Abstract
Although human tumours are shaped by the genetic evolution of cancer cells, evidence also suggests that they display hierarchies related to developmental pathways and epigenetic programs in which cancer stem cells (CSCs) can drive tumour growth and give rise to differentiated progeny1. Yet, unbiased evidence for CSCs in solid human malignancies remains elusive. Here we profile 4,347 single cells from six IDH1 or IDH2 mutant human oligodendrogliomas by RNA sequencing (RNA-seq) and reconstruct their developmental programs from genome-wide expression signatures. We infer that most cancer cells are differentiated along two specialized glial programs, whereas a rare subpopulation of cells is undifferentiated and associated with a neural stem cell expression program. Cells with expression signatures for proliferation are highly enriched in this rare subpopulation, consistent with a model in which CSCs are primarily responsible for fuelling the growth of oligodendroglioma in humans. Analysis of copy number variation (CNV) shows that distinct CNV sub-clones within tumours display similar cellular hierarchies, suggesting that the architecture of oligodendroglioma is primarily dictated by developmental programs. Subclonal point mutation analysis supports a similar model, although a full phylogenetic tree would be required to definitively determine the effect of genetic evolution on the inferred hierarchies. Our single-cell analyses provide insight into the cellular architecture of oligodendrogliomas at single-cell resolution and support the cancer stem cell model, with substantial implications for disease management.
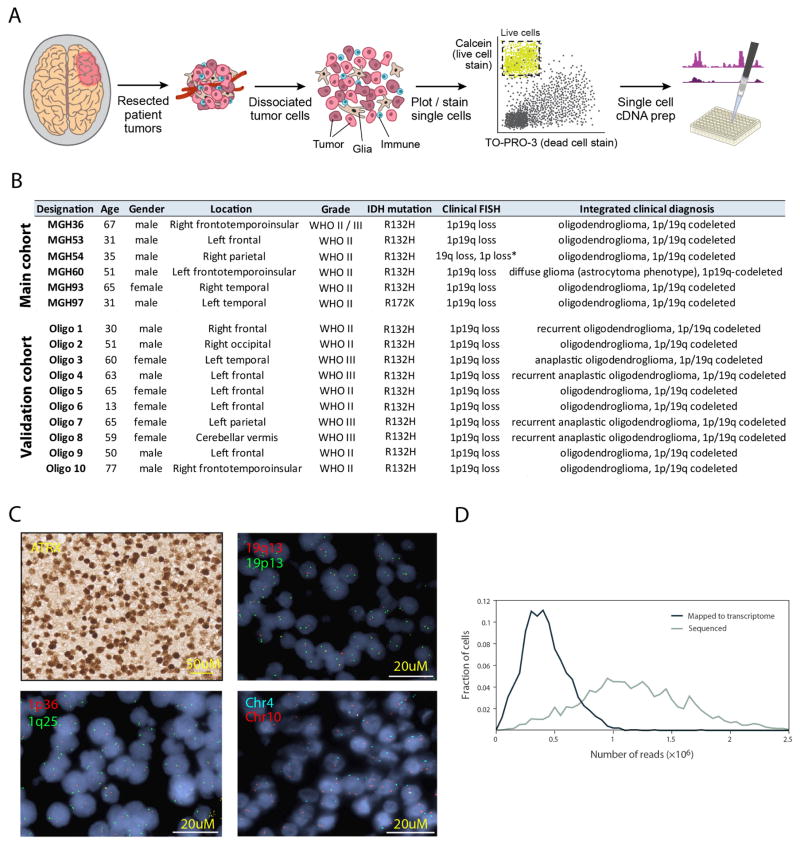
Intra-tumoural heterogeneity contributes to therapy failure and cancer progression1. Although branched genetic evolution of cancer cells is a key determinant of tumour heterogeneity, non-genetic programs such as those associated with the self-renewal of tissue stem cells and their differentiation into specialized cell types contribute further to tumour functional heterogeneity. In human gliomas, candidate CSCs have been functionally isolated in high-grade (WHO grade III–IV) lesions2. However, functional approaches such as in vivo orthotopic xenotransplantation in mice or in vitro sphere formation assays have generated controversy, as they identify candidate CSCs through selection in xenogeneic environments that are very different from the native tumour milieu and only provide limited genetic characterization of putative CSCs. In addition, it remains unknown if gliomas contain CSCs early in their development—as grade II lesions—a question central to our understanding of the initial steps of gliomagenesis3. Thus, it is critical to develop a framework that allows the analysis of cellular programs at single-cell resolution and across different genetic clones in human tumours in situ at each stage of clinical progression. We focused on oligodendroglioma, an incurable glioma characterized by mutations in IDH1 or IDH2 and co-deletion of chromosome arms 1p and 19q4. We performed single-cell RNA-seq5 (scRNA-seq) from six untreated grade II oligodendrogliomas, in which IDH1 or IDH2 mutation and 1p/19q co-deletion were confirmed (Extended Data Fig. 1a–c). Overall, we analysed 4,347 cells that passed quality controls (Methods; Extended Data Fig. 1d). Three tumours were analysed more deeply (MGH36, MGH53 and MGH54, with analysis of 791 to 1,229 cells per tumour) and three (MGH60, MGH93 and MGH97) were profiled at medium depth (430 to 598 cells analysed).
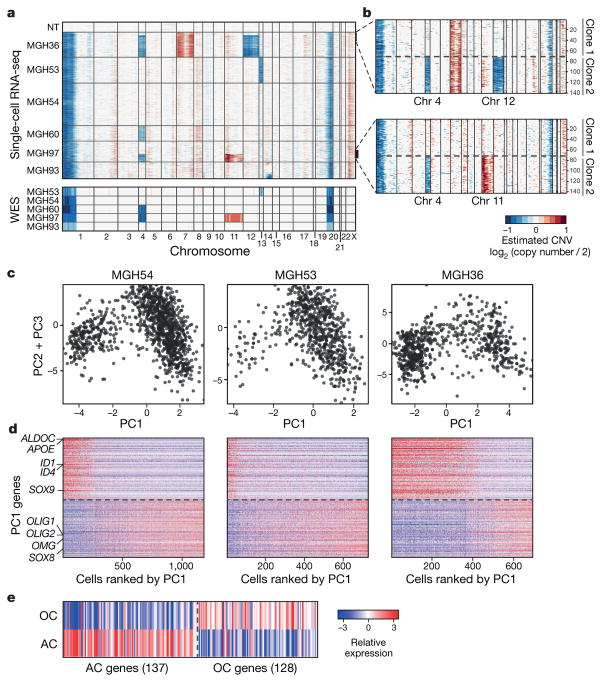
We distinguished malignant from non-malignant cells by estimating CNV from the average expression of genes in large chromosomal regions within each cell6 (Fig. 1a; Methods). Each tumour contained a large majority of cells with the 1p/19q co-deletion, as well as some cases of tumour-specific CNVs, which were validated by fluorescence in situ hybridization (FISH) and by whole-exome sequencing (WES) (Fig. 1a and Extended Data Fig. 1c). In two tumours (MGH36 and MGH97), CNV analysis identified two sub-clones (Fig. 1a, b).
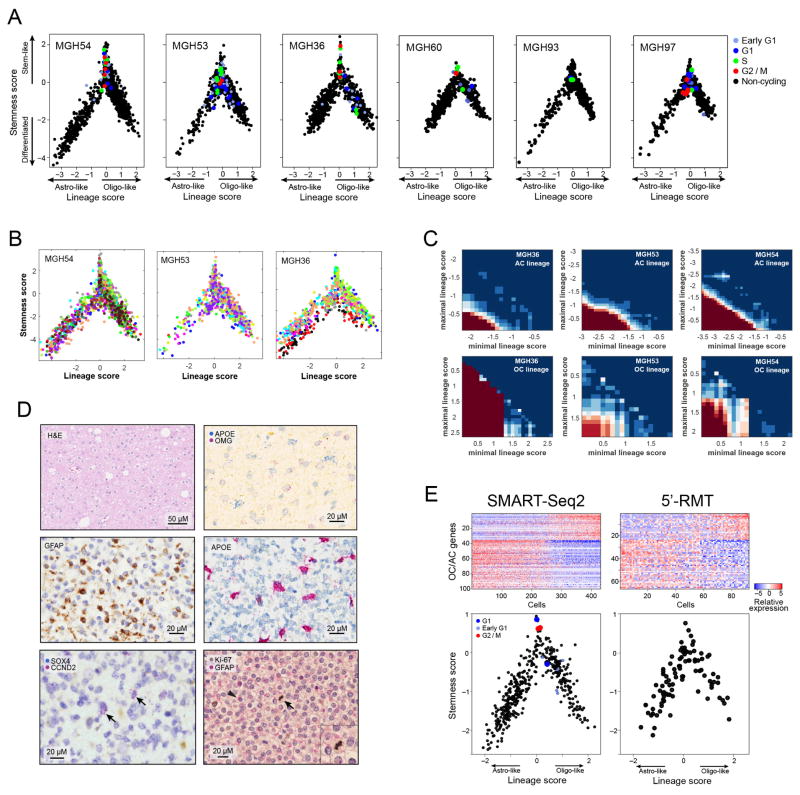
Figure 1. Single-cell RNA-seq of cancer and non-cancer cells in six oligodendrogliomas.
a, CNV profiles inferred from scRNA-seq (top) and DNA whole-exome sequencing (WES) (bottom) of oligodendrogliomas. Cells (rows, n = 4,347) are ordered from non-tumoural cells (NT, n = 303) to cancer cells (n = 4,044), ordered into six oligodendrogliomas. b, In MGH36 and MGH97, cells are ordered by CNVs, with zoomed in view shown. c, PCA of malignant cells. Shown are PC1 (x axis) versus PC2 and PC3 (y axis) scores of cells from three tumours based on a single combined PCA. d, Astrocyte-like and oligodendrocyte-like signatures. Relative expression of genes correlated most positively (bottom) or negatively (top) with PC1, in cancer cells from each of the three tumours (marked as in c), ranked by PC1 scores. Selected astrocyte (AC) and oligodendrocyte (OC) marker genes are highlighted. e, Relative expression of the mice orthologues of the genes shown in d (log2-ratio of the respective cell type compared to the average of oligodendrocyte, astrocyte, OPC and neurons)9.
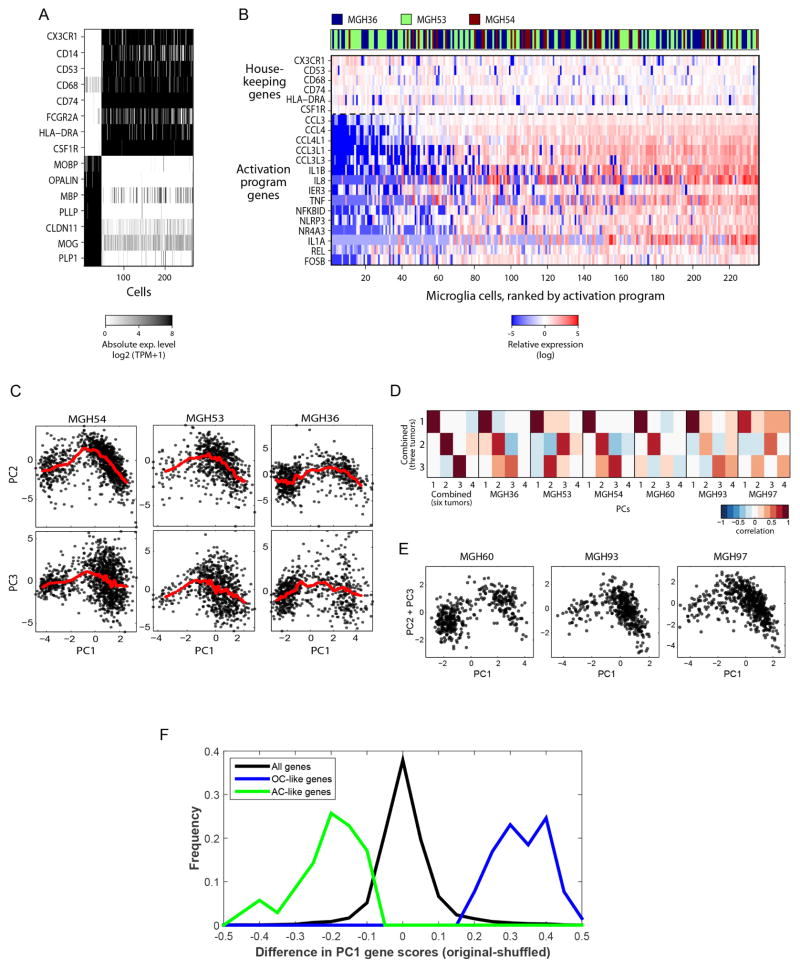
Another 303 cells lacked detectable CNVs and clustered by gene expression into subsets that expressed microglia and mature oligodendrocyte markers, respectively (Extended Data Fig. 2a). There was significant variation between the microglia cells, with a set of pro-inflammatory cytokines (IL1A, IL1B, IL8 and TNF), chemokines (CCL3, CCL4) and early response genes coordinately expressed by ~ 80% of the microglia (Extended Data Fig. 2b). This program differs from canonical macrophage M1 and M2 responses7, suggesting an unknown microglia program that may be glioma-specific.
We next examined cancer cell heterogeneity from the three tumours with the largest cell numbers. A combined principal component analysis (PCA) (Methods) identified two prominent groups of cells, corresponding to low and high PC1 scores (Fig. 1c) and expressing distinct lineage markers of astrocytes and oligodendrocytes, respectively. These results were highly consistent across all six tumours, and were not accounted for by technical or batch effects (Extended Data Fig. 2c–f and Supplementary Note 1). In each tumour, cells with high PC1 scores were strongly associated with the high expression of 137 genes, including oligodendrocytic markers (for example, OLIG1, OLIG2, OMG), and with the low expression of 128 genes, including astrocytic markers (for example, APOE, ALDOC, SOX9) (Fig. 1d, e and Supplementary Table 1)8,9. Cells with low PC1 scores had the opposite patterns of expression. This suggests that oligodendrogliomas are primarily composed of two subpopulations of glial cells, and this mirrors the histopathology4.
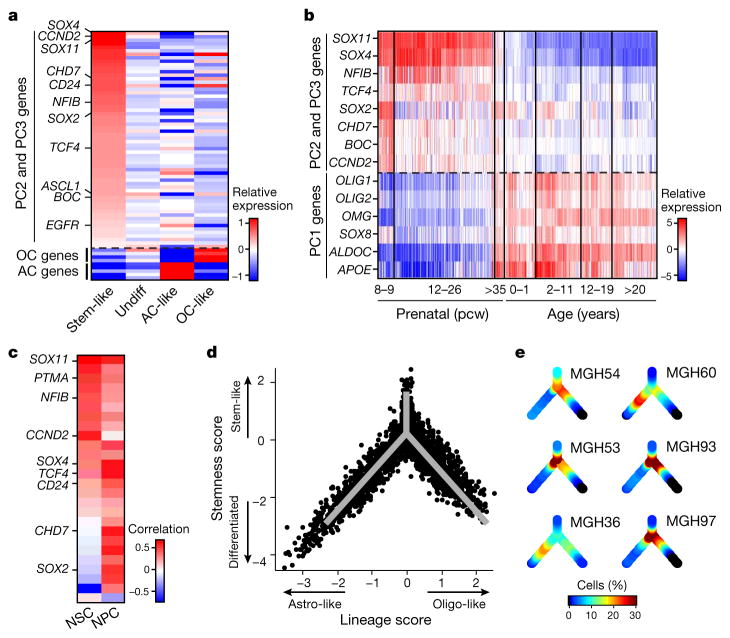
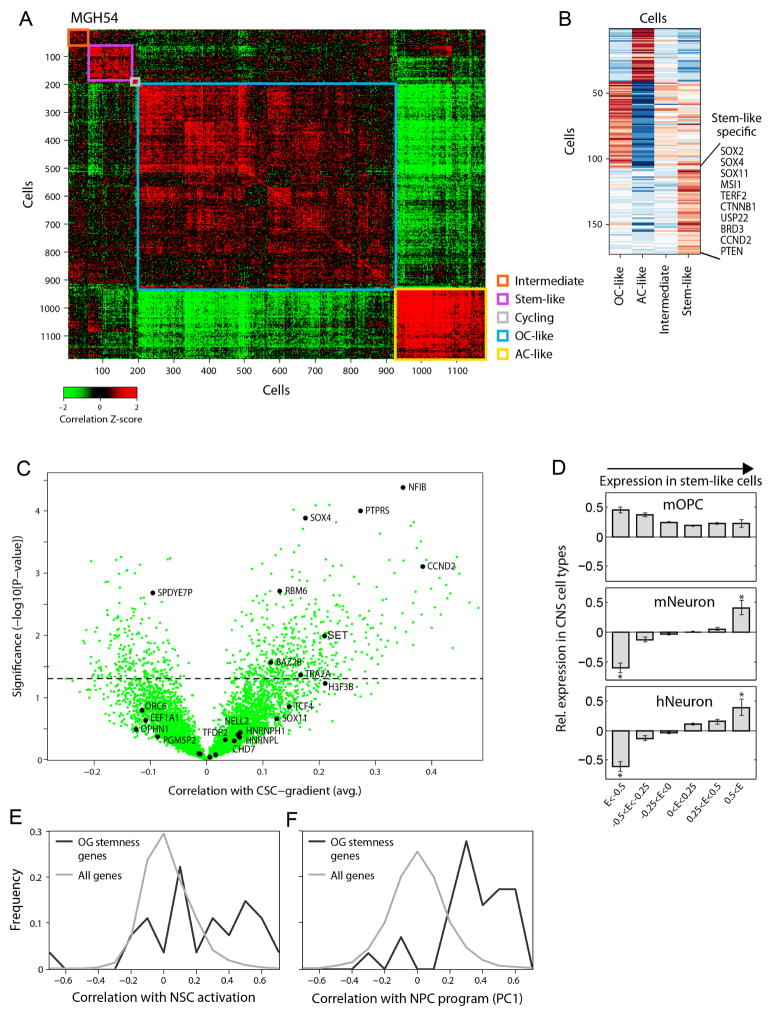
Cells with high PC2 and PC3 scores had intermediate PC1 scores (Fig. 1c and Extended Data Fig. 2c, e), suggesting a lack of differentiation, and prompting us to explore additional programs. A total of 63 genes were associated with high PC2 and PC3 (Fig. 2a, Supplementary Table 1 and Methods), and several lines of evidence suggest that they represent a ‘stemness’ program. The 20 highest-ranking genes include SOX4, SOX11 and SOX2, neurodevelopmental transcription factors critical to neural stem cells and glioma CSCs10–12. Additional signature genes with important roles in neurogenesis and in glioma CSCs included NFIB, ASCL1, CHD7, CD24, BOC and TCF4 (refs 6, 10–14). Similar results were obtained by hierarchical clustering, which showed a distinct cluster of cells preferentially expressing PC2- and PC3-associated genes (Extended Data Fig. 3a, b). Several of these genes were identified by scRNA-seq in primary glioblastoma CSCs (Extended Data Fig. 3c, P = 1.5 × 10−4, hypergeometric test). Expression of PC2- and PC3-associated regulators was highest in prenatal human brain and dropped significantly after birth, suggesting a role in early neural development (Allen Brain Atlas15, Fig. 2b, P = 8 × 10−18, t-test). Similarly, PC2- and PC3-associated genes were preferentially expressed in single cells from fetal human brain (P = 0.006, hypergeometric test)16. On the basis of these lines of evidence, we separated cells with intermediate PC1 values into ‘undifferentiated’ (low PC2 and PC3) and ‘stem/progenitors’ (high PC2 and PC3) cells (Fig. 2a).
Figure 2. Stemness expression program and a developmental hierarchy in oligodendroglioma.
a, Average relative expression of genes most highly correlated with PC2 and PC3 (top), and selected astrocyte and oligodendrocyte genes (bottom), in stem-like cells, undifferentiated cells, oligodendrocyte-like and astrocyte-like cells (Methods). Genes were sorted by relative expression in stem-like cells. b, Stemness genes are preferentially expressed in early human brain development. Relative expression of PC2 and PC3 putative stemness genes (top) and oligodendrocyte and astrocyte marker genes (bottom) across 524 human brain samples (Allen Brain Atlas). Samples are ordered in columns by age, from prenatal (left) to adult (right). c, The stemness program is correlated to mouse activated NSC and human NPCs. Pearson correlation coefficients between the expression of PC2 and PC3 genes (rows) and expression programs of mouse NSC activation19 (left) and human NPCs (right) across single cells from the respective datasets (Extended Data Figs 3e, f and 4). d, Inferred developmental hierarchy in oligodendroglioma cells (n = 4,044). Lineage and stemness scores (Methods) of malignant cells from the six tumours. Grey lines indicate the ‘backbone’ used in e and Extended Data Fig. 8b. e, Colour-coded density of cells (fraction of cells within a Euclidean distance of 0.3) from each tumour across the backbone of the hierarchy.
Oligodendrogliomas are thought to arise from transformation of oligodendrocyte progenitor cells (OPCs)17. However, PC2 and PC3 genes were not preferentially expressed in OPCs; instead, they were preferentially expressed in cells of neuronal lineage9,18 (Extended Data Fig. 3d) and upregulated upon activation of tri-potent mouse neural stem cells19 (NSCs) (Fig. 2c, Extended Data Fig. 3e; P = 3 × 10−6, t-test).
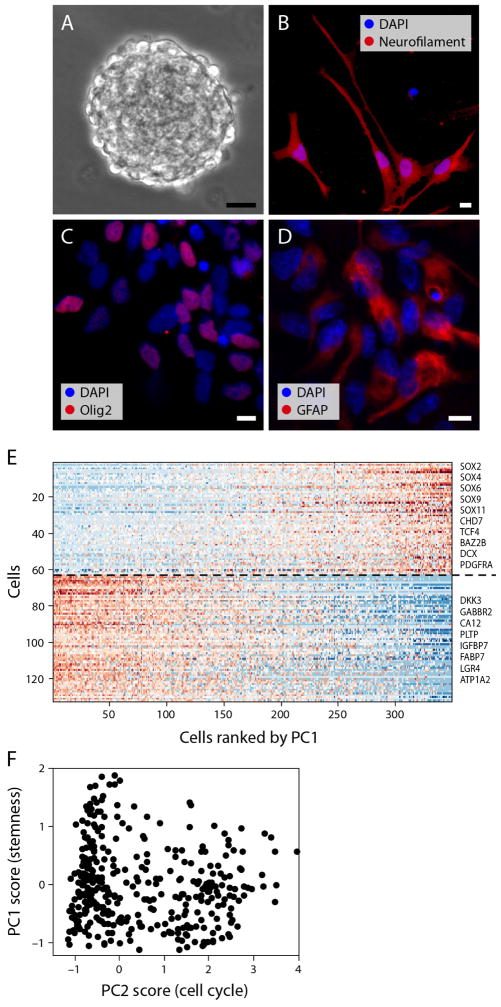
To further examine if the stemness program is associated with tri-potent stem/progenitor cells, we profiled human neural progenitor cells (NPCs) by scRNA-seq (Extended Data Fig. 4a–d). PCA of the NPC profiles identified an expression program highly similar to the PC2 and PC3 associated program of tumour cells (Fig. 2c, Extended Data Figs 3f and 4e, f and Supplementary Table 2; P = 2 × 10−35, t-test). Thus, a common program is shared by subsets of putative oligodendroglioma stem cells and normal NPCs and NSCs. Together, our analysis reveals three main expression patterns recapitulating oligodendrocytic and astrocytic differentiation, and stem/progenitor programs of early neural development.
To assign a cellular state to each tumour cell precisely, we defined differentiation and stemness scores (Methods). Plotting these scores across the cells of all six tumours revealed similarity to normal development (Fig. 2d), with a transition from stem/progenitor programs into differentiation along two glial lineages. Notably, the same architecture was observed in each of the six tumours and also found when tested with a different method for scRNA-seq (Fig. 2e, Extended Data Fig. 5a, e, Methods). Statistical analysis of the lineage scores suggests the existence of intermediate states for each lineage (Extended Data Fig. 5c and Supplementary Note 2).
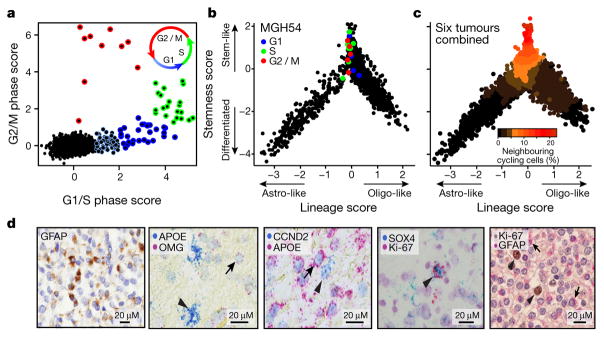
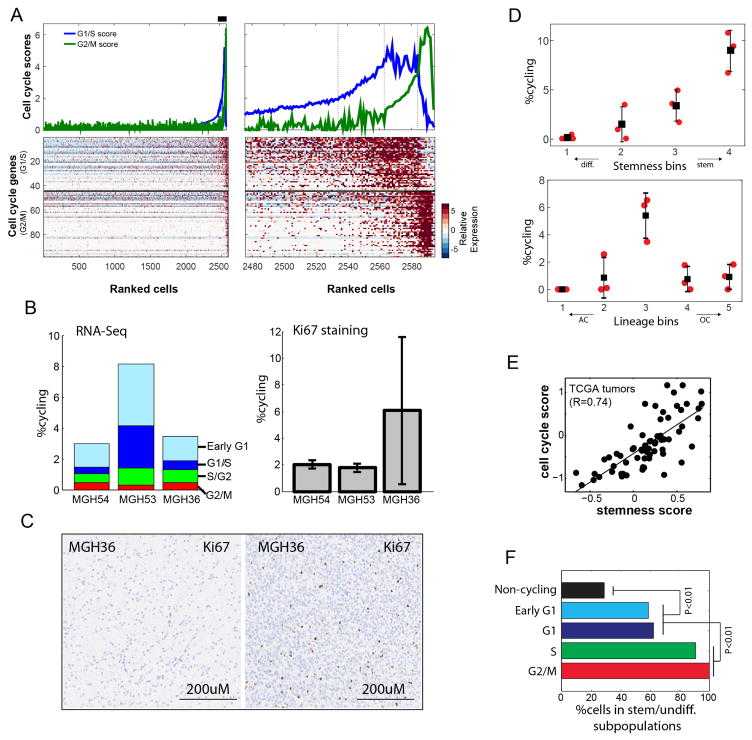
To assess how tumour cell proliferation and self-renewal may relate to developmental programs, we next scored each cell for the expression of signatures for the G1/S and G2/M phases (Methods)20,21. We found a small proportion of cells in each tumour (1.5–8%) that were proliferating, consistent with Ki-67 staining, and we estimated the cell-cycle phase of proliferating cells (Fig. 3a, Extended Data Fig. 6a–c and Supplementary Table 3). Almost all proliferating cancer cells were confined to the stem/progenitor and undifferentiated subpopulation of the tumour (Fig. 3b, c, Extended Data Fig. 6d and Supplementary Table 3), suggesting that this is the compartment fuelling the growth of oligodendroglioma in humans. We confirmed these patterns in tumours by both RNA in situ hybridization and immunohistochemistry with markers of astrocytes (GFAP and APOE), oligodendrocytes (OMG), stem/progenitor cells (SOX4, CCND2) and cell proliferation (Ki-67) in tissue staining across the original six tumours and in a validation cohort of ten additional tumours (Fig. 3d, Extended Data Figs 5d and 8c and Supplementary Table 3). Additionally, there is a strong correlation between our cell-cycle and stem/progenitor signatures across 69 bulk oligodendroglioma samples in The Cancer Genome Atlas22 (Extended Data Fig. 6e). Finally, the enrichment of cell-cycle signatures among stem/progenitor and undifferentiated cells was even more striking for cells inferred to be in G2/M phases compared to those in G1 phase (Extended Data Fig. 6f), possibly reflecting a short G1 phase in stem cells23.
Figure 3. Cycling cells are enriched among oligodendroglioma stem/progenitor cells.
a, Classification of cells (n = 4,044) to non-cycling (black) and categories of cycling cells (colour-coded by approximated phase as per inset) based on the relative expression of gene-sets associated with G1/S (x axis) and G2/M (y axis). b, c, Only stem/progenitor cells are cycling. b, Hierarchy plot, as in Fig. 2d, for MGH54 cells (n = 1,174), with cycling cells colour-coded as in a. c, Hierarchy plot for the six tumours, with each cell colour-coded based on the fraction of neighbouring cells (within Euclidean distance of 0.3) that are cycling. d, Immunohistochemistry for astrocytic marker (GFAP) in MGH54, with expression in subset of cells (left). In situ RNA hybridization shows mutually exclusive expression of astrocytic (APOE, arrowhead) and oligodendrocytic (OMG, arrow) markers, and of stem/progenitor (CCND2, arrowhead) and APOE (arrow) markers, but co-expression of stemness (SOX4) and cell cycle (Ki-67) markers (arrowhead) (middle). Double immunohistochemistry for GFAP (red, arrows) and Ki-67 (brown, arrowheads), showing mutual exclusivity (right).
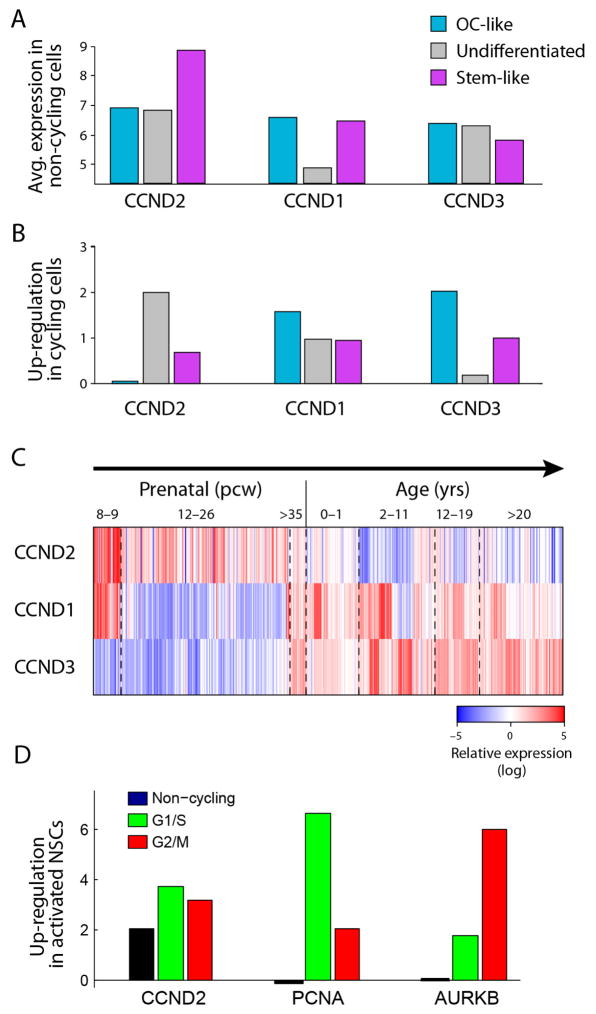
Although cycling cells were highly enriched among stem/progenitors, their frequency was low (~ 10%) even in that compartment; accordingly, the PC2 and PC3 program did not include a signature for the cell cycle, except for CCND2 (Fig. 2a), a gene controlling cell cycle and self-renewal of glioma CSCs24. CCND2 was highly expressed both in cycling cells and in non-cycling stem/progenitor cells (Extended Data Fig. 7a, b), consistent with CCND2 priming cells for the cell cycle23. Interestingly, stem/progenitor tumour cells preferentially express CCND2, whereas differentiated cells express CCND1 and CCND3, mirroring their expression patterns in normal neural development (Extended Data Fig. 7c). Furthermore, CCND2 was upregulated in activated mouse NSCs before these cells enter the cell cycle (Extended Data Fig. 7d). These results suggest CCND2 has a role in both normal and malignant stem cell programs.
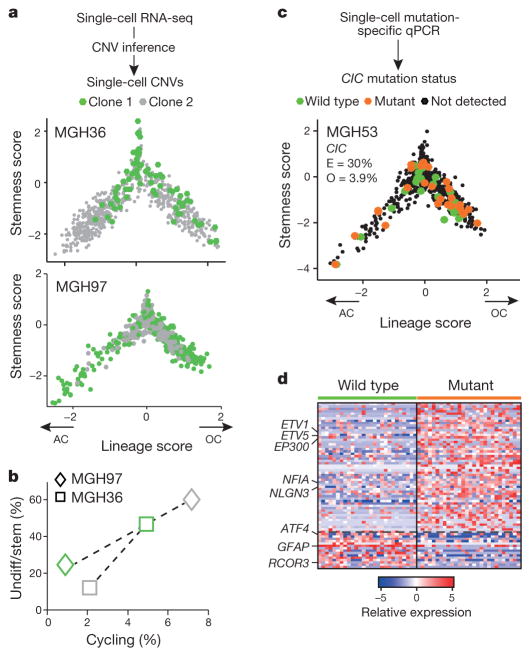
Finally, we explored the role of genetic events in shaping cellular identity, devising two approaches to obtain genetic information from scRNA-seq and classify cells into tumour subclones. First, we used the inferred CNVs in each cell (Fig. 1a, b). Second, we defined subclonal point mutations from bulk DNA whole-exome sequencing, using ABSOLUTE25, and identified these mutations in the RNA-seq reads of individual cells, albeit with limited sensitivity (Methods).
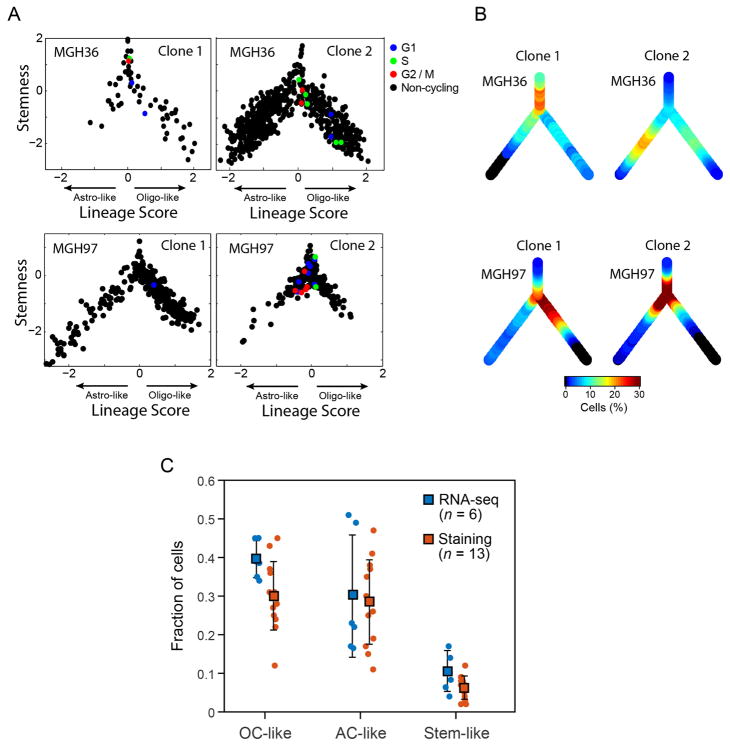
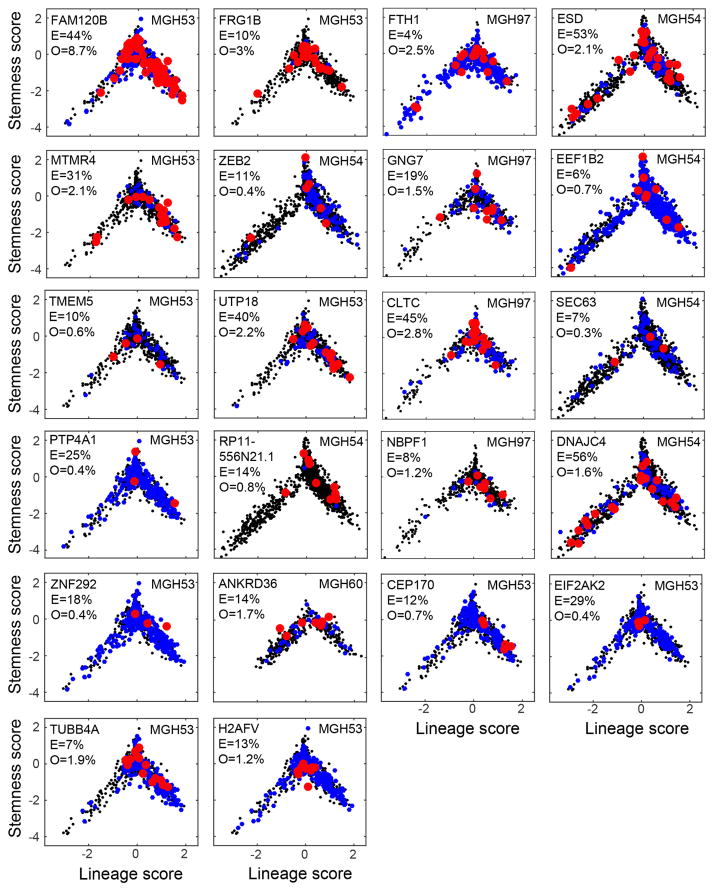
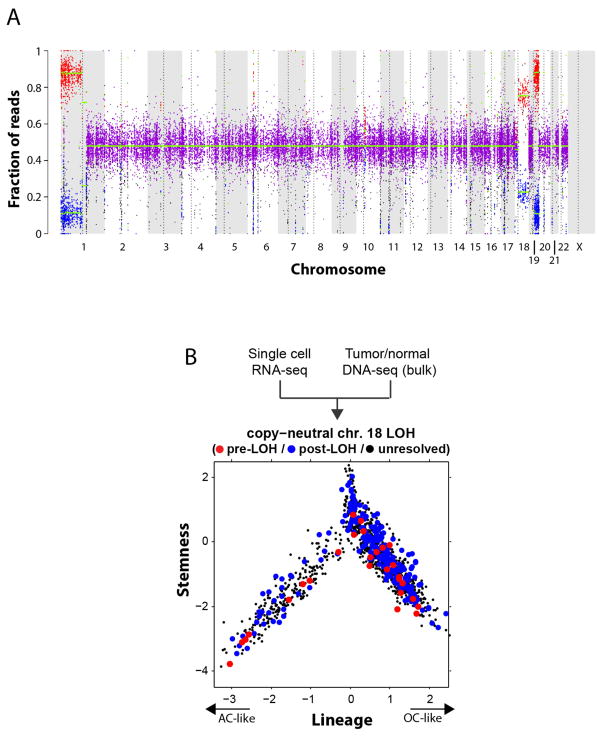
In both analyses, we found genetic subclones that span all three of the compartments, although the genetic information obtained with these two approaches is partial and is not sufficient to reconstruct a full phylogenetic tree. We observed the same three sub-population architectures in distinct CNV subclones in MGH36 and in MGH97 (Fig. 1b), with cycling stem/progenitor cells and two lineages of differentiated non-cycling cells (Fig. 4a and Extended Data Fig. 8). Similarly, examining the distribution of expression states for cells harbouring subclonal point mutations, we found that 22 subclonal point mutations (Extended Data Fig. 9) and a subclonal loss-of-heterozygosity event (Extended Data Fig. 10) are not significantly restricted to particular developmental states and often span all three states. Thus, the three compartments exist in different genetic subclones.
Figure 4. Intra-tumoural genetic heterogeneity and association with gene expression states.
a–d, Cells were classified into genetic subclones based on CNVs (a, b) or CIC point mutation status (c, d), and examined for differences in gene expression states. a, Two CNV clones (green and grey) in MGH36 and MGH97 mapped to the cellular hierarchy defined by lineage (x axis) and stemness (y axis) scores. b, Percentages of cycling cells (x axis) and of stem/progenitor cells (y axis) in clone 1 (green) and clone 2 (grey) of MGH36 (square) and MGH97 (diamond). c, Cells were classified using mutation-specific qPCR as wild-type CIC (green), mutant CIC (orange) or CIC status not detected (black) and mapped to the cellular hierarchy. The fraction of mutant CIC cells as observed by qPCR (O) and as expected by ABSOLUTE (E) is indicated. d, An expression signature for mutant CIC cells. Shown is a heatmap of relative expression levels for CIC-dependent genes (rows) in mutant CIC cells (right) and wild-type CIC cells (left). Selected gene names are indicated.
Although most subclonal mutations were of unknown functional relevance, we identified a subclonal mutation of CIC (~ 30% frequency in MGH53), a known tumour suppressor in oligodendroglioma26. RNA-seq reads detected the CIC mutation only in 7 MGH53 cells. We therefore designed a mutation-sensitive qPCR testing approach and were able to identify 28 mutant CIC cells (including all cells detected by RNA-seq) and 27 wild-type CIC cells (Fig. 4c). Notably, we identified a signature of expression changes between the mutant CIC and wild-type cells (Fig. 4d, Supplementary Table 5), including increased expression of ETV1 and ETV5 (ref. 27) in mutant CIC cells8. Despite these specific transcriptional changes, mutant CIC and wild-type CIC cells spanned all three subpopulations (Fig. 4c). Thus, many subclonal mutations can accrue within the hierarchy (but not drive it), although without a comprehensive phylogenetic reconstruction, we cannot categorically rule out a genetic influence.
Although CNV subclones in MGH36 and MGH97 included cells from all three tumour compartments, they differed in their relative distributions (Fig. 4a, b and Extended Data Fig. 8). Clone 1 of MGH36 displayed a higher frequency of stem/progenitors, whereas clone 2 displayed higher frequency of astrocyte-like cells (P < 10−9, Fisher’s exact test). Similarly, clone 2 of MGH97 contained a higher frequency of stem/progenitors (P < 10−16). Furthermore, the frequencies of cycling cells were higher in clone 1 of MGH36 and clone 2 of MGH97, consistent with their increased frequencies of stem/progenitor cells. Thus, genetic evolution may modulate patterns of self-renewal and differentiation.
In conclusion, our results highlight the fact that there is a subpopulation of undifferentiated cells in oligodendroglioma that possess stem cell expression signatures and enriched proliferative potential. Thus, the most primitive and undifferentiated population of cancer cells might be the main source of proliferating cells in oligodendroglioma. Although we cannot rule out an influence from genetic mutations, many subclonal events span all three states, consistent with this architecture being primarily dictated by non-genetic developmental programs. A caveat of our work is that because grade II oligodendroglioma cells do not grow in xenotransplantation, we could not functionally validate the stem/progenitor program, and instead we infer its function from the inverse association with differentiation programs, the enriched proliferation and the similarity to normal stem/progenitors. Our single-cell profiles suggest that oligodendroglioma stem/progenitor cells more closely resemble a primitive tri-potent neural cell type, such as NSC or NPC than a more committed glial progenitor like an OPC17,28. By providing the genome-wide transcriptional signature of cancer stem/progenitor cells in oligodendroglioma, our work delineates cellular programs that represent promising targets to affect tumour growth. Further studies will be needed to functionally validate our findings, interrogate their generality across other glioma subtypes, and investigate opportunities for clinical translation.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment
Tumour dissociation
Patients at the Massachusetts General Hospital consented preoperatively to take part in the study in all cases following the Institutional Review Board Protocol 1999P008145. Fresh tumours were collected at time of resection and the presence of malignant cells was confirmed in frozen sections on adjacent, representative pieces of tissue. Fresh tumour tissue was minced with a scalpel and enzymatically dissociated using a gentle papain-based brain tumour dissociation kit (Miltenyi Biotec). Large pieces of debris were removed with a 100 μm strainer, and dissociated cells were layered carefully onto a 5 ml density gradient (Lympholyte-H, Cedar Lane labs), which was centrifuged at 2,000 r.p.m. for 10 min at room temperature to pellet dead cells and red blood cells. The interface containing live cells was saved and used for staining and flow cytometry. Viability was measured using trypan blue exclusion, which confirmed > 90% cell viability.
Fluorescence-activated cell sorting
For primary tumour sorting, tumour cells were blocked in 1% bovine serum albumin in Hanks buffered saline solution (BSA/HBSS), and then stained first with CD45-Vioblue direct antibody conjugate (Miltenyi Biotec) for 30 min at 4 °C. Cells were washed with cold PBS, and then resuspended in 1 ml of BSA/HBSS containing 1 μM calcein AM (Life Technologies) and 0.33 μM TO-PRO-3 iodide (Life Technologies) to co-stain for 30 min before sorting. Fluorescence-activated cell sorting was performed on FACSAria Fusion Special Order System (Becton Dickinson) using 488 nm (calcein AM, 530/30 filter), 640 nm (TO-PRO-3, 670/14 filter), and 405 nm (Vioblue, 450/50 filter) lasers. Fluorescence-minus-one controls were included with all tumours, as well as heat-killed controls in early pilot experiments, which were crucial to ensure proper identification of the TO-PRO-3 positive compartment and ensure sorting of the live cell population. Standard, strict forward scatter height versus area criteria were used to discriminate doublets and gate only singlets. Viable cells were identified by staining positive with calcein AM but negative for TO-PRO-3. Single cells were sorted into 96-well plates containing cold buffer TCL buffer (Qiagen) containing 1% β-mercaptoethanol, snap frozen on dry ice, and then stored at − 80 °C before whole transcriptome amplification, library preparation and sequencing.
Whole-transcriptome amplification, library construction, sequencing, and processing
Libraries from isolated single cells were generated based on the Smartseq2 protocol (Picelli 2014) with the following modifications. RNA from single cells was first purified with Agencourt RNAClean XP beads (Beckman Coulter) before oligo-dT primed reverse transcription with Maxima reverse transcriptase and locked TSO oligonucleotide, which was followed by 20 cycle PCR amplification using KAPA HiFi HotStart ReadyMix (KAPA Biosystems) with subsequent Agencourt AMPure XP bead purification as described. Libraries were tagmented using the Nextera XT Library Prep kit (Illumina) with custom barcode adapters (sequences available upon request). Libraries from 384 cells with unique barcodes were combined and sequenced using a NextSeq 500 sequencer (Illumina).
We also analysed 96 cells from MGH60 with an alternative protocol that incorporates random molecular tags (RMTs, also known us unique molecular identifiers, or UMIs) in order to control for PCR amplification bias, as described previously29 and we obtained similar results.
Paired-end, 38-base reads were mapped to the UCSC hg19 human transcriptome using Bowtie with parameters “-q–phred33-quals -n 1 -e 99999999 -l 25 -I 1 -X 2000 -a -m 15 -S -p 6”, which allows alignment of sequences with single base changes, such as point mutation in the IDH1 gene. Expression values were calculated from SAM files using RSEM v1.2.3 in paired-end mode using parameters “–estimate-rspd–paired end -sam -p 6”, from which TPM values for each gene were extracted.
Immunohistochemistry
Haematoxylin and eosin and single antibody staining (GFAP, Ki-67) was done by the clinical pathology laboratory at the Massachusetts General Hospital per routine protocol. For GFAP and Ki-67 double immunohistochemistry, paraffin-embedded sections were mounted on glass slides, deparaffinized in xylene, treated with 0.5% peroxide in methanol, and rehydrated. Antigen retrieval was done using sodium citrate-based, heat-induced antigen retrieval at pH 6.0. The Dako EnVision G/2 double stain system was used for blocking, staining, and development using rabbit anti-Ki67 antibody (Abcam ab15580 at 1:300) and mouse anti-GFAP antibody (Dako M0761 at 1:100).
RNA in situ hybridization
Human tissue was obtained from the Massachusetts General Hospital according to an Institutional Review Board-approved protocol (1999P008145) and informed consent was obtained from all patients. ViewRNA technology (Affymetrix) was used for manual format RNA in situ hybridization. Tissue sections mounted on glass slides were stored at − 80 °C until they were used for hybridization. Slides were baked at 60 °C for 1 h, then denatured at 80 °C for 3 min, deparaffinized with Histoclear and ethanol dehydration. RNA targets in dewaxed sections were unmasked by treating with pre-treatment buffer at 95 °C for 10 min and digested with 1:100 dilution protease at 40 °C for 10 min, followed by fixation with 10% formalin for 5 min at room temperature. Probe concentrations were 1:40 for both type 1 (red) and type 6 (blue) probe sets, except that the ApoE probe was used at 1:80 dilution. The probe was incubated on sections for 2 h at 40 °C and then washed serially. Affymetrix Panomics probes included ApoE (type 6, catalogue number VA6-16904 and type 1, catalogue number VA1-18265), OMG (type 1, catalogue number VA1-18161), Sox4 (type 6, catalogue number VA6-18162), CCND2 (type 6, catalogue number VA6-18266), Ki-67 (type 1, catalogue number VA1-11033). Signal was amplified using PreAmplifier mix QT for 25 min at 40 °C followed by Amplifier mix QT for 15 min at 40 °C, and then signal was hybridized with labelled probe at 1:1,000 dilution for 15 min at 40 °C. Colour was developed using Fast Blue substrate for Type 6 probes and Fast Red substrate for Type 1 probes for 30 min at 40 °C. Tissue was counterstained with Gill’s haematoxylin for 25 s at room temperature followed by mounting with ADVANTAGE mounting media (Innovex). For quantification of compartments by ISH, at least 1,000 cells were counted in representative areas of the tumours.
Fluorescent in situ hybridization (FISH)
The probes used in this study consisted of centromeric (CEP) and locus-specific identifiers (LSI) probes. CEP probes included: CEP2 (2p11.1-q11.1, spectrum orange), CEP4 (4p11-q11, spectrum aqua), CEP9 (9p11-q11, spectrum aqua), CEP12 (12p11.1-q11, spectrum green), CEP17 (17p11.1-q11.1, spectrum aqua) and Y (Yp11.1-q11.1, spectrum green) all obtained from Abbott Molecular, Inc. (Des Plaines, IL). LSI probes were1p36/1q25 and 19q13/19p13 dual-colour probe set (Abbott), and bacterial artificial chromosome RP11-351D16 (10q11.21, spectrum red or green; CHORI, Oakland, CA).
FISH was performed as described previously30. Briefly, 5-μm sections of formalin-fixed, paraffin-embedded tumour material were deparaffinized, hydrated, and pretreated with 0.1% pepsin for 1 h. Slides were then washed in 2× saline-sodium citrate buffer (SSC), dehydrated, air dried, and co-denatured at 80 °C for 5 min with a three-colour probe panel and hybridized at 37 °C overnight using the Hybrite Hybridization System (Abbott). Two 2-min post-hybridization washes were performed in 2× SSC/0.3%NP40 at 72 °C followed by one 1-min wash in 2× SSC at room temperature. Slides were mounted with Vectashield containing 4′,6-diamidino-2-phenylindole (Vector, Burlingame, CA, USA). Entire sections were observed with an Olympus BX61 fluorescent microscope equipped with a charge-coupled device camera and analysed with Cytovision software (Applied Imaging, Santa Clara, CA).
Human NPC culturing
Human NPCs were dissociated from the subventricular zone of 19 week fetal tissue and resulting neurospheres were expanded in a 1:1 mixture of DMEM/F12 and Neurobasal A (Invitrogen), supplemented with B27 lacking vitamin A, EGF, FGF, and heparin. Single live NPCs were isolated by FACS from a passage 8 culture and sorted into 96-well plates containing Buffer TCL (Qiagen) + 1% β-mercaptoethanol. For differentiation assays, NPCs were plated in chamber slides coated with poly-d-lysine and laminin, and proliferation media was exchanged over a period of 3 days with base media supplemented with either 1% FBS, 1% FBS + 60 ng ml−1 T3, or FBS + 100 nM trans-retinoic acid and 10 ng ml−1 NT3. Multipotency was confirmed by indirect immunofluorescence after 7 days of differentiation with GFAP (Abcam ab53554), Olig2 (Millipore AB9610), and Neurofilament (Aves).
Single-cell RNA-seq data processing
Expression levels were quantified as Ei,j = log2(TPMi,j/10 + 1), where TPMi,j refers to transcript-per-million for gene i in sample j, as calculated by RSEM31. TPM values are divided by 10, since we estimate the complexity of single-cell libraries in the order of 100,000 transcripts and would like to avoid counting each transcript ~ 10 times, as would be the case with TPM, which may inflate the difference between the expression level of a gene in cells in which the gene is detected and those in which it is not detected.
For each cell, we quantified two quality measures: the number of genes for which at least one read was mapped, and the average expression level of a curated list of housekeeping genes. We then conservatively excluded all cells with either fewer than 3,000 detected genes or an average housekeeping expression (E, as defined above) below 2.5. For the remaining cells we calculated the aggregate expression of each gene as log2(average(TPMi,1...n)+1), and excluded genes with an aggregate expression below 4, leaving a set of 8,008 analysed genes. For the remaining cells and genes, we defined relative expression by centering the expression levels, Eri,j = Ei,j-average[Ei,1...n]. Centring was performed within each tumour separately in order to decrease the impact of inter-tumoural variability on the combined analysis across tumours.
CNV estimation
Initial CNVs (CNV0) were estimated by sorting the analysed genes by their chromosomal location and applying a moving average to the relative expression values, with a sliding window of 100 genes within each chromosome, as previously described6. To avoid considerable impact of any particular gene on the moving average, we limited the relative expression values to [− 3,3] by replacing all values above 3 by 3, and replacing values below − 3 by − 3. This was performed only in the context of CNV estimation. For visualization purposes, in order to include the two chromosomes with fewest analysed genes (chromosome 18 and 21 with 105 and 75 genes, respectively), we extended the moving average to include up to 50 genes from the flanking chromosomes (for example, the first window in chromosome 18 consisted of the last 50 genes of chromosome 17 and the first 50 genes of chromosome 18, whereas the 51 through 56 windows in that chromosome consisted only of chromosome 18 genes). This initial analysis is based on the average expression of genes in each cell compared to the other cells and therefore does not have a proper reference which is required to define the baseline. However, we detected a cluster of cells that have higher values at chromosome 1p and 19q, which we know are deleted in the six tumours, and that have consistent ‘CNV patterns’ across the genome, despite the fact that they originate from all three tumours. We thus defined these as the non-cancer cells and used the average CNV estimate at each gene across these cells as the baseline. The non-cancer cells included both microglia and oligodendrocytes, which differed in gene expression patterns and therefore also in CNV estimates (for example, the MHC region in chromosome 6 had consistently higher values in microglia than in oligodendrocytes and cancer cells). We therefore defined two baselines, as the average of all microglia and the average of all oligodendrocytes, and based on these the maximal (BaseMax) and minimal (BaseMin) baseline at each genomic position. The final CNV estimate of cell i at position j was defined as:
Principal component analysis
We performed principal component analysis (PCA) for the relative expression values of all cancer cells (as defined by CNV analysis) from the three tumours combined. The covariance matrix used for PCA was generated using an approach outlined in ref. 32 to decrease the weight of less reliable ‘missing’ values in the data. The basis of this approach is that due to the limited sensitivity of single-cell RNA-seq, many genes are not detected in particular cells despite being expressed. This is particularly pronounced for genes expressed at low levels, and for cells with low library complexity (that is, for which relatively few genes are detected), and results in non-random patterns in the data, whereby cells may cluster based on their complexity and genes may cluster based on their expression levels, rather than ‘true’ co-variation. To mitigate this effect, we assign weights to missing values, such that the weight of Ei,j is proportional to the expectation that gene i will be detected in cell j given the average expression of gene i and the total complexity (number of detected genes) of cell j.
To further verify that the PCA results are not driven by library complexity, we compared the PCA results to those of shuffled data. We iteratively swapped the expression of individual genes between pairs of cells with similar complexities, swapping each gene in each cell at least once. In that way we shuffled the data and removed the biological clustering, but maintained the distribution of complexities across cells, as well as the distribution of expression levels for each gene. PCA over the shuffled data defined the complexity-based effect, as evident by a Pearson correlation of 0.96 between the PC1 cell scores and their complexities (in the original data this correlation is only 0.41). We then compared PC1 gene scores between the original and the shuffled data (Extended Data Fig. 2f). Although PC1 gene scores of most genes are comparable between the two analyses, the loadings of the oligodendrocyte and astrocyte gene sets (Supplementary Table 1) were highly affected. Oligodendrocyte genes were originally associated with highly positive PC1 scores, and their scores are significantly decreased upon shuffling (97% of the oligodendroglial genes were among the 5% genes with the most decreased loadings, P < 10−32); similarly, astrocytic genes were originally associated with negative PC1 scores, and their scores are significantly increased upon shuffling (all astrocytic genes were among the 5% of genes with the most increased loadings, P < 10−32). As a result, none of the genes with highest and lowest PC1 scores (after shuffling) overlap with our oligodendroglial and astrocytic gene sets. Thus, complexity does not account for the association of PC1 with the differentiation programs. Similarly, complexity clearly does not account for the PC2 and PC3 stemness program, as PC2 cell scores are positively correlated with complexity (R = 0.27), whereas PC3 cell scores are negatively correlated with complexity (R = − 0.24) and stemness genes were defined as those associated with both PC2 and PC3.
PC1-associated genes and lineage scores
The top correlated genes with PC1 scores (across all tumour cells) were defined as PC1-associated genes. We focused on the genes with an absolute correlation value above 0.35, but note that other thresholds gave similar results (not shown). Of those genes, the subset that was differentially expressed by at least threefold between oligodendrocyte (OC) and astrocyte (AC) mouse cells9, and for which the two comparisons were consistent (that is, PC1-positively correlated genes with higher OC expression, and PC1-negatively correlated genes with higher AC expression) were defined as the OC and AC lineage gene sets. Lineage scores were then calculated as the average relative expression of the lineage gene set minus the average relative expression of a control gene set, that is, Lini,j = average[Er(Gj,i)] − average[Er(Gjcont,i)], in which Lini,j is the score of cell i to lineage j, Gj is the gene set for lineage j and Gjcont is a control gene set for lineage j. The control gene set was defined by first binning all 8,008 analysed genes into 25 bins of aggregate expression levels and then, for each gene in the lineage gene set, randomly selecting 100 genes from the same expression bin. In this way, the control gene set has a comparable distribution of expression levels to that of the lineage gene set and the control gene set is 100-fold larger, such that its average expression is analogous to averaging over 100 randomly selected gene sets of the same size as the lineage gene set. The final lineage score of each cell was defined as the maximal score over the two lineages, LINi = max(Lini OC, Lini AC). For visualization purposes where the two lineage scores are shown in a single axis, we first assigned random scores within (0–0.15) to all cells with LIN < 0, to avoid having many overlapping cells at x = 0. Second, we assigned negative scores to the cells with higher AC than OC scores (that is, a cell with AC and OC scores of 0.1 and 1, respectively, would be assigned a lineage score of 1, wheresa a cell with AC and OC scores of 1 and 0.1 would be assigned a lineage score of −1).
PC2 and PC3 associated genes and stemness scores
Both PC2 and PC3 were associated with intermediate values of PC1 (Extended Data Fig. 2c) and therefore with presumably less differentiated cells, and both were correlated with a shared set of genes, but distinguished by their correlation with cell ‘complexity’. We considered their sum as a potential stemness program. To detect potential stem-related genes, we chose the top 100 most positively correlated genes with PC2 + PC3 scores across all cancer cells from the three tumours. The 100 candidate genes were then restricted to (1) genes that are positively correlated with both PC2 and PC3, which primarily excluded ribosomal protein genes that were only correlated with PC2; (2) genes for which the average relative expression among the stem-like cells was above average. Stemness scores for each cell, stem(i), were then defined as the average relative expression of the stemness gene-set (Gstem) minus the average of a control gene set ( ) and minus the lineage score of cell i:
Assignment of cells to four subpopulations: stem/progenitor-like, undifferentiated, OC-like and AC-like
Cells were scored for the three programs defined above (two lineage scores and a stemness score) and assigned to the subpopulation that corresponds to their highest scoring program, if the maximal score was above 0.5 and was higher by 0.5 than the score for the other programs. Cells in which the maximal score did not pass these thresholds were assigned to the undifferentiated subpopulation, for which we did not detect a specific expression program. We note that the expression programs are continuous and thus it is difficult to assign every cell to a discrete subpopulation. Nevertheless, most cells are highly biased towards one of the three states, and the overall estimates are consistent between analysis of single-cell RNA-seq data and tissue staining experiments (Extended Data Fig. 8c and Supplementary Table 3). Furthermore, very few cells (~ 1% on average, and 5% at most) scored for two programs simultaneously (with the same threshold of 0.5, Supplementary Table 3).
Cell cycle analysis
Analysis of single-cell RNA-seq in human (293T) and mouse (3T3) cell lines20, and in mouse haematopoietic stem cells21 revealed in each case two prominent cell cycle expression programs that overlap considerably with genes that are known to function in replication and mitosis, respectively, and that have also been found to be expressed at G1/S phases and G2/M phases, respectively, in bulk samples of synchronized HeLa cells33. We thus defined a core set of 43 G1/S and 55 G2/M genes that included those genes that were detected in the corresponding expression clusters in all four datasets from the three studies described above (Supplementary Table 2). As expected, the genes in each of those expression programs were highly co-regulated in a small fraction of the oligodendroglioma cells, such that some cells expressed only the G1/S or the G2/M programs and other cells expressed both programs (Extended Data Fig. 6a). Plotting the average expression of these programs revealed an approximate circle (Fig. 3a), which we hypothesize describes the progression along the cell cycle. Putative cycling cells were identified by at least a twofold upregulation and a t-test P value < 0.01 for either the G1/S or the G2/M gene set compared to the average of all cells. Although we cannot confidently define the regions that correspond to each phase of the cell cycle in an automatic way, we manually defined four regions in the apparent circle and assigned them to approximate cell cycle phases.
Analysis of whole-exome DNA sequencing data
Output from Illumina software was processed by the Picard processing pipeline to yield BAM files containing aligned reads (bwa version 0.5.9, to the NCBI Human Reference Genome Build hg19) with well-calibrated quality scores34,35. Sample contamination by DNA originating from a different individual was assessed using ContEst36. Somatic single nucleotide variations (sSNVs) were then detected using MuTect37. Following this standard procedure, we filter sSNVs by (1) removing potential DNA oxidation artefacts38; (2) removing events seen in sequencing data of a large panel of ~ 8,000 TCGA normal samples; (3) realigning identified sSNVs with NovoAlign (http://www.novocraft.com) and performing an additional iteration of MuTect with the newly aligned BAM files. sSNVs were finally annotated using Oncotator39. Sample purity and ploidy, as well as Cancer Cell Fraction (CCF) of identified sSNVs were determined by ABSOLUTE25. Genome-wide copy-ratio profiles were inferred using CapSeg. Read depth at capture targets in tumour samples was calibrated to estimate copy ratio using the depths observed in a panel of normal genomes. Next, we performed allelic copy analysis using reference and alternate counts at germline heterozygous SNP sites.
Mutation calling in single cells
sSNVs that were identified by WES were examined in single-cell RNA-seq data by the mpileup command of SAMtools40. The fraction of cells in which we identified these mutations was, on average, only 1.3% of the expected fraction estimated by ABSOLUTE. This low sensitivity primarily reflects the low coverage of the RNA-seq reads over the transcriptome of single cells. Accordingly, sensitivity was correlated with the expression levels of the genes that harbour the mutations, and reached 20.4% for the top 10% most highly expressed genes. Sensitivity was also affected by heterozygosity and allele-specific expression, as in some heterozygote mutant cells we might only sequence the wild-type allele.
We used a targeted sequencing approach to increase our sensitivity for three specific mutations in MGH54 which were identified by WES but detected in very few cells by single-cell RNA-seq. We designed primers flanking these three mutations (in ZEB2, EEF1B2 and DNAJC4), PCR-amplified single cell cDNAs (frozen stocks of product from the pre-amplification reaction of the Smart-seq2 protocol) and sequenced the amplified material. This approach was applied for 1,056 cells from MGH54. Mutant cells were defined as those with at least 50 reads that mapped to the mutant allele as defined by WES, and for which the fraction of mutant reads was at least 20% of all reads and fivefold higher than the overall rate of mutant reads (in order to exclude a low rate of mutant reads due to PCR or sequencing errors). The mutations detected by this criteria were highly consistent with those identified from single-cell RNA-seq (P < 10−5, hypergeometric test) and uncovered 19 additional mutant calls (three for ZEB2, three for EEF1B2 and 13 for DNAJC4).
We next focused on the 23 subclonal mutations for which (1) the estimated clonal fraction by ABSOLUTE was at most 60%; (2) at least three cells were identified as harbouring the mutation; and (3) at least one cell was identified as having a wild-type allele of the mutant gene. For each of those 23 mutations we plotted the lineage and stemness scores of all mutant cells to examine their distribution of expression states (Fig. 4 and Extended Data Fig. 9). Note that for these mutations we detected on average 9.4% of the expected fraction by ABSOLUTE.
To estimate the frequency of false-positive errors we defined, for each mutation that is detected by WES and analysed by RNA-seq mutation calling, (i) ‘expected mutations’: the number of events in which we find the exact mutation reported by WES; and (ii) ‘false mutations’, the number of events in which we find a mismatch in the same exact site but to a different base than expected by WES (there are 2 such possible bases). This approach focuses on the exact genomic context of the real mutations to obtain a reliable estimate of the false positive rate. This estimate is half the number of false mutations divided by the number of expected mutations (given 4 bases, one of which is wild type, there are two types of false mutations but only one type of expected mutations). The result of this analysis was an estimated average false positive rate of 0.85%, suggesting that the confidence of each detected mutation is, on average, higher than 99%. Accordingly, even in the most extreme case (for example, ZEB2) where only a single mutant cell is detected in one of the compartments of the hierarchy, we still have a 99% confidence that the mutation is represented in that compartment.
Mutation-detecting qPCR and analysis of CIC mutations
To detect CIC mutations in single cells from MGH53, we performed qPCR using SuperSelective PCR primers, which are highly specific to single base changes due to a loop-out sequence adjacent to the mutant base (http://legacy.labroots.com/user/webinars/details/id/95). The following qPCR primers were designed to target the c.4543 C > T, p.1515 R > C mutation on CIC cDNA which had been identified as subclonal in MGH53 via whole-exome sequencing analysis. Wild-type-specific forward primer: 5′-CCCTCCAAGGTTTGTCTGCAGccattcGAGGTGC-3′; mutant-specific forward primer: 5′-CCCTCCAAGGTTTGTCTGCAGccattcGAGGTGT-3′; universal reverse primer: 5′-tcgGGCAGCCTGCATGATCTT-3′.
The specificity of the single cell qPCR primers was validated by two approaches: (1) qPCR on artificial templates differing by only the mutant base; and (2) qPCR on cDNA of single MGH53 tumour cells for which RNA-seq already detected mutant or wild-type reads. These positive control reactions were highly consistent between duplicates and with the mutation status as inferred from RNA-seq: qPCR identified 7 out of 7 mutant cells and 12 out of 15 wild-type cells, while the remaining three cells had no qPCR signal, and therefore all qPCR signal was consistent with RNA-seq data. We also took advantage of the fact that CIC is located on chr19q which is deleted in MGH53 cancer cells and therefore each cell only contains one CIC allele (loss-of-heterozygosity, LOH). Thus, in a single MGH53 cancer cell, we expect evidence of either mutant or wild-type CIC, but not both. Indeed, all cells with a signal in the positive control assay showed a difference in Ct values of at least 5 between mutant and wild-type reactions, consistent with LOH.
cDNA was taken from frozen stocks of product from the preamplification reaction of the Smartseq2 protocol. 1 μl from each well of cDNA was used as template for a second round of Smartseq2 preamplification and bead purification in order to increase overall signal downstream. qPCR was performed with the Fast Plus EvaGreen qPCR Master Mix Low Rox (Biotium 31014-1) according to the manufacturer’s instructions with the sole modification of adding EDTA to a final reaction concentration of 1.6 mM to enhance primer selectivity. Cp ≥ 33 were considered negative signal; Cp < 33 was considered positive signal.
We performed SuperSelective qPCR on cDNA from 467 single MGH53 tumour cells. Of these, 61 cells had signal in both replicates for either mutant or wild type primers, but never for both. These were used to define 28 mutant CIC cells and 27 wild-type CIC cells, after excluding 6 cells which did not pass the single cell RNA-seq quality control filters.
To identify genes regulated by the CIC mutation, we compared the 28 mutant CIC cells and 27 wild-type CIC cells and identified genes with at least twofold average expression difference and P < 0.01 (before correction for multiple hypothesis testing) based both on a permutation test and a t-test. To further filter the list of differentially expressed genes we also compared the mutant CIC cells to the 671 unresolved cells (in which we did not detect signal for either mutant or wild-type alleles by qPCR and by RNA-seq). As the fraction of CIC mutants was estimated as 30% by ABSOLUTE, we expect the unresolved cells to be a mixture of about one-third mutant CIC cells and about two-thirds wild-type CIC cells, and thus CIC-regulated genes should also differ between this mixture and the CIC mutants but to a lesser extent; we used a threshold of 1.5-fold difference between the average expression in CIC mutants and in unresolved cells. The resulting set of differentially expressed genes is given in Supplementary Table 5. We simulated this analysis with 1,000 randomly selected sets of cells (to replace the mutant CIC and wild-type CIC cells) and found an average of only five upregulated genes by the same criteria, suggesting a false discovery rate lower than 0.1 for the genes upregulated by the CIC mutation.
Data availability
Data generated for this study are available through the Gene Expression Omnibus (GEO) under accession number GSE70630.
Extended Data
Extended Data Figure 1. Single-cell RNA-seq analysis of human oligodendroglioma samples.
a, Experimental workflow. b, Clinical information of the main and validation patient cohorts analysed in this study. Asterisk indicates a borderline result of chromosome 1p loss based on clinical testing. c, ISH (top left) and FISH (all other panels) in a representative tumour (MGH36). All our cases retain ATRX protein expression by ISH (top left) and show loss of chromosomes arms 1p (bottom left) and 19q (top right) by FISH. In addition, tumour-specific CNVs identified by single-cell RNA-seq were confirmed by FISH (for example, loss of chromosome 4 in MGH36, bottom right panel). d, Distributions of the total number of sequenced paired-end reads per cell (grey) and of paired-end reads that were mapped to the transcriptome and used to quantify gene expression (black).
Extended Data Figure 2. Diversity of expression programs in oligodendroglioma.
a, Two populations of non-cancer cells identified in oligodendroglioma. Selected genes that are differentially expressed among the two populations of non-cancer cells that lack CNVs (Fig. 1b, top), including markers of microglia (top) and oligodendrocytes (bottom). b, Expression programs in microglia cells from three tumours. The heat map shows relative expression of genes (rows) across microglia cells (columns). Above the dashed line are microglia markers expressed in all microglia cells and below the line are the genes of a microglia activation program, which is variably expressed, and includes cytokines, chemokines, early response genes and other immune effectors. This latter gene set might reflect a microglia activation program that could either be a general microglia program or potentially specific to the context of oligodendroglioma. Microglia cells (n = 235) (columns) are rank ordered by their relative expression of the activation program. The tumour of origin of each cell is colour-coded as indicated in the top row. c, PC2 and PC3 are associated with intermediate values of PC1. PC1 scores are shown along with PC2 (top) and PC3 (bottom) scores for cells in each of the three tumours profiled at high depth. The red line indicates local weighted regression (LOWESS) with a span of 5%, which demonstrates that PC2 and PC3 values tend to be highest in intermediate values of PC1 and to decrease in either high PC1 (that is, oligodendrocyte-like cells) or low PC1 (that is, astrocyte-like cells). d, Consistency of PCA across tumours. Shown are the Pearson correlations in gene loadings (over all analysed genes) between the top three PCs in PCA of the three tumours profiled at high depth (y axis, as shown in Fig. 1) and the top four PCs in alternative PCA of either all six tumours (left), as well as of PCA of each individual tumour (right). PC1–3 are highly consistent between the three-tumour and six-tumour PCAs (R > 0.9); PC1 is highly consistent (R > 0.8) between the three-tumour analysis and all other analysis. e, PC1 (x axis) and PC2 plus PC3 (y axis) scores of malignant cells from each of the three tumours profiled at intermediate depth, showing consistent patterns with those shown in Fig. 1d. f, Distribution of differences in PC1 loadings between the original PCA and the shuffled PCA (see description in the Methods section for principal component analysis) for all genes (black), oligodendrocyte-like (OC-like) genes (blue) and astrocyte-like (AC-like) genes (green). This analysis demonstrates that oligodendrocyte-like and astrocyte-like gene sets are highly skewed in the original PCA and their loadings are not recapitulated by shuffled data reflecting the effect of complexity.
Extended Data Figure 3. The stemness program in oligodendroglioma.
a, Cell–cell correlation matrix based on all analysed genes across all malignant cells in MGH54 (n = 1,174). Cells are ordered by average linkage hierarchical clustering, and coloured boxes indicate distinct clusters. Clusters are marked based on the identity of differentially expressed genes as OC-like (blue), AC-like (yellow), cycling (pink) stem-like (purple) and intermediate cells that do not score highly for any of those expression programs (orange). b, Most differentially expressed genes. Shown is the average expression in each of the OC-like, AC-like, stem-like and intermediate cell clusters (columns) of differentially expressed genes (rows) defined by comparing cells from each of the OC-like, AC-like and stem-like clusters to cells from the remaining clusters with a t-test. Similar genes are highlighted as in Fig. 1 (OC-like: OMG, OLIG1, OLIG2, SOX8; AC-like: ALDOC, APOE, SOX9; Stem-like: SOX4, SOX11, CCND2, SOX2). Stem-like genes also include CTNNB1, USP22, and MSI1. c, Overlap with human GBM stemness program. We previously6 identified a GBM stemness program and determined the association of each gene with that program by the correlation between the expression of that gene and the average expression of the stemness program’s genes across individual cells (‘CSC gradient’) in each of five GBM tumours. Shown is the average correlation (x axis) of each analysed gene (green dots) across the five cases and the P values of those correlations as determined with a t-test (y axis). Genes identified in the oligodendroglioma stemness program (this work) are marked in black and are significantly enriched for the GBM stemness genes (1.5 × 10−4, hypergeometric test), defined as those with P < 0.05 and an average correlation above 0.1. d, Preferential expression of the oligodendroglioma stemness program in neurons but not in OPCs. Genes expressed in the oligodendroglioma single cells were divided into six bins (bars) based on their relative expression (log2-ratio) in stem-like cells with high PC2/3 and intermediate PC1 scores compared to all other cells. Each panel shows for each bin the average relative expression in each of three normal brain cell types (y axis) based on data from the Barres laboratory RNA-seq database9,18: mice oligodendrocyte progenitor cells (mOPC, top), mouse neurons (mNeurons, middle), and human neurons (hNeurons, bottom). Relative expression of each gene in each cell type was defined as the log2-ratio between the respective cell type divided by the average over AC, OC and neurons. Error bars denote standard error as defined by bootstrapping. Asterisks denote bins with significantly different relative expression (in the respective normal cell type) compared to all genes expressed in oligodendroglioma, based on P < 0.001 (by t-test) and average expression change of at least 30%. e, Correlation with mouse activated NSC program. Shown is the distribution of correlation values (x axis) of either all genes (grey) or genes from the oligodendroglioma stemness program (black) with the expression program of mice NSC activation states, as previously quantified by ‘pseudotime’, across single mouse NSCs19. The average correlation of the NSC activation program genes with oligodendroglioma stemness genes is significantly higher than with all other genes (P = 3 × 10−6; t-test). f, Correlation with human NPC program. Shown is the distribution of correlation values (x axis) of either all genes (grey) or genes from the oligodendroglioma stemness program (black) with an expression program of human NPCs identified by PCA (Extended Data Fig. 4). Each gene’s correlation to the average expression of the NPC program genes was calculated across single human NPCs. The average correlation with oligodendroglioma stemness genes is significantly higher than with all other genes (P = 2 × 10−35, t-test).
Extended Data Figure 4. Analysis of human NPCs.
a–d, Differentiation potential of human SVZ NPCs. Human SVZ NPCs isolated from 19-week-old fetuses form neurospheres in culture (a), and can be differentiated to neuronal (neurofilament, b), oligodendrocytic (OLIG2, c), or astrocytic (GFAP, d) lineages in vitro. Scale bars, 25 μm (a), 10 μm (b–d). We note that although OLIG2 can represent different cell types, it is expressed at a low level in the fetal NPCs before differentiation (an average log2(TPM + 1) of 0.82, compared to a threshold of 4 that we use to define expressed genes in our analysis, and with zero cells with expression above this threshold). Thus, the undifferentiated NPCs do not express OLIG2, and we interpret the expression of OLIG2 as a sign of oligodendroglial lineage differentiation. e, f, Single-cell RNA-seq analysis of NPCs. e, NPCs have an expression program similar to the oligodendroglioma stemness program. Heat map shows the expression of genes (rows) most positively (top) or negatively (bottom) correlated with PC1 of a PCA of RNA-seq profiles for 431 single NPCs, across NPC cells (columns) rank ordered by their PC1 scores. Selected genes are indicated, and a full list of correlated genes for PC1 and PC2 is given in Supplementary Table 2. f, NPC cell scores for PC1 (y axis) and PC2 (x axis). PC2 correlated genes are associated with the cell cycle. Cells with the highest PC1 scores tend to be non-cycling (low PC2 score), indicating that while the stemness program is coupled to the cell cycle in oligodendroglioma, it is decoupled from the cell cycle in NPCs.
Extended Data Figure 5. Developmental hierarchy in oligodendroglioma.
a, Shown are plots as in Fig. 2d for each of the six tumours with cycling cells coloured as in Fig. 3. b, Lineage and stemness scores for three tumours with high-depth profiling, coloured based on sequencing batches, demonstrating the lack of considerable batch effects. c, For each of the three tumours profiled at high depth (horizontal panels) and for the two lineages (vertical panels), we calculated the significance of co-expression among sets of AC-related (top panels) or OC-related (bottom panels) genes within limited ranges of lineage scores (between the value of the x axis and that of the y axis). Significance was calculated by comparison of average co-expression to that of 100,000 control gene-sets with similar number of genes and distribution of average expression levels, and is indicated by colour. The significant co-expression patterns within limited ranges of lineage scores suggest that variability of lineage scores in these ranges cannot be driven by noise alone, and implies the existence of multiple states within each lineage, presumably reflecting intermediate differentiation states (see Supplementary Note 2). d, Characterization of tumour subpopulations by histopathology and tissue staining. Top/middle panels denote two predominant lineages of AC-like and OC-like cells. Shown are MGH53 with haematoxylin and eosin (H&E, top left), immunohistochemistry for OLIG2 (oligodendrocyte marker, top right) and GFAP (astrocyte marker, middle left), as well as in situ RNA hybridization for astrocytic markers ApoE (apolipoprotein E, astrocytic lineage, middle right), with patterns similar to GFAP immunohistochemistry. Bottom panels denote stem-like cells and association with cell cycle. In situ RNA hybridization for the stem/progenitor markers SOX4 and CCND2 (bottom left) and the proliferation marker Ki-67 (bottom right) in MGH36 identifies cells positive for both markers (arrows). Immunohistochemistry for GFAP (arrowhead, bottom right) and Ki-67 (arrow, bottom right) shows mutually exclusive expression patterns. e, Consistency of MGH60 hierarchy between the full-length SMART-Seq2 protocol used throughout this work (left panels), and an alternative protocol (right panels) in which only the 5′-ends of transcripts are analysed while incorporating random molecular tags (RMTs, also known us unique molecular identifiers, or UMIs) that decrease the biases of PCR amplification. Top panels: PC1 reflects an AC-like and OC-like distinction. Shown are heatmaps of the AC-like and OC-like specific genes (rows, as defined in Supplementary Table 1 and restricted to genes with average expression log2(TPM + 1) > 4 in each data set) with cells ordered by their PC1 score. Bottom, lineage (x axis) and stemness (y axis) scores (defined as in Fig. 2d).
Extended Data Figure 6. Cell-cycle analysis.
a, High expression of G1/S and G2/M gene sets in a subset of cycling cells. Shown are the average expression (top panels, lines) or the expression of all individual genes (bottom, heat maps) of the G1/S and G2/M gene sets, in all cells (n = 2,594) (left) or only among the putative cycling cells (n = 119) (right) from the three tumours profiled at high-depth ordered by cell cycle expression. Dashed lines (top right) separates the four inferred phases of cycling cells, corresponding to light blue, blue, green and red in Fig. 3a, respectively. b, Estimated fraction of cycling cells (y axis) in each of 3 tumours (x axis) based on single cell RNA-seq (left; different phases marked by colour code as in Fig. 3a) or Ki-67 immunohistochemistry (right). c, Variation in cycling cells between regions of the same tumour. Shown is Ki-67 immunohistochemistry in two regions in MGH36. Such regional variability in proliferation complicates direct comparisons as done in b. d, Cycling cells are enriched in stem-like and undifferentiated cells compared to differentiated cells. Shown is the percentage of cycling cells (y axis) in four bins based on stemness scores (top) or lineage scores (bottom). Black squares and error-bars correspond to the mean and standard deviation of the percentages in the three tumours profiled at high depth (MGH36, MGH53, MGH54), and red circles denote the percentages in individual tumours. Bins in left panel were defined as stemness scores below − 1.5 (n = 711), between − 1.5 and 0.5 (n = 1,100), between − 0.5 and 0.5 (n = 939), and above 0.5 (n = 274), respectively. The first two bins are significantly depleted with cycling cells, while the last two bins are significantly enriched (P < 0.05, hypergeometric test). Bins in left panel were defined as AC score above 1 (n = 503), AC score between 0.5 and 1 (n = 1,013), AC and OC scores below 0.5 (n = 1,130), OC score between 0.5 and 1 (n = 855), and OC score above 1 (n = 597), respectively. The third bin is significantly enriched with cycling cells, while the four other bins are significantly depleted (P < 0.05, hypergeometric test). e, Correlation between the average expression of cell cycle (y axis) and that of stemness genes (x axis) across molecularly defined oligodendrogliomas (by IDH mutation, chromosome 1p and 19q co-deletion, and absence of P53 and ATRX mutations) profiled by TCGA (n = 69) with bulk RNA-seq. Average expression was defined by centring the log2-transformed RSEM gene quantifications. Also shown are the linear least-square regression and Pearson correlation coefficient. f, Specific enrichment of S/G2/M cells compared to G1 cells among stem-like or undifferentiated cells. Shown is the proportion (y axis) of each marked category of cells among the stem-like or undifferentiated subpopulations. Significant enrichments are marked (P < 0.01, hypergeometric test).
Extended Data Figure 7. CCND2 is associated with both cycling and non-cycling stem/progenitor cells.
a, CCND2, but not CCND1 or CCND3, is upregulated in non-cycling stem-like oligodendroglioma cells. Shown are the average expression levels (y axis, log-scale) of three cyclin D genes (x axis) in non-cycling cells classified as OC-like cells (light blue), undifferentiated cells (grey) and stem-like cells (purple). CCND2 is approximately fourfold higher in stem-like non-cycling cells than in OC-like and undifferentiated cells (P < 0.001 by permutation test). Conversely, CCND1 and CCND3 are expressed at comparable levels in stem-like and OC-like cells. b, Upregulation of cyclin D genes in cycling cells compared to non-cycling cells. As in a but for up regulation (log2-ratio) in cycling cells vs. non-cycling cells. CCND2 levels further increase in cycling undifferentiated and stem-like cells but not in OC-like cells, whereas CCND1 and CCND3 levels increase in OC-like cycling cells more than in undifferentiated and stem-like cycling cells. c, Distinct expression patterns of cyclin D genes in human brain development. Shown are the expression patterns of three cyclin D genes (rows) in human brain samples at different points in pre- and post-natal development, sorted by age (columns) from the Allen Brain Atlas15. CCND2 is associated with prenatal samples, whereas CCND1 and CCND3 are expressed mostly in childhood and adult samples. d, CCND2 is upregulated in activated versus quiescent NSCs19, both among cycling and non-cycling cells. Activated NSCs were partitioned into non-cycling cells (black) and cycling cells in the G1/S (green) or G2/M (red) phases (Methods). Expression difference (y axis) for each of three genes (x axis) was quantified for each of these subsets as the log2-ratio of the average expression in the respective subset versus the quiescent NSCs, and was significant for each of the three subsets (P < 0.05 by permutation test). Although CCND2 (left) is induced in both cycling and non-cycling activated NSCs, two canonical cell cycle genes (PCNA, middle; and AURKB, right) are not induced in non-cycling genes but were induced preferentially in G1/S and G2/M cells, respectively.
Extended Data Figure 8. Distribution of cellular states in distinct genetic clones of MGH36 and MGH97.
a, Stemness (y axis) and lineage (x axis) score plots for MGH36 (top) and MGH97 (bottom), each separated into clone 1 (left) and clone 2 (right) as determined by CNV analysis (Fig. 1a, b). Cycling cells are coloured as in Fig. 3, with G1/S cells in blue, S/G2 cells in green, and G2/M cells in red. b, Colour-coded density of cells across the cellular hierarchy as shown in Fig. 2e, for the two clones (left: clone 1, right: clone 2) in each of the two tumours (top: MGH36, bottom: MGH97). c, The fraction of cells assigned to the different tumour compartments (y axis, Methods) based on either single-cell RNA-seq (blue) or RNA in situ hybridization (orange). Circles denote individual tumours; squares denote average of all tumours; error bars denote standard deviation across tumours, showing general agreement between scRNA-Seq and IHC estimates.
Extended Data Figure 9. Subclonal mutations tend to span the cellular hierarchy.
Each panel shows lineage (x axis) and stemness (y axis) scores of cells in which we ascertained by single cell RNA-seq a mutant (red), a wild-type (blue) or none (black) of the alleles. Included are mutations for which at least three cells were identified as mutants and that were identified by WES as subclonal (fraction < 60%). The gene names, tumour name, ABSOLUTE-derived fraction of mutant cells (E, expected fraction) and the fraction of cells detected as mutant by RNA-seq (O, observed) are also indicated within each panel. We note that identification of a wild-type allele (blue) does not imply a wild-type cell because mutations may be heterozygous, and thus cells could contain both alleles while only one may be detected by single-cell RNA-seq. The observed fraction of mutations (O) is much lower than expected (E) due to limited coverage of the single-cell RNA-seq data, as well as due to heterozygosity. The vast majority of mutations (20 of 22) are distributed across the hierarchy and span multiple compartments. Two remaining mutations (H2AFV and EIF2AK2) appear more restricted to the ‘undifferentiated’ region (intermediate lineage and stemness scores), which could reflect our limited detection rate of mutant cells and/or a bias of the mutation to a particular region. To test the significance of potential biases in the distribution of mutations we calculated, for each mutation, a Euclidean distance among all pairs of mutant cells (based on their lineage and stemness scores), and compared the average pairwise distances among mutant cells to that among randomly selected subsets of the same number of cells. None of the mutations were significant with a false discovery rate (FDR) of 0.1, although this could reflect our limited statistical power and we cannot exclude a potential bias. The apparent bias of mutant cells to the OC lineage over the AC lineage (that is, positive versus negative lineage scores) reflects the lower frequencies of AC-like cells compared to OC-like cells in MGH53 and MGH54 (MGH53: 17% AC versus 39% OC; MGH54: 23% AC vs. 45% OC); this bias is also observed for the detection of wild-type alleles (blue) demonstrating that there is no bias against mutation detection in the AC lineage.
Extended Data Figure 10. Loss-of-heterozygosity event in MGH54 reveals two clones that span the cellular hierarchy.
a, Chromosome 18 loss of heterozygosity (LOH) in MGH54. Allelic fraction analysis of MGH54 SNPs from WES shows an imbalance (red and blue dots) in the frequency of alternative alleles in chromosome 1p, 19q, as well as chromosome 18, despite the normal copy number at this chromosome (Fig. 1a). This is consistent with an LOH event in which presumably one copy of chromosome 18 was deleted, and the other copy amplified. The weaker imbalance compared to chromosomes 1p and 19q further suggests that this is a subclonal event. b, Each of two clones defined by chromosome 18 LOH status spans the full hierarchy. Shown are the lineage (x axis) and stemness (y axis) scores for each cell from MGH54 (n = 1,174) classified as pre-LOH (red), post-LOH (blue) and unresolved (black) based on RNA-seq reads that map to SNPs in the minor (that is, deleted) chromosome. Both the pre- and post-LOH clones span the different tumour subpopulations. Pre-LOH cells were defined as all cells with reads that map to minor alleles in chromosome 18; post-LOH cells were defined as all cells with reads that map to at least five different major alleles, but no reads that map to minor alleles in chromosome 18; all other cells were defined as unresolved.
Supplementary Material
Acknowledgments
We thank L. Gaffney for graphic support. This work was supported by grants from the National Brain Tumor Society (to M.L.S. and D.N.L.), the Smith Family Foundation (to M.L.S.), NIH-NCI SPORE on brain cancer Career Enhancement Project and Developmental Research Project (to M.L.S.), the Broad Institute Broadnext10 program (to M.L.S. and O.R.R.), the American Cancer Society (to M.L.S.) and start-up funds from the MGH department of Pathology. A.S.V. was supported by the NIH R25 fellowship (NS065743) and research grants from the American Brain Tumor Association and Neurosurgery Research and Education Foundation. I.T. was supported by a Human Frontier Science Program fellowship and a Rothschild fellowship. A.R. was supported by funds from the Howard Hughes Medicine Institute, the Klarman Cell Observatory, STARR cancer consortium, NCI grant 1U24CA180922, by the Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute, the Ludwig Center and the Broad Institute. A.R. is a scientific advisory board member for ThermoFisher Scientific and Syros Pharmaceuticals and a consultant for Driver Group. Flow cytometry and sorting services were supported by shared instrumentation grant 1S10RR023440-01A1. M.M. was supported by the California Institute of Regenerative Medicine (CIRM) grants RB4-06093 and RN3-06510 and the Virginia and D.K. Ludwig Fund for Cancer Research.
Footnotes
Reviewer Information Nature thanks P. Dirks, J. Rich and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author Contributions I.T., A.S.V., A.R. and M.L.S. conceived the project, designed the study, and interpreted results. A.S.V., C.H., L.E.E. and C.N. collected single cells and generated single-cell sequencing data. I.T. performed computational analyses. J.M.Fra, K.Y. and G.G. provided support for genomic and genetic analyses. J.M.Fis, C.R. and K.J.L. designed and performed qPCR experiments. C.C.L. and R.M. provided flow cytometry expertise. C.M. and M.M. developed normal human cell cultures used in the study. N.D., N.R., M.N.R., M.L.O. and A.J.I. performed in situ hybridization and FISH experiments. A.P.P., A.A.P., D.G., B.I., J.N., R.S., M.G.F., B.V.N., D.P.C., W.T.C., R.L.M., M.P.F., O.R.R., T. R.G., B.E.B. and D.N.L. provided experimental and analytical support. I.T., A.R. and M.L.S. wrote the manuscript with feedback from all authors.
Supplementary Information is available in the online version of the paper.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumors of the Central Nervous System. 4. IARC; 2016. [DOI] [PubMed] [Google Scholar]
- 5.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 6.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butovsky O, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nature Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rousseau A, et al. Expression of oligodendroglial and astrocytic lineage markers in diffuse gliomas: use of YKL-40, ApoE, ASCL1, and NKX2-2. J Neuropathol Exp Neurol. 2006;65:1149–1156. doi: 10.1097/01.jnen.0000248543.90304.2b. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng W, et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Ikushima H, et al. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Suvà ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rheinbay E, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Reports. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugiarto S, et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin J, et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Research Network. Comprehensive integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange C, Calegari F. Cdks and cyclins link G1 length and differentiation of embryonic, neural and hematopoietic stem cells. Cell Cycle. 2010;9:1893–1900. doi: 10.4161/cc.9.10.11598. [DOI] [PubMed] [Google Scholar]
- 24.Koyama-Nasu R, et al. The critical role of cyclin D2 in cell cycle progression and tumorigenicity of glioblastoma stem cells. Oncogene. 2013;32:3840–3845. doi: 10.1038/onc.2012.399. [DOI] [PubMed] [Google Scholar]
- 25.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettegowda C, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padul V, Epari S, Moiyadi A, Shetty P, Shirsat NV. ETV/Pea3 family transcription factor-encoding genes are overexpressed in CIC-mutant oligodendrogliomas. Genes Chromosom Cancer. 2015;54:725–733. doi: 10.1002/gcc.22283. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohapatra G, et al. Glioma test array for use with formalin-fixed, paraffin-embedded tissue: array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. J Mol Diagn. 2006;8:268–276. doi: 10.2353/jmoldx.2006.050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos AH, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study are available through the Gene Expression Omnibus (GEO) under accession number GSE70630.