Abstract
In contrast to enveloped viruses, the mechanisms involved in membrane penetration by nonenveloped viruses are not as well understood. In these studies, we determined the relationship between adenovirus (Ad) capsid disassembly and the development of membrane lytic activity. Exposure to low pH or heating induced conformational changes in wild-type Ad but not in temperature-sensitive Ad (ts1) particles that fail to escape the early endosome. Wild-type Ad but not ts1 particles permeabilized model membranes (liposomes) and facilitated the cytosolic delivery of a ribotoxin. Alterations in wild-type Ad capsids were associated with the exposure of a pH-independent membrane lytic factor. Unexpectedly, this factor was identified as protein VI, a 22-kDa cement protein located beneath the peripentonal hexons in the viral capsid. Recombinant protein VI and preprotein VI, but not a deletion mutant lacking an N-terminal amphipathic α-helix, possessed membrane lytic activity similar to partially disassembled virions. A new model of Ad entry is proposed based on our present observations of capsid disassembly and membrane penetration.
Virus infection of host cells requires that these microbes gain access to the cytoplasm and/or nucleus to accomplish subsequent steps in their replication cycle. Situated between the virus and its final destination are the cell plasma or endosomal membranes. While the molecular processes involved in membrane penetration by enveloped viruses have been analyzed in some detail, analogous events in cell entry by nonenveloped viruses are currently the focus of intense interest. Studies of several nonenveloped animal viruses such as poliovirus (17), reovirus (6), and rotavirus (8) have revealed some general features of membrane penetration by naked viruses. First, the capsids of nonenveloped virions are assembled as immature stable structures that are rendered metastable upon proteolytic processing of capsid proteins (17). During infection, these mature metastable virions undergo conformational changes in response to external stimuli, including binding to cell receptor (46), proteolytic digestion (42, 44), altered divalent cation concentrations (9, 22, 32), or acidification in endosomes (8, 30, 37). Conformational changes often expose hydrophobic residues that subsequently interact with and reorganize the membrane bilayer, enabling cytosolic translocation of the viral genome, transcription complex, or partially disassembled virus capsid.
The relatively large size (90-nm-diameter) and complex architecture of adenovirus (Ad), a nonenveloped virus with icosahedral symmetry (T = 25), has hampered a detailed analysis of its structure, as well as its precise interactions with host cells. The Ad capsid is composed primarily of 240 homotrimeric hexon capsomers with 12 penton capsomers located at each of the 12 fivefold axes of symmetry. Each penton consists of the homotrimeric fiber protein protruding from the homopentameric penton base. Other so-called minor capsid components, including proteins IIIa, VI, VIII, and IX, are thought to act primarily as cement proteins that help to stabilize the viral particle. While the location in the capsid and precise role in the Ad life cycle of these capsid components have yet to be fully elucidated, protein VI was recently shown to act as a shuttling adaptor for hexon import into the nucleus (53). Ad assembly occurs in the nucleus following cleavage of preproteins TP, IIIa, VI, VII, VIII, and μ by the virally encoded 23K cysteine protease, a process that may confer a metastable capsid structure. Subsequent infection of host cells by newly assembled mature virions is initiated by the binding of the Ad fiber protein to the coxsackie B and Ad receptor (CAR) (4) or CD46 (12, 27, 34, 41, 54). Interaction of the penton base with αv integrins triggers virus internalization via clathrin-coated pits (50).
Internalization of Ad is thought to trigger capsid disassembly, starting with the loss of the fiber protein at the cell surface (26). Subsequent disassembly of the capsid occurs in endosomes and appears to require acidification (14). Acid-induced conformational changes in the capsid are thought to be required for membrane penetration, allowing cytosolic translocation of the partially uncoated virion containing the DNA, core proteins, hexon, and protein VIII (14). However, the precise relationship between Ad capsid disassembly and the development of membrane lytic activity has not been established.
Further insights into the role of Ad capsid alterations in membrane penetration have been obtained by analyses of a temperature-sensitive mutant of Ad type 2 (Ad2), ts1. When propagated at the nonpermissive temperature, ts1 capsids contain only two to three copies of the 23K protease (compared to ∼10 copies in wild-type [WT] virions) (1). This defect results in the arrest of the capsid in the immature state, containing uncleaved preproteins (3, 48). Although ts1 can still bind the CAR receptor and undergo internalization via αv integrins, this mutant virus fails to escape endosomes (13), suggesting a potential link between capsid structural proteins and membrane penetration.
To help clarify the role of capsid disassembly in membrane penetration, we compared the structural stability of Ad5 and ts1 particles as a function of pH. We also examined the ability of these different viruses to disrupt model membranes under conditions similar to those required to initiate capsid disassembly. These studies demonstrated unambiguously that partial uncoating of the capsid occurs prior to membrane penetration. Furthermore, protein VI, an internal capsid protein exposed during capsid disassembly, exhibits pH-independent membrane lytic activity. An N-terminal domain in protein VI with predicted amphipathic α-helical structure was also shown to be required for membrane lytic activity. These findings are discussed in light of an emerging theme for how nonenveloped viruses overcome the barrier of the host cell membrane.
MATERIALS AND METHODS
Cell culture and virus propagation.
HEK293 and A459 cells were maintained in Dulbecco's complete modified Eagle's medium (DMEM) supplemented with 10 mM HEPES, 2 mM glutamine, 1 mM pyruvate, 0.1 mM nonessential amino acids, 100 U of penicillin G/ml, 0.3 mg of gentamicin/ml, and 10% fetal bovine serum. An Ad5 vector containing an enhanced green fluorescent protein expression cassette was propagated in the HEK293 cells by infection with 200 particles per cell. Propagation of the Ad temperature-sensitive mutant H2ts1 (hereafter referred to ts1) (3, 48) in A459 cells was performed at the nonpermissive temperature of 39.5°C. Cells were infected with 200 particles of ts1/cell that had been previously grown at the permissive temperature of 33°C. Infected cells were harvested after nearly complete cytopathic effect, generally 48 to 60 h postinfection. Virus was purified by cesium chloride density gradient centrifugation as previously described (54) and stored at −80°C in 40 mM Tris, 150 mM NaCl, 10% glycerol, and 1 mM MgCl2 (pH 8.2).
Analysis of membrane penetration in intact cell cultures.
A549 lung epithelial cells were plated in 96-well plates (catalogue no. 3904; Costar) at a density of 10,000 cells/well for 24 h prior to infection. One hour prior to infection, cells were washed once with DMEM without cysteine or methionine and supplemented with 10 mM HEPES, 2 mM glutamine, and 0.5% bovine serum albumin (BSA) with penicillin and streptomycin (DMEM−) and incubated in DMEM− for 1 h at 37°C. The medium was then removed, and cells were incubated with DMEM− containing 0.1 mg of α-sarcin (Sigma)/ml and various amounts of Ad5 or ts1 particles, ranging from 1 to 10−3 μg/ml for 2 h at 37°C. The effect of endosome acidification on α-sarcin entry was also assessed by infection of cells with Ad5 in the presence of 0.3 μM bafilomycin A1 (Sigma), an inhibitor of vacuolar H+-ATPase. The cells were then washed once with DMEM− and incubated with DMEM− containing 0.1 μCi of [35S]l-methionine (Amersham Biosciences) per well for an additional 2 h at 37°C. The cells were then washed twice with 100 μl of 20 mM Tris-buffered saline (pH 7.4), incubated with 50 μl of ice-cold 5% trichloroacetic acid (TCA) at 4°C for 1 h, then centrifuged at 1,000 × g for 15 min at 4°C, and washed twice with cold 100% ethanol. The pelleted material was then dissolved overnight in 5 μl of 1% sodium dodecyl sulfate (SDS)-0.1 N NaOH at 4°C on a rotary shaker. The resulting solutions were neutralized with 1 μl of 0.6 N HCl, and 34 μl of MicroScint 20 liquid scintillation cocktail (Packard) was added to each well with vortexing. Radioactivity was measured with a Topcount (Packard) 96-well liquid scintillation counter.
Measurements of Ad capsid stability and disassembly.
The thermal stability of the Ad capsid after exposure to various pH conditions was measured by the accessibility of the viral DNA to a fluorescent intercalating dye, TOTO-1 (Molecular Probes), as previously described with minor modifications (31). Briefly, 100 μg of Ad5 or ts1/ml was incubated with 60 nM TOTO-1 at various pH values ranging from 4.5 to 7.5 in buffers containing of 50 mM acetate (pH 4.5 to 5.5), morpholineethanesulfonic acid (MES) (pH 6.0 to 6.5), or HEPES (pH 7.0 to 8.0) with 100 mM NaCl. The fluorescence emission of TOTO-1 was monitored as a function of temperature with an ABI Prism 7900HT real-time PCR machine (Applied Biosystems) programmed to measure fluorescence (λex 488; λem 540) every 2.5°C between 20 and 70°C. Samples were equilibrated at each temperature for 2 min prior to fluorescence measurement.
To measure capsid disassembly, Ad5 or ts1 particles were dialyzed against either 50 mM acetate, 100 mM NaCl (pH 5.0), or 50 mM HEPES-100 mM NaCl (pH 7.4) as a control. A 0.5-ml aliquot of a 100-μg/ml solution was then placed in 1.5-ml microcentrifuge tubes precoated with 3% (wt/vol) BSA and then incubated at various temperatures between 25 and 60°C for 10 min before being loaded onto a Nycodenz (Sigma) step density gradient consisting of 1 ml of 80% (wt/vol) Nycodenz and 1.5 ml of 40% Nycodenz gradients prepared in acetate or HEPES buffers. Gradients were centrifuged in an SW60 rotor for 2 h at 180,000 × g, and the partially disassembled virions were collected from the 40 to 80% interface and analyzed for protein content by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Simply Blue staining. Gel images were obtained with an AlphaImager 2200 (Alpha Innotech), and densitomety was performed with Scion Image (Scion Corporation). The densities of several Ad capsid protein bands were normalized to that of the major core antigen, protein VII.
Liposome preparation and analysis of membrane disruption.
Liposomes composed of bovine liver phosphatidylcholine, bovine liver phosphatidylethanolamine, bovine brain phosphatidylserine, and entrapping 100 mM sulforhodamine B (SulfoB; Molecular Probes) were prepared as previously described (5). The liposomes containing entrapped SulfoB were separated from unentrapped material with a Sephadex G-75 column preequilibrated with an elution buffer consisting of 5 mM HEPES and 150 mM NaCl (pH 7.0). The phospholipid concentration of the liposomes was determined by a phosphate assay modified from reference 11.
Ad-mediated membrane disruption was assessed by the dequenching of SulfoB fluorescence upon its release from the liposomes. Fluorescence intensity was monitored using a Tecan Genios microplate reader equipped with 535/20 nm excitation and 585/20 nm emission filters, respectively. Liposomes were diluted to a final phospholipid concentration of 2 μM in 100 μl of buffer in 96-well plates (catalogue no. 3904; Costar). Various amounts of virus or recombinant Ad proteins in 5 mM HEPES (pH 7.0) were added to the liposomal solutions preequilibrated at 37°C, and the kinetics of membrane disruption were monitored by the increase in SulfoB fluorescence. One hundred percent dye release was determined by adding Triton X-100 to liposomes at a final concentration of 0.2% (wt/vol). The percentage of SulfoB release was calculated by the equation % SulfoB released = 100 × [(Fmeas − F0)/(Ftx100 − F0)], where Fmeas is the maximum fluorescence intensity measured for each sample, F0 is the fluorescence intensity in the absence of virus or protein, and Ftx100 is the fluorescence intensity in the presence of 0.2% Triton X-100.
Electron microscopy.
Purified protein VI was mixed with liposomes at a 1:1 weight ratio (0.4 mg/ml of sprotein) in HEPES-buffered saline (pH 7.4) and incubated at room temperature for 30 min. Copper grids (300 mesh; EM Sciences) were subjected to glow discharge in the presence of butylamine and immediately floated on the 10 μl of sample for 1 min. The grids were then rinsed twice with 50 mM HEPES (pH 7.4) before incubation with 2% phosphotungstate (pH 7.0) for 1 min. Samples were rinsed once with 50 mM HEPES, the excess solution was removed by being blotted on Whatman paper, and the sample was then examined with a Phillips CM100 electron microscope (EM) with an accelerating voltage of 80 V and a magnification of ×21,000.
Immunodepletion analyses.
To identify the protein(s) responsible for the membrane-disrupting activity, Ad5 virions were incubated at 45°C for 10 min, and the partially disassembled virions were separated from free capsomers with a Nycodenz step gradient as described above. Isolated disrupted viral particles and free capsomers were collected and dialyzed against 5 mM HEPES-buffered saline before membrane-disrupting activity was assessed as described above. Immunodepletion of membrane lytic activity was performed by incubation of free capsomers with affinity-purified antibodies directed against the Ad5 penton base or protein VI in a total volume of 100 μl of PBS at 4°C. After 1 h, 10 μl of Gammabind Plus resin (Amersham Biosciences) was added, and the samples were incubated on a rocking plate at 4°C for an additional 4 h. The resin was then pelleted by centrifugation at 10,000 × g for 2 min, and the amount of membrane lytic activity remaining in the immunodepleted supernatant was measured by the liposome-dye release assay as described above.
Sequence analysis and generation of recombinant protein VI molecules.
Prediction of protein VI secondary structure was performed with the PSIPRED program. (18, 25). Identification of potential membrane interacting domains within protein VI was performed with Membrane Protein Explorer software employing the hydropathy scale of Wimley and White (52). Helical wheel projections were made with this software, while an alignment of protein VI sequences from different human and nonhuman Ad serotypes was performed with the ClustalW algorithm.
cDNA encoding preprotein VI (pVI), protein VI, and a truncated protein VI lacking residues 34 to 54 (VIΔ54) was amplified from pAdeasy-1 (Clontech) by standard procedures and then cloned into the NdeI and BamHI sites of the pET15b expression vector (Novagen, Madison, Wis.) that contains an enterokinase cleavage site positioned between the N-terminal His6 tag and the N terminus of the recombinant proteins. The 5′ and 3′ primers, containing an NdeI restriction site (underlined) and an enterokinase cleavage site (italics) (in the 5′ primers) or a BamHI restriction site (underlined) (in the 3′ primers) were used as follows: for pVI, 5′-GG AAA TTC CAT ATG GAA GAC ATC AAC and 5′-AA ACC GGA TCC TCA GAA GCA TCG TCG; for protein VI, 5′-GG AAA TTC CAT ATG GAC GAC GAC GAC AAG GCC TTC AGC TGG GGC and 5′-AAA GGA TCC TCA CAG ACC CAC GAT GCT; and for VIΔ54, 5′-AAA CAT ATG GAC GAC GAC GAC AAG TAT GGC AGC AAG GCC and 5′-AAA GGA TCC TCA CAG ACC CAC GAT GCT.
Recombinant proteins were expressed in BL21(DE3) cells (Invitrogen, Carlsbad, Calif.). For expression of pVI and protein VI under conditions that reduce cell lysis, cells were grown at 37°C in Luria-Bertani broth to an optical density at 600 nm of 1 to 1.2. Cells were then brought to 25°C, the NaCl concentration was increased to 300 mM, and protein expression was induced by the addition of 0.5 mM IPTG (isopropyl-ā-d-thiogalactopyranoside) for 2 h. For expression of VIΔ54 in BL21(DE3) cells, the cells were grown at 37°C in Luria-Bertani broth to an optical density at 600 nm of 0.8 before 0.5 mM IPTG was added. In all cases, the cells were induced for 2 h, pelleted, and washed once in PBS. The pellets were then resuspended in Bugbuster Protein extraction reagent supplemented with 20 U of Benzonase (Novagen)/ml, 0.5 mg of lysozyme (Sigma)/ml, and a protease inhibitor cocktail (catalogue no. P8849; Sigma). After 15 min at room temperature, the cell debris was pelleted at 16,000 × g for 15 min at 4°C. Recombinant proteins were purified with nitrilotriacetic acid-Ni2+ agarose with the manufacturer's protocol (QIAGEN). Hexon was purified from Ad5-infected HEK293 cells as previously described (53).
RESULTS
Low pH destabilizes the Ad5 but not the ts1 capsid.
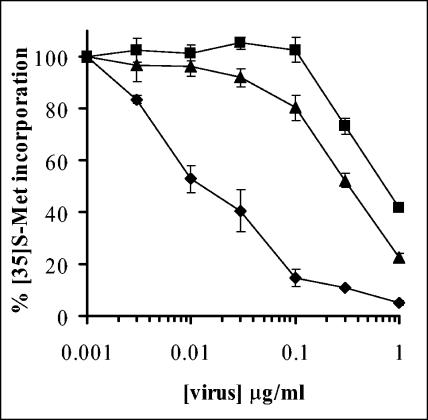
To gain a better understanding of the role of the Ad capsid in membrane penetration, we compared the ability of WT Ad5 and ts1 particles to facilitate the cytosolic translocation of the ribosomal toxin α-sarcin. This approach was previously used to study membrane penetration by several enveloped (30) and nonenveloped (8, 9) viruses. Cointernalization of virus particles and α-sarcin into endosomes allows cytosolic entry of the toxin upon membrane penetration by the virus. α-Sarcin entry restricts protein synthesis by specific hydrolysis of rRNA (33), as measured by reduced incorporation of [35-S]methionine into newly synthesized proteins. As shown in Fig. 1, Ad5 particles caused a dose-dependent increase in membrane penetration, as indicated by α-sarcin inhibition of protein synthesis. Approximately 50% inhibition of protein synthesis was observed at 0.02 μg/ml (∼200 particles/cell). The Ad concentration required for 50% inhibition of protein synthesis was increased 40 fold (∼8,000 particles/cell) when cells were treated with bafilomycin A1, an inhibitor of vacuolar H+-ATPase (Fig. 1), suggesting a requirement for endosome acidification in Ad membrane penetration. ts1 particles grown at the nonpermissive temperature of 39.5°C exhibited a reduced capacity to promote α-sarcin entry, requiring ∼30-fold-higher particle concentrations (∼6,000 particles/cell) than Ad5. Since ts1 capsids have significantly reduced membrane lytic activity, we were compelled to investigate whether the structural differences in Ad5 and ts1 Ad particles might be related to the observed differences in membrane lytic activity.
FIG. 1.
Cytosolic translocation of the ribotoxin α-sarcin by Ad5 or ts1 particles. Sarcin entry into A549 cells was performed in the presence of various amounts of Ad5 (♦), Ad5 plus 0.3 uM bafilomycin (▪) or ts1 particles (▴). Toxin activity was assessed by inhibition of [35S]methionine incorporation into cellular proteins. Less than 10% inhibition of protein synthesis was observed with the highest amount of Ad5 in the absence of α-sarcin.
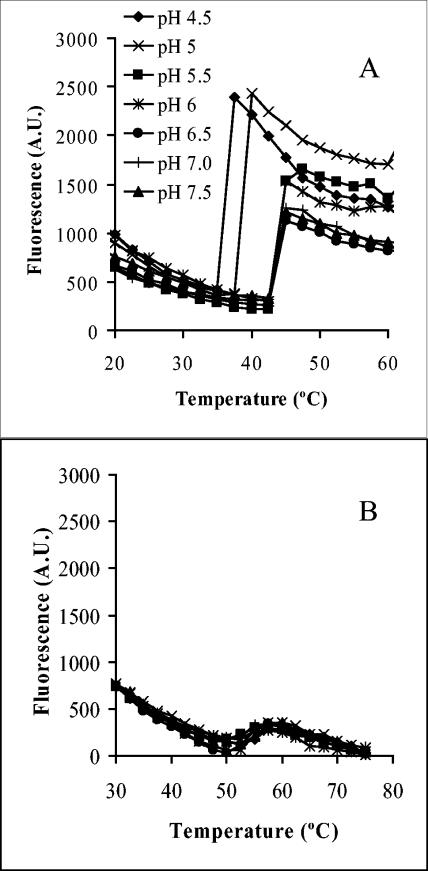
Previous studies suggested that the Ad capsid undergoes partial disassembly prior to membrane disruption (14) and that Ad membrane penetration occurs from within acidified endosomal compartments. Thus, we compared the thermal stability of Ad5 to ts1 capsids at various pH values as measured by the accessibility of Ad DNA to the bis-intercalating dye TOTO-1. When Ad5 was incubated at pH values ranging from 5.5 to 7.5, a structural transition in the capsid occurred when these particles were heated above 42.5°C, as indicated by an increase in TOTO-1 fluorescence emission (Fig. 2A). At pH values below 5.5, the Ad5 capsid was significantly destabilized, as indicated by the pronounced increase in TOTO-1 fluorescence occurring at or below 37.5°C (Fig. 2A). In parallel studies, we observed that ts1 particles were actually more stable than Ad5 capsids (Fig. 2B). A small structural transition was observed in the ts1 capsid above 50°C, and the overall fluorescence intensity was significantly reduced compared to Ad5 (Fig. 2B). Moreover, reduced pH appeared to have little or no destabilizing effect on the ts1 capsid.
FIG. 2.
Analysis of Ad capsid stability as a function of pH and temperature. The stability of Ad5 (A) or ts1 particles (B) was assessed at various pH levels and temperatures by monitoring the accessibility of the viral DNA to TOTO-1, a bis-intercalating fluorophore.
Partial disassembly of the Ad5 capsid occurs below pH 5.5.
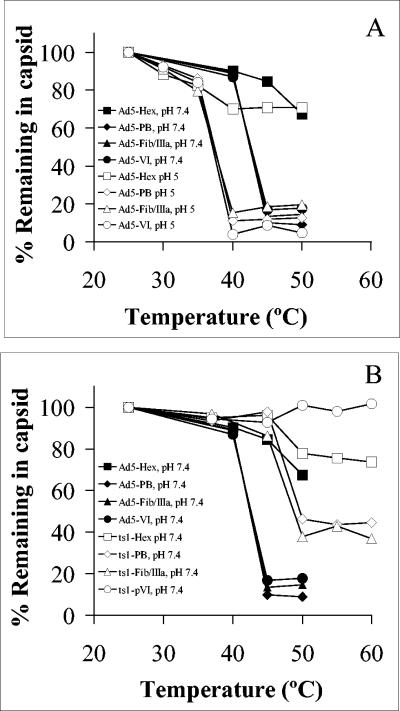
To more precisely define the structural alterations in the Ad capsid occurring during exposure to low pH, we examined the integrity of low-pH or heat-treated Ad capsid proteins, following sedimentation on 40 to 80% Nycodenz step gradients. Proteins associated with the gradient-purified capsids were quantitated by SDS-PAGE and densitometry. Proteins II through VII were clearly resolved by SDS-PAGE, with the exception of the fiber and protein IIIa, which comigrated as a single band (data not shown). Ad5 particles incubated at pH 7.4 and increasing temperature (>40°C) exhibited a significant loss of the vertex proteins located at the fivefold axis of symmetry, including the penton base, fiber/IIIa, and protein VI (Fig. 3). At pH 5.0, partial disassembly of Ad5 occurred when the virus was heated above 35°C, in agreement with the TOTO-1 accessibility experiments discussed above (Fig. 3A). Examination of both heat- and low pH-treated Ad capsids by negative-stain transmission EM confirmed that these treatments dissociated the vertices of the Ad capsid as previously described (data not shown) (31).
FIG. 3.
Examination of thermally induced capsid disassembly. Ad5 at pH 7.4 (A and B, solid symbols) or pH 5.0 (A, open symbols) or ts1 particles at pH 7.4 (B, open symbols) were heated to various temperatures and then subjected to density gradient centrifugation to separate partially disassembled virions from soluble capsomers. Individual capsid proteins associated with the treated virions were analyzed by SDS-PAGE and Simply Blue staining and then quantitated by scanning densitometry. Densities were normalized to that of the major core protein, protein VII.
In contrast to Ad5, dissociation of capsid proteins from ts1 particles was substantially reduced. Densitometric analysis indicated that only 10 to 20% remained associated with Ad5 capsids when heated to 40 to 50°C, while 40 to 95% of these proteins remained with ts1 particles when heated to even higher temperatures (Fig. 3B). These findings are consistent with the prior observation that ts1 capsids have decreased TOTO-1 accessibility and strongly suggest that disassembly of this mutant virus is much less efficient than WT Ad5. Of particular interest, pVI remained almost entirely associated with the partially disassembled ts1 capsid.
Partially disassembly of the Ad5 capsid is required for membrane disruption.
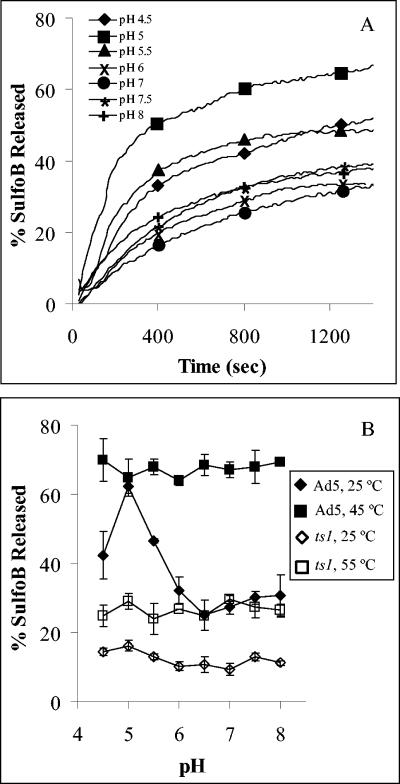
In further studies, we sought to correlate the observed structural changes in Ad capsids with Ad-mediated membrane disruption with model membranes (liposomes). The integrity of liposomal membranes was monitored by the release of an entrapped fluorescent dye, SulfoB, whose fluorescence emission is insensitive to pH in the range of 4 to 9. The dye is entrapped at concentrations that result in significant self-quenching of fluorescence emission. Upon release, the dye is effectively diluted, allowing fluorescence dequenching. When Ad5 was incubated with liposomes at 37°C, a saturable and time-dependent release of SulfoB was observed (Fig. 4A). The initial rate of SulfoB release was pH dependent and correlated well with the pH dependence of maximum dye release (Fig. 4B). Although Ad membrane disruption was observed at all pH values examined, maximal release occurred near pH 5.0. By comparison, ts1 produced significantly less membrane-disrupting activity at pH 4.5 to 8 (Fig. 4B).
FIG. 4.
Ad-mediated disruption of model membranes (liposomes) containing an entrapped fluorophore. (A) Ad5 particles (75 μg/ml) were incubated with liposomes (2 μg/ml) at different pH levels, and the release of SulfoB was monitored over time as described in Materials and Methods. (B) The amount of SulfoB released from liposomes incubated with Ad5 or ts1 particles was measured at different pHs, following preincubation of virions at 25 or 45°C (for WT) or 55°C (for ts1).
Unexpectedly, we found that partial disassembly of Ad5 by applying heat prior of addition to liposomes generated a membrane lytic activity that was independent of pH (Fig. 4B). Thus, disassembly of the Ad capsid, either by lowering pH or elevating temperature, was necessary for membrane disruption. In contrast, pretreatment of the more stable ts1 virus at 55°C for 10 min produced only a minor increase in membrane disruption. These findings indicated that the increased structural stability of mutant Ad particles correlated with decreased membrane lytic activity.
Immunodepletion of protein VI from dissociated capsomers removes membrane lytic activity.
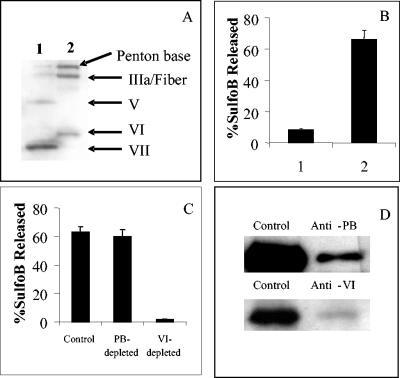
Further studies were next undertaken to determine whether a particular capsid protein in WT Ad5 was responsible for pH-independent membrane disruption. Ad5 was partially disassembled by being heated to 45°C, and the dissociated capsomers were separated from the partially uncoated virions on a Nycodenz step gradient. The top fraction (supernatant) contained the dissociated hexon, penton base, IIIa, fiber, and protein VI. The banded fraction, composed of partially disassembled virions, was largely depleted of these proteins, although they retained the majority of the hexon and the core proteins V and VII (Fig. 5A). Incubation of these different virion fractions with liposomes at 37°C and pH 7.4 indicated that the membrane lytic activity was predominantly associated with the free vertex proteins (supernatant) (Fig. 5B). Somewhat to our surprise, we found that that protein VI was responsible for the majority of membrane lytic activity (Fig. 5C) as indicated by immunodepletion with affinity-purified antibodies (Fig. 5D and data not shown).
FIG. 5.
Identification of the Ad capsid protein associated with pH-independent membrane disruption. (A) Immunoblot of partially disassembled Ad5 capsids (lane 1) and the soluble capsomer fraction (lane 2). Individual Ad proteins were detected with a polyclonal anti-Ad antibody. (B) Liposome disruption by virion (lane 1) and supernatant fraction (lane 2) was analyzed by the SulfoB release assay. (C) The supernatant fraction was immunodepleted with a control or anti-penton base (PB) or protein VI antibodies, followed by immunoblot analysis. (D) The supernatant- or PB- or protein VI-depleted fractions were analyzed for liposome disruption at pH 7.4 by the SulfoB release assay.
Recombinant protein VI possesses pH-independent membrane lytic activity.
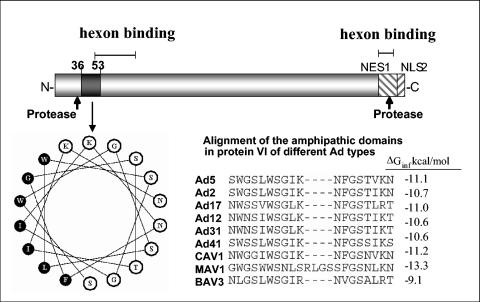
Protein VI has been proposed to be located underneath the peripentonal hexons in intact virions, existing as a trimer of dimers (43). Further analysis of the primary structure of protein VI revealed that cleavage of pVI by the virus-encoded 23K protease gives rise to the mature protein VI containing a predicted N-terminal amphipathic α-helix (residues 36 to 53) (Fig. 6). This domain is highly conserved among distinct Ad protein VI molecules from both human and nonhuman viruses. Importantly, it possesses many similarities to amphipathic helices of class I viral fusion peptides, with hydrophobic residues being composed of large bulky aromatic or aliphatic residues and several glycine residues throughout the helix length. Even in instances where residues vary among different viral serotypes, the amphipathic nature and physicochemical properties of the helix are maintained. The free energy of partitioning of this helix into the interfacial region of lipid membranes was calculated using the Wimley-White hydropathy scale, assuming a helical structure (52). For Ad5, ΔGinterfacial is −11.1 kcal/mol, with the majority of helices from different Ad types falling within the range of −9.1 to −13.3 kcal/mol (Fig. 6). For comparison, the predicted ΔGinterfacial for the influenza hemagglutinin fusion peptide is −7.5 kcal/mol, which is in good agreement with the experimentally observed value of −7.6 kcal/mol (19).
FIG. 6.
Schematic representation of Ad protein VI. Sites for cleavage by the 23K cysteine protease, hexon binding, and nuclear export (NES1) and import (nuclear localization signal 2 [NLS2]) are indicated. A predicted amphipathic α-helical domain is located between residues 36 to 53. A helical wheel diagram of this region is shown with the hydrophobic residues in black and the hydrophilic residues in white. An alignment of protein VI sequences corresponding to the amphipathic helical domain of human and nonhuman Ads is shown. Predicted ΔGinterfacial values for the partitioning of these domains into lipid membranes are provided for comparison.
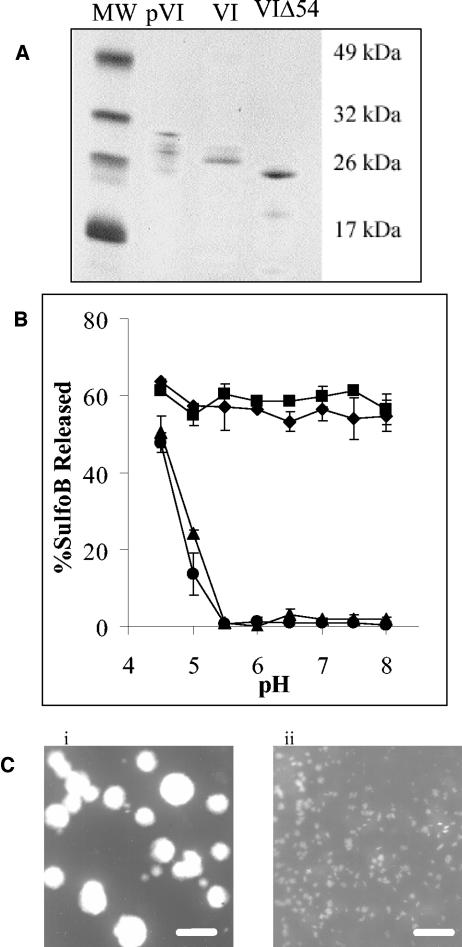
To substantiate the role of protein VI in membrane disruption and to identify its putative membrane-reactive domain, we generated recombinant forms of protein VI containing an N-terminal His6 tag. Both the mature and preprotein forms of protein VI were capable of disrupting liposomal membranes in a pH-independent manner, similar to heated virions (Fig. 7B). The membrane lytic activity occurred with or without removal of the His6 tag (data not shown). In contrast, deletion of the amphipathic sequence from protein VI (VIΔ54) resulted in the complete loss of membrane lytic activity at pH values above 5, similar to that observed with an irrelevant protein (BSA) (Fig. 7B). Since no other amphipathic sequences are predicted to be present in protein VIΔ54, we concluded that the membrane-disrupting activity occurring below pH 5.0 was due to a pH-dependent conformational change in the protein that exposes a hydrophobic surface, capable of interacting nonspecifically with the lipid membrane. To gain further insights into the mechanism of protein VI membrane disruption, we examined the morphological changes in the lipid membranes, following incubation with this protein by EM. Untreated liposomes ranged in size from 200 to 500 nm (Fig. 7Ci). In striking contrast, liposomes treated with a weight equivalent of protein VI resulted in vesicles or membrane fragments reduced in size to 20 to 100 nm in diameter (Fig. 7Cii). While the exact mechanism responsible for this membrane alteration remains to be determined, the dramatic change in membrane morphology is likely associated with the ability of Ad particles to disrupt endosomes.
FIG. 7.
Analysis of the membrane lytic activity recombinant protein VI molecules. (A) SDS-PAGE of recombinant preprotein VI, protein VI, and a truncated protein VI (VIΔ54), following purification on nitriloacetic acid-Ni2+ agarose columns. (B) Liposome disruption by equivalent amount of pVI ♦, VI ▪, VIΔ54 ▴, or BSA • as a control was assessed at various pH levels by SulfoB release. (C) EMs of untreated liposomes (i) or liposomes treated with an equivalent weight of protein VI at pH 7.4 (ii) obtained at a magnification of ×21,000 after being negatively stained with 2% phosphotungstate. Bar, 500 nm.
DISCUSSION
Uncoating of nonenveloped viruses, and Ad in particular, is triggered by a variety of environmental stimuli that initiate penetration of endocytic vesicles. While endosomal acidification has previously been observed to promote Ad infection (37), the relationship between uncoating and membrane penetration has been largely unexplored. Here, we demonstrate that reduced pH destabilizes the Ad capsid, resulting in dissociation of proteins from the capsid vertex. The pH-dependent capsid disassembly liberates protein VI, and this promotes membrane disruption. Membrane disruption is also dependent upon exposure of a predicted N-terminal amphipathic α-helix in protein VI that is presumably buried within a hexon-protein VI interface. Consistent with this cell entry model, the ts1 mutant virus, which fails to undergo significant capsid destabilization at low pH as measured by increased accessibility of the viral DNA to the dye TOTO-1 and does not release preprotein VI from the capsid upon disassembly, also fails to escape endosomes (13) or to deliver a ribotoxin into the cytoplasm of host cells (Fig. 1).
Earlier studies had investigated the kinetics of Ad uncoating by coimmunoprecipitating capsid proteins from infected cells with anti-hexon antibodies (14). These studies reported that during the first 15 min after synchronous Ad internalization, the fiber, penton base, and proteins IIIa, VI, VIII, and IX were lost from the capsid. This observation is consistent with our data, showing that low pH destabilizes the Ad5 capsid. However, a more precise molecular description of Ad capsid disassembly and its role in virus entry has been lacking. It was recently reported that the thermally induced disassembly of Ad5 corresponded to a partial loss of protein tertiary structure, as measured by changes in intrinsic tryptophan fluorescence (31). Since ∼70% of tryptophan residues of all Ad proteins are present in the hexon protein, this observation suggests that subtle changes in the hexon tertiary structure occur during capsid disassembly.
The availability of a mutant Ad that is capable of binding and internalization but not membrane penetration provided a useful tool to investigate the link between virus uncoating and membrane penetration. Of particular interest was the fact that several of the preproteins present in the ts1 capsid are associated with hexon-including proteins IIIa, VI, and VIII. Since capsid disassembly corresponds with minor alterations in hexon tertiary structure, the enhanced stability of the ts1 capsid may be related to altered protein interactions between hexon and preproteins compared to WT Ad. Additional insight into this pH-induced capsid instability may eventually be gained by a detailed structural analysis of Ad5 and ts1 capsids.
Previous studies demonstrated the pH dependence of Ad-mediated disruption of cell membranes (37, 38), purified cell membrane preparations (35), or liposomes (5). Maximal membrane lytic activity was generally observed between pH 5 and 6; however, the precise role of low pH in membrane disruption was not addressed. By comparing the membrane-disrupting activity of Ad5 and ts1 particles, we showed that the low pH dependence of Ad membrane disruption was related to disassembly of the capsid rather than a direct requirement for membrane lytic activity. In fact, protein VI-mediated disruption of model membranes was observed to be pH independent. Furthermore, dissociation of protein VI from the capsid appears to be a requirement for efficient membrane disruption, since disassembly of the ts1 capsid at elevated temperatures failed to dissociate preprotein VI or to generate significant membrane lytic activity.
An unanticipated finding was the identification of protein VI as the membrane lytic protein for Ad, since previous reports suggested that the penton base mediates membrane penetration (36, 39, 40). The interpretation of the data from these earlier studies may have been confounded by the fact that the penton base also mediates Ad internalization via interaction with αv integrins (50). Thus, the observed inhibitory effects of anti-penton base antiserum may have been incorrectly attributed to the ability to restrict membrane disruption rather than to limit virus internalization. Alternatively, penton base interaction with integrin αvβ5 may serve to promote virus capsid disassembly in the low-pH environment of the endosome rather than having a direct role in membrane disruption (49). Intact Ad particles (but not purified recombinant penton base) were shown to permeabilize cell membranes (49). Greber and coworkers also reported that protein VI is degraded during Ad entry into the cytosol, suggesting that its release from the capsid is accompanied by digestion by host cell enzymes or the Ad protease (13, 14). Protein VI degradation was reportedly inhibited by integrin antagonists (e.g., RGD peptides), consistent with the notion that integrin binding to the penton base facilitates capsid disassembly and subsequent release of protein VI.
The identification of protein VI as a membrane lytic factor adds to its expanding functional repertoire in the Ad life cycle, including facilitating hexon nuclear import (53), stimulating 23K protease activity during Ad assembly (21), and providing structural stability to the inner capsid (43). A full understanding of protein VI functions will undoubtedly require increased knowledge of its structure and precise location in the capsid. Unfortunately, little structural information on protein VI is currently available. Different images generated from the electron density of the Ad capsid obtained by cryo-EM and the crystal structure of the hexon protein have been used to tentatively assign the location of protein VI to a region beneath the peripentonal hexons (43). This observation, along with limited biochemical data (47), suggests that protein VI exists as a trimer of dimers (43) with a stoichiometry of 360 monomers per virion (30 per vertex region). Amino acid residues 48 to 74 and 235 to 239 in protein VI have been identified as necessary for binding to the hexon (23, 24).
The N terminus of the mature protein VI is predicted to contain a four-helix bundle, while the majority of the remaining protein is predicted to be largely unstructured. The first helix appears to be responsible for pH-independent membrane disruption, since its deletion substantially reduced activity. Although the nature of the interaction between protein VI and lipid membranes remains to be determined, two possibilities exist. The helix either could lie relatively parallel to the lipid bilayer in the interfacial region, as observed for fusion peptides of the influenza hemagglutinin (15), or could span the lipid bilayer. In the latter case, this might lead to the formation of pore structures. Given that both the preprotein and protein VI possess identical membrane lytic activity, we propose that the former case is more likely, since the thermodynamic penalty for translocating the N-terminal 33 residues proximal to the amphipathic helix through the lipid bilayer would likely be a significant barrier to pore formation.
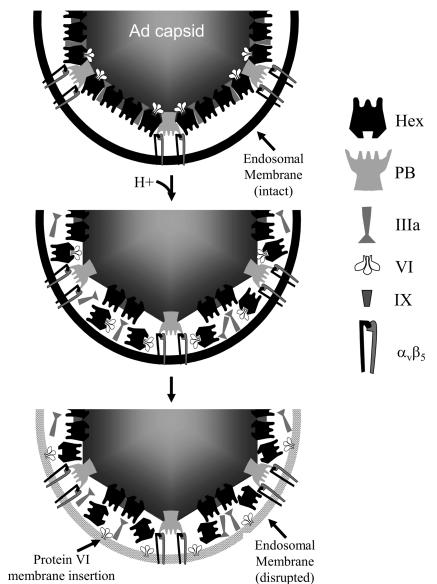
Ad entry bears similarities with entry by other nonenveloped viruses. In general, the assembly of immature virions is accompanied by a transition to a metastable state by proteolytic processing of the capsid, by either autocatalysis (e.g., poliovirus) (2, 16) or endogenous host proteases (e.g., reovirus and rotavirus) (7, 10, 20, 28). In the case of Ad, immature particles (e.g., ts1) are initially formed as stable particles that are rendered metastable by cleavage of proteins IIIa, VI, VII, VIII, and μ. Proteolytic cleavage of Ad preprotein VI by the 23K protease results in the generation of an N-terminal amphipathic helix in the mature form of the protein. Upon subsequent infection by these newly formed viral particles, an environmental stimulus triggers a conformational change in the capsid, resulting in release of capsid components and membrane penetration (e.g., poliovirus, reovirus) (7, 17, 29, 44-46, 51). Ad possesses similarities with these schemes in that the reduced endosomal pH triggers a conformational change in the capsid. This conformational change results in release of peripentonal proteins and exposure of protein VI, which can mediate membrane disruption. Generally, the membrane lytic factor of nonenveloped viruses is also shielded from the external environment, perhaps to avoid nonproductive interactions with membranes at the wrong time and place. Based on our present findings as well as previous studies, we propose a new scheme for Ad-mediated membrane penetration (Fig. 8). While further studies are needed to test this scheme, the identification of protein VI as a key membrane lytic factor fills an important gap in our understanding of cell entry by this complex viral pathogen.
FIG. 8.
Hypothetical model of Ad-mediated endosomal membrane disruption. (Top) Penton base binding to αv integrins triggers Ad uptake into endosomes, coincident with the loss of the fiber protein. (Middle) Endosome acidification induces partial disassembly of the capsid, resulting in the removal of the peripentonal hexons, penton base, IIIa, and VI. (Bottom) Protein VI dissociates from peripentonal hexons and associates with the endosome, resulting in membrane disruption and release of partially disassembled particles into the cytoplasm.
Acknowledgments
We thank Kelly White and Joan Gauspohl for their help in preparation of the manuscript. We would also like to acknowledge Phoebe Stewart for helpful discussions.
We further acknowledge financial support from the NIH (AI007354 to C.M.W., HL054352 and EY011431 to G.R.N., and AI055729 to L.G.).
REFERENCES
- 1.Anderson, C. W. 1990. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177:259-272. [DOI] [PubMed] [Google Scholar]
- 2.Basavappa, R., R. Syed, O. Flore, J. P. Icenogle, D. J. Filman, and J. M. Hogle. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 3:1651-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begin, M., and J. Weber. 1975. Genetic analysis of adenovirus type 2. I. Isolation and genetic characterization of temperature-sensitive mutants. J. Virol. 15:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal, R., P. Seth, M. C. Willingham, and I. Pastan. 1986. pH-dependent lysis of liposomes by adenovirus. Biochemistry 25:2231-2237. [DOI] [PubMed] [Google Scholar]
- 6.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 76:9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran, K., J. S. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The delta region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chemello, M. E., O. C. Aristimuño, F. Michelangeli, and M.-C. Ruiz. 2002. Requirement for vacuolar H+-ATPase activity and Ca2+ gradient during entry of rotavirus into MA104 cells. J. Virol. 76:13083-13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuadras, M. A., C. F. Arias, and S. Lopez. 1997. Rotaviruses induce an early membrane permeabilization of MA104 cells and do not require a low intracellular Ca2+ concentration to initiate their replication cycle. J. Virol. 71:9065-9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowling, W., E. Denisova, R. LaMonica, and E. R. Mackow, 2000. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 74:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiske, C. H., and Y. Subbarow. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66:374-389. [Google Scholar]
- 12.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 13.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 14.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 15.Han, X., J. H. Bushweller, D. S. Cafiso, and L. K. Tamm. 2001. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8:715-720. [DOI] [PubMed] [Google Scholar]
- 16.Hindiyeh, M., Q. H. Li, R. Basavappa, J. M. Hogle, and M. Chow. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 73:9072-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y., X. Han, and L. K. Tamm. 2003. Thermodynamics of fusion peptide-membrane interactions. Biochemistry 42:7245-7251. [DOI] [PubMed] [Google Scholar]
- 20.Ludert, J. E., A. A. Krishnaney, J. W. Burns, P. T. Vo, and H. B. Greenberg. 1996. Cleavage of rotavirus VP4 in vivo. J. Gen. Virol. 77:391-395. [DOI] [PubMed] [Google Scholar]
- 21.Mangel, W. F., W. J. McGrath, D. L. Toledo, and C. W. Anderson. 1993. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361:274-275. [DOI] [PubMed] [Google Scholar]
- 22.Martin, S., M. Lorrot, M. A. El Azher, and M. Vasseur. 2002. Ionic strength- and temperature-induced KCa shifts in the uncoating reaction of rotavirus strains RF and SA11: correlation with membrane permeabilization. J. Virol. 76:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews, D. A., and W. C. Russell. 1994. Adenovirus protein-protein interactions: hexon and protein VI. J. Gen. Virol. 75:3365-3374. [DOI] [PubMed] [Google Scholar]
- 24.Matthews, D. A., and W. C. Russell. 1995. Adenovirus protein-protein interactions: molecular parameters governing the binding of protein VI to hexon and the activation of the adenovirus 23K protease. J. Gen. Virol. 76:1959-1969. [DOI] [PubMed] [Google Scholar]
- 25.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, M. Y., K. Boucke, M. Suomalainen, R. P. Stidwill, and U. F. Greber. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsson, M., S. Karlsson, and X. Fan. 2004. Functionally distinct subpopulations of cord blood CD34+ cells are transduced by adenoviral vectors with serotype 5 or 35 tropism. Mol. Ther. 9:377-388. [DOI] [PubMed] [Google Scholar]
- 28.Odegard, A. L., K. Chandran, X. Zhang, J. S. L. Parker, T. S. Baker, and M. L. Nibert. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78:8732-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier, I., L. Ouzilou, M. Arita, A. Nomoto, and F. Colbere-Garapin. 2003. Characterization of the poliovirus 147S particle: new insights into poliovirus uncoating. Virology 305:55-65. [DOI] [PubMed] [Google Scholar]
- 30.Perez, L., and L. Carrasco. 1994. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J. Gen. Virol. 75:2595-2606. [DOI] [PubMed] [Google Scholar]
- 31.Rexroad, J., C. M. Wiethoff, A. P. Green, T. D. Kierstead, M. O. Scott, and C. R. Middaugh. 2003. Structural stability of adenovirus type 5. J. Pharm. Sci. 92:665-677. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz, M. C., A. Charpilienne, F. Liprandi, R. Gajardo, F. Michelangeli, and J. Cohen. 1996. The concentration of Ca2+ that solubilizes outer capsid proteins from rotavirus particles is dependent on the strain. J. Virol. 70:4877-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindler, D. G., and J. E. Davies. 1977. Specific cleavage of ribosomal RNA caused by alpha sarcin. Nucleic Acids Res. 4:1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seth, P. 1994. Adenovirus-dependent release of choline from plasma membrane vesicles at an acidic pH is mediated by the penton base protein. J. Virol. 68:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seth, P., D. Fitzgerald, H. Ginsberg, M. Willingham, and I. Pastan. 1984. Evidence that the penton base of adenovirus is involved in potentiation of toxicity of Pseudomonas exotoxin conjugated to epidermal growth factor. Mol. Cell. Biol. 4:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seth, P., D. J. Fitzgerald, M. C. Willingham, and I. Pastan. 1984. Role of a low-pH environment in adenovirus enhancement of the toxicity of a Pseudomonas exotoxin-epidermal growth factor conjugate. J. Virol. 51:650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seth, P., I. Pastan, and M. C. Willingham. 1985. Adenovirus-dependent increase in cell membrane permeability. J. Biol. Chem. 260:9598-9602. [PubMed] [Google Scholar]
- 39.Seth, P., M. C. Willingham, and I. Pastan. 1984. Adenovirus-dependent release of 51Cr from KB cells at an acidic pH. J. Biol. Chem. 259:14350-14353. [PubMed] [Google Scholar]
- 40.Seth, P., M. C. Willingham, and I. Pastan. 1985. Binding of adenovirus and its external proteins to Triton X-114. Dependence on pH. J. Biol. Chem. 260:14431-14434. [PubMed] [Google Scholar]
- 41.Sirena, D., B. Lilienfeld, M. Eisenhut, S. Kalin, K. Boucke, R. R. Beerli, L. Vogt, C. Ruedl, M. F. Bachmann, U. F. Greber, and S. Hemmi. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 78:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spendlove, R. S., M. E. McClain, and E. H. Lennette. 1970. Enhancement of reovirus infectivity by extracellular removal or alteration of the virus capsid by proteolytic enzymes. J. Gen. Virol. 8:83-94. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, P. L., S. D. Fuller, and R. M. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12:2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tillotson, L., and A. J. Shatkin. 1992. Reovirus polypeptide σ3 and N-terminal myristoylation of polypeptide μ1 are required for site-specific cleavage to μ1C in transfected cells. J. Virol. 66:2180-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang, S. K., B. M. McDermott, V. R. Racaniello, and J. M. Hogle. 2001. Kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J. Virol. 75:4984-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Oostrum, J., and R. M. Burnett. 1985. Molecular composition of the adenovirus type 2 virion. J. Virol. 56:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber, J. 1976. Genetic analysis of adenovirus type 2. III. Temperature sensitivity of processing viral proteins. J. Virol. 17:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin αvβ5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, G. J., E. L. Nason, C. S. Hardy, D. H. Ebert, J. D. Wetzel, B. V. Venkataram Prasad, and T. S. Dermody. 2002. A single mutation in the carboxy terminus of reovirus outer-capsid protein σ3 confers enhanced kinetics of σ3 proteolysis, resistance to inhibitors of viral disassembly, and alterations in σ3 structure. J. Virol. 76:9832-9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 53.Wodrich, H., T. Guan, G. Cingolani, D. Von Seggern, G. Nemerow, and L. Gerace. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22:6245-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, E., S. A. Trauger, L. Pache, T. M. Mullen, D. J. von Seggern, G. Siuzdak, and G. R. Nemerow. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J. Virol. 78:3897-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]