Abstract
Purpose
Inherited retinal dystrophies are a significant cause of vision loss and are characterized by the loss of photoreceptors and the retinal pigment epithelium (RPE). Mutations in approximately 250 genes cause inherited retinal degenerations with a high degree of genetic heterogeneity. New techniques in next-generation sequencing are allowing the comprehensive analysis of all retinal disease genes thus changing the approach to the molecular diagnosis of inherited retinal dystrophies. This review serves to analyze clinical progress in genetic diagnostic testing and implications for retinal gene therapy.
Methods
A literature search of PubMed and OMIM was conducted to relevant articles in inherited retinal dystrophies.
Results
Next-generation genetic sequencing allows the simultaneous analysis of all the approximately 250 genes that cause inherited retinal dystrophies. Reported diagnostic rates range are high and range from 51% to 57%. These new sequencing tools are highly accurate with sensitivities of 97.9% and specificities of 100%. Retinal gene therapy clinical trials are underway for multiple genes including RPE65, ABCA4, CHM, RS1, MYO7A, CNGA3, CNGB3, ND4, and MERTK for which a molecular diagnosis may be beneficial for patients.
Conclusion
Comprehensive next-generation genetic sequencing of all retinal dystrophy genes is changing the paradigm for how retinal specialists perform genetic testing for inherited retinal degenerations. Not only are high diagnostic yields obtained, but mutations in genes with novel clinical phenotypes are also identified. In the era of retinal gene therapy clinical trials, identifying specific genetic defects will increasingly be of use to identify patients who may enroll in clinical studies and benefit from novel therapies.
Keywords: inherited retinal dystrophy, retinal gene therapy, genetic sequencing, retinitis pigmentosa
Inherited retinal dystrophies can be classified as to whether they cause degeneration of rod and/or cone photoreceptors.1 These retinal disorders are associated with a wide degree of genetic heterogeneity. Specifically, mutations in many genes can lead to a single clinical phenotype. In the Retinal Information Network (http://www.sph.uth.tmc.edu/RetNet/), there are approximately 250 retinal disease–causing genes. Supplemental Digital Content (see Table 1, http://links.lww.com/IAE/A538) shows a list of genes causing inherited retinal dystrophies based on information contained within the Retinal Information Network, the Online Mendelian Inheritance in Man (OMIM) database, the genetic eye disease gene list, and the National Ophthalmic Disease Genotyping and Phenotyping Network.2–5 Many of the genes listed in this table lead to a similar retinal dystrophy phenotype.6–8 Additionally, there are likely to be clinical phenotypes associated with genetic mutations that have not yet been identified. Current diagnostic testing including fundoscopic examination, visual field testing, optical coherence tomography, dark adaptation, fundus autofluorescence, and electroretinography rarely leads to the prediction of a specific genetic mutation; therefore, genetic sequencing is necessary to identify the causative retinal disease gene.
Traditional methods of genetic sequencing include Sanger single-gene sequencing; however, it is both costly and time consuming to screen all retinal dystrophy genes. Arrayed primer extension genotyping microarrays are available; however, they only test for known variants, and thus are limited to detecting known mutations.9 Recently, next-generation sequencing using targeted panels of all known and candidate retinal dystrophy genes has been developed and yields high diagnostic rates.3 This sequencing technology shows promise for the identification of retinal disease–causing genes and the identification of patients who may benefit from inclusion in retinal gene therapy clinical trials. A summary of ongoing retinal gene therapy clinical trials available through the National Institutes of Health registry (www.clinicaltrials.gov) and preclinical gene therapy studies are displayed in Supplemental Digital Content (see Table 1, http://links.lww.com/IAE/A538).10–33
Retinal Gene Therapy Clinical Trials
65 kDa Retinal Pigment Epithelium–Specific Protein–Associated Leber Congenital Amaurosis
Leber congenital amaurosis is an inherited retinal degeneration that causes visual loss early in childhood through dysfunction of both rod and cone photoreceptors. Full-field electroretinography typically shows severely decreased to absent cone and rod responses. A Phase III clinical trial of subretinally delivered 65 kDa retinal pigment epithelium–specific protein (RPE65) (OMIM 180069) in patients with RPE65-associated Leber congenital amaurosis was recently completed. Although the data have not yet been published in a peer-review journal, it was reportedly successful at improving both sensitivity to light and functional vision34,35 (ClinicalTrials.gov identifier: NCT00999609).
In this trial, patients with a confirmed mutation in RPE65 were randomized and those in the intervention group underwent a three-port vitrectomy, and an adeno-associated viral vector containing RPE65 was delivered subretinally in both eyes. This virus served as a delivery vehicle allowing the production of the RPE65 protein in the RPE. The 20 intervention patients showed a functional improvement in vision with a 1.9 specified lux level improvement in bilateral mobility testing at 1 year.34 Over one third of the patients in the treatment group (7 of 20) demonstrated a 15-letter improvement in the first eye treated at 1 year compared with none in the control group.35 Thirteen of the 20 intervention patients were able to complete the mobility test at 1 year at 1 lux, similar to a low light condition compared with none of the control patients. Intervention patients also showed a statistically significant improvement in full-field light sensitivity threshold testing for white light compared with control patients. Given the success of this randomized controlled Phase III trial, Spark Therapeutics of Philadelphia, PA plans to submit a Biologics Licensing Application for Food and Drug Administration approval, and if approved, will be the first gene therapy available in the United States.
Stargardt Macular Dystrophy
In addition to RPE65-associated Leber congenital amaurosis, a Phase II/III clinical trial is in progress for ATP-binding cassette, subfamily A, member 4 (ABCA4)-associated Stargardt macular dystrophy (OMIM 601691), the most common juvenile macular dystrophy. Stargardt disease is characterized by progressive accumulation of lipofuscin material in the RPE with associated macular atrophy. Subretinally delivered ABCA4 using StarGen, a lentiviral viral vector, is being tested in patients with Stargardt macular dystrophy (ClinicalTrials.gov identifiers: NCT01367444 and NCT01736592). The advantage of the lentiviral vector as compared with the adeno-associated viral vector is that it has a larger genetic carrying capacity, which allows it to package the ABCA4 gene, which is 6.8 kb. Proof-of-concept studies in a mouse model of Stargardt disease using ABCA4-based gene therapy showed improved disease phenotypes with decreased lipofuscin accumulation.36 The human early phase study completion date is expected in 2017, and if the results are positive, it would allow the initiation of a Phase III gene therapy clinical trial for Stargardt disease.
Choroideremia
Choroideremia is an X-linked retinal dystrophy that primarily affects men and is caused by mutations in the CHM gene (OMIM 300390). Clinically, patients present with loss of night vision and peripheral visual constriction that eventually affects central visual acuity. A Phase I/II clinical trial of subretinally delivered CHM in six patients with choroideremia was recently completed and showed the therapy to be safe (ClinicalTrials.gov identifier: NCT01461213).37 One third of the patients in the treatment group (2 of 6) had more than 15-letter improvement at 3.5 years compared with none in the control group.38 The other 4 patients had good baseline visual acuity, and thus limited potential for gains in visual function.38 Based in part on these results, multiple Phases I and II clinical studies enrolling patients with confirmed mutations in CHM are being initiated or ongoing (ClinicalTrials.gov identifiers: NCT02341807, NCT02077361, NCT02553135, NCT02671539, and NCT02407678).
MYO7A-Associated Usher Syndrome Type I
MYO7A-associated Usher syndrome Type I is an inherited retinal dystrophy characterized by congenital sensory hearing loss and progressive retinitis pigmentosa (OMIM 276903). Preclinical work showed subretinal delivery of Myo7A to be effective at preventing light-induced retinal degeneration in a mouse model of disease and to be safe in rhesus macaques.39 Based on this work, a Phase I/II clinical study using UshStat in patients with MYO7A-associated Usher syndrome is currently underway (ClinicalTrials.gov identifier: NCT01505062).
X-linked Retinoschisis
Retinoschisin (RS1)-associated X-linked retinoschisis is an inherited retinal dystrophy characterized by schisis of the neurosensory retina (OMIM 300839). It is a common inherited cause of decreased central visual acuity in men. Preclinical gene delivery of human RS1 in a mouse model of the disease led to improvements in retinal morphology and retinal function as measured by electroretinography testing.40 Based on these results, two Phase I/II clinical trials of intravitreally delivered RS1 for X-linked retinoschisis are underway (ClinicalTrials. gov identifiers: NCT02317887 and NCT02416622).
MERTK-Associated Retinitis Pigmentosa
Retinitis pigmentosa is a characterized by loss of rod photoreceptors leading to night blindness and constricted peripheral vision. Over time, progressive loss of rod photoreceptors causes cone photoreceptors to be affected resulting in decreased central visual acuity. MER Proto-Oncogene, Tyrosine Kinase (Mertk) (OMIM 604705) gene replacement therapy in a rat model of disease resulted in both functional and structural retina preservation.41
Based on this proof-of-concept study, a phase I trial of subretinally administered MERTK for MERTK-associated retinitis pigmentosa took place in six patients and demonstrated peak gains of greater than three lines of vision in one third (two of six) patients.42 Visual gains were maintained in one patient and declined in the other patient at 2 years. While progression of disease cannot be excluded, visual decline may have been associated with the development of a posterior subcapsular cataract from the vitrectomy, as spectral-domain optical coherence tomography showed stable central macular thickness.42 The primary outcome of safety was demonstrated in this Phase I clinical trial (ClinicalTrials.gov identifier: NCT01482195). Further studies will be needed to assess the efficacy of MERTK gene replacement therapy.
ND4-Associated Leber Hereditary Optic Neuropathy
Leber hereditary optic neuropathy is a maternally inherited mitochondrial optic neuropathy resulting in optic nerve dysfunction and visual loss. A Phase I/II clinical trial of intravitreally administered NADH dehydrogenase subunit 4 Complex I (ND4) was performed in nine patients with Leber hereditary optic neuropathy due to point mutations of guanine to adenine at position 11,778 (ClinicalTrials.gov identifier: NCT01267422). At 9 months follow-up, 6 of 9 patients had an improvement in visual acuity equal to or greater than 0.3 logarithm of the minimum angle of resolution with enlargement of visual fields.43 Ongoing Phase I (ClinicalTrials.gov identifier: NCT02161380) and Phase I/II (ClinicalTrials.gov identifier: NCT02064569) studies for Leber hereditary optic neuropathy will provide further data on the safety and efficacy of intravitreal ND4 gene therapy for Leber hereditary optic neuropathy.
CNGA3 and CNGB3-Linked Achromatopsia
Achromatopsia is a cone dystrophy, which manifests clinically as color blindness, photophobia, and decreased visual acuity. Electroretinography reveals reduced to absent physiologically measurable cone responses. It has been reported that approximately 25% of cases of achromatopsia are due to mutations in the α-subunit of the cone cGMP-gated channel (CNGA3).44 Subretinal delivery of CNGA3 to a sheep model of achromatopsia improved cone responses on electroretinography testing as well as behavioral maze testing.16 Phase I/II clinical studies of subretinally injected CNGA3 and CNGB3 using adeno-associated viral vector are being initiated to assess the safety and efficacy of this treatment in patients (ClinicalTrials.gov identifier: NCT02610582 and NCT02599922).
Genetic and Clinical Heterogeneity Underlying Inherited Retinal Dystrophies
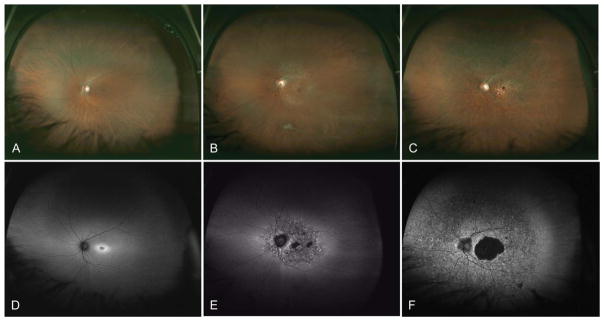
Currently approximately 250 genes have been identified that cause inherited retinal dystrophies.5 Inherited retinal degenerations show significant genetic heterogeneity, which rarely allows a genetic diagnosis to be determined based on clinical phenotype alone. For example, more than 100 different genes have been associated with retinitis pigmentosa.45 Twenty-three genes have been associated with Leber congenital amaurosis. Furthermore, mutations in a many genes such as RPE65, LRAT, MERTK, SPATA7, and TULP1 can all cause either Leber congenital amaurosis or early-onset retinitis pigmentosa, which makes single-gene genetic testing for these conditions both costly and time consuming. Genetic heterogeneity also exists for cone dystrophies, cone–rod dystrophies, and macular dystrophies. In the Retinal Information Network database, 25 genes have been implicated as causing cone–rod dystrophy. As an example, mutations in ABCA4 can cause macular dystrophy with degeneration of foveal cones (Figure 1A), cone dystrophy (Figure 1B), cone–rod dystrophy (Figure 1C), and retinitis pigmentosa (rod–cone dystrophy).
Fig. 1.
Variety of retinal dystrophies associated with homozygous ABCA4 mutations. Images in A and autofluorescence in D show an area of parafoveal hyperautofluorescence associated with a macular dystrophy. Images in B and autofluorescence in E show a small central area of hypoautofluorescence corresponding to RPE atrophy with surrounding flecks of hyperautofluorescence associated with a cone dystrophy. Images in C and autofluorescence in F show a central area of hypoautofluorescence in the macula with peripheral areas of punctate hyperautofluorescence and hypoautofluorescence associated with a cone–rod dystrophy.
Fundus autofluorescence demonstrates the variety of retinal pathologies resulting from genetic mutations in ABCA4. In Figure 1D, the retina of a patient with Stargardt macular dystrophy shows parafoveal hyperautofluorescence corresponding to an increase in lipofuscin in the RPE.46 Fundus autofluorescence of a patient with ABCA4-associated cone dystrophy shows a small central area of hypoautofluorescence corresponding to RPE atrophy with surrounding flecks of hyperautofluorescence (Figure 1E). Fundus autofluorescence of a patient with ABCA4-associated cone–rod dystrophy demonstrates a central area of hypoautofluorescence in the macula with peripheral areas of punctate hyperautofluorescence and hypoautofluorescence (Figure 1F). Of note, given the sensitivity of fundus autofluorescence for detecting subtle retinal pathology in the RPE, performing a fluorescein angiogram to examine for the lack of RPE autofluorescence (dark choroid) is unnecessary for the clinical diagnosis. These clinical examinations show the wide variety of clinical phenotypes that may result from a mutation in ABCA4.
Additionally, mutations in USH2A can cause both Usher syndrome with associated hearing loss and USH2A-associated retinitis pigmentosa without hearing loss.47 Mutations in CRB1 can result in Leber congenital amaurosis, retinitis pigmentosa, and cone–rod dystrophy.48 The genetic heterogeneity makes predicting the genetic mutation based on the clinical phenotype rarely possible. While single-gene sequencing is occasionally able to determine the predicted genetic defect, this technology is laborious and not feasible to perform for all known retinal dystrophy genes. The development of targeted next-generation sequencing of all retinal degeneration genes is a major advance for the molecular diagnosis of inherited retinal dystrophies, and is yielding high diagnostic yields.3 Furthermore, the identification of genetic defects identifies patients who may benefit from inclusion in retinal gene therapy clinical trials as well as genome editing using CRISPR technology described below.
Recently, a new approach using nucleases such as Cas9 has enabled targeted, site-specific genome modifications in mammalian organisms. This would represent an advance over retinal gene therapy for three reasons. First, it would allow genome editing of genes, which are too large to be contained within adeno-associated viral vectors. Second, genome editing would enable both autosomal-dominant and autosomal-recessive inherited retinal dystrophies to be targeted. Third, CRISPR gene editing technology would allow autologous induced pluripotent stem cells to be differentiated into photoreceptors, genetically modified, and transplanted subretinally to potentially reverse loss of vision. This gene editing approach was successfully able to reverse the phenotype in patient-derived photoreceptors with a point mutation in the gene CEP290.49 Editas Medicine of Cambridge, MA hopes to perform a human clinical trial using this technology for CEP290-associated Leber congenital amaurosis by 2017. If successful, it would potentially be able to be applied to many additional inherited retinal dystrophies.
Comprehensive Genetic Testing for Inherited Retinal Dystrophies
Given the recent increase in knowledge about genes associated with inherited retinal dystrophies, how can a retinal specialist translate this into clinical practice? For patients with a family history of a known genetic mutation, performing single-gene sequencing is a logical approach to confirm the molecular diagnosis. For example, if a sibling has a known mutation in ABCA4, performing Sanger single-gene sequencing will validate the genetic diagnosis. However, as in most cases, if the genetic mutation is unknown, the use of single-gene sequencing is not feasible to perform routinely for all approximately 250 known genes associated with inherited retinal dystrophies.
Over the past few years, comprehensive next-generation sequencing has been developed and is currently being used for the molecular diagnosis of inherited retinal dystrophies at multiple institutions. This approach uses high-throughput genetic sequencing in parallel of all known genes associated with inherited retinal dystrophies. Specifically, the patient’s genomic DNA is fragmented into small pieces. An RNA probe set specific for the target inherited retinal dystrophy genes and pathogenic noncoding regions is used to capture DNA fragments using streptavidin beads. Finally, selected regions are amplified and sequenced using specifically designed polymerase chain reaction primers.3
In a recently described approach, a genetic eye disease sequencing panel was generated toward all known retinal disease genes. This genetic eye disease sequencing panel yielded a high diagnostic rate of 51% in 192 patients with inherited retinal degenerations.3 The sensitivity was 97.9% and specificity 100% for variant detection.3 Next-generation sequencing data were shown to have equal quality to Sanger sequencing.50 A similar next-generation sequencing approach targeted toward a different panel of 105 retinal dystrophy genes yielded a detection rate of 50%.51 In patients for whom the disease-causing mutation was not identified, whole-exome sequencing led to the identification of pathogenic mutations in five of eight cases.51 In one case unsolved by whole-exome sequencing, whole-genome sequencing led to the identification of a disease-causing intronic mutation.51
Once genetic defects are identified by next-generation sequencing, Sanger single-gene sequencing is able to confirm the mutation. As new retinal disease genes are identified, they are continually added to the next-generation sequencing panel. Of note, blood samples may be shipped at room temperature and insurance companies are increasingly providing coverage.
In other recent studies, next-generation sequencing using a comprehensive retinal dystrophy panel of genes yielded a higher diagnostic rate than using a narrower panel of genes. In 50 patients with retinitis pigmentosa or cone–rod dystrophy, using a next-generation sequencing panel of 73 inherited retinal dystrophy genes yielded a diagnostic yield of 25%.52 Using a more comprehensive panel that includes all known and candidate 254 retinal dystrophy genes led to a diagnostic yield of 57%,53 which supports the use of a nonbiased, comprehensive sequencing approach.
Conclusion
Given ongoing retinal gene therapy clinical trials for inherited retinal degenerations, the identification of the disease-causing gene allows patients to potentially benefit from inclusion in clinical studies. As inherited retinal dystrophies are genetically heterogeneous, the use of high-throughput next-generation sequencing yields high diagnostic rates. These sequencing platforms are currently clinically available for retinal specialists to use on a Clinical Laboratories Improvement Amendments-certified basis. As retinal gene therapies become available for patients with inherited retinal dystrophies such as for RPE65, patients will increasingly benefit from genetic diagnoses.
Supplementary Material
Acknowledgments
This work was supported by the NIH/NEI K08 EY026652 Mentored Clinical Scientist Award. The author serves as a consultant to Achillion Pharmaceuticals and receives research funding from Astellas Pharmaceuticals.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the this work was supported by the NIH/NEI K08 EY026652 Mentored Clinical Scientist Award. This article on the journal’s Web site (www.retinajournal.com).
References
- 1.den Hollander AI, Black A, Bennett J, Cremers FP. Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies. J Clin Invest. 2010;120:3042–3053. doi: 10.1172/JCI42258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz KE, Reeves MJ, Tumminia SJ, Brooks BP. eyeGENE (R): a novel approach to combine clinical testing and researching genetic ocular disease. Curr Opin Ophthalmol. 2012;23:355–363. doi: 10.1097/ICU.0b013e32835715c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consugar MB, Navarro-Gomez D, Place EM, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2015;17:253–261. doi: 10.1038/gim.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM. org: Online Mendelian Inheritance in Man (OMIM), an online catalog of human genes and genetic disorders. Nucleic Acids Research. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daiger SP, Rossiter BF, Greenberg J, et al. Data services and software for identifying genes and mutations causing retinal degeneration. Invest Ophthalmol Vis Sci. 1998;39:S295. [Google Scholar]
- 6.Boon CJ, den Hollander AI, Hoyng CB, et al. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog Retin Eye Res. 2008;27:213–235. doi: 10.1016/j.preteyeres.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Boon CJ, Klevering BJ, Leroy BP, et al. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog Retin Eye Res. 2009;28:187–205. doi: 10.1016/j.preteyeres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Schorderet DF, Escher P. NR2E3 mutations in enhanced S-cone sensitivity syndrome (ESCS), Goldmann-Favre syndrome (GFS), clumped pigmentary retinal degeneration (CPRD), and retinitis pigmentosa (RP) Hum Mutat. 2009;30:1475–1485. doi: 10.1002/humu.21096. [DOI] [PubMed] [Google Scholar]
- 9.Jaakson K, Zernant J, Kulm M, et al. Genotyping microarray (gene chip) for the ABCR (ABCA4) gene. Hum Mutat. 2003;22:395–403. doi: 10.1002/humu.10263. [DOI] [PubMed] [Google Scholar]
- 10.Zhong H, Eblimit A, Moayedi Y, et al. AAV8(Y733F)-mediated gene therapy in a Spata7 knockout mouse model of Leber congenital amaurosis and retinitis pigmentosa. Gene Ther. 2015;22:619–627. doi: 10.1038/gt.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng WT, Dyka FM, Dinculescu A, et al. Stability and safety of an AAV vector for treating RPGR-ORF15 x-linked retinitis pigmentosa. Hum Gene Ther. 2015;26:593–602. doi: 10.1089/hum.2015.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi VW, Bigelow CE, McGee TL, et al. AAV-mediated RLBP1 gene therapy improves the rate of dark adaptation in Rlbp1 knockout mice. Mol Ther Methods Clin Dev. 2015;2:15022. doi: 10.1038/mtm.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao H, Gorbatyuk MS, Rossmiller B, et al. Long-term rescue of retinal structure and function by rhodopsin RNA replacement with a single adeno-associated viral vector in P23H RHO transgenic mice. Hum Gene Ther. 2012;23:356–366. doi: 10.1089/hum.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlichtenbrede FC, da Cruz L, Stephens C, et al. Long-term evaluation of retinal function in Prph2Rd2/Rd2 mice following AAV-mediated gene replacement therapy. J Gene Med. 2003;5:757–764. doi: 10.1002/jgm.401. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho LS, Xu J, Pearson RA, et al. Long-term and age-dependent restoration of visual function in a mouse model of CNGB3-associated achromatopsia following gene therapy. Hum Mol Genet. 2011;20:3161–3175. doi: 10.1093/hmg/ddr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banin E, Gootwine E, Obolensky A, et al. Gene augmentation therapy restores retinal function and visual behavior in a sheep model of CNGA3 achromatopsia. Mol Ther. 2015;23:1423–1433. doi: 10.1038/mt.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boye SL, Peterson JJ, Choudhury S, et al. Gene therapy fully restores vision to the all-cone NrlGucy2e mouse model of leber congenital amaurosis-1. Hum Gene Ther. 2015;26:575–592. doi: 10.1089/hum.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellissier LP, Quinn PM, Alves CH, et al. Gene therapy into photoreceptors and Muller glial cells restores retinal structure and function in CRB1 retinitis pigmentosa mouse models. Hum Mol Genet. 2015;24:3104–3118. doi: 10.1093/hmg/ddv062. [DOI] [PubMed] [Google Scholar]
- 19.Nishiguchi KM, Carvalho LS, Rizzi M, et al. Gene therapy restores vision in rd1 mice after removal of a confounding mutation in Gpr179. Nat Commun. 2015;6:6006. doi: 10.1038/ncomms7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ku CA, Chiodo VA, Boye SL, et al. Viral-mediated vision rescue of a novel AIPL1 cone-rod dystrophy model. Hum Mol Genet. 2015;24:670–684. doi: 10.1093/hmg/ddu487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burnight ER, Wiley LA, Drack AV, et al. CEP290 gene transfer rescues Leber congenital amaurosis cellular phenotype. Gene Ther. 2014;21:662–672. doi: 10.1038/gt.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch S, Sothilingam V, Garcia Garrido M, et al. Gene therapy restores vision and delays degeneration in the CNGB1(−/−) mouse model of retinitis pigmentosa. Hum Mol Genet. 2012;21:4486–4496. doi: 10.1093/hmg/dds290. [DOI] [PubMed] [Google Scholar]
- 23.Dinculescu A, Min SH, Deng WT, et al. Gene therapy in the rd6 mouse model of retinal degeneration. Adv Exp Med Biol. 2014;801:711–718. doi: 10.1007/978-1-4614-3209-8_89. [DOI] [PubMed] [Google Scholar]
- 24.Dai X, Han J, Qi Y, et al. AAV-mediated lysophosphatidylcholine acyltransferase 1 (Lpcat1) gene replacement therapy rescues retinal degeneration in rd11 mice. Invest Ophthalmol Vis Sci. 2014;55:1724–1734. doi: 10.1167/iovs.13-13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz NM, Yuan Y, Leehy BD, et al. Modifier genes as therapeutics: the nuclear hormone receptor Rev Erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS One. 2014;9:e87942. doi: 10.1371/journal.pone.0087942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low BE, Krebs MP, Joung JK, et al. Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6N mice via TALEN-mediated homology-directed repair. Invest Ophthalmol Vis Sci. 2014;55:387–395. doi: 10.1167/iovs.13-13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lheriteau E, Petit L, Weber M, et al. Successful gene therapy in the RPGRIP1-deficient dog: a large model of cone-rod dystrophy. Mol Ther. 2014;22:265–277. doi: 10.1038/mt.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo S, Mullins RF, Dumitrescu AV, et al. Subretinal gene therapy of mice with Bardet-Biedl syndrome type 1. Invest Ophthalmol Vis Sci. 2013;54:6118–6132. doi: 10.1167/iovs.13-11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molday LL, Djajadi H, Yan P, et al. RD3 gene delivery restores guanylate cyclase localization and rescues photoreceptors in the Rd3 mouse model of Leber congenital amaurosis 12. Hum Mol Genet. 2013;22:3894–3905. doi: 10.1093/hmg/ddt244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Collin RW, Cremers FP, et al. Expression of wild-type Rp1 protein in Rp1 knock-in mice rescues the retinal degeneration phenotype. PLoS One. 2012;7:e43251. doi: 10.1371/journal.pone.0043251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guziewicz KE, Zangerl B, Komaromy AM, et al. Recombinant AAV-mediated BEST1 transfer to the retinal pigment epithelium: analysis of serotype-dependent retinal effects. PLoS One. 2013;8:e75666. doi: 10.1371/journal.pone.0075666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh MS, Broadgate S, Mathur R, et al. Hypotrichosis and juvenile macular dystrophy caused by CDH3 mutation: a candidate disease for retinal gene therapy. Sci Rep. 2016;6:23674. doi: 10.1038/srep23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiting RE, Jensen CA, Pearce JW, et al. Intracerebroventricular gene therapy that delays neurological disease progression is associated with selective preservation of retinal ganglion cells in a canine model of CLN2 disease. Exp Eye Res. 2016;146:276–282. doi: 10.1016/j.exer.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. [Accessed June 8, 2016];Spark Therapeutics Investor Relations. [Press Release]. Available at: http://ir.sparktx.com.
- 35.Maguire A. Phase 3 trial of AAV2-hRPE65v2 (SPK-RPE65) to treat RPE65 mutation-associated inherited retinal dystrophies. Paper presented at: American Academy of Ophthalmology 2015 Annual Meeting, Retina Subspecialty Day; November 13–14, 2015; Las Vegas, NV. [Google Scholar]
- 36.Kong J, Kim SR, Binley K, et al. Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy. Gene Ther. 2008;15:1311–1320. doi: 10.1038/gt.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLaren RE, Groppe M, Barnard AR, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards TL, Jolly JK, Groppe M, et al. Visual acuity after retinal gene therapy for choroideremia. N Engl J Med. 2016;374:1996–1998. doi: 10.1056/NEJMc1509501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zallocchi M, Binley K, Lad Y, et al. EIAV-based retinal gene therapy in the shaker1 mouse model for usher syndrome type 1B: development of UshStat. PLoS One. 2014;9:e94272. doi: 10.1371/journal.pone.0094272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min SH, Molday LL, Seeliger MW, et al. Prolonged recovery of retinal structure/function after gene therapy in an Rs1h-deficient mouse model of x-linked juvenile retinoschisis. Mol Ther. 2005;12:644–651. doi: 10.1016/j.ymthe.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Deng WT, Dinculescu A, Li Q, et al. Tyrosine-mutant AAV8 delivery of human MERTK provides long-term retinal preservation in RCS rats. Invest Ophthalmol Vis Sci. 2012;53:1895–1904. doi: 10.1167/iovs.11-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghazi NG, Abboud EB, Nowilaty SR, et al. Treatment of retinitis pigmentosa due to MERTK mutations by ocular sub-retinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum Genet. 2016;135:327–343. doi: 10.1007/s00439-016-1637-y. [DOI] [PubMed] [Google Scholar]
- 43.Wan X, Pei H, Zhao MJ, et al. Efficacy and safety of rAAV2-ND4 treatment for Leber’s hereditary optic neuropathy. Sci Rep. 2016;6:21587. doi: 10.1038/srep21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wissinger B, Gamer D, Jagle H, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–737. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chizzolini M, Galan A, Milan E, et al. Good epidemiologic practice in retinitis pigmentosa: from phenotyping to biobanking. Curr Genomics. 2011;12:260–266. doi: 10.2174/138920211795860071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Ruckmann A, Fitzke FW, Bird AC. Distribution of fundus autofluorescence with a scanning laser ophthalmoscope. Br J Ophthalmol. 1995;79:407–412. doi: 10.1136/bjo.79.5.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet. 2000;66:1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrenberg M, Pierce EA, Cox GF, Fulton AB. CRB1: one gene, many phenotypes. Semin Ophthalmol. 2013;28:397–405. doi: 10.3109/08820538.2013.825277. [DOI] [PubMed] [Google Scholar]
- 49.Morgan L, Maeder SS, Burnight ER, et al. Therapeutic correction of an LCA-causing splice defect in the CEP290 gene by CRISPR/cas-mediated genome editing. American Society of Gene & Cell Therapy (ASGCT) 18th Annual Meeting; Saturday; May 16, 2015.2015. [Google Scholar]
- 50.Sikkema-Raddatz B, Johansson LF, de Boer EN, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–1042. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]
- 51.Weisschuh N, Mayer AK, Strom TM, et al. Mutation detection in patients with retinal dystrophies using targeted next generation sequencing. PLoS One. 2016;11:e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shanks ME, Downes SM, Copley RR, et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet. 2013;21:274–280. doi: 10.1038/ejhg.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Audo I, Bujakowska KM, Leveillard T, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis. 2012;7:8. doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.