Significance
DNA methylation is generally considered an epigenetic mark for transcriptional gene silencing. In this work, we generated loss-of-function mutant alleles of SlDML2. We characterized the mutant fruits that failed to ripen and discovered that SlDML2 is required for the demethylation and activation of genes important for fruit ripening, including genes involved in fruit pigment and flavor synthesis, ethylene synthesis and signaling, and cell wall hydrolysis. Unexpectedly, we found that SlDML2-mediated DNA demethylation is also necessary for fruit ripening-induced repression of hundreds of genes involved in photosynthesis and cell wall synthesis and organization. Our study has therefore revealed a broad and critical role of DNA methylation as an activation mark for the expression of many genes in a eukaryotic organism.
Keywords: DNA demethylase, 5-mC DNA glycosylase, DNA methylation, epigenetic regulation, gene silencing
Abstract
DNA methylation is a conserved epigenetic mark important for genome integrity, development, and environmental responses in plants and mammals. Active DNA demethylation in plants is initiated by a family of 5-mC DNA glycosylases/lyases (i.e., DNA demethylases). Recent reports suggested a role of active DNA demethylation in fruit ripening in tomato. In this study, we generated loss-of-function mutant alleles of a tomato gene, SlDML2, which is a close homolog of the Arabidopsis DNA demethylase gene ROS1. In the fruits of the tomato mutants, increased DNA methylation was found in thousands of genes. These genes included not only hundreds of ripening-induced genes but also many ripening-repressed genes. Our results show that SlDML2 is critical for tomato fruit ripening and suggest that active DNA demethylation is required for both the activation of ripening-induced genes and the inhibition of ripening-repressed genes.
DNA methylation is a conserved epigenetic modification that is generally associated with inactive transcription in plants and mammals. As such, DNA methylation plays important roles in many biological processes, such as genome stability, gene imprinting, development, and response to the environment (1–3). In contrast to mammals, in which DNA methylation predominantly occurs at cytosines in the symmetric CG sequence context, plants commonly have methylation in the asymmetrical CHH sequence context (H = A, C, or T), as well as in the symmetrical CG and CHG contexts (1, 2). In plants, cytosines in all sequence contexts can be de novo methylated through the well-known RNA-directed DNA methylation pathway (RdDM), in which 24-nt siRNAs guide the DNA methyltransferase domains rearranged methyltransferase 2 (DRM2) to methylate target loci (4). DNA methylation can be maintained during replication; mCG and mCHG are maintained by the DNA methyltransferases DNA methyltransferase 1 (MET1) and chromomethylase 3 (CMT3), respectively, whereas mCHH is maintained by CMT2 and RdDM (4, 5).
Cytosine methylation levels are dynamically regulated by DNA methylation and demethylation reactions (3, 6). DNA methylation can be lost either because of failure in maintaining methylation after replication (i.e., passive DNA demethylation) or because of active removal by enzymes (i.e., active DNA demethylation). Previous studies have identified and characterized several enzymes important for active DNA demethylation in Arabidopsis (7–11). The ROS1 family of bifunctional 5-methylcytosine DNA glycosylases/lyases, often referred to as DNA demethylases, initiate active DNA demethylation by removing the methylcytosine base from the DNA backbone, resulting in a single nucleotide gap that can be filled with an unmethylated cytosine through a base excision repair pathway (7, 8, 12, 13). Several enzymes acting downstream of ROS1, such as the 3′ DNA phosphatase ZDP, AP endonuclease-like protein APE1L, and DNA ligase I (AtLIG1), have also been identified (9–11). Because the ROS1 family of enzymes function in the first step of the DNA demethylation pathway, their targeting to specific genomic sequences determines the patterns of DNA methylation removal. Recent studies revealed a histone acetyltransferase complex that regulates DNA demethylation by facilitating the targeting of ROS1 (14–17).
Because they prevent DNA hypermethylation, enzymes and regulatory factors involved in DNA demethylation are regarded as antisilencing factors. Expression of some transgenes, endogenous genes, and transposable elements (TEs) is reduced in ros1 mutants because of DNA hypermethylation (14, 16, 18, 19). In Arabidopsis, ROS1 preferentially targets TEs (20). When ROS1-targeted TEs are located in the promoter regions of genes, ROS1 can protect gene expression by demethylating the adjacent TEs. In Arabidopsis, ros1 mutants have an abnormal epidermal cell organization due to hypermethylation of a TE located in the promoter region of EPIDERMAL PATTERNING FACTOR 2, which encodes a negative regulator of stomata formation (21). In addition, Arabidopsis DNA demethylase mutants show enhanced susceptibility to fungal infection due to hypermethylation of TEs located in pathogen-responsive genes (22). These studies have demonstrated that, by demethylating nearby TEs, active DNA demethylation is critical for gene activation or for the prevention of silencing.
To date, active DNA demethylation in plants has been studied mostly in Arabidopsis. Arabidopsis mutants defective in DNA demethylation have been important in understanding the mechanism and functions of active DNA demethylation. Despite the many advantages of Arabidopsis, the use of this model plant is limited by its lack of some agronomically important processes, such as fiber growth in cotton and ripening of fleshy fruits, such as tomato. Tomato (Solanum lycopersicum) is an economically important crop and a model for studying fleshy fruit ripening. Fruit ripening is associated with distinct physiological, biochemical, and structural changes, such as the accumulation of sugars, flavor volatiles, and pigments, and the hydrolysis of cell walls (23). These changes are driven by phytohormones and developmental factors. Ethylene is one of the most important factors promoting ripening, especially for climacteric fruits, which show a rapid increase in ethylene level during ripening (23). In addition, several transcription factors (TFs), including RIPENING INHIBITOR (RIN), NON-RIPENING (NOR), and COLORLESS NON-RIPENING (CNR), have been identified as major ripening regulators. These regulators function upstream of both ethylene-dependent and -independent ripening pathways by directly and indirectly regulating the expression of many ripening-related genes (24–26). Evidence has recently emerged that, in addition to ethylene and the TFs, DNA methylation is another key regulator of fruit ripening (27). Furthermore, a recent study revealed that DNA methylation is associated with chilling-induced flavor loss in tomato fruits (28). Tomato fruits undergo a dramatic loss in DNA methylation during ripening (27). The application of a DNA methylation inhibitor facilitated ripening, and RNAi-mediated down-regulation of putative DNA demethylases inhibited fruit ripening in tomato, suggesting that active DNA demethylation plays an important role in regulating fruit ripening (27, 29). It remains unclear, however, how active DNA demethylation contributes to ripening and ripening-associated changes in DNA methylation patterns.
Tomato contains four putative DNA demethylases (SlDML1 to -4) according to sequence homology with the Arabidopsis DNA demethylases, and two of them, SlDML1 and SlDML2, are most closely related to the Arabidopsis ROS1 (AtROS1) (29, 30). A previous study used RNAi against the conserved HhH-GPD domain to investigate the function of SlDMLs in tomato (29). The RNAi lines showed an altered expression of all four SlDMLs, with SlDML2, the most abundant DNA demethylase in fruits, showing the most dramatic reduction in expression. Fruits of the RNAi plants displayed a delayed ripening phenotype, suggesting critical roles of the SlDMLs in fruit ripening. However, because the RNAi lines were not specific to a particular SlDML, it is unclear which SlDML(s) is required for fruit ripening. In addition, the genome-wide effect of active DNA demethylation during fruit ripening has not been determined thus far.
In the present study, we generated two mutant alleles of SlDML2 in the tomato cultivar (cv.) Micro-Tom using CRISPR/Cas9-mediated gene editing. To determine the genome-wide effect of SlDML2 on DNA demethylation during fruit ripening, we compared the DNA methylomes of WT and sldml2 mutant fruits. We found that SlDML2 is responsible for the demethylation of as many as 29,764 genomic regions, which are preferentially distributed in chromosomal arms, and is required for virtually all ripening-induced DNA demethylation. Our transcriptome analysis of sldml2 mutant fruits suggested that SlDML2 is necessary for the activation of hundreds of ripening-related genes, such as RIN, and genes involved in ethylene and pigment synthesis and cell wall hydrolysis. Unexpectedly, our analysis also revealed that SlDML2-mediated DNA demethylation is required for the repression of hundreds of genes during fruit ripening. These repressed genes are involved in processes such as photosynthesis and cell wall synthesis, and their repression correlates with DNA hypomethylation in their promoter regions. Thus, our study documents a broad silencing role of DNA demethylation in gene regulation.
Results
Generation of Stable Loss-of-Function Mutant Alleles of SlDML2 Using the CRISPR/Cas9 Gene-Editing System.
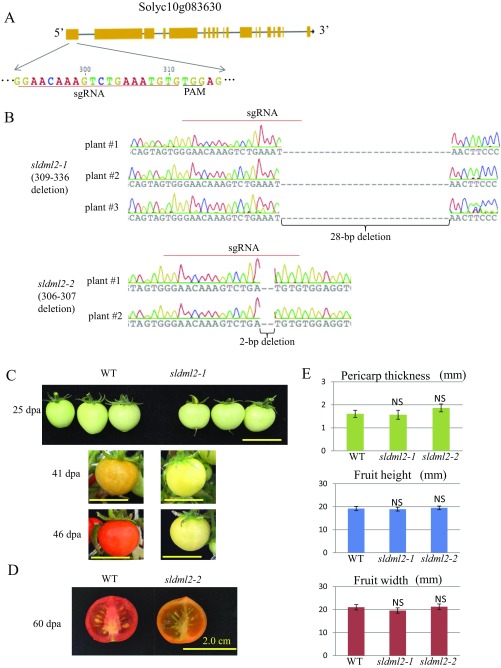
During tomato fruit ripening, DNA demethylation occurs in numerous genomic regions (27). In tomato, there are four annotated 5-mC DNA glycosylase/DNA demethylase genes; one of them, SlDML2, is highly induced during fruit ripening (29, 30). A recent study using RNAi lines of the demethylases suggested that SlDMLs-mediated active DNA demethylation is important for fruit ripening (29). To better define the role of SlDML2 in active DNA demethylation and fruit ripening, we generated stable loss-of-function sldml2 mutants using the CRISPR/Cas9 gene-editing system. A single guide RNA (sgRNA) was designed to specifically target the first exon of SlDML2 (Fig. S1A). We cloned the sgRNA sequence into a binary vector that contains sgRNA and Cas9 expression cassettes (31), and the resulting construct was transformed into WT tomato cv. Micro-Tom using Agrobacterium infection of leaf explants.
Fig. S1.
Generation and characterization of tomato sldml2 mutants. (A) Diagram showing the sequence of single guide RNA (sgRNA) for targeting the first exon of SlDML2. PAM, protospacer adjacent motif. (B) Genotyping of mutations in sldml2-1 and sldml2-2. T0 plants were genotyped by sequencing genomic regions flanking the sgRNA. Three sldml2-1 plants (Upper) had a homozygous 28-bp deletion, and two sldml2-2 plants (Lower) had a homozygous 2-bp deletion in the first exon of SlDML2. The red line indicates the position of the sgRNA. (C) Fruit phenotypes of the WT and sldml2-1 at 25 dpa, 41 dpa, and 46 dpa. (Scale bars: 2 cm.) (D) Photograph of the inside of fruits of the WT and sldml2-2 at 60dpa. Scale bars: 2 cm. (E) Fruit sizes of the WT, sldml2-1, and sldml2-2 plants. Fruit height and width and the thickness of pericarp were measured at 46 dpa. Fruits of sldml2-1 and sldml2-2 were not significantly different from those of the WT (n = 6–8; *P value < 0.01, two-tailed t test; NS, not significant).
A total of 30 transgenic plants regenerated from tissue culture were genotyped through direct sequencing of PCR products from genomic DNA flanking the target site. We found 2- to 56-bp deletion mutations in 18 plants. Three of the plants carried the same homozygous 28-bp deletion, and two of the plants had the same homozygous 2-bp deletion in the first exon of SlDML2 (Fig. S1B). Both the 28-bp and 2-bp deletion mutations are predicted to cause premature stop codons in the first exon of SlDML2. We did not find any off-target editing events in any of the other three DNA demethylase genes. We refer to the mutant with the 28-bp deletion as sldml2-1 and that with the 2-bp deletion as sldml2-2 (Fig. S1B).
The sldml2 Mutations Inhibit Fruit Ripening.
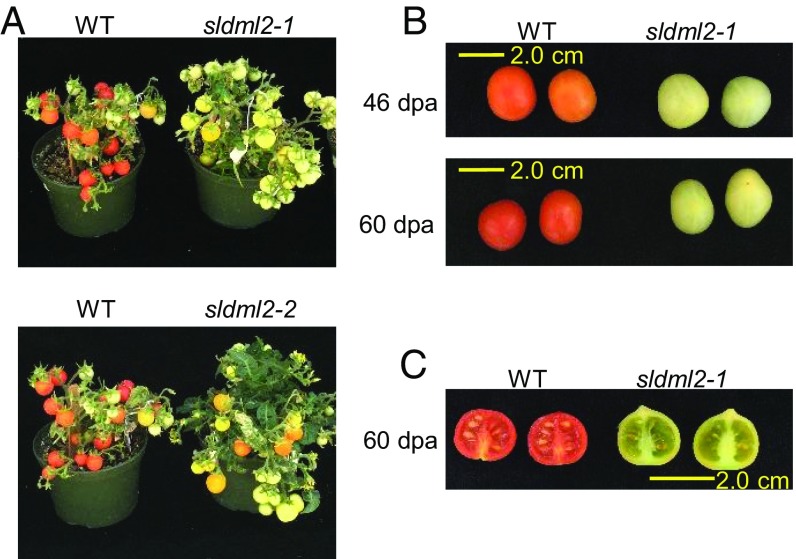
To exclude the effect of the tissue culture process, we also regenerated from tissue culture the WT plants that were used as controls for the sldml2 mutants in this study. Compared with fruit ripening in the WT plants, fruit ripening in the sldml2-1 mutants was dramatically inhibited (Fig. 1A). We compared fruits of the WT and sldml mutants at four stages: 25 d after pollination (dpa), 41 dpa, 46 dpa, and 60 dpa. Fruits of the WT turned red by 46 dpa, but fruits of the sldml2-1 mutants remained green at all stages (Fig. 1B and Fig. S1C). The sldml2-2 mutation had a similar inhibitory effect on fruit ripening (Fig. 1A).
Fig. 1.
Fruit-ripening phenotypes of tomato sldml2 mutants. (A) Plants of the WT (cv. Micro-Tom), sldml2-1, and sldml2-2 at the same stage. All plants were from the T0 generation. (B) Fruits of the WT and sldml2-1 at 46 dpa and 60 dpa. (C) Photograph of the inside of fruits of the WT and sldml2-1 at 60 dpa.
The fruits of the sldml2-1 and sldml2-2 mutants contained seeds although they seemed to contain fewer seeds than the WT fruits (Fig. 1C and Fig. S1D). At 60 dpa, fruits of the WT and sldml2 mutants were similar in height and in the width and thickness of the pericarp (Fig. S1E), which is consistent with the fact that the induction of SlDML2 expression occurs after the breaker stage, when fruits have already reached full size (29). Our results are consistent with the previous study that used RNAi lines (29) and demonstrate that SlDML2 is critical for tomato fruit ripening.
The sldml2 Mutations Cause Genome-Wide DNA Hypermethylation.
To investigate the genetic function of SlDML2 in DNA demethylation during fruit ripening, we performed whole-genome bisulfite sequencing to generate single-base resolution maps of DNA methylation in the following fruits: WT at 25 dpa (WT-25dpa), sldml2-1 at 25 dpa (sldml2-1-25dpa), WT at 46 dpa (WT-46dpa), and two biological replicates of sldml2-1 at 46 dpa (sldml2-1-46dpa). Over 95% of the genomic cytosines were covered in each sample, and each methylome was sequenced with >10.7-fold coverage per DNA strand (Table S1). The coverage and depth of these methylomes were higher than those of published methylomes of Solanum lycopersicum cv. Ailsa Craig (AC) (27).
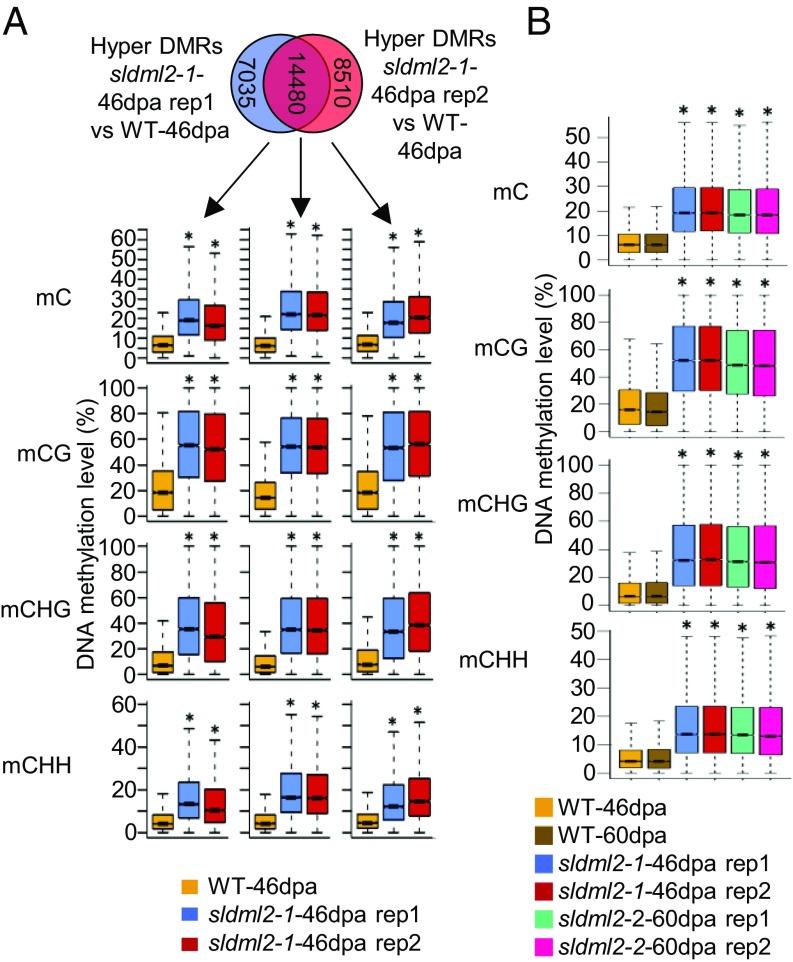
To begin to identify SlDML2 targets in fruits, we compared the methylome of WT-46dpa with those of two biological replicates of sldml2-1-46dpa. Our analysis identified 21,515 hyper-DMRs (Table S2) and 6,643 hypo-DMRs in sldml2-1-46dpa replication 1 (rep1), and 23,026 hyper-DMRs (Table S3) and 6,689 hypo-DMRs in sldml2-1-46dpa rep2. The overwhelmingly higher numbers of hyper-DMRs than hypo-DMRs in the sldml2 mutant are consistent with the presumed function of SlDML2 in DNA demethylation. To investigate the role of SlDML2 in DNA demethylation, we focused on hyper-DMRs in the sldml2 mutant. There were 14,480 hyper-DMRs shared between the two biological replicates of sldml2-1-46dpa (Fig. 2A). For hyper-DMRs unique to sldml2-1-46dpa rep1, we also observed increased DNA methylation in sldml2-1-46dpa rep2, and, similarly, for hyper-DMRs unique to sldml2-1-46dpa rep2, we also observed increased DNA methylation in sldml2-1-46dpa rep1 (Fig. 2A), suggesting that the number of overlapped hyper-DMRs was underestimated because of the artificial cutoff used in defining the DMRs. We therefore used the combined hyper-DMRs (29,764 hyper-DMRs, using bedtools merge) of the two biological replicates as SlDML2 targets in the following analysis.
Fig. 2.
sldml2 mutations cause genome-wide DNA hypermethylation in ripening tomato fruits. (A) DNA methylation levels of sldml2-1 hyper-DMRs in fruits at 46 dpa. (Upper) Hyper-DMRs in two biological replicates of sldml2-1-46dpa relative to WT-46dpa were identified and compared using a Venn diagram. (Lower) Boxplot analysis of mC, mCG, mCHG, and mCHH levels of different groups of the hyper-DMRs. (B) Boxplots showing DNA methylation levels at combined sldml2-1 hyper-DMRs in WT-46dpa, WT-60dpa, two biological replicates of sldml2-1-46dpa, and two biological replicates of sldml2-2-60dpa. WT-46dpa served as control for sldml2-1-46dpa, and WT-60dpa served as control for sldml2-2-60dpa (*P value < 2.2e−16, one-tailed t test).
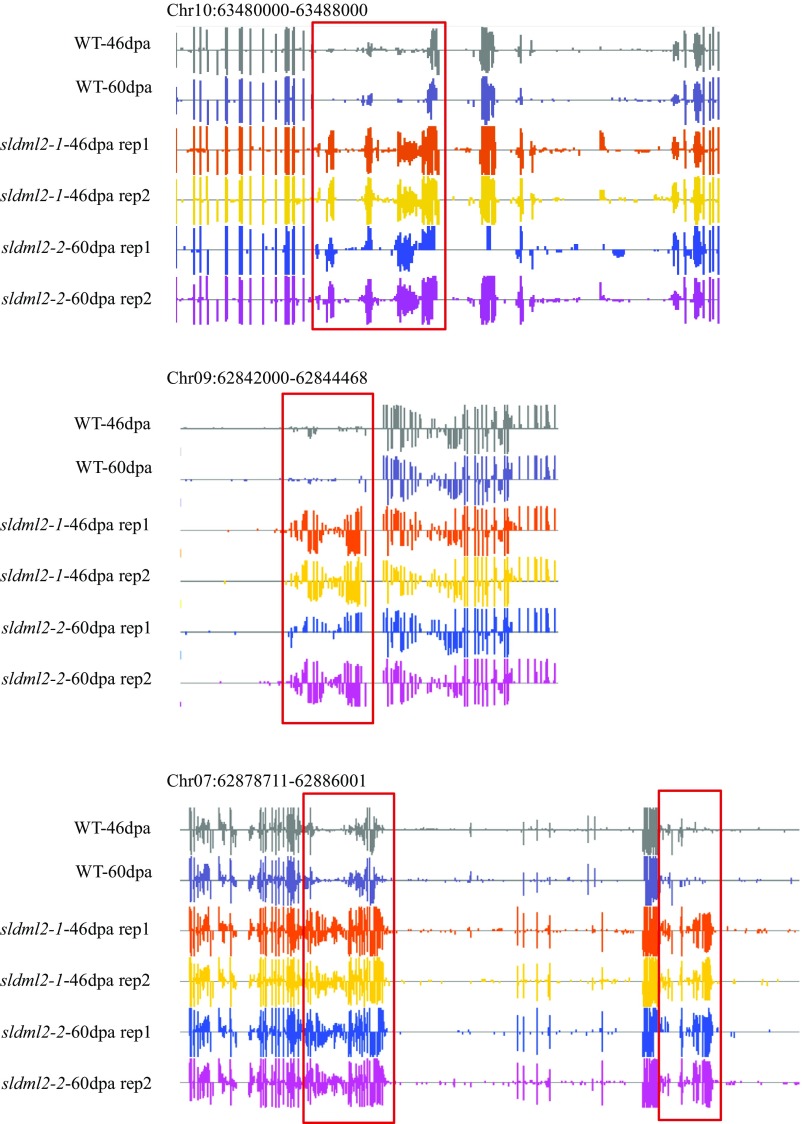
The sldml2 hyper-DMRs contain 1.18 million hyper-DMCs and cover a total length of 17.67 Mb of the genomic sequence (with an average length of 594 bp per DMR). As shown in Fig. 2A and consistent with features of AtROS1 targets in Arabidopsis (20), DNA hypermethylation in sldml2 mutant fruits occurred in all three sequence contexts: mCG, mCHG, and mCHH. To confirm the function of SlDML2 in DNA demethylation, we examined DNA methylation levels of the other mutant allele of SlDML2, sldml2-2, and found that the identified SlDML2 targets in sldml2-1 have similar increases in mCG, mCHG, and mCHH levels in sldml2-2 at 60 dpa compared with the WT at 60 dpa (Fig. 2B). Several examples of sldml2 hyper-DMRs are shown in Fig. S2. Even though the methylation level of sldml2-1 was measured at 46 dpa and the methylation level of sldml2-2 was measured at 60 dpa, the similar increases in DNA methylation in the two mutants are consistent with the inference that the hyper-DMRs of sldml2-1 represent the genuine targets of SlDML2.
Fig. S2.
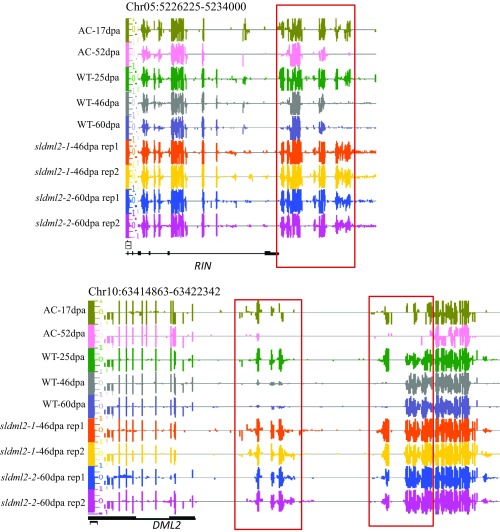
DNA methylation levels of several sldml2 hyper-DMRs (boxed regions) in WT-46dpa, WT-60dpa, sldml2-1-46dpa, and sldml2-2-60dpa. Shown are screenshots from the Integrative Genome Browser (IGB) display of whole-genome bisulfite sequencing data. Vertical bars on each track indicate DNA methylation levels. Bisulfite sequencing results of two replicates of sldml2-1-46dpa and sldml2-2-60dpa are displayed.
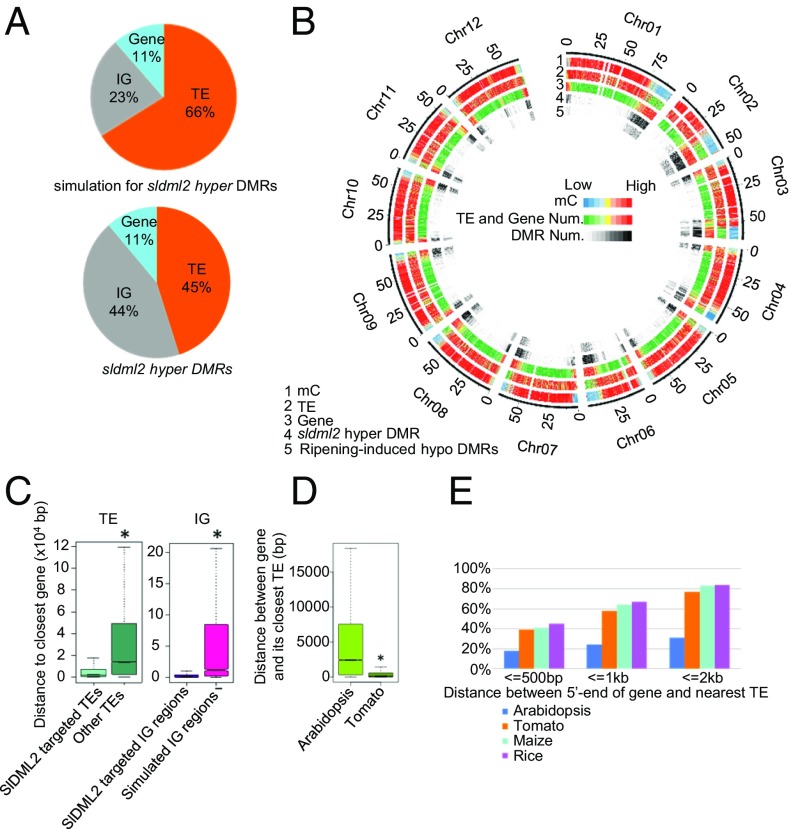
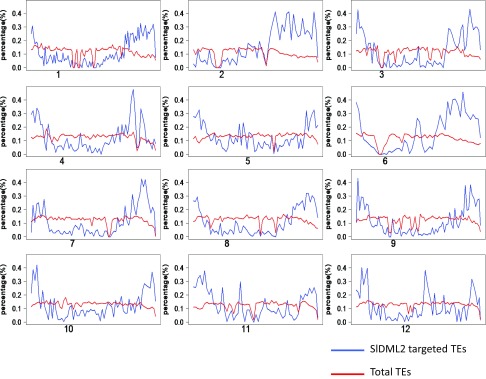
In Arabidopsis, ROS1 preferentially targets TEs (20). Among the simulated genomic regions in tomato, 66.1% are in TEs, 22.5% in intergenic regions (IGs), and 11.4% in genic regions. Among the SlDML2 targets, 45.0% are in TEs, 43.7% in IGs, and 11.3% in genic regions (Fig. 3A). These data suggest that SlDML2 preferentially targets IG regions and TEs in tomato. In both Arabidopsis and tomato, DNA methylation and TEs are mostly distributed in pericentromeric regions (27) (Fig. 3B). However, in contrast to AtROS1 targets that are concentrated around pericentromeric regions (20), sldml2 hyper-DMRs aggregate in chromosomal arms (Fig. 3B). These results indicate that SlDML2 may preferentially target TEs located in euchromatin regions in tomato. This conclusion is also supported by analysis of the density of SlDML2-targeted TEs and total TEs across chromosomes (Fig. S3). We found that SlDML2-targeted TEs are closer to genes than are TEs that are not targeted by SlDML2 (Fig. 3C). Similarly, SlDML2-targeted IG regions are also closer to genes than are simulated IG regions (Fig. 3C). These results suggest that, during fruit ripening, SlDML2 may have a function similar to that of AtROS1 in preventing gene silencing by demethylating nearby TEs or highly methylated intergenic regions. However, in contrast to AtROS1, which targets TEs in both euchromatic and heterochromatic regions (20), SlDML2 preferentially targets TEs in euchromatic regions.
Fig. 3.
Characterization of the DNA methylomes of ripening sldml2 mutant fruits. (A) The compositions of hyper-DMRs (combined hyper-DMRs from two biological replicates of sldml2-1-46dpa) in the sldml2-1 mutant and simulated genomic regions. (B) Density plots of DNA methylation, transposable elements (TEs), genes, sldml2-1 hyper-DMRs, and ripening-induced hypo-DMRs. Chromosome numbers and coordinates (Mb) are indicated on the outer rim. (C) Boxplots showing distances between SlDML2-targeted or nontargeted TEs (Left) and IG regions (Right) and their nearest protein-coding genes. (D) Boxplot showing the distances between genes and their nearest TEs in Arabidopsis and tomato. (E) Percentages of genes that contain TEs in their 500-bp, 1,000-bp, and 2,000-bp upstream sequences in Arabidopsis, tomato, maize, and rice (*P value < 2.2e−16, one-tailed Wilcoxon rank sum test).
Fig. S3.
Chromosomal distribution of SlDML2-targeted TEs and total TEs. Chromosome numbers are indicated at the bottom of each graph, and the centromere positions are indicated by the positions of chromosome numbers on the x axis.
In both Arabidopsis and tomato, TEs are concentrated in pericentromeric regions, and genes are preferentially located in chromosomal arms (Fig. 3B). The content of TEs is higher in the tomato genome than in the Arabidopsis genome. Approximately 20.9% of the Arabidopsis genome but ∼54.7% of the tomato genome consists of TEs. As shown in Fig. 3D, the distance between a gene and its closest TE is much less in tomato than in Arabidopsis. About 18%, 24%, and 31% of Arabidopsis genes have TEs within 500 bp, 1,000 bp, and 2,000 bp of the 5′ end of genes, respectively, but these percentages are ∼39%, 58%, and 77% in tomato (Fig. 3E). The percentages are 41%, 64%, and 83% in rice and 45%, 67%, and 84% in maize (Fig. 3E). These results suggest that, compared with Arabidopsis genes, genes in tomato and the other crops are more likely to be affected by the methylation status of nearby TEs. Consequently, the DNA demethylases likely play more important roles in regulating gene expression in these crop plants than in Arabidopsis.
SlDML2 Is Required for Ripening-Induced DNA Demethylation.
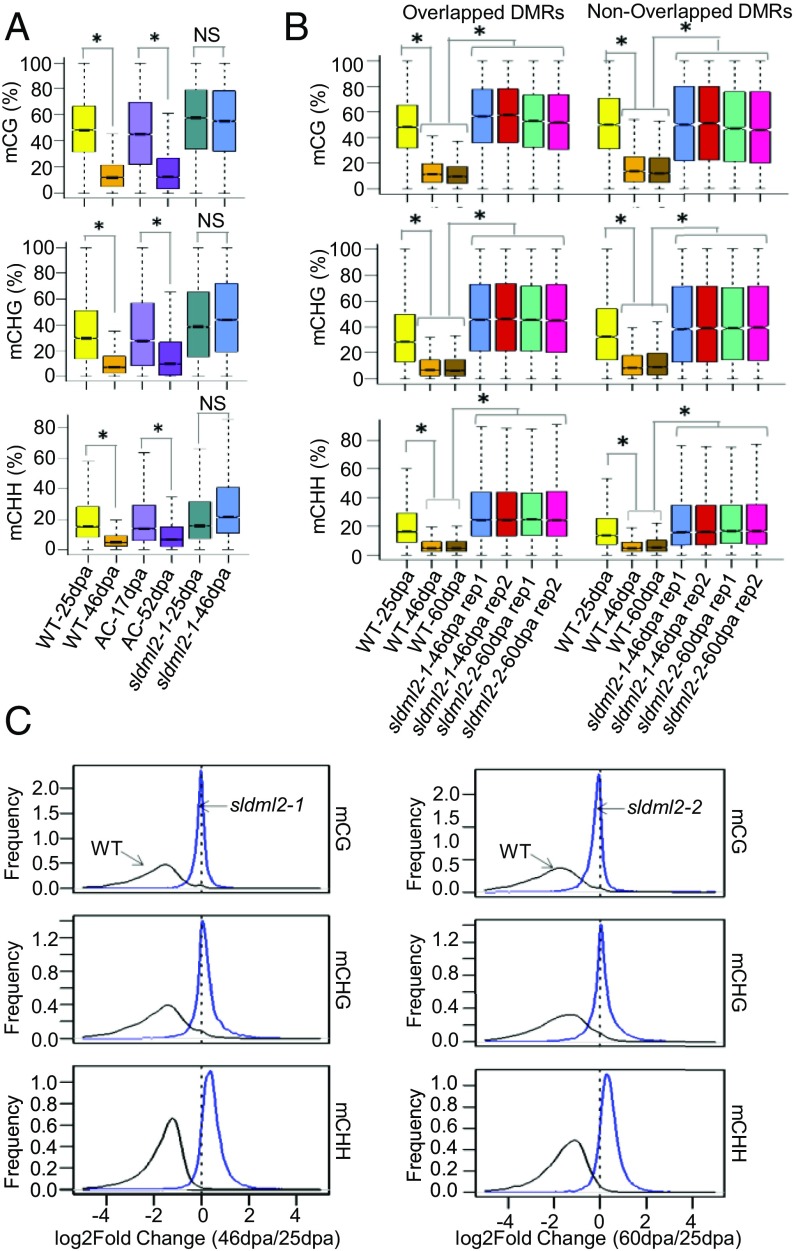
We profiled DNA methylation changes during fruit ripening in tomato cv. Micro-Tom. We compared the methylomes of WT-25dpa and WT-46dpa and identified a total of 16,412 differentially methylated regions (hereafter referred to as ripening-induced DMRs), among which 3,306 are hypermethylated and 13,106 are hypomethylated in WT-46dpa relative to WT-25dpa (Table S4). The DNA methylation changes occurred in all three sequence contexts (Fig. 4A). These 13,106 hypo-DMRs are still hypomethylated in WT-60dpa, suggesting that the hypomethylation status is maintained during later stages of ripening (Fig. 4B). In the sldml2-1 mutant, the DNA methylation levels at these hypo-DMRs do not decrease from 25 dpa to 46 dpa, suggesting that SlDML2 is required for the fruit ripening-induced DNA demethylation (Fig. 4A). We examined the DNA methylation levels of these DMRs in tomato cv. AC (27) and found that the hypo-DMRs in cv. Micro-Tom are also hypomethylated in AC during fruit ripening (from 17 dpa to 52 dpa) (Fig. 4A), which suggests that ripening-induced DNA demethylation is conserved in the two tomato varieties. In contrast, the hyper-DMRs in cv. Micro-Tom are not hypermethylated in cv. AC during fruit ripening (Fig. S4A), suggesting that the ripening-associated increase in DNA methylation differs between the two varieties.
Fig. 4.
SlDML2 is required for ripening-induced DNA demethylation. (A) Boxplots showing mCG, mCHG, and mCHH levels at ripening-induced hypo-DMRs in WT-25dpa, WT-46dpa, AC-17dpa, AC-52dpa, sldml2-1-25dpa, and sldml2-1-46dpa (average of two replicates) (*P value < 2.2e−16, one-tailed t test; NS, not significant). (B) Boxplots showing mCG, mCHG, and mCHH levels at ripening-induced hypo-DMRs that are overlapped (Left) or not overlapped (Right) with sldml2-1 hyper-DMRs in WT-25dpa, WT-46dpa, WT-60dpa, two biological replicates of sldml2-1-46dpa, and two biological replicates of sldml2-2-46dpa. For mCG, mCHG and mCHH levels, WT-25dpa, two biological replicates of sldml2-1-46dpa, and two biological replicates of sldml2-2-46dpa were significantly higher than WT-46dpa and WT-60dpa (*P value < 2.2e−16, one-tailed t test). (C) The distribution of logtwofold-change (46dpa/25dpa) of mCG, mCHG, and mCHH levels in sldml2-1 mutant and the WT (Left), and the distribution of logtwofold-change (60dpa/25dpa) in the sldml2-2 mutant and the WT (Right). A peak at 0 corresponds to no change in DNA methylation during ripening.
Fig. S4.
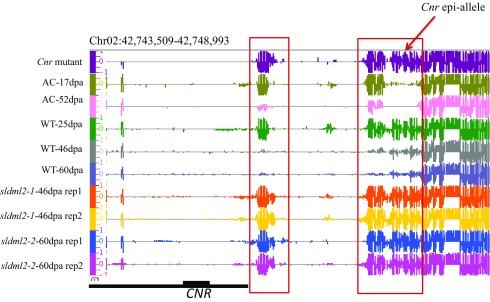
SlDML2 is required for ripening-induced DNA hypomethylation but not for ripening-induced DNA hypermethylation. (A) mCG, mCHG, and mCHH levels of ripening-induced hyper-DMRs in WT-25dpa, WT-46dpa, sldml2-1-46dpa rep1, sldml2-1-46dpa rep2, AC-17dpa, and AC-52dpa (*P value < 1e−10, one-tailed t test; NS, not significant). (B) DNA methylation levels at several ripening-induced hypo-DMRs (boxed areas) in AC-17dpa, AC-52dpa, WT-25dpa, WT-46dpa, WT-60dpa, and two biological replicates of sldml2-1-46dpa and sldml2-2-60dpa. The red arrow indicates the location of the Cnr epi-allele.
To further investigate the role of SlDML2 in ripening-induced DNA demethylation, we compared the ripening-induced hypo-DMRs with sldml2 hyper-DMRs and found that 8,176 of the 13,106 ripening-induced hypo-DMRs overlap with the sldml2 hyper-DMRs. As shown in Fig. 4B, the 8,176 shared ripening-induced hypo-DMRs have increased DNA methylation levels in sldml2-1 and sldml2-2 mutants compared with the WT at the same stage, and the data were consistent for the two biological replicates of both mutants. For the 4,930 nonoverlapped hypo-DMRs, DNA methylation was also higher in the sldml2-1 and sldml2-2 mutants than in the WT (Fig. 4B). This result suggested that, although these 4,930 ripening-induced hypo-DMRs do not overlap with sldml2 hyper-DMRs (perhaps because of the artificial cutoff used to define the DMRs), their methylation levels are still controlled by SlDML2 during ripening. Several examples of ripening-induced hypo-DMRs, such as those at the promoters of RIN, CNR, and DML2, are shown in Fig. S4B. The methylation patterns at these DMRs are consistent with our analysis above, which showed that, during ripening, they are hypomethylated in the WT but not in sldml2 mutants. To further assess the effect of SlDML2 dysfunction on ripening-induced DNA demethylation, we compared the DNA methylation changes in ripening-induced hypo-DMRs from the immature to the ripe stage in the WT and in the sldml2-1 and sldml2-2 mutants. During ripening, DNA methylation decreased in the WT in all three sequence contexts, as shown in Fig. 4A and also indicated by the left-skewed peaks in Fig. 4C. This decrease in DNA methylation, however, did not occur in the fruits of sldml2-1 (Fig. 4A). The failure in DNA demethylation in both sldml2-1 and sldml2-2 mutants is also indicated by the peak at “0” in Fig. 4C. Interestingly, the peaks of mCHG and mCHH in sldml2-1 and sldml2-2 mutants were skewed to the right of 0, indicating that non-CG methylation, particularly mCHH in sldml2 mutants, was slightly increased from the immature stage to 46 or 60 dpa (Fig. 4C). The increase in DNA methylation during ripening was not compromised by SlDML2 dysfunction (Fig. S4). These results demonstrate that SlDML2 is responsible for virtually all ripening-induced DNA demethylation and but not for ripening-induced hyper-DNA methylation.
SlDML2-Mediated DNA Demethylation also Has Silencing Functions.
DNA methylation is generally associated with inactive transcription, especially when it occurs in the promoter regions of genes (1–3). Enzymes and regulators involved in active DNA demethylation are also known as antisilencing factors because of their function in preventing DNA hypermethylation and in therefore restraining transcriptional gene silencing (3, 14). In sldml2-1, there are 29,764 hyper-DMRs relative to the WT, and 47.3% (14,074) of them are located within 1 kb of 12,902 genes. Expression of these hyper-DMR–associated genes is potentially regulated by SlDML2-mediated DNA demethylation.
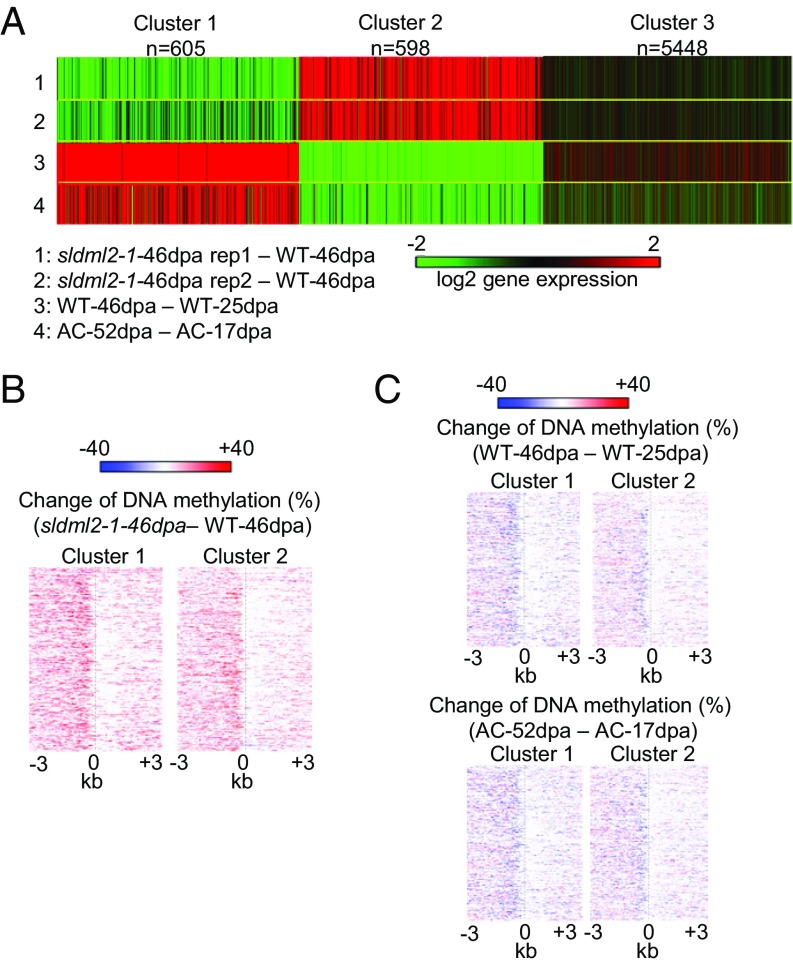
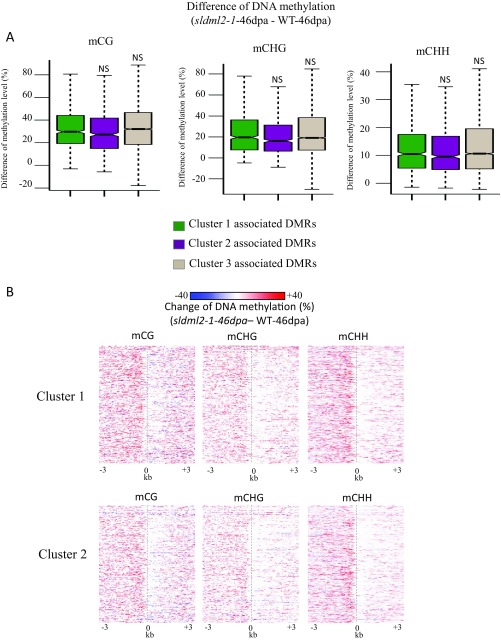
To determine the role of SlDML2 in the regulation of gene expression during tomato fruit ripening, we generated gene expression profiles for WT-25dpa, WT-46dpa, sldml2-1-25dpa, and two biological replicates of sldml2-1-46dpa. Among the 12,902 hyper-DMR-associated genes, 6,651 were expressed in at least one sample and were used for the following analysis. For each gene, we calculated the expression change in WT-46dpa vs. WT-25dpa [log2 (WT-46dpa/WT-25dpa)] and in sldml2-1-46dpa vs. WT-46dpa [log2 (sldml2-1-46dpa/WT-46dpa)]. Using k-means clustering, we assigned the 6,651 hyper-DMR–associated genes to three clusters: cluster 1 included 605 genes that were up-regulated during ripening in the WT but not in sldml2 (Table S5); cluster 2 included 598 genes that were silenced during ripening in the WT but the silencing was compromised in sldml2 (Table S6); and cluster 3 included 5,448 genes that were not obviously influenced by ripening or SlDML2 dysfunction (Fig. 5A). To investigate whether the hyper-DMRs that are associated with cluster 3 genes might be weaker SlDML2 targets (and therefore less able to cause expression changes) than those associated with cluster 1 and 2 genes, we examined the DNA methylation levels of cluster 1, 2, and 3 genes in sldml2-46dpa and WT-46dpa. We found that the increases in DNA methylation were similar in cluster 1, 2, and 3 genes (Fig. S5A), suggesting that the DNA methylation increases at cluster 3 genes are not sufficient to cause gene expression changes in sldml2.
Fig. 5.
SlDML2-mediated DNA demethylation correlates with ripening-induced gene activation, as well as with ripening-induced gene repression. (A) The k-means clustering of sldml2-1 hyper-DMR–associated gene expression. Genes associated with sldml2-1 (46 dpa) hyper-DMRs were subjected to the clustering analysis. (B) DNA methylation changes in cluster 1 and cluster 2 genes in sldml2-1-46dpa vs. WT-46dpa. The sequences flanking cluster 1 and 2 genes were aligned, and DNA methylation levels for each 100-bp interval were plotted. The dashed line marks transcription start sites. (C) DNA methylation changes in cluster 1 and cluster 2 genes in ripe fruits compared with immature fruits. (Upper) DNA methylation differences between WT-46dpa and WT-25dpa. (Lower) DNA methylation differences between AC-52dpa and AC-17dpa.
Fig. S5.
Cluster 1, 2, and 3 genes are hypermethylated in sldml2-1-46dpa vs. WT-46dpa. (A) Boxplots showing changes in DNA methylation levels at different groups of DMRs in sldml2-1-46dpa compared with WT-46dpa (NS, not significant, one-tailed Wilcoxon rank sum test). (B) Changes in DNA methylation levels of cluster 1 and cluster 2 genes in sldml2-1-46dpa relative to WT-46dpa. The sequences flanking cluster 1 and 2 genes are aligned, and mCG, mCHG, and mCHH levels for each 100-bp interval are plotted. The dashed line marks the positions of transcription start sites.
Consistent with the general notion that DNA hypermethylation is associated with inactive transcription, the expression level of the 605 cluster 1 genes was negatively correlated with their DNA methylation level. Cluster 1 genes were hypermethylated in sldml2 vs. WT-46dpa (Fig. 5B and Fig. S5A) and had reduced expression levels in sldml2 vs. WT-46dpa (Fig. 5A). The data were consistent between the two biological replicates of sldml2-1-46dpa (Fig. 5A). For both cluster 1 and cluster 2 genes, the hypermethylation mainly occurred in the genomic regions upstream of the transcription start site (Fig. 5B). Compared with mCG and mCHG changes, mCHH changes in the upstream regions were more concentrated near the transcription start site of the genes (Fig. S5B). By comparing the expression profiles of immature and ripe fruits, we found that cluster 1 genes were up-regulated during fruit ripening in both Micro-Tom and AC varieties (Fig. 5A) and that their promoter regions lost DNA methylation in both varieties (Fig. 5C). These results suggest that SlDML2-mediated DNA demethylation is required for active transcription of the ripening-induced genes and that SlDML2 has an antisilencing function for the genes.
In addition to cluster 1 genes, we also identified 598 genes (cluster 2) that were repressed during ripening in the WT but remained active in the sldml2 mutant (Fig. 5A). As shown in Fig. 5B, the promoters of cluster 2 genes had increased DNA methylation in sldml2 mutants, but their expression levels were consistently higher in the two biological replicates of sldml2-46dpa than in WT-46dpa (Fig. 5A). These results demonstrate that the expression of cluster 2 genes is positively correlated with DNA methylation and that the DNA hypermethylation in sldml2 is important for these genes to remain active in the sldml2 mutants. Cluster 2 genes were repressed during fruit ripening in both cv. Micro-Tom and cv. AC (Fig. 5A), and this repression was correlated with a decrease in the DNA methylation level during ripening in both varieties (Fig. 5C). Our data demonstrate that a large number of genes are repressed during ripening and that SlDML2-mediated DNA demethylation is critical for this repression. Thus, SlDML2 has a silencing function for these ripening-repressed genes.
Functions of SlDML2-Activated and -Repressed Genes.
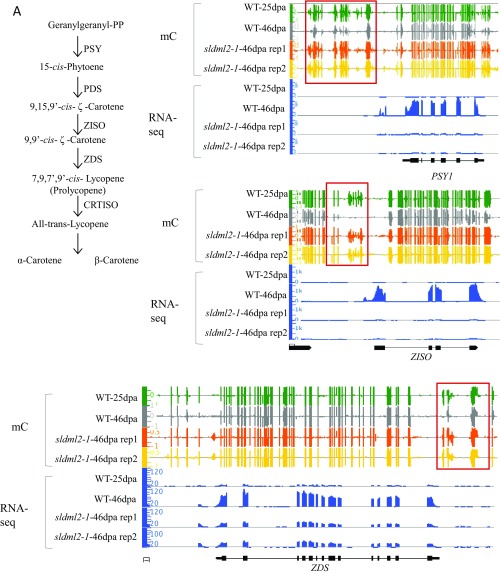
To understand why SlDML2 is critical for tomato fruit ripening, we performed Gene Ontology (GO) term enrichment analysis of cluster 1 and cluster 2 genes. For cluster 1, genes involved in flavonoid biosynthesis and carotenoid biosynthesis were highly enriched (Table S7). We examined the promoter methylation and expression pattern of genes in the carotenoid biosynthesis pathway, including PSY1, Z-ISO, ZDS, and CRTISO, and found that all of them are regulated by SlDML2-mediated DNA demethylation during ripening because all of them were hypermethylated and silenced in the sldml2 mutant (Fig. S6A).
Fig. S6.
Expression and DNA methylation levels of several ripening-induced and ripening-repressed genes in WT-25dpa, WT-46dpa, and two biological replicates of sldml2-1-46dpa fruits. (A) IGB screenshots of DNA methylation levels and expression (normalized RNA-seq reads) of several ripening-induced genes, including several genes in the pigment biosynthesis pathway (pathway shown Top Left), and RIN and CNR. (B) IGB screenshots of DNA methylation levels and expression (normalized RNA-seq reads) of two ripening-repressed cell wall organization genes.
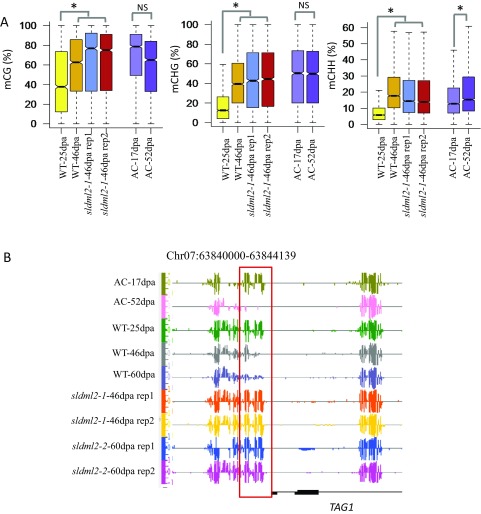
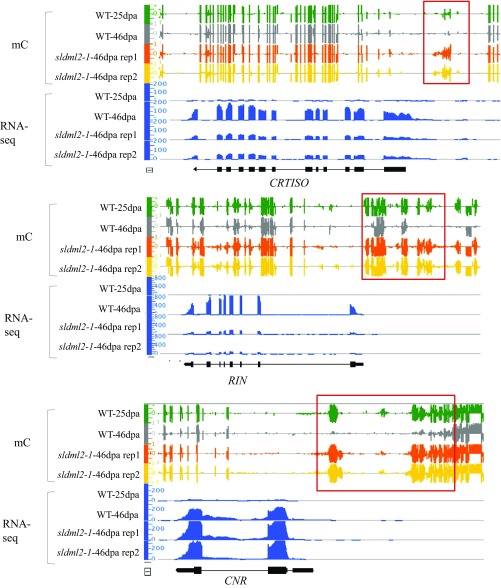
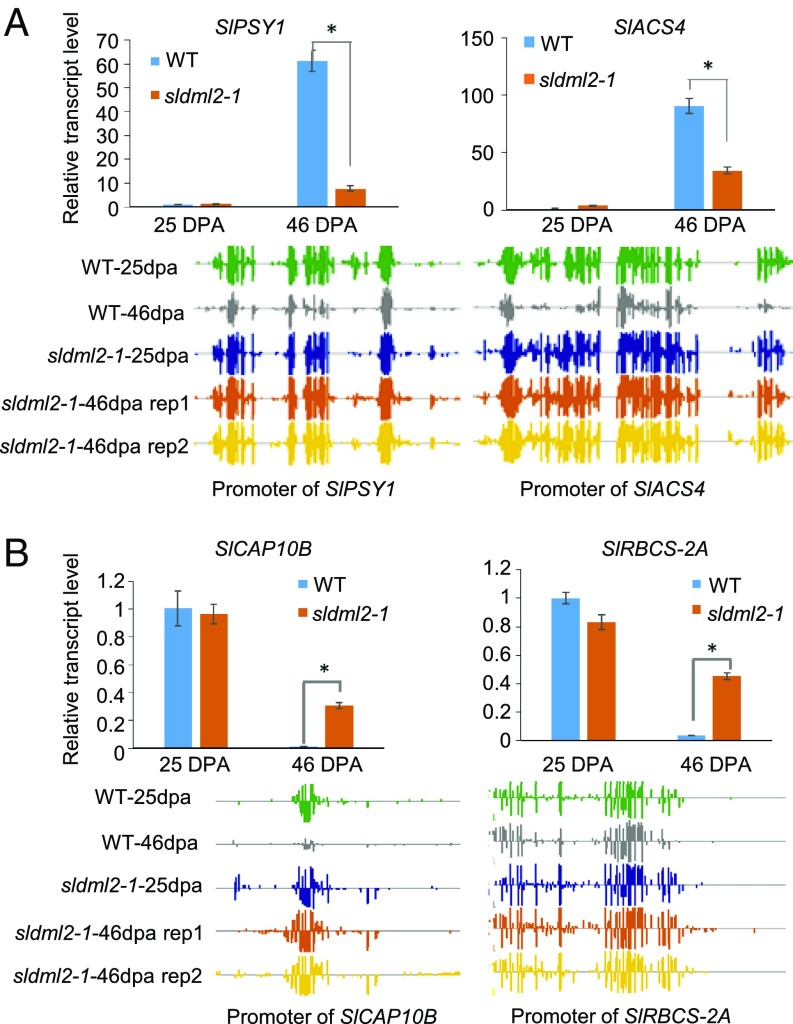
Many other genes known to be important for fruit ripening were also found in cluster 1, and these included PG2a and PL (which are involved in cell wall degradation); ACS, ACO, and ETR (which are involved in ethylene biosynthesis or signaling); and genes encoding ripening-associated transcription factors, such as RIN (Fig. S6A). A previous study showed that three ripening-related genes, RIN, NOR, and PSY1, are hypermethylated and silenced during ripening in the RNAi lines of DNA demethylases (29). In agreement with this previous finding, these three genes were found in cluster 1 (Table S5). We performed the reverse transcription quantitative PCR (RTqPCR) to examine the expression patterns of PSY1 and ACS4 in fruits of sldml2-1 mutant and the WT. Consistent with the RNA-seq results, PSY1 and ACS4 were highly induced in WT fruits during ripening, but their induction was significantly less in the sldml2-1 mutant than in the WT at 46 dpa (Fig. 6A, Upper). As shown in Fig. 6A, Lower, the promoters of these two genes were demethylated in the WT but not in the sldml2 mutants during ripening. These data show that SlDML2 is required for ripening-induced demethylation and expression of genes involved in ripening-related processes, including pigment synthesis, flavonoid synthesis, and fruit softening.
Fig. 6.
Expression and methylation levels of representative cluster 1 and cluster 2 genes in fruits of the WT and sldml2 mutants. RT-qPCR measurements to validate the expression of two cluster 1 genes, SlPSY1 and SlACS4 (A), and two cluster 2 genes, SlCAP10B and SlRBCS-2A (B), in WT and sldml2-1 mutant fruits at 25 dpa and 46 dpa. EF1α was used as the internal control. The DNA methylation levels of promoter regions of these four genes are shown with screenshots of Integrative Genome Browser (IGB) display of whole-genome bisulfite sequencing data, where each vertical bar represents an mC and the height of the bar indicates methylation level. Error bars indicate SD, n = 3 (*P value < 0.01, two-tailed t test).
CNR is an important transcription factor for tomato fruit ripening, and its expression is known to be regulated by DNA methylation (25). RIN binds to the CNR promoter in a DNA demethylation-dependent manner (32). Intriguingly, in our study, ripening-induced expression of CNR was not silenced by the sldml2 mutation (Fig. S6A) even though the CNR promoter was hypermethylated in sldml2 mutants (Figs. S4B and S6A). This observation suggests that promoter hypermethylation is not sufficient to cause the silencing of CNR.
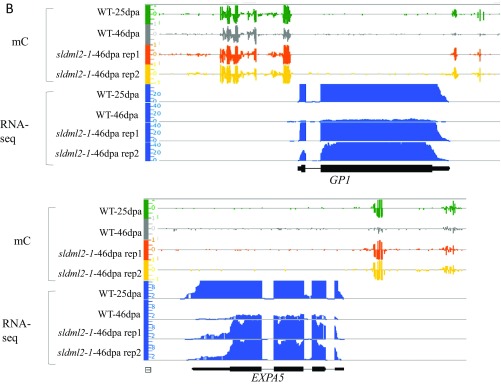
Our GO enrichment analysis showed that cluster 2 genes involved in photosynthesis and cell wall organization or biogenesis are highly enriched (Table S8). We tested the expression of CAP10B (encoding a chlorophyll a/b-binding protein) and RBCS-2A (encoding a Rubisco small subunit) by RT-qPCR. The results validated the RNA-seq data in that the expression of these genes was completely inhibited in the WT-46dpa but not in the sldml2 mutant (Fig. 6B, Upper). Integrated genome browser (IGB) screenshots clearly show that the promoter regions of these genes were demethylated in the WT but not in the sldml2 mutants during fruit ripening (Fig. 6B, Lower). Similarly, the sldml2 mutation also disrupted ripening-induced promoter demethylation and silencing of GP1 (encoding a regulator of polygalacturonase) and EXPA5 (encoding an expansin) (Fig. S6B).
Fruit development has three stages: cell division, cell enlargement, and ripening. Fruit photosynthesis has been suggested to be important in early fruit development by contributing to fruit growth and production of starch that is turned into soluble sugars during ripening (33). As fruits ripen, however, an important transition takes place: The photosynthetically active chloroplasts differentiate into chromoplasts. Chromoplasts no longer contain chlorophylls but instead produce the lycopene and carotene necessary for fruit color. This transition is coupled with a decreased expression of both nuclear- and plastid-encoded genes involved in photosynthesis. Our analysis showed that SlDML2 regulates at least 23 genes involved in photosynthesis during ripening, including 6 genes encoding chlorophyll a/b binding proteins and 8 genes encoding proteins of photosystem 1 and 2 reaction centers (Table S6). These genes were repressed during ripening, and this repression coincided with hypomethylation of their promoter. However, the promoter hypomethylation and ripening-induced repression were disrupted or diminished in sldml2 mutants (Fig. 6B), suggesting a critical role of SlDML2 in the regulation of the chloroplast-to-chromoplast transition during ripening.
In addition to genes involved in photosynthesis, 22 genes involved in cell wall biogenesis were enriched in cluster 2, and these included genes encoding cellulose synthase, expansins, and xyloglucan endotransglucosylases/hyrolases (XETs) (Table S6). In the first stage of fruit development, enzymes involved in cell wall synthesis, such as cellulose synthase, are active. As fruits shift to the second stage, cell walls are loosened via expansins and XETs to allow the cells to expand. As the fruit ripens, genes involved in cell wall biogenesis and organization are no longer required for fruit development and are thus repressed. Our results revealed that these cluster 2 cell wall-related genes are silenced in ripe fruits because of promoter hypomethylation and that the silencing depends on SlDML2-mediated DNA demethylation (Fig. 5 A and B and Fig. S6B). Thus, SlDML2 is required not only for the activation of genes important for ripening but also for the silencing of genes that are needed for the early stages of fruit development, but that must be repressed for ripening to occur.
Zhong et al. (27) showed that genomic regions that are hypomethylated during ripening are enriched with RIN-binding sites. RIN has both transcription activation and transcription repression activities (34). Previous studies identified 137 RIN activation targets and 109 RIN repression targets (34). We found that previously identified RIN activation targets are enriched in cluster 1 genes whereas RIN repression targets are enriched in cluster 2 genes (Table S9).
Discussion
Cytosine methylation is generally thought to be associated with inactive transcription (i.e., with silencing), especially when the methylation occurs in the promoter region of genes. Therefore, components involved in DNA demethylation are often referred to as antisilencing factors (3). In Arabidopsis, dysfunction of the 5-mC DNA glycosylase/demethylase ROS1 enhances the transcriptional silencing of some transgenes, endogenous genes, and TEs due to DNA hypermethylation (14, 16, 19). RNAi of ROS1 orthologs in cotton and tomato also reduce the expression of several genes due to promoter DNA hypermethylation (29, 35). Consistent with the expected role of the tomato ROS1 ortholog SlDML2 as an antisilencing factor, sldml2 mutations caused the silencing of hundreds of genes (cluster 1 genes in Fig. 5A) in tomato fruits due to DNA hypermethylation. Surprisingly, we also found that SlDML2-mediated DNA demethylation is associated with the repression of hundreds of genes (cluster 2 genes in Fig. 5A) during tomato fruit ripening. Thus far, there are only a few reports of DNA demethylation causing transcriptional silencing. In Arabidopsis, the expression of ROS1 is regulated by both ROS1-mediated active DNA demethylation and DNA methylation pathways, including RNA-directed DNA methylation (RdDM) (36). In met1 and RdDM mutants, AtROS1 is silenced due to promoter hypomethylation whereas, in ros1 mutants with point mutations, the AtROS1 transcript level is enhanced due to promoter hypermethylation (36). Thus, ROS1 expression is promoted by DNA methylation and antagonized by DNA demethylation. In tomato, we discovered that hundreds of genes are repressed by DNA hypomethylation during ripening. The expression of these cluster 2 genes was higher in the fruits of sldml2 mutants than in those of the WT. These results have revealed a broad role of DNA methylation in promoting gene expression. The mechanisms underlying the antisilencing role of DNA methylation and the silencing role of DNA demethylation at the cluster 2 genes are unknown. It is possible that a transcriptional activator is recruited to the promoters of the cluster 2 genes by a methyl-DNA–binding protein. It is also possible that DNA methylation prevents the binding of a transcriptional repressor to these genes.
Like AtROS1, the tomato SlDML2 also regulates its own promoter DNA methylation (Fig. S4B). However, contrary to AtROS1, SlDML2 promoter demethylation is correlated with its transcriptional induction during ripening. It would be interesting to determine whether any of the other SlDML genes may be regulated by DNA methylation like AtROS1 and may thus function like AtROS1 as a “methylstat” to maintain DNA methylation homeostasis (36). AtROS1 antagonizes RdDM in Arabidopsis (20). A recent study reported that RdDM targets in tomato are located in gene-rich euchromatin regions (37), which is similar to the distribution of SlDML2 targets (Fig. 3B), suggesting a potential antagonism between SlDML2 and RdDM in tomato as well. The increase in mCHH levels in sldml2 fruits at ripening-induced hypo-DMRs (Fig. 4C) may be caused by enhanced RdDM in the mutant because ROS1 dysfunction in Arabidopsis causes enhanced RdDM (20). RdDM in tomato might also be important for the DNA methylation-dependent expression of cluster 2 genes in immature fruits.
Tomato fruits undergo genome-wide loss of DNA methylation during ripening (27). By characterizing the DNA methylomes of sldml2 mutant fruits, we found that ∼30,000 genomic regions require SlDML2 for demethylation during fruit ripening. In fact, SlDML2 was critical for virtually all ripening-induced demethylation (Fig. 4). Our analyses showed that, among the SlDML2-targeted genes, cluster 1 is enriched in genes involved in the synthesis of pigments and flavor compounds, as well as in ethylene biosynthesis and signaling, and cell wall hydrolysis. These genes are well-known to be required for fruit ripening. In contrast, cluster 2 is enriched with genes involved in photosynthesis and cell wall organization. These cluster 2 genes have important functions in the growth of immature fruits but are no longer needed after ripening and therefore must be repressed during ripening. Despite the genome-wide role of SlDML2 in fruit ripening, many highly methylated genomic regions remain methylated. It would be useful to investigate how SlDML2 is preferentially targeted to genes relevant to fruit ripening. Given that so many genes are affected by SlDML2-mediated demethylation, the tomato fruit would be an excellent system for studying the mechanism underlying the targeting of DNA demethylases to genes.
The mechanism and function of active DNA demethylation has been studied mostly in Arabidopsis. AtROS1, the predominant DNA demethylase in vegetative tissues in Arabidopsis, targets more than 6,000 genomic regions for demethylation (20). Most of these genomic regions are in TEs or intergenic regions. Only a small number of genes are hypermethylated and silenced in Arabidopsis ros1 mutants, and the mutants do not show dramatic phenotypes in growth or development (7, 14, 19, 21). Like ROS1 in Arabidopsis (20), SlDML2 in tomato preferentially targets highly methylated TEs and intergenic regions near genes (Fig. 3C). However, unlike in Arabidopsis, the expression of thousands of genes is impaired by DNA demethylase dysfunction in tomato, and the sldml2 mutants display dramatic phenotypes in fruit ripening. Clearly, the role of active DNA demethylation in regulating gene expression is broader in tomato than in Arabidopsis. One reason for this difference is that the tomato genome has a higher content of TEs and other repeats and that more tomato genes than Arabidopsis genes contain highly methylated TEs and other repeats in their promoters (Fig. 3 D and E). Therefore, the expression of these tomato genes is more likely to be regulated by DNA demethylation. Rice genes tend to have mCHH methylation in their promoter regions (38). Our analysis showed that, like the tomato genome, rice and maize genomes also have higher percentages of genes with highly methylated TEs in their promoters compared with the Arabidopsis genome (Fig. 3E). These results suggest that active DNA demethylation has a broader effect on gene expression in crops and other plants with large genomes than in Arabidopsis.
Our DNA methylome analysis showed that ripening-induced demethylation is abolished at promoters of several well-known fruit-ripening genes, such as PSY1, RIN, and CNR in sldml2 mutants (Fig. 6A and Figs. S4B and S6A). The CNR promoter is a direct RIN-binding target (27, 32). CNR expression is substantially reduced in the Cnr epiallele, which is thought to be caused by DNA hypermethylation at the CNR promoter region (25, 27). Interestingly, in Micro-Tom WT fruit, we observed a high level of DNA methylation at the CNR promoter, and the methylation was lost during ripening in a SlDML2-dependent manner (Figs. S4B and S6A). However, even though RIN expression was silenced and both RIN and CNR promoters were hypermethylated in sldml2 mutant fruits (Figs. S4B and S6A), CNR expression was even higher in the mutant fruits than in WT fruits at the same stage (Fig. S6A). In the RNAi lines for SlDMLs, induction of CNR is also not fully suppressed but is instead delayed for several days during ripening (29). These observations suggest that DNA hypermethylation is not sufficient to silence CNR and that RIN binding is not required for CNR expression. It may be worthwhile to reexamine the role of DNA hypermethylation at the Cnr epiallele: e.g., by using CRISPR/dCas9 to target SlDML2 to specifically demethylate the CNR promoter in the epiallele.
In summary, our results demonstrate that SlDML2 is critical for tomato fruit ripening because it mediates all ripening-induced DNA demethylation. This genome-wide demethylation is important not only for activating the expression of ripening factors but also for silencing hundreds of genes that are no longer needed and must be repressed when the fruit is ripe.
Materials and Methods
Plant Materials.
All plant materials used in this study were first generation (T0) plants regenerated from callus. Tomato cv. Micro-Tom was obtained from Ball. Tomato plants were grown in a greenhouse at Purdue University, West Lafayette campus. Tomato tissue culture was performed according to the established protocol (39). All tissue culture steps were conducted at room temperature with a photoperiod of 16 h light and 8 h dark.
CRISPR/Cas9 Gene Editing.
A 19-bp sgRNA oligo (5′- GAACAAAGTCTGAAATGTG-3′) targeting the first exon of SLDML2 was cloned into the psgR-Cas9-At vector (31), and the cassette including chimeric RNA driven by the AtU6 promoter and Cas9 driven by the AtUBQ1 promoter was subcloned into the pCAMBIA1300 binary vector. The construct was transformed into tomato cv. Micro-Tom using Agrobacterium infection of leaf explants.
Whole-Genome Bisulfite Sequencing and Analysis.
Genomic DNA was extracted from tomato fruits using a DNeasy Plant Maxi Kit (Qiagen) and then was used for library construction using Illumina's standard DNA methylation analysis protocol and the NEB Ultra II DNA Library Prep Kit. The samples were sequenced at the Genomics Core Facility of the Shanghai Centre for Plant Stress Biology, Chinese Academy of Sciences with Illumina HiSeq2500.
For data analysis, low-quality sequences (q < 20) were trimmed using trim in BRAT-BW (40), and clean reads were mapped to the reference genome using BRAT-BW, allowing two mismatches. The reference genome version is SL2.50 (ftp://ftp.solgenomics.net/tomato_genome/assembly/build_2.50/) (41).
To remove potential PCR duplicates, the remove-dupl command of BRAT-BW was used. DMRs were identified using the same method as in ref. 20. In brief, only cytosines with at least 4× coverage in all libraries were considered. A sliding-window approach with a 200-bp window sliding at 50-bp intervals was used to identify DMRs. Fisher's exact test was performed for methylated vs. unmethylated cytosines for each context within each window, with false discovery rates (FDRs) estimated using a Benjamini–Hochberg adjustment of Fisher's P values calculated in the R environment. Windows with an FDR ≤ 0.05 were considered for further analysis, and windows within 100 bp of each other were merged into larger regions. Regions were then adjusted to shrink to the first and last differentially methylated cytosines (DMCs). A cytosine was considered a DMC if it showed at least a twofold change in methylation percentage in the mutant. The regions were then filtered to include only those with at least 10 DMCs and with at least a twofold change in the arithmetic mean of methylation percentage of all cytosines.
RNA Analysis.
Total RNA was extracted with TRIzol reagent (Ambion) from pericarps of fruits.
For reverse transcription, 1 μg of RNA and oligo dT primers were used to synthesize cDNA in a 20-μL reaction using the qScript cDNA SuperMix kit (Quanta). Real-time PCR was then carried out on a CFX96 Real-Time PCR Detection System (Bio-Rad) with PerfeCTa SYBR Green FastMix (Quanta). EF1α was used as an internal control. Primers used in q-PCR reactions are listed in Table S10.
For RNA-seq, the libraries were constructed and sequenced at the Genomics Core Facility of the Shanghai Centre for Plant Stress Biology, Chinese Academy of Sciences, with an Illumina HiSeq2500.
For RNA-seq data processing, quality control was checked using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc). RNA-Seq reads were trimmed using the fastx_trimmer command FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit/index.html) with parameter “-f 14 -l 80” before alignment. The trimmed reads were aligned to the tomato genome using TopHat2 (42). The ITAG2.4 annotation gff3 file was supplied as the “-G” option to TopHat2. The program featureCounts (43) was used to count the mapped fragments for each gene. The output count table was used as the input for edgeR (44) to compute the logtwofold change of each gene.
DMR-associated genes were defined as genes with DMRs within 1 kb. For gene clustering, we used only DMR-associated genes that had transcripts per kilobase million (TPM) ≥ 5 in at least one sample. The input data had two columns: logtwofold-change WT-46dpa vs. WT-25dpa and logtwofold-change sldml2-46dpa vs. WT-46dpa. The Kmeans function of R was used for clustering.
The heatmap was generated using Matrix2png (45).
Data Deposition.
The data discussed in this publication have been deposited in the NCBI's Gene Expression Omnibus and are accessible through the GEO Series accession number GSE94903. Previously published data used in this study were from Zhong et al. (27).
Supplementary Material
Acknowledgments
This work was supported by the Chinese Academy of Sciences and by US NIH Grants R01GM070795 and R01GM059138 (to J.-K.Z.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE94903).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705233114/-/DCSupplemental.
References
- 1.He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J-K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 5.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Zhu J-K. Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol. 2012;77:161–173. doi: 10.1101/sqb.2012.77.014936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 8.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Duan C-G, Zhu X, Qian W, Zhu J-K. A DNA ligase required for active DNA demethylation and genomic imprinting in Arabidopsis. Cell Res. 2015;25:757–760. doi: 10.1038/cr.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis, and correction (2015) 11:e1005198. PLoS Genet. 2015;11:e1004905. doi: 10.1371/journal.pgen.1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez-Macías MI, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agius F, Kapoor A, Zhu J-K. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morales-Ruiz T, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian W, et al. Regulation of active DNA demethylation by an α-crystallin domain protein in Arabidopsis. Mol Cell. 2014;55:361–371. doi: 10.1016/j.molcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang Z, et al. The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol Cell. 2015;57:971–983. doi: 10.1016/j.molcel.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan CG, et al. A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 2017;27:226–240. doi: 10.1038/cr.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, et al. Methyl-CpG-binding domain protein MBD7 is required for active DNA demethylation in Arabidopsis. Plant Physiol. 2015;167:905–914. doi: 10.1104/pp.114.252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu J-K. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 20.Tang K, Lang Z, Zhang H, Zhu JK. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat Plants. 2016;2:16169. doi: 10.1038/nplants.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamuro C, et al. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le T-N, et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014;15:458. doi: 10.1186/s13059-014-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- 24.Vrebalov J, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 25.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 26.Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol. 2007;10:283–289. doi: 10.1016/j.pbi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, et al. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc Natl Acad Sci USA. 2016;113:12580–12585. doi: 10.1073/pnas.1613910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu R, et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci USA. 2015;112:10804–10809. doi: 10.1073/pnas.1503362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao D, et al. Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene. 2014;550:230–237. doi: 10.1016/j.gene.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Mao Y, et al. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6:2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 2011;157:1568–1579. doi: 10.1104/pp.111.181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cocaliadis MF, Fernández-Muñoz R, Pons C, Orzaez D, Granell A. Increasing tomato fruit quality by enhancing fruit chloroplast function: A double-edged sword? J Exp Bot. 2014;65:4589–4598. doi: 10.1093/jxb/eru165. [DOI] [PubMed] [Google Scholar]
- 34.Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–386. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin X, et al. A potential role for CHH DNA methylation in cotton fiber growth patterns. PLoS One. 2013;8:e60547. doi: 10.1371/journal.pone.0060547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei M, et al. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA. 2015;112:3553–3557. doi: 10.1073/pnas.1502279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gouil Q, Baulcombe DC. DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 2016;12:e1006526. doi: 10.1371/journal.pgen.1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zemach A, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107:18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H-J, Uchii S, Watanabe S, Ezura H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006;47:426–431. doi: 10.1093/pcp/pci251. [DOI] [PubMed] [Google Scholar]
- 40.Harris EY, Ponts N, Le Roch KG, Lonardi S. BRAT-BW: Efficient and accurate mapping of bisulfite-treated reads. Bioinformatics. 2012;28:1795–1796. doi: 10.1093/bioinformatics/bts264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlidis P, Noble WS. Matrix2png: A utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.