Significance
The spread of multidrug-resistant bacteria constitutes a significant unmet medical need. Fluoroquinolone antibiotics have been compromised by resistance mutations in their targets: DNA gyrase and topoisomerase IV. Using biochemical and genetic techniques, we have identified and characterized a class of antibacterials which transforms DNA gyrase into toxic DNA-cleavage complexes, similar to fluoroquinolones, but with a distinct mechanism of action. X-ray crystallography shows that the inhibitors access a previously unexploited pocket in gyrase, leading to their activity against fluoroquinolone-resistant bacteria and providing a strategy to target bacterial topoisomerases.
Keywords: antibiotic, topoisomerase, drug discovery
Abstract
A paucity of novel acting antibacterials is in development to treat the rising threat of antimicrobial resistance, particularly in Gram-negative hospital pathogens, which has led to renewed efforts in antibiotic drug discovery. Fluoroquinolones are broad-spectrum antibacterials that target DNA gyrase by stabilizing DNA-cleavage complexes, but their clinical utility has been compromised by resistance. We have identified a class of antibacterial thiophenes that target DNA gyrase with a unique mechanism of action and have activity against a range of bacterial pathogens, including strains resistant to fluoroquinolones. Although fluoroquinolones stabilize double-stranded DNA breaks, the antibacterial thiophenes stabilize gyrase-mediated DNA-cleavage complexes in either one DNA strand or both DNA strands. X-ray crystallography of DNA gyrase–DNA complexes shows the compounds binding to a protein pocket between the winged helix domain and topoisomerase-primase domain, remote from the DNA. Mutations of conserved residues around this pocket affect activity of the thiophene inhibitors, consistent with allosteric inhibition of DNA gyrase. This druggable pocket provides potentially complementary opportunities for targeting bacterial topoisomerases for antibiotic development.
The antibiotic era, heralded by the advent of penicillin, has led to a long-standing improvement in global human health. The wide range of antibacterial agents discovered from the 1940s to the early 1960s enabled efficient treatment of an extensive range of infections and was a key contributor to many aspects of modern clinical care, such as wound care, surgery, and organ transplantation. However, the rise of multidrug-resistant (MDR) strains of pathogens, combined with the limited number of novel antibacterials discovered since the 1960s, has resulted in a marked rise in the rate of untreatable infections. The issue is especially acute for MDR Gram-negative pathogens, which are exerting a significant strain on health systems worldwide (1, 2). Efforts to identify and optimize novel mechanism-of-action antibacterials to cope with the current limited pipeline are thus of high importance. Furthermore, many multinational pharmaceutical companies have withdrawn from antibacterial research due to, among other reasons, a perceived lack of commercial return of investment (3). Recently, several calls for renewed and concerted efforts in antibiotic discovery have been made (4, 5). One response to this urgent need is the New Drugs for Bad Bugs (ND4BB) suite of projects launched by the Innovative Medicines Initiative (IMI), a public–private partnership (6). We [GlaxoSmithKline (GSK)] have identified a series of DNA gyrase inhibitors that bind to a site on the gyrase enzyme and have determined that the mode of action both is distinct from known DNA gyrase antibacterials and has no cross-resistance to currently used topoisomerase-targeting antibacterials. This series was further developed in a collaborative effort within the IMI-funded European Gram-negative Antibacterial Engine (ENABLE) project (6), an academic–industry open-innovation partnership for antibacterial discovery that is a component of the ND4BB platform.
Bacterial type II DNA topoisomerases [DNA gyrase and DNA topoisomerase (topo) IV] are among the few clinically validated targets for antibacterial therapy (7, 8). These enzymes modulate bacterial chromosome topology by performing transient double-strand DNA cleavage via a covalent link between a tyrosine side chain of the enzyme and a 5′ phosphate of DNA, followed by strand passage and religation (9, 10), thereby either relaxing supercoils, decatenating, or introducing negative supercoils in DNA. The highly successful fluoroquinolone (FQ) class of antibiotics, exemplified by ciprofloxacin (Fig. 1D), target the DNA-cleavage active site located at the “DNA gate” of these enzymes and interfere with DNA religation, thereby converting the enzyme–DNA complex into a “cleavage complex” that can release highly lethal DNA double-strand breaks (11). This phenomenon is commonly referred to as “poisoning” and is thought to underlie the efficiency and, ultimately, the clinical success of the FQ antibiotics (11, 12). Balanced dual targeting of these highly related bacterial enzymes is attractive from a drug discovery perspective due to the resulting lower propensity for the development of antibiotic resistance. Unfortunately, despite this dual targeting, the rise of clinical resistance to FQs, most commonly by mutation in the conserved GyrA S83 and D87 amino acid residues (Escherichia coli numbering), has compromised the use of this important class of antibiotics, especially in infections caused by Gram-positive and -negative pathogens (13) and in MDR tuberculosis (14).
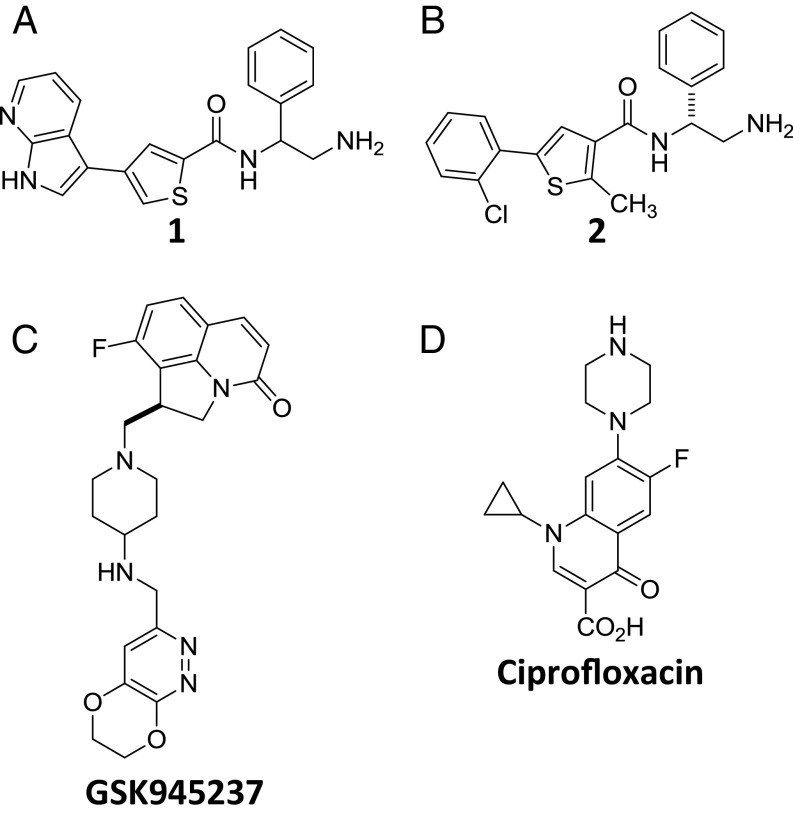
Fig. 1.
Chemical structures of compounds. (A) Compound 1. (B) Compound 2. (C) The NBTI GSK945237. (D) The fluoroquinolone ciprofloxacin.
Two new classes of antibacterial agents targeting bacterial topoisomerases triazaacenaphthylenes and spiropyrimidinetriones are currently in clinical trials. In the Novel Bacterial type II Topoisomerase Inhibitor (NBTI) class (15, 16), the triazaacenaphthylene gepotidacin is in phase II clinical trials (17). Gepotidacin inhibits these enzymes by bridging the DNA and topoisomerase at a site midway between the two DNA-cleavage sites (15). This NBTI-binding site is different from the FQ-binding sites, and gepotidacin retains minimum inhibitory concentration (MIC) activity against FQ-resistant strains (16). The spiropyrimidinetriones, exemplified by ETX0914, also in phase II trials, bind the topoisomerase complex at a site that overlaps with that of FQs, but remains effective against FQ-resistant strains (18, 19). In contrast to the FQ, triazaacenaphthylene, and spiropyrimidinetrione classes, which all bind to the DNA gate, the aminocoumarins, represented by novobiocin, target the gyrase ATPase domain. Besides the FQs, novobiocin represents the only other gyrase inhibitor class that has been approved for use in the clinic although it was later withdrawn (for other classes of topoisomerase inhibitors, see ref. 7). The focus of our work was the identification of a new druggable pocket to target gyrase, which has the potential to deliver new antibacterials without cross-resistance to fluoroquinolones. Here, we describe the identification of a series of DNA gyrase inhibitors with broad-spectrum antibacterial activity. Although not drug candidates in their own right, characterization of these inhibitors has led further to the discovery of a previously unexploited site for inhibition of gyrase. Unlike other topoisomerase II poisons, these inhibitors do not make direct contact with DNA, indicating an unforeseen allosteric mechanism for DNA-cleavage complex stabilization. We also present biochemical data suggesting that this mechanism is distinct from the poisoning of DNA gyrase by the FQs. We discuss the potential implications for further drug-targeting endeavors and for the mechanism of DNA cleavage by DNA gyrase.
Results
Discovery and Characterization of DNA Gyrase Inhibitor Compound 1.
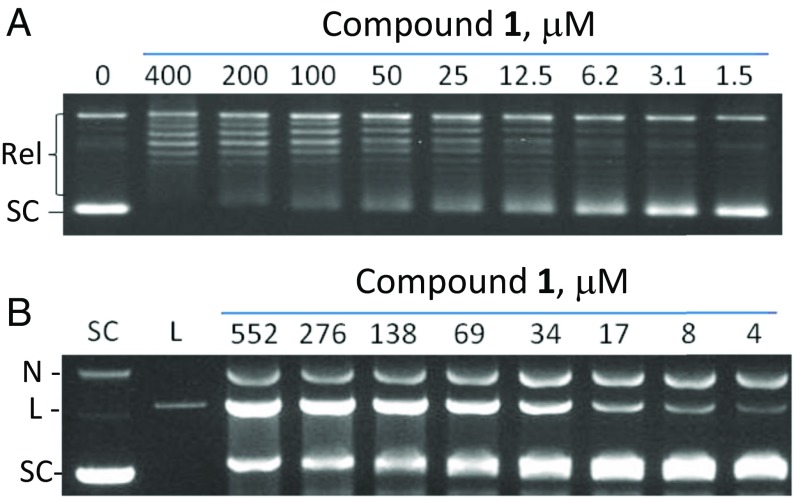
High-throughput screening (HTS) efforts have generally had limited success in identifying antibacterial hits (20, 21). However, an HTS of the GSK compound library measuring the DNA-dependent ATPase activity of E. coli DNA gyrase, and confirmation of hits using biochemical and genetic tools, led to the identification of the thiophene 1 [N-(2-amino-1-phenylethyl)-4-(1H,2H,3H-pyrrolo[2,3-β]pyridin-3-yl)thiophene-2-carboxamide] (Fig. 1A) as a weak (IC50 = 5 µg/mL) inhibitor of DNA gyrase. This compound inhibited the ability of gyrase to introduce negative supercoils in relaxed DNA (Fig. 2A and SI Appendix, Table S3) and stabilized gyrase-mediated DNA-cleavage complex formation in vitro, consistent with a gyrase poisoning mechanism (Fig. 2B). Beyond activity at the target level, compound 1 was shown to have MICs against both Gram-positive (e.g., Staphylococcus aureus and Streptococcus pneumoniae) and Gram-negative (e.g., E. coli and Pseudomonas aeruginosa) bacterial pathogens, with significantly greater activity against strains in which genes for major efflux systems (22) had been inactivated (Table 1). To further evaluate the mechanism of action of compound 1, we monitored the incorporation of radiolabeled, pathway-specific precursors (23) in an efflux-deficient strain of E. coli. In this assay, compound 1 primarily inhibited the incorporation of 14C-thymidine, similar to the behavior of another DNA-synthesis inhibitor, ciprofloxacin (SI Appendix, Fig. S1).
Fig. 2.
Characterization of activity of compound 1 against DNA gyrase. (A) Inhibition of E. coli DNA gyrase supercoiling activity by compound 1. Relaxed (Rel) pBR322 DNA was incubated with gyrase and different concentrations of compound and separated by agarose gel electrophoresis. (B) Stabilization of the cleavage complex with E. coli DNA gyrase by compound 1. Supercoiled pBR322 DNA was incubated with gyrase and different concentrations of compound 1. Cleaved DNA complexes were trapped with SDS, followed by proteinase K digestion, and DNA was separated by agarose gel electrophoresis. L, linearized DNA; N, nicked DNA; SC, supercoiled DNA.
Table 1.
Antimicrobial activity of thiophene inhibitors
| Bacteria isolate | MIC, μg/mL | ||
| Compound 1 | Compound 2 | Ciprofloxacin | |
| S. aureus OXFORD | 32 | 8 | 0.125 |
| S. pneumoniae ERY2 | 1 | 2 | 2 |
| E. coli 7623 | 64 | 8 | ≤0.008 |
| E. coli 7623 ΔtolC | 4 | ≤0.125 | ≤0.008 |
| P. aeruginosa PAO1 | >128 | >128 | 0.125 |
| P. aeruginosa PAO322 Δ(mexAB-oprM) Δ(mexCD-oprJ) Δ(mexEF-oprN) | 16 | 8 | ≤0.008 |
| A. baumannii BM4454 | NT | 4 | >8 |
| A. baumannii BM4652 ΔadeABC ΔadeIJK | NT | 0.25 | 2 |
| K. pneumoniae 1161486 | NT | 64 | 0.063 |
| K. pneumoniae 1161486 GyrA S83I ParC S80I (FQr) | NT | 32 | 8 |
| E. coli TOP10 ΔtolC | 4 | 0.125 | 0.0005 |
| E. coli W4753 ΔtolC GyrA S83L D87N ParC S80I ParE S458A (FQr) | NT | 0.063 | 8 |
| E. coli TOP10 ΔtolC GyrB E793K | 16 | 4 | 0.0005 |
MICs were determined against WT strains, efflux knockout mutants, fluoroquinolone-resistant strains, and a laboratory-generated, thiophene-resistant GyrB E793K mutant of E. coli. A. baumannii, Acinetobacter baumannii; E. coli, Escherichia coli; FQr, fluoroquinolone-resistant strain; K. pneumoniae, Klebsiella pneumoniae; NT, not tested; P. aeruginosa, Pseudomonas aeruginosa; S. aureus, Staphylococcus aureus; S. pneumoniae, Streptococcus pneumoniae.
The Compound-Binding Pocket Is Remote from DNA.
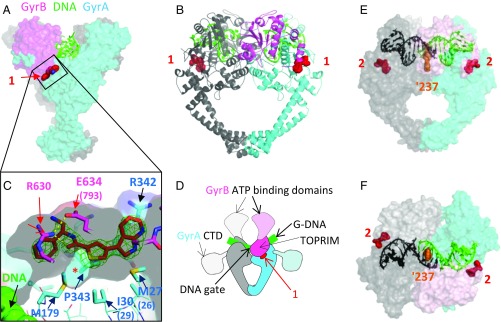
In parallel with the biochemical and genetic studies, we explored cocrystallization of the inhibitor with DNA gyrase. We have found that the S. aureus DNA gyrase crystallography platform can be used to gain structural information on compounds that target both Gram-positive and Gram-negative organisms (15, 19, 24). Using this crystallographic platform, we obtained a 1.98-Å structure of compound 1 complexed to an S. aureus DNA gyrase core fusion [lacking the gyrase A (GyrA) C-terminal domain, the gyrase B (GyrB) Greek-key domain, and the GyrB N-terminal domain] and DNA (Fig. 3 and SI Appendix, Table S1 and Fig. S2). These structural data showed that compound 1 bound to the protein at a site remote from the DNA-cleavage site and devoid of any direct DNA contacts. As far as we know, neither of these features has been previously reported in topoisomerase II poisons (15, 19, 25, 26). The binding pocket lies at the interface between the GyrB topoisomerase-primase (TOPRIM) domain (27) and GyrA winged helix domain (WHD) and opens as a groove toward the outer side of the enzyme (Fig. 3 A–D). The residues contacting the inhibitor are conserved across key pathogenic bacteria and, importantly, are not found in human topoisomerase IIα or IIβ (Fig. 4A and SI Appendix, Table S2).
Fig. 3.
Crystal structures of compounds 1 and 2 with S. aureus DNA gyrase and DNA. (A) Compound 1 binds in a pocket between the GyrA (cyan) and GyrB (pink) subunits in the complex with DNA (green) and S. aureus GyrB27A56. (B) Orthogonal view of complex with protein shown in schematic and one GyrB-GyrA subunit in pink and cyan and the other in gray. Carbon atoms of compound 1 are in red. (C) An enlarged view of the ligand binding site looking from GyrB (R630 and E634 are near the viewer). A semitransparent surface on the protein is shown gray on the inside (view from GyrB) and colored on the outside. Note that the front clipping plane cuts through the near surface at one point—allowing a clearer view of the “hydrophobic cavity” under the compound (indicated with *). GyrA P343 is behind the far surface of the pocket. An Fo-Fc ligand omit map at 3.5 sigma (green mesh) is shown. Note that DNA is remote from the compound binding site. (D) Orientation schematic showing the location of the compound 1 binding site (same view and color scheme as in B). The dashed delineated domains are absent from the crystallographic core construct, and their approximate orientation is indicated. (E and F) Two orthogonal views of the 2.22-Å crystal structure of compound 2 with S. aureus DNA gyrase, DNA, and GSK945237 (orange spheres, an NBTI used to cocrystallize and aid crystallography) (Materials and Methods). The protein subunits are shown with semitransparent surface so that the thiophene (red spheres) can be seen. The DNA is colored black/green to indicate the positions of the four base pair staggered breaks.
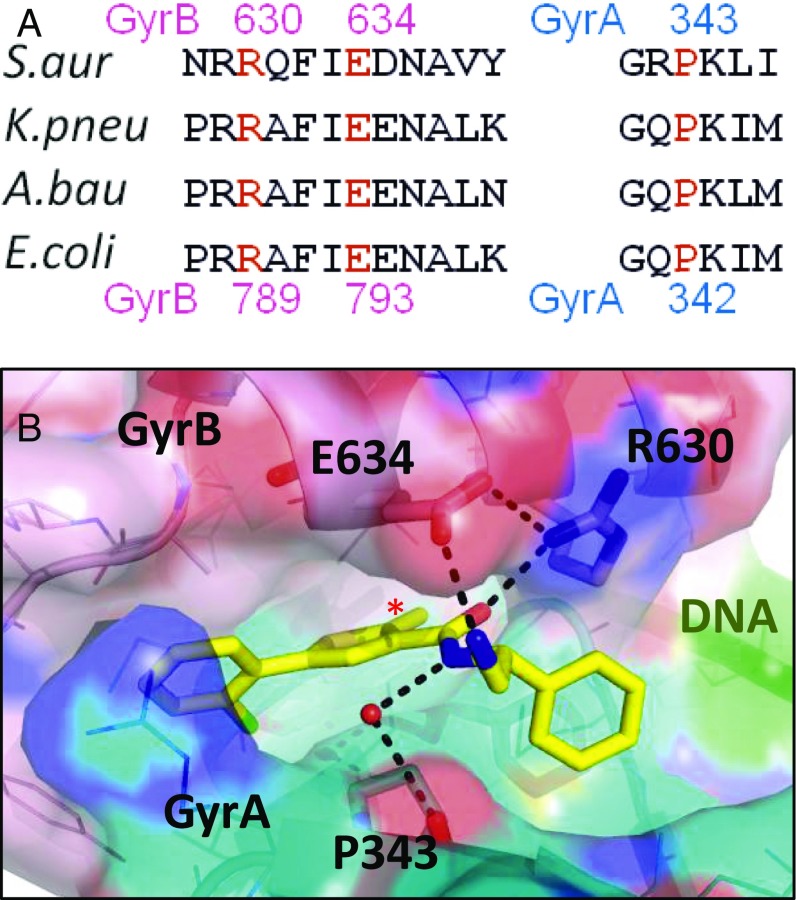
Fig. 4.
Polar interactions at the thiophene binding pocket of compound 2. (A) Sequence alignment with S. aureus residue numbers above and E. coli numbers below. (B) View of crystal structure of compound 2 with S. aureus DNA gyrase and DNA. Carbon atoms of compound 2 are in yellow, GyrB carbons in light pink, and GyrA in cyan. A semitransparent (40%) surface is shown, with GyrB R630 and E634, and GyrA P343 shown in stick (other residues as lines). Direct contacts between compound and amino acid residues including via a water molecule (red sphere) are shown by dotted lines. GyrB E634 in S. aureus is equivalent to E793 in E. coli. The hydrophobic cavity is filled by a methyl group (*).
Structure-Guided Optimization of Compound 1 Produced a More Active Lead Compound 2.
Examination of the contacts between compound 1 and the protein in the cocrystal structure revealed a small hydrophobic cavity (Fig. 3C). We hypothesized that filling this cavity would improve inhibitor binding and activity. Because the position of the sulfur atom in compound 1 would not allow appropriate substitution, we prepared a number of regioisomeric analogs both with and without methyl groups designed to fill the cavity. In parallel, we discovered that the azaindole of compound 1 could be replaced by a 2-chlorophenyl group.
This chemistry campaign resulted in lead compound 2 (Fig. 1B), which possessed increased activity against E. coli DNA gyrase (SI Appendix, Table S3), as well as improved MIC against E. coli strains (up to ∼32-fold compared with inhibitor 1) (Table 1). A crystal structure of compound 2 bound to the same gyrase core supported the original hypothesis that an added methyl group would pack more tightly against the protein (Figs. 3 E and F and 4B and SI Appendix, Fig. S2). Additional optimization of compound 2 was attempted within the IMI ENABLE consortium, however work on the series was discontinued due to toxicity findings.
Compounds 1 and 2 Do Not Show Cross-Resistance with Fluoroquinolones.
Importantly, compounds 1 and 2 showed no cross-resistance to high level, FQ-resistant E. coli and Klebsiella pneumoniae strains carrying the most common mutations found in clinical strains (Table 1). In contrast, ciprofloxacin showed up to a 16,000-fold MIC increase with FQ-resistant mutants compared with WT. The mechanism of action of both compounds 1 and 2 was further confirmed as DNA gyrase inhibition by isolation and identification of E. coli mutants resistant to compound 1 and its analogs with mutations that mapped to this enzyme (Table 1 and SI Appendix, Table S4), and through biochemical characterization of these resistant-mutant proteins (SI Appendix, Fig. S3 and Table 2).
Table 2.
Relative target activity of compound 2 and ciprofloxacin against E. coli WT and thiophene-resistant mutant gyrase protein
| Compound | IC50 fold increase | |||
| WT | GyrB E793K | GyrA M26A | GyrA I29V | |
| 2 | 1 | >100 | ∼5 | ∼5 |
| Ciprofloxacin | 1 | <5 | 1 | 1 |
The propensity for target-mediated resistance to these inhibitors in E. coli, Acinetobacter baumannii, and K. pneumoniae was found to be modest (frequency of spontaneous resistance at four times above the MIC was ∼1.8 × 10−8). The resistant mutants in E. coli generated in these experiments conferred up to eightfold MIC increase to compound 1 and ∼32-fold MIC increase to compound 2 compared with the parent strain (Table 1 and SI Appendix, Table S4). In contrast, ciprofloxacin showed no (E793K mutant) (Table 1) or little (up to fourfold) (SI Appendix, Table S4) cross-resistance to the thiophene series-resistant mutants generated. Genetic characterization of these mutants by PCR and DNA sequencing revealed that several had altered amino acids that were located in or close to the thiophene-binding pocket (SI Appendix, Tables S2 and S4 and Fig. 4).
Laboratory-Generated Resistant Mutants Confirmed the Biological Relevance of the Thiophene Compound-Binding Site.
Mapping the laboratory-generated resistant mutants revealed that three of these mutants, E. coli GyrB-E793K, GyrB-R789, and GyrA-P342 (equivalent to E634, R630, and P343 in S. aureus, respectively) directly contact compounds 1 and 2 in the crystal structures (Figs. 3C and 4). Furthermore, resistant mutants selected to compound 2 in an A. baumannii efflux-deficient strain mapped to gyrase residues located in or adjacent to this binding pocket (SI Appendix, Table S4), which supported the relevance of the structure in understanding the mode of action of these compounds and the biochemical validity of this binding pocket.
To obtain independent, biochemical confirmation of the relevance of this binding site, we overexpressed, purified, and characterized DNA gyrase that carries a mutation in a key residue for inhibitor binding (E. coli GyrB-E793K) (Fig. 4). Using in vitro gyrase supercoiling inhibition assays, compound 2 displayed >100-fold less activity against this mutant enzyme compared with the WT enzyme (Table 2 and SI Appendix, Fig. S3). In contrast, the mutant enzyme showed only limited resistance to ciprofloxacin treatment (less than fivefold relative to WT), consistent with the lack of cross-resistance observed in MIC testing (Table 1). Similarly, we also designed and prepared other mutant enzymes based on analysis of the structure (Fig. 3C), namely E. coli GyrA-M26A and GyrA-I29V, equivalent to S. aureus GyrA M27 and I30, respectively. These mutants displayed no change in IC50 to ciprofloxacin and an approximately fivefold increase in IC50 to compound 2 compared with WT (Table 2).
Compounds 1 and 2 Stabilize Both Single- and Double-Strand DNA-Cleavage Complexes.
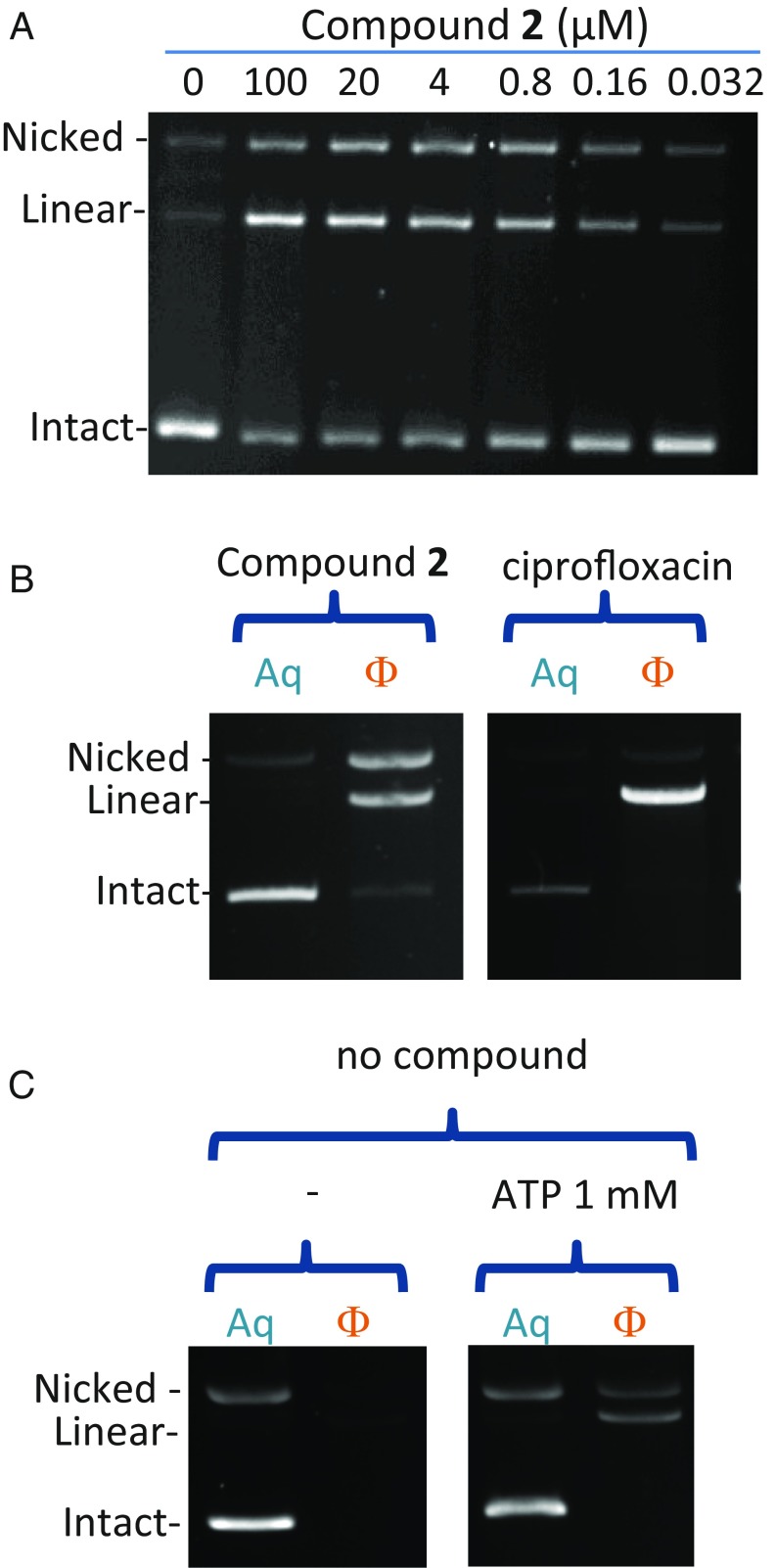
Because compounds 1 and 2 bind away from the site of DNA cleavage, we wanted to confirm that the DNA cleavage detected in our experiments was due to the stabilization of the covalently linked gyrase–DNA-cleavage complexes. After denaturing the polypeptide chains using SDS, the chains can be harvested from the phenol-buffer interface along with any DNA to which they are covalently linked. The uncleaved DNA remains in the aqueous layer whereas DNA covalently attached to the denatured protein is enriched at the phenol-buffer interface and can be recovered in the phenol fraction (28). As expected, ciprofloxacin strongly induced double-stranded DNA cleavage as shown by linear plasmid DNA extractible in the phenol (Fig. 5 B, Right). Compound 2 induced a mixture of double- and single-strand cleavage, both extractible in the phenol phase (Fig. 5 A and B, Left and SI Appendix, Fig. S4), confirming that compound 2 stabilizes a genuine cleavage complex in which the gyrase enzyme is covalently linked to cleaved DNA. When DNA gyrase is submitted to the same procedure of trapping the cleavage complexes followed by phenol enrichment in the presence of only ATP, a lower baseline level of single- and double-stranded cleavage is observed (Fig. 5C).
Fig. 5.
Compound 2 induces a mixture of single- and double-strand DNA cleavage. (A) DNA-cleavage activity of compound 2 with WT E. coli gyrase showing single- and double-stranded breaks (labeled as nicked and linear DNA, respectively). (B) Both single- and double-strand cleavage induced by compound 2 are trapped at a phenol-buffer interface (Left) and so is the cleavage induced by ciprofloxacin (Right). Aq, DNA harvested from the aqueous layer; Φ, DNA harvested from the phenol-buffer interface. Reactions run without ATP. (C) ATP induces phenol-extractible, single- and double-strand DNA cleavage by the unpoisoned E. coli DNA gyrase. Approximately five times more enzyme was used in the reactions compared with B.
A similar experiment to that shown in Fig. 5A was performed with varying the concentrations of ciprofloxacin (SI Appendix, Fig. S5). Concentrations of compound 2 (0.8 μM) and ciprofloxacin (0.16 μM) that produced the same overall extent of cleavage (i.e., the same amount of remaining intact plasmid) (SI Appendix, Fig. S5) gave different levels of single- and double-stranded DNA. Compound 2 stabilized 28% single-stranded DNA and 29% double-stranded DNA whereas ciprofloxacin stabilized 3% single-stranded DNA and 54% double-stranded DNA (SI Appendix, Fig. S5). These results show that compound 2 stabilizes a higher level of nicking compared with ciprofloxacin, which stabilizes mostly double-strand cleavage. It is noteworthy that, to achieve the same level of cleavage, roughly fivefold more of compound 2 is needed compared with ciprofloxacin, despite the two compounds having similar IC50 (SI Appendix, Fig. S5 and Discussion). It is possible that some of the compound 2–gyrase–DNA ternary complexes do not result in cleavage when trapped with SDS.
The DNA-Cleavage Sites Stabilized by Compound 2 Differ from Those Stabilized by Ciprofloxacin.
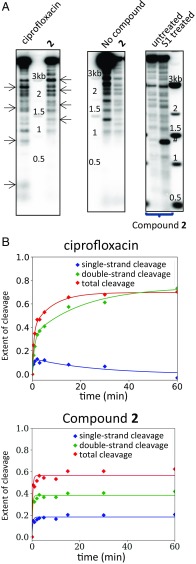
Upon low-resolution mapping of the DNA-cleavage sites induced by compound 2 on plasmid pBR322, we found that the cleavage sites observed in the presence of compound 2 colocalized with cleavage sites trapped with the uninhibited enzyme, albeit with a different intensity (Fig. 6 A, Middle). This result is consistent with the crystal structure of compound 2 (Fig. 3 E and F), which shows that the compound does not make contact with the DNA. In contrast, the cleavage site pattern induced by ciprofloxacin differs from that of the uninhibited enzyme (Fig. 6 A, Left), presumably because the ciprofloxacin–gyrase complex has preferred DNA sequences that may be different from the intrinsic DNA sequence preference of DNA gyrase alone (29, 30). Our results (Fig. 6A) suggest that compound 2 does not change the preferred cleavage sites of the enzyme whereas ciprofloxacin has different preferred cleavage sites.
Fig. 6.
A distinct cleavage mechanism for compound 2 compared with ciprofloxacin. (A) Mapping of cleavage induced by ciprofloxacin and compound 2 on plasmid pBR322. E. coli DNA gyrase cleavage products were digested by EcoRI, separated by electrophoresis, transferred onto a nitrocellulose membrane, and probed with a short PCR fragment hybridizing next to the EcoRI site, on one side only. (Left) The cleavage pattern differs between ciprofloxacin and compound 2. Arrows point at specific differences between the two compounds. (Center) Cleavage observed in the absence and presence of compound 2, respectively, showing similar cleavage patterns. Six reactions were pooled for the “No compound” control to increase the signal. (Right) Treatment by nuclease S1 (which degrades single-stranded DNA and converts nicked plasmids into linear forms) (SI Appendix, Fig. S10) does not change the profile. #, This band is strictly nuclease S1-dependent and occurs with a much higher intensity with the uninhibited enzyme (SI Appendix, Fig. S10). (B) Kinetics of DNA cleavage in the presence of E. coli DNA gyrase and ciprofloxacin or compound 2. Cleavage reactions were allowed to continue for the times indicated before trapping and separation of the cleaved products by electrophoresis, followed by quantification. Data were fitted using a simple kinetic scheme involving a single-strand intermediate for ciprofloxacin and to a single exponential for compound 2; this line is indicative only.
Conversion of single-strand DNA breaks into double-strand DNA breaks using nuclease S1 allowed the single- and double-stranded DNA breaks stabilized by compound 2 to be visualized on the same gel (Fig. 6 A, Right). We found that nuclease S1 treatment merely increased the intensity of the cleavage profile for compound 2 and did not produce new cleavage sites (i.e., new double-stranded break sites) (Fig. 6 A, Right), which suggests that the single-strand cleavage occurs at the same sites as the double-strand cleavage, consistent with the hypothesis that they are two products of the same event.
Kinetics of Cleavage Complex Formation Differs Between Compound 2 and Ciprofloxacin.
We analyzed the kinetics of formation of the cleavage complexes stabilized by compound 2 (Fig. 6B) and found that maximum cleavage is achieved within 2 min with compound 2 and that this timing is not influenced by ATP (SI Appendix, Fig. S6). Importantly, the timing of the appearance of the single- and double-strand cleavages is identical, and the ratio between the two remains the same throughout the experiment, which is in contrast to ciprofloxacin, which induces double-strand cleavage with slower kinetics and through a detectable single-strand intermediate, as reported previously (31). This rate of cleavage with ciprofloxacin is further slowed when ATP is absent (SI Appendix, Fig. S6).
Compound 2 Does Not Inhibit ADPNP-Induced Relaxation of Positively Supercoiled DNA.
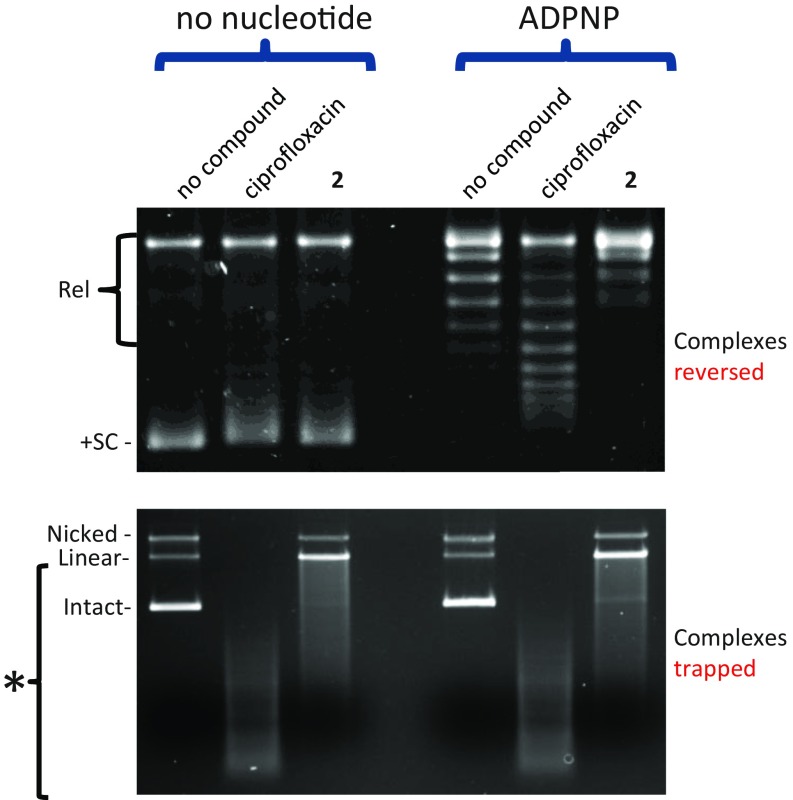
When DNA gyrase acts on positively supercoiled DNA, ATP binding results in efficient DNA capture, strand passage (due to the presence of positive plectonemes), and, ultimately, relaxation of the substrate DNA (32, 33). In the presence of the nonhydrolyzable ATP analog 5′-adenylyl-β,γ-imidodiphosphate (ADPNP), DNA gyrase can perform only one round of relaxation on positively supercoiled DNA (32) (no-compound controls in Fig. 7, Upper), and the amount of nicked or linear DNA does not change noticeably after relaxation of the positively supercoiled DNA (no-compound controls in Fig. 7, Lower). In the presence of ciprofloxacin, the positively supercoiled DNA is not fully relaxed (Fig. 7, Upper), and multiple cleavage complexes are trapped on the DNA plasmid substrate so it runs as a smear (Fig. 7, Lower). In the presence of compound 2 and ADPNP, the positively supercoiled DNA substrate is fully relaxed (Fig. 7, Upper), but much of the substrate DNA plasmid has been converted into a linear or nicked form (Fig. 7, Lower), suggesting that, although the DNA has passed through the DNA gate, to formally relax the positively supercoiled DNA, the DNA is still stabilized in a cleaved form.
Fig. 7.
Compound 2 does not inhibit ADPNP-induced relaxation of positively supercoiled DNA by E. coli DNA gyrase. Reactions containing gyrase, the indicated compounds (at 4 µM) ± nucleotide were carried out with positively supercoiled DNA (see Materials and Methods for details). (Upper) After relaxation, cleavage complexes were removed from the DNA by sequential addition of EDTA and proteinase K to the reaction (this procedure fully reverses the cleavage induced by both compounds—tested independently on relaxed DNA) (SI Appendix, Fig. S7). Rel, relaxed DNA, +SC, positively supercoiled DNA. (Lower) After relaxation, cleavage complexes were trapped on the DNA by the addition of SDS to identical reactions as in Upper. The gel in Lower was run in the presence of 0.5 µg/mL ethidium bromide (see Materials and Methods for details). Asterisk denotes multiple cleavage events resulted in the smearing of the DNA substrate on the gel.
Fig. 7 shows that compound 2 does not inhibit ADPNP-induced relaxation of positively supercoiled pBR322 whereas ciprofloxacin partially blocks this process. These results are consistent with compound 2 blocking the resealing of the DNA only after strand passage and not inhibiting the operation of the DNA gate whereas the data with ciprofloxacin (which binds in the cleaved DNA) suggest inhibition of strand passage.
Discussion
In this study, we detail the discovery and characterization of a class of bacterial DNA gyrase inhibitors. Compound 1, identified via a DNA gyrase-targeted HTS, was shown to have modest antibacterial activity, albeit activity against WT Gram-negative pathogens was limited due to penetration and/or efflux effects. Compound 1 was shown to inhibit the supercoiling activity of DNA gyrase and to stabilize DNA gyrase-dependent DNA cleavage. A crystal structure showed that compound 1 bound to a previously unknown site on gyrase remote from the catalytic DNA-cleavage site. The structural information was used to design a modified inhibitor with a tighter fit inside the pocket, resulting in compound 2, which had improved activity at both the target and whole cell level. Binding to this site was confirmed to be relevant for inhibition of DNA gyrase by the isolation and characterization of mutations in the binding site, which showed resistance to compounds 1 and 2, but not to ciprofloxacin. DNA gyrase enzymes with mutations in the binding site were expressed and tested in vitro, showing decreased susceptibility to compounds 1 and 2 but not to ciprofloxacin, further confirming the relevance of this binding site. In contrast to ciprofloxacin, compound 2 does not change the preferred cleavage pattern of the uninhibited enzyme nor does it inhibit the opening of the DNA gate.
Compound 2 inhibits negative supercoiling by E. coli DNA gyrase as efficiently as ciprofloxacin (SI Appendix, Table S3) and is less efficient in stabilizing cleavage than ciprofloxacin (SI Appendix, Fig. S5). This apparent discrepancy can be explained by assuming that compound 2 has a high affinity for binding to gyrase (similar to that of ciprofloxacin) but that DNA is not cleaved in every ternary complex formed by compound 2 with the enzyme and DNA. In other words, we propose that a pool of compound 2–enzyme–DNA complexes remain uncleaved whereas the ciprofloxacin–enzyme–DNA complexes are primarily cleaved. Compound 2 was also shown to stabilize both single- and double-strand DNA breaks (Fig. 5 A and B and SI Appendix, Figs. S4 and S5). The mixture of single- and double-strand DNA-cleavage complexes observed distinguishes this class of compounds mechanistically from the FQs. We found that ATP can also induce a mixture of single- and double-strand breaks with the uninhibited enzyme, albeit at much lower levels (Fig. 5C). In addition, compound 2 induces a rapidly appearing (relative to ciprofloxacin) cleavage profile similar to that observed with ATP-stimulated cleavage, with single- and double-strand breaks arising together at the same sites on DNA, which suggests that all strand breaks with compound 2 arise from the trapping of specific and homogeneous cleavage complexes. Altogether, these results point toward the idea that the thiophene compounds 1 and 2 allosterically stabilize a DNA cleavage-competent conformation of gyrase that resembles the cleavage-competent conformation transiently adopted by the enzyme during its normal ATP-catalyzed strand passage catalytic cycle. This cleavage-competent conformation presumably affords some flexibility to the DNA gate consistent with the observations that (i) the DNA gate opening is unimpaired by binding of compound 2 and (ii) a mixture of intact DNA and single- and double-strand cleaved DNA molecules within the ternary complex is formed by compound 2.
A structural hypothesis for the action of thiophene compounds 1 and 2 can be found by considering two structures of A. baumannii topo IV: apo and in a complex with DNA and moxifloxacin (an FQ related to ciprofloxacin). The thiophene compound-binding pocket does not exist in a 2.2-Å apo A. baumannii crystal structure due to a hinge movement around R33 of the ParC subunit (25), which moves the TOPRIM domain relative to the WHD (SI Appendix, Figs. S8 and S9). In apo bacterial type IIA topoisomerases, R33 is at the C-terminal end of the Aα1 helix (SI Appendix, Fig. S8), and, in many apo-bacterial topoisomerase structures, the N terminus of the Aα1 helix is disordered, as are the Bα9 and Bα10 helices (e.g., ref. 15). The thiophene compound-binding pocket described here is not observed in any apo bacterial topoisomerase II structure that we are aware of. In complexes of bacterial topoisomerases with DNA, R33 is no longer in the Aα1 helix (SI Appendix, Fig. S8), and the peptide plane between residues 32 and 33 has rotated so that R33 has phi–psi angles outside the normal regions of the Ramachandran plot (15). In eukaryotic type IIA topoisomerases, the Aα1 helix does not act as a hinge; this difference may be due to the insertion of an extra amino acid in bacterial sequences, just before the conserved arginine (GyrA residue R33 in alignment in SI Appendix, Table S2). The residues lining the thiophene compound-binding pocket are conserved among bacteria, perhaps because the pocket needs to open and close during the bacterial catalytic cycle, as observed when the two A. baumannii topo IV structures are compared (SI Appendix, Figs. S8 and S9) (25). This finding suggests that the opening and closing of this Aα1 hinge pocket may be coupled to the binding, cleavage, religation, and release of DNA from the DNA gate. The relative motion of the TOPRIM domain and WHD within a covalently fused subunit (SI Appendix, Figs. S8 and S9) can result in the opening and closing of the Aα1 hinge pocket, and blocking this motion with inhibitors, such as compounds 1 and 2, can stabilize cleavage complexes.
We speculate that interfering with this movement inhibits religation by favoring a DNA cleavage-prone conformation. The residues lining the pocket are almost completely conserved in key bacterial pathogens (SI Appendix, Table S2); the only exception to this residue conservation is in Mycobacterium tuberculosis gyrase, where the residue equivalent to E634 (in S. aureus) is a threonine. Recent crystal structures of M. tuberculosis gyrase with FQs (34) show that the pocket described here is present; however, the distinct shape of the pocket suggests that different compounds may be required to exploit this pocket in M. tuberculosis gyrase.
Clinically proven targets for antibacterials are few in number, and endeavors to find new ones making use of genomic information have proven challenging (20, 21). Producing new molecules against well-validated targets continues to be a successful strategy for delivering new antibacterials for clinical trials. Although we were unable to optimize the current series into a developable drug, the thiophene-binding pocket described here constitutes a unique way of targeting DNA gyrase, corrupting the enzyme into a DNA-cleavage complex with toxic results for the pathogen. Its mechanism of action bypasses existing target-mediated resistance to gyrase-targeting antibiotics currently in clinical use. Moreover, we have found that the frequency of resistance was low, suggesting either that the residues involved in the binding of the thiophenes are important for the enzyme function and/or that the pocket is small enough that the number of mutable residues is limited. These features make this allosteric pocket a possible target for the design and development of antibacterials with a potential broad-spectrum against Gram-negative bacteria.
Materials and Methods
Chemistry.
Compounds 1 and 2 were synthesized as described in SI Appendix, Supporting Materials and Methods.
Crystallography.
Proteins were purified and complexes were prepared and crystallized and structures were determined using methods as previously described (19, 24). The X-ray data collection (35) and refinement statistics for the two thiophene structures described in this paper are shown in SI Appendix, Table S1. More details of the crystallization and structure determinations are given below. Structural figures were drawn with Pymol (36).
Compound 1.
For crystallization, 1 µL of complex [0.035 mM GyrB27A56(GKdel/Tyr123Phe) dimer, 0.174 mM 20-12p-8 DNA duplex (24), 2.2 mM compound 1, 4.3 mM MnCl2, 4.3% DMSO, 17 mM Hepes, 87 mM Na2SO4, pH 7.0] was mixed with 1 µL of crystallization buffer (11% PEG 5000 MME, 150 mM Bis-Tris, pH 6.2). The 1.98-Å structure was determined by molecular replacement and refined using Refmac (37) and Phenix-refine (38); the structure was rebuilt with coot (39). The structure was twinned with refined twin fractions of 0.76/0.24. The structure contained two complexes in the asymmetric, each with two molecules of compound 1 bound. The central four base pairs of DNA, between the two DNA-cleavage sites, were largely disordered, as in a binary complex, with the same doubly nicked DNA duplex (19). The complex with compound 1 was in a very similar cell to a previously determined structure of S. aureus DNA gyrase with ciprofloxacin and DNA (PDB ID code 2XCT; cell P21 89.0 123.2 170.4, beta = 90.25° compared with compound 1 cell, P21 89.4 120.7 168.9, beta = 90.0°), suggesting that compound 1 and ciprofloxacin can stabilize similar conformations of the enzyme.
Compound 2 (soaked into GSK945237 cocrystal).
In all our previous complex structures of S. aureus DNA gyrase with DNA and a variety of DNA-gate inhibitors (15, 19, 40), the thiophene class inhibitor-binding pocket was present. Therefore, a soakable crystal system for determining thiophene crystal structures was developed by cocrystallizing with an NBTI (namely, GSK945237) (41) and then soaking in compound 2. For crystallization, 1 µL of complex [0.047 mM GyrB27A56(GKdel) dimer, 0.072 mM 20-12p-8 DNA duplex, 0.36 mM GSK945237, 2.7 mM MnCl2, 3.6% DMSO, 18.6 mM Hepes, 54 mM Na2SO4, pH 7.0] was mixed with 1 µL of crystallization buffer (7–11% PEG 5000 MME, 130 to 190 mM Bis-Tris, pH 6.2) and streak seeded before covering with paraffin oil. A hexagonal cocrystal with GSK945237 was transferred into a cryo-buffer containing compound 2 (15% glycerol, 20% PEG 5000 MME, 110 mM BisTris, pH 6.2, 2 mM compound 2) and soaked for 4 h at room temperature before being frozen in liquid nitrogen. The structure was refined using Refmac (37) and Phenix-refine (38) from related structures in the same space group (15), and rebuilding was carried out with coot (39). The structure contains one complex in the asymmetric with two molecules of compound 2 bound, and one GSK945237 sitting on the internal twofold axis of the complex. The central four base pairs of DNA, between the two DNA-cleavage sites, were well-ordered, with the left-hand side of GSK945237 sitting between the central bases of the DNA as in other NBTI structures (15, 40). Tyr-639 from the GyrB subunit was displaced from the thiophene-hinge pocket by the binding of compound 2.
In Vitro Topoisomerase Assays.
DNA cleavage and topoisomerase activity assays were performed as described (19). For cleavage site mapping, the DNA material from a cleavage assay was purified by phenol extraction and ethanol precipitation in the presence of 150 mM NaCl to remove the SDS. The pellet was resuspended in water and digested by EcoRI (New England Biolabs) according to the manufacturer’s instructions. The samples were then ethanol-precipitated and analyzed by electrophoresis on a 1% agarose gel containing 0.5 µg/mL ethidium bromide, followed by DNA transfer onto a Hybond-N membrane (GE Life Sciences) according to the manufacturer’s recommendations. The DNA was detected by hybridization to a 100-bp PCR fragment (oligo forward, AGCTTTAATGCGGTAGGCATAGG; oligo reverse, TGGCGATGCTGTCGGAATG) labeled by random priming in the presence of 32P-labeled dATP (Thermofisher random priming labeling kit) according to the manufacturer’s instructions. Nuclease S1 (New England Biolabs) treatments were carried out according to the manufacturer’s instructions just after the first ethanol precipitation. Subsequently, an additional phenol extraction and a PCR purification procedure (Qiagen) were performed before the EcoRI digestion. An aliquot of the untreated (undigested by EcoRI) sample was analyzed separately on an agarose gel to assess the conversion of the nicked plasmid into a linear form (compared with no S1 treatment).
For the ADPNP-induced relaxation of positively supercoiled DNA, 250 ng of positively supercoiled pBR322 (see below) was incubated in the presence of ∼4 µg of A2B2 E. coli DNA gyrase using the same buffer as for the cleavage assays. To this reaction, 1 mM ADPNP (Sigma) and/or 4 µM either compound 2 or ciprofloxacin was added. The mixture was incubated for 30 min at 37 °C and either trapped by SDS as in the cleavage assay (treatment 1) (SI Appendix, Fig. S7) or treated by 8 mM EDTA followed by the addition of 20 µg of proteinase K (treatment 3) (SI Appendix, Fig. S7). Treatment 3 resulted in the complete reversal of all cleavage complexes (SI Appendix, Fig. S7), revealing the topology of the substrate DNA plasmid (Fig. 7). Treatment 1 resulted in the entrapment of the cleavage complexes in the form of irreversible DNA cleavage that could be visualized by an agarose gel containing ethidium bromide (Fig. 7, Lower).
Preparation of Positively Supercoiled DNA.
Relaxed pBR322 was incubated for 30 min at 37 °C with 4 µg of A2B2 E. coli DNA gyrase in a cleavage assay in the absence of ATP. The enzyme constrained positive writhing through its binding to the DNA substrate. The intrinsic relaxation activity of the enzyme removed the compensatory negative writhing, thereby introducing net positive supercoiling. The reaction was stopped with treatment 1 (above). The DNA was purified by ethanol precipitation, followed by a PCR purification column (Qiagen). The DNA was eluted in ultrapure water. The handedness of the supercoiling was ascertained by migration on a chloroquine agarose gel.
Purification of E. coli DNA Gyrase.
GyrA and GyrB subunits were expressed in E. coli and purified separately as described previously (42).
MIC Assays.
Antibacterial MIC assays were determined according to Clinical and Laboratory Standards Institute guidelines (43).
Resistant Mutant Isolation.
Bacterial cells were plated onto Müller–Hinton agar plates containing 2× or 4× above the MIC of the thiophene inhibitor. After 48 h incubation at 37 °C, resistant colonies were purified on compound containing plates and then on plates without compound. The quinolone resistance-determining region or full length of gyrA/B was amplified by PCR from the resistant mutants, and DNA was sequenced using the following forward and reverse primer pairs (all 5′ to 3′): for E. coli gyrA, GAACTCACCTTCCAGATCCCA and TCGGCGCATGACGGATCAGTTC; and gyrB, CGCTGGACTGGCTGGTGAAAGAG and GCCCTTTTCTTATAGCTTCTTGC; for K. pneumoniae gyrA, CGATTTCGTCGACAACTATG and GGTTATGTGGCGGTATGTTG; and gyrB, TCTCCATCCAGCGCTATAAAGG and AGCAGCGGATTCCTCTTTTAGC; for A. baumannii gyrA, GGAAGATAACTACGACGGTT and GCCATACCTACAGCAATACC; and gyrB, AAAGGTCTGGGCGAGATGAATGC and CTAAAGCCGCACATGAAATCAGC. Mutations in thiophene-resistant transformants were confirmed by PCR and DNA sequence analysis.
Supplementary Material
Acknowledgments
We thank Steve Baker and Mike Hann (GSK) and Diarmaid Hughes (Uppsala) for critical reading of the manuscript. The research leading to these results received funding from the Innovative Medicines Initiative Joint Undertaking under Grant Agreement 115583, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and the in-kind contributions of European Federation of Pharmaceutical Industries and Associations (EFPIA) companies. The ENABLE project is also financially supported by contributions from academic and small-medium enterprise (SME) partners. Work in the A.M. laboratory is also supported by Grant BB/J004561/1 from BBSRC (United Kingdom) and the John Innes Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5NPK and 5NPP).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700721114/-/DCSupplemental.
References
- 1.World Health Organization 2017 Global priority list of antiobiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Available at www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. Accessed May 1, 2017.
- 2.CDC . Antibiotic Resistance Threats in the United States, 2013. CDC; Atlanta: 2016. [Google Scholar]
- 3.Payne DJ, Miller LF, Findlay D, Anderson J, Marks L. Time for a change: Addressing R&D and commercialization challenges for antibacterials. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140086. doi: 10.1098/rstb.2014.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien S. Meeting the societal need for new antibiotics: The challenges for the pharmaceutical industry. Br J Clin Pharmacol. 2015;79:168–172. doi: 10.1111/bcp.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.President’s Council of Advisors on Science and Technology 2014 Report to the President on Combating Antibiotic Resistance, 2014. Available at https://obamawhitehouse.archives.gov/sites/default/files/microsites/ostp/PCAST/pcast_amr_jan2015.pdf. Accessed February 1, 2017.
- 6.Kostyanev T, et al. The Innovative Medicines Initiative’s New Drugs for Bad Bugs programme: European public-private partnerships for the development of new strategies to tackle antibiotic resistance. J Antimicrob Chemother. 2016;71:290–295. doi: 10.1093/jac/dkv339. [DOI] [PubMed] [Google Scholar]
- 7.Chan PF, Huang J, Bax BD, Gwynn MN. Recent developments in inhibitors of bacterial type IIA topoisomerases. In: Gualerzi CO, Brandi L, Fabbretti A, Pon CL, editors. Antibiotics: Targets, Mechanisms and Resistance. Wiley, Weinheim; Germany: 2013. pp. 263–297. [Google Scholar]
- 8.Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl Microbiol Biotechnol. 2011;92:479–497. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush NG, Evans-Roberts K, Maxwell A. DNA Topoisomerases. Ecosal Plus. 2015;6:1–34. doi: 10.1128/ecosalplus.esp-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoeffler AJ, Berger JM. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 11.Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper DC, Jacoby GA. Mechanisms of drug resistance: Quinolone resistance. Ann N Y Acad Sci. 2015;1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aldred KJ, Blower TR, Kerns RJ, Berger JM, Osheroff N. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proc Natl Acad Sci USA. 2016;113:E839–E846. doi: 10.1073/pnas.1525055113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bax BD, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 16.Biedenbach DJ, et al. In vitro activity of gepotidacin, a novel triazaacenaphthylene bacterial topoisomerase inhibitor, against a broad spectrum of bacterial pathogens. Antimicrob Agents Chemother. 2016;60:1918–1923. doi: 10.1128/AAC.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. A dose-ranging study evaluating the efficacy, safety, and tolerability of GSK2140944 in the treatment of uncomplicated urogenital gonorrhea caused by Neisseria gonorrhoeae. Available at https://clinicaltrials.gov/ct2/show/NCT02294682?term=GSK2140944&rank=8. Accessed February 2, 2017.
- 18.Basarab GS, et al. Responding to the challenge of untreatable gonorrhea: ETX0914, a first-in-class agent with a distinct mechanism-of-action against bacterial Type II topoisomerases. Sci Rep. 2015;5:11827. doi: 10.1038/srep11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan PF, et al. Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin. Nat Commun. 2015;6:10048. doi: 10.1038/ncomms10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 21.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido H, Pagès JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King AC, Wu L. Macromolecular synthesis and membrane perturbation assays for mechanism of action studies of antimicrobial agents. Curr Protoc Pharmacol. 2009;47:7.1–7.23. doi: 10.1002/0471141755.ph13a07s47. [DOI] [PubMed] [Google Scholar]
- 24.Srikannathasan V, et al. Crystallization and initial crystallographic analysis of covalent DNA-cleavage complexes of Staphyloccocus aureus DNA gyrase with QPT-1, moxifloxacin and etoposide. Acta Crystallogr F Struct Biol Commun. 2015;71:1242–1246. doi: 10.1107/S2053230X15015290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlkonig A, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat Struct Mol Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 26.Laponogov I, et al. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Modular structure of the full-length DNA gyrase B subunit revealed by small-angle X-ray scattering. Structure. 2007;15:329–339. doi: 10.1016/j.str.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnoldi E, Pan XS, Fisher LM. Functional determinants of gate-DNA selection and cleavage by bacterial type II topoisomerases. Nucleic Acids Res. 2013;41:9411–9423. doi: 10.1093/nar/gkt696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 31.Kampranis SC, Maxwell A. The DNA gyrase-quinolone complex: ATP hydrolysis and the mechanism of DNA cleavage. J Biol Chem. 1998;273:22615–22626. doi: 10.1074/jbc.273.35.22615. [DOI] [PubMed] [Google Scholar]
- 32.Bates AD, O’Dea MH, Gellert M. Energy coupling in Escherichia coli DNA gyrase: The relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry. 1996;35:1408–1416. doi: 10.1021/bi952433y. [DOI] [PubMed] [Google Scholar]
- 33.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci USA. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blower TR, Williamson BH, Kerns RJ, Berger JM. Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2016;113:1706–1713. doi: 10.1073/pnas.1525047113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schrödinger, LLC (2015) The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.8.
- 37.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miles TJ, et al. Novel hydroxyl tricyclics (e.g., GSK966587) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg Med Chem Lett. 2013;23:5437–5441. doi: 10.1016/j.bmcl.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Miles TJ, et al. Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg Med Chem Lett. 2016;26:2464–2469. doi: 10.1016/j.bmcl.2016.03.106. [DOI] [PubMed] [Google Scholar]
- 42.Maxwell A, Howells AJ. 1999. Overexpression and purification of bacterial DNA gyrase. DNA Topoisomerase Protocols, Part 1: DNA Topology and Enzymes, Methods in Molecular Biology, eds Bjornsti M-A, Osheroff N (Humana, Totoway, NJ), Vol 94, pp 135–144.
- 43.Clinical and Laboratory Standards Institute 2009. Methods for Dilution Antimicrobial Susceptibility Tests For Bacteria That Grow Aerobically: Approved Standard. (CLSI, Wayne, PA), 8th Ed, Publication M07-A8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.