Abstract
The catalytic core domain (CCD) of human immunodeficiency virus type 1 (HIV-1) integrase (IN) harbors the enzyme active site and binds viral and chromosomal DNA during integration. Thirty-five CCD mutant viruses were constructed, paying particular attention to conserved residues in the Phe139-Gln146 flexible loop and abutting Ser147-Val165 amphipathic alpha helix that were implicated from previous in vitro work as important for DNA binding. Defective viruses were typed as class I mutants (specifically blocked at integration) or pleiotropic class II mutants (additional particle assembly and/or reverse transcription defects). Whereas HIV-1P145A and HIV-1Q146K grew like the wild type, HIV-1N144K and HIV-1Q148L were class I mutants, reinforcing previous results that Gln-148 is important for DNA binding and uncovering for the first time an important role for Asn-144 in integration. HIV-1Q62K, HIV-1H67E, HIV-1N120K, and HIV-1N155K were also class I mutants, supporting findings that Gln-62 and Asn-120 interact with viral and target DNA, respectively, and suggesting similar integration-specific roles for His-67 and Asn-155. Although results from complementation analyses established that IN functions as a multimer, the interplay between active-site and CCD DNA binding functions was unknown. By using Vpr-IN complementation, we determined that the CCD protomer that catalyzes integration also preferentially binds to viral and target DNA. We additionally characterized E138K as an intramolecular suppressor of Gln-62 mutant virus and IN. The results of these analyses highlight conserved CCD residues that are important for HIV-1 replication and integration and define the relationship between DNA binding and catalysis that occurs during integration in vivo.
An essential step in the retroviral life cycle is the integration of the cDNA made by reverse transcription into a cell chromosome. Integration is mediated by the viral integrase (IN) protein, a specialized DNA recombinase that catalyzes two distinct endonucleolytic reactions during the early phase of infection. The first reaction, 3′ processing, occurs shortly after reverse transcription is completed. During 3′ processing, human immunodeficiency virus type 1 (HIV-1) IN removes a GT dinucleotide from each end of linear viral cDNA. IN performs its second reaction, DNA strand transfer, after finding a suitable chromosome target for integration. During DNA strand transfer, IN uses the oxygen moieties at the processed 3′ ends to cut the target DNA at two 5′ phosphates that are separated by 5 bp, which concomitantly joins the 3′ ends to the chromosome. The resulting DNA recombination intermediate, with viral 5′ ends unattached to the chromosome, is likely repaired by cellular enzymes (see references 16 and 34 for reviews).
Partial proteolysis (24), deletion mutagenesis (9, 70), and functional complementation (23, 69) analyses revealed that HIV-1 IN is comprised of three domains, the N-terminal domain, catalytic core domain (CCD), and C-terminal domain (CTD) (reviewed in reference 29). The CCD (residues 50 to 212 of the 288-residue protein) harbors a triad of invariant carboxylate amino acids (HIV-1 residues Asp-64, Asp-116, and Glu-152, which form the D,D-35-E motif) that comprises the enzyme active site (7, 20, 21, 24, 46, 49, 67). Results of IN-DNA photo-cross-linking (28, 40, 43), footprinting (19), and in vitro enzyme assays (4, 26, 33, 38, 43, 67) revealed other conserved CCD residues that likely contact viral (28, 33, 40, 41, 43) and target (4, 19, 38, 40, 67) DNA during integration. Specifically, Lys-159, Tyr-143, and Gln-148 cross-linked to viral DNA substrates (28, 43) and IN enzymes with alterations at Lys-159 or Gln-148 were defective in in vitro integration assays (33, 43, 67, 68). Peptides spanning residues 49 to 69 (40) and 51 to 64 (28) cross-linked to viral DNA and IN proteins altered at Gln-62 were sensitive to the concentration of salt in in vitro assays (26, 33), suggesting that Gln-62 also binds HIV-1 DNA (13, 28, 33). Since IN enzymes carrying substitutions of Ser-119 (38) or Asn-120 (67) displayed altered patterns of DNA strand transfer, these residues likely contact target DNA during integration. Since Lys-159, Gln-148, and Tyr-143 interacted with 1-(5-chloroindol-3-yl)-3-hydroxy-(2 H-tetrazol-5-yl)-propenone (5CITEP) in an IN-inhibitor cocrystal structure, it was suggested that 5CITEP might inhibit IN activity by interfering with critical IN-DNA contacts that occur during integration (36).
Mixtures of two defective IN mutant proteins can display 3′ processing and DNA strand transfer activities, suggesting that IN normally functions as a multimer (23, 69). Since mutants carrying changes in two different active-site residues failed to complement each other, the D,D-35-E active-site residues belong to the same complementation group (69). In other words, integration requires the same protomer within the active multimer to donate all three active-site residues. The organization of the active site to viral and target DNA-binding functions within the CCD, however, has not been probed by complementation. Although three-dimensional structures of IN-DNA complexes have not been solved, models based on protein fragment structures (14, 71), photo-cross-linking (31, 41), and/or computer simulations (2, 18, 32, 58, 59, 76) have been proposed. Most models posit that the CCD that donates the active site also binds the viral end and the target DNA to which that end will integrate; however, other arrangements are plausible (32, 41). To probe the relationships between these CCD functions, mutants altered for catalytic and presumed viral and target DNA binding activities were tested in complementation assays. This was done in the context of HIV-1 infection by fusing one mutant partner to the virion accessory protein Vpr and testing that fusion in trans to defective mutant virus (30, 73). A prerequisite for this strategy was the identification of viral mutants that supported relatively low levels of inherent infectivity. Thus, we began by characterizing a relatively large panel of CCD mutants in assays for HIV-1 spread.
Numerous investigators have analyzed HIV-1 mutants altered at IN active-site residues (3, 10, 25, 27, 39, 47, 48, 54, 55, 57, 60, 62, 63, 65, 72, 74, 75). The resulting class I replication-defective mutants were specifically blocked at integration as evidenced by (i) wild-type (WT) levels of cDNA synthesis in acutely infected cells and (ii) two long terminal repeat (2-LTR)-containing DNA circles that were elevated compared to WT levels, likely due to transient increases in unintegrated DNA pools (3, 25, 39, 48, 54, 57, 60, 62, 72, 75; see reference 22 for a review). In contrast, CCD mutants with changes in potential DNA binding residues have not been systematically analyzed, and unlike active site mutants, wherein any of a number of different missense mutations rendered HIV-1 inactive, the identity of the amino acid substitution had a relatively large impact on the virus replication phenotype. For example, whereas HIV-1K159Q displayed WT replication kinetics (63) and HIV-1E157A/K159A transduced cells 46% as efficiently as the WT (72), HIV-1K159P replicated with an approximate 2-week delay compared to the WT (10), and HIV-1K159E and HIV-1K156E/K159E failed to replicate over a 20-day observation period (43). As another example, HIV-1N120L and HIV-1N120E transduced cells at only about 0.06% of the WT level under conditions where HIV-1N120G supported about 3.3% of the WT transduction activity (48). Cells infected with HIV-1K159E, HIV-1K156E/K159E (43), or HIV-1N120L (48) contained elevated levels of 2-LTR circles, which defined these mutants as class I (reviewed in reference 22). In contrast to the integration-specific class I phenotype, class II IN mutant viruses display particle assembly and/or reverse transcription defects (22).
In this study, 22 CCD residues were targeted by site-directed mutagenesis, yielding 35 mutant viruses. In addition to their potential role in DNA binding, residues were targeted due to 5CITEP binding (36), degree of sequence conservation and surface exposure in crystal structures, and/or previous determination as essential for integration. Our results categorize conserved CCD residues as either dispensable or important for HIV-1 replication and classify defective viruses as either class I or class II. Of note, defective viruses altered at putative DNA contacts were class I mutants, highlighting the integration-specific roles of these residues in HIV-1 replication. Results of Vpr-IN complementation assays indicated that the active CCD protomer within the IN multimer interacts with both viral and target DNA during integration. Revertant mutant viruses were screened for the presence of second-site mutations, and E138K was characterized as an effective suppressor of Gln-62 mutant viruses and IN. Our results help to define the residues and regions of the IN CCD that engage viral and target DNA during integration.
MATERIALS AND METHODS
Plasmids.
Mutations were incorporated into HIV-1NL4-3 molecular clones pNL4-3 (1) or pNL43/XmaI (6) using PCR-directed mutagenesis as previously described (25, 51). Previously described mutants were HIV-1D116N (25), HIV-1F185K (42), HIV-1K156E, HIV-1K159E, HIV-1K156E/K159E (43), HIV-1D64N/D116N, and HIV-11-212 (57). The bacterial expression vector pINSD.His (17) was mutagenized as previously described (26). Plasmid pINSD.His(Q62A) encoding recombinant His-tagged INQ62A has been described (26).
The envelop (Env)-deleted single-round vector pNLX.Luc(R−) was derived from pNLXΔenvCAT (52) as follows. Plasmid pNLX.Luc was generated by amplifying the gene for firefly luciferase (Luc) from pGL3-Basic (Promega Corp., Madison, Wis.) with ClaI- and XhoI-tagged primers, digested, and ligated to ClaI, XhoI-digested pNLXΔenvCAT. To make pNLX(Vpr−), pNL43/XmaI was digested with EcoRI, filled in with Klenow, and religated. Plasmid pNLX.Luc(R−) was built by swapping the 1.6-kb PflMI-NheI fragment from pNLX(Vpr−) for the corresponding fragment in pNLX.Luc. IN mutant derivatives of pNLX.Luc(R−) were built by swapping altered 1.8-kb AgeI-PflMI fragments from pNL4-3 or pNL43/XmaI-based plasmids for the corresponding fragment in pNLX.Luc(R−). The HIV-1NL4-3 Env expression vector pNLXE7 was previously described (52).
Plasmid pRL2P-Vpr-IN (73) encoding HIV-1YU2-derived Vpr fused to HIV-1SG3 IN was a generous gift from J. Kappes (University of Alabama). Mutations were introduced into pRL2P-Vpr-IN using QuikChange mutagenesis as recommended by the manufacturer (Stratagene, La Jolla, Calif.). DNA sequencing was used to verify the regions of all plasmids that were generated by PCR.
Cells, viruses, and infections.
HeLa and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented to contain 10% fetal calf serum (FCS), 100 IU of penicillin per ml, and 0.1 μg of streptomycin sulfate per ml. Jurkat, CEM-12D7, and C8166 T-cells were grown in RPMI 1640 containing 10% FCS, 100 IU of penicillin/ml, and 0.1 μg of streptomycin/ml (RPMI-FCS).
Viruses for spreading-infection and 3′ processing assays were derived from HeLa or 293T cells by transfection in the presence of calcium phosphate as previously described (11, 25, 57). The concentration of virus in cell supernatants was determined using a 32P-based assay for reverse transcriptase (RT) activity (25, 57).
Single-round viruses carrying the HIV-1NL4-3 Env for real-time quantitative PCR (RQ-PCR) assays were derived by cotransfecting 293T cells with pNLX.Luc(R−) and pNLXE7 at the ratio of 30:1 using FuGENE 6 (Roche Molecular Biochemicals, Indianapolis, Ind.). Resulting cell supernatants were treated with 40 U of TURBO DNase (Ambion, Austin, Tex.)/ml for 1 h at 37°C to degrade the bulk of plasmid DNA remaining after transfection. Single-round viruses for Vpr-IN complementation studies were assembled by cotransfecting 293T cells with pNLX.Luc(R−), pRL2P-Vpr-IN, and pNLXE7 at the ratio of 30:15:1, using calcium phosphate. Background infectivity measurements of IN mutant viruses in the absence of added Vpr-IN were made by substituting pcDNA3 (Invitrogen, Carlsbad, Calif.) for pRL2P-Vpr-IN during transfection. All single-round viral stocks were normalized for RT content prior to infection. To analyze Vpr-IN incorporation, 293T cells were transfected with pNLX(Vpr−) and pRL2P-Vpr-IN in a 2:1 ratio by using FuGENE6.
For assays of viral spread, Jurkat or CEM-12D7 cells (2 × 106) were infected with WT or IN mutant HIV-1NL4-3 (106 RT cpm) for 18 h at 37°C in 0.5 ml of medium unless otherwise noted. The following day, cells were washed with serum-free RPMI 1640 and plated in 5 ml of RPMI-FCS. Cells were split at regular intervals, at which time aliquots of cell supernatants were stored at −20°C for RT assays. CEM-12D7 cells (5 × 106) were infected with 106 RT cpm of WT or IN mutant HIV-1NL4-3 for 2-LTR circle PCR analyses as previously described (6).
For RQ-PCR assays, Jurkat cells (4 × 106 per well of a six-well plate) were infected with DNase-treated WT or IN mutant single-round HIV-1NLX.Luc(R-) (5 × 106 RT cpm) by spinoculation for 2 h as previously described (52). Spun cells were washed three times with serum-free RPMI 1640 before plating in 4 ml of RPMI-FCS. For Vpr-IN complementation assays, Jurkat cells (2 × 106) were infected with single-round viruses (5 × 105 RT cpm in 0.5 ml) for 18 h at 37°C, at which time culture volumes were expanded by the addition of 5 ml of RPMI-FCS.
Analysis of HIV-1 proteins.
Transfected HeLa cells were radiolabeled with 35S-Met and 35S-Cys as previously described (26). Labeled cells and viruses were lysed and processed for immunoprecipitation, and the resulting immunoprecipitates were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and fluorography as previously described (25, 26).
For Western blot analysis of viral particles, 293T cell supernatants were pelleted through 20% sucrose cushions for 120 min at 4°C and 27,000 rpm in a Beckman SW28 rotor. Pellets were lysed in 25 μl of radioimmunoprecipitation assay buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS) and normalized for p24 protein content (Alliance HIV-1 p24 ELISA kit; New England Nucleotides, Boston, Mass.), and 2.5 μg of total p24 was analyzed by Western blotting using polyclonal anti-IN antibodies (15).
Analysis of HIV-1 DNA synthesis. (i) 2-LTR circle PCR.
Total DNA was extracted from CEM-12D7 cells 18 h postinfection (hpi), and 10 μl of cell extract was analyzed for levels of HIV-1 2-LTR circles by nested PCR as previously described (6).
(ii) RQ-PCR.
Total DNA was extracted from infected Jurkat cells 7 and 24 hpi by using the DNeasy kit (QIAGEN, Valencia, Calif.), and 10 μl was amplified by PCR in a 30-μl volume as described previously (51, 54). The PCR primers and Taqman probe were designed to detect total viral cDNA after the second template switch of reverse transcription, and HIV-1 signals were normalized to total cellular DNA using endogenous retrovirus 3 as described previously (51, 54). Parallel infections with Env− viruses were performed to control for residual levels of plasmid DNA that may have resisted DNase treatment, and the resulting PQ-PCR values were subtracted from those obtained with Env+ viruses.
Analysis of intracellular 3′ processing activity.
To determine 3′ processing levels, WT and IN mutant preintegration complexes were obtained from cytoplasmic extracts of infected C8166 cells as described previously (11-13). Following deproteinization and precipitation with ethanol, HIV-1 cDNA was cleaved with HaeIII and HindIII and separated by denaturing PAGE. Following electrotransfer, the blot was probed by indirect end labeling as described previously (11, 12). Results were detected by autoradiography and quantified by densitometry (IS-1000 Digital Imaging System; Alpha Innotech Corp., San Leandro, Calif.).
Cloning and sequencing of revertant viruses.
Supernatants were prepared from Jurkat cells infected with WT and revertant viruses by Hirt fractionation as previously described (57). Viral sequences amplified from Hirt supernatants by PCR were molecularly cloned as described previously (57).
Expression and purification of recombinant IN protein and in vitro integration assays.
INWT and INQ62A proteins were purified from insoluble fractions of bacterial lysates as previously described (26). INQ62A/E138K and INE138K were similarly purified and refolded into CN buffer {20 mM HEPES (pH 7.6), 1 mM EDTA, 0.2 M NaCl, 10% (wt/vol) glycerol, 1 mM dithiothreitol, 15 mM [(3-cholamidopropyl)-dimethylammonio]-l-propanesulfonate [CHAPS]}. The N-terminal hexahistidine tag used for protein purification was cleaved from IN using thrombin as previously described (26).
The 5′ end of oligonucleotide AE144 (5′-TTTTAGTCAGTGTGGAAAATCTCTAGCAGT) was radiolabled with T4 polynucleotide kinase as described previously (17). After heat inactivation of the kinase, the complementary single strand AE143 (5′-ACTGCTAGAGATTTTCCACACTGACTAAAA) was annealed and the 30-bp DNA substrate was separated from unincorporated nuclide as described previously (17).
In vitro integration reaction mixtures (16 μl) contained 25 mM morpholinepropanesulfonic acid (pH 7.2), 0.1 mg of bovine serum albumin/ml, 10 mM β-mercaptoethanol, 10% glycerol, 25 nM substrate DNA, 0.4 mM CHAPS, 7 mM NaCl, 7.5 mM MnCl2, and 0.3 μM IN. The concentration of NaCl in the reactions was varied as appropriate. Reactions were terminated after 60 min at 37°C and analyzed by electrophoresis on denaturing polyacrylamide sequencing gels as previously described (17). Results were visualized by autoradiography and quantitated using densitometry.
Vpr-IN complementation.
Cells harvested 48 hpi were washed with phosphate-buffered saline (Mediatech, Hamilton, Va.) and lysed with 75 μl of passive lysis buffer (Promega Corp., Madison, Wis.). Lysates frozen on dry ice and thawed at 37°C were centrifuged at 18,730 × g for 15 min at 4°C. Supernatants (20 μl) were tested in duplicate for Luc activity using the Luciferase Assay system (Promega Corp.) and an EG&G Berthold Microplate LB 96-V luminometer and Microlite 1 flat-bottom microtiter plate (Thermo Labsystems, Franklin, Mass.). Luc activity was normalized to the total cellular protein concentration as determined by the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, Calif.). Background levels of activity derived from mock infections with Env− HIV-1NLX.Luc(R−) were subtracted from WT and IN mutant values.
RESULTS
Mutagenesis strategy.
Twenty-two CCD residues were targeted for mutagenesis, and the resulting mutant viruses were initially assayed for spread in T-cell lines. A major focus of this study was to analyze the phenotypes of viruses mutated at residues proposed to bind DNA, and 16 of the 22 residues fell into this category (Table 1). Overall, residues were chosen for mutagenesis based on one or more of six different selection criteria: the residue (i) was implicated in binding to either viral or target DNA via in vitro activity (33, 43, 67, 68) or photo-cross-linking (28, 40, 41, 43, 56) assays and/or through molecular modeling (58), (ii) interacted with 5CITEP in an IN-inhibitor structure (36), (iii) displayed a high degree of conservation (>95% identity at that position) among a collection of 120 HIV-1 strains (45), (iv) was relatively well conserved among divergent retroviruses (24), (v) was surface exposed in crystal structures (14, 21, 35, 71), and (vi) was previously identified as essential for HIV-1 replication (10, 43, 47, 48, 63, 72). Table 1 lists the 22 residues, their degrees of sequence conservation, the ways in which the different selection criteria applied, and the resulting 35 mutant viruses. Because of this tabulation, selection details will for the most part be sidestepped in text. Yet some examples are noteworthy. Gln-148 was of interest, since this highly conserved residue was implicated in viral DNA (28, 33) and inhibitor (36) binding, and mutant proteins displayed <10 to 54% of INWT levels of 3′ processing and DNA strand transfer activities in in vitro assays (33, 67, 68). Asn-155 was targeted because it too was highly conserved among HIV-1 strains and divergent retroviruses, was implicated via molecular modeling as a potential DNA-binding residue (58), and contacted 5CITEP in the cocrystal structure (36), and mutant proteins displayed only 3 to 18% of INWT levels of 3′ processing, DNA strand transfer, and disintegration activities (33) (Table 1). Neither Gln-148 nor Asn-155 was previously analyzed in assays for HIV-1 infectivity. Since Gln-148 was predictably involved in DNA binding, previously characterized DNA binding mutants HIV-1K156E, HIV-1K159E, and HIV-1K156E/K159E (43) were included for comparison (Table 1). Since active-site residue Glu-152 was also shown to interact with HIV-1 cDNA (33), various Glu-152 mutants were constructed.
TABLE 1.
Targeted amino acid residues and mutagenesis strategy
| Residue | Conservationa | Criteriab | Reference(s) | Mutation(s) analyzed |
|---|---|---|---|---|
| Gln-53c | 100/33 | iii | cond | Q53K |
| Asp-55c | 98/10 | iii, vi | 72 | D55A, D55S, D55K |
| Gln-62 | 100/76 | i, iii, iv, v | 14, 28, 33, 40, 71 | Q62K |
| His-67 | 100/71 | i, iii, iv, v | 14, 40, 41, 58, 71 | H67E, H67Q/K71E |
| Lys-71 | 99/48 | iii, v | 14, 71 | K71E |
| His-114 | 100/38 | i, iii, v | 14, 58, 71 | H114E |
| Asn-117 | 100/90 | i, iii, iv, v | 14, 33, 58, 71 | N117K |
| Asn-120 | 100/38 | i, iii, v, vi | 14, 33, 48, 67, 71 | N120L, N120K, N120L/Q148K |
| Lys-136 | 31/24 | i, v, vi | 14, 19, 56, 71, 72 | K136A |
| Glu-138 | 98/10 | i, iii, v | 14, 19, 35, 71 | E138K |
| Tyr-143 | 99/86 | i, iii, iv, v | 28, 35, 36, 40 | Y143G |
| Asn-144 | 100/71 | iii, iv, v | 35 | N144K |
| Pro-145 | 100/95 | i, iii, iv, v | 14, 35, 40 | P145A |
| Gln-146 | 100/81 | i, iii, iv, v | 14, 35, 40, 58 | Q146K |
| Gln-148 | 100/62 | i, ii, iii, iv, v | 14, 28, 33, 35, 36, 67, 68 | Q148L, Q148K |
| Glu-152 | 99/100 | i, ii, iii, iv, v, vi | 10, 14, 35, 36, 47, 63, 71, 72 | E152D, E152Q, E152K |
| Ser-153 | 94/10 | i, v | 14, 58, 71 | S153R |
| Asn-155 | 100/76 | i, ii, iii, iv, v | 14, 33, 36, 58, 71 | N155E, N155L, N155K |
| Lys-156 | 97/5 | i, ii, iii, v, vi | 14, 19, 33, 36, 43, 71 | K156E |
| Lys-159 | 99/100 | i, ii, iii, iv, v, vi | 14, 19, 33, 36, 43, 71 | K159E, K156E/K159E |
| Asn-184 | 100/95 | iii, iv | con | N184D, N184L |
| Arg-199 | 99/38 | iii, v, vi | 14, 71, 72 | R199A, R199E |
The first number is the percentage at which the residue is found at that position in a collection of 120 HIV-1 strains (45), and the second number is percent identity among 21 different retroviruses (24).
The following criteria were utilized: i, implicated via in vitro and/or in silico analyses in DNA binding (19, 28, 33, 40, 41, 43, 56, 58, 67, 68); ii, identified as binding to 5CITEP in an IN-drug cocrystal structure (36); iii, highly conserved (>95%) among HIV-1 isolates; iv, conserved in >50% of retroviral INs; v, surface exposed in crystal structures (14, 35, 71); vi, mutant viruses previously reported as noninfectious (10, 43, 47, 48, 63, 72).
Neither Gln-53 nor Asp-55 was visible in X-ray structures.
con, conservation. The only selection criterion was degree of sequence conservation.
Although Asn-144 has not been implicated in DNA binding, this well-conserved residue was targeted because it resides within the Phe139-Gln146 flexible loop region that cross-linked to DNA substrates (40). Of the four loop residues targeted here (Tyr-143, Asn-144, Pro-145, and Gln-146), Tyr-143 cross-linked to viral DNA (28). Certain other residues, such as Asp-55 and Lys-136, were targeted because previous mutants were replication defective (72), yet neither residue was well conserved among divergent retroviruses (Table 1). In particular, Lys-136 was conserved in only 31% of HIV-1 strains, with Gln and Thr predominating at this position approximately 42 and 17% of the time, respectively (45). Mutations were introduced into the NL4-3 strain of HIV-1 (1).
Replication profiles of IN mutant viruses.
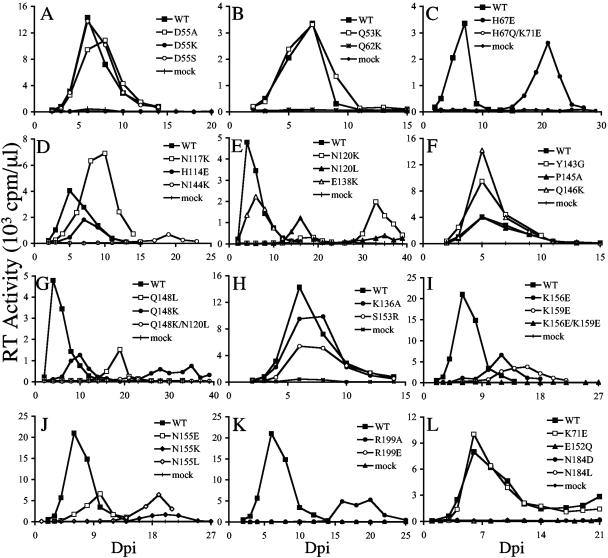
Jurkat and CEM-12D7 T-cells infected with 106 RT cpm (an approximate multiplicity of infection [MOI] of 0.04; see references 52 and 57) supported peak levels of HIV-1NL4-3 replication 4 to 6 days postinfection (dpi) (Fig. 1). Since substituting Ala for Asp-55 rendered HIV-1R7-3 inactive (72), we were a bit surprised to find that NL4-3-based HIV-1D55A replicated similarly to the WT (Fig. 1A). Because of this, a similar substituent, Ser, as well as nonconservative Lys, were tested at this position. Whereas HIV-1D55S grew like the WT, HIV-1D55K was defective (Fig. 1A).
FIG. 1.
Replication kinetics of WT and IN mutant viruses. (A) Aliquots of infected Jurkat T-cell supernatants were assayed for RT activity at the indicated time points. (B and C) Supernatants of CEM-12D7 cells were assayed for RT activity at the indicated times. (D through L) RT profiles of WT and IN mutant viruses in Jurkat cells. Results are representative of a minimum of two independent sets of infections.
Approximately one-third of the viruses were classified as WT because their replication peaks were reached at the same day or at most 2 days delayed from HIV-1NL4-3. In addition to HIV-1D55A and HIV-1D55S, these included the following: HIV-1Q53K (Fig. 1B), HIV-1H114E (Fig. 1D), HIV-1E138K (Fig. 1E), HIV-1Y143G, HIV-1P145A, and HIV-1Q146K (Fig. 1F), HIV-1K136A and HIV-1S153R (Fig. 1H), and HIV-1K71E (Fig. 1L). A few of the mutants, including HIV-1N117K (Fig. 1D), HIV-1Q148K (Fig. 1G), HIV-1K156E (Fig. 1I), and HIV-1N155E (Fig. 1J), displayed moderate replication delays that ranged from 4 days to 1 week. The following IN mutants replicated with more-serious delays that ranged from 1 to 3 weeks compared to WT replication in repeat experiments: HIV-1H67E (Fig. 1C), HIV-1N144K (Fig. 1D), HIV-1N120L and HIV-1N120K (Fig. 1E), HIV-1Q148L (Fig. 1G), HIV-1K159E (Fig. 1I), HIV-1N155K, HIV-1N155L (Fig. 1J), and HIV-1R199A (Fig. 1K). Numerous mutants in addition to HIV-1D55K were categorized as replication defective, since the cells they infected failed to yield any evidence of virus spread over 2 months of observation. These included the following: HIV-1Q62K (Fig. 1B), HIV-1H67Q/K71E (Fig. 1C), HIV-1Q148K/N120L (Fig. 1G), HIV-1K156E/K159E (Fig. 1I), HIV-1R199E (Fig. 1K), HIV-1E152Q, HIV-1N184D, and HIV-1N184L (Fig. 1L), HIV-1E152D, and HIV-1E152K (data not shown). Table 2 summarizes mutant viral replication profiles and phenotypic classifications.
TABLE 2.
Summary of IN mutant viral phenotypes
| Mutation | Replicationa | Mutant phenotypeb |
|---|---|---|
| Q53K | ++ | NAc |
| D55A | ++ | NA |
| D55S | ++ | NA |
| D55K | − | II |
| Q62K | − | I |
| H67E | ± | Id |
| H67Q/K71E | − | I |
| K71E | ++ | NA |
| H114E | ++ | NA |
| N117K | + | NDe |
| N120L | ± | Id |
| N120K | ± | Id |
| N120L/Q148K | − | I |
| K136A | ++ | NA |
| E138K | ++ | NA |
| Y143G | ++ | NA |
| N144K | ± | Id |
| P145A | ++ | NA |
| Q146K | ++ | NA |
| Q148L | ± | Id |
| Q148K | + | Id |
| E152D | − | I |
| E152Q | − | I |
| E152K | − | II |
| S153R | ++ | NA |
| N155E | + | ND |
| N155L | ± | Id |
| N155K | ± | Id |
| K156E | + | If |
| K159E | + | If |
| K156E/K159E | − | If |
| N184D | − | II |
| N184L | − | II |
| R199A | ± | IId |
| R199E | − | II |
++, replication peak detected either the same day or at most 2 days delayed from the WT; +, replication delayed 4 days to 1 week from the WT; ±, peak of virus replication delayed 1 to 3 weeks from the WT; −, replication undetected over 2 months of observation. Values are representative of a minimum of two independent infections.
Phenotypic classification based on results in Fig. 1 through 3. Whereas class I mutants are defined as replication-defective viruses that yield 2-LTR circle levels in excess of WT HIV-1 following acute infection, class II mutants display pleiotropic defects at the step(s) of virus assembly and/or reverse transcription (see reference 22 for a review).
NA, not applicable, as these viruses replicated similarly to the WT.
Although the class I and class II designations are usually reserved for completely dead viruses, here they indicate whether the reason(s) for the replication delays shown in Fig. 1 was primarily due to a specific block in integration (class I phenotype) or whether reduced levels of reverse transcription (class II) may have also contributed.
ND, not determined.
As determined in reference 43.
Defective IN mutant classification: (i) DNA synthesis.
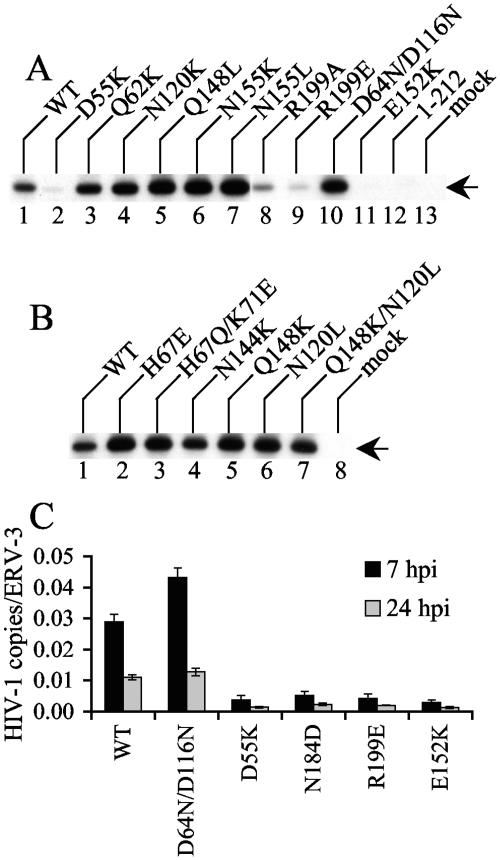
The results in Fig. 1 and Table 2 revealed that the majority of IN mutants (24 of 35) were defective, since either they failed to grow or their replication peaks were delayed minimally 4 days from the WT peak. We next investigated the replication defects for the majority of these viruses. Since cells infected with integration-defective class I mutants contain more 2-LTR circles than WT-infected controls and pleiotropic class II IN mutants are defective for reverse transcription (reviewed in 22), levels of WT and IN mutant 2-LTR circles after acute infection were analyzed. As expected (54, 57), the class I active-site mutant control strain HIV-1D64N/D116N yielded more 2-LTR circles than WT HIV-1NL4-3 at 18 hpi (Fig. 2A, compare lane 10 to lane 1). Two of the novel active-site mutants, HIV-1E152D and HIV-1E152Q, also yielded elevated levels of 2-LTR circles (Table 2; also data not shown). Unexpectedly, the other novel active-site mutant (HIV-1E152K) behaved more like the class II CTD deletion mutant strain HIV-11-212 (57), since both yielded minimal levels of 2-LTR circles that were detected only upon prolonged autoradiographic exposure (Fig. 2A, lanes 11 to 13; also data not shown). Thus, HIV-1E152K seemed defective for DNA synthesis. In contrast, most of the other replication-defective mutants, including HIV-1Q62K, HIV-1N120K, HIV-1Q148L, HIV-1N155K, HIV-1N155L (Fig. 2A), HIV-1H67E, HIV-1H67Q/K71E, HIV-1N144K, HIV-1Q148K, HIV-1N120L, and HIV-1Q148K/N120L (Fig. 2B), formed more 2-LTR circles than the WT, which classified these as class I mutant viruses (Table 2). HIV-1D55K (Fig. 2A, lane 2), HIV-1R199E (lane 9), HIV-1N184L, and HIV-1N184D (data not shown) yielded less 2-LTR circular DNA than the WT, indicating that like HIV-1E152K, these mutants were defective for reverse transcription. Although HIV-1R199A also yielded fewer 2-LTR circles than the WT (Fig. 2A, lane 8), we note that repeat experiments revealed that these levels were consistently above those detected for HIV-1D55K, HIV-1R199E, HIV-1E152K, and HIV-11-212 (Fig. 2; also data not shown).
FIG. 2.
WT and IN mutant viral DNA synthesis. (A and B) CEM-12D7 cells infected with the indicated viruses were lysed 18 hpi, and 2-LTR circles were detected by nested PCR as previously described (6). The arrow indicates the 170-bp circle junction product. Results are representative of those obtained in a minimum of two independent sets of infections. (C) Jurkat cells infected with the indicated single-round viruses were lysed at the indicated time points, and total HIV-1 DNA levels quantified by RQ-PCR were normalized to the cellular endogenous retrovirus 3 marker as previously described (51, 54). Error bars represent the variation between duplicate sets of RQ-PCR assays. Results are representative of those obtained in four independent sets of infections. hpi, hours postinfection.
A subset of the mutants that yielded reduced levels of 2-LTR circles was analyzed for overall HIV-1 levels by RQ-PCR. Primers and Taqman probe were designed to detect products after the second template switch of reverse transcription, and single-round Env− viruses were employed to restrict the spread of the WT control during the 24-h experiment. Since reverse transcription peaks approximately 7 to 8 hpi (12, 13, 44), DNA levels were initially analyzed at 7 hpi. WT HIV-1NLX.Luc(R-) and the class I mutant HIV-1D64N/D116N.Luc(R−) supported similar levels of reverse transcription (Fig. 2C). In contrast, HIV-1D55K.Luc(R-), HIV-1N184D.Luc(R-), HIV-1R199E.Luc(R-), and HIV-1E152K.Luc(R-) yielded levels that were reduced approximately five- to 10-fold from the WT in repeat experiments (Fig. 2C; also data not shown). To address whether these were absolute or kinetic differences in DNA synthesis, lysates were prepared at 24 hpi. Similar to the results at 7 hpi, HIV-1D55K.Luc(R-), HIV-1N184D.Luc(R−), HIV-1R199E.Luc(R−), and HIV-1E152K.Luc(R-) DNA levels were significantly lower (approximately five- to 10-fold) than those for WT HIV-1NLX.Luc(R−) and HIV-1D64N/D116N.Luc(R−) at 24 hpi (Fig. 2C). Based on this, we conclude that the lower levels of 2-LTR circles detected for class II mutant viruses in Fig. 2A were primarily due to overall reductions in DNA synthesis.
(ii) Mutant virus assembly.
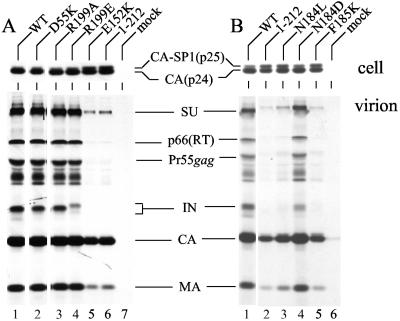
In addition to the common reverse transcription defect, certain class II IN mutants are defective for the relatively late steps of particle assembly and release (8; reviewed in reference 22). To address whether any of the DNA synthesis-defective mutants displayed late event defects, virus-producing cells were radiolabeled with [35S]Met and [35S]Cys, cell and viral lysates were immunoprecipitated with AIDS patients' antisera, and HIV-1 proteins were analyzed after SDS-PAGE and fluorography. Assembly-release mutants were identified as viruses containing levels of HIV-1 proteins that were significantly lower than their intracellular levels of gene expression.
As previously reported (26, 42), the HIV-1F185K class II mutant was released poorly from cells (Fig. 3B, compare lane 5 to lane 1). As predicted from previous analyses of IN deletion mutants (3, 8, 25), the CTD truncation mutant HIV-11-212 also released poorly in this assay (Fig. 3A, lane 6 and Fig. 3B, lane 2). Despite these disproportionately low levels of HIV-1F185K and HIV-11-212 virion proteins, the mutants were expressed in cells as efficiently as WT HIV-1NL4-3 (Fig. 3A, upper panel, lanes 1 and 6; upper panel of Fig. 3B, lanes 1, 2, and 5). Whereas HIV-1E152K (Fig. 3A lower panel, lane 5) and HIV-1N184L (Fig. 3B, lane 3) also displayed release defects, HIV-1D55K, HIV-1R199A, HIV-1R199E, and HIV-1N184D were released from cells similarly to the WT (Fig. 3). We note that assembly-defective class II mutants expressed an increase in the ratio of the Gag processing intermediate capsid-spacer peptide 1 (p25) to mature capsid (p24) (Fig. 3A and B, upper panels), a phenotype associated with a variety of maturation-defective HIV-1 mutants (reference 50 and references therein).
FIG. 3.
IN mutant viral assembly and release. (A) HeLa cells were transfected with the indicated plasmid clones, and radioimmunoprecipitated cell (upper panel) and virion (lower panel) lysates were analyzed by SDS-PAGE. (B) Cells transfected with the indicated viral clones were analyzed as for panel A. The migration positions of HIV-1 proteins are indicated. CA-SP1, capsid-spacer peptide 1; CA, capsid; SU, surface; MA, matrix. The minor amount of capsid in lane 6 of Fig. 3B was caused by spillover during gel loading. Similar assembly-release profiles were observed in an independent set of radioimmunoprecipitation assays.
Target DNA binding-defective phenotype of HIV-1N120K.
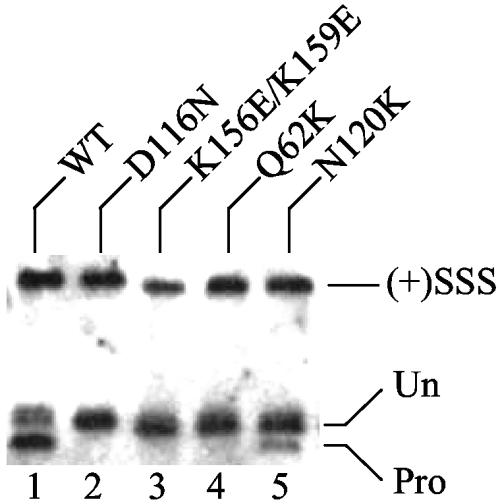
A major goal of this study was to test IN mutants defective for distinct CCD functions for their abilities to complement each other during infection, and identifying numerous class I mutant viruses (Table 2) afforded this possibility. Since recombinant Asn-120 mutant proteins (67) as well as proteins altered at the adjacent Ser-119 residue (38) displayed novel DNA strand transfer patterns in vitro and HIV-1N120K was a class I mutant (Fig. 1E and 2A), the virus was predictably defective for target DNA binding. To investigate this hypothesis, preintegration complexes isolated from acutely infected C8166 cells were quantified for in vivo levels of IN 3′ processing activity. If HIV-1N120K was defective for target DNA binding, it should in theory support higher levels of 3′ processing activity than class I mutants that were primarily defective for catalysis or viral DNA binding. Indeed, this was the observed phenotype. Whereas HIV-1NL4-3 processed about 70% of its U5 end, active-site mutant HIV-1D116N and viral DNA binding mutants HIV-1K156E/K159E and HIV-1Q62K failed to detectably process their ends (<3% of U5 processed) (Fig. 4, lanes 1 to 4). In contrast, HIV-1N120K processed about 17% of its end (lane 5). Thus, since (i) HIV-1N120K, HIV-1Q62K, HIV-1K156E/K159E, and HIV-1D116N were class I mutant viruses (Fig. 2A) (25, 43) and (ii) HIV-1N120K processed substantially more of its DNA than HIV-1D116N, HIV-1K156E/K159E, or HIV-1Q62K (Fig. 4), we inferred that HIV-1N120K was partially defective for target DNA interactions in vivo. This set the stage to test CCD mutants that were predictably defective for viral DNA binding, target DNA binding, or catalysis in complementation assays. Since HIV-1N120K was about fourfold defective for 3′ processing activity, we noted that efficient trans complementation of target DNA binding might yield 10 to 25% of WT function.
FIG. 4.
WT and class I IN mutant 3′ processing in cells. The structures of WT and IN mutant U5 plus strands are shown. (+)SSS, cleaved plus-strand strong-stop DNA; Un, 105-nucleotide nonprocessed U5 end; Pro, 103-nucleotide 3′ processing product. Similar levels of WT and mutant 3′ processing were observed at U3 minus-strand ends (data not shown).
Vpr-IN complementation.
A subset of the more deleterious changes that yielded viruses predictably defective for viral DNA binding (Q62K and K156E/K159E), target DNA binding (N120K), or catalysis (D64N/D116N) were introduced into the single-round Luc reporter virus HIV-1NLX.Luc(R−), and Q62K and N120K were additionally introduced into pRL2P-Vpr-IN (73). Since Vpr-INV165A efficiently complemented active site mutant HIV-1 (5, 53), it served as a positive control. Since HIV-1V165A is a CCD class II mutant virus (51, 53), class II mutant fusion proteins Vpr-INR199E and Vpr-IND55K were also constructed.
As predicted from their poor performances in spreading-infection assays, HIV-1Q62K.Luc(R−), HIV-1K156E/K159E.Luc(R−), and HIV-1N120K.Luc(R−) infectivities were near the detection limit of the Luc assay, similar to the case with active-site mutant HIV-1D64N/D116N.Luc(R−) [approximately 0.05 to 0.3% of that for WT HIV-1NLX.Luc(R−)] (Table 3). Vpr-INWT restored 10 to 30% of HIV-1NLX.Luc(R−) function to HIV-1Q62K.Luc(R−), HIV-1K156E/K159E.Luc(R−), HIV-1N120K.Luc(R−), and HIV-1D64N/D116N.Luc(R−) (Table 3). Mutant Vpr-IN activity was reported as percent Vpr-INWT function. As expected (5, 53), Vpr-INV165A efficiently complemented active-site mutant HIV-1D64N/D116N.Luc(R−), and HIV-1Q62K.Luc(R−) was also efficiently complemented by Vpr-INV165A (Table 3). Although Vpr-INV165A also complemented HIV-1K156E/K159E.Luc(R−) and HIV-1N120K.Luc(R−), these levels were approximately 5 to 10% of the levels observed with HIV-1D64N/D116N.Luc(R−) and HIV-1Q62K.Luc(R−) (Table 3). In contrast to the relatively robust activity of Vpr-INV165A, Vpr-INQ62K did not complement HIV-1D64N/D116N.Luc(R−), HIV-1Q62K.Luc(R−), HIV-1K156E/K159E.Luc(R−), or HIV-1N120K.Luc(R−) (Table 3). Vpr-INN120K behaved similarly to Vpr-INQ62K: although Vpr-INN120K weakly complemented HIV-1Q62K.Luc(R−) (approximately 3% of Vpr-INWT), we noted that the reciprocal arrangement yielded negligible activity (Table 3). Based on this, we concluded that Q62K, D64N/D116N, K156E/K159E, and N120K belong to the same functional complementation group. We therefore infer that D,D-35-E active site, viral DNA binding, and target DNA binding functions of the CCD are supplied by the same protomer within the active IN multimer during integration.
TABLE 3.
Complementation of IN mutant viruses with Vpr-INa
| IN mutants | % WT activityb | Vpr-IN
|
|||||
|---|---|---|---|---|---|---|---|
| WTc | Complementation relative to Vpr-INWTd
|
||||||
| V165A | Q62K | N120K | R199E | D55K | |||
| Q62K | 0.32 (0.09) | 30.9 (19.4) | 181 (17.3) | 0.1 (0.1) | 3.1 (0.8) | 30.4 (15.7) | 51.2 (24.9) |
| K156E/K159E | 0.05 (0.00) | 17.5 (1.9) | 7.9 (1.0) | 0.4 (0.4) | 1.3 (0.7) | 0.7 (0.2) | 12.3 (0.1) |
| D64N/D116N | 0.09 (0.01) | 9.6 (2.0) | 132 (26.7) | 0.5 (0.2) | 0.7 (0.2) | 10.1 (1.8) | 25.3 (12.0) |
| N120K | 0.08 (0.05) | 20.7 (11.5) | 12.5 (0.3) | 0.8 (0.9) | 1.7 (1.6) | 5.4 (5.0) | 17.9 (8.0) |
Values represent an average of a minimum of two infections.
Luc activity of the indicated IN mutant virus relative to that of WT HIV-1NLX.Luc(R−) with standard deviations in parentheses.
Luc activity of the indicated IN mutant viruses complemented with Vpr-INWT relative to WT HIV- 1NLX.Luc(R−) with standard deviations in parentheses.
Percent activity with standard deviations in parentheses.
Vpr-INR199E and Vpr-IND55K were also tested with the various class I mutant viruses. Since Vpr-INR199A complemented active-site mutant virus (30, 53), it was not surprising that Vpr-INR199E complemented HIV-1D64N/D116N.Luc(R−) (Table 3). Although Vpr-INR199E complemented HIV-1Q62K.Luc(R−) and HIV-1N120K.Luc(R−) to levels similar to that observed with HIV-1D64N/D116N.Luc(R−), it failed to function with HIV-1K156E/K159E.Luc(R−) (Table 3). Vpr-IND55K was more promiscuous than Vpr-INR199E in this assay, since it displayed between 12% and approximately 50% of Vpr-INWT activity with the different class I mutant viruses (Table 3). Although Vpr-INQ62K efficiently complemented HIV-1V165A.Luc(R−) (53), we note that neither Vpr-IND55K nor Vpr-INR199E complemented HIV-1V165A.Luc(R−) (data not shown).
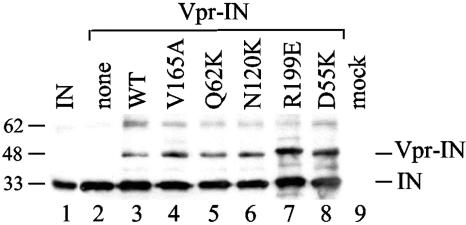
Our results demonstrate that CCD class II mutant Vpr-INs effectively complemented CCD class I mutant viruses under conditions where analogous class I mutant proteins failed to function (Table 3). Our ability to assign various CCD functions to the same IN protomer during postentry integration therefore relied on efficient incorporation of class I and class II Vpr-IN proteins during virus assembly. Although there was little reason to expect differential packaging of different mutant classes, virus particles generated by cotransfecting Vpr-IN expression vectors with HIV-1NLX(Vpr−) were pelleted through sucrose and normalized for p24 content, and Western blots were probed with anti-IN antibodies. Whereas the only reactive HIV-1NLX(Vpr−) band comigrated with recombinant IN protein (Fig. 5, compare lane 2 to lane 1), trans-complemented particles contained an additional ∼45-kDa product indicative of uncleaved Vpr-IN (lanes 3 to 8) (30, 61, 73). Although class II mutants Vpr-IND55K and Vpr-INR199E were incorporated somewhat more efficiently than class I Vpr-INQ62K and Vpr-INN120K, the level of class I mutant incorporation was indistinguishable from that of Vpr-INV165A and Vpr-INWT (Fig. 5). Based on this, we conclude that CCD class I mutant Vpr-INs do not function with CCD class I mutant viruses due to the lack of postviral entry phenotypic complementation.
FIG. 5.
Vpr-IN incorporation into HIV-1 particles. Lane 1, 50 ng of recombinant IN protein; lane 2, HIV-1NLX(Vpr−) cotransfected with empty vector; lanes 3 to 8, cotransfections with indicated Vpr-IN expression vector; lane 9, mock-transfected cell lysate. The migrations positions of Vpr-IN and IN are indicated on the right, and those of molecular mass standards are indicated on the left. Since the weakly reactive ∼62-kDa band in lanes 3 to 8 comigrated with dimeric recombinant IN (not shown), we speculate this was cleaved, dimeric INSG3.
Selection and characterization of IN revertant viruses.
The experiments in the previous section probed the organization of the CCD through the use of functional complementation, and the results indicated that multiple functions, including metal-based catalysis, viral DNA binding, and target DNA binding, were accomplished by the same protomer within the active IN multimer. The interplay between different residues in the CCD was also analyzed by selecting and characterizing revertant mutant viruses.
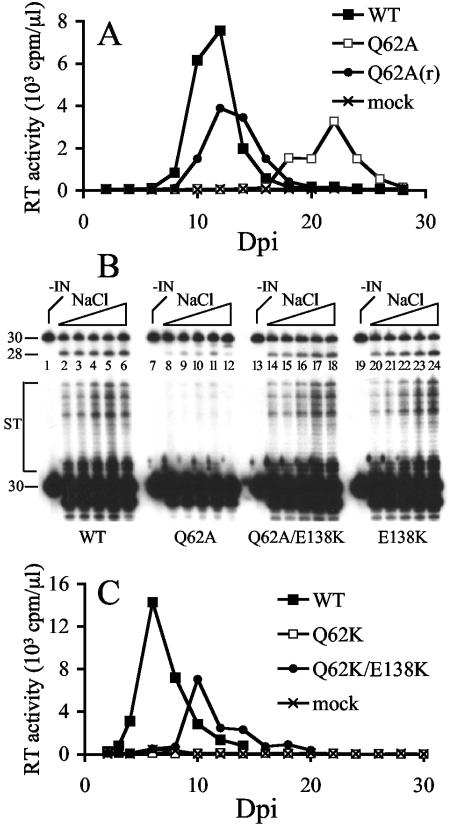
Although some class I mutants, such as HIV-1H67E and HIV-1N120K, showed growth that was significantly delayed compared to that of the WT, their titers eventually approached those of the WT (Fig. 1C and E). We considered two possibilities for these observations. First, the replication delay was a direct consequence of the activity of the mutant IN protein. In other words, replication occurred in the face of the original mutation, and the delay was due to the time required to amass sufficient infection events to drive replication throughout the culture. In the second case, the original mutation, although sufficient to support an initial low level of integration, was insufficient to drive virus spread, and thus, other mutations were required to revert the IN to a more active form. In this case the delay was due to the time required for the initial infection to generate the required mutation(s) through subsequent rounds of reverse transcription and integration. To distinguish between these two scenarios, viruses derived from the peaks of HIV-1H67E (Fig. 1C) and HIV-1K159E (Fig. 1I) replication were passed onto fresh cells. If the first model was true, the passaged viruses would grow with delays similar to those seen in the original infections. If on the other hand additional mutations had occurred, the passaged viruses would replicate at increased rates. Since HIV-1Q62A also grew with an approximate 2-week delay compared to the WT (26), it was included in this analysis. Using a similar selection scheme, we previously determined that a second CCD substitution (T125A) effectively suppressed the deleterious effects of P109S (64).
Jurkat cells infected with passaged HIV-1H67E and HIV-1K159E revealed cytopathic evidence of viral spread 9 dpi (data not shown). Since HIV-1H67E and HIV-1K159E reached their original growth peaks 21 and 16 dpi, respectively (Fig. 1C and I), we reasoned that the passaged viruses had accumulated new changes. To investigate this, infected cells were lysed by Hirt extraction, and HIV-1 DNA in Hirt supernatants was sequenced after PCR amplification. Whereas HIV-1NL4-3 uses an AAG codon for Lys-159, GAA was engineered in HIV-1K159E. The revertant virus harbored AAA at this position, indicating back reversion of the mutation to restore WT HIV-1NL4-3. This was not unexpected for a substitution such as Lys→Glu wherein a single nucleotide change would restore INWT function. HIV-1H67E contained GAG in place of CAT, and the revertant virus altered this to AAG, indicating that a Lys at position 67 was sufficient for near-INWT function. Due to what were considered relatively uninformative back reversions of K159E and H67E changes, these revertant viruses were not studied further.
Similar to the results with HIV-1H67E and HIV-1K159E, passaged HIV-1Q62A(r) (r for revertant) spread significantly faster than the starting mutant strain (Fig. 6A). Hirt supernatant DNA prepared from HIV-1Q62A(r)-infected cells at day 14 postinfection was PCR amplified, and IN coding regions were molecularly cloned prior to sequencing. In one of four cases, the mutant GCG codon underwent two reversional substitutions back to the WT CAG. However, in the other three cases, the mutant GCG codon persisted and a G-to-A transition occurred at nucleotide 4641 of HIV-1NL4-3. This altered the GAA encoding IN residue Glu-138 to the same E138K change that we had included in our starting mutant pool (Table 1 and Fig. 1E). The entire IN coding regions of two of the HIV-1Q62A(r) molecular clones were sequenced, which revealed the G-to-A change as the only new alteration in each case.
FIG. 6.
Identification and characterization of a HIV-1Q62A revertant virus. (A) Jurkat cells (5 × 106) infected with 5 × 105 RT cpm of HIV-1Q62A produced from (▪) transiently transfected HeLa cells (26) or (•) HIV-1Q62A(r) obtained from a prior infection of Jurkat cells at 19 dpi were monitored for RT activity at the indicated time points. WT HIV-1NL4-3 was derived from transfected HeLa cells. (B) In vitro 3′ processing (upper panel) and DNA strand transfer (lower panel) activities of WT and IN mutant proteins. IN was omitted from the reaction in lane 1. The reactions in lanes 2 to 6 contained 7, 14, 28, 56, and 112 mM NaCl, respectively (the triangle represents the increase in NaCl concentration). The reactions in lanes 7 to 12, 13 to 18, and 19 to 24 were the same as in lanes 1 to 6 except for the identity of the IN protein, as indicated beneath the gel. The migration positions of the 3′ processing substrate and product are marked 30 and 28, respectively. ST, products of strand transfer. Whereas the gel in the upper panel was exposed to X-ray film for 15 min, the lower panel was exposed for 8 h. (C) Jurkat cells (2 × 106) infected with the indicated molecularly cloned viruses (106 RT cpm) were monitored for RT activity at the indicated times.
To investigate whether the E138K change impacted INQ62A function, the G-to-A substitution was incorporated into INWT and INQ62A bacterial expression constructs, and purified INWT, INQ62A, INQ62A/E138K, and INE138K proteins were analyzed in in vitro integration assays. Previous studies revealed that INQ62A supported approximately 5 to 20% of INWT 3′ processing and DNA strand transfer activities in reactions containing 25 mM NaCl but substantially less activity (<1 to 3% of INWT) in the presence of 80 to 90 mM NaCl (26, 33). INWT converted between 30 and 40% of a 30-bp U5 substrate to the nicked 3′ processing product, and this level of activity persisted through a range of NaCl concentration that spanned from 7 to 112 mM (Fig. 6B, upper panel, lanes 1 to 6). Consistent with previous reports, INQ62A converted less of the U5 substrate (8 to 24%) to product than INWT under low-salt conditions (Fig. 6B, upper panel, compare lanes 8 to 11 to lanes 2 to 5) and only about 5% of the substrate under higher-salt conditions (compare lane 12 to lane 6). As predicted from the near-WT replication profile of HIV-1E138K (Fig. 1E), the activity of the INE138K enzyme mirrored that of INWT (Fig. 6B, compare lanes 20 to 24 to lanes 2 to 6). The activity profile of INQ62A/E138K also mirrored that of INWT. In this case between 15 and 40% of substrate was converted to product, and most notably, INQ62A/E138K displayed the WT level of IN 3′ processing activity under high-salt conditions (Fig. 6B, upper panel, compare lane 18 to lanes 12 and 6). Longer autoradiographic exposure revealed that the E138K change also restored efficient DNA strand transfer activity to the defective INQ62A enzyme (Fig. 6B, lower panel).
The phenotype of E138K-containing virus was next analyzed. Since HIV-1Q62K was completely defective (Fig. 1B), E138K was tested in the stringent background of HIV-1Q62K rather than the HIV-1Q62A strain from which it originated. HIV-1Q62K/E138K was observed to replicate, although with an approximately 4-day delay from WT replication in repeat experiments (Fig. 6C; also data not shown). Thus, E138K is a bona fide second site suppressor mutation that can revert the deleterious effects of different changes (Ala or Lys) at Gln-62.
DISCUSSION
CCD DNA binding and HIV-1 replication.
Although results of in vitro photo-cross-linking (28, 40, 41, 43, 56), integration (33, 43, 67, 68), and in silico analyses (58) have implicated a variety of CCD residues as potentially important for DNA binding (Table 1), just some of these, including Gln-62 (26), Asn-117 (63), Asn-120 (48), Lys-136 (26, 72), Tyr-143 (63, 66, 72), Ser-153 (63), Lys-156 (43, 72), and Lys-159 (10, 43, 63, 72), were previously analyzed for their roles in HIV-1 replication. Whereas some strains, including HIV-1K159Q (63), HIV-1K156A (72), and HIV-1K136E (26), displayed WT replication profiles, other mutants, for example, HIV-1K159P (10), HIV-1K156E, HIV-1K159E (43), and HIV-1K136A (72), were either severely delayed (10) or completely defective (43, 72). To obtain a comprehensive view of the roles of 16 confirmed or proposed CCD DNA-binding residues in HIV-1 replication, 25 mutant viruses were generated (Table 1) and tested in side-by-side infections (Fig. 1). Several conclusions can be drawn from the results. In contrast to active-site mutations wherein a single amino acid substitution is sufficient to render HIV-1 replication defective, almost all singly substituted viral DNA-interacting mutants retained some capacity to replicate, with the lone exception of HIV-1Q62K (Fig. 1B to 1J). Since the other purported DNA binding residues that were tested by nonconservative substitution retained some replication capacity (Table 2), we infer that Gln-62 plays a particularly important role in viral DNA end recognition during integration. By analogy, His-67 (Fig. 1C), Asn-120 (Fig. 1E), Gln-148 (Fig. 1G), Lys-159 (Fig. 1I), and Asn-155 (Fig. 1J) also play important roles in HIV-1 replication. Whereas single missense mutants derived from these residues replicated to some degree, we note that double mutants HIV-1H67Q/K71E (Fig. 1C), HIV-1Q148K/N120L (Fig. 1G), and HIV-1K156E/K159E (Fig. 1I) were completely defective. In contrast, since HIV-1Y143G grew as did the WT (Fig. 1F) and HIV-1N117K showed just marginally delayed growth (Fig. 1D), we conclude that neither Asn-117 nor Tyr-143 played a significant role in HIV-1 replication under these conditions. We note that Tyr-143 was suggested to play a particularly important role during macrophage infection (66), a cell type that was not tested here. Similarly, we were unable to ascertain a role for Lys-136 in HIV-1 replication (Fig. 1H) (26). Since Wiskerchen and Muesing (72) previously reported that the K136A change rendered HIV-1R7-3 replication defective, Lys-136 could play a strain-dependent role in replication. The same may hold true for Asp-55, since we determined that NL4-3-based HIV-1D55A grew as did WT (Fig. 1A) yet R7-3-based HIV-1D55A was replication defective (72). Other aspects of tissue culture infections could also contribute to disparate results. For example, whereas CEM-12D7 cells infected at an approximate MOI of 10−3 failed to support replication of HIV-1K156E or HIV-1K159E over 20 days of observation (43), Jurkat cells infected at an MOI of 0.04 revealed HIV-1K156E and HIV-1K159E replication peaks at 12 and 16 dpi, respectively (Fig. 1I).
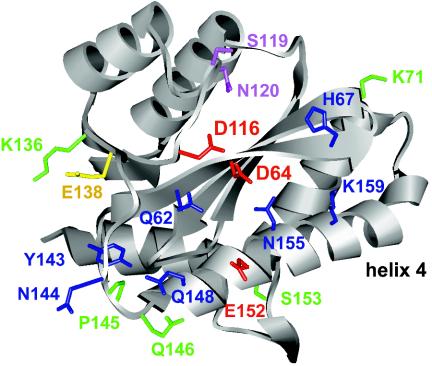
Our results help in understanding conserved residues and structural features of the CCD Phe139-Gln146 flexible loop and abutting Ser147-Val165 amphipathic alpha-helix, which are likely important for DNA binding during HIV-1 replication. In terms of the alpha-helix, previous in vitro work indicated that the polar face of the amphipathic structure interacted with viral DNA (76), and our work with mutant viruses supports these findings. For example, Gln-148, Glu-152, Asn-155, and Lys-159 reside on the same side of the helix (Fig. 7, upward face of helix 4 as drawn).
FIG. 7.
Structural summary of CCD residues important and dispensable for HIV-1 replication. The B monomer of the IN CCD dimer from reference 35 is shown (Protein Data Bank identifier 1BIS). The following color code was used: red, active-site residues (Asp-64, Asp-116, and Glu-152); blue, residues that cross-linked to viral DNA substrates (Tyr-143, Gln-148, and Lys-159; references 28 and 43) and/or when mutated yielded class I IN mutant viruses (see Fig. 1 and 2) (43); green, residues that when mutated yielded viruses displaying WT replication kinetics; yellow, Glu-138; magenta, residues that predictably interact with target DNA (38, 67). The amphipathic alpha-helix is labeled helix 4.
Since class I mutant HIV-1N155K (Fig. 1J) grew significantly slower than HIV-1K156E (Fig. 1I) and Asn-155 contacted 5CITEP in the cocrystal structure (36), we speculate that Asn-155 interacts with HIV-1 cDNA during integration. This conclusion is in agreement with the results of a molecular modeling study that proposed that Asn-155 might bind DNA (58). Ser-153, which like other helix 4 residues was well conserved among HIV-1 isolates (Table 1), instead jutted down from the underside of the helix (Fig. 7). Whereas Ser-153 was dispensable for HIV-1 replication, alterations of the conserved residues positioned atop helix 4 yielded class I phenotypic viruses (Table 2).
The Phe139-Gln146 flexible loop cross-linked to DNA substrates (40), and Tyr-143 was subsequently shown to cross-link to the adenosine at the 5′ end of the nontransferred U5 minus-strand (28). The other loop residues targeted here, Asn-144, Pro-145, and Gln-146, were similar to Tyr-143 in that they were well conserved among HIV-1 strains and divergent retroviruses (Table 1). However, out of these four highly conserved residues, only Asn-144 played a significant role in HIV-1 replication (Fig. 1D and F). Since HIV-1N144K synthesized more 2-LTR circles than the WT (Fig. 2B), our results indicate that Asn-144 plays a specific role in integration. However, it is unclear if this includes DNA binding. From the structure-based pattern of required residues, one can deduce an extended pocket that encompasses the approximate lower half of the molecule, as drawn in Fig. 7, to which viral DNA might bind. Asn-144 seems to jut away from this pocket. However, it should be noted that due to its flexibility, the loop is generally not observed in CCD structures, and the precise locations of loop side chains differ dramatically among the few structures where they are observed (7, 35, 37). Despite these limitations, Gln-146 may be dispensable for HIV-1 replication because it points down toward the underside of helix 4 (Fig. 7).
Although results of an in vitro footprinting assay indicated that DNA strongly protected Glu-138 from proteolysis (19), HIV-1E138K grew similarly to the WT, indicating that Glu-138 does not play an important role in HIV-1 replication under these conditions (Fig. 1E). Interestingly, E138K was identified as an intramolecular mutation that suppressed the adverse affects of Q62A and Q62K changes (Fig. 6). In the CCD structure, Glu-138 is positioned toward oppositely charged Lys-136, noticeably away from Gln-62 (Fig. 7). Because of this, we speculate that substituting Lys for Glu-138 would redirect the mutant side chain away from Lys-136, which might orient it toward Gln-62 and in doing so directly compensate for a loss of DNA binding incurred through Gln-62 substitutions.
The roles of other conserved CCD residues in HIV-1 replication.
Five residues, Gln-53, Asp-55, Lys-71, Asn-184, and Arg-199, that were neither implicated in DNA binding nor part of the Phe139-Gln146 flexible loop, were also targeted in this study. Although Gln-53 was invariant among HIV-1 strains (Table 1), HIV-1Q53K grew as did the WT (Fig. 1B), and thus, our assay system did not reveal a role for Gln-53 in HIV-1 replication. The same can be said for highly conserved Lys-71 (Table 1 and Fig. 1L). In contrast, HIV-1D55K, HIV-1N184D, HIV-1N184L, and HIV-1R199E were replication defective (Fig. 1A, K, and L). Since each of these mutants synthesized significantly less cDNA than the WT (Fig. 2 and data not shown) and HIV-1N184L was released less efficiently than the WT from HeLa cells (Fig. 3B), these viruses were typed as pleiotropic class II mutants. The results of Asp-55 mutagenesis are reminiscent of our previous findings with Phe-185 (26). In that paper we determined that HIV-1F185H replicated as did the WT and that HIV-1F185K, HIV-1F185A, and HIV-1F185L were replication-defective class II mutants (26; see reference 22 for a review). Somewhat surprisingly, HIV-1E152K was also a class II mutant (Fig. 2A and 3A; Table 2). Since HIV-1E152D and HIV-1E152Q were class I viruses (Table 1), these results demonstrate that different substitutions of the same residue can yield either the class I or class II mutant phenotype.
Functional organization of the HIV-1 IN CCD.
Vpr-IN complementation was used to probe the functional relationship between catalysis and viral and target DNA binding that occurs during integration. For this we needed viral mutants for each of these functions that supported relatively low levels of inherent infectivity. By characterizing a large collection of CCD mutants in assays for viral spread, HIV-1Q62K, HIV-1K156E/K159E, and HIV-1N120K were determined to meet these requirements. Since Vpr-INQ62K complemented HIV-1V165A.Luc(R−) (53) but failed to complement either HIV-1K156E/K159E.Luc(R−) or HIV-1D64N/D116N.Luc(R−), our results indicate that different DNA-binding residues, such as Gln-62 and Lys-159, originate from the same functional protomer and that the active CCD binds to the viral DNA end that it functionally integrates. This result enforces previous IN-DNA models, which invariably predicted that the active CCD also bound to the viral DNA that it processed and joined to target DNA (2, 14, 18, 31, 32, 41, 58, 59, 71). Despite this “cis” requirement for active-site and CCD viral DNA-binding functions to reside within the same protomer, we note that the CTD's viral DNA end binding function is donated by a separate IN molecule (14, 31).
HIV-1N120K was a class I mutant (Fig. 2A, lane 4) that processed significantly more of its 3′ ends than other class I mutants, including active-site mutant HIV-1D116N and viral DNA-binding mutants HIV-1Q62K and HIV-1K156E/K159E (Fig. 4). Based on these results and previous observations that recombinant IN proteins altered at Asn-120 or the adjacent Ser-119 displayed altered patterns of DNA strand transfer (38, 67), we inferred that HIV-1N120K was in part defective for target DNA binding in vivo. Since Vpr-INQ62K did not complement HIV-1N120K.Luc(R-) and Vpr-INN120K failed to appreciably complement HIV-1Q62K.Luc(R-), HIV-1K156E/K159E.Luc(R−),or HIV-1D64N/D116N.Luc(R−), our results indicate that the active protomer within the IN multimer that binds and processes viral DNA also binds the chromosomal DNA into which it integrates. Although we inferred from in vitro studies that Gln-62, Asn-120, and Lys-159 directly contact DNA, we have not extensively characterized biochemical properties of recombinant proteins and because of this cannot rule out that disruption of other IN functions (for example, multimerization) contributed to the class I mutant phenotypes described here.
In conclusion, our results prioritized the importance of a number of conserved CCD residues that were previously implicated in DNA binding for their roles in HIV-1 replication. Together with the results of Vpr-IN complementation, our results were used to refine the regions of the active CCD protomer that interact with viral and target DNA during integration (Fig. 7). Precise structural delineation of the active IN-DNA complex is expected to aid the design of inhibitors that block HIV-1 replication before the virus has the chance to integrate into chromosomal DNA.
Acknowledgments
We thank J. Kappes for his generation donation of plasmid DNA, M. Farzan for his help in constructing Fig. 7, and Y. Liu for her technical assistance during the early phase of this project.
This work was supported by National Institutes of Health grant AI39394. Core facilities were supported by a Center for AIDS Research grant (AI28691) and the Dana-Farber Cancer Institute/Harvard Cancer Center.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adesokan, A. A., V. A. Roberts, K. W. Lee, R. D. Lins, and J. M. Briggs. 2004. Prediction of HIV-1 integrase/viral DNA interactions in the catalytic domain by fast molecular docking. J. Med. Chem. 47:821-828. [DOI] [PubMed] [Google Scholar]
- 3.Ansari-Lari, M. L., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 4.Appa, R. S., C.-G. Shin, P. Lee, and S. A. Chow. 2001. Role of the non-specific DNA binding region and α helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 276:45848-45855. [DOI] [PubMed] [Google Scholar]
- 5.Bouyac-Bertoia, M., J. D. Dvorin, R. A. M. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 6.Brown, H. E. V., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73:9011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bujacz, G., J. Alexandratos, Z. L. Qing, C. Clement-Mella, and A. Wlodawer. 1996. The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant. FEBS Lett. 398:175-178. [DOI] [PubMed] [Google Scholar]
- 8.Bukovsky, A., and H. Göttlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon, P. M., W. Wilson, E. Byles, S. M. Kingsman, and A. J. Kingsman. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J. Virol. 68:4768-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H., and A. Engelman. 2000. Characterization of a replication-defective human immunodeficiency virus type 1 att site mutant that is blocked after the 3′ processing step of retroviral integration. J. Virol. 74:8188-8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H., and A. Engelman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., S.-Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 14.Chen, J. C.-H., J. Krucinski, L. J. W. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cherepanov, P., W. Pluymers, A. Claeys, P. Proost, E. De Clercq, and Z. Debyser. 2000. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 14:1389-1399. [DOI] [PubMed] [Google Scholar]
- 16.Craigie, R. 2002. Retroviral DNA integration, p. 613-630. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 17.Craigie, R., A. B. Hickman, and A. Engelman. 1995. Integrase, p. 53-71. In J. Karn (ed.), HIV: a practical approach, vol. 2. Oxford University Press, Oxford, United Kingdom.
- 18.De Luca, L., A. Pedretti, G. Vistoli, M. L. Barreca, L. Villa, P. Monforte, and A. Chimirri. 2003. Analysis of the full-length integrase-DNA complex by a modified approach for DNA docking. Biochem. Biophys. Res. Commun. 310:1083-1088. [DOI] [PubMed] [Google Scholar]
- 19.Dirac, A. M. G., and J. Kjems. 2001. Mapping DNA-binding sites of HIV-1 integrase by protein footprinting. Eur. J. Biochem. 268:743-751. [DOI] [PubMed] [Google Scholar]
- 20.Drelich, M., R. Wilhelm, and J. Mous. 1992. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology 188:459-468. [DOI] [PubMed] [Google Scholar]
- 21.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 22.Engelman, A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411-426. [DOI] [PubMed] [Google Scholar]
- 23.Engelman, A., F. D. Bushman, and R. Craigie. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman, A., Y. Liu, H. Chen, M. Farzan, and F. Dyda. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol. 71:3507-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Englund, G., T. S. Theodore, E. O. Freed, A. Engelman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral DNA end binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esposito, D., and R. Craigie. 1999. HIV integrase structure and function. Adv. Virus Res. 52:319-333. [DOI] [PubMed] [Google Scholar]
- 30.Fletcher, T. M., III, M. A. Soares, S. McPhearson, H. Hui, M. Wiskerchen, M. A. Muesing, G. M. Shaw, A. D. Leavitt, J. D. Boeke, and B. H. Hahn. 1997. Complementation of integrase function in HIV-1 virions. EMBO J. 16:5123-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao, K., S. L. Butler, and F. Bushman. 2001. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 20:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao, K., S. Wong, and F. Bushman. 2004. Metal binding by the D,DX35E motif of human immunodeficiency virus type 1 integrase: selective rescue of Cys substitutions by Mn2+ in vitro. J. Virol. 78:6715-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerton, J. L., S. Ohgi, M. Olsen, J. DeRisi, and P. O. Brown. 1998. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J. Virol. 72:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goff, S. P. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 35.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral design. Proc. Natl. Acad. Sci. USA 96:13040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenwald, J., V. Le, S. L. Butler, F. D. Bushman, and S. Choe. 1999. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry 38:8892-8898. [DOI] [PubMed] [Google Scholar]
- 38.Harper, A. L., L. M. Skinner, M. Sudol, and M. Katzman. 2001. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J. Virol. 75:7756-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 40.Heuer, T. S., and P. O. Brown. 1997. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry 36:10655-10665. [DOI] [PubMed] [Google Scholar]
- 41.Heuer, T. S., and P. O. Brown. 1998. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry 37:6667-6678. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and C-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo crosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, S., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuiken, C. L., B. Foley, E. Freed, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, and S. Wolinksy. 2002. HIV sequence compendium 2002. Report no. LA-UR 03-3564. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 46.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. G. Mack, and A. M. Skalka. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leavitt, A. D., L. Shiue, and H. E. Varmus. 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase function in vitro. J. Biol. Chem. 268:2113-2119. [PubMed] [Google Scholar]
- 50.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. USA 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Limón, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Limón, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu, R., A. Limón, E. Devroe, P. A. Silver, P. Cherepanov, and A. Engelman. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu, R., N. Nakajima, W. Hofmann, M. Benkirane, K. Teh-Jeang, J. Sodroski, and A. Engelman. 2004. Simian virus 40-based replication of catalytically inactive human immunodeficiency virus type 1 integrase mutants in nonpermissive T cells and monocyte-derived macrophages. J. Virol. 78:658-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masuda, T., V. Planelles, P. Krogstad, and I. S. Y. Chen. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazumder, A., N. Neamati, A. A. Pilon, S. Sunder, and Y. Pommier. 1996. Chemical trapping of ternary complexes of human immunodeficiency virus type 1 integrase, divalent metal, and DNA substrates containing an abasic site. J. Biol. Chem. 271:27330-27338. [DOI] [PubMed] [Google Scholar]
- 57.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perryman, A. L., and J. A. McCammon. 2002. Autodocking dinucleotides to the HIV-1 integrase core domain: exploring possible binding sites for viral and genomic DNA. J. Med. Chem. 45:5624-5627. [DOI] [PubMed] [Google Scholar]
- 59.Podtelezhnikov, A. A., K. Gao, F. D. Bushman, and J. A. McCammon. 2003. Modeling HIV-1 integrase complexes based on their hydrodynamic properties. Biopolymers 68:110-120. [DOI] [PubMed] [Google Scholar]
- 60.Poon, B., and I. S. Y. Chen. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Priet, S., J.-M. Navarro, G. Querat, and J. Sire. 2003. Reversion of the lethal phenotype of an HIV-1 integrase mutant virus by overexpression of the same integrase mutant protein. J. Biol. Chem. 278:20724-20730. [DOI] [PubMed] [Google Scholar]
- 62.Saenz, D. T., N. Loewen, M. Peretz, T. Whitwam, R. Barraza, K. G. Howell, J. M. Holmes, M. Good, and E. M. Poeschla. 2004. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J. Virol. 78:2906-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin, C.-G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taddeo, B., F. Carlini, P. Verani, and A. Engelman. 1996. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J. Virol. 70:8277-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taddeo, B., W. A. Haseltine, and C. M. Farnet. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J. Virol. 68:8401-8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Gent, D. C., A. A. M. O. Groeneger, and R. H. A. Plasterk. 1992. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl. Acad. Sci. USA 89:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Gent, D. C., A. A. M. O. Groeneger, and R. H. A. Plasterk. 1993. Identification of amino acids in HIV-1 integrase involved in site-specific hydrolysis and alcoholysis of viral DNA termini. Nucleic Acids Res. 21:3373-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Gent, D. C., C. Vink, A. A. M. O. Groeneger, and R. H. A. Plasterk. 1993. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 12:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vink, C., A. A. M. O. Groeneger, and R. H. A. Plasterk. 1993. Identification of the catalytic and DNA-binding region of human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 21:1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, J.-Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 75.Wu, Y., and J. W. Marsh. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zargarian, L., M. S. Benleumi, J. G. Renisio, H. Merad, R. G. Maroun, F. Wieber, O. Mauffret, H. Porumb, F. Troalen, and S. Fermandjian. 2003. Strategy to discriminate between high and low affinity bindings of human immunodeficiency virus, type 1 integrase to viral DNA. J. Biol. Chem. 278:19966-19973. [DOI] [PubMed] [Google Scholar]