Abstract
Virus-infected plants often display developmental abnormalities that include stunting, leaf curling, and the loss of apical dominance. In this study, the helicase domain of the Tobacco mosaic virus (TMV) 126- and/or 183-kDa replicase protein(s) was found to interact with the Arabidopsis Aux/IAA protein PAP1 (also named IAA26), a putative regulator of auxin response genes involved in plant development. To investigate the role of this interaction in the display of symptoms, a TMV mutant defective in the PAP1 interaction was identified. This mutant replicated and moved normally in Arabidopsis but induced attenuated developmental symptoms. Additionally, transgenic plants in which the accumulation of PAP1 mRNA was silenced exhibit symptoms like those of virus-infected plants. In uninfected tissues, ectopically expressed PAP1 accumulated and localized to the nucleus. However, in TMV-infected tissues, PAP1 failed to accumulate to significant levels and did not localize to the nucleus, suggesting that interaction with the TMV replicase protein disrupts PAP1 localization. The consequences of this interaction would affect PAP1's putative function as a transcriptional regulator of auxin response genes. This is supported by gene expression data indicating that ∼30% of the Arabidopsis genes displaying transcriptional alterations in response to TMV contain multiple auxin response promoter elements. Combined, these data indicate that the TMV replicase protein interferes with the plant's auxin response system to induce specific disease symptoms.
Developmental abnormalities, including leaf curling, stunting, and the loss of apical dominance, are some of the most common and economically important symptoms associated with virus-induced plant diseases (29). The majority of these disease symptoms occur only when newly emerged tissue becomes infected, suggesting that interference in developmental processes is a primary cause of disease. Many of these disease responses are likely derived from the ability of specific virus components to interact with and disrupt the function of specific host components. Unfortunately, the host components and pathways through which viruses induce disease remain poorly characterized.
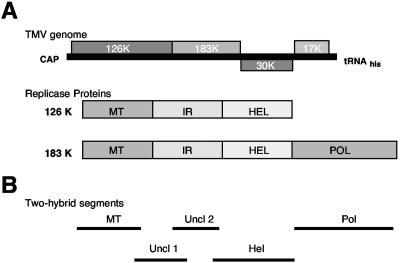
Tobacco mosaic virus (TMV) is the type member of the genus Tobamovirus and serves as a model for studying virus-host interactions. TMV is a positive-stranded RNA virus that encodes at least four proteins (22) (Fig. 1A). The two largest open reading frames (ORFs) encode 126- and 183-kDa replicase proteins, the larger produced via read-through of an amber stop codon (40). Homology studies indicate that the 126-kDa replicase protein ORF encodes a methyltransferase domain (MT) implicated in viral RNA capping and a helicase domain (HEL) involved in double-stranded RNA unwinding (17, 25). The read-through portion of the 183-kDa replicase protein ORF encodes the RNA-dependent RNA polymerase domain (POL) (34). A 30-kDa protein required for cell-to-cell movement and the 17.5-kDa capsid protein are produced from subgenomic mRNAs (15, 30, 38).
FIG. 1.
TMV genome organization and two-hybrid constructs. (A) Diagrammatic representations of the TMV genome and replicase proteins. 126K, 126-kDa; IR, inverted repeat. (B) Replicase segments used in two-hybrid screens covering the methyltransferase (MT), helicase (Hel), polymerase (Pol), and uncharacterized domains (Uncl 1 and 2).
During infection, TMV induces a specific set of disease symptoms in Arabidopsis thaliana ecotype Shahdara. These symptoms include stunting, necrosis of the inoculated leaf, loss of apical dominance, and leaf curling. Numerous changes in specific TMV genes have been identified as conferring either attenuated or severe disease symptoms (13). For example, amino acid substitutions in the coat protein have been linked to chlorosis, while specific mutations in the replicase protein can disrupt its function as an RNA silencing suppressor, resulting in reduced accumulation of virus and milder disease symptoms (3, 10, 16, 35). However, the specific molecular mechanisms through which these TMV components affect the disease process remain unidentified. In other virus-host systems, more specific links between virus and host components involved in the display of symptoms have been made. For example, suppression of RNA silencing conferred by the HC-Pro protein of turnip mosaic virus affects the accumulation of specific micro-RNAs involved in the control of plant developmental pathways and results in the display of specific disease symptoms (32). For geminiviruses, the AL1 replication protein interacts with a host-encoded cell cycle regulator protein to modulate virus tissue specificities and associated disease symptoms (33). Thus, virus-induced diseases appear to have multiple causes.
In this study, an interaction between the TMV 126- or 183-kDa replicase protein(s) and an A. thaliana auxin/indole-3-acetic acid (Aux/IAA) protein, phytochrome-associated protein 1 (PAP1/IAA26; At3g16500), is described. In general, Aux/IAA proteins, such as PAP1, are thought to function as negative regulators of auxin response factor (ARF) proteins that in turn control the transcriptional activity of primary auxin response genes involved in various aspects of plant development, including cell division, cell expansion, and apical dominance (41, 48). The nuclear localization of Aux/IAA proteins and their ability to heterodimerize with ARF DNA binding proteins support their function as transcription factors (1). Furthermore, the stability of Aux/IAA proteins is modulated by the plant hormone auxin, providing a sensitive method for the spatial and temporal control of their function (48). Interaction with the TMV replicase protein was found to disrupt PAP1 localization and corresponded with the inappropriate expression of auxin-regulated genes and the appearance of disease symptoms.
MATERIALS AND METHODS
Two-hybrid assays.
Five overlapping bait sequences covering the TMV 126- or 183-kDa ORF(s) were cloned into the Saccharomyces cerevisiae bait vector pLexA-NLS as previously described (24). An Arabidopsis leaf and root cDNA library, CD4-10, which was derived from A. thaliana ecotype Nossen and cloned into the yeast prey vector pACT-GAL4, was obtained from the Arabidopsis Biological Resource Center, Columbus, Ohio. Each of the five TMV 126-183/LexA bait constructs was cotransformed with the Arabidopsis/GAL4 prey library into S. cerevisiae strain L40. Transformants displaying bait to prey interactions were selected on minimal medium lacking histidine, leucine, tryptophan, and uracil at 25°C, a temperature previously shown to promote the interaction of TMV helicase sequences (24). Interacting transformants were assayed for β-galactosidase activity on nitrocellulose filter lifts, frozen at −80°C for 5 min, and soaked in a 4% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) solution containing 0.1% Triton X-100 and Z buffer (1 M Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol [pH 7.0]). Yeast colonies turning blue within 30 min were selected for further analysis.
Interaction-positive Arabidopsis/GAL4 prey plasmids were shuttled into Escherichia coli by the method of Ward (53). Positive interacting Arabidopsis clones were retransformed with the interacting TMV 126-183/LexA replicase clone to confirm the maintenance of the interaction and with an empty pLexA-NLS clone or one encoding the noninteracting Arabidopsis protein ETR1 (8) to screen for false interactions. β-Galactosidase activity was quantified in liquid culture as previously described (24, 39). Positive Arabidopsis/GAL4 prey clones were then sequenced for identification. Full-length PAP1 and IAA10 ORFs were obtained by reverse transcription PCR using mRNA derived from leaves of ecotype Shahdara. Full-length ORFs were cloned into the GAL4/prey plasmid and analyzed in yeast for their interaction with TMV HEL/LexA.
PAP1-replicase interaction assays.
Full-length PAP1 was cloned into the expression vector pTrcHis-A (Invitrogen, Carlsbad, Calif.) to produce an ORF containing an N-terminal hexahistidine tag. PAP1 protein was then expressed and purified via Ni-affinity columns (Amersham Biosciences, Piscataway, N.J.) as previously described (25). Full-length TMV 126- or 183-kDa protein was generated by in vitro translation. Purified TMV virions (3 μg) were incubated for 15 min in 0.01 M Tris-HCl (pH 8.0) pelleted by centrifugation at 65,000 rpm in a Beckman TLA100.3 rotor for 20 min and resuspended in water. Treated virions were added directly to an mRNA-dependent rabbit reticulocyte lysate system (Promega, Madison, Wis.) containing 250 μCi of l-[35S]methionine per ml and incubated for 90 min at 30°C. Translation reaction mixtures were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a PhosphorImager and quantified using the program ImageQuant (Molecular Dynamics, Sunnyvale, Calif.).
For overlay assays, purified PAP1 protein was immobilized onto nitrocellulose sheets. Sheets were then blocked for 2 h at 4°C with 5% (wt/vol) nonfat dry milk in Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 6.8], 200 mM NaCl). Blocked sheets were incubated overnight in TBS containing 5% nonfat dry milk and equivalent levels of 35S-labeled 126- or 183-kDa protein, either wild type or V1087I. Sheets were then washed three times with TBS and dried, and bound 126- or 183-kDa protein was visualized with a PhosphorImager.
RNAi construct and plant transformation.
A derivative of the Agrobacterium binary transformation vector pBI121 (Clontech, Palo Alto, Calif.) was used to construct a PAP1-specific RNAi (RNA interference) silencing vector. Within the pBI121 polylinker, complementary PAP1 sequences nucleotides (nt) 1 to 500 were cloned onto opposite sides of a 102-nt spacer containing an EF1α intron (At5g60390, nt 961 to 1061). Transcription of this construct, derived from the 35S cauliflower mosaic virus promoter, produces a 500-bp PAP1-specific double-stranded RNA. Agrobacterium transformations were performed on 5-week-old A. thaliana Shahdara plants as previously described (9). Transformants were selected on 1× Murashige and Skoog agar containing 30 mg of kanamycin per liter. Integration of the PAP1-RNAi construct was confirmed by PCR analysis of genomic DNA extracted from leaves (14).
Endogenous PAP1 mRNA levels were quantified in both T0 and T1 lines by real-time PCR. Total RNA was extracted from the leaves of 4-week-old PAP1-RNAi transformed and nontransformed plants using the RNeasy Plant Miniprep kit (QIAGEN, Valencia, Calif.). cDNA was generated from 1 μg of isolated RNA pretreated with RQ1 DNase (Promega) and reverse transcribed in a SuperScript first-strand synthesis system (Invitrogen) per the manufacturer's instructions. Quantitative real-time PCR (QRT-PCR) was preformed using Platinum qPCR supermix-UDG reagents (Invitrogen). Each 20-μl QRT-PCR mixture contained 10 μM concentrations of both labeled LUX PAP1 primer (CACGCTTTCATCTGTGAAGAGACTGCG5G) and unlabeled PAP1 primer (TTGCTTACTGCATCCAAATGTCAA) designed using LUX designer software (Invitrogen), carboxy-x-rhodamine reference dye (0.5 μl), cDNA (1.5 μl), and sterile distilled H2O (4.5 μl). QRT-PCR was performed on a GeneAMP 5700 sequence detection system (Applied Biosystems, Foster City, Calif.) as follows: (i) 2 min at 50°C; (ii) 10 min at 95°C; (iii) 40 cycles, with 1 cycle consisting of 15 s at 95°C and 1 min at 60°C. Relative expression levels of PAP1 were normalized to those of the housekeeping gene EF1α, and fold expression levels were determined using the comparative ΔCt method (31). Expression levels of EF1α were determined as described above using 1.5 μl of cDNA and EF1α-specific LUX-labeled (GACTGCCCACACCTCTCACATTGCAG5C) and unlabeled (TCCTTACCAGAACGCCTGTCA) primers.
Identification and construction of a TMV helicase mutant.
TMV-V1087I was previously created by random mutagenesis using hydroxylamine treatment of the TMV helicase/LexA bait construct (24). Yeast cotransformations with mutant TMV helicase/LexA and PAP1/GAL4 constructs were done as described above. Stability and expression of HEL-LexA fusion proteins were confirmed by Western immunoblotting using anti-LexA antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.). The noninteracting TMV-V1087I helicase coding sequence, SacII (nt 2654) to BamHI (nt 3333), was cloned into a similarly digested recombinant full-length infectious TMV clone, pSNC004 (12, 43, 49). DNA sequencing was performed to confirm the presence of the mutation within the viral helicase sequence. Infectious RNA transcripts derived from the mutant full-length virus construct were generated in vitro as described previously and used to inoculate leaves of Nicotiana benthamiana and A. thaliana ecotype Shahdara plants (43).
Plant growth conditions and virus assay.
A. thaliana ecotype Shahdara and N. benthamiana plants were grown and maintained in growth chambers under a 12-h photoperiod at 24°C. Four-week-old A. thaliana ecotype Shahdara plants and 4- to 5-week-old N. benthamiana plants were used for virus inoculations. Mature rosette leaves of A. thaliana ecotype Shahdara plants were dusted with carborundum (Fisher Scientific Company, Pittsburgh, Pa.) and mechanically inoculated with 10 μg of purified wild-type TMV (WT-TMV) or TMV-V1087I. The youngest leaves of N. benthamiana plants were similarly treated with carborundum and inoculated with 5 μg of WT-TMV and TMV-V1087I. Control plants were similarly dusted with carborundum and mock inoculated with distilled water.
Virus accumulation and movement were monitored by Western blotting and tissue print immunoblotting to detect the virus capsid protein as previously described (11). Arabidopsis protoplasts were derived from leaf tissues as previously described and inoculated by electroporation with 5 μg of purified viral RNA (11, 44). The accumulation of viral RNA in inoculated protoplasts was determined by QRT-PCR using TMV-specific LUX-labeled (CACTCTGGATGCAGCAATCAGGCAGAGGGGGGG) and unlabeled (AGCGGCATAGCACGTATGGA) primers. A QRT-PCR standard curve derived from known amounts of viral RNA was used to determine the concentration of virus.
For transient-expression studies, inoculated A. thaliana ecotype Shahdara leaves were used at 12 days postinoculation, and systemic leaves were used at 3 weeks postinoculation. Inoculated N. benthamiana leaves were used 4 to 6 days postinoculation, and systemic leaves were used 7 to 9 days postinoculation. Viral loads within these tissues were monitored by immunodot blots using TMV coat protein-specific antiserum (11).
PAP1 transient-expression constructs and assays.
The expression vector pCMC1100, containing the cauliflower mosaic virus 35S promoter, served as the parental plasmid for all transient-expression constructs (37). The enhanced green fluorescent protein (EGFP) ORF (Clontech) was PCR modified to contain a 5′-end BsiWI site and 3′-end PstI site and inserted into similarly cut pCMC1100, creating pCMC-GFP. PCR amplification was also used to engineer the PAP1 ORF with a unique 5′-end BspHI site and a 3′-end BsiWI site minus the termination codon. The modified PAP1 ORF was cloned into similarly digested pCMC-GFP, placing the PAP1 ORF in frame with GFP and creating pPAP1-GFP. IAA10 was likewise cloned into pCMC-GFP using PCR-engineered 5′-end KpnI and 3′-end BsiWI restriction sites to create pIAA10-GFP.
pCMC-GFP, pPAP1-GFP, and pIAA10-GFP DNA was introduced into N. benthamiana and A. thaliana Shahdara leaf cells by particle bombardment. The bombardment method was performed essentially by the method of Figueira et al. (19). Briefly, 2 μg of plasmid DNA was ethanol precipitated onto 0.5 mg of tungsten particles (1.3 μm in diameter; Bio-Rad, Hercules, Calif.). The DNA-coated particles were resuspended in 95% ethanol by sonication in a Brandon 2200 ultrasonic cleanser (Branson Equipment, Shelton, Conn.). The nucleic acid-tungsten mixture was loaded onto plastic filter screens (Gelman Sciences, Ann Arbor, Mich.) and dried. The coated screens were mounted into the particle inflow gun (20, 45) and bombarded into leaf tissue using a 50-ms pulse of helium (50 lb/in2). Bombarded leaf tissues were incubated for 12 to 16 h at room temperature and mounted on glass microscope slides in distilled water under coverslips. The tissue was imaged for GFP fluorescence using a Zeiss LSM 510 laser-scanning confocal microscope system with a 10× dry 0.8-numerical-aperture lens and a 63× 1.2-numerical-aperture water immersion lenses (Carl Zeiss Inc., Thornwood, N.Y.). Images were modified using Zeiss LSM Imager Examiner software, version 3.0 and processed for printing with Adobe Photoshop (Grand Prairie, Tex.).
Auxin leaf treatment and expression.
Rosette leaves were excised from 4- to 5-week-old A. thaliana ecotype Shahdara plants and vacuum infiltrated with water or water plus 50 μM IAA. Infiltrated leaves were incubated in the same solutions for 90 min in the dark, and total RNA was extracted using the RNeasy Plant Miniprep kit (QIAGEN). RNA expression levels of At4g38850 (SAUR-AC1), At5g21010, At1g19350, At5g02160, and At3g17790 were quantified via QRT-PCR as described above. Gene-specific LUX primers (Invitrogen) were At5g21010 (CACGACGGGTCGCATCAATTCGG and TCGCTATGTGCTTCCCTATACCC), At1g19350 (GACTCGTTCCTCTTCTTCATTCCCGAGC and TCCTGAGGAAAGGGAAGATTGTG), At5g02160 (CACATTTCACCATCACCGAACAATGG and GCGACAACTACGGAGGAAGAAGA), At3g17790 (GACGAATTGTGTATCTTCACCACCTTCGC and ACTAACGGAACCGTCGCTTT), and At4g38850 (GACCGAAGAGGATTCATGGCGGC and AAGTATGAAACCGGCACCACAT). Relative expression levels were determined as described above for RNAi analysis.
RESULTS
The TMV replicase protein interacts with the Aux/IAA protein PAP1.
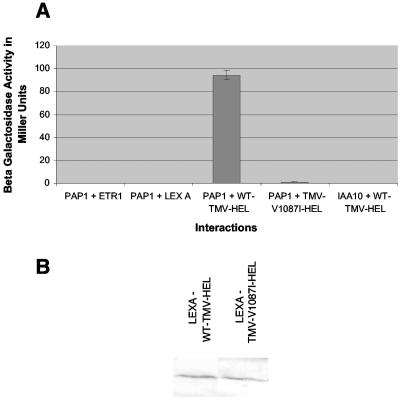
A yeast two-hybrid approach was used to identify Arabidopsis host proteins capable of interacting with TMV replicase sequences (18, 24). The entire TMV 126- or 183-kDa ORF was divided into five overlapping segments covering the MT (amino acids [aa] 1 to 376), HEL (aa 814 to 1211), POL (aa 1205 to 1613), and uncharacterized (aa 369 to 615 and aa 589 to 820) domains (Fig. 1B). All five segments were used as “bait” to screen a library of cDNA “prey” constructs derived from Arabidopsis leaf and root tissues. Only the “bait” construct covering the TMV HEL domain (aa 814 to 1211) was found to interact with clones from the cDNA library. Specifically, three cDNA clones displayed a strong interaction with the TMV HEL domain as measured by β-galactosidase activity (Fig. 2A). Sequence analysis of these clones revealed that all were identical, encoding nearly all (nt 3 to 789) of the 810-nt ORF of PAP1/IAA26 (At3g16500), a member of the Aux/IAA family of auxin-regulated transcription factors.
FIG. 2.
TMV helicase interactions. (A) Quantification of β-galactosidase activity between PAP1 and ETR1, a noninteracting control protein; PAP1 and LEXA, the empty vector; PAP1 and WT-TMV HEL, encoding the helicase domain of the viral replicase; PAP1 and TMV-V1087I HEL, a mutant TMV helicase; and IAA10, an Aux/IAA family member, with WT-TMV HEL. (B) Western immunoblot assay comparing the accumulation of WT-TMV HEL and TMV-V1087I HEL proteins in yeast.
Sequence comparisons between the two-hybrid PAP1 cDNA, derived from A. thaliana ecotype Nossen, and PCR-amplified cDNA from the ecotype Shahdara as well as expressed sequence tag and genomic sequences available for A. thaliana ecotype Columbia show 100% identity at the protein level. Furthermore, a full-length PAP1 cDNA derived from ecotype Shahdara and cloned into the yeast prey vector also interacted with the TMV helicase domain in a manner similar to that of the original PAP1 library clone (data not shown).
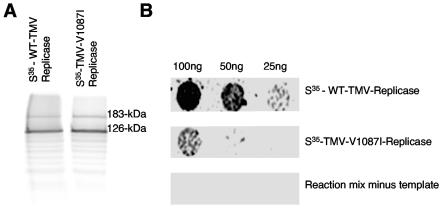
To assess this interaction further, an in vitro protein-protein assay was used to evaluate the ability of PAP1 to interact with the full-length viral replicase proteins. In this assay, purified histidine-tagged PAP1 protein was immobilized on nitrocellulose sheets and used to capture 35S-labeled full-length replicase proteins translated directly from purified virions. Results indicate that PAP1 is capable of binding full-length wild-type TMV replicase (Fig. 3).
FIG. 3.
Interaction of PAP1 with full-length TMV replicase. (A) 35S-labeled 126- and 183-kDa TMV replicase proteins. (B) PAP1 overlay assays using 25, 50, or 100 ng of 35S-labeled full-length WT-TMV replicase or TMV-V1087I replicase proteins.
A TMV helicase mutation disrupts the PAP1 interaction and confers attenuated disease symptoms.
To investigate the roles of PAP1 in virus replication and disease development, a series of previously characterized TMV helicase mutations were screened via two-hybrid assay for the ability to interact with PAP1 (24). One mutant helicase, containing a V-to-I substitution at position 1087 in the TMV replicase protein displayed a significantly reduced ability to interact with PAP1 in the two-hybrid system (Fig. 2A). Western immunoblots for the detection of the helicase-LexA fusion protein in yeast extracts indicate that the mutation at position 1087 does not affect protein expression or stability (Fig. 2B). However, extending the incubation time for the two-hybrid interaction assay from 10 min to 1 h resulted in detectable levels of β-galactosidase activity for PAP1 and V1087I helicase (data not shown). In addition, the V1087I mutation similarly disrupted the ability of the full-length viral replicase protein to interact with purified PAP1 protein in vitro (Fig. 3B). These findings suggest that the V1087I mutation greatly reduces the interaction between the TMV HEL domain and PAP1 but does not totally abolish the interaction.
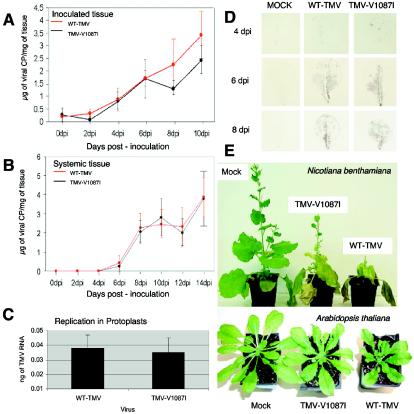
A recombinant TMV containing this mutation, TMV-V1087I, was subsequently tested for its ability to replicate and move in both Arabidopsis and tobacco (Fig. 4A and B). Within tobacco protoplasts, TMV-V1087I was previously shown to replicate at levels similar to those of the wild-type virus (24). Similar levels of replication were also observed in Arabidopsis leaf protoplasts infected with either WT-TMV or TMV-V1087I (Fig. 4C). Immunoblots monitoring virus accumulation and spread demonstrated that TMV-V1087I moves from cell to cell and systemically at rates and levels similar to those of WT-TMV (Fig. 4D).
FIG. 4.
Biological characterization of helicase mutant TMV-V1087I. (A) Accumulation of WT-TMV and TMV-V1087I in inoculated Arabidopsis leaf tissue. CP, coat protein; dpi, days postinoculation. (B) Virus accumulations in systemic leaf tissues. (C) WT-TMV and TMV-V1087I replication in Arabidopsis leaf protoplasts. Results from QRT-PCR-amplified TMV genomic products were compared against similarly amplified known TMV RNA standards to determine virus concentrations. (D) Tissue print immunoblots showing the cell-to-cell spread of WT-TMV and TMV-V1087I infection foci at 4, 6, and 8 days postinoculation (dpi) in A. thaliana ecotype Shahdara. MOCK, mock inoculated. (E) Attenuation of disease symptoms caused by TMV helicase mutation V1087I in N. benthamiana or A. thaliana Shahdara plants. Plants were inoculated with water (mock), TMV-V1087I, or WT-TMV, and photos were taken 2 weeks postinoculation.
Although the replication and movement of TMV-V1087I were not affected by its inability to interact with PAP1, this recombinant virus consistently induced milder disease symptoms than the wild-type virus did. Most significantly, the loss of apical dominance, characteristic of a WT-TMV infection, was reduced in plants infected with TMV-V1087I (Fig. 4E and 5C). Plants infected with TMV-V1087I also displayed a near-normal rosette pattern compared to the pattern of wild-type TMV-infected plants. Thus, the V1087I mutation functions to uncouple the display of specific disease symptoms from virus replication.
FIG. 5.
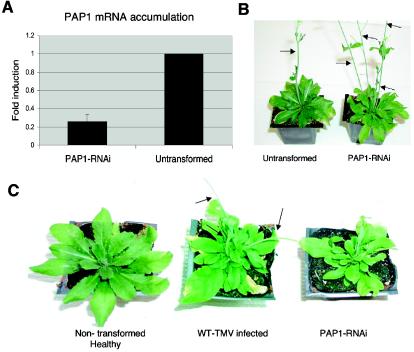
Characterization of PAP1-silenced RNAi plants. (A) QRT-PCR analysis of the levels of PAP1 mRNA accumulated. Expression levels were normalized to those of an internal control, EF1α. (B) Photo showing the development of multiple floral bolts (arrows) in PAP1-silenced plants. (C) Comparison of the phenotypes of nontransformed healthy and WT-TMV-infected plants to the phenotype of a PAP1-silenced plant. Arrows mark the multiple floral bolts produced in the TMV-infected plant.
The separation of symptom attenuation and virus replication is significant, since the TMV replicase also functions as a suppressor of gene silencing, and mutations that disrupt suppressor activity reduce virus accumulation and spread, which in turn results in the attenuation of disease symptoms (16, 35). Additionally, mutations that disrupt replicase suppressor activity all map outside the tobamovirus helicase domain (16, 35). Thus, the location of the V1087I mutation in the helicase domain of the viral replicase and the ability of TMV-V1087I to replicate and move at levels similar to those of the wild-type virus indicate that this mutation does not significantly affect the suppressor function of the replicase protein.
Transgenic RNAi plants with reduced levels of PAP1 mRNA display symptoms like those of virus-infected plants.
An RNAi construct was used to transgenically silence the expression of PAP1 mRNA in A. thaliana ecotype Shahdara. This construct produces a double-stranded RNA containing nt 1 through 500 of the PAP1 coding sequence. T0 and T1 plants from six independent transformants were identified as having significantly reduced levels of PAP1 mRNA (Fig. 5A). PAP1 RNAi-silenced plants were slightly stunted in appearance and displayed a loss in apical dominance compared to nontransformed control plants. Most notably, PAP1-silenced plants produced multiple shoot apexes, disrupting the rosette patterning of leaves and resulting in the appearance of numerous floral bolts (Fig. 5B and C). The phenotype produced by the RNAi suppression of PAP1 is similar to that observed for WT-TMV-infected plants.
RNAi-suppressed PAP1 plants also were found to accumulate TMV in both inoculated and systemically infected tissues at levels comparable to those of nontransformed Arabidopsis plants (data not shown). Thus, reduction in the accumulation of PAP1 does not adversely affect TMV replication or spread.
PAP1 is inhibited in its ability to accumulate and localize to the nucleus in TMV-infected cells.
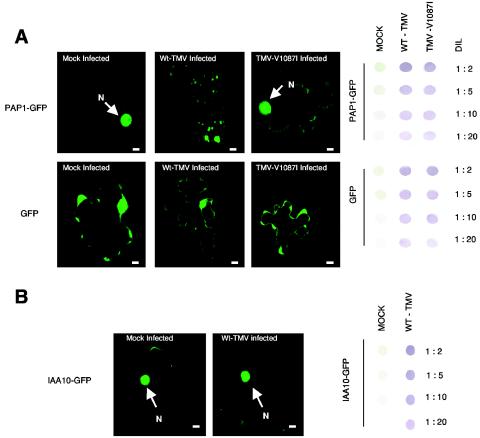
To confirm TMV's ability to interfere with PAP1 function in vivo, a PAP1-GFP fusion construct was transiently expressed in either mock-inoculated, WT-TMV-infected, or TMV-V1087I-infected tobacco and Arabidopsis leaf tissues. Relative levels of virus infectivity for both wild-type and mutant virus were monitored in these tissues by immunodot blot assays (Fig. 6A). An unmodified GFP construct was also utilized to demonstrate that transient expression of GFP is not altered in TMV-infected tissues (Fig. 6A).
FIG. 6.
Transient expression of PAP1-GFP in N. benthamiana leaf tissues. (A) Fluorescent images of cells expressing a PAP1-GFP fusion protein or GFP alone in noninfected (mock-infected), WT-TMV-infected, or TMV-V1087I-infected tissue. Bars, 10 μm. Immunodot blots showing dilutions (DIL) of leaf tissue homogenate used to monitor virus levels in PAP1-GFP-transformed leaf tissues are shown to the right of the fluorescent cell images. (B) Fluorescent cell images of IAA10-GFP in either mock- or TMV-infected tissue.
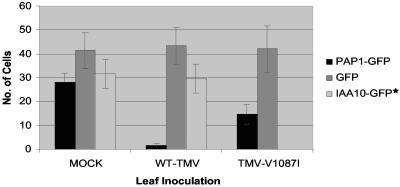
In mock-inoculated tissues, the PAP1-GFP fusion protein was found to localize predominantly in the nucleus (Fig. 6A). By comparison, in WT-TMV-infected tissues, only a few cells displayed detectable levels of PAP1-GFP fluorescence, indicating a reduction in the stability or accumulation of PAP1-GFP (Fig. 7). In addition, localization of PAP1-GFP in TMV-infected tissues appeared primarily as faint fluorescent cytoplasmic inclusions that were not localized in the nucleus (Fig. 6A). In contrast, the localization of PAP1-GFP in TMV-V1087I-infected tissues showed significantly higher numbers of cells that displayed nuclear localized fluorescence, similar to what is observed in uninfected tissues (Fig. 6A and 7). However, the number of cells within TMV-V1087I-infected tissues that displayed PAP1-GFP fluorescence in the nucleus was half of that observed in uninfected tissues, suggesting that TMV-V1087I interfered with PAP1-GFP accumulation or localization but to a lesser degree than for wild-type TMV (Fig. 7). This finding is consistent with the reduced ability of the V1087I mutant helicase to interact with PAP1.
FIG. 7.
Expression and localization totals of GFP, PAP1-GFP, and IAA10-GFP constructs in noninfected (mock-infected), WT-TMV-infected, and TMV-V1087I-infected tissues. GFP bars represent the numbers of cells displaying detectable levels of GPF fluorescence within a 15-mm2 leaf area at 16 h posttransformation. PAP1-GFP and IAA10-GFP bars represent the numbers of cells displaying detectable levels of nucleus-localized GFP fluorescence. Values are the mean numbers of cells ± standard errors of the means (error bars) for six independent bombardment transformations. Note that IAA10-GFP was not tested in TMV-V1087I-infected tissues (IAA10-GFP*).
To further confirm that TMV interference in PAP1 localization was specific to its interaction with the replicase protein, a second member of the Aux/IAA family, IAA10 (At1g04100), was cloned and analyzed. IAA10 shares 41% sequence identity with PAP1 but does not interact with the TMV HEL domain in two-hybrid assays (Fig. 2A). The expression of IAA10 as a GFP fusion protein in either TMV-infected or mock-inoculated tissue resulted in similar levels of fluorescence accumulated in both the nucleus and cytoplasm (Fig. 6B and 7). Thus, TMV does not affect the localization of a noninteracting Aux/IAA family member.
Transcriptionally altered Arabidopsis genes contain AuxREs within their promoters.
Previously performed microarray studies of both inoculated and systemically infected Arabidopsis leaf tissues identified 68 genes displaying transcriptional alterations in response to infection by TMV (23). Microarrays used in these experiments contained cDNAs representing approximately one-third of the Arabidopsis genome. An analysis of the 2,000 nt immediately upstream of the translational start codon for each of these genes revealed that 20 contained two or more TGTCTC auxin-responsive elements (AuxREs) (see Table S1 in the supplemental material). This element is present in the promoters of primary and early auxin response genes that are under the transcriptional control of ARF and Aux/IAA proteins (28, 50). The presence of multiple AuxREs has been correlated with increased alterations in gene expression, both up and down, in response to auxin (47, 52).
The effect of TMV on the localization of PAP1 suggests that the transcription of specific AuxRE-containing genes should be similarly altered in response to either a TMV infection or auxin treatment. To test this possibility, 4 of the 20 TMV-altered AuxRE genes were selected for further studies (Table 1). Additionally, SAUR-AC1, a known auxin-induced gene that is not affected by TMV infection was used as a positive control (21, 23). Upon auxin treatment, all four TMV-altered AuxRE genes showed reduced levels of transcription with two genes, At5g02160 and At3g17790 having reductions of greater than fourfold from the control (Table 1). Thus, the expression trends for these genes were similar in both auxin-treated and TMV-infected tissues. Variations in the levels of gene repression observed for these genes may be due to differences between the auxin treatment that presumably affects all Aux/IAA family members and TMV infection that specifically targets PAP1. Alternatively, TMV may affect other regulatory pathways that contribute to the transcriptional profile of these genes in a manner not replicated by auxin treatment. The nonsynchronous nature of a TMV infection may also impact observed transcriptional levels. However, combined, these data suggest that ∼30% of the genes displaying transcriptional alterations in response to TMV infection may be linked to an auxin response transcription factor, such as PAP1.
TABLE 1.
Auxin response of AuxRE-containing TMV-altered genes
| AFGC gene model IDa | Protein nameb | No. of AuxREc | Fold expression in TMV infectiond | Fold expression after 50 μM IAA treatmente |
|---|---|---|---|---|
| At5g02160 | Putative protein | 3 | −1.9 | −4.1 ± 1.7 |
| At1g19350 | Unknown protein | 3 | −2.0 | −1.8 ± 1.1 |
| At3g17790 | Acid phosphatase type 5 | 2 | −2.3 | −6.6 ± 4.0 |
| At5g21010 | Putative stress protein | 3 | −3.5 | −1.5 ± 1.2 |
| At4g38850 | SAUR-AC1 | 1 | −1.1f | 5.1 ± 2.1 |
AFGC, Stanford University's Arabidopsis Functional Genomics Consortium; ID, identification.
Gene functions based on sequence homologies.
Number of TGTCTC and GAGACA sequences within 2 kb upstream of the start codon.
cDNA microarray fold expression values averaged from three independent biological replicates of TMV-infected tissues. Fold changes at or above 1.6 and at or below −1.9 occur above the 95% confidence interval used to identify transcriptionally altered genes (23).
QRT-PCR fold expression values and standard errors averaged from four independent auxin leaf treatments. Data were normalized against the expression levels of EF1α.
Microarray fold changes for SAUR-AC1 were not above the 95% confidence interval cutoff in TMV-infected tissues.
DISCUSSION
The etiology of disease remains one of the least understood areas of virology. Of particular importance is the identification of host components and pathways that directly interact with or are affected by the infecting virus. In this study, domains from the TMV replicase protein were assessed for their ability to interact with a library of Arabidopsis host proteins. The virus replicase protein was selected for this study, because it is an essential component of the infection process and previously has been implicated in disease development (5). Of the five replicase segments used to screen for interacting Arabidopsis proteins, only the segment covering the TMV helicase domain yielded putative interacting clones. Of these cDNA clones, only those encoding the PAP1 ORF displayed levels of β-galactosidase activity indicative of a strong protein-protein interaction. In vitro interaction assays established the ability of the full-length viral replicase protein to interact with PAP1. Additional genetic and localization studies demonstrated a role for this interaction in disrupting the nuclear localization of PAP1. Specifically, the reduced ability of TMV-V1087I to interact with PAP1 corresponded with reduced disruptions in PAP1 localization and attenuated disease symptoms. Correspondingly, RNAi disruption of PAP1 produced a plant phenotype with similarities to the virus-induced disease response. Combined, these findings indicate that the interaction of the TMV replicase with PAP1 modulates the display of disease symptoms.
PAP1 encodes a 30-kDa member of the Aux/IAA family of early auxin-responsive proteins. PAP1, like other Aux/IAA proteins, contains four conserved domains involved in nuclear localization (domains I and II), protein destabilization (domain II), and dimerization (domains III and IV) (36, 51). At this time, the model for auxin signaling suggests that in the absence of auxin, Aux/IAA proteins form heterodimers with ARF proteins and repress their ability to modulate auxin response genes. In the presence of auxin, Aux/IAA proteins dissociate from ARF proteins and are targeted for degradation via the Skp1/Cullin/F-box subunit-containing E3 ubiquitin ligase complex, SCFTIR1 (26, 48, 54). TIR1 encodes the F-box component of this complex and interacts directly with Aux/IAA proteins to promote their ubiquitination and degradation via the proteasome. In the absence of Aux/IAA proteins, ARF proteins function as transcriptional activators or repressors, binding the AuxRE TGTCTC within the promoters of primary auxin response genes (27). During normal plant development, the stability of Aux/IAA proteins is regulated by an auxin concentration gradient emanating from the shoot apex. Disrupting the function of Aux/IAA proteins or the genes controlling their stability results in numerous developmental abnormalities, including the loss of apical dominance, alterations in leaf development, and changes in floral promotion (41, 42).
On the basis of the present model for auxin signaling, we hypothesize that during a TMV infection, interaction with the viral replicase promotes the destabilization and/or inappropriate sequestration of PAP1, thus interfering with its function. Disruption of PAP1 function directed by the TMV replicase protein would occur independently of the plant's auxin gradient, resulting in the activation of ARFs and alterations in the transcription levels of specific auxin response genes. Consistent with this possibility, a significant portion (∼30%) of the transcriptionally altered genes in TMV-infected leaf tissues contained multiple AuxREs within their promoter sequences (23). Furthermore, experimental results indicate that TMV-altered AuxRE genes display auxin-induced expression trends similar to those observed in TMV-infected tissues (Table 1). Microarray results also indicate that other genes containing AuxRE promoter sequences, including members of several primary auxin response gene families, such as SAUR (see SAUR-AC1 results in Table 1), GH3, and other Aux/IAA proteins do not display transcriptional alteration in response to TMV (23). Thus, the regulation of TMV-altered AuxRE genes (Table 1) appears specific and not part of a genome-wide disruption in auxin sensing. Specificity in the effect of TMV on the auxin response system is further demonstrated by the inability of TMV to alter the localization of IAA10, a non-replicase-interacting Aux/IAA family member. Combined, these data support a link between TMV-altered AuxRE genes and the disruption of PAP1 stability or localization by TMV. Thus, TMV-altered AuxRE genes are candidates for additional studies directed at determining their role in the development of disease symptoms.
The induction of disease symptoms is likely to be complex, involving multiple interactions between host and pathogen components. PAP1 is only 1 of 29 predicted members of the Aux/IAA family of auxin-responsive transcription factors and shares between ∼26 and ∼67% sequence homology with the other members. Although IAA10, which has 41% homology with PAP1, did not interact with the TMV helicase, it is possible that other more closely related Aux/IAA members interact in a PAP1-like fashion. In addition, recent studies have determined that several auxin regulatory components, including ARF8, ARF10, and TIR1, are targets for micro-RNA (miRNA) regulation (6, 7, 32). The ability of virus-encoded RNA-silencing suppressors to interfere with the miRNA-directed regulation of such components has also been correlated with the appearance of symptom-like developmental defects (7, 32). Thus, interaction of the TMV replicase with PAP1 likely represents only one avenue through which plant viruses can disrupt the auxin signaling pathway. The ability of TMV-V1087I to induce developmental symptoms, albeit reduced in severity, supports a role for other viral processes and interactions in the development of disease symptoms. At this time, we are investigating this possibility as well as the potential contributions of other Aux/IAA family members in the display of disease symptoms.
Interestingly, silencing PAP1 mRNA did not produce a detectable effect on virus replication or movement. Similarly, TMV-V1087I, a helicase mutant with reduced ability to interact with PAP1, replicated and spread at levels similar to those of the wild-type virus. Thus, the interaction between the TMV HEL domain and PAP1 is not rate limiting for virus function. This type of nonessential interaction may account for the differences between diseased and tolerant host responses. Both diseased and tolerant hosts show similar levels of susceptibility to a pathogen; however, only the diseased host displays significant damage (2). In addition, disease and tolerant phenotypes in both host and pathogen backgrounds are heritable characteristics, suggesting the involvement of specific host-pathogen interactions. In fact, virus-induced disease severity often does not correlate with the ability of an infecting virus to replicate at high levels or spread rapidly within a specific host (29). Therefore, nonessential interactions, such as the one between TMV replicase and PAP1, may play significant roles in determining disease severity.
Combined, these experiments suggest that the TMV replicase protein disrupts PAP1 function. One possibility is that this interaction destabilizes PAP1 through a ubiquitin-mediated process similar to the auxin-directed degradation of other Aux/IAA proteins. While virus-directed protein degradation has not been established as a disease mechanism in plants, it has been directly linked to disease development in several animal virus systems (4). For example, human papillomavirus (HPV) E6 protein directs the degradation of the cellular tumor suppressor protein p53 as well as several membrane-associated guanylate kinases, contributing directly to the malignant progression of HPV-associated cancers (46). Alternatively, the TMV replicase protein may simply sequester PAP1 protein and prevent its ability to localize to the nucleus. The precise mechanism by which TMV disrupts PAP1 function remains to be determined.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Science Foundation (IBN-0113536) and the United States Department of Agriculture (200301163).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abel, S., P. W. Oeller, and A. Theologis. 1994. Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91:326-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrios, N. G. 1996. Plant pathology. Academic Press, London, United Kingdom.
- 3.Banerjee, N., J. Y. Wang, and M. Zaitlin. 1995. A single nucleotide change in the coat protein gene of tobacco mosaic virus is involved in the induction of severe chlorosis. Virology 207:234-239. [DOI] [PubMed] [Google Scholar]
- 4.Banks, L., D. Pim, and M. Thomas. 2003. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 28:452-459. [DOI] [PubMed] [Google Scholar]
- 5.Bao, Y., S. A. Carter, and R. S. Nelson. 1996. The 126- and 183-kilodalton proteins of tobacco mosaic virus, and not their common nucleotide sequence, control mosaic symptom formation in tobacco. J. Virol. 70:6378-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, E., J. Wuyts, P. Rouze, and Y. Van de Peer. 2004. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101:11511-11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, K. L., P. B. Larsen, X. Want, and C. Chang. 1998. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 95:5401-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clough, S. J., and A. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735-743. [DOI] [PubMed] [Google Scholar]
- 10.Culver, J. N. 2002. Tobacco mosaic virus assembly and disassembly: determinants in pathogenicity and resistance. Annu. Rev. Phytopathol. 40:287-308. [DOI] [PubMed] [Google Scholar]
- 11.Dardick, C. D., S. Golem, and J. N. Culver. 2000. Susceptibility and symptom development in Arabidopsis thaliana to tobacco mosaic virus is influenced by virus cell-to-cell movement. Mol. Plant-Microbe Interact. 12:247-251. [DOI] [PubMed] [Google Scholar]
- 12.Dawson, W. O., D. L. Beck, D. A. Knorr, and G. L. Grantham. 1986. cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc. Natl. Acad. Sci. USA 83:1832-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson, W. O. 1999. Tobacco mosaic virus virulence and avirulence. Philos. Trans. R. Soc. Lond. B 354:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1:19-21. [Google Scholar]
- 15.Deom, C. M., M. J. Oliver, and R. N. Beachy. 1987. The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science 237:389-394. [DOI] [PubMed] [Google Scholar]
- 16.Ding, X. S., J. Liu, N.-H. Cheng, A. Folimonov, Y.-M. Hou, Y. Bao, C. Katagi, S. A. Carter, and R. S. Nelson. 2004. The tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant-Microbe Interact. 17:583-592. [DOI] [PubMed] [Google Scholar]
- 17.Dunigan, D. D., and M. Zaitlin. 1990. Capping of tobacco mosaic virus RNA. Analysis of viral-coded guanylyltransferase-like activity. J. Biol. Chem. 265:7779-7786. [PubMed] [Google Scholar]
- 18.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature (London) 340:245-246. [DOI] [PubMed] [Google Scholar]
- 19.Figueira, A., S. Golem, S. P. Goregaoker, and J. N. Culver. 2002. A nuclear localization signal and a membrane association domain contribute to the cellular localization of the tobacco mosaic virus 126-kDa replicase protein. Virology 301:81-89. [DOI] [PubMed] [Google Scholar]
- 20.Finer, J. J., P. Vain, M. W. Jones, and M. D. McMullin. 1992. Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 11:323-328. [DOI] [PubMed] [Google Scholar]
- 21.Gil, P., Y. Liu, V. Orbovi, E. Verkamp, K. L. Poff, and P. J. Green. 1994. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 104:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goelet, P., G. P. Lomonossoff, P. J. G. Butler, M. E. Akam, M. J. Gait, and J. Karn. 1982. Nucleotide sequence of tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 79:5818-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golem, S., and J. N. Culver. 2003. Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 16:681-688. [DOI] [PubMed] [Google Scholar]
- 24.Goregaoker, S. P., D. J. Lewandowski, and J. N. Culver. 2001. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320-328. [DOI] [PubMed] [Google Scholar]
- 25.Goregaoker, S. P., and J. N. Culver. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray, W. M., S. Kepinski, D. Rouse, O. Leyser, and M. Estelle. 2001. Auxin regulates SCF(TIRI)-dependent degradation of AUX/IAA proteins. Nature (London) 414:271-276. [DOI] [PubMed] [Google Scholar]
- 27.Hagen, G., and T. Guilfoyle. 2002. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49:373-385. [PubMed] [Google Scholar]
- 28.Higo, K., Y. Ugawa, M. Iwamoto, and T. Korenaga. 1999. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hull, R. 2002. Matthews' plant virology. Academic Press, London, United Kingdom.
- 30.Hunter, T., T. Hunt, J. Knowland, and D. Zimmern. 1976. Messenger RNA for the coat protein of tobacco mosaic virus. Nature (London) 260:759-764. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, M. R., K. Wang, J. B. Smith, M. J. Heslin, and R. B. Diasio. 2000. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal. Biochem. 278:175-184. [DOI] [PubMed] [Google Scholar]
- 32.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 33.Kong, L.-J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 35.Kubota, K., S. Tsuda, A. Tamai, and T. Meshi. 2003. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 77:11016-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liscum, E., and J. W. Reed. 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49:387-400. [PubMed] [Google Scholar]
- 37.McCabe, D. E., W. F. Swain, B. J. Martinel, and P. Christou. 1988. Stable transformation of soybean (Glycine max) by particle acceleration. Bio/Technology 6:923-926. [Google Scholar]
- 38.Meshi, T., Y. Watanabe, T. Saito, A. Sugimoto, T. Maeda, and Y. Okada. 1987. Function of the 30kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J. 6:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Pelham, H. R. B. 1978. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature (London) 272:469-471. [DOI] [PubMed] [Google Scholar]
- 41.Reed, J. W. 2001. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6:420-425. [DOI] [PubMed] [Google Scholar]
- 42.Rogg, L. E., J. Lasswell, and B. Bartel. 2001. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13:465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shivprasad, S., G. P. Pogue, D. J. Lewandowski, J. Hidalgo, J. Donson, L. K. Grill, and W. O. Dawson. 1999. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255:312-323. [DOI] [PubMed] [Google Scholar]
- 44.Simon, A. E., X. H. Li, J. E. Lew, R. Stange, C. Zhang, M. Polacco, and C. D. Carpenter. 1992. Susceptibility and resistance of Arabidopsis thaliana to turnip crinkle virus. Mol. Plant-Microbe Interact. 5:496-503. [Google Scholar]
- 45.Takeuchi, Y., M. Dotson, and N. T. Keen. 1992. Plant transformation: a simple particle bombardment device based on flowing helium. Plant Mol. Biol. 18:835-839. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, M., D. Pim, and L. Banks. 1999. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 18:7690-7700. [DOI] [PubMed] [Google Scholar]
- 47.Tian, Q., N. J. Uhlir, and J. W. Reed. 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiwari, S. B., X.-J. Wang, G. Hagen, and T. J. Guilfoyle. 2001. Aux/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turpen, T. H., S. J. Reini, Y. Charoenvit, S. L. Hoffman, V. Fallarme, and L. K. Grill. 1995. Malarial epitopes expressed on the surface of recombinant tobacco mosaic virus. Bio/Technology 13:53-57. [DOI] [PubMed] [Google Scholar]
- 50.Ulmasov, T., Z. B. Liu, G. Hagen, and T. J. Guilfoyle. 1995. Composite structure of auxin response elements. Plant Cell 7:1611-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulmasov, T., G. Hagen, and T. J. Guilfoyle. 1999. Dimerization and DNA binding of auxin response factors. Plant J. 19:309-319. [DOI] [PubMed] [Google Scholar]
- 52.Ulmasov, T., G. Hagen, and T. J. Guilfoyle. 1999. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward, A. C. 1990. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 18:5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zenser, N., A. Ellsmore, C. Leasure, and J. Callis. 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98:11795-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.