Abstract
Prevention of virus infections is a major objective in agriculture and human health. One attractive approach to the prevention is inhibition of virus replication. To demonstrate this concept in vivo, an artificial zinc finger protein (AZP) targeting the replication origin of the Beet severe curly top virus (BSCTV), a model DNA virus, was created. In vitro DNA binding assays indicated that the AZP efficiently blocked binding of the viral replication protein (Rep), which initiates virus replication, to the replication origin. All of the transgenic Arabidopsis plants expressing the AZP showed phenotypes strongly resistant to virus infection, and 84% of the transgenic plants showed no symptom. Southern blot analysis demonstrated that BSCTV replication was completely suppressed in the transgenic plants. Since the mechanism of viral DNA replication is well conserved among plants and mammals, this approach could be applied not only to agricultural crop protection but also to the prevention of virus infections in humans.
Various DNA viruses are known to cause severe infectious diseases in both plants and mammals, including humans (11). For many of these infectious diseases, we have yet to find an effective prevention or treatment. Therefore, new methodologies for the prevention of virus infections in both agricultural crops and humans have been vigorously sought for a long time.
The basic mechanism of DNA virus replication is well conserved among plants and mammals (11). After infection, a viral DNA binding protein expressed from its genome binds to the replication origin of the genome and then initiates replication alone or in cooperation with another viral protein(s). For example, the large T antigen is the only simian virus 40 (SV40) protein required for viral DNA replication in SV40 (13). It binds tightly and specifically to the SV40 origin to initiate replication. Among plant DNA viruses, the geminiviruses constitute a large family. Members of the geminivirus family possess a circular single-stranded DNA (ssDNA) genome encapsidated within geminate icosahedral virions (reference 18 and references cited therein). Several lines of evidence independently support the hypothesis that geminivirus double-stranded DNA (dsDNA) produced within infected cells serves as a replicative intermediate in a rolling-circle replication mechanism (22). Beet curly top virus (BCTV) is a member of this family that has an unusually wide dicotyledonous host range and induces severe diseases (21). Mutational analysis of the seven BCTV genes has indicated that the replication protein known as Rep in BCTV is the only virus-encoded protein absolutely required for BCTV DNA replication (2, 6, 8, 19, 20). The Rep protein binds to the direct repeat sequence in the viral replication origin and induces nicking in the stem-loop of the origin for initiation of DNA replication.
I developed a method for the rational design of artificial zinc finger proteins (AZPs) by using a nondegenerate recognition code table (17). This design method can be used to target diverse DNA sequences, and it enables the creation of millions of AZPs in a high-throughput manner, as well as a huge combinatorial library of AZPs. The AZPs designed by this method showed both high affinities and high selectivities. In particular, six-finger AZPs bound to 19-bp DNA targets with extremely high affinities (i.e., apparent dissociation constants of <3 pM), including the Rep binding site (e.g., a six-finger AZP binding to the Rep binding site designated AZP-A4 in reference 17). In this report, the six-finger AZP was applied to prevention of DNA virus infection. If binding of a replication protein to its replication origin can be blocked with AZP, it should be possible to inhibit virus replication, which will result in prevention of virus infection. The principle is here demonstrated by using Beet severe curly top virus (BSCTV), which is one of the BCTV strains causing the severest damage, as a model DNA virus. Since this virus is known to induce severe symptoms in Arabidopsis thaliana (12), it is easy to evaluate the ability of AZP to prevent viral infection of living organisms by observing phenotypes of A. thaliana.
MATERIALS AND METHODS
Construction of a Rep expression vector.
DNA encoding the BSCTV Rep protein (residues 1 to 176) was amplified from the pCFH vector (American Type Culture Collection) with primers 5′-AGCCCGATTCGAATTCCCTTTTTACAAAAAAGCCAAAAATTTTTTC-3′ and 5′-GAGGAAAAGGAAGCTTTTAAAGATCTGGAGGAGGAAGAAAAATAGCC-3′ and cloned into the EcoRI/HindIII sites of the pET-21a vector (Novagen).
Protein overexpression and purification of AZP and Rep.
The design of the AZP (designated AZP-A4 in reference 17) for inhibition of Rep binding and construction of the E. coli expression vector have been reported previously (16, 17). The AZP was overexpressed and purified as previously described (16, 17). The Rep protein was also overexpressed and purified essentially as previously described (17), except that the protein was eluted with 200 mM NaCl buffer. All purified proteins were >95% homogeneous, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The protein concentration was determined with a Protein Assay ESL kit (Roche Molecular Biochemicals).
DNA binding assays.
A 26-bp synthetic DNA duplex consisting of the sequence 5′-(TA)4TTGGGTGCTTTGGGTGCTC(TA)4-3′ was labeled by a Klenow fill-in reaction with [α-32P]dATP and [α-32P]dTTP. Purified AZP and Rep protein were incubated on ice in 10 mM Tris-HCl (pH 7.5)-100 mM NaCl-5 mM MgCl2-0.1 mM ZnCl2-0.05% bovine serum albumin-10% glycerol containing the labeled probe (1 fmol/10 μl of buffer) and 0.1 μg of poly(dA-dT)2. The order of addition of these proteins to the labeled probe and the incubation time for each binding experiment are described in detail in the legend to Fig. 1. The probe-protein complexes and the free probe were separated on a 6% nondenaturing polyacrylamide gel (45 mM Tris-borate) by electrophoresis at 140 V for 2 h at 4°C. The radioactive signals were recorded on X-ray films.
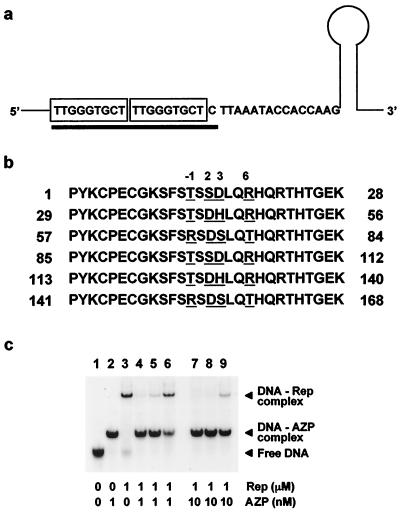
FIG. 1.
Inhibition of Rep binding to direct repeats by AZP. (a) Rep binding site in the BSCTV replication origin. The two boxes indicate the direct repeats recognized by the Rep protein. The underlined 19-bp region is the target site of the designed six-finger AZP to block Rep binding. Only the sense strand is shown. (b) Amino acid sequence of the AZP. The underlined amino acids were selected from the nondegenerate recognition code table (17) for recognition of the 19-bp target shown in panel a. The numbers at the top are positions relative to the first amino acid of each recognition helix. (c) Lanes: 1, 32P-labeled probe containing direct repeats; 2, band shift in the presence of 1 nM AZP; 3, band shift in the presence of 1 μM Rep; 4 to 6 and 7 to 9, band shifts in the presence of Rep (1 μM) together with 1 and 10 nM AZP, respectively. In lanes 4 and 7, Rep was added to the binding mixture after incubation of the probe with AZP for 30 min. In lanes 5 and 8, Rep and AZP were mixed together with the probe. In lanes 6 and 9, AZP was added to the binding mixture after incubation of the probe with Rep for 30 min.
Preparation of transgenic Arabidopsis plants.
The AZP expression cassette was prepared as previously described (17). The expression cassette encodes a nuclear localization signal from the SV40 large T antigen, the AZP, and a FLAG epitope tag (in that order from the amino terminus) under the control of a cestrum yellow leaf curling virus promoter (9). The expression cassette was cloned between the left and right borders of a T-DNA vector, pNOV3510, containing a protoporphyrinogen IX oxidase (PPO) selection marker gene (7). The transgenic Arabidopsis plants were prepared from the Columbia (Col) ecotype with the above-described T-DNA vector by the floral-dip method (3) with Agrobacterium tumefaciens strain GV3101. The inoculation medium contained 0.005% Silwet L-77. PPO selection was conducted in accordance with a modified version of a previously reported method (3). The selection plates contained 0.5× Murashige and Skoog salts (pH 5.8), 0.8% agar, 1× GM vitamins (1 μg of thiamine HCl, 0.5 μg of pyridoxine HCl, 0.5 μg of nicotinic acid, and 10 mg of myoinositol/ml), 1% sucrose, and 12.5 nM butafenacil. The transformants were identified as butafenacil-resistant seedlings that produced green leaves and well-established roots. Positive transformants were transferred onto soil and grown in a greenhouse (20°C, 18 h of light-6 h of dark) until seeds were harvested.
Agroinoculation with BSCTV.
A. tumefaciens strain GV3101 containing pAbar-CFH was incubated at 30°C until the optical density at 600 nm was 1.5. After brief centrifugation of 1 ml of the culture, the resulting pellet was resuspended in 1 ml of the inoculation medium. A short primary inflorescence (less than 1 cm tall) of each 3- to 5-week-old plant was cut, and the suspension was injected into the center of each cut stem with a 23-gauge needle. The injected plants were covered with a plastic dome to keep them moist and incubated at room temperature overnight. The next day, the plants were transferred to a growth chamber or a greenhouse (22°C, 16 h of light-8 h of dark) and grown until symptoms appeared. This method consistently produced severe symptoms in all 44 wild-type (WT) Col ecotype plants.
Total DNA isolation.
Total DNA was isolated from agroinoculated A. thaliana plants with a DNeasy Maxi Kit (QIAGEN). The yield was 25 to 35 mg/g of frozen plant tissue.
Southern blot analysis.
A 200-bp digoxigenin (DIG)-labeled probe specific to the BSCTV genome was prepared from pCFH by PCR with DIG-11-dUTP (Roche Molecular Biology) and primers 5′-CCATCGACCTGAAATTCACCCCAGTCGATG-3′ and 5′-GGGGAACCACATCTGCATGCCCTTATTCAA-3′. Two micrograms of each isolated DNA sample and 50 ng of EcoRI-digested pCFH were separated on a 0.8% agarose gel containing 5 μg of ethidium bromide per ml. The digested pCFH plasmid was used as a size marker to show a linear dsDNA form of the BSCTV genome. After a picture was taken under UV irradiation, DNA bands were transferred onto a Nytran SuPerCharge membrane with a TurboBlotter (Schleicher & Schuell). After hybridization with the DIG-labeled probe, DNA bands corresponding to BSCTV were visualized with anti-DIG-AP and CDP-Star in accordance with the accompanying protocols (Roche Molecular Biology).
RESULTS
Inhibition of Rep binding to the replication origin of BSCTV by AZP.
To examine whether or not the six-finger AZP can block the Rep protein of BSCTV from binding to the direct repeats, gel shift assays were performed. The Rep protein is known to bind to dsDNA of the direct repeats (i.e., 5′-TTGGGTGCTTTGGGTGCT-3′) in the viral replication origin (Fig. 1a). In a previous study (17), a six-finger AZP (designated AZP-A4 in reference 17) was designed (Fig. 1b) and shown to bind to a 19-bp target (5′-TTGGGTGCTTTGGGTGCTC-3′) containing the direct repeats (underlined) with extremely high affinity (i.e., with an apparent dissociation constant of <3 pM). First, to test whether or not the AZP bound to the direct repeats can block Rep binding, the Rep protein was added to a premixture of the AZP and the DNA probe containing the direct repeats. This experimental condition seems to simulate BSCTV infection in transgenic Arabidopsis plants constitutively expressing the AZP. In actual virus infection of transgenic plants, it is likely that the direct repeats are preoccupied by the AZP before Rep binding because there is a time lag between generation of the dsDNA form of the viral genome by endogenous enzymes and appearance of the Rep protein (expressed from the dsDNA) in nuclei. In lanes 4 and 7 of Fig. 1c, the AZP was first mixed with the DNA probe to final concentrations of 1 and 10 nM, respectively, where all of the probe molecules were bound to the AZP (Fig. 1c, lane 2). After incubation for 30 min on ice, Rep protein was added to the binding reaction mixture to a final concentration of 1 μM. As shown in lane 4, the AZP bound to the direct repeats efficiently prevented Rep protein binding even at a concentration of 1 nM. Moreover, the AZP could completely inhibit Rep protein binding at a concentration of 10 nM (lane 7). Next, I examined how efficiently the AZP prevents Rep binding when both the AZP and Rep are mixed with the DNA probe at the same time. During coincubation with 1 nM AZP, a small amount of Rep protein still bound to the direct repeats (lane 5). However, 10 nM AZP completely inhibited Rep binding again (lane 8). Finally, I examined whether or not the AZP can eliminate Rep bound to the direct repeats. In this experiment, the Rep protein was first mixed with the DNA probe. After incubation for 30 min, the AZP was added to the binding reaction mixture. As shown in lanes 6 and 9 of Fig. 1c, it seemed to be difficult for the AZP to completely eliminate Rep protein bound to the target at concentrations of ≤10 nM. These results suggest that the presence of AZP in plants before invasion of virions will achieve more efficient inhibition of Rep binding to the replication origin in vivo.
Infection of BSCTV in transgenic Arabidopsis plants expressing AZP.
To examine the ability of the AZP to inhibit the DNA replication of BSCTV in vivo, the AZP expression construct was introduced into A. thaliana by transformation with A. tumefaciens strain GV3101 (3). The construct possesses a mutated PPO-encoding gene as a selection marker (7). By PPO selection of ∼4,000 T1 seeds harvested from the transgenic plants, eight butafenacil-resistant seedlings were obtained and grown in a greenhouse. Among them, seven T1 plants were confirmed by PCR to possess the AZP-encoding region in each of the genomes and by reverse transcription-PCR to express the AZP gene (data not shown). Each of the T2 lines obtained from two randomly chosen T1 plants (designated 1 and 2, respectively) were further grown to obtain their T3 seeds. For the following first infection experiment, one of each of the T2 lines (designated lines 1-1 and 2-1) that were obtained from T1 lines 1 and 2, respectively, was used to obtain a young T3 plant. The resulting T3 plants were designated 1-1A and 2-1A, respectively. WT and the T3 transgenic plants, 1-1A and 2-1A, were infected with BSCTV by Agrobacterium-mediated inoculation (agroinoculation) with strain GV3101(pAbar-CFH). The pAbar-CFH construct is an infectious clone containing 1.5 copies of the BSCTV genome between the left and right borders of the T-DNA vector. The conventional agroinoculation method is injection of an Agrobacterium inoculum containing a viral genome into leaves or crowns (2, 12). However, WT Arabidopsis plants agroinoculated under my experimental conditions did not show a consistent phenotype. Therefore, an alternative injection method was developed. Namely, a short primary inflorescence (≤1 cm tall) was cut and an Agrobacterium inoculum containing the pAbar-CFH construct was injected into the center of the cut stem. All of the WT plants agroinoculated by this new injection method consistently showed severe symptoms such as curling and stunting of inflorescences, deformation of floral structures, leaf curling and deformation, vein swelling, accumulation of anthocyanins, and finally death. A typical infected WT plant 2.5 weeks after agroinoculation is shown in Fig. 2a (the plant on the left). In contrast, the T3 plants, 1-1A and 2-1A, clearly showed strong resistance to viral infection. The 1-1A plant did not show any symptoms and grew identically to a healthy WT plant (the plant on the right in Fig. 2a). The 2-1A plant, shown in Fig. 2b (the plant on the right), was almost identical to a healthy WT plant except for one gently curling inflorescence. A magnified image of the inflorescence is shown in Fig. 2c. The shape was slightly similar to that of inflorescences observed consistently in infected WT plants (compare with Fig. 2d). In infected WT plants, inflorescences were short, thick, curling, and severely deformed. Severely deformed floral structures and anthocyanin accumulation were also observed, as shown in Fig. 2d.
FIG. 2.
Photographs of WT and AZP-transgenic A. thaliana plants agroinoculated with strain GV3101(pAbar-CFH). (a) Agroinoculated WT (left) and T3 transgenic 1-1A (right) plants. (b) Agroinoculated WT (left) and T3 transgenic 2-1A (right) plants. (c) Magnified image of the gently curling inflorescence of the 2-1A plant indicated by the white rectangular frame in panel b. (d) Magnified image of a typical inflorescence of an agroinoculated WT plant.
Southern blot analysis of virus replication in AZP-expressing transgenic lines.
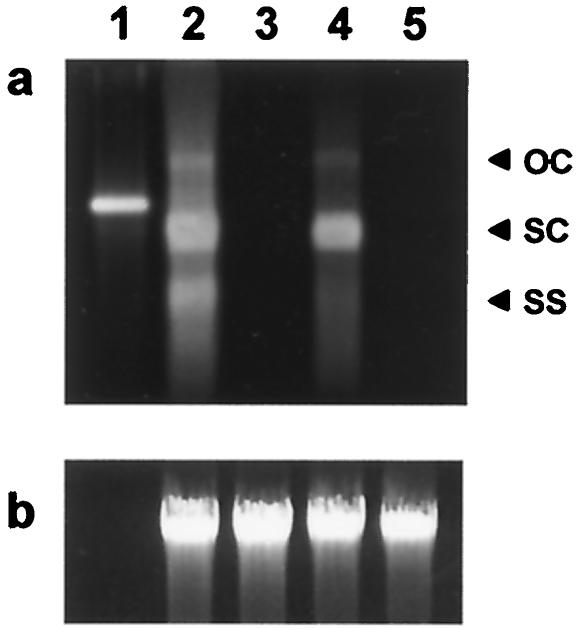
Virus replication in T3 transgenic plants 1-1A and 2-1A, which showed strong resistance to viral infection (Fig. 2), and an infected WT plant was examined at the molecular level by Southern blot analysis. Total DNA was isolated from each agroinoculated plant and a noninfected WT plant that served as a control. From each plant, 2 μg of total DNA was separated on a 0.8% agarose gel to obtain open circular, supercoiled, and single-stranded forms of the genome length progeny viral DNA. As shown in lane 2 of Fig. 3a, all three forms were detected in the infected WT plant with severe symptoms. In contrast, no form of the progeny viral DNA was detected in the 1-1A plant (lane 3 of Fig. 3a), as expected from the symptom-free phenotype (Fig. 2a). The 2-1A plant was divided into two parts, half containing the gently curling inflorescence and the remaining half with no symptoms, and total DNA was isolated from each half. In the first half, a much reduced but significant amount of the supercoiled form was detected (lane 4 of Fig. 3a). On the other hand, no form of the progeny viral DNA was detected at all in the symptom-free half, as shown in lane 5 of Fig. 3a. This result indicates at the molecular level that the virus infection observed in the curling inflorescence of the 2-1A plant had not spread throughout the transgenic plant.
FIG. 3.
Southern blot analysis of total DNA isolated from agroinoculated WT and T3 transgenic 1-1A and 2-1A plants. (a) DNA bands probed with the DIG-labeled PCR product (200 bp) of the BSCTV genome. Lanes: 1, 50 ng of pCFH digested with EcoRI; 2, 2 μg of total DNA isolated from a whole agroinoculated WT plant; 3, 2 μg of total DNA isolated from a whole agroinoculated 1-1A plant; 4, 2 μg of total DNA isolated from the half of an agroinoculated 2-1A plant that had gently curling inflorescence, indicated by the white frame in Fig. 2b; 5, 2 μg of total DNA isolated from the remaining half of the agroinoculated 2-1A plant, in which no symptoms were observed. (b) Ethidium bromide-stained gel image of total DNA used for the Southern blot shown in panel a. This photograph was taken before processing of the Southern blot. OC, open circular DNA; SC, supercoiled DNA; SS, ssDNA.
Infection of BSCTV in 116 transgenic plants expressing AZP.
More T3 transgenic plants derived from the nine T2 lines (i.e., four T2 lines from T1 line 1 and five T2 lines from T1 line 2) were infected with BSCTV. As summarized in Table 1, all of these lines showed strong resistance to viral infection. Of 116 transgenic plants agroinoculated, 97 transgenic plants showed no symptoms of infection. Thus, 84% of the transgenic plants were completely resistant to viral infection. Fourteen transgenic plants displayed only minor symptoms. After further incubation (more than 4 weeks after agroinoculation), the minor symptoms observed in the 14 transgenic plants had not developed further (Fig. 4): the number of curling inflorescences did not increase, and rosette leaves of the transgenic plants were identical to those of noninfected, healthy WT plants. The roots were also identical to those of noninfected WT plants (data not shown). Therefore, it is most likely that the virions present in the curling inflorescences did not spread beyond the infected curling inflorescences in the transgenic plants. In contrast, in WT plants, agroinoculated viruses spread to other tissues, including rosette leaves, and the infected plants finally died (Fig. 4). Finally, no phenotype indicating toxicity derived from AZP expression was observed in any of the 116 transgenic plants.
TABLE 1.
Resistance of AZP-expressing A. thaliana to BSCTV infection
| Lineb | No. of plantsa with:
|
|||
|---|---|---|---|---|
| No symptomsc | Minor symptomsd | Mild symptomse | Severe symptomsf | |
| 1-1 | 12 | 3 | 0 | 0 |
| 1-2 | 17 | 2 | 0 | 0 |
| 1-3 | 14 | 1 | 0 | 0 |
| 1-4 | 18 | 3 | 0 | 0 |
| 2-1 | 8 | 2 | 0 | 0 |
| 2-2 | 14 | 1 | 3 | 0 |
| 2-3 | 3 | 0 | 0 | 0 |
| 2-4 | 5 | 1 | 0 | 0 |
| 2-5 | 6 | 1 | 2 | 0 |
| WT | 0 | 0 | 0 | 44 |
Each value is the number of WT or T3 transgenic plants showing each symptom after agroinoculation with BSCTV.
The line numbers correspond to the T2 lines. T3 transgenic plants used for the agroinoculation were obtained from these T2 lines.
Same phenotypes as healthy, noninfected WT Col ecotype plants.
Same phenotypes as healthy, noninfected WT Col ecotype plants, except for gentle curling of a couple of tops of inflorescences.
Plants were as tall as WT plants, but all tops of inflorescences were curled or deformed. Some stems in some plants were thicker than WT stems. No symptom were observed on rosette leaves.
Observed symptoms included curling and stunting of all inflorescences, deformation of floral structures, short and thick stems, leaf curling and deformation, vein swelling, and accumulation of anthocyanins.
FIG. 4.
Transgenic plants 4 weeks after agroinoculation. (a) Images of whole plants. (b) Images of rosette leaves. In both panels, a noninfected WT plant, an infected WT plant, a 1-3A plant with minor symptoms, and a 1-3B plant with no symptoms are shown from the left to the right. T3 transgenic plants 1-3A and 1-3B were obtained from T2 line 1-3.
DISCUSSION
Various antiviral drugs have been developed and used for the treatment of viral infections. Their main targets are viral enzyme molecules, including viral thymidine kinases and DNA polymerases. However, the emergence of virus mutants that inactivate antiviral drugs is always a formidable problem (1). Viruses with high mutation rates have escaped the attacks of drugs by mutation of an amino acid(s) in viral enzymes, resulting in rapid emergence of drug-resistant virus mutants. Therefore, it is very important to develop new agents (or approaches) with different modes of action. In this study, binding of a viral protein to its replication origin was targeted in order to prevent viral infections. One advantage of this approach is the absence or reduced possibility of rapid emergence of virus mutants resistant to blocking of viral protein binding by AZP. For DNA viruses to escape AZP attack, there are two choices: inactivation of the AZP and mutation of viral genomes. Since no gene for an enzyme capable of inactivating AZPs by chemical modification (e.g., phosphorylation) has been identified in DNA viruses (because the AZPs are artificial proteins), rapid evolution of viruses to inactivate AZPs seems to be unlikely. In order to escape by mutation of existing viral genomes, the mutation of both amino acids of the viral replication proteins and DNA sequences of the binding sites of the emerging mutant proteins in their replication origins must happen at the same time because either mutation alone will dramatically reduce the efficiency of viral DNA replication, which is unfavorable for viruses. However, the possibility of the occurrence of synchronized double mutations should be very low. Therefore, the rapid emergence of virus mutants resistant to this approach is less likely than that of resistance to conventional antiviral drugs, such as nucleoside analogues. For inhibition of a viral protein's binding to its replication origin, a dominant negative replication protein composed of its DNA binding domain has been used. For example, the dominant negative Rep protein of the Tomato golden mosaic virus reduced DNA replication ∼50% in vitro (14). One drawback of this approach is the requirement of much larger amounts of the dominant negative protein for efficient inhibition of DNA replication because the affinities of WT and dominant negative replication proteins for the replication origin are the same. As an alternative approach to the inhibition of virus replication, repression of viral promoters with pyrrole-imidazole polyamides (5) or engineered zinc finger proteins (15) has been reported. Although this approach was shown to be efficient in vitro, its in vivo efficacy has not been reported.
Transgenic plants expressing the six-finger AZP showed clear resistance to BSCTV infection. In all 116 of the transgenic plants agroinoculated, no phenotype suggesting that the expression of AZP is toxic to A. thaliana was observed. For example, as shown in Fig. 3 and 4, the 97 transgenic plants with no symptoms were identical to noninfected WT plants; these plants were as large as noninfected WT plants, and no deformation was observed in their stems and leaves. Seeds were harvested from the transgenic plants and grown to healthy T4 plants (data not shown). The minor symptom of curling inflorescences was observed in 14 transgenic plants, and the viral progenies were detected in the infected tissues (Fig. 3, lane 4). In plants, it is well known that viruses spread to other tissues through plasmodesmata (4). However, in the transgenic plants in this study, the spread seemed to be restricted to (or around) the infected tissues. Four weeks post agroinoculation, when infected WT plants were dead (Fig. 4), the number of curling inflorescences per plant had not increased and the leaves did not show any clear phenotype, such as curling and accumulation of anthocyanins, in the transgenic plants with minor symptoms (Fig. 4). Southern blot analysis also supported the restriction of infection (compare lane 5 to lane 4 in Fig. 3a). Thus, symptoms did not develop in the transgenic plants even when topical infection occurred.
In this study, BSCTV DNA was introduced into plants via agroinoculation. Agroinoculation with a T-DNA vector containing tandemly repeated copies (generally 1.5 copies) of viral genomic DNA has been extensively used for plant viral infection experiments, including analysis of viral DNA replication. In agroinoculation, ssDNA between the right and left borders (containing tandem copies of the viral genomic DNA) in the double-stranded T-DNA is released from the vector in Agrobacterium, the ssDNA forms a complex with virulence proteins in Agrobacterium, and then the complex moves from Agrobacterium into a plant cell. After reaching a plant cell nucleus, the complex is converted to linear dsDNA by endogenous enzymes (agroinoculation has also been used for Agrobacterium-mediated transient gene expression; for an example, see reference 23) and/or integrated into a plant genome (24). In either case, the Rep protein produced from the dsDNA (linear T-DNA or a T-DNA integrated genome) binds to the replication origin in the dsDNA, and then the virus replication is initiated by a rolling-circle mechanism to generate the unit length progeny genome. During agroinoculation as described above, the T-DNA containing 1.5 tandem copies of the viral genomic DNA exists as an ssDNA entirely covered by virulence proteins in a plant cell (nucleus). The double-stranded T-DNA exists only in Agrobacterium. Since AZP cannot pass through Agrobacterium membranes, AZP (a dsDNA binder) has no opportunity to interact with the T-DNA during agroinoculation. Therefore, it is unlikely that AZP inhibits the process of agroinoculation, thereby generating the resistance phenotypes. Moreover, results obtained with 14 transgenic plants with minor symptoms support this hypothesis. If AZP worked only in the agroinoculation step, severe symptoms should have developed in the plants. However, as described above, the transgenic plants with minor symptoms, in which unit length viral progeny DNA was detected, did not show any further development of symptoms or spread of viral progenies even 4 weeks after agroinoculation, when WT plants were dead.
One issue of this approach is how to deal with variants of viruses. For example, >100 variants in the Geminiviridae family have been identified so far (http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_gemin.htm). Each virus contains its own replication origin. It is not practically possible to generate a single transgenic plant expressing AZPs against all of the variants. However, as shown in this study, this approach could be very powerful for inhibition of DNA replication of a single DNA virus. Therefore, the approach can be useful in a particular geographic region, where one specific DNA virus causes severe damage to a significant crop(s). Expression of two or more AZPs in a single transgenic plant may expand the practical potential of this approach. Use of plant internal ribosomal entry sites (10) or development of expression systems of several genes from a single mRNA is required for that.
In this study, it was demonstrated in A. thaliana that blocking the binding of a viral replication protein to its replication origin with AZP prevents DNA virus infection in vivo. Thus, this method would be applicable to agricultural crop protection against DNA virus infection. Moreover, because the basic mechanism of DNA virus replication is well conserved between plants and mammals, this antiviral approach with AZP could also be used for prevention of infections of humans by DNA viruses such as human papillomavirus and herpes simplex virus.
Acknowledgments
I thank Fumiaki Katagiri and Yi Tao for technical advice on Agrobacterium transformation of A. thaliana, Kimberly Campbell for maintenance of A. thaliana plants, Kiyoshi Tachikawa for help with Rep protein purification, Sheng Quan for pAbar-CFH, and Joel Kreps for pNOV3510.
REFERENCES
- 1.Balzarini, J., L. Naesens, and E. De Clercq. 1998. New antivirals—mechanism of action and resistance development. Curr. Opin. Microbiol. 1:535-546. [DOI] [PubMed] [Google Scholar]
- 2.Briddon, R. W., J. Watts, P. G. Markham, and J. Stanley. 1989. The coat protein of beet curly top virus is essential for infectivity. Virology 172:628-633. [DOI] [PubMed] [Google Scholar]
- 3.Clough, S. J., and A. F. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735-743. [DOI] [PubMed] [Google Scholar]
- 4.Deom, C. M., M. Ladidot, and R. N. Beachy. 1992. Plant virus movement proteins. Cell 69:221-224. [DOI] [PubMed] [Google Scholar]
- 5.Dickinson, L. A., R. J. Gulizia, J. W. Trauger, E. E. Baird, D. E. Mosier, J. M. Gottesfeld, and P. B. Dervan. 1998. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl. Acad. Sci. USA 95:12890-12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frischmuth, S., T. Frischmuth, J. R. Latham, and J. Stanley. 1993. Transcriptional analysis of the virion-sense genes of the geminivirus beet curly top virus. Virology 197:312-319. [DOI] [PubMed] [Google Scholar]
- 7.Hanin, M., S. Volrath, A. Bogucki, M. Briker, E. Ward, and J. Paszkowski. 2001. Gene targeting in Arabidopsis. Plant J. 28:671-677. [DOI] [PubMed] [Google Scholar]
- 8.Hormuzdi, S. G., and D. M. Bisaro. 1993. Genetic analysis of beet curly top virus: evidence for three virion sense genes involved in movement and regulation of single- and double-stranded DNA levels. Virology 193:900-909. [DOI] [PubMed] [Google Scholar]
- 9.Horn, T., L. Stavolone, P. T. De Haan, H. T. Ligon, and M. Kononova. 2001. Cestrum yellow leaf curling virus promoters. PCT WO 01/73087 A1. Patent pending.
- 10.Jaag, H. M., L. Kawchuk, W. Rohde, R. Fischer, N. Emans, and D. Prüfer. 2003. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. USA 100:8939-8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knipe, D. M., and P. M. Howley (ed.). 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Lee, S., D. C. Stenger, D. M. Bisaro, and K. R. Davis. 1994. Identification of loci in Arabidopsis that confer resistance to geminivirus infection. Plant J. 6:525-535. [DOI] [PubMed] [Google Scholar]
- 13.Li, J. J., and T. J. Kelly. 1984. Simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. USA 81:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orozco, B. M., L.-J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds, L., C. Ullman, M. Moore, M. Isalan, M. J. West, P. Clapham, A. Klug, and Y. Choo. 2003. Repression of the HIV-1 5′ LTR promoter and inhibition of HIV-1 replication by using engineered zinc finger transcription factors. Proc. Natl. Acad. Sci. USA 100:1615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sera, T. 2002. Zinc finger domain recognition code and uses thereof. PCT WO 02/08286 A2. Patent pending.
- 17.Sera, T., and C. Uranga. 2002. Rational design of artificial zinc finger proteins using a nondegenerate recognition code table. Biochemistry 41:7074-7081. [DOI] [PubMed] [Google Scholar]
- 18.Stanley, J. 1991. Molecular determinants of geminivirus pathogenesis. Semin. Virol. 2:139. [Google Scholar]
- 19.Stanley, J., and J. R. Latham. 1992. A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 190:506-509. [DOI] [PubMed] [Google Scholar]
- 20.Stanley, J., J. R. Latham, M. S. Pinner, I. Bedford, and P. G. Markham. 1992. Mutational analysis of the monopartite geminivirus beet curly top virus. Virology 191:396-405. [DOI] [PubMed] [Google Scholar]
- 21.Stenger, D. C., D. Carbonaro, and J. E. Duffus. 1990. Genomic characterization of phenotypic variants of beet curly top virus. J. Gen. Virol. 71:2211-2215. [DOI] [PubMed] [Google Scholar]
- 22.Stenger, D. C., G. N. Revington, M. C. Stevenson, and D. M. Bisaro. 1991. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. USA 88:8029-8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai, T. H., D. Dahlbeck, E. T. Clark, P. Gajiwala, R. Pasion, M. C. Whalen, R. E. Stall, and B. J. Staskawicz. 1999. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96:14153-14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzfira, T., J. Li, B. Lacroix, and V. Citovsky. 2004. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 20:375-383. [DOI] [PubMed] [Google Scholar]