.
By performing interspecies heterokaryon assays and by using specific inhibitors of transcription, we have demonstrated that p17 is a nucleocytoplasmic shuttling protein. The capacity of GFP-p17(104-146) to translocate into the nuclei of rodent cells in the heterokaryon assay demonstrated that the shuttling ability of p17 is maintained on its C-terminal 43-residue fragment. In spite of being able to shuttle back and forth between the nucleus and the cytoplasm, p17 accumulates in the nuclei of actively transcribing cells, suggesting that nuclear import dominates over nuclear export at a steady state. On the other hand, our observation that leptomycin B does not prevent the cytoplasmic accumulation of p17 in transcriptionally arrested cells or the transfer of p17 to the nuclei of rodent cells in heterokaryon assays (data not shown) suggests that the route of p17 nuclear export is unrelated to the classical CRM1-mediated pathway. In support of this suggestion, we were unable to find a canonical leucine-rich NES motif within the C-terminal fragment of p17 that might target the CRM1 export pathway in the presence of Ran-GTP (40). CRM1-independent nuclear export has also been reported for several viral proteins, including human adenovirus E4orf6 (7), the human cytomegalovirus transactivating protein pUL69 (28), and herpesvirus 1 ICP27 (52), and for cellular proteins such as importin α (18), the cytoplasmic signaling mediators Smad2 and Smad3 (19), the translation initiation factor eIF-5A (27), and several RNA-binding proteins (35).
The intracellular distribution of p17 is transcription dependent.
An important observation emerging from the results of this study is the dependence of the intracellular distribution of p17 on the transcriptional state of the cell. Whereas in actively transcribing cells p17 accumulates within the nucleus, an inhibition of transcription induces a cytoplasmic redistribution of p17. Our finding that cycloheximide alone does not perturb the nuclear accumulation of p17 suggests both that ongoing protein synthesis is not required for the redistribution effect induced by transcriptional inhibitors and that the effect caused by transcriptional inhibitors is a direct result of the transcriptional block, not an indirect result of an inhibition of protein synthesis due to the transcriptional block. Therefore, the avian reovirus nonstructural p17 protein should be added to the small but growing list of proteins that exhibit dynamic transcription-dependent trafficking between the nucleus and the cytoplasm. The regulated localization of shuttling proteins is achieved in many cases by reversible phosphorylation (23), but this does not appear to be the mechanism that controls the intracellular distribution of p17, since we were unable to detect phosphorylation in either nuclear or cytoplasmic p17 (data not shown).
The nucleocytoplasmic shuttling properties of p17 are similar to those reported for several proteins that participate in a wide variety of nuclear processes as well as in mRNA export and stability. For example, hnRNP A1 has a 38-residue M9 shuttling domain which is glycine rich and has only a single essential basic residue (32). The hnRNP K protein contains a 39-residue nuclear shuttling signal which induces both nuclear import and export and probably contacts the NPC directly (33). Finally, the HuR protein possesses a 32-residue shuttling sequence designated HNS that does not appear to correspond to any previously described class of shuttling motifs (11). These shuttling sequences differ from the classical unidirectional NLS and NES signals in that only the former are coupled with bidirectional nuclear import and export activities. Like p17, these proteins accumulate predominantly in the nuclei of actively transcribing cells, their nuclear export does not rely on CRM1, and their shuttling is controlled by the activity of RNA polymerase II. Furthermore, the export receptors for these proteins are unknown, and their import- and export-inducing signals could not be separated by extensive mutagenesis. Accordingly, it is possible that the mechanism which governs the nucleocytoplasmic transport of p17 is related to those described for the above-mentioned shuttling proteins and is fundamentally different from those that govern the import and export of classical NLS- and NES-containing proteins.
Implications of nucleocytoplasmic shuttling on p17 activity.
In contrast to the avian reovirus nonstructural protein p17, most of the reported nucleocytoplasmic shuttling proteins encoded by viruses that assemble in the cytoplasm are structural proteins (17), and thus their shuttling is justified on the basis that they must return to the cytoplasm after performing their nuclear functions in order to participate in viral assembly. Therefore, our finding that p17, being a nonstructural protein, is constantly moving in and out of the nucleus suggests that this protein performs specific duties in both the nucleus and the cytoplasm, as has been reported for several RNA-binding proteins. The shuttling properties of these proteins have been reported to be important for controlling their activities in several nuclear processes as well as for modulating the functions of mRNAs in the cytoplasm (35, 50). Alternatively, the regulated nucleocytoplasmic shuttling of p17 might be used to control the activity of p17 within the nucleus and/or the cytoplasm.
Why should a cytoplasmic virus need a nucleus-targeted protein? The presence of p17 in the nuclei of avian reovirus-infected cells implies a potential role in the regulation of nuclear processes during virus replication. Nuclear p17 may avoid triggering the host immune response by blocking nuclear signaling pathways. This strategy might disrupt the interferon-mediated response and may be one of the reasons that avian reoviruses are so resistant to the antiviral effects of interferons (9, 29). Another possibility is that nuclear p17 decreases cellular transcription to induce host cell shutoff or to divert biosynthetic resources from the nucleus to the cytoplasm, the site of avian reovirus replication. Finally, p17 may activate the transcription of specific genes to enhance viral replication. A potential role of p17 in transcription was reinforced by our recent finding that this protein has specific double-stranded DNA binding activity (our unpublished data) and by its dependence on transcription for nuclear accumulation.
The activities of several transcription factors or cofactors, including the human immunodeficiency virus type 1 Rev protein (12), signal transducer and activator of transcription (STAT1) (34), the adenomatosis polyposis coli protein (36), the tumor protein p53 (54), the breast cancer 1 early onset (BRCA1) protein (44), certain members of the SOX family of development transcription factors (51), and several members of the SMAD family of signaling mediators (41), are modulated by shuttling in and out of the nucleus. Likewise, it is also possible that the accumulation of p17 in the nuclei of actively transcribing cells is caused by an association with a DNA target and that the inhibition of transcription might lead to DNA dissociation and the activation of export, either by unmasking a functional NES or by promoting its binding to a protein containing a transcription-dependent and CRM1-independent export activity. The generation of a cell line that stably expresses p17 would surely help to elucidate the effects of this viral protein on cellular metabolism.
Acknowledgments
We are grateful to Laboratorios Intervet (Salamanca, Spain) for providing specific-pathogen-free embryonated eggs. We thank Salvador Blanco Turnes and Lois Hermo Parrado for lab maintenance and technical support and assistance. We thank Aaron Shatkin for a critical reading of the manuscript.
This research was supported by grants from the Spanish Ministry of Ciencia y Tecnología (BMC2001-2839) and from the Xunta de Galicia (PGIDIT02PXIC20301PN).
REFERENCES
- 1.Belli, B. A., and C. E. Samuel. 1991. Biosynthesis of reovirus-specified polypeptides: expression of reovirus S1-encoded sigma 1NS protein in transfected and infected cells as measured with serotype specific polyclonal antibody. Virology 185:698-709. [DOI] [PubMed] [Google Scholar]
- 2.Bodelon, G., L. Labrada, J. Martinez-Costas, and J. Benavente. 2001. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology 290:181-191. [DOI] [PubMed] [Google Scholar]
- 3.Bodelon, G., L. Labrada, J. Martinez-Costas, and J. Benavente. 2002. Modification of late membrane permeability in avian reovirus-infected cells: viroporin activity of the S1-encoded nonstructural p10 protein. J. Biol. Chem. 277:17789-17796. [DOI] [PubMed] [Google Scholar]
- 4.Ceruzzi, M., and A. J. Shatkin. 1986. Expression of reovirus p14 in bacteria and identification in the cytoplasm of infected mouse L cells. Virology 153:35-45. [DOI] [PubMed] [Google Scholar]
- 5.Dang, C. V., and W. M. Lee. 1988. Identification of the human c-Myc protein nuclear translocation signal. Mol. Cell. Biol. 8:4048-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawe, S., and R. Duncan. 2002. The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J. Virol. 76:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan, R., J. Corcoran, J. Shou, and D. Stoltz. 2004. Reptilian reovirus: a new fusogenic orthoreovirus species. Virology 319:131-140. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, M. N., C. S. Eidson, J. Brown, and S. H. Kleven. 1983. Studies on interferon induction and interferon sensitivity of avian reoviruses. Avian Dis. 27:927-936. [PubMed] [Google Scholar]
- 10.Ernst, H., and A. J. Shatkin. 1985. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc. Natl. Acad. Sci. USA 82:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, X. C., and J. A. Steitz. 1998. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95:15293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 14.Grlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 15.Grlich, D., and I. W. Mattaj. 1996. Nucleocytoplasmic transport. Science 271:1513-1518. [DOI] [PubMed] [Google Scholar]
- 16.Grande, A., and J. Benavente. 2000. Optimal conditions for the growth, purification and storage of the avian reovirus S1133. J. Virol. Methods 85:43-54. [DOI] [PubMed] [Google Scholar]
- 17.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hood, J. K., and P. A. Silver. 1998. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem. 273:35142-35146. [DOI] [PubMed] [Google Scholar]
- 19.Inman, G. J., F. J. Nicolas, and C. S. Hill. 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol. Cell 10:283-294. [DOI] [PubMed] [Google Scholar]
- 20.Joklik, W. K. 1983. The Reoviridae. Plenum Publishing Corp., New York, N.Y.
- 21.Joklik, W. K. 1985. Recent progress in reovirus research. Annu. Rev. Genet. 19:537-575. [DOI] [PubMed] [Google Scholar]
- 22.Jordon, L. E., and H. D. Mayor. 1962. The fine structure of reovirus, a new member of the icosahedral series. Virology 17:597-599. [Google Scholar]
- 23.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 24.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 25.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lipowsky, G., F. R. Bischoff, P. Schwarzmaier, R. Kraft, S. Kostka, E. Hartmann, U. Kutay, and D. Gorlich. 2000. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19:4362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Costas, J., C. Gonzalez-Lopez, V. N. Vakharia, and J. Benavente. 2000. Possible involvement of the double-stranded RNA-binding core protein σA in the resistance of avian reovirus to interferon. J. Virol. 74:1124-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Costas, J., A. Grande, R. Varela, C. Garcia-Martinez, and J. Benavente. 1997. Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J. Virol. 71:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCutcheon, A. M., T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus M3 gene sequences and conservation of coiled-coil motifs near the carboxyl terminus of the μNS protein. Virology 10:16-24. [DOI] [PubMed] [Google Scholar]
- 32.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 33.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mowen, K., and M. David. 2000. Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol. 20:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 36.Neufeld, K. L., F. Zhang, B. R. Cullen, and R. L. White. 2000. APC-mediated downregulation of beta-catenin activity involves nuclear sequestration and nuclear export. EMBO Rep. 1:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 38.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 39.Olson, V. A., J. A. Wetter, and P. D. Friesen. 2002. Baculovirus transregulator IE1 requires a dimeric nuclear localization element for nuclear import and promoter activation. J. Virol. 76:9505-9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ossareh-Nazari, B., C. Gwizdek, and C. Dargemont. 2001. Protein export from the nucleus. Traffic 2:684-689. [DOI] [PubMed] [Google Scholar]
- 41.Reguly, T., and J. L. Wrana. 2003. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 13:216-220. [DOI] [PubMed] [Google Scholar]
- 42.Richardson, W. D., B. L. Roberts, and A. E. Smith. 1986. Nuclear location signals in polyoma virus large-T. Cell 44:77-85. [DOI] [PubMed] [Google Scholar]
- 43.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a sigma1s-null mutant. J. Virol. 72:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, J. A., and B. R. Henderson. 2000. Identification of a functional nuclear export sequence in BRCA1. J. Biol. Chem. 275:38589-38596. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sarkar, G., J. Pelletier, R. Bassel-Duby, A. Jayasuriya, B. N. Fields, and N. Sonenberg. 1985. Identification of a new polypeptide coded by reovirus gene S1. J. Virol. 54:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapouri, M. R., M. Arella, and A. Silim. 1996. Evidence for the multimeric nature and cell binding ability of avian reovirus sigma 3 protein. J. Gen. Virol. 77:1203-1210. [DOI] [PubMed] [Google Scholar]
- 48.Shmulevitz, M., and R. Duncan. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 19:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shmulevitz, M., Z. Yameen, S. Dawe, J. Shou, D. O'Hara, I. Holmes, and R. Duncan. 2002. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation. J. Virol. 76:609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 51.Smith, J. M., and P. A. Koopman. 2004. The ins and outs of transcriptional control: nucleocytoplasmic shuttling in development and disease. Trends Genet. 20:4-8. [DOI] [PubMed] [Google Scholar]
- 52.Soliman, T. M., and S. J. Silverstein. 2000. Herpesvirus mRNAs are sorted for export via Crm1-dependent and -independent pathways. J. Virol. 74:2814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 54.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tourís-Otero, F., J. Martínez-Costas, V. N. Vakaria, and J. Benavente. 2004. Avian reovirus nonstructural protein μNS forms viroplasm-like inclusions and recruits protein σNS to these structures. Virology 319:94-106. [DOI] [PubMed] [Google Scholar]
- 56.Turpin, P., B. Ossareh-Nazari, and C. Dargemont. 1999. Nuclear transport and transcriptional regulation. FEBS Lett. 452:82-86. [DOI] [PubMed] [Google Scholar]