Significance
Resistance to endocrine treatment remains a significant clinical obstacle. ESR1 mutations were found to be the mechanism of endocrine resistance in a substantial number of patients with metastatic ER-positive breast. However, these mutations are primarily linked to aromatase inhibitor resistance and are not strongly associated with tamoxifen resistance. Herein, we show that tamoxifen treatment promotes a RUNX2–ER complex, which mediates an altered ER cistrome that facilitates the up-regulation of SOX9. We show that up-regulation of SOX9, an embryonic transcription factor with key roles in metastases, is a driver of endocrine resistance in the setting of tamoxifen treatment. Our data provide putative targets for the development of new strategies to treat tamoxifen-resistant breast cancer.
Keywords: estrogen receptor, breast cancer, cistrome, endocrine resistance, SOX9
Abstract
The estrogen receptor (ER) drives the growth of most luminal breast cancers and is the primary target of endocrine therapy. Although ER blockade with drugs such as tamoxifen is very effective, a major clinical limitation is the development of endocrine resistance especially in the setting of metastatic disease. Preclinical and clinical observations suggest that even following the development of endocrine resistance, ER signaling continues to exert a pivotal role in tumor progression in the majority of cases. Through the analysis of the ER cistrome in tamoxifen-resistant breast cancer cells, we have uncovered a role for an RUNX2–ER complex that stimulates the transcription of a set of genes, including most notably the stem cell factor SOX9, that promote proliferation and a metastatic phenotype. We show that up-regulation of SOX9 is sufficient to cause relative endocrine resistance. The gain of SOX9 as an ER-regulated gene associated with tamoxifen resistance was validated in a unique set of clinical samples supporting the need for the development of improved ER antagonists.
Approximately 70% of breast cancers are hormone receptor-positive (HR+) and express estrogen receptor (ERα), progesterone receptor, or both. ERα is a nuclear receptor that is a key driver of tumor development and progression and is the most important therapeutic target in HR+ breast cancers. Therapies targeting ERα signaling include aromatase inhibitors that inhibit ERα by blocking the synthesis of estrogen in peripheral tissues and are effective in postmenopausal women or in combination with ovarian suppression in premenopausal women. Another type of endocrine therapy is the use of pure antiestrogens, also called selective estrogen receptor degraders (SERDs), which do not have agonistic activity and cause ERα degradation. A third class of endocrine therapy consists of the selective estrogen receptor modulators (SERMs), such as tamoxifen (TAM), which bind to ERα and competitively inhibit estrogen binding in the breast. The efficacy of TAM in the treatment of breast cancer patients was first confirmed in clinical trials conducted over 30 y ago (1–4), and TAM remains an important drug in the adjuvant and metastatic setting of HR+ disease.
Despite the known efficacy of endocrine treatments, endocrine resistance remains an important clinical challenge. A significant number of patients with early stage disease will develop disease recurrence after adjuvant endocrine treatment, and in metastatic disease, the majority of patients will eventually develop resistance (5). Loss of ERα expression is seen in 10–15% of patients who develop resistance to endocrine treatment. Thus, in the majority of these cases, ERα continues to be expressed (6–8). Moreover, ligand-independent ERα activity is a key feature underlying the mechanism of endocrine resistance in multiple preclinical studies. These mechanisms include increased ERα coactivator interactions (9) and cross-talk between ERα and growth factor receptor pathways, such as the human epidermal growth factor receptor 2 (HER2) and insulin-like growth factor receptor 1 (IGF1R) (10, 11). This cross-talk leads to the activation of the PI3K signaling pathway, which facilitates ligand-independent activation of ERα. In addition, we and other groups have detected ESR1 ligand binding domain (LBD) mutations in ∼20% of patients with metastatic HR+ disease. These mutations confer constitutive ligand-independent activity and resistance to estrogen deprivation (12). Also, recently reported is a YAP1–ESR1 translocation, whereby the ER LBD is lost, leading to ligand-independent growth and endocrine resistance (13). More recently up-regulation of FOXA1, a pioneer transcription factor for ERα in breast cancer, has been implicated as a mechanism of endocrine resistance (14). Taken together, these preclinical studies suggest that the ERα transcriptional activity remains an important factor in the majority of cases exhibiting endocrine resistance.
A number of clinical trials also support the notion that ERα is a pivotal driver of endocrine-resistant breast cancer and continues to be an important therapeutic target. In the metastatic setting, ∼30% of patients who progress on an aromatase inhibitor respond to fulvestrant (15, 16). Additionally, increasing the dose of fulvestrant resulted in improved disease-free survival and overall survival (17). Furthermore, there is evidence that the combination of fulvestrant with an aromatase inhibitor is superior to an aromatase inhibitor alone in first-line treatment for metastatic disease (18). In early-stage disease, it has been demonstrated that prolonging endocrine treatment or enhancing endocrine blockade with the combination of ovarian suppression with an aromatase inhibitor or TAM in premenopausal women can improve clinical outcomes in certain patients (19, 20), suggesting that more effective inhibition of ERα signaling may also prevent the development of endocrine resistance.
Upon ERα activation, ERα is recruited to thousands of sites across the genome, defining its cistrome. This process is highly organized through epigenetic events that restrict the recruitment of the receptor to a subset of its potential binding sites in a cell type-specific manner (21–24). In addition to the lineage-specific transcriptional program, the ERα cistrome is also dictated by the specific stimuli; as an example, the transcriptional response to growth factor stimulation is different from that of estrogen and regulates genes that are overexpressed in HR+ breast cancers that overexpress ERBB2, which may explain endocrine resistance in this setting (25). Moreover, the ERα cistrome is heterogeneous in HR+ breast tumors, and distinct binding sites are associated with clinical outcomes (26). Collectively, based on the evidence that the ERα transcriptional activity is a key driver in endocrine resistance, the ERα cistrome is cell type- and stimuli-specific, and the ERα cistrome is linked to clinical outcomes, we hypothesized that altered ERα binding to the genome and the resulting changes of its transcriptional program constitute a fundamental mechanism of endocrine resistance. To test this hypothesis, we studied the alterations in the ERα cistrome in breast cancer cell line models of endocrine resistance. Given the recent clinical data supporting the continuation of TAM treatment up to 10 y and the combination of ovarian suppression with TAM in subsets of premenopausal patients (19, 27, 28), our study focused on a model of TAM and estrogen deprivation resistance.
In the present study, we demonstrate that with the acquisition of TAM resistance, the transcription factor RUNX2 is up-regulated and in complex with ERα induces a distinct ERα cistrome, which regulates the transcription of a set of genes that promote a metastatic phenotype. We show that the distinct ERα–RUNX2 binding sites are also increased in metastatic HR+ patient samples compared with early-stage tumors. Furthermore, the stem cell transcription factor SOX9 is induced by the ERα–RUNX2 complex and is sufficient for the development of resistance to estrogen deprivation and TAM.
Results
TAM-Resistant Cell Growth Is ERα-Dependent.
Using an established TAM-resistant (TAMR) cell line model derived from MCF7 cells that were grown in long-term estrogen-deprived (LTED) conditions and TAM (14, 29), we assessed the contribution of ERα to cell proliferation. These studies were performed in the presence of TAM, as removal of TAM in this model abrogates cell growth (Fig. S1). The TAM growth dependency in TAMR cell line models has been demonstrated previously in preclinical studies (30), and in the clinic, there is evidence of tumor regression with TAM withdrawal in a subset of patients with metastatic disease (31). We first confirmed that ERα is expressed in TAMR cells and, in fact, detected higher levels of ERα compared with the parental MCF7 cells (Fig. 1A). ERα reduction by siRNA inhibited cell proliferation in parental and TAMR cells (Fig. 1A). In addition, fulvestrant treatment resulted in a dose-dependent reduction in ERα levels and growth inhibition (Fig. 1B). Thus, cell proliferation in TAMR remains dependent on ERα.
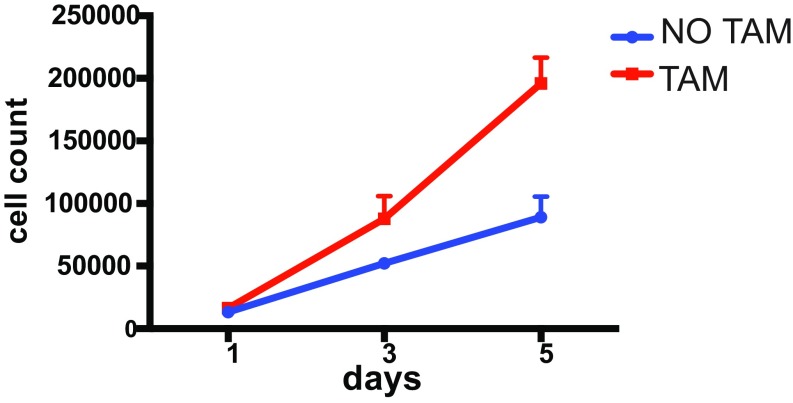
Fig. S1.
TAMR proliferation counted on day 1, 3, and 5 in WM (white medium) in the presence (TAM) or absence of TAM (No TAM).
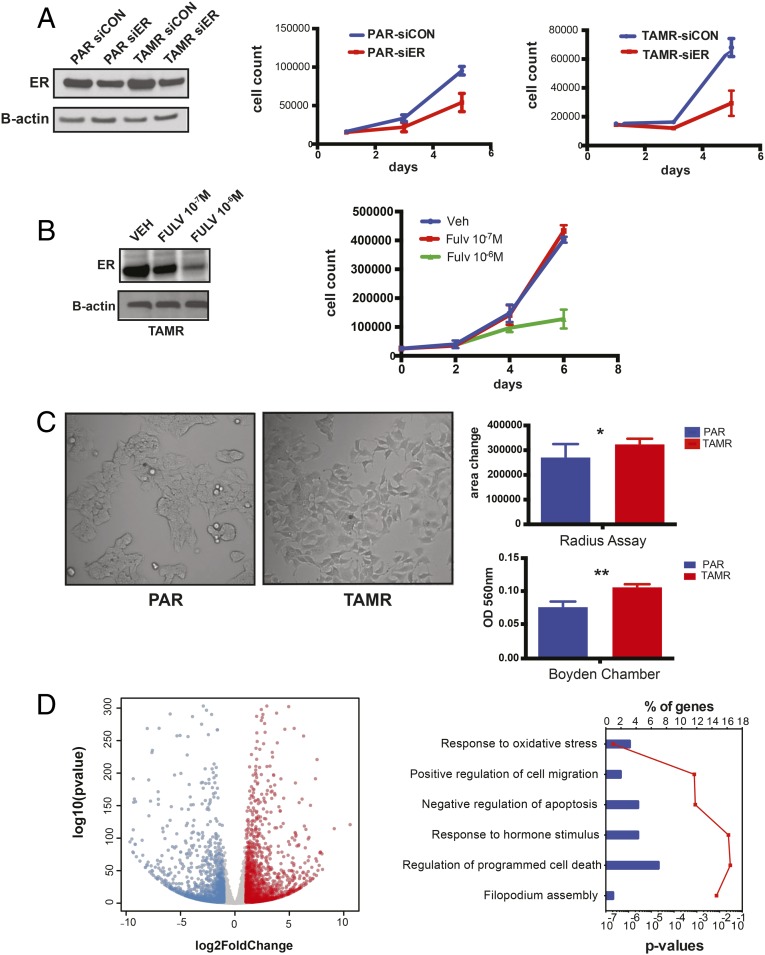
Fig. 1.
TAMR cell proliferation is ERα-dependent and increased migratory capacity with the acquisition of TAM resistance. (A) Cell proliferation analyzed by cell counting on days 1, 3, and 5 after down-regulation of ERα with siRNA in TAMR and PAR (parental) cells. As controls, TAMR and PAR cells were transfected with a siCON, and ERα down-regulation was confirmed by Western blot. Western blot also confirms ERα expression in TAMR. B-actin was used as a loading control. (B) Western blot for ERα and cell proliferation curves after treatment of TAMR cells with fulvestrant (FULV) 10−7 M and 10−6 M or vehicle (Veh) control. (C) Brightfield microscopy picture of 2D culture of PAR and TAMR cells. Magnification, 20×. Shown are the radius assay and Boyden chamber assay testing PAR cell migration in FM (full medium) and TAMR in hormone-depleted white medium. *P = 0.02; **P = 0.003. Error bars denote SEM; figure represents the results of three replicates. (D) Volcano plot of the RNA-seq differential gene expression between PAR and TAMR cells using DE-seq2, log twofold change >1 or <−1, and P <0.05. The red dots are the genes that are significantly up-regulated in TAMR cells, and blue dots are the genes down-regulated. Functional annotation of genes up-regulated in TAMR cells. Top overrepresented gene categories from GO using DAVID are shown here. P values are represented by a red line and % of genes by blue bars.
We further characterized the TAMR cells and observed that these cells acquired morphological changes consisting of cellular elongation with a mesenchymal-like appearance and a more dispersed growth pattern compared with the parental cells (Fig. 1C). The TAMR cells also develop filopodia, which are plasma membrane protrusions known to be important for cell migration (32, 33). In line with the morphological changes, the TAMR cells exhibit a significantly increased migratory capacity compared with the parental cells, as demonstrated by the radius assay and a Boyden chamber (Fig. 1C). Furthermore, differential gene expression by RNA-sequencing (RNA-seq) analysis revealed 1,092 genes up-regulated and 855 genes down-regulated in TAMR compared with parental cells [using DESeq. (32) with a log2FC > 1 or < –1]. Functional annotation of the up-regulated genes with DAVID (33) showed an enrichment of Gene Ontology (GO) terms that included response to oxidative stress, positive regulation of cell migration, negative regulation of apoptosis, response to hormone stimulus, regulation of programmed cell death, and filopodium assembly (P < 0.001). These functions are consistent with the morphological changes and indicative of a metastatic phenotype.
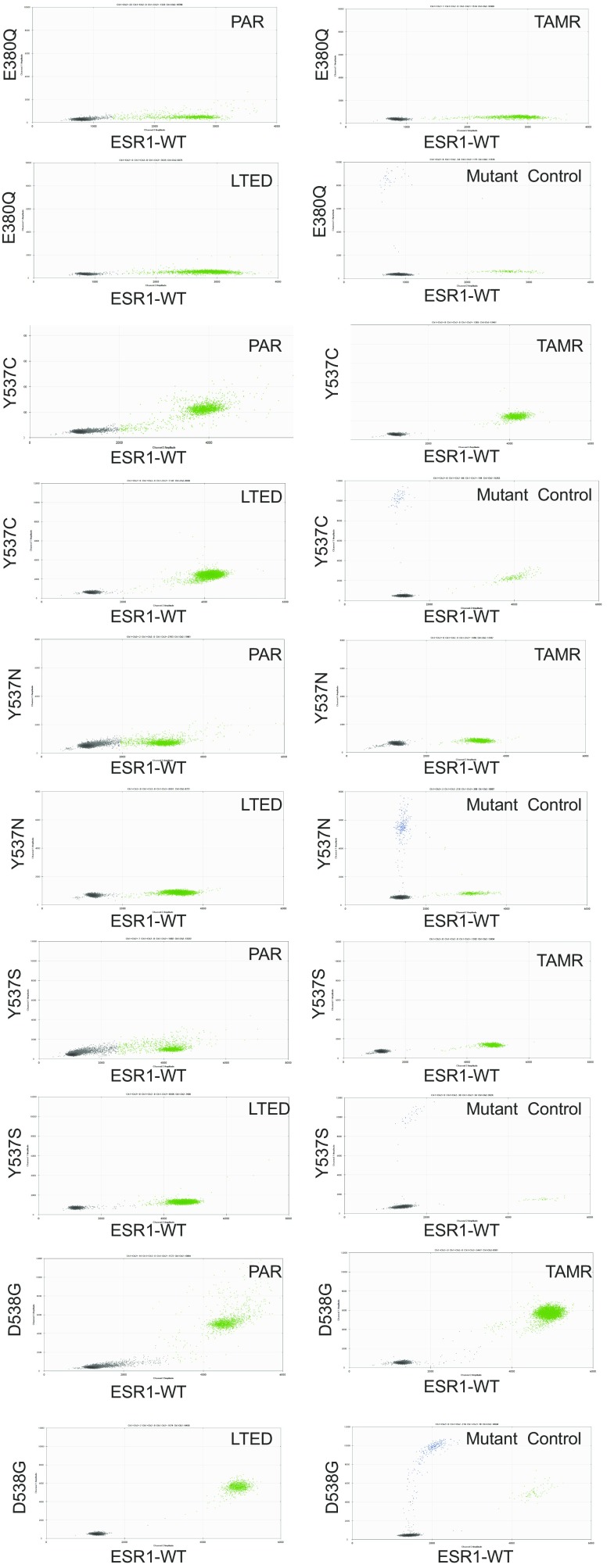
Because the TAMR cells grow under the selective pressure of estrogen deprivation and TAM treatment and remain ER-dependent, we tested the cells for the presence of the most common ligand-binding ESR1 mutations (Y537S, Y537N, Y537C, D538G, and E380Q). Using droplet digital PCR, we did not detect these mutations (Fig. S2), confirming that these mutations do not contribute to endocrine resistance in these cells and suggesting that the mechanism of resistance is due to other alterations in the ERα transcriptional axis. In addition, we did not detect other mutations or splice variations in ESR1 in the TAMR cells by RNA-seq.
Fig. S2.
Genomic DNA samples of MCF7 cell lines were screened for ESR1 E380Q, Y537C, Y537N, Y537S, and D538G mutations by droplet digital PCR in PAR (parental), LTED (long-term estrogen deprived), and TAMR cells. Mutant plasmid DNA was serially diluted into ESR1 WT plasmid DNA (1,000, 250, and 63 mutant copies into 10,000 WT copies per reaction) as the positive control.
TAM Resistance Leads to the Redistribution of ERα–Chromatin Binding.
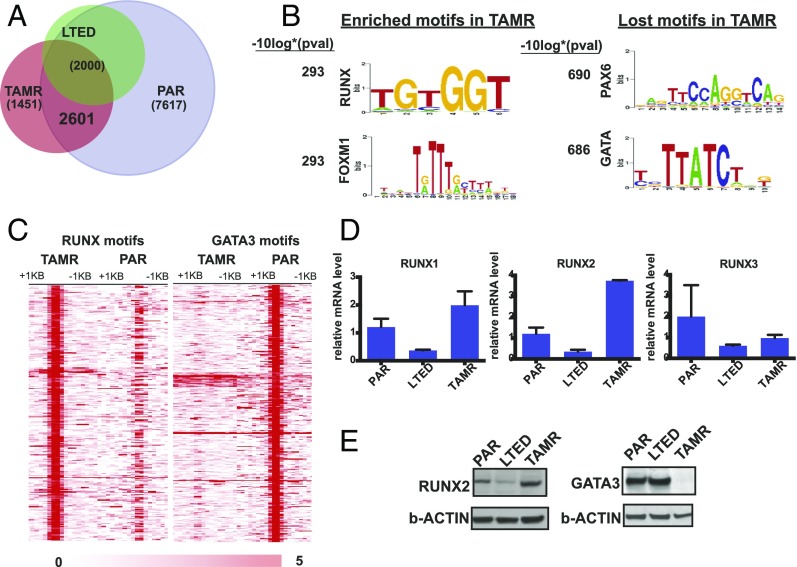
To determine whether there is differential ERα recruitment after the acquisition of resistance to TAM and estrogen deprivation, we compared the ERα cistromes of parental cells after estradiol stimulation with TAMR and LTED cells (also derived from MCF7 cells) in the absence of ligand stimulation (Fig. 2A). While the LTED cells without ligand stimulation had a relatively small number of binding sites (Q value of <0.001) and 95% of these peaks overlapped with the parental cistrome, the TAMR cells had a higher number of estradiol (E2)-independent binding sites (Q value <0.001) and 37% of these binding sites were unique to the TAMR cells compared with the parental cells. In addition, over 60% of the parental ligand-stimulated binding sites were lost in the TAMR cells without E2 stimulation. Of interest, GATA motifs were enriched in the ERα binding events depleted in the TAMR cells [–log10 (P value) = 686] (Fig. 2 B and C). This finding was shown in previous work (26) and explained here by the loss of GATA3 protein expression with the acquisition of TAM resistance (Fig. 2E). GATA3 is a transcription factor that promotes mammary luminal differentiation and a key determinant of luminal-type breast cancers (34). A link between GATA3 and ERα that promotes HR+ tumor development is well established; GATA3 and ERα participate in a positive feedback loop in which each transcription factor stimulates the expression of the other and GATA3 is also a pioneer factor and required for ERα binding at sites that lack active histone modifications (35, 36). On the other hand, GATA3 was shown to suppress epithelial–mesenchymal transition (EMT) and metastases and is associated with a favorable outcome in HR+ breast cancers (34, 37). Consistent with the latter role of GATA3, we detected loss of GATA3 expression and consequently loss of ER binding at sites enriched in GATA motifs in the TAMR cells, which have an increased migratory capacity and EMT phenotype.
Fig. 2.
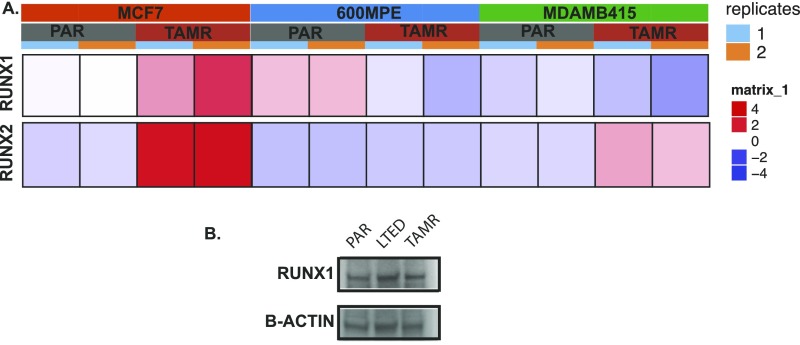
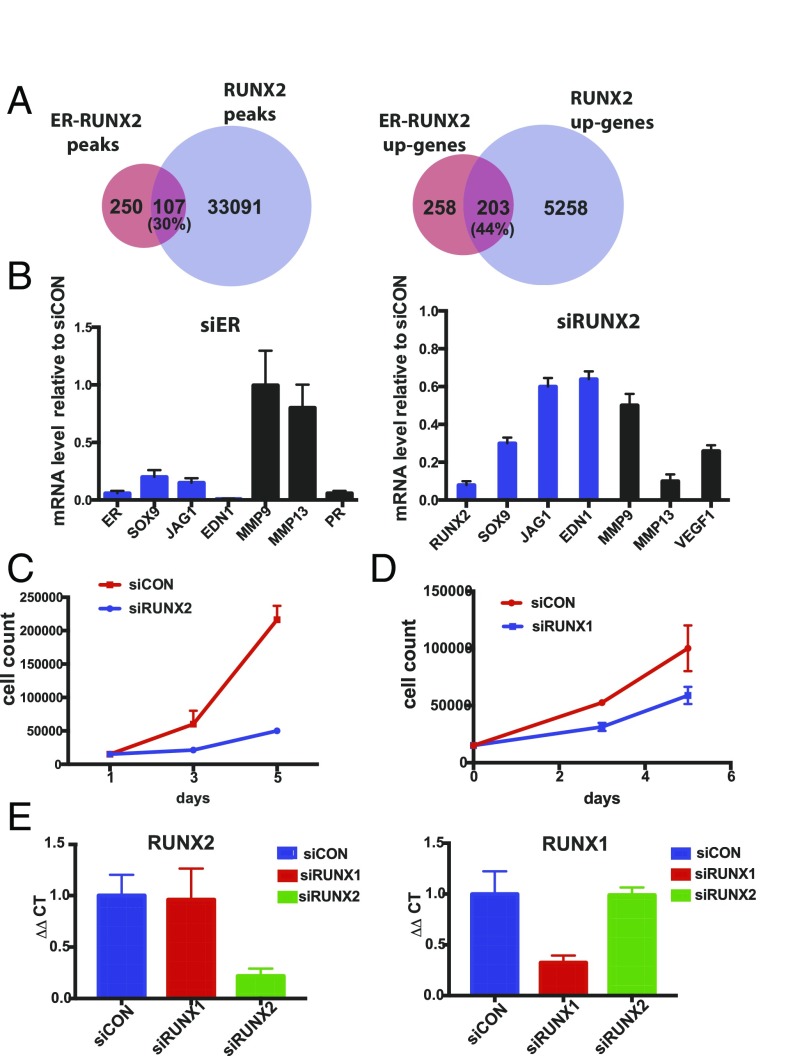
Redistribution of ERα chromatin binding with the acquisition of TAM resistance. (A) Venn diagram showing the overlap of ERα binding in TAMR and LTED cells without E2 stimulation and PAR (parental cells) with E2 stimulation. (B) Top motifs enriched and lost in the TAMR cells compared with parental cells. (C) Heatmap showing clustered ERα binding signals enriched in RUNX and GATA in TAMR and parental cells. The windows represent ±1-kb regions from the center of the ERα binding events. The color scale shows relative enrichment based on raw signal. (D) Relative mRNA levels of the RUNX transcription factors determined by RT-PCR calculated by ΔΔCT in parental, LTED, and TAMR cells. (E) Immunoblots for RUNX2 and GATA3 using cell lysates of PAR, LTED, and TAMR cells.
Motif analysis of the sites that were gained following the acquisition of TAM resistance revealed that these sites were enriched for several motifs, with the RUNX motif being highly significant (–log10 P value = 293) (Fig. 2 B and C). The RUNX transcription factors consist of three family members that are master regulators of differentiation in distinct tissues (Runx1-hematopoeisis, Runx2-bone, Runx3-neuronal/gastrointestinal). The RUNX transcription factors also function as tumor suppressors or oncogenes in a cell context-dependent manner (38). Because the three RUNX transcription factors share a common motif (TGTGGT consensus), we looked at the expression level of the three transcription factors in the parental and TAMR cells. Although RUNX3 levels were down-regulated in TAMR compared with the parental cells, RUNX1 and RUNX2 expression levels were increased in TAMR cells (Fig. 2D). We next performed RNA-seq to evaluate the transcript levels of RUNX1 and RUNX2 in TAMR compared with matched parental cells in two other cell line models, MDAMB415 and 600MPE. RUNX2 levels were up-regulated in both TAMR cell lines compared with the matched parental cells. In contrast, RUNX1 levels were decreased in the MDAMB415 TAMR and 600MPE TAMR cells compared with the parental cells (Fig. S3A). In addition, we were able to confirm up-regulation of RUNX2 and not RUNX1 in TAMR cells at the protein level (Fig. 2F and Fig. S3B). Collectively, the up-regulation of RUNX2 in a number of cell lines, the established role of RUNX2 in bone metastases and EMT (39–42), and emerging data on the association between RUNX2 and poor outcomes in HR+ breast cancer (43, 44) led us to focus our initial studies on RUNX2.
Fig. S3.
(A) Heat maps of RUNX1 and RUNX2 in TAMR and PAR cells (MDAMB415 and 600MPE) determined by RNA-seq. (B) Immunoblot for RUNX1 in PAR (parental), LTED (long-term estrogen deprived), and TAMR cells.
The ERα–RUNX Regulated Genes Are Enriched in Genes Associated with Metastases and Poor Outcomes in Breast Cancer.
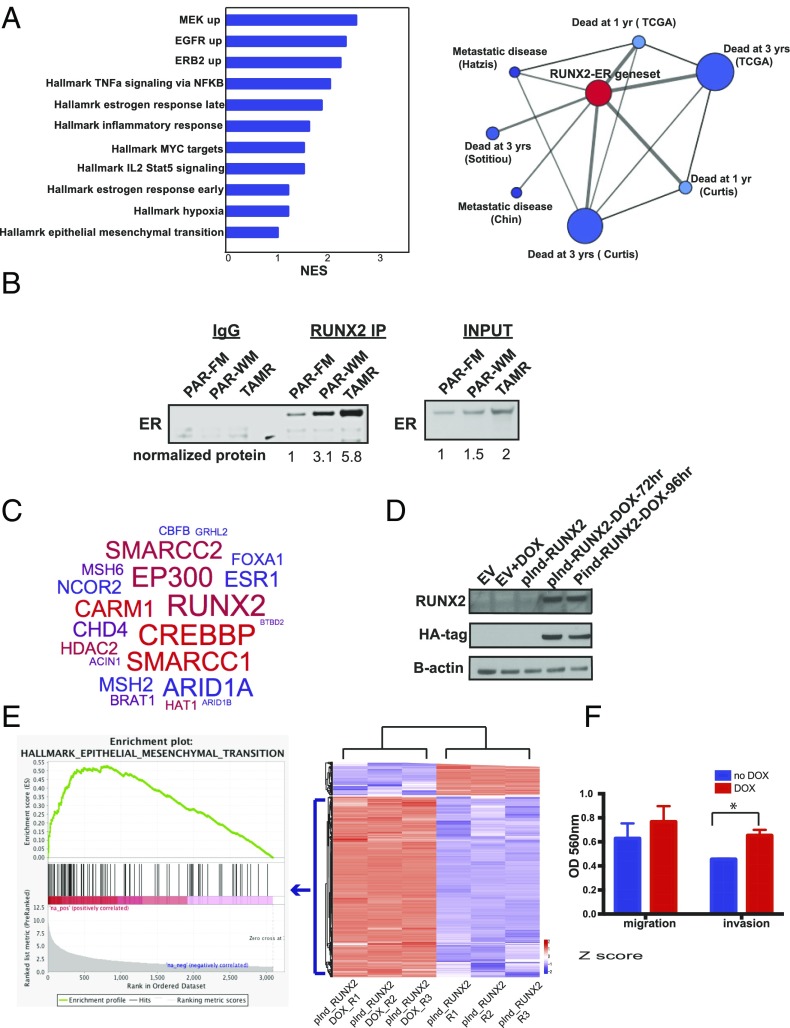
To determine which genes are uniquely up-regulated by ERα transcriptional activity specifically at sites of the RUNX motif in TAMR cells, we used binding and expression target analysis (BETA) to integrate the TAMR-unique ERα–RUNX binding sites with the genes differentially expressed between parental and TAMR cells as determined by RNA-seq (45). We identified 461 genes that are up-regulated by ERα–RUNX after the acquisition of TAM resistance with a rank product of <0.01. Ranked gene set enrichment analysis (GSEA) (46) identified signatures up-regulated in MCF7 cells after the expression of MEK [normalized enrichment score (NES) = 2.5, q = 0.015], ERBB2 (NES = 2.3, q = 0.091), and EGFR (NES = 2.3, q = 0.071) as the top scoring gene sets enriched in the 461 ERα–RUNX2 up-regulated genes in TAMR. These results suggest that the ERα–RUNX transcriptional activity is involved in the cross-talk between ERα and the receptor tyrosine kinase signaling pathways, which is a key mechanism of ligand-independent activation of ERα and endocrine resistance. This finding is supported by previous studies that have shown that RUNX2 is a downstream mediator of the PI3K/AKT pathway (47, 48). In addition, other gene sets that were highly scored in the ERα–RUNX2 up-regulated genes include signatures of TNFα signaling, IL2–STAT5 signaling, and EMT, which are important signatures of the metastatic phenotype (Fig. 3A).
Fig. 3.
The ERα binding sites unique to TAMR cells and enriched in RUNX2 motifs are associated with poor outcomes in breast cancer, and endogenous RUNX2 and ERα interact to mediate transcriptional changes that promote EMT. (A) Ranked GSEA of the genes determined to be up-regulated by ERα–RUNX2 in TAMR cells based on BETA analysis. NES, normalized enrichment score; q < 0.25. Oncomine Concepts Map analysis (Compendia Bioscience) was used to compare the ER–RUNX2 gene signature in TAMR cells against published gene signatures from primary breast cancers. This analysis showed significant correlations between the ER–RUNX2-induced genes and gene expression signatures of poor outcome. The association between the molecular concepts of different gene signatures is represented as a graph using Cytoscape (www.cytoscape.org). A node represents a gene set, and significantly associated (q < 0.2) sets were connected by an edge. The node of the ER–RUNX2 gene set is in red, and the nodes of poor outcomes are in blue. The size of the nodes is proportional to the number of overlapping genes between the corresponding gene set and the ERα–RUNX2 gene set, and the thickness of the edges that connect between the nodes is proportional to the rank of the association significance. (B) Immunoprecipitation of endogenous RUNX2 using nuclear extracts and immunoblotting for endogenous ERα with IgG and input controls. FM, full medium; PAR, parental; TAMR, tamoxifen resistant; WM, white medium. Normalized protein quantification was done using ImageJ (imageJ.nih.gov). (C) RUNX2 RIME results in TAMR cells depicted in a word cloud. (D) Western blot for RUNX2 and HA confirming stable DOX-inducible expression of HA-tagged RUNX2 in MCF7 parental cells after treatment with DOX. EV, empty vector. (E) Heat map of a K-means 2 clustering of the top 1,000 genes differentially expressed between the DOX-inducible RUNX2 MCF7 cells with DOX treatment (pInd-RUNX2_DOX) or no DOX treatment (pInd-RUNX2). R1–R3, replicates 1–3. (Left) An enrichment plot from GSEA showing the Hallmark EMT gene set, which is the gene set most significantly enriched after the induction of RUNX2 expression. (F) Results of a Boyden chamber assay comparing migration and invasion in MCF7 cells with and without DOX induction of RUNX2. *P < 0.05. Error bars represent the SEM of three replicates.
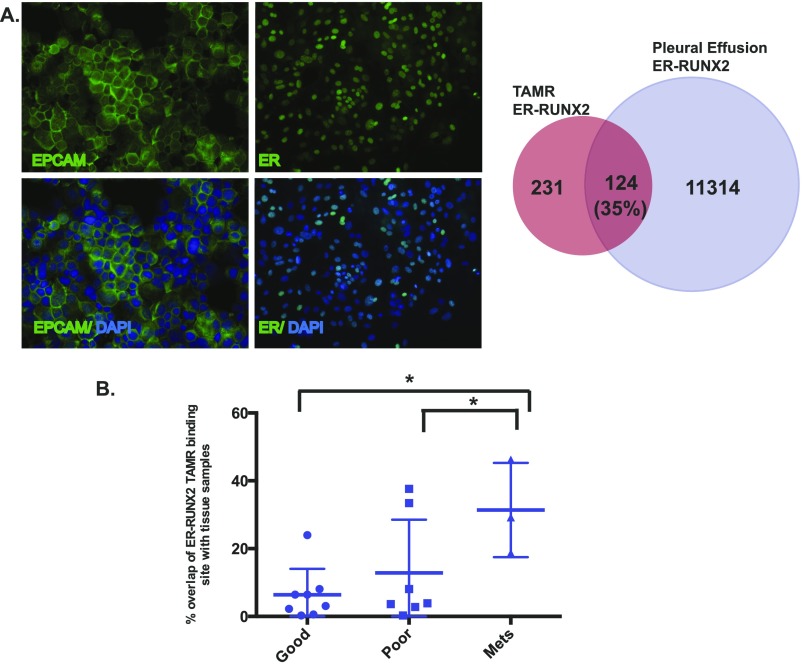
To address the relevance of the TAMR ERα–RUNX transcriptional activity in breast tumors, we established the correlation between the TAMR ERα–RUNX-regulated genes and breast tumor expression signatures using Oncomine Concepts Map analysis (49). Significant correlations were revealed between the TAMR ERα–RUNX2-regulated genes and gene expression signatures of poor outcome including death and metastatic disease (Fig. 3A). We also determined the ERα cistrome of ER+ cancer cells isolated from a metastatic pleural effusion from a patient that had progressive metastatic disease after TAM treatment. A significant proportion of the ERα binding sites in this tumor contained a RUNX motif, and there was a significant overlap between the ERα–RUNX cistrome detected in the TAMR cells and that from this clinical case of TAMR metastatic breast cancer (P < 3e-7, random permutation) (Fig. S4A). In addition, we compared the ERα–RUNX binding sites that were gained in the TAMR cells to published data mapping ER binding sites in ER+ primary tumors of good and poor outcomes and in metastatic ER+ tumors (26). We found that the overlap between the TAMR ERα–RUNX2 binding sites and ER binding sites in the metastatic tumors was significantly higher than the overlap between the TAMR ERα–RUNX binding sites and ERα binding sites in good (P = 0.01) and poor outcome primary tumors (P = 0.04) (Fig. S4B). Thus, the TAMR-unique ERα cistrome associated with RUNX motifs supports transcriptional changes that promote metastases and is associated with breast tumors of poor outcome and metastases.
Fig. S4.
(A) Immunofluorescence staining of cells isolated from a malignant pleural effusion. Staining is for epcam and DAPI (Left) and ERα and DAPI (Right). Magnification, 20×. Venn diagram showing the overlap between the ER–RUNX2 binding sites enriched in the TAMR cells and the pleural effusion metastatic cells in ER binding sites with a RUNX motif within ±1 kb from the summit. (B) Comparison of overlap between the ERα–RUNX2 binding sites in TAMR cells with published ERα binding in ER+ breast cancer tissue samples including primary breast cancers of good prognosis and poor prognosis and metastatic samples. Unpaired Student’s t test was used to calculate the P values. *P < 0.05.
A RUNX2–ER Interaction Occurs with the Acquisition of TAM Resistance and Regulates Genes That Mediate EMT and Metastases.
Because we detected the enrichment of RUNX motifs in the TAMR-unique ERα cistrome, we performed a protein immunoprecipitation study to examine whether there is a direct interaction between ERα and RUNX2. Indeed, when using a RUNX2 antibody for the immunoprecipitation and blotting for ER, we detected a RUNX2–ERα interaction. This interaction was detectable in the parental MCF7 cells, mildly increased after 3 days of estrogen deprivation, and was greatest with the acquisition of TAM and estrogen deprivation resistance (Fig. 3B). This was not merely due to the increase in ERα levels, as the ratio between the amount of ER that coprecipitated with RUNX2 comparing the TAMR to parental cells was higher than the ratio of the level of ERα in TAMR to parental cells in the input determined by the normalized relative density. (The ratio is 5.8 in the immuneprecipitated samples and 2 in the input samples.) To validate this interaction and to identify other proteins that interact with RUNX2 in the context of chromatin binding, we used the RIME approach [rapid immunoprecipitation mass spectrometry (MS) of endogenous proteins] (50). As expected, RUNX2 and CBFβ, a known RUNX2 heterodimer partner, were among the most significantly enriched proteins after removing the nonspecific proteins also found in the IgG control (Fig. 3C, Table S1, and Fig. S5). Other interacting proteins identified included ER and known ER coregulators such as AIB1 (NCOA3), P300, CARM1 (51–53), and the ER pioneer factors FOXA1 and AP2γ (21, 54). In addition, GRHL2, which was recently found to bind to FOXA1, was also detected (55). Furthermore, the RIME assay for RUNX2 failed to identify RUNX2 or significant interacting proteins in the parental cells, which is in keeping with the low RUNX2 levels observed in these cells.
Table S1.
RIME results
| Description | Score | Coverage | Unique peptides | No. of peptides |
| Nuclear receptor coactivator 3 OS = Homo sapiens GN = NCOA3 PE = 1 SV = 1 - [NCOA3_HUMAN] | 1,771.35 | 34.3 | 35 | 35 |
| CREB-binding protein OS = Homo sapiens GN = CREBBP PE = 1 SV = 3 - [CBP_HUMAN] | 557.7 | 14.1 | 11 | 17 |
| Histone acetyltransferase p300 OS = Homo sapiens GN = EP300 PE = 1 SV = 2 - [EP300_HUMAN] | 526.46 | 10.31 | 9 | 15 |
| SWI/SNF complex subunit SMARCC1 OS = Homo sapiens GN = SMARCC1 PE = 1 SV = 3 - [SMRC1_HUMAN] | 346.66 | 11.58 | 5 | 8 |
| AT-rich interactive domain-containing protein 1A OS = Homo sapiens GN = ARID1A PE = 1 SV = 3 - [ARI1A_HUMAN] | 285.15 | 6.96 | 12 | 12 |
| SWI/SNF complex subunit SMARCC2 OS = Homo sapiens GN = SMARCC2 PE = 1 SV = 1 - [SMRC2_HUMAN] | 280.76 | 10.54 | 8 | 10 |
| Runt-related transcription factor 2 OS = Homo sapiens GN = RUNX2 PE = 1 SV = 2 - [RUNX2_HUMAN] | 242.29 | 28 | 7 | 8 |
| Histone-arginine methyltransferase CARM1 OS = Homo sapiens GN = CARM1 PE = 1 SV = 3 - [CARM1_HUMAN] | 224.05 | 23 | 8 | 8 |
| Estrogen receptor OS = Homo sapiens GN = ESR1 PE = 1 SV = 2 - [ESR1_HUMAN] | 200.46 | 10.6 | 4 | 4 |
| Chromodomain-helicase-DNA-binding protein 4 OS = Homo sapiens GN = CHD4 PE = 1 SV = 2 - [CHD4_HUMAN] | 185.26 | 6.83 | 5 | 5 |
| DNA mismatch repair protein Msh2 OS = Homo sapiens GN = MSH2 PE = 1 SV = 1 - [MSH2_HUMAN] | 140.27 | 7.7 | 4 | 4 |
| Nuclear receptor corepressor 2 OS = Homo sapiens GN = NCOR2 PE = 1 SV = 2 - [NCOR2_HUMAN] | 135.24 | 3.3 | 6 | 6 |
| Histone deacetylase 2 OS = Homo sapiens GN = HDAC2 PE = 1 SV = 2 - [HDAC2_HUMAN] | 109.10 | 6.4 | 2 | 2 |
| Hepatocyte nuclear factor 3-alpha OS = Homo sapiens GN = FOXA1 PE = 1 SV = 2 - [FOXA1_HUMAN] | 101.11 | 21.4 | 4 | 4 |
| DNA mismatch repair protein Msh6 OS = Homo sapiens GN = MSH6 PE = 1 SV = 2 - [MSH6_HUMAN] | 89.7 | 1 | 1 | 1 |
| BRCA1-associated ATM activator 1 OS = Homo sapiens GN = BRAT1 PE = 1 SV = 2 - [BRAT1_HUMAN] | 78.79 | 5 | 2 | 4 |
| Histone acetyltransferase type B catalytic subunit OS = Homo sapiens GN = HAT1 PE = 1 SV = 1 - [HAT1_HUMAN] | 62.77 | 7.4 | 2 | 2 |
| Core-binding factor subunit beta OS = Homo sapiens GN = CBFB PE = 1 SV = 2 - [PEBB_HUMAN] | 52.89 | 13.3 | 3 | 3 |
| Apoptotic chromatin condensation inducer in the nucleus OS = Homo sapiens GN = ACIN1 PE = 1 SV = 2 - [ACINU_HUMAN] | 40.35 | 1 | 1 | 1 |
| Grainyhead-like protein 2 homolog OS = Homo sapiens GN = GRHL2 PE = 1 SV = 1 - [GRHL2_HUMAN] | 34.62 | 10 | 4 | 4 |
| AT-rich interactive domain-containing protein 1B OS = Homo sapiens GN = ARID1B PE = 1 SV = 2 - [ARI1B_HUMAN] | 31.79 | 1 | 1 | 1 |
| BTB/POZ domain-containing protein 2 OS = Homo sapiens GN = BTBD2 PE = 1 SV = 1 - [BTBD2_HUMAN] | 23.95 | 3 | 1 | 1 |
Fig. S5.
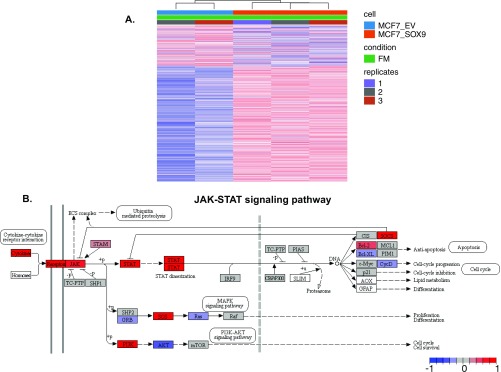
(A) Heat map of unsupervised analysis of RNA-seq from control MCF7 parental cells (EV, empty vector) and MCF7 parental cells stably overexpressing SOX9. (B) Graph depicting the genes up-regulated in the JAK–STAT pathway with up-regulation of SOX9 in MCF7 cells. Graph was rendered by Pathview (82).
Because we confirmed an interaction between ERα and RUNX2 in TAMR cells, we hypothesized that with the acquisition of TAM resistance, RUNX2 is up-regulated and interacts with ERα and the ER–RUNX2 complex is in part responsible for the reprogramming of the ERα cistrome and transcriptional activity in TAM resistance. To test this hypothesis, we generated MCF7 cells stably expressing doxycycline (DOX)-inducible HA-tagged RUNX2 (Fig. 3D). Globally, RNA-seq analysis revealed a large number of differentially expressed transcripts when comparing cells without and with DOX treatment (FDR < 0.01 and log2FC > 1 or < –1). Ranked GSEA revealed that the genes up-regulated by the induction of RUNX2 expression are most significantly enriched in the Hallmark gene set of EMT (NES = 2.06, q-val = 0.002), consistent with the metastatic phenotype we observed in the TAMR cells (Fig. 3E). Furthermore, we saw that overexpression of RUNX2 led to a significant increase in invasion and a trend toward increased migration (Fig. 3F). Next, we performed RUNX2 ChIP-sequencing (ChIP-seq) in the MCF7 cells after RUNX2 induction using an HA antibody (Fig. 4A). We compared the RUNX2 cistrome to the TAMR-unique ER–RUNX2 binding sites and found that 30% of the TAMR-unique binding sites overlapped with RUNX2 binding sites. This overlap is statistically significant (P < 3e-7, random permutation). Moreover, we found that 44% of the 461 genes up-regulated by the RUNX2–ERα transcriptional activity overlapped with the RUNX2-specific up-regulated genes defined by integrating the RUNX2 ChIP-seq and the differential gene expression induced by RUNX2 expression in the DOX-inducible cell line applying BETA (46). To test if the overlapping RUNX2–ERα up-regulated genes are coregulated by both transcription factors, we looked at the expression levels of three of the overlapping genes that are known to have key functions in metastases (SOX9, EDN1, JAG1) after knockdown of ERα or RUNX2. By RT-PCR we confirmed that all three genes are regulated by both ERα and RUNX2. In contrast, MMP9 and MMP13 (39, 40), which are published RUNX2 target genes and are within our list of RUNX2 up-regulated genes that do not overlap with the RUNX2–ERα up-regulated genes, are down-regulated after RUNX2 knockdown but not ER knockdown (Fig. 4B). Lastly, we demonstrated that down-regulation of RUNX2 abrogated proliferation (Fig. 4C). Because there is a recently published small molecule (56) that inhibits the RUNX–CBFβ binding and we showed that RUNX1 is expressed in the MCF7–TAMR cells, we tested the effect of RUNX1 down-regulation by siRUNX1 and showed that down-regulation of RUNX1 also had a growth inhibitory effect, suggesting that a nonselective RUNX inhibitor could be effective in this setting. Additionally, we confirmed that siRUNX1 was specific to RUNX1 (Fig. 4E). Taken together, these results imply that with the acquisition of TAM resistance, ERα interacts with RUNX2 and these two transcription factors coregulate a unique set of genes that mediate proliferation and a metastatic phenotype.
Fig. 4.
The RUNX2 cistrome highly overlaps with the ER–RUNX2 cistrome. (A) Venn diagram (Left) depicts the overlap between the ERα binding sites with a RUNX2 motif in TAMR cells and RUNX2 binding sites in MCF7 cells with DOX-inducible expression of RUNX2. Venn diagram (Right) showing the overlap between the genes up-regulated by the ERα–RUNX2 complex in TAMR cells and genes up-regulated by RUNX2 in MCF7. Both gene sets were determined by integrating ChIP-seq and RNA-seq applying BETA (45). (B) Relative mRNA levels of genes regulated by the ERα–RUNX2 complex after transfection of TAMR cells with either siERα, siRUNX2, or siControl (siCON) and extraction of RNA on day 3 after transfection. Shown here are the relative mRNA levels after siERα knockdown or siRUNX2 knockdown compared with siControl. (C) Cell proliferation analyzed by cell counting on days 1, 3, and 5 after down-regulation of RUNX2 with siRNA. (D) Cell proliferation analyzed by cell counting on days 1, 3, and 5 after down-regulation of RUNX1 with siRUNX1 in TAMR cells. As controls, TAMR cells were transfected with a siCON. (E) Relative mRNA levels of RUNX2 (Left) and RUNX1 (Right) in control MCF7 cells and after down-regulation of RUNX1 and RUNX2.
SOX9 Is a ERα–RUNX2 Target Gene and Is Sufficient for TAM Resistance.
SOX9 is a transcription factor that regulates stem and progenitor cells in adult tissues (57, 58). It is up-regulated in basal cell breast cancers and confers a tumor-initiating stem/progenitor cell and metastatic phenotype in breast cancer cells (57, 59). Because we found that SOX9 is one of the target genes of the ERα–RUNX2 complex, we next turned our attention to the role of SOX9 in TAM resistance.
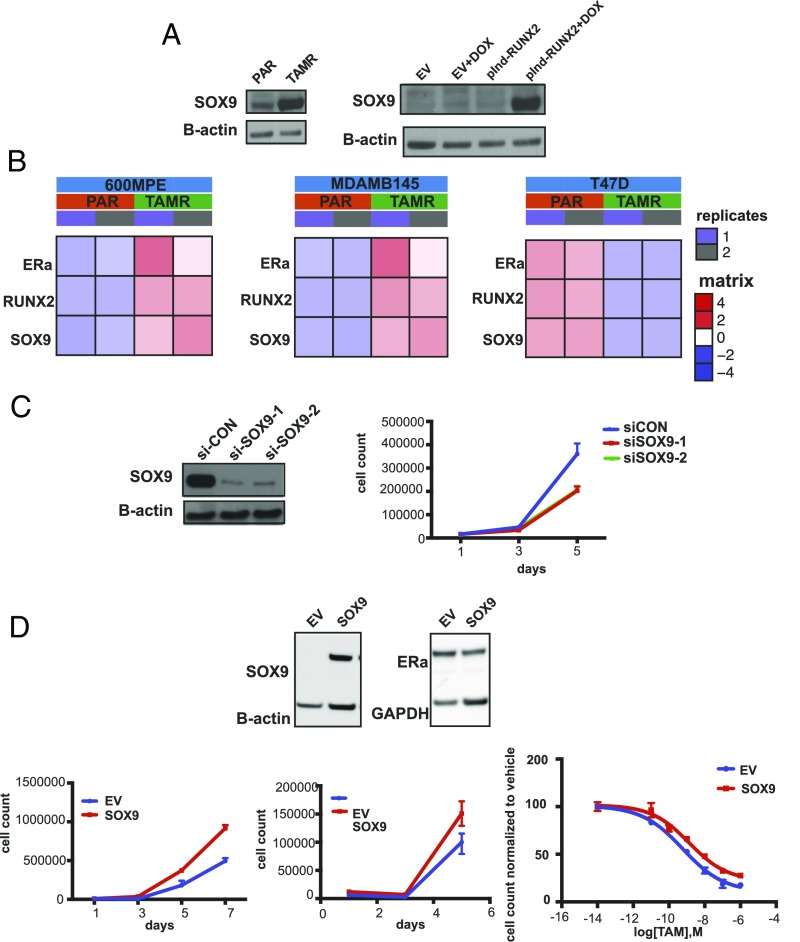
We first confirmed the up-regulation of SOX9 in TAMR cells compared with parental cells and after the induction of RUNX2 expression at the protein level (Fig. 5A). We also tested other ER+ cell line models of TAM resistance and found that similar to the MCF7 cells, in TAMR cell lines in which ERα expression is maintained (MDAMB415 and 600MPE), RUNX2 and SOX9 are up-regulated after the development of TAM resistance. In contrast, in the T47D cell line, ERα is suppressed after the development of TAM resistance and RUNX2 and SOX9 are both down-regulated (Fig. 5B). We next determined that TAMR cell growth is dependent on SOX9 by knockdown of SOX9 in TAMR cells (Fig. 5C). Subsequently, we showed that overexpression of SOX9 in parental cells, which does not affect ER levels, leads to a growth advantage in full medium and estrogen-deprived conditions compared with cells expressing an empty vector (EV) (Fig. 5D). Furthermore, dose–response studies showed that overexpression of SOX9 leads to relative resistance to TAM with a 2.3-fold increase in the TAM IC50 in the SOX9-overexpressing cells compared with the EV-expressing cells with a P value close to significant (EV TAM IC50 = 6 × 10−10 M, SOX9 overexpression TAM IC50 = 1.4 × 10−9 M, P = 0.054) (Fig. 5D). We analyzed the transcriptional changes after SOX9 overexpression in PAR MCF7 cells and identified 277 genes that were up-regulated compared with PAR control (EV) (Fig. S4A) (FDR < 0.01). The top ranked gene sets enriched in the PAR cells overexpressing SOX9 were genes down-regulated during apoptosis in breast cancer cell lines (NES = 1.94) and genes associated with acquired endocrine resistance in breast tumors expressing ER and ERBB2 (NES = 1.55). In addition, Kegg Pathways analysis with GAGE (general applicable gene set enrichment for pathway analysis) (60) revealed that the top ranked pathway enriched in the SOX9 overexpressing PAR cells was the JAK–STAT pathway (Fig. S5). In summary, we show that SOX9 is up-regulated in TAMR by the ERα–RUNX2 complex, TAMR cell proliferation is dependent on SOX9, and SOX9 up-regulation is sufficient to cause resistance to estrogen deprivation and decreased sensitivity to TAM treatment.
Fig. 5.
SOX9 is up-regulated in TAMR and mediates resistance to estrogen deprivation and TAM. (A) Western blot for SOX9 in parental (PAR) and TAMR MCF7 cells (Left) and in MCF7 cells infected with an EV control and a DOX-inducible RUNX2 expression vector with and without DOX treatment. (B) Heat maps of mRNA levels of ESR1, RUNX2, and SOX9 in parental (PAR) and TAMR cell lines derived from 600MPE, MDAMB415, and T47D cell lines. (C) Cell proliferation analyzed by cell counting on days 1, 3, and 5 in TAMR cells after SOX9 silencing with two different siRNAs or with an siRNA control (si-CON). Western blot for SOX9 in TAMR cells showing successful knockdown of SOX9. (D, Top) Western blot of SOX9 in parental MCF7 cells after stable expression of SOX9 and an EV. Western blot shows stable ERα expression after expression of SOX9 in MCF7 parental cells. Cell proliferation analyzed by cell counting on days 1, 3, and 5 in MCF7 parental cells after SOX9 expression or an EV control in (Top Left) full medium or (Top Right) white medium. Dose–response curves for TAM treatment in MCF7 cells expressing SOX9 and an EV as control. TAM IC50 in SOX9-overexpressing cells is 1.4 × 10−9 M and in control cells expressing an EV 6 × 10−10 M.
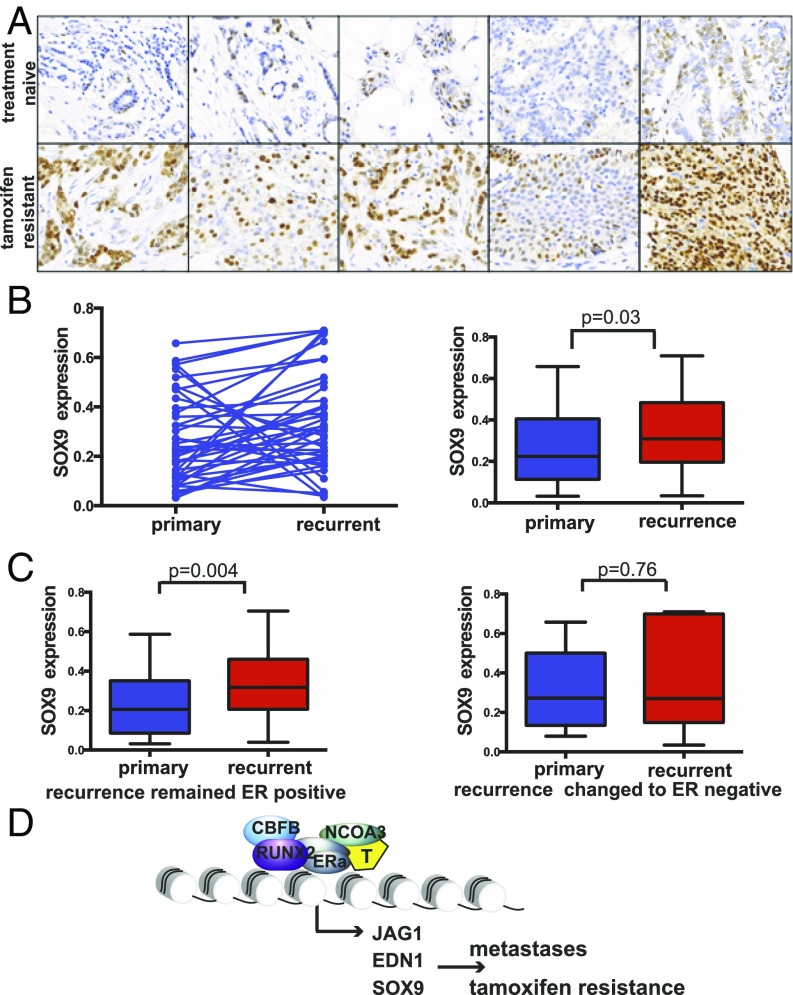
To validate our preclinical findings, we examined a unique set of clinical samples from patients that developed local recurrences during or after adjuvant treatment with TAM. This sample set included 42 pairs of primary ERα+ breast cancers and the matched recurrent lesions. Comprehensive clinical data were available for 39 of the patients (Table S2). The majority of the recurrent lesions remained ERα+ (67%), and the majority of the patients developed the recurrence while on TAM treatment (80%). For the 20% of patients that developed the recurrence after TAM treatment, the average time between completion of TAM and disease recurrence was 35 mo. The average duration of TAM treatment was 34 mo (ranging between 6 and 83 mo). We were able to examine the expression of SOX9 by IHC in the 42 tumor pairs (Fig. 6A). SOX9 showed strong nuclear staining of breast cancer epithelial cells in the ERα+ primary and recurrent tumors. Taking into account the staining intensity and percentage of positive epithelial cells, there was variation in the level of SOX9 expression between tumors. Nonetheless, there was a significant increase in the expression of SOX9 after the development of TAM resistance [expression in treatment-naïve tumors, mean ± SE, 0.268 ± 0.028; expression in TAMR-recurrent tumors, mean ± SE, 0.342 ± 0.031, P value of 0.033 (t = –2.207)] (Fig. 6B). Furthermore, we performed a subset analysis and looked at the pairs in which the recurrent disease remained ERα+ (n = 28) and found that in this subgroup SOX9 expression was significantly up-regulated after the development of TAM resistance (P value of 0.004), whereas in the recurrent tumors with loss of ERα expression (n = 14), SOX9 expression was not up-regulated (Fig. 6C). These clinical results show a correlation between TAM resistance and SOX9 expression in ERα+ tumors and support the notion that SOX9 is an ERα transcriptional target gene in the setting of TAM resistance.
Table S2.
Patient characteristics
| Sample no. | ER status of recurrence | HER2 status of primary | HER2 status of recurrence | Duration of TAM treatment, months | Timing of recurrence, months |
| 1 | NEG | NEG | NEG | 12 | 11 |
| 2 | POS | NEG | NEG | 46 | 146 |
| 3 | POS | NEG | NEG | 52 | 52 |
| 4 | POS | NEG | NEG | 12 | 12 |
| 5 | NEG | NEG | NEG | 15 | 15 |
| 6 | NEG | POS | POS | 25 | 27 |
| 7 | NEG | NEG | NEG | 20 | 21 |
| 8 | POS | NEG | NEG | 83 | 83 |
| 9 | NEG | NEG | NEG | 13 | 13 |
| 10 | POS | NEG | NEG | 11 | 11 |
| 11 | NEG | NEG | NEG | 45 | 45 |
| 12 | POS | NEG | NEG | 15 | 15 |
| 13 | POS | POS | POS | 26 | 27 |
| 14 | POS | POS | POS | 21 | 22 |
| 15 | POS | NEG | NEG | 52 | 53 |
| 16 | POS | NEG | NEG | 33 | 35 |
| 17 | NEG | NEG | NEG | 8 | 10 |
| 18 | POS | NEG | NEG | 68 | 69 |
| 19 | POS | NEG | NEG | 6 | 6 |
| 20 | POS | NEG | POS | 17 | 17 |
| 21 | POS | NEG | NEG | 69 | 69 |
| 22 | POS | NEG | NEG | 51 | 52 |
| 23 | NEG | POS | POS | 14 | 13 |
| 24 | POS | UNK | NEG | 62 | 119 |
| 25 | NEG | POS | NEG | 15 | 15 |
| 26 | NEG | NEG | NEG | 8 | 12 |
| 27 | POS | NEG | NEG | 22 | 54 |
| 28 | NEG | NEG | NEG | 42 | 43 |
| 29 | NEG | NEG | NEG | 36 | 37 |
| 30 | NEG | NEG | NEG | 30 | 30 |
| 31 | POS | NEG | NEG | 54 | Unk |
| 32 | NEG | UNK | NEG | 23 | 23 |
| 33 | POS | NEG | UNK | 25 | 49 |
| 34 | POS | UNK | UNK | 70 | 69 |
| 35 | POS | NEG | NEG | 60 | 90 |
| 36 | POS | UNK | NEG | 60 | 101 |
| 37 | UNK | UNK | UNK | 23 | 23 |
| 38 | POS | NEG | NEG | 54 | 98 |
| 39 | POS | NEG | NEG | 18 | 22 |
Fig. 6.
TAM resistance in clinical samples is associated with SOX9 expression. (A) Representative figures of immunohistochemistry staining for SOX9 showing an increase in nuclear staining in the recurrent TAMR samples. Magnification, 40×. (B) SOX9 expression in ER+ primary and recurrent breast cancer samples. The scatter plot shows that in a number of cases there was a decrease in SOX9 expression after the development of TAM resistance, but in the majority of the cases (67%) there was an increase and overall there was a significant increase. (C) SOX9 expression levels in the recurrent tumors that remained ER+ and the recurrent tumors that changed to ER– and their matched primary tumors. (D) Scheme of the ERα–RUNX2 model in TAM resistance.
Discussion
Our results demonstrate that differential recruitment of ERα resulting in an aberrant transcriptional network is a fundamental mechanism of resistance to TAM treatment. Furthermore, as endocrine resistance is commonly associated with the development of distant recurrences (61), our results indicate that the aberrant ERα transcriptional axis also has a key role in driving an EMT and prometastatic program. More specifically, we found that after long-term exposure to TAM and estrogen deprivation, ERα–chromatin binding is enriched in RUNX motifs mediated by a direct interaction between ERα and RUNX2. Our RIME data suggest that in addition to RUNX2 and known ERα coregulators, the SWI/SNF chromatin-remodeling complex, which we have previously shown to be important for ERα activity and is emerging as an important cancer driver (62), may be involved in RUNX2–ERα chromatin binding.
In this study, we found that other transcription factor binding motifs of potential interest were enriched in the TAMR ERα DNA binding sites. These include the FOXM1 motif, which was enriched in the TAMR ERα unique binding sites, consistent with published studies showing that FOXM1 interacts with ER and can also promote TAM resistance (63, 64). Furthermore, we demonstrated the loss of transcription factors essential for the ER transcription axis in luminal-type breast cancers, such as GATA3. Thus, multiple alterations in the ERα cistrome orchestrate resistance to TAM and disease progression, underscoring the therapeutic implications of better targeting of ERα. Our study also depicts the complexity of endocrine resistance and the fact that there are multiple mechanisms of resistance. As an example, in the TAMR cells derived from T47D cells, ERα is down-regulated and RUNX2 and SOX9 are not up-regulated, implying that the mechanism of resistance is different in this model and likely due to the loss of ERα expression. Likewise, in the clinic, there are multiple mechanisms of endocrine resistance, and studies to identify biomarkers of the specific drivers of resistance will be important.
A RUNX1–ERα interaction has been described in the literature in a triple-negative breast cancer cell line engineered to ectopically express ERα (65). RUNX1 has been shown to function as both a tumor suppressor and an oncogene in breast cancer, and recently, point mutations in RUNX1 and CBFB have been detected in ER+ breast cancers by next-generation sequencing studies (66–68). Similar to the cell- and context-dependent activity of RUNX1, the RUNX2–ERα complex appears to be distinct in a cell- and context-dependent manner. A RUNX2–ERα interaction has been described previously in COS7 cells transfected with an expression vector encoding ERα (69). Although in the COS7 cell type ERα repressed RUNX2 transcriptional activity in a ligand-dependent manner, here we show an interaction between endogenous RUNX2 and ERα proteins in an ERα+ endocrine-resistant breast cancer cell line, and these two transcription factors in concert mediate transcription of genes that promote EMT and metastases. In addition, we also show that down-regulation of RUNX1 has a moderate inhibitory effect on the TAMR cells, suggesting that RUNX1 likely also has a role in endocrine resistance in this model and future studies are needed for further evaluation.
A number of studies have shown that RUNX2 can increase invasiveness, but these studies were in triple-negative breast cancer models (70–72). Likewise, in clinical samples of primary breast cancers, RUNX2 is expressed and found to be prognostic mainly in triple-negative breast cancers (44, 73). However, in a more recent study, RUNX2 level in metastatic ER+ breast cancers was associated with poor outcomes (74). In line with these clinical results, our RUNX2–ER gene signature correlated with gene signatures of metastases and poor outcomes. Our findings have important clinical implications, as a small-molecule inhibitor of the RUNX–CBFβ complex was recently identified (56). One of the genes regulated by the ERα–RUNX2 complex of particular significance is SOX9. SOX9 is a master regulator of embryonic stem cells and more recently was shown to have key roles in the development of metastases in triple-negative and HER2-positive breast cancer. Here, we show that SOX9 is expressed in ERα-positive breast cancers, increases with progression, and plays an important role in endocrine resistance.
In this study, we were also able to extend our findings to endocrine-resistant clinical samples. We found a significant overlap between the ERα–RUNX2 DNA binding sites unique to the TAMR cell line model and the ERα binding sites in ER+ breast cancer clinical samples. Furthermore, we found that the overlap between the TAMR-unique ERα–RUNX2 DNA binding sites and metastatic ERα binding sites was higher compared with the overlap with the ER binding sites in primary treatment-naïve tumor samples. We also detected an association between the ERα–RUNX2 coregulated genes that are up-regulated in TAMR and gene sets of poor prognosis in breast cancer. Lastly, we show that up-regulation of SOX9 expression is linked to TAM resistance in clinical samples. Taken together, these results support the notion that in a subset of breast cancers with acquired TAM resistance, ERα activity is maintained but reprogrammed, leading to an aberrant ERα cistrome. In part, the aberrant chromatin binding is due to the noncanonical activation of ERα in complex with RUNX2. This complex results in the activation of a set of genes that include SOX9 and promote endocrine resistance and metastases. These results highlight the importance of developing improved ERα antagonists as well as agents that target other key proteins in complex with ER and key ER transcriptional targets, such as RUNX2 and SOX9, respectively.
Methods
Cell Culture and Proliferation Assays.
The TAMR and LTED cells were derived from MCF7 cells (MCF7L originally from Marc Lippman’s laboratory, University of Miami, Miami) using methods previously reported (29). Other TAMR cells were derived from 600MPE (originally from Joe Gray’s laboratory, Oregon Health and Science University, Portland, OR) and MDAMB415 (purchased from ATCC). All of the cells were authenticated and regularly tested for mycoplasma contamination. The MCF7 cells were maintained in RPMI/1640 or supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin (P/S). The endocrine-resistant cells were kept in phenol-red free medium supplemented with 10% heat-inactivated charcoal-stripped (CS)-FBS and 1% P/S. The TAMR cells were grown with the addition of 100 nM 4-OH-TAM (H7904, Sigma). All cells were incubated at 37 °C in 5% CO2.
For proliferation assays, the breast cancer cells were plated in 24-well plates (2.5 × 104 per well). At indicated time points, the cells were trypsinized and collected. The number of viable cells was determined by Trypan blue exclusion staining and directly assessed with a hemacytometer using independent triplicates.
Cell Migration–Invasion Assay.
The Radius assay and Boyden Chamber assays were performed following the manufacturer’s instructions (Cell Biolabs).
Western Blotting and Protein Immunoprecipitation.
For Western blot analysis, cells were lysed in 50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Nonidet P-40, and 1% Triton X-100 supplemented with protease inhibitors and subjected to SDS/PAGE. Antibodies used were as follows: ERα (sc-543, Santa Cruz Biotechnology), GATA3 (607102, Biolegend), RUNX2 (D130-3, MBL), SOX9 (Ab5535, EMD Millipore), Beta-Actin (Sigma), GAPDH (Santa Cruz Biotechnology), and HA (Ab9110, Abcam). Protein immunoprecipitation was carried out as previously described (75).
ChIP-Seq.
Chromatin immunoprecipitation (ChIP) experiments were conducted as described previously (76). ChIP-seq reads were aligned to the hg19 genome assembly using Bowtie (77), and ChIP-seq peaks were called using MACS 2.0 (78, 79). Regions of enrichment comparing ChIP and input control signal exceeding q < 0.01 were called as peaks. Read densities were calculated for each peak in reads per million per nucleotide (RPM), which were used for comparison of cistromes across samples. We used SeqPos, available at www.cistrome.org, for motif analysis. Correlation between RNA-seq differential gene expression and transcription factor binding based on ChIP-seq was performed with the BETA basic algorithm (45).
All of the ChIP-seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE86538).
RNA-Seq.
Total RNA was isolated using an RNeasy Mini Kit (Qiagen). RNA-seq libraries were made using the TruSeq RNA Sample Preparation Kit (Illumina) adapted for use on the Sciclone (Perkin-Elmer) liquid handler. Samples were sequenced on an Illumina Nextseq500, and the 75 bp-long reads were aligned to hg19 with STAR aligner. Cufflinks was used to generate the expression value (RPKM) for each gene (80), and the differential expression analysis was performed using the DEseq method. For GO, we used the DAVID website. GSEA was performed using the online tool from the Broad Institute. All of the RNA-seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE86538).
SiRNA Transfection.
SiRNAs were chemically synthesized by Dharmacon Inc. A nonspecific siRNA duplex was used as the negative control. For transfection, cells were seeded in complete medium without antibiotics the day before the experiment. After 24 h, cells were transfected with 50 nM of siRNA, using Lipofectanmine 2000 transfection reagent (Invitrogen), according to the manufacturer’s protocol.
Lentiviral Infection.
For the Dox-inducible RUNX2 MCF7 cells, RUNX2-HA tag cDNA (GeneCopoeia) was transferred to the pInducer 20 destination vector (5) using the Gateway system (Invitrogen). Lentivirus was produced in 293T cells to infect cells in media containing polybrene (8 μg/mL). Cells were selected after the infection with G418.
RIME.
The RIME experiment using the TAMR and parental MCF7 cells was conducted as published previously (81). The RUNX2 antibody (D130-3, MBL) was used for the experiments, and as controls, an IgG antibody (SC-2025, Santa Cruz Biotechnology) was used. MS was performed using an LTQ Velos-Orbitrap MS (Thermo Scientific) coupled to an Ultimate RSLCnano-LC system (Dionex). Raw MS data files were processed using Proteome Discoverer v.1.3 (Thermo Scientific). Processed files were searched against the SwissProt human database using the Mascot search engine version 2.3.0.
Patient Tissue Analysis.
A total of 42 matched pairs of ER+ breast cancer tumors from patients treated with TAM, including treatment-naïve primary tumors and local recurrent tumors that developed TAM treatment, were used. The tissue samples were evaluated on two tissue microarrays (TMAs) obtained from consented patients at the Royal Marsden Hospital, London. More details of these tissue samples were previously published. Immunohistochemical staining of SOX9 was performed on 4-µm sections of the TMA, using the Bond Refine Detection System following the manufacturer’s protocols on the Leica Bond III automated immunostainer. The sections were automatically deparaffinized, and antigen retrieval was done with EDTA buffer (pH 9.0) and processed for 20 min. The slides were incubated with the antibody against SOX9 (8G5, Mouse monoclonal, D130-3, MBL) at a dilution of 1:500. The sections were then treated according to the streptavidin–biotin–peroxidase complex method (Bond Polymer Refine Detection, Leica Microsystems) with diaminobenzidine (DAB) as a chromogen and counterstained with hematoxylin. Incubation with the biotinylated universal secondary antibody was then performed. Visualization was performed with DAB as the chromogen substrate. Testis tissue was used as positive controls for SOX9. Omission of the primary antibody was used as a negative control. Once stained, the TMA slides were scanned on the Olympus BX-51 W1 microscope using Vectra 2.0.8 software (Perkin-Elmer). Cores that were disrupted or contained insufficient tissue were eliminated from the analysis. Following the standard bright-field TMA scanning protocol, a chromogenic spectral library was composed using the spectra of both the counterstain (hematoxylin) and the immunostain (DAB). Tissue segmentation algorithms were subsequently constructed using inForm V2.0.2 (Perkin-Elmer). Initially a training set comprising three classes of tissue was created (i.e., tumor, stroma, and other). Representative regions of interest (ROIs) for each of these classes were marked on 15–20 images from the TMAs. The software was trained on these areas and tested to determine how accurately it could differentiate between the tissue classes. Cell segmentation algorithms were then constructed for the cell nucleus. Cell segmentation algorithms identified nuclei as pixels above the minimum signal value. The spectral library and algorithms were then run on all samples. Poorly segmented cores were manually corrected via touchscreen editing following pathology review. Data generated from every cell per core, where expression of SOX9 was observed, was used to calculate a core mean intensity value for subsequent statistical analysis.
Statistical Analysis.
Statistical analyses were performed using unpaired Student’s t tests, and P values less than 0.05 were considered statistically significant. Error bars in figures represent SEM. For the patient tissue sample analysis, a two-sided (paired Student's t test) was performed.
Study Approval.
The pleural effusion was collected with patient consent and Dana Farber Cancer Institute/Harvard Cancer Center Institutional Review Board approval (protocol 12–259). The tissue samples for the generation of the TMAs were obtained from consented patients at the Royal Marsden Hospital, London.
SI Methods
ChIP-Seq.
Chromatin from ∼1 × 107 fixed cells was sonicated to a size range of 200–300 bp. Solubilized chromatin was subjected to immunoprecipitation with the ER antibody, Ab10 (Lab Vision corporation) and SC-543 (Santa Cruz Biotechnology), or HA antibody (Ab9110, Abcam) bound to protein A and protein G beads (Life Technologies). A fraction of the sample was not exposed to antibody in order to be used as control (input). The samples were reversed cross-linked, treated with proteinase K, and DNA was extracted. DNA sequencing libraries were prepared using the ThruPLEX-FD Prep Kit (Rubicon Genomics). Libraries were sequenced using 50-bp reads on the Illumina platform at the Dana-Farber Cancer Institute.
ChIP-seq data for the patient tumor samples were downloaded from GSE32222. For the pleural effusion sample, the pleural effusion was collected with patient consent and DFCI/HCC IRB approval (protocol 12–259). Epithelial cells were isolated after a Percoll gradient to remove the red blood cells and dead cells using BerEp4 beads (Dynal). Immunofluorescence was done as previously published using the ERα antibody (sc-543, Santa Cruz Biotechnology) and Epcam antibody (Abcam-Ab71916).
Overlap significance was calculated based on 104 random permutations. In each permutation, peaks from the ChIP-seq datasets were randomly shuffled through the whole hg19 genome. Then count overlaps with the specific peaks in the respective cistromes were calculated after 104 permutations, and we determined the significance of the mean value of empirical random distribution of overlapped peaks (P < 0.001).
RT-PCR.
Total RNA was isolated using the RNeasy kit (Qiagen). cDNA was prepared with the high capacity RT kit (Applied Biosystems), and PCR was carried out with SYBR Green (Qiagen). The following primers were used in this study:
ESR1 forward: 5′-CAAGCCCATGGAACATTTCT-3′;
ESR1 reverse: 5′-TCCCTTGGATCTGATGCAGT-3′;
RUNX2 forward: 5′-AACCCTCAGCCTCAGAGTCA-3′;
RUNX2 reverse: 5′-CAATGGATCCCAGGTATTGG-3′;
SOX9 forward: 5′-AGTACCCGCACTTGCACAAC-3′;
SOX9 reverse: 5′-CGTTCTTCACCGACTTCCTC-3′;
EDN1 forward: 5′-AGTACCCGCACTTGCACAAC-3′;
EDN1 reverse: 5′-CGTTCTTCACCGACTTCCTC-3′;
JAG1 forward: 5′-GTCCCACTGGTTTCTCTGGA-3′;
JAG1 reverse: 5′-GGCAGTGGTCTTTCAGGTGT-3′;
EDN1 forward: 5′-TCTCTGCTGTTTGTGGCTTG-3′;
EDN1 reverse: 5′-GATGTCCAGGTGGCAGAAGT-3′;
MMP9 forward: 5′-GAGACCGGTGAGCTGGATAG-3′;
MMP9 reverse: 5′-TACACGCGAGTGAAGGTGAG-3′;
MMP13 forward: 5′-GGTTCCTGATGTGGGTGAAT-3′;
MMP13 reverse: 5′-CAATGCCATCGTGAAGTCTG-3′;
GAPDH forward: 5′-CGAGA TCCCTCCAAAA TCAA-3′; and
GAPDH reverse: 5′-TTCACACCCATGACGAACAT-3′.
Droplet Digital-PCR.
TaqMan PCR mixtures were assembled from a Mastermix (Bio-Rad). The PCR product was subsequently loaded onto the Automated Droplet Generator (Bio-Rad). The final product was read on the QX-100 droplet reader (Bio-Rad). Analysis of the droplet digital-PCR data was performed with QuantaSoft analysis software (Bio-Rad) that accompanied the droplet reader. The following primers were used: for the ESR1 E380Q assay, forward 5′-TGGATTTGACCCTCCATGATCAG-3′, reverse 5′-AGACGAGACCAATCATCAGGATCT-3′; probe sequences are 5′-VIC-CCACCTTCTAGAATGTG-MGB-NFQ-3′ and 5′-FAM-CCACCTTCTACAATGTG-MGB-NFQ-3′; for ESR1 Y537C assay, forward 5′-CAGCATGAAGTGCAAGAACGT-3′, reverse 5′-TGGGCGTCCAGCATCTC-3′; probe sequences are 5′-VIC-TGCCCCTCTATGACCTG-MGB-NFQ-3′ and 5′-FAM-CCCCTCTGTGACCTG-MGB-NFQ-3′; for the ESR1 Y537N assay, forward 5′-CTGTACAGCATGAAGTGCAAGAAC-3′, reverse 5′-TGGGCGTCCAGCATCTC-3′; probe sequences are 5′-VIC-TGGTGCCCCTCTATGAC-MGB-NFQ-3′ and 5′-FAM-TGCCCCTCAATGAC-MGB-NFQ-3′; for the ESR1 Y537S assay, forward 5′-CAGCATGAAGTGCAAGAACGT-3′ and reverse 5′-TGGGCGTCCAGCATCTC-3′; probe sequences are 5′-VIC- CCCCTCTATGACCTGC-MGB-NFQ-3′ and 5′-FAM-CCCTCTCTGACCTGC-MGB-NFQ-3′; for the ESR1 D538G assay, forward 5′-CAGCATGAAGTGCAAGAACGT-3′ and reverse 5′-TGGGCGTCCAGCATCTC-3′; probe sequences are 5′-VIC-CCCCTCTATGACCTGCT-MGB-NFQ-3′ and 5′-FAM-CCCTCTATGGCCTGCT-MGB-NFQ-3′.
Acknowledgments
This study was supported in part by grants from Susan G. Komen for the Cure (to M. Brown), Breast Cancer SPORE Career Development Award (to R.J.), and Royal Marsden National Institute for Health Research Biomedical Research Centre (to M.D.); the Breast Cancer Research Foundation (R.S.); the National Cancer Institute Dan L. Duncan Comprehensive Cancer Center Grant P30CA125123 (to R.S.); Susan G. Komen for the Cure Promise Grant PG12221410 (to R.S.); a Stand Up 2 Cancer Dream Team Translational Research Grant (SU2C-AACR-DT0409); the Cancer Prevention Research Institute of Texas (CPRIT Award RP140102) and Baylor College of Medicine Comprehensive Cancer Training Program (CDA); and the Department of Defense Breakthrough Award W81XWH-14-1-0326 (to X.F.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE86538).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620993114/-/DCSupplemental.
References
- 1.Lerner HJ, Band PR, Israel L, Leung BS. Phase II study of tamoxifen: Report of 74 patients with stage IV breast cancer. Cancer Treat Rep. 1976;60:1431–1435. [PubMed] [Google Scholar]
- 2.Morgan LR, Jr, et al. Therapeutic use of tamoxifen in advanced breast cancer: Correlation with biochemical parameters. Cancer Treat Rep. 1976;60:1437–1443. [PubMed] [Google Scholar]
- 3.Ingle JN, et al. Randomized clinical trial of diethylstilbestrol versus tamoxifen in postmenopausal women with advanced breast cancer. N Engl J Med. 1981;304:16–21. doi: 10.1056/NEJM198101013040104. [DOI] [PubMed] [Google Scholar]
- 4.Ingle JN, et al. Randomized trial of bilateral oophorectomy versus tamoxifen in premenopausal women with metastatic breast cancer. J Clin Oncol. 1986;4:178–185. doi: 10.1200/JCO.1986.4.2.178. [DOI] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 6.Lindström LS, et al. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–2608. doi: 10.1200/JCO.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- 7.Curigliano G, et al. Should liver metastases of breast cancer be biopsied to improve treatment choice? Ann Oncol. 2011;22:2227–2233. doi: 10.1093/annonc/mdq751. [DOI] [PubMed] [Google Scholar]
- 8.Gong Y, Han EY, Guo M, Pusztai L, Sneige N. Stability of estrogen receptor status in breast carcinoma: A comparison between primary and metastatic tumors with regard to disease course and intervening systemic therapy. Cancer. 2011;117:705–713. doi: 10.1002/cncr.25506. [DOI] [PubMed] [Google Scholar]
- 9.Torres-Arzayus MI, Zhao J, Bronson R, Brown M. Estrogen-dependent and estrogen-independent mechanisms contribute to AIB1-mediated tumor formation. Cancer Res. 2010;70:4102–4111. doi: 10.1158/0008-5472.CAN-09-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massarweh S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 11.Miller TW, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010;120:2406–2413. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Reports. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X, et al. FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc Natl Acad Sci USA. 2016;113:E6600–E6609. doi: 10.1073/pnas.1612835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perey L, et al. Clinical benefit of fulvestrant in postmenopausal women with advanced breast cancer and primary or acquired resistance to aromatase inhibitors: Final results of phase II Swiss Group for Clinical Cancer Research Trial (SAKK 21/00) Ann Oncol. 2007;18:64–69. doi: 10.1093/annonc/mdl341. [DOI] [PubMed] [Google Scholar]
- 16.Ingle JN, et al. North Central Cancer Treatment Group Trial N0032 Fulvestrant in women with advanced breast cancer after progression on prior aromatase inhibitor therapy: North Central Cancer Treatment Group Trial N0032. J Clin Oncol. 2006;24:1052–1056. doi: 10.1200/JCO.2005.04.1053. [DOI] [PubMed] [Google Scholar]
- 17.Di Leo A, et al. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2010;28:4594–4600. doi: 10.1200/JCO.2010.28.8415. [DOI] [PubMed] [Google Scholar]
- 18.Mehta RS, et al. Combination anastrozole and fulvestrant in metastatic breast cancer. N Engl J Med. 2012;367:435–444. doi: 10.1056/NEJMoa1201622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies C, et al. Adjuvant Tamoxifen: Longer Against Shorter (ATLAS) Collaborative Group Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet. 2013;381:805–816. doi: 10.1016/S0140-6736(12)61963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goss PE, et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349:1793–1802. doi: 10.1056/NEJMoa032312. [DOI] [PubMed] [Google Scholar]
- 21.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 23.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krum SA, et al. Unique ERalpha cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22:2393–2406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupien M, et al. Growth factor stimulation induces a distinct ER(alpha) cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24:2219–2227. doi: 10.1101/gad.1944810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis PA, Regan MM, Fleming GF. Adjuvant ovarian suppression in premenopausal breast cancer. N Engl J Med. 2015;372:1673. doi: 10.1056/NEJMc1502618. [DOI] [PubMed] [Google Scholar]
- 28.Burstein HJ, et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: American Society of Clinical Oncology clinical practice guideline update on ovarian suppression. J Clin Oncol. 2016;34:1689–1701. doi: 10.1200/JCO.2015.65.9573. [DOI] [PubMed] [Google Scholar]
- 29.Morrison G, et al. Therapeutic potential of the dual EGFR/HER2 inhibitor AZD8931 in circumventing endocrine resistance. Breast Cancer Res Treat. 2014;144:263–272. doi: 10.1007/s10549-014-2878-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottardis MM, Jiang SY, Jeng MH, Jordan VC. Inhibition of tamoxifen-stimulated growth of an MCF-7 tumor variant in athymic mice by novel steroidal antiestrogens. Cancer Res. 1989;49:4090–4093. [PubMed] [Google Scholar]
- 31.Agrawal A, Robertson JF, Cheung KL. Clinical relevance of “withdrawal therapy” as a form of hormonal manipulation for breast cancer. World J Surg Oncol. 2011;9:101. doi: 10.1186/1477-7819-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W, et al. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 35.Eeckhoute J, et al. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 36.Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013;23:12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouros-Mehr H, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blyth K, Cameron ER, Neil JC. The RUNX genes: Gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 39.Pratap J, et al. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selvamurugan N, Kwok S, Partridge NC. Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth factor-beta1-stimulated collagenase-3 expression in human breast cancer cells. J Biol Chem. 2004;279:27764–27773. doi: 10.1074/jbc.M312870200. [DOI] [PubMed] [Google Scholar]
- 41.Li XQ, et al. ITGBL1 is a Runx2 transcriptional target and promotes breast cancer bone metastasis by activating the TGFβ signaling pathway. Cancer Res. 2015;75:3302–3313. doi: 10.1158/0008-5472.CAN-15-0240. [DOI] [PubMed] [Google Scholar]
- 42.Sharp JA, Waltham M, Williams ED, Henderson MA, Thompson EW. Transfection of MDA-MB-231 human breast carcinoma cells with bone sialoprotein (BSP) stimulates migration and invasion in vitro and growth of primary and secondary tumors in nude mice. Clin Exp Metastasis. 2004;21:19–29. doi: 10.1023/b:clin.0000017167.17065.61. [DOI] [PubMed] [Google Scholar]
- 43.El-Gendi SM, Mostafa MF. Runx2 expression as a potential prognostic marker in invasive ductal breast carcinoma. Pathol Oncol Res. 2016;22:461–470. doi: 10.1007/s12253-015-0018-5. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Zhang B, Liu B, Xie Y, Cao X. Combined Runx2 and Snail overexpression is associated with a poor prognosis in breast cancer. Tumour Biol. 2015;36:4565–4573. doi: 10.1007/s13277-015-3101-3. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pande S, et al. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J Cell Physiol. 2013;228:1784–1792. doi: 10.1002/jcp.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen-Solal KA, Boregowda RK, Lasfar A. RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol Cancer. 2015;14:137. doi: 10.1186/s12943-015-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhodes DR, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammed H, et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 51.Anzick SL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 52.Hanstein B, et al. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 54.Tan SK, et al. AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011;30:2569–2581. doi: 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Reports. 2016;17:2715–2723. doi: 10.1016/j.celrep.2016.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Illendula A, et al. Small molecule inhibitor of CBFβ-RUNX binding for RUNX transcription factor driven cancers. EBioMedicine. 2016;8:117–131. doi: 10.1016/j.ebiom.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jo A, et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malladi S, et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell. 2016;165:45–60. doi: 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metzger-Filho O, et al. Patterns of recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: Results from international breast cancer study group trials VIII and IX. J Clin Oncol. 2013;31:3083–3090. doi: 10.1200/JCO.2012.46.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30:356–363. doi: 10.1016/j.tig.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergamaschi A, et al. The forkhead transcription factor FOXM1 promotes endocrine resistance and invasiveness in estrogen receptor-positive breast cancer by expansion of stem-like cancer cells. Breast Cancer Res. 2014;16:436. doi: 10.1186/s13058-014-0436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, Balasubramanian S. Genome-wide mapping of FOXM1 binding reveals co-binding with estrogen receptor alpha in breast cancer cells. Genome Biol. 2013;14:R6. doi: 10.1186/gb-2013-14-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stender JD, et al. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Browne G, et al. Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J Cell Physiol. 2015;230:2522–2532. doi: 10.1002/jcp.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janes KA. RUNX1 and its understudied role in breast cancer. Cell Cycle. 2011;10:3461–3465. doi: 10.4161/cc.10.20.18029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khalid O, et al. Modulation of Runx2 activity by estrogen receptor-alpha: Implications for osteoporosis and breast cancer. Endocrinology. 2008;149:5984–5995. doi: 10.1210/en.2008-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Owens TW, et al. Runx2 is a novel regulator of mammary epithelial cell fate in development and breast cancer. Cancer Res. 2014;74:5277–5286. doi: 10.1158/0008-5472.CAN-14-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taipaleenmäki H, et al. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75:1433–1444. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tandon M, Chen Z, Pratap J. Runx2 activates PI3K/Akt signaling via mTORC2 regulation in invasive breast cancer cells. Breast Cancer Res. 2014;16:R16. doi: 10.1186/bcr3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDonald L, et al. RUNX2 correlates with subtype-specific breast cancer in a human tissue microarray, and ectopic expression of Runx2 perturbs differentiation in the mouse mammary gland. Dis Model Mech. 2014;7:525–534. doi: 10.1242/dmm.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.El-Gendi SM, Mostafa MF. Runx2 expression as a potential prognostic marker in invasive ductal breast carcinoma. Pathol Oncol Res. 2016;22:461–470. doi: 10.1007/s12253-015-0018-5. [DOI] [PubMed] [Google Scholar]
- 75.Ni M, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohammed H, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Reports. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]