Abstract
Triple-negative breast cancers (TNBCs) are defined as tumors that are negative for estrogen, progesterone and HER-2 receptor. At a percentage of 10–20% TNBCs represent a minority in all breast cancers. However, because of the poor prognosis this particular subtype, triple negative disease accounts for a disproportionate number of metastatic cases and breast cancer deaths. Identification of its subtypes is essential for understanding the biological characteristics and clinical behavior of TNBC, as well as for developing personalized treatments. This review will focus on the great progress that has been made in the past few years on identifying new targets in TNBC subtypes and a variety of new treatment options that are on the verge of routine clinical application.
Key Words: Triple-negative, Breast cancer, Subtypes, Biological activity
Introduction
Triple-negative breast cancers (TNBCs) represent a heterogeneous breast cancer subtype with a poor prognosis. These cancers are defined by the absence of estrogen receptors (ER) and progesterone receptors (PR), and the absence of HER2 overexpression. The definition of negative ER status by immunohistochemistry (IHC) is not concordant in the literature, with some definitions considering ER expression to be significant only if at least 10% of tumor cells express the receptors. However, the St. Gallen guidelines [1], the American Society of Clinical Oncology [2], and the American College of Pathology [3] have defined TNBC as breast cancer with less than 1% of tumor cells expressing ER and PR via IHC. It is widely accepted that TNBC is a very inhomogeneous group. This diversity is further highlighted by the high prevalence of rare histopathological subtypes, such as metaplastic (90%), medullary (95%), adenoid cystic (90–100%), and apocrine (40–60%) carcinomas. Some common markers have been identified, such as basal cytokeratin (CK) 5/6 and epidermal growth factor receptor (EGFR) [4]. On the other hand, the molecular differentiation shows also a wide range of heterogeneity. New studies have now refined the understanding of TNBCs.
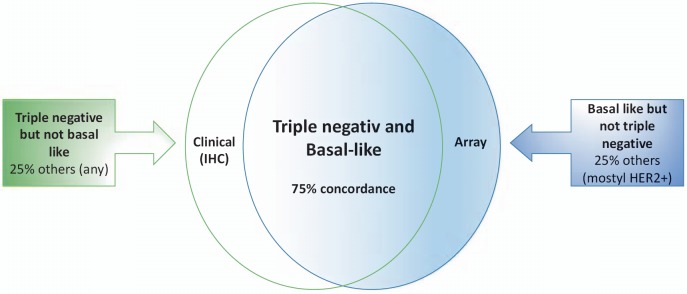
The basal-like subtype was discovered more than a decade ago by first-generation cDNA microarrays [5]. These tumors are often referred to as TNBCs because most basal-like tumors are typically negative for ER, PR, and HER2. This subtype of breast cancer is characterized by a gene expression profile that is similar to that of the basal-myoepithelial layer of the normal breast [5]. However, approximately 75% of TNBCs are basal-like, with the other 25% comprising all other mRNA subtypes [6]. These subtypes include mostly HER2+ breast cancer (fig. 1). 25% of all TNBCs lack ER, PR, and HER2 in IHC but do not exhibit the features of the basal-like subtype. The Cancer Genome Atlas (TCGA) Research Network analyzed primary breast cancers using 6 platforms, including genomic DNA copy number arrays, DNA methylation, exome sequencing, messenger RNA arrays, microRNA sequencing, and reverse phase protein arrays [7]. By integrating information across platforms, the authors were able to examine the genomic heterogeneity of tumors. The TCGA analysis revealed that the most frequent loss-of-function and gain-of-function alterations in TNBC involve genes associated with DNA damage repair and phosphatidylinositol 3-kinase (PI3K) signaling pathways, respectively. Alterations in DNA damage repair genes include loss of TP53, RB1, and BRCA1 function. Aberrant activation of the PI3K pathway occurs due to loss of negative regulators such as the lipid phosphatases PTEN or INPP4B [8], [9] or activating mutations in PIK3CA, along with other genes in the PI3K/TOR signaling network [10], [11]. Another study sequenced and analyzed 104 TNBC tumors at the time of diagnosis and confirmed the high rate of TP53 mutations; however, they showed that 12% of cases did not have somatic mutations in any established ‘driver’ genes, suggesting that primary TNBCs are mutationally heterogeneous from the outset [12].
Fig. 1.
Microarray analysis and immunohistochemistry show 75% concordance for basal-like and triple-negative breast cancer (TNBC), respectively. Approximately 25% of TNBCs are not basal-like on gene expression array. Similarly, there are basal-like breast cancers that are not triple-negative, which also represent approximately 25% of cases. Therefore, in clinical trials looking at basal-like biology and using the triple-negative phenotype to identify patients, the potential exists for misclassification.
BRCA1-Associated TNBC
Patients with a BRCA1 mutation develop tumors with many similarities to basal-like sporadic breast tumors, including greater likelihood of being high-grade, ER/PR-negative, HER2-negative, and of having a high frequency of TP53 mutations. Basal keratins are expressed by both sporadic basal-like tumors and tumors with BRCA1 mutations, and both groups cluster together in gene expression profiling [13]. BRCA1 breast cancers share features with a subset of sporadic tumor, indicating a similar etiology. Hallmarks of this ‘BRCAness’ include basal-like phenotype (associated with the BRCA1 phenotype but not with the BRCA2 phenotype), ER-negativity, EGFR expression, c-MYC amplification, TP53 mutations, loss of RAD51 focus formation, extreme genomic instability, and sensitivity to DNA-crosslinking agents [14]. Apart from germline or somatic BRCA1 mutations, BRCA1 hypermethylation and/or loss of heterozygosity may give rise to a BRCA1-like molecular profile in wild-type TNBC [15,16]. DNA damage response is the cellular reaction to exogenous and endogenous genotoxic injuries that may produce DNA single-strand breaks and DNA double-strand breaks. If the repair process is not executed correctly, the DNA injuries result in mutations and chromosomal aberrations which alter the cellular behavior and lead to cancer and tumor progression independent of BRCA mutational status. Frequent loss of several other genes involved in BRCA1-dependent homologous recombination (HR) repair has been demonstrated in basal-like/triple-negative cancer, most likely contributing to BRCA1-like features [17]. Due to innovative treatment options, information about the BRCA1-like or ‘BRCAness’ status may have important clinical implications: A number of studies have shown that homologous recombination deficiency (HRD) sensitizes the tumor to DNA-damaging agents such as platinum compounds, or to poly(ADP-ribose)polymerase (PARP) inhibitors, or their combination [18,19,20]. Accordingly, biomarkers to identify and select patients with BRCA1-like (‘BRCAness’) signatures are urgently required. Identification of patients with tumors deficient in homologous repair or HRD-like behavior moves cancer treatment towards individualized therapies.
Tumor-Infiltrating Lymphocytes in TNBC
Approximately 20% of TNBCs classify as immunomodulatory and are highly enriched in immune cell makers and signaling. Tumors that have more than 50% lymphocytic infiltrate are considered lymphocyte-predominant breast cancer and have the best prognosis [21]. In TNBC, without treatment, the presence of tumor-infiltrating lymphocytes (TILs) is correlated with improved overall survival, increased metastasis-free survival, and decreased distant recurrence [22]. TILs can predict improved pathological complete response (pCR) to neoadjuvant chemotherapy [23]. Additionally, presence of TILs in residual TNBC after neoadjuvant chemotherapy is also prognostic for better metastases-free and overall survival [24]. TILs emerge as a robust prognostic biomarker of the host antitumor immune response in ‘immunogenic’ breast cancer subtypes, especially TNBC. In TNBC, the increase in immune infiltrate with high levels of TILs predicts not only response to chemotherapy but also, as a consequence, better survival [25].
Molecular Subtyping of TNBC
The first attempt to molecularly distinguish TNBC subtypes was a direct comparison of 374 TNBC samples extracted from 14 datasets where investigators sought to determine the relationship between the PAM50 intrinsic and TNBC molecular subtypes. The majority of the TNBC samples were indeed classified as basal-like (80.6%) followed by HER2 (0.2%), normal-like (14.6%), luminal B (3.5%), and luminal A (1.1%) by PAM50 [26].
Using gene expression analyses, distinct TNBC subtypes have been recently identified, each displaying a unique biology. In this pivotal study, Lehman et al. [27] analyzed gene expression profiles from 21 breast cancer data sets with a total of 3,247 breast cancers and identified 587 TNBC cases representing 18% of all breast cancers. The 6 TNBC subtypes in this study included 2 basal-like (BL1 and BL2), 1 immunomodulatory (IM), 1 mesenchymal (M), 1 mesenchymal stem-like (MSL), and 1 luminal androgen receptor (LAR) subtype, the last being characterized by androgen receptor (AR) signaling. Masuda et al. [28] confirmed the classification of Lehmann et al. [27] in an independent analysis, and they classified TNBC with high correlation into 7 subtypes (BL1, BL2, M, IM, MSN, LAR) including 1 unstable subtype (UNS).
From the progress in TNBC subtyping evolved an increasing need to identify clinically relevant subtypes. Using RNA and DNA profiling, Burstein et al. [29] was able to distinguish 4 stable molecularly defined TNBC subtypes: luminal-AR (LAR), mesenchymal (MES), basal-like immune-suppressed (BLIS), and basal-like immune-activated (BLIA). MES, BLIS, and BLIA are characterized by distinct clinical prognoses, with BLIS tumors having the worst and BLIA tumors having the best outcome. DNA analysis demonstrated subtype-specific gene amplifications, suggesting the possibility of using in situ hybridization techniques to identify these TNBC subsets. These results also demonstrated subtype-specific molecular expression, thereby enabling TNBC subtype classification based on molecules that are being expressed as opposed to molecules that are not being expressed.
Due to the complexity of the varying histological landscape of tumor specimens, a very recent study by Lehman et al. [30] used histopathological quantification and laser-capture microdissection to determine that transcripts in the previously described IM and MSL subtypes were contributed from infiltrating lymphocytes and tumor-associated stromal cells, respectively. Therefore, the new classification takes into account the contribution of transcripts from normal stromal and immune cells in the tumor environment and has refined TNBC molecular subtypes from 6 (TNBCtype) into 4 (TNBCtype-4) tumor-specific subtypes (BL1, BL2, M, and LAR).
Very recently, an expression algorithm reduced to 101 genes was describedwith the power to subtype TNBC tumors similarly to the original 2,188-gene expression algorithm and predict patient outcomes [31].
Distinct Subtypes According to Molecular Profiling
Basal-Like Subtypes (BL1 and BL2)
The BL1 subtype (table 1) is characterized by enrichment of cell cycle and cell division components and pathways. The proliferation pathways in the BL1 subtype are accompanied by elevated DNA damage response (ATR/BRCA) pathways. The proliferative nature of this subtype was further supported by the observation of high Ki67 mRNA expression and nuclear Ki67 staining by ICH (> 70%). The BL2 subtype (table 1) involves growth factor signaling pathways (EGF, NGF, MET, Wnt/β-catenin, and IGF1R) as well as glycolysis and gluconeogenesis. Likewise, the BL2 subtype is uniquely enriched in growth factor receptors such as EGFR, MET, and EPHA2 [27].
Table 1.
Subtypes of triple negative breast cancer (TNBC) based on analysis of gene expression profiles
| TNBCtype | TNBCtype-4 | Subtypes of TNBC |
|---|---|---|
| (Lehman et al. [27]) | (Lehman et al. [30]) | (Burstein et al. [29]) |
| Basal-like 1 (BL1) | basal-like1 (BL1) | basal-like immune-suppressed (BLIS) |
| Basal-like 2 (BL2) | basal-like 2 (BL2) | |
| Immunomodulatory (IM) | basal-like immune-activated (BLIA) | |
| Mesenchymal (M) | mesenchymal (M) | mesenchymal (MES) |
| Mesenchymal stem-like (MSL) | ||
| Luminal androgen receptor (LAR) | luminal androgen receptor (LAR) | luminal androgen receptor (LAR) |
Burstein et al. [29] subclassified the basal-like subtype also in 2 basal-like clusters. The BLIS subtype exhibits downregulation of B cell, T cell, and natural killer cell immune-regulating pathways and cytokine pathways. This subtype has the worst prognosis and low expression of molecules controlling antigen presentation, immune cell differentiation, and innate and adaptive immune cell communication. However, this cluster uniquely expresses multiple SOX family transcription factors. The second basal-like subtype in their analysis is similar to the immunomodulatory subtype (IM) described by Lehmann et al. [27].
Nearly all of the cell lines with known mutations in BRCA1 and BRCA2 have gene expression patterns that correlate with the basal-like subtype, which is in agreement with the current view that BRCA-mutant tumors display a basal-like phenotype [32].
Immunomodulatory Subtype
The IM subtype (table 1) is enriched for factors involved in immune cell processes. These processes include immune cell signaling. Immune signaling genes within the IM subtype substantially overlap with a gene signature for medullary breast cancer, a rare, histologically distinct form of TNBC that despite its high-grade histology is associated with a favorable prognosis. In the classification by Burstein et al. [29], the other basal-like subtype, BLIA, exhibits an upregulation of immune regulation pathways. Contrary to BLIS, tumors of this subtype display upregulation of genes controlling B cell, T cell, and natural killer cell functions. This subtype has the best prognosis, exhibits activation of STAT transcription factor-mediated pathways, and has high expression of STAT genes [29].
Mesenchymal and Mesenchymal Stem-Like Subtypes
The M subtype (table 1) is heavily enriched in components and pathways involved in cell motility, extracellular receptor interaction, and cell differentiation pathways. The MSL subtype (table 1) shares enrichment of genes for similar biological processes with the M subtype, including cell motility, cellular differentiation, and growth pathways. However, unique to the MSL are genes representing components and processes linked to growth factor signaling pathways that include inositol phosphate metabolism, EGFR, PDGF, calcium signaling, G-protein coupled receptor, and ERK1/2 signaling as well as ABC transporter and adipocytokine signaling. Additionally, Burstein et al. [29] showed in their analysis that this subtype expresses genes normally exclusive to osteocytes (OGN) and adipocytes (ADIPOQ, PLIN1), and important growth factors (IGF-1) are also highly expressed in this type of TNBC.
Luminal Androgen Receptor Subtype
The LAR subtype (table 1) is the most differential among the TNBC subtypes. This subtype is ER-negative, but gene ontologies are heavily enriched in hormonally regulated pathways including steroid synthesis, porphyrin metabolism, and androgen/estrogen metabolism. Tumors exhibit AR, ER, prolactin, and ErbB4 signaling, but ERα-negative IHC staining. Gene expression profiling demonstrates expression of ESR1 (the gene encoding ERα), and other estrogen-regulated genes (PGR, FOXA, XBP1, GATA3). Thus, these ‘ER-negative’ tumors demonstrate molecular evidence of ER activation. This may be because 1% of these tumor cells express low levels of ER protein, defining them as ‘ER-negative’ by IHC analysis [29]. AR mRNA is highly expressed, on average at 9-fold greater than all other subtypes. Tumors within the LAR group also expressed numerous downstream AR targets and coactivators. AR expression by IHC in the LAR subtype is more than 10-fold higher compared to other TNBC subtypes. All these observations suggest that the LAR TNBC subtype is composed of AR-driven tumors [27].
Impact of Different Subtypes on Response to Chemotherapy and Prognosis
Various studies have analyzed the response to neoadjuvant chemotherapy according to molecular subtypes of TNBC. In a retrospective analysis, it was shown for the first time that the TNBC subtype can serve as an independent predictor of pCR status in patients who receive current standard chemotherapy regimens [28]. 130 TNBC cases treated with neoadjuvant anthracyclines and/or taxanes were retrospectively classified into subtypes as follows: BL1, 21 patients; BL2, 8 patients; M, 26 patients; IM, 27 patients; MSL, 13 patients; LAR, 20 patients; and UNS, 15 patients. There was no statistically significant difference in treatment regimens between subtypes. The overall pCR response was 28%, and interestingly, subtype-specific responses differed substantially. The BL1 subtype achieved the highest pCR rate (52%), and the BL2, LAR, and MSL subtypes were found to have the lowest response rates (0, 10, and 23%, respectively).
The most recent paper investigated the response rate according to TNBC subtypes classified by either PAM50 or TNBCtype-4 [30]. PAM50 subtyping of tumors in to basal and non-basal subtypes did not result in significant differences in pCR. TNBCtype-4 subtyping did not result in significant differences in pCR for TNBC patients treated with neoadjuvant chemotherapy; the pCR incidence for the subtypes displayed shows similar trends to previous studies, with BL1 displaying the greatest response and BL2 and LAR showing lower pCR. However, compared to all other subtypes, BL1 patients had significantly higher pCR (49 vs. 31%; p = 0.0441).
Distant relapse-free survival (DRFS) was evaluated in the same cohort to determine if chemotherapy responses to neoadjuvant AC-T (anthracycline-cyclophosphamide and taxane) resulted in differences in survival within PAM50 and TNBCtype-4 subtypes. Despite having better pCR to neoadjuvant chemotherapy (34 vs. 11%), TNBC patients had significantly worse DRFS compared to non-TNBC. However, TNBC patients that responded to chemotherapy and achieved a pCR clearly had a far better DRFS compared to those patients that did not, with 95% of patients surviving 7 years after treatment compared to a median survival of 2.7 years. While there were no differences in DRFS between basal and non-basal PAM50 subtypes (p = 0.41), stratification by TNBCtype-4 trended towards significance (p = 0.09), with BL2 patients displaying the worst outcome with a median survival of 2.4 years compared to a median survival for unselected TNBC of greater than 7 years. In contrast, the BL1 subtype displayed the highest pCR (49%) and also the best long-term DRFS with 72% of patients relapse-free at 7-year follow-up.
Treatment Options in Different Subtypes
Basal-Like Subtype: Genomic Instability as Therapeutic Target
Among the breast tumors, the triple-negative type is characterized by some form of genomic instability easily identified by alterations in chromosome number and structure. Cells have evolved a complex DNA damage response (DDR) to maintain genomic integrity. The most deleterious lesion, double-strand breaks, are repaired by either HR (homologous recombination) or with non-homologous end joining. A subset of familial and sporadic breast, ovarian, and pancreatic cancers are deficient in HR repair, resulting from mutations in the breast cancer susceptibility proteins BRCA1 and BRCA2 [14]. The observation that a majority of BRCA1 mutation carriers develop TNBC rather than other forms of breast cancer led investigators to evaluate neoadjuvant platinum compounds in unselected TNBC. In a trial of neoadjuvant platinum, gemcitabine, and iniparib, HR deficiency predicted residual cancer burden scores of 0 or I (RCB 0/I) and pCR. HR deficiency remained a significant predictor of RCB 0/I when adjusted for clinical variables [33]. In 2 other trials of neoadjuvant cisplatin therapy, HR deficiency predicted RCB 0/I and pCR. In a multivariable model of RCB 0/I, HR deficiency retained significance when clinical variables were included. When restricted to BRCA1/2 non-mutated tumors, response was higher in patients with high HRD scores: RCB 0/I p = 0.062, pCR p = 0.063 in the neoadjuvant platinum, gemcitabine, and iniparib trial; RCB 0/I p = 0.0039, pCR p = 0.018 in the neoadjuvant cisplatin trials. This study showed an overall pCR of 22% and associated both BRCA-mutated tumors and those with decreased BRCA1 expression with cisplatin sensitivity [33]. Further evidence of the activity of platinum agents in TNBC comes from the GeparSixto trial that compared neoadjuvant paclitaxel, anthracycline, and bevacizumab +/− carboplatin, and found that the addition of carboplatin improved pCR from 38 to 59% [34].
PARP facilitates cell recovery from DNA damage. BRCA can also repair double-strand DNA breakage. PARP inhibitors may be particularly useful in BRCA-mutated or HR-deficient breast cancer. Various early-phase clinical trials have assessed the activity of PARP inhibitors in cohorts of patients with TNBC. Most responses have been seen in patients with BRCA germline mutations, with often significant and long-lasting clinical benefit. Several PARP inhibitors are currently being investigated in clinical trials. Olaparib, veliparib, and rucaparib are showing considerable potential. Olaparib is the most extensively studied in breast cancer to date. It is currently approved for recurrent BRCA-mutant ovarian cancer. A phase I study of olaparib for human subjects was initiated 2005. Since then, a number of phase II trials have been conducted. Tutt et al. [35] reported the results of a trial which enrolled 54 patients with germline BRCA mutations and metastatic breast cancer previously treated with a median of 3 prior chemotherapy lines. 2 dose cohorts were studied, and 29 of those enrolled had TNBC. The overall response rate (ORR) was 41% in the cohort assigned to 400 mg olaparib twice daily. However, responses were not limited to TNBC. Also patients with ER-positive and HER2-positive cancers benefited equally from the treatment. The authors concluded that responsiveness to PARP inhibition in breast cancer was not a feature of TNBC but of BRCA-mutated breast cancer.
A recently published trial with nearly 300 patients with a variety of BRCA-mutated cancers included 62 patients with breast cancer [36]. 12.9% of these BRCA-mutated breast cancer patients responded to the treatment with olaparib, and 47% showed disease stabilization. The relatively low response rate in this study compared to the data from Tutt et al. [35] might have been due to previous exposure to platinum compounds in 47.6% of the patients. Currently, a large number of clinical trials are investigating olaparib as monotherapy or in combination with chemotherapy or other targeted agents.
Veliparib has also been extensively investigated in breast cancer, although almost entirely in combination with chemotherapy. The agent was mostly used as chemosensitizer to induce synthetic lethality in the setting of BRCA mutation. The combination of veliparib with the alkylating agent temozolomide in a xenograft model utilizing a variety of tumors including breast cancer showed a significant anti-metastatic effect, even in tumors resistant to temozolomide alone [37]. In the clinical setting, the combination of veliparib and temozolomide was tested in 41 women with advanced TNBC in a single-arm phase II study. In the subgroup with BRCA mutations, the ORR and clinical benefit rate were 37.5 and 62.5%, respectively [38]. However, the most promising combination in TNBC is veliparib and carboplatinum. In the I-SPY 2 trial, the PARP inhibitor and the platinum compound had an 88% predicted probability of success in a phase III trial in terms of pCR when added to standard neoadjuvant therapy [39].
Recent studies have shown that rucaparib effectively treats patients with platinum-sensitive, relapsed, high-grade ovarian cancer harboring a BRCA mutation. A phase II study published in 2016 showed also considerable activity in BRCA-mutated TNBC. 44% of the breast cancer patients harboring a BRCA mutation experienced a 12-week progression-free survival (PFS) period under rucaparib monotherapy [40]. In April 2015, rucaparib received a U.S. Food and Drug Administration (FDA) Breakthrough Therapy designation.
Targeting the LAR Subtype
The LAR subtype is characterized by a luminal gene expression profile and is enriched for expression of the AR an its gene targets. In addition to AR dependency, all LAR TNBC cell lines analyzed harbor an activating mutation in the kinase domain of PIK3CA (H1047R) and display greater sensitivity to PIK3CA inhibitors versus models of other subtypes [27]. In TNBC, the prevalence of AR positivity is approximately 10–20% [41]. The same observation was made for BRCA1-mutated TNBCs [42]. Antiandrogens have been studied for advanced AR-positive breast cancer. The antiandrogen enzalutamide has shown activity in a subset of women with advanced TNBC whose tumors express the AR. Gucalp et al. [43] reported from a phase II trial that the AR was expressed in 12% of TNBC. The 6-month clinical benefit rate was 19% for bicalutamide, and the median PFS appeared to be 12 weeks. Another phase II single-arm trial was presented in 2015 at the annual ASCO meeting [44]. In the largest trial so far with 118 women with AR-positive TNBC, more than 50% received enzalutamide as either first- or second-line therapy for metastatic disease. Patients were treated with 160 mg enzalutamide daily until disease progression. The trial met its primary endpoint of clinical benefit at 16 weeks of therapy. Of the 75 patients who could be evaluated, 35% achieved a clinical benefit. There were 2 complete responses and 7 partial responses. The clinical benefit rate at ≥ 24 weeks was 29%. Median PFS was 14.7 weeks. The 47% of study patients whose tumors stained positive for AR expression and who had an AR-related gene signature had better outcomes, including improved overall survival, compared with those whose tumors were AR-positive but did not have the gene signature. Patients who were positive for this gene signature had a median PFS of 16 weeks compared with 8 weeks in patients whose tumors lacked the gene signature. However, AR by IHC is not perfect for predicting who is going to respond. The authors also found that even those tumors with really low AR expression levels had excellent responses. The combination of AR expression with the gene signature will allow to enrich the population that appears to truly benefit from enzalutamide. The most common drug-related adverse events were fatigue (34%), nausea (25%), decreased appetite (13%), diarrhea (10%), and hot flushes (10%). The only high-grade adverse event that occurred in at least 5% of patients was fatigue (5%).
Immune Checkpoint Inhibitors for the IM Subtype
The description of an immunomodulatory subtype of TNBC characterized by elevated expression of genes involved in antigen processing and T cell functions suggests a likely option for immunotherapy against this disease. The key targets of cancer immunotherapy include immune checkpoint inhibitors or antagonists, such as immune checkpoint antagonists of the PD-1/PD-L1 axis and CTLA4. In breast cancer, the reported rates of PD-L1 in carcinoma cells vary between studies due to differences in sample size, sampling format (e.g., tissue microarray vs. whole section), and the method used to detect PD-L1 expression [45, 46]. PD-L1 protein expression has been observed in 15.8–30% in studies, and in situ mRNA hybridization was detected in PDL1 mRNA in 55–60% of tissue microarrays of primary breast cancers [47]. PD-L1 expression in tumor cells or its presence in the tumor microenvironment has been correlated with high levels of TILs, and it has been positively associated with triple-negative status in breast cancer [48]. Furthermore, high PD-L1 and TILs have also been correlated with pCR after neoadjuvant chemotherapy and with improved clinical outcomes in TNBC [48]. The potential value of PD-L1 could be its use as a target for PD-L1 axis-directed therapies, but PD-L1 expression status in tumor tissues does not seem to be an appropriate predictive biomarker to select patients for treatment with either anti-PD-1 or anti-PD-L1 antibodies, because PD-L1 is a dynamic marker that changes over time [49]. As opposed to mutated genes in cancers that permanently mark a tumor, the immune response is dynamic and changes rapidly. Moreover, in a phase I trial, approximately 10% of patients were deemed to be PD-L1-negative but did have clinical responses to anti PD-L1 therapy [50].
Pembrolizumab, a monoclonal anti PD-1, was tested in a phase Ib clinical trial in metastatic PD-L1-positive TNBC. It showed a preliminary ORR of 18.5% in heavily pretreated TNBC [51]. The safety profile of the antibody was very acceptable.
Another anti-PD-L1 antibody, atezolizumab or MPDL3280A, has also shown promising activity in TNBC. In a phase I study with atezolizumab, 21 evaluable patients with metastatic PD-L1-positive TNBC (37 PD-L1-positive from 54 initially tested patients) achieved an ORR according to RECIST of 19% [50]. 3 patients with PD-L1-positive TNBC experienced pseudo-progression, continued treatment, and finally demonstrated responses.
Preliminary results of a combination of atezolizumab plus nab-paclitaxel chemotherapy in metastatic TNBC (mTNBC) have been reported [52], and a phase III trial with this combination as first-line therapy in untreated mTNBC has been opened. Multiple additional immune checkpoint receptors and their ligands are prime targets for blockade, such as CTLA-4 (cytotoxic T lymphocyte antigen-4). The monoclonal antibody tremelimumab, which inhibits the CTLA-4 pathway, was evaluated in hormone-positive breast cancer and has shown certain activity [53].
Conclusion
TNBC continuous to be a heterogeneous disease. The identification of several specific subtypes characterized by different biologic pathways and various sensitivities to chemotherapy is instrumental in delivering a more personalized therapy for TNBC.
Disclosure Statement
None of the authors declare any conflict of interest.
References
- 1.Gnant M, Thomssen C, Harbeck N. St. Gallen/Vienna 2015: a brief summary of the consensus discussion. Breast Care (Basel) 2015;10:124–130. doi: 10.1159/000430488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denduluri N, Somerfield MR, Eisen A, et al. Selection of optimal adjuvant chemotherapy regimens for human epidermal growth factor receptor 2 (HER2)-negative and adjuvant targeted therapy for HER2-positive breast cancers: an American Society of Clinical Oncology guideline adaptation of the Cancer Care Ontario Clinical Practice Guideline. J Clin Oncol. 2016;34:2416–2427. doi: 10.1200/JCO.2016.67.0182. [DOI] [PubMed] [Google Scholar]
- 3.Wolff AC, Hammond MEH, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 4.Rakha EA, El-Sayed ME, Green AR, et al. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 9.Gewinner C, Wang ZC, Richardson A, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 11.Abramson VG, Lehmann BD, Ballinger TJ, Pietenpol JA. Subtyping of triple-negative breast cancer: implications for therapy. Cancer. 2015;121:8–16. doi: 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matros E, Wang ZC, Lodeiro G, et al. BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res Treat. 2005;91:179–186. doi: 10.1007/s10549-004-7603-8. [DOI] [PubMed] [Google Scholar]
- 14.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 16.Wei M, Grushko TA, Dignam J, et al. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 2005;65:10692–10699. doi: 10.1158/0008-5472.CAN-05-1277. [DOI] [PubMed] [Google Scholar]
- 17.Weigman VJ, Chao H-H, Shabalin AA, et al. Basal-like breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res Treat. 2012;133:865–880. doi: 10.1007/s10549-011-1846-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vollebergh MA, Lips EH, Nederlof PM, et al. An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann Oncol. 2011;22:1561–1570. doi: 10.1093/annonc/mdq624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerrato A, Morra F, Celetti A. Use of poly ADP-ribose polymerase (PARP) inhibitors in cancer cells bearing DDR defects: the rationale for their inclusion in the clinic. J Exp Clin Cancer Res. 2016;35:179. doi: 10.1186/s13046-016-0456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muvarak NE, Chowdhury K, Xia L, et al. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents – a potential therapy for cancer. Cancer Cell. 2016;30:637–650. doi: 10.1016/j.ccell.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani H, Mori-Shiraishi K, Nakajima M, Ueki H. Defining lymphocyte-predominant breast cancer by the proportion of lymphocyte-rich stroma and its significance in routine histopathological diagnosis. Pathol Int. 2015;65:644–651. doi: 10.1111/pin.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanton SE, Adams S, Disis ML. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: a systematic review. JAMA Oncol. 2016;2:1354–1360. doi: 10.1001/jamaoncol.2016.1061. [DOI] [PubMed] [Google Scholar]
- 23.Mao Y, Qu Q, Chen X, et al. The prognostic value of tumor-infiltrating lymphocytes in breast cancer: a systematic review and meta-analysis. PLoS ONE. 2016;11:e0152500. doi: 10.1371/journal.pone.0152500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Martínez E, Gil GL, Benito AC, et al. Tumor-infiltrating immune cell profiles and their change after neoadjuvant chemotherapy predict response and prognosis of breast cancer. Breast Cancer Res. 2014;16:488. doi: 10.1186/s13058-014-0488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Teijido P, Cabal ML, Fernández IP, Pérez YF. Tumor-infiltrating lymphocytes in triple negative breast cancer: the future of immune targeting. Clin Med Insights Oncol. 2016;10:31–39. doi: 10.4137/CMO.S34540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple negative breast cancer subtypes. J Pathol. 2014;232:142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda H, Baggerly KA, Wang Y, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19:5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann BD, Jovanović B, Chen X, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS ONE. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ring BZ, Hout DR, Morris SW, et al. Generation of an algorithm based on minimal gene sets to clinically subtype triple negative breast cancer patients. BMC Cancer. 2016;16:143. doi: 10.1186/s12885-016-2198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefansson OA, Jonasson JG, Johannsson OT, et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11:R47. doi: 10.1186/bcr2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telli ML, Timms KM, Reid J, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Minckwitz G, Schneeweiss A, Loibl S, et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol. 2014;15:747–756. doi: 10.1016/S1470-2045(14)70160-3. [DOI] [PubMed] [Google Scholar]
- 35.Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palma JP, Wang Y-C, Rodriguez LE, et al. ABT-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res. 2009;15:7277–7290. doi: 10.1158/1078-0432.CCR-09-1245. [DOI] [PubMed] [Google Scholar]
- 38.Isakoff SJ, Overmoyer B, Tung NM, et al. A phase II trial of the PARP inhibitor veliparib (ABT888) and temozolomide for metastatic breast cancer. J Clin Oncol. 2010;28((suppl)) abstr 1019; meetinglibrary.asco.org/content/43191-74. [Google Scholar]
- 39.Rugo HS, Olopade OI, DeMichele A, et al. Adaptive randomization of veliparib-carboplatin treatment in breast cancer. N Engl J Med. 2016;375:23–34. doi: 10.1056/NEJMoa1513749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drew Y, Ledermann J, Hall G, et al. Phase 2 multicentre trial investigating intermittent and continuous dosing schedules of the poly(ADP-ribose) polymerase inhibitor rucaparib in germline BRCA mutation carriers with advanced ovarian and breast cancer. Br J Cancer. 2016;114:723–730. doi: 10.1038/bjc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niemeier LA, Dabbs DJ, Beriwal S, et al. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol. 2010;23:205–212. doi: 10.1038/modpathol.2009.159. [DOI] [PubMed] [Google Scholar]
- 42.Pristauz G, Petru E, Stacher E, et al. Androgen receptor expression in breast cancer patients tested for BRCA1 and BRCA2 mutations. Histopathology. 2010;57:877–884. doi: 10.1111/j.1365-2559.2010.03724.x. [DOI] [PubMed] [Google Scholar]
- 43.Gucalp A, Tolaney S, Isakoff SJ, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic breast cancer. Clin Cancer Res. 2013;19:5505–5512. doi: 10.1158/1078-0432.CCR-12-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traina TA, Miller K, Yardley DA, et al. Results from a phase 2 study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in advanced AR+ triple-negative breast cancer (TNBC). J Clin Oncol. 2015;33((suppl)) abstr 1003; meetinglibrary.asco.org/content/150040-156. [Google Scholar]
- 45.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittendorf EA, Philips AV, Meric-Bernstam F, et al. PD-L1 Expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–370. doi: 10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schalper KA, Velcheti V, Carvajal D, et al. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res. 2014;20:2773–2782. doi: 10.1158/1078-0432.CCR-13-2702. [DOI] [PubMed] [Google Scholar]
- 48.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 50.Emens LA, Braiteh FS, Cassier P, et al. Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer. Cancer Res. 2015;75(suppl):PD1–6. [Google Scholar]
- 51.Nanda R, Chow LQM, Dees EC, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams S, Diamond JR, Hamilton EP, et al. Phase Ib trial of atezolizumab in combination with nab-paclitaxel in patients with metastatic triple-negative breast cancer (mTNBC). J Clin Oncol. 2016;34((suppl)) abstr 1009; meetinglibrary.asco.org/content/169304-176. [Google Scholar]
- 53.Vonderheide RH, LoRusso PM, Khalil M, et al. Tremelimumab in combination with exemestane in patients with advanced breast cancer and treatment-associated modulation of inducible costimulator expression on patient T cells. Clin Cancer Res. 2010;16:3485–3494. doi: 10.1158/1078-0432.CCR-10-0505. [DOI] [PubMed] [Google Scholar]