Significance
We found that the yeast Hsp90 chaperone system is greatly impaired in naa10Δ cells, which cannot N-terminally acetylate a majority of normally N-terminally acetylated proteins, including Hsp90 and its cochaperones. Hsp90 clients, including Chk1, Kar4, Tup1, Gpd1, Ste11, and even the Hsp90 chaperone (Hsc82) itself, became short-lived substrates of the Arg/N-end rule pathway in naa10Δ cells. Ubr1 targets the Chk1 kinase through its internal degron. Interactions of Hsp90 with Chk1 could be detected in wild-type but not in naa10Δ cells. These results revealed a major role of N-terminal acetylation in the Hsp90-mediated protein homeostasis and showed that a number of Hsp90 clients are previously unknown substrates of the Arg/N-end rule pathway.
Keywords: Naa10, Chk1, Ubr1, Ufd4, protein degradation
Abstract
We found that the heat shock protein 90 (Hsp90) chaperone system of the yeast Saccharomyces cerevisiae is greatly impaired in naa10Δ cells, which lack the NatA Nα-terminal acetylase (Nt-acetylase) and therefore cannot N-terminally acetylate a majority of normally N-terminally acetylated proteins, including Hsp90 and most of its cochaperones. Chk1, a mitotic checkpoint kinase and a client of Hsp90, was degraded relatively slowly in wild-type cells but was rapidly destroyed in naa10Δ cells by the Arg/N-end rule pathway, which recognized a C terminus-proximal degron of Chk1. Diverse proteins (in addition to Chk1) that are shown here to be targeted for degradation by the Arg/N-end rule pathway in naa10Δ cells include Kar4, Tup1, Gpd1, Ste11, and also, remarkably, the main Hsp90 chaperone (Hsc82) itself. Protection of Chk1 by Hsp90 could be overridden not only by ablation of the NatA Nt-acetylase but also by overexpression of the Arg/N-end rule pathway in wild-type cells. Split ubiquitin-binding assays detected interactions between Hsp90 and Chk1 in wild-type cells but not in naa10Δ cells. These and related results revealed a major role of Nt-acetylation in the Hsp90-mediated protein homeostasis, a strong up-regulation of the Arg/N-end rule pathway in the absence of NatA, and showed that a number of Hsp90 clients are previously unknown substrates of the Arg/N-end rule pathway.
Proteins called chaperones promote the in vivo protein folding and counteract misfolding, maladaptive protein aggregation, and other perturbations of proteostasis (1–4). Hsp90 is an ATP-dependent chaperone system that mediates the conformational maturation and stabilization of many proteins. Referred to as Hsp90 clients, these proteins include kinases, transcriptional regulators, and many other proteins that comprise at least 20% of a cellular proteome. The diversity of Hsp90 clients and the broad range of processes that involve Hsp90 underlie its necessity for cell viability in all eukaryotes (5–11). Cochaperones of the ∼90-kDa Hsp90 comprise, in mammals, ∼30 distinct proteins. They participate in the delivery of clients to Hsp90 and contribute to related processes, including ATP hydrolysis by Hsp90 (12). Because of its ability to modulate protein conformations, the Hsp90 system can either exacerbate or counteract the fitness-reducing effects of mutations in cellular proteins. This property of Hsp90 affects the standing genetic variation and thereby influences the impact of natural selection on rates and directions of evolution (13, 14).
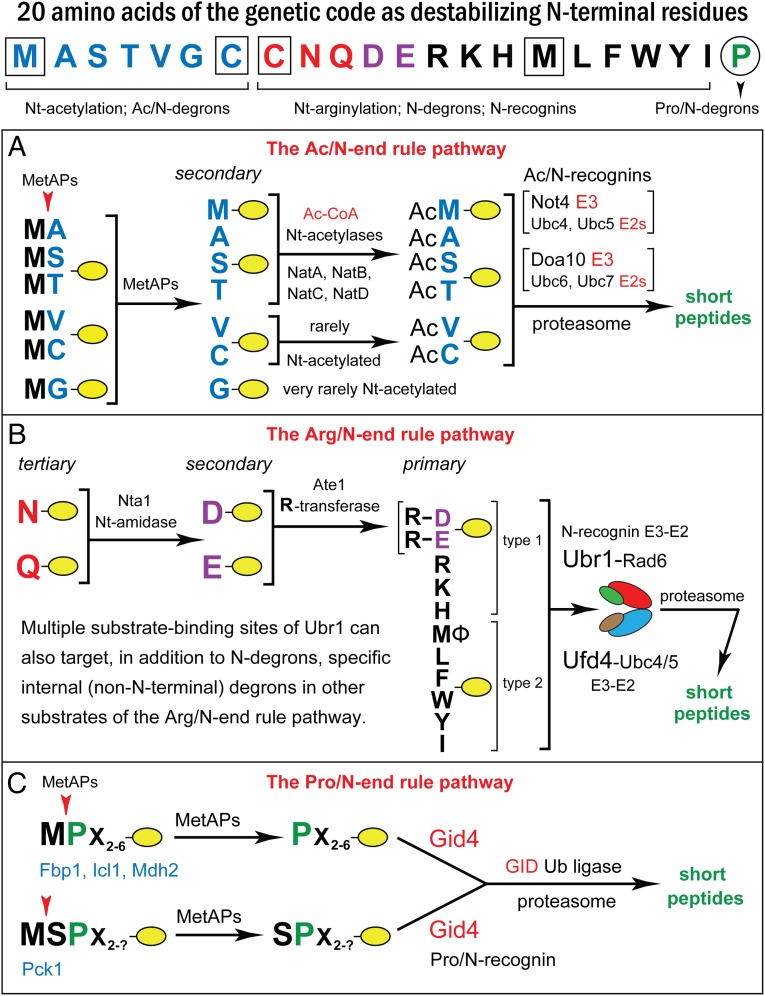
The N-end rule pathways comprise a set of proteolytic systems whose unifying feature is their ability to recognize proteins containing N-terminal (Nt) degradation signals called N-degrons, thereby causing the degradation of these proteins by the proteasome or autophagy in eukaryotes and by the proteasome-like ClpAP protease in bacteria (Fig. 1) (15–26). The main determinant of an N-degron is a destabilizing Nt-residue of a protein. Many N-degrons become active through their enzymatic Nt-acetylation, Nt-formylation, Nt-deamidation, Nt-arginylation, or Nt-leucylation. Studies over the last three decades have shown that all 20 amino acids of the genetic code can act, in cognate sequence contexts, as destabilizing Nt-residues. Consequently, many, possibly most, proteins in a cell are conditionally short-lived N-end rule substrates, either as full-length proteins or as protease-generated natural protein fragments (21, 27). Recognition components of N-end rule pathways are called “N-recognins.” In eukaryotes, N-recognins are E3 ubiquitin (Ub) ligases that can target N-degrons (Fig. 1).
Fig. 1.
The N-end rule pathways. N-terminal residues at the top of the diagram are indicated by single-letter abbreviations. Twenty DNA-encoded amino acids are arranged to delineate three sets of N-degrons in S. cerevisiae, corresponding to three N-end rule pathways. N-terminal Met is cited twice, because it can be recognized by the Ac/N-end rule pathway (as Nt-acetylated N-terminal Met) and by the Arg/N-end rule pathway (as unacetylated N-terminal Met). N-terminal Cys is also cited twice, because it can be recognized by the Ac/N-end rule pathway (as Nt-acetylated Cys) and by the Arg/N-end rule pathway (as an oxidized, Nt-arginylatable N-terminal Cys, denoted as Cys* and formed in multicellular eukaryotes but apparently not in unstressed S. cerevisiae). (A) The Ac/N-end Rule pathway. (B) The Arg/N-end rule pathway. (C) The Pro/N-end rule pathway. See the Introduction for references and brief descriptions of these pathways.
Regulated degradation of proteins or their fragments by N-end rule pathways has been shown to mediate a wide range of processes, including the sensing of oxygen, nitric oxide (NO), heme, and short peptides; the control of subunit stoichiometry in protein complexes; the elimination of misfolded proteins; the degradation of proteins retrotranslocated to the cytosol from other compartments; the regulation of apoptosis and repression of neurodegeneration; the regulation of DNA repair, transcription, replication, and chromosome cohesion/segregation; the regulation of G proteins, cytoskeletal proteins, autophagy, gluconeogenesis, peptide import, meiosis, immunity, circadian rhythms, fat metabolism, cell migration, cardiovascular development, spermatogenesis, and neurogenesis; the functioning of organs, including the brain, muscle, testis, and pancreas; and the regulation of leaf and shoot development, oxygen/NO sensing, and many other processes in plants (refs. 15–17, 21–24, 28, and the references therein).
Eukaryotes contain three N-end rule pathways. One of them, the Arg/N-end rule pathway, targets unacetylated Nt-residues (Fig. 1B) (18, 29–31). N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, Ile, and Met (if Met is followed by a bulky hydrophobic residue) are directly recognized by N-recognins (15, 27, 32). In contrast, N-terminal Asn, Gln, Glu, and Asp (as well as Cys, under some conditions) are destabilizing because of the enzymatic deamidation of N-terminal Asn and Gln and the arginylation of N-terminal Asp, Glu, and (oxidized) Cys (33–37).
In the yeast Saccharomyces cerevisiae, the Arg/N-end rule pathway is mediated by the Ubr1 N-recognin, a 225-kDa RING-type E3 Ub ligase and a part of the targeting complex comprising the Ubr1–Rad6 and Ufd4–Ubc4/5 E3-E2 holoenzymes (Fig. 1B) (38). Two substrate-binding sites of Ubr1 recognize N-degrons, whereas its other binding sites recognize internal degrons in proteins such as Cup9 or a variety of misfolded proteins (39–43). Thus, physiological substrates of the Arg/N-end rule pathway comprise not only proteins bearing N-degrons but also proteins containing internal degrons.
Another N-end rule pathway is the Ac/N-end rule pathway. It recognizes Nα-terminally acetylated (Nt-acetylated) residues (Fig. 1A) (20, 27, 28, 44). N-degrons and E3 Ub ligases of the Ac/N-end rule pathway are called Ac/N-degrons and Ac/N-recognins, respectively. At least 60% of S. cerevisiae proteins and ∼90% of human proteins are cotranslationally and irreversibly Nt-acetylated by ribosome-associated Nt-acetylases (Fig. 1A) (45, 46). In examined cases, the presence of the Nt-Ac group in a subunit of a protein complex increases the thermodynamic stability of the complex, because affinity-enhancing contacts by the Nt-Ac group cause a slower dissociation of the Nt-acetylated subunit (as compared with its unacetylated counterpart) from the rest of the complex (47–50). Many, possibly most, Nt-acetylated proteins contain Ac/N-degrons, whose regulation includes their steric shielding in cognate protein complexes (44).
The third eukaryotic N-end rule pathway, the Pro/N-end rule pathway, is mediated by the GID Ub ligase. Gid4, a subunit of GID that functions as the Pro/N-recognin, targets proteins that bear the N-terminal Pro residue or a Pro at position 2, in addition to adjoining (and also required) sequence motifs (Fig. 1C) (21). Physiological substrates of the Pro/N-end rule pathway include gluconeogenic enzymes, which are long-lived in cells deprived of glucose but are selectively destroyed upon return to glucose-replete conditions (21, 51, 52).
We show here that the yeast Hsp90 chaperone system is greatly impaired in naa10Δ cells, which lack the NatA Nt-acetylase and therefore cannot Nt-acetylate a majority of normally Nt-acetylated proteins. By exploring this finding, we identified extensive connections between Hsp90 and the Arg/N-end rule pathway. These and related results revealed a major role of Nt-acetylation in the Hsp90-mediated protein homeostasis, a strong up-regulation of the Arg/N-end rule pathway in the absence of NatA, and also showed that a number of Hsp90 clients are previously unknown substrates of the Arg/N-end rule pathway.
Results
Promoter Reference Technique.
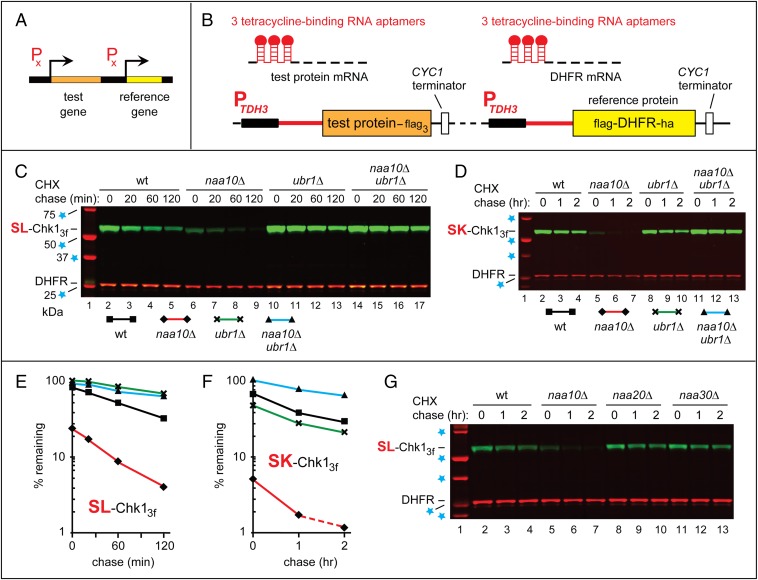
Degradation assays used the promoter reference technique (PRT) (Fig. 2 A and B) (21). In this method, one measures, during a chase, the ratio of a test protein to a long-lived reference protein, the mouse dihydrofolate reductase (DHFR). The reference protein and test proteins were coexpressed on a low-copy plasmid from two identical PTDH3 promoters containing additional DNA elements developed by others (53). Once transcribed into an mRNA, these elements form 5′-RNA aptamers that can bind to tetracycline (Tc), thereby repressing translation of that mRNA in cis (Fig. 2B). This PRT design improves degradation assays in two independent ways: by providing a reference protein and by allowing chases that involve a gene-specific (Tc-mediated) repression of translation.
Fig. 2.
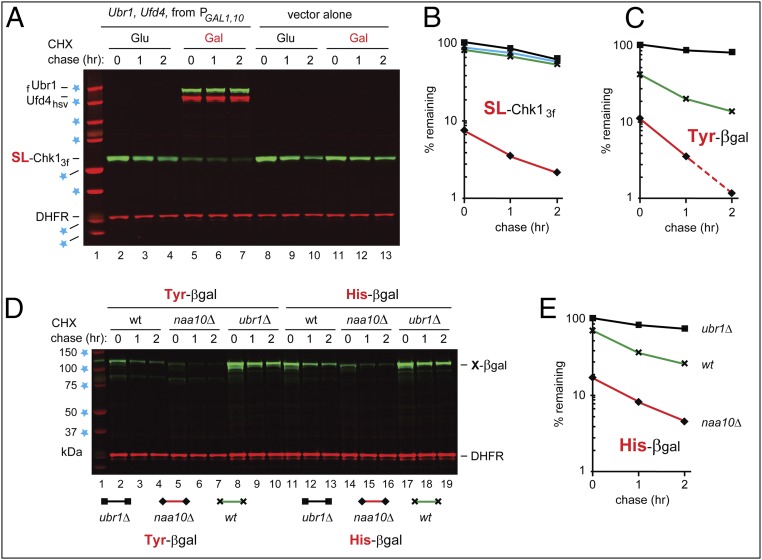
The PRT and degradation of Chk1 in naa10Δ cells. (A and B) The PRT. See the main text for a discussion. (C) Lane 1, kDa markers. CHX-chases, using PRT with wild-type SL-Chk13f, were performed at 30 °C for 0, 20, 60, and 120 min, with wild-type (lanes 2–5), naa10Δ (lanes 6–9), ubr1Δ (lanes 10–13), and naa10Δ ubr1Δ (lanes 14–17) S. cerevisiae. Extracts were prepared from cells withdrawn at the indicated times of a chase. Proteins in an extract were fractionated by SDS/PAGE, followed by immunoblotting with anti-flag and anti-HA antibodies. (D) As in C, but CHX-chases, with the SK-Chk13f mutant, were for 0, 1, and 2 h, with wild-type (lanes 2–4), naa10Δ (lanes 5–7), ubr1Δ (lanes 8–10), and naa10Δ ubr1Δ (lanes 11–13) S. cerevisiae. (E and F) Quantification of data in C and D, respectively. For curve designations, see the keys below the immunoblots in C and D. All degradation assays in this study were performed at least twice, yielding results that differed by less than 10%. (G) As in C, but CHX-chases, with wild-type SL-Chk13f, were for 0, 1, and 2 h, with wild-type (lanes 2–4), naa10Δ (lanes 5–7), naa20Δ (lanes 8–10), and naa30Δ (lanes 11–13) S. cerevisiae. In C, D, and G, lane 1 shows kDa markers and blue stars denote 25-, 37-, 50-, and 75-kDa proteins.
Chk1 Kinase Is Short-Lived in naa10Δ Cells.
This work began with a finding about the S. cerevisiae mitotic checkpoint kinase Chk1 (SL-Chk1, bearing N-terminal Ser-Leu) (54). C-terminally flag-tagged wild-type SL-Chk13f was relatively long-lived in wild-type cells but became short-lived in naa10Δ (ard1Δ) cells, which lacked the NatA Nt-acetylase (Fig. 2 C and E and SI Appendix, Fig. S2 A, C, and D).
The accelerated degradation of SL-Chk13f in naa10Δ cells was manifested, in part, by much lower prechase (time-zero) levels of SL-Chk13f in these cells (Fig. 2 C and E and SI Appendix, Fig. S2 A, C, and D). 35S-pulse-chases with SL-Chk13f produced similar results (SI Appendix, Fig. S2B). We could ascribe the decreased prechase levels of SL-Chk13f in naa10Δ cells to its faster degradation (rather than to a decreased synthesis of Chk1), inasmuch as these assays used PRT and its built-in reference protein (Fig. 2B). As shown earlier, prechase effects analogous to those in naa10Δ vs. wild-type cells stem from the accelerated destruction of nascent and/or newly formed proteins (15, 27, 44). This aspect of SL-Chk13f degradation was particularly prominent in naa10Δ cells (Fig. 2 C and E and SI Appendix, Fig. S2 A, C, and D).
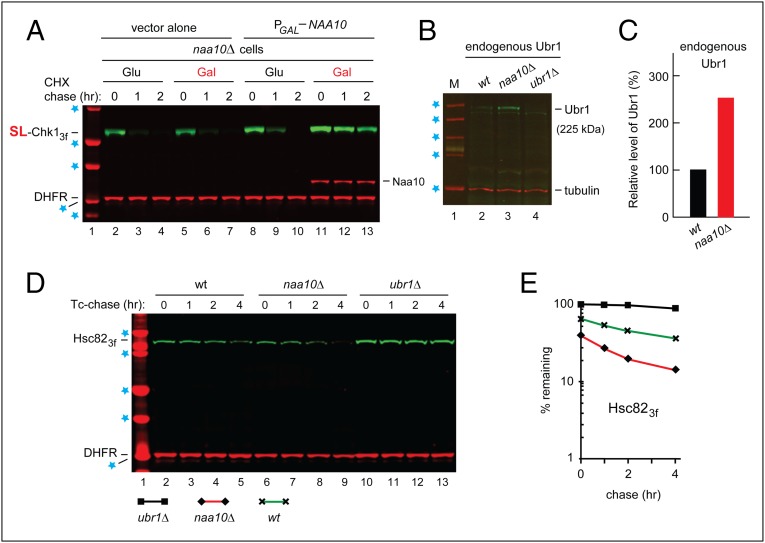
The faster degradation of SL-Chk13f in naa10Δ cells was NatA-specific: This effect was absent in naa20Δ cells (which lacked the NatB Nt-acetylase) and in naa30Δ cells (which lacked the NatC Nt-acetylase) (Fig. 2G). An ectopic expression of NatA (Naa10) in naa10Δ cells (monitored by antibody to Naa10) restored the relative stability of SL-Chk13f (Fig. 3A).
Fig. 3.
Degradation of Hsc82 by the Arg/N-end rule pathway in naa10Δ cells. (A) CHX-chases, using PRT (Fig. 2B), with wild-type SL-Chk13f, for 0, 1, and 2 h, with naa10Δ S. cerevisiae carrying a high-copy plasmid that expressed Naa10 from the PGAL promoter. Lane 1, kDa markers. Blue stars denote 20-, 25-, 37-, 50-, and 75-kDa markers, respectively. Lanes 2–4 and 5–7, cells (containing vector alone) in glucose and galactose medium, respectively. Lanes 8–10 and 11–13 are as in lanes 2–4 and 5–7, respectively, but with cells containing the PGAL-NAA10 plasmid. The presence of Naa10 was verified directly, using an affinity-purified antibody to Naa10 (bands in lanes 11–13, above the bands of DHFR). (B) Immunoblotting-based comparison of the levels of endogenous (untagged) Ubr1 in wild-type naa10Δ and ubr1Δ S. cerevisiae (with ubr1Δ cells as a negative control), using an affinity-purified antibody to Ubr1 (see the main text), with immunoblots of tubulin as a loading control. Blue stars denote 50-, 75-, 100-, 150-, and 250-kDa markers, respectively. (C) Quantification of the data in B, with subtraction of the background-level signal in lane 4 (ubr1Δ cells) and with the level of Ubr1 in wild-type cells taken as 100%. (D) Degradation of Hsc82 in naa10Δ cells. Blue stars denote 25-, 37-, 50-, 75-, and 100-kDa markers, respectively. Tc-chases, using PRT (Fig. 2B) with wild-type Hsc823f, were for 0, 1, 2, and 4 h, with wild-type (lanes 2–5), naa10Δ (lanes 6–9), and ubr1Δ (lanes 10–13) S. cerevisiae. Extracts were prepared from cells withdrawn at the indicated times of a chase. Proteins in an extract were fractionated by SDS/PAGE, followed by immunoblotting with anti-flag and anti-HA antibodies. (E) Quantification of data in D. For curve designations, see the keys below the immunoblot in D. All chases in this study were performed at least twice, yielding results that differed by less than 10%.
Chk1 as a Substrate of the Arg/N-End Rule Pathway in naa10Δ Cells.
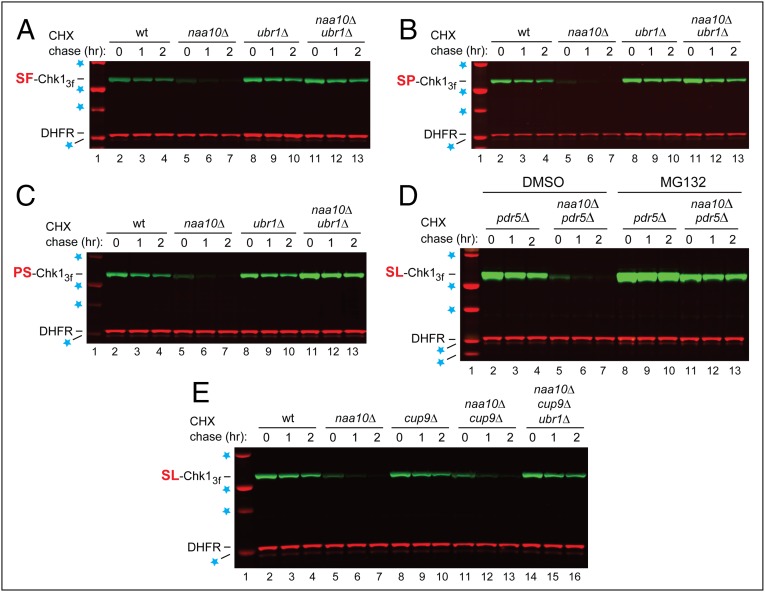
The E3 Ub ligases Ubr1 and Ufd4 are a part of the targeting apparatus of the Arg/N-end rule pathway (Fig. 1B). Remarkably, the ablation of either Ubr1 or Ufd4 abrogated the accelerated degradation of SL-Chk13f in naa10Δ cells. SL-Chk13f was as stable in ubr1Δ naa10Δ and ufd4Δ naa10Δ double mutants as it was in wild-type cells, despite the absence of the NatA (Naa10) Nt-acetylase in double mutants and in contrast to the rapid degradation of SL-Chk13f in single-mutant naa10Δ cells (Figs. 2 C and E and SI Appendix, Fig. S2 A, C, and D). As would be expected (Fig. 1B), the degradation of SL-Chk13f required the proteasome (Fig. 4D).
Fig. 4.
Degradation of Chk1 mutants by the Arg/N-end rule pathway. (A) As in Fig. 2D, but with the SF-Chk13f mutant. CHX-chases, using PRT (Fig. 2B), with SF-Chk13f, for 0, 1, and 2 h, with wild-type (lanes 2–4), naa10Δ (lanes 5–7), ubr1Δ (lanes 8–10), and naa10Δ ubr1Δ (lanes 11–13) S. cerevisiae. Blue stars are as in Fig. 2D. (B) As in A but with the SP-Chk13f mutant. (C) As in A but with the PS-Chk13f mutant. (D) Lanes 2–4, CHX-chase, using PRT (Fig. 2B), with wild-type SL-Chk13f, for 0, 1, and 2 h, in pdr5Δ cells. Lanes 5–7 are as in lanes 2–4 but in pdr5Δ naa10Δ cells. Lanes 8–10 and 11–13 are as in lanes 2–4 and 5–7, respectively, but in the presence of 50 μM MG132, a proteasome inhibitor. The absence of the Pdr5 multidrug transporter was necessary to make S. cerevisiae sensitive to MG132. Blue stars denote 20-, 25-, 37-, 50-, and 75-kDa markers. (E) CHX-chases, using PRT (Fig. 2B), with wild-type SL-Chk13f, for 0, 1, and 2 h, with wild-type (lanes 2–4), naa10Δ (lanes 5–7), cup9Δ (lanes 8–10), naa10Δ cup9Δ (lanes 11–13), and naa10Δ cup9Δ ubr1Δ (lanes 14–16) S. cerevisiae. Blue stars denote 25-, 37-, 50-, and 75-kDa markers.
Degradation of Chk1 in naa10Δ Cells Does Not Involve the Cup9 Regulon.
Cup9, a transcriptional repressor of a regulon containing ∼50 genes, is conditionally destroyed by the Arg/N-end rule pathway through an internal degron of Cup9 (39). It was conceivable that the tripartite Cup9–Tup1–Ssn6 repressor complex (39) might dissociate more rapidly in the absence of NatA-mediated Nt-acetylation, because of the “affinity” effect of Nt-Ac groups (described in the Introduction). This effect would lead to a faster degradation of Cup9 by the Arg/N-end rule pathway in naa10Δ cells and, consequently, to an induction of the Cup9 regulon, whose functions are incompletely understood. We therefore asked whether the faster degradation of SL-Chk13f in naa10Δ cells might be mediated by the Arg/N-end rule pathway indirectly, through a faster destruction of Cup9 and the resulting up-regulation of a member of the Cup9 regulon (e.g., an uncharacterized Ub ligase) that would accelerate the degradation of SL-Chk13f in naa10Δ cells. This model was found to be incorrect, inasmuch as SL-Chk13f remained relatively long-lived in cup9Δ cells, in which the Cup9 regulon becomes derepressed (Fig. 4E) (39).
Endogenous Chk1 and Its Targeting by the Arg/N-End Rule Pathway.
To address the degradation of endogenous SL-Chk1, we replaced the chromosomal CHK1 gene with a DNA segment that expressed wild-type SL-Chk1 C-terminally tagged with myc (SL-Chk19m) from the endogenous PCHK1 promoter. The prechase (time-zero) level of SL-Chk19m in wild-type cells was ∼2.5-fold lower than in ubr1Δ cells (which lacked the Arg/N-end rule pathway), confirming that a large fraction of the endogenous SL-Chk19m was destroyed by the Arg/N-end rule pathway in wild-type cells (SI Appendix, Fig. S3 C and D). Moreover, the prechase level of endogenous SL-Chk19m was further decreased in naa10Δ cells, becoming ∼4.7-fold lower than in ubr1Δ cells (SI Appendix, Fig. S3 C and D). These findings were qualitatively similar to the results with (moderately) overexpressed SL-Chk13f (Fig. 2 C and E and SI Appendix, Fig. S2 A, C, and D).
Degradation of Chk1 in naa10Δ Cells Does Not Involve an N-Degron.
Might a small unacetylated Nt-residue (such as Ser) followed by a bulky hydrophobic residue (such as Leu) function, in the contexts of SL-Chk13f and naa10Δ cells, as a previously unknown N-degron? To address this question, we constructed position-1, 2 mutants of SL-Chk13f. Both wild-type SL-Chk13f and its mutants SD-Chk13f and SF-Chk13f were predicted substrates of the NatA Nt-acetylase. In contrast, the SK-Chk13f and SR-Chk13f mutants were unlikely to be Nt-acetylated in wild-type cells, because a position-2 basic residue impedes Nt-acetylation in yeast. The PS-Chk13f and SP-Chk13f mutants, which contained Pro at positions 1 and 2, respectively, were not Nt-acetylated in wild-type cells (45).
The results with XZ-Chk13f proteins (X = S, P; Z = L, D, F, K, R, P), including wild-type SL-Chk13f, were essentially indistinguishable. All these proteins were degraded relatively slowly in wild-type cells, became much shorter-lived in naa10Δ cells, and required the Arg/N-end rule pathway for their degradation in naa10Δ cells (Figs. 2 C–F and 4 A–C and SI Appendix, Fig. S2 E and F). We concluded that XZ-Chk13f (X = S, P; Z = L, D, F, K, R, P), including wild-type SL-Chk13f, were targeted by the Arg/N-end rule pathway in naa10Δ cells through an internal degron, which was largely inactive in wild-type cells (see below).
Hsp90 and Cochaperones as Substrates of the NatA Nt-Acetylase.
Approximately 60% of proteins in the S. cerevisiae proteome are predicted substrates of NatA (Naa10) (SI Appendix, Fig. S1) (45). A parsimonious explanation of the accelerated degradation of SL-Chk13f in naa10Δ (NatA-lacking) cells is that in wild-type cells SL-Chk13f is shielded from the Arg/N-end rule pathway by a protective entity that becomes impaired in the absence of Nt-acetylation by NatA. Given the moderate overexpression of SL-Chk13f in our assays (Fig. 2B), a protective entity had to be relatively abundant. The leading candidate was Hsp90, inasmuch as SL-Chk1 is a putative client of Hsp90 (55).
Hsc82 and Hsp82 are two similar S. cerevisiae Hsp90 chaperones (5, 8). Remarkably, our survey of the yeast Hsp90 system showed that not only Hsc82 and Hsp82, but also their cochaperones Aha1, Cdc37, Cns1, Cpr6, Cpr7, Hch1, Pih1, Ppt1, Rvb1, Rvb2, Sba1, Sse1, Sse2, Sti1, and Tah1 were either identified or predicted substrates of the NatA Nt-acetylase (SI Appendix, Figs. S1 and S5).
A bias this high was, a priori, an unlikely finding, because only ∼60% of the S. cerevisiae proteome are either identified or predicted substrates of NatA (45). It is unknown whether the observed near-exclusive preference for NatA by the Hsp90 system (SI Appendix, Figs. S1 and S5) is functionally adaptive, i.e., whether this bias was caused by natural selection. The alternative interpretation is that this pattern resulted largely from a “fluctuation” in a quasi-neutral genetic drift. In either case, these results (SI Appendix, Fig. S5) were consistent with the relevance of Hsp90 to degradation of SL-Chk13f in naa10Δ cells (Fig. 2 C and E and SI Appendix, Fig. S2 A, C, and D).
Faster Degradation of Chk1 in Cells Containing Lower Levels of Hsp90.
If destabilization of SL-Chk13f in naa10Δ cells is caused by a lower Hsp90 activity, a decreased level of Hsp90 would be expected to accelerate the degradation of SL-Chk13f in Naa10+ cells. S. cerevisiae Hsp90 is encoded by the constitutive HCS82 and the stress-inducible HSP82, whose single nulls (but not a double null) yield viable cells (8). Immunoblotting-based comparisons of the levels of Hsc82+Hsp82 in wild-type cells vs. hsc82Δ, hsp82Δ, and other mutants are shown in SI Appendix, Fig. S3 D and E. We also found that SL-Chk13f was indeed destabilized in either hsc82Δ cells or hsp82Δ cells but not as strongly as in naa10Δ cells (SI Appendix, Fig. S3 A and B).
Ablation of Hsp90 Cochaperones Can Destabilize Chk1.
We also found that SL-Chk13f was shorter lived in Naa10+ cells lacking the cochaperones Sba1, Sse1, Sti1, or Tah1 (SI Appendix, Fig. S6 A and B). These assays used Tc-PRT chases (Fig. 2 A and B). Tc is at most weakly toxic, in contrast to highly toxic cycloheximide (CHX). The basal degradation of SL-Chk13f was faster in Tc-chases than in CHX-chases (e.g., compare Fig. 2 C and E with SI Appendix, Fig. S6C). The cause of this difference was traced to the in vivo degradation of the Ubr1 N-recognin (a study of this aspect of the Arg/N-end rule pathway is under way). Although chases that used CHX (at present the standard translation inhibitor for degradation assays in the Ub field) produced results qualitatively similar to those with Tc, all chases at later stages of the present study were done using Tc, for the reason above.
Hsp90 Itself and Other Proteins as Substrates of Ubr1 in naa10Δ Cells.
We also examined other, quasi-randomly chosen non-Chk1 proteins for their degradation in naa10Δ cells vs. ubr1Δ cells. The Ubr1 targets thus identified were Kar4, a transcriptional regulator of pheromone responses; Tup1, a transcriptional corepressor; Gpd1, glycerol-3-phosphate dehydrogenase; Ste11, a kinase regulator of pheromone responses; and also, remarkably, Hsc82, the main Hsp90 chaperone (Fig. 3 D and E and SI Appendix, Figs. S3 E and F, S6D, and S7). Tup1 and Ste11 are clients of Hsp90 (56); Kar4 and Gpd1 may also be Hsp90 clients; and Hsc82 is itself an Hsp90.
The illuminating result that the Hsp90 (Hsc82) chaperone becomes vulnerable to the Arg/N-end rule pathway in naa10Δ cells (Fig. 3 D and E, and SI Appendix, Fig. S7 C and D) was in agreement with a technically independent finding, that the level of Hsc82+Hsp82 in naa10Δ cells was approximately twofold lower than in wild-type cells (SI Appendix, Fig. S4 D and E). Together, these findings strongly suggested that newly formed Hsc82 and Hsp82 are themselves clients of mature Hsc82/Hsp82, requiring the (unimpaired) Hsp90 system for their conformational maturation and protection from the Arg/N-end rule pathway. The Ubr1-mediated degradation of Hsc82 (Fig. 3 D and E and SI Appendix, Figs. S4 D and E and S7 C and D) would account, at least in part, for the decreased protection, in naa10Δ cells, of Hsp90 clients such as SL-Chk13f (Fig. 2 C and E).
As already observed with SL-Chk13f, one aspect of the Ubr1-dependent patterns of degradation of Kar4, Tup1, Gpd1, Ste11, and Hsc82 was a significant increase in the prechase (time-zero) level of a test protein in ubr1Δ cells (which lacked the Arg/N-end rule pathway), as compared with, e.g., naa10Δ cells. Particularly clear examples of this increase were Hsc82, Ste11, and Gpd1 (Fig. 3 D and E and SI Appendix, Figs. S6D and S7).
A parsimonious interpretation of the relative flatness of decay curves in some of these cases (e.g., SI Appendix, Fig. S6D), with the major change being a time-zero (prechase) alteration, is that “young” molecules of a protein (either cotranslationally or shortly after translation) are particularly vulnerable to the Arg/N-end rule pathway. The subsequent folding and quaternary structure formation (for example, mature Gpd1 is a homodimer) would make these proteins either partly or completely resistant to the Ubr1-mediated degradation through the shielding of a cognate degron and/or through a higher resistance to the unfolding of a targeted protein by the 26S proteasome. Conformational maturation of the above (and other) proteins can be partly reversible and takes place, at least in part, through their (protective) interactions with the Hsp90 system. Thus impairment of Hsp90 in naa10Δ cells increases the vulnerability of some Hsp90 clients, such as conditional Ubr1 substrates, to attacks by the Arg/N-end rule pathway.
An extensively analyzed precedent for the non–first-order (nonexponential) degradation kinetics by the Arg/N-end rule pathway was X–β-galactosidase (X–β-gal) bearing a destabilizing N-terminal residue (18). These first (engineered) Arg/N-end rule substrates were vulnerable to the targeting by Ubr1 cotranslationally and/or shortly after translation. The subsequent formation of an X–β-gal homotetramer led to the nearly (but not quite) complete cessation of the degradation of a “mature” X–β-gal tetramer by the Arg/N-end rule pathway (15, 18, 19). Later studies have shown that a non–first-order degradation kinetics is also a frequent aspect of proteolysis by the Ub system at large, resulting, primarily, from the same causes of time-dependent protein folding and formation of protein complexes (44, 57–59).
As shown below, Ubr1 recognizes an internal, C terminus-proximal degron of SL-Chk1. It is likely (but remains to be verified) that Kar4, Tup1, Gpd1, Ste11, and Hsc82 also bear Ubr1-specific internal degrons (as distinguished from N-degrons). Overall, ∼60% of 11 examined yeast proteins were identified as substrates of the Arg/N-end rule pathway in naa10Δ cells. Given this percentage and the quasi-random selection of proteins for these experiments, it is possible, indeed likely, that roughly a half of natural Hsp90 clients, i.e., ∼10% of the cellular proteome, will eventually be identified as conditional (Hsp90 state-dependent) substrates of the Arg/N-end rule pathway.
Overexpression of Ubr1–Ufd4 Destabilizes Chk1 in Wild-Type Cells.
Natural levels of Ubr1 are rate-limiting in the Arg/N-end rule pathway (15). We therefore asked whether co-overexpression of Ubr1 and Ufd4, two interacting Ub ligases of the Arg/N-end rule pathway (Fig. 1B), might accelerate the degradation of SL-Chk13f in wild-type cells, despite the presence of the NatA Nt-acetylase. Remarkably, we found that they did so (Fig. 5 A and B). This result strongly suggested that the rate of degradation of an Hsp90 client such as SL-Chk13f is determined by a dynamic partitioning of SL-Chk13f between a protected Hsp90-bound state and a “free” state vulnerable to the Arg/N-end rule pathway.
Fig. 5.
Overexpression of the Arg/N-end rule pathway destabilizes Chk1. (A) CHX-chases, using PRT (Fig. 2B), with wild-type SL-Chk13f, for 0, 1, and 2 h, in wild-type S. cerevisiae carrying a high-copy plasmid that expressed both flag-tagged Ubr1 (fUbr1) and Hsv-tagged Ufd4 (Ufd4hsv) from the bidirectional PGAL1,10 promoter. Lanes 2–4 and 5–7: cells containing the PGAL1,10-UBR1/UFD4 plasmid in glucose and galactose medium, respectively. Lanes 8–10 and 11–13 are as in lanes 2–4 and 5–7, respectively, but with cells containing a vector alone. The presence of fUbr1 and Ufd4hsv in lanes 5–7 was verified directly, using anti-flag and anti-Hsv antibodies. Blue stars denote 20-, 25-, 37-, 50-, 75-, 100-, 150-, and 250-kDa markers. (B) Quantification of data in A. Rhombuses, squares, triangles, and crosses denote, respectively, the decay curves of SL-Chk13f in wild-type cells overexpressing Ubr1-Ufd4 (in galactose), or containing vector alone (in glucose), or containing the repressed Ubr1-Ufd4 plasmid (in glucose), or containing vector alone (in galactose). (C) Quantification of data in D, lanes 2–10. For curve designations, see the keys below the immunoblot in D. (D) Lane 1, kDa markers. CHX-chase, using PRT (Fig. 2B), for 0, 1, and 2 h, with wild-type (lanes 2–4), naa10Δ (lanes 5–7), and ubr1Δ (lanes 8–10) S. cerevisiae expressing Tyr–β-gal, derived from Ub–Tyr–β-gal. Lanes 11–19 are as in lanes 2–10 but with His–β-gal, derived from Ub–His–β-gal. Blue stars indicate 37-, 50-, 75-, 100-, and 150-kDa markers. (E) Quantification of data in D, lanes 11–19. For curve designations, see the keys below the immunoblot in D. All chases in this study were performed at least twice, yielding results that differed by less than 10%.
The Endogenous Arg/N-End Rule Pathway Is Up-Regulated in naa10Δ Cells.
We also asked whether the Arg/N-end rule pathway might become naturally up-regulated in naa10Δ cells, which exhibited the accelerated degradation of SL-Chk13f (Fig. 2 C and E and SI Appendix, Fig. S3 A, C, and D). We began by comparing, using an affinity-purified antibody to S. cerevisiae Ubr1 (60), the levels of Ubr1 in wild-type vs. naa10Δ cells, with ubr1Δ cells as a negative control (Fig. 3 B and C). Despite the high sensitivity of this antibody [it could detect, by immunoblotting, down to ∼2 ng of Ubr1 (60)], the levels of endogenous Ubr1 in wild-type cells [500–1,000 Ubr1 molecules per haploid cell in exponential growth (38)] were low enough to make these measurements technically challenging. Nevertheless, we reproducibly observed an ∼2.5-fold increase of the endogenous, untagged Ubr1 in naa10Δ cells as compared with wild-type cells (Fig. 3 B and C).
In a conceptually independent set of assays, we used X–β-gal (in which X = His or Tyr) as Ub fusion-based reporters of the Arg/N-end rule pathway (15, 18, 19). N-terminal His and Tyr are recognized by Ubr1 through its type-1 site (this site binds to basic Nt-residues) and type-2 site (this site binds to bulky hydrophobic Nt-residues), respectively (Fig. 1B). In agreement with the higher levels of Ubr1 in naa10Δ cells (Fig. 3 B and C), the degradation of His–β-gal and Tyr–β-gal was much faster in naa10Δ cells than in wild-type cells (see Fig. 7 C–E). A strong up-regulation of the Arg/N-end rule pathway in the absence of NatA provides a glimpse of still unexplored circuits that regulate the levels of pathway’s components and the activity of this proteolytic system.
Fig. 7.
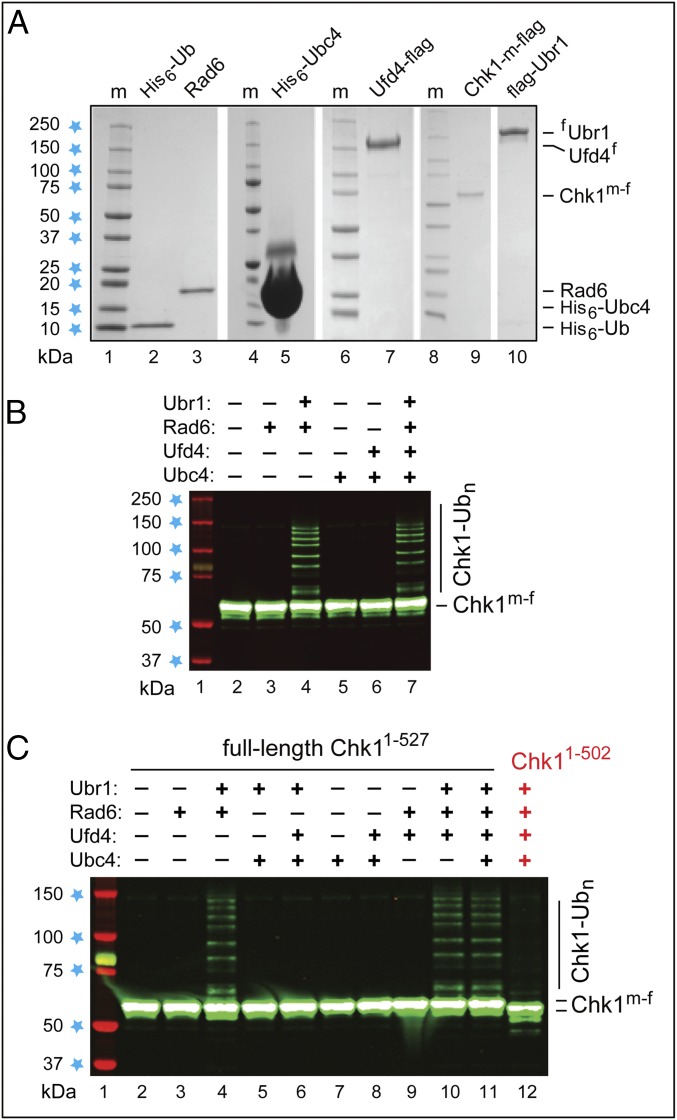
Degron-specific polyubiquitylation of Chk1 in a defined in vitro system. (A) Coomassie-stained SDS/PAGE patterns of purified S. cerevisiae His6-Ub, Rad6, His6-Ubc4, Ufd4f, full-length SL-, and fUbr1. (B) Lane 1, kDa markers. Lanes 2–7, SL- incubated in the presence of ATP, Ub, Uba1, and other purified proteins as indicated. Immunoblotting with anti-flag antibody. (C) As in B but independent Chk1 polyubiquitylation assays that included, in addition, the assay with C-terminally truncated SL- (lane 12).
Mapping the Chk1 Degron by Degradation Assays.
Having determined that the targeting of SL-Chk13f did not involve an N-degron (Figs. 2 C–F and 4 A–C, and SI Appendix, Fig. S2 E and F), we mapped an internal degron of Chk1 by examining its C-terminally truncated derivatives (SI Appendix, Fig. S8). Except for SL- and SL-, the truncated derivatives of SL- could not be expressed in reference-based PRT assays at readily detectable levels in either wild-type or ubr1Δ cells (SI Appendix, Fig. S8), suggesting their rapid (and Ubr1-independent) destruction, likely as the result of their misfolding. The absence, in SL-, of only 25 C-terminal residues of wild-type SL- abolished a decreased prechase level of Chk1 in naa10Δ cells and also nearly sufficed to eliminate the dependence of degradation on Ubr1 (SI Appendix, Fig. S6 A and B). SL- yielded similar results, except that the effect of Ubr1 was now completely gone (SI Appendix, Fig. S6 A and C). Thus, a degron recognized by Ubr1 is located largely within the last 25 residues of the 527-residue Chk1.
Mapping the Chk1 Degron by Two-Hybrid Binding Assays.
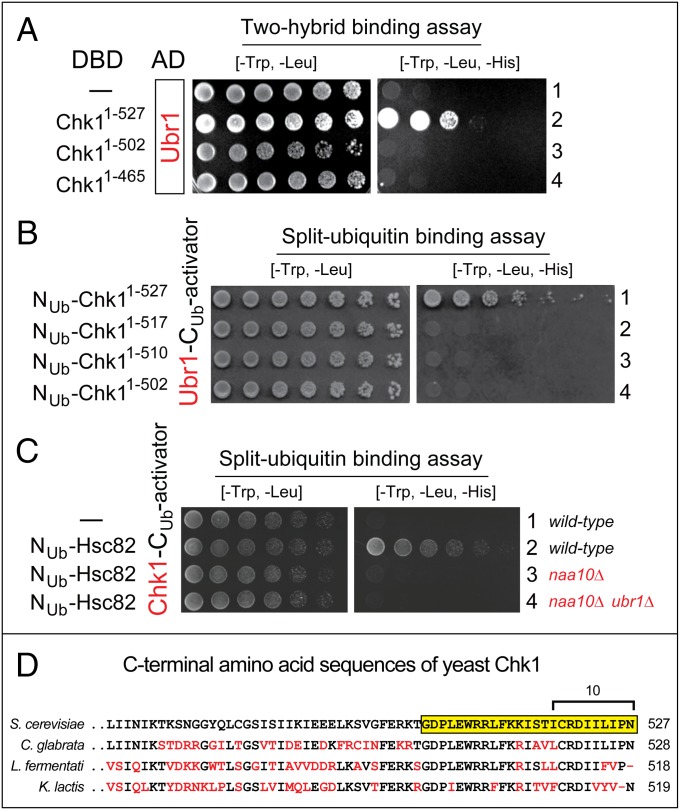
We recently used two-hybrid assays (61) to map the recognition of a degron by its cognate E3 Ub ligase (21). Application of this method to interactions between Chk1 and the Ubr1 E3 is shown in Fig. 6A. Controls included immunoblotting to verify the expression of two-hybrid fusions and also confirming that binding-positive fusions did not stay positive in assays with one of two fusions (e.g., SI Appendix, Fig. 6 A, 1). In agreement with truncation-degradation assays (SI Appendix, Fig. S6), the in vivo interaction between Ubr1 and the 527-residue Chk1 could be abrogated by deleting the last 25 residues of Chk1 (Fig. 6A).
Fig. 6.
Mapping Chk1 degron by two-hybrid and split-ubiquitin assays. (A) Two-hybrid binding assays with Ubr1 vs. Chk1. In both two-hybrid and split-Ub assays, the expression of HIS3 (the ultimate readout of both assays), in otherwise His− cells, was a function of affinity between test proteins. (A–C, Left) Images of His-containing plates, on which all yeast strains grew. (Right) His-lacking plates, on which only His+ cells grew. A two-hybrid-based Ubr1 fusion bearing the activation domain (AD) was examined vs. full-length Chk11–527 and its truncated derivatives bearing the DNA-binding domain (DBD). (A, 1) Ubr1 alone (negative control). (A, 2) Full-length Chk11–527 vs. Ubr1. (A, 3) Chk11–502 vs. Ubr1. (A, 4) Chk11–465 vs. Ubr1. (B) Split-Ub binding assays with Ubr1 vs. Chk1. Split-Ub–based fusions (Materials and Methods) of Ubr1 vs. Chk11–527, Chk11–517, Chk11–510, and Chk11–502. (C) Split-Ub binding assays with full-length Chk1 vs. full-length Hsc82 in wild-type, naa10Δ, and naa10Δ ubr1Δ cells. C1, Chk1 alone (negative control). (D) Alignment of C-terminal regions of Chk1 from the indicated yeast species. The last 25 residues of S. cerevisiae Chk1 are highlighted in yellow. The last 10 residues of Chk1 (found to be essential for the binding to Ubr1; see C, 2) are marked as well. Residues of Chk1 in species other than S. cerevisiae that differed from those of S. cerevisiae Chk1 are in red.
Mapping the Chk1 Degron by Split-Ubiquitin Binding Assays.
In addition and independently, Ubr1–Chk1 interactions were also analyzed by the split-Ub assay. In this method, test proteins are expressed as fusions, respectively, to a C-terminal half of Ub (Cub) and to its mutant N-terminal half (Nub). Interactions between test proteins would reconstitute a quasi-native Ub moiety from Cub and mutant Nub. The resulting cleavage of a Cub-containing fusion by deubiquitylases downstream from the Ub moiety serves as a readout of this technique (62, 63).
In agreement with truncation-degradation and two-hybrid assays (Fig. 6A and SI Appendix, Fig. S6), Ubr1 could bind, in split-Ub assays, to the full-length SL-Chk11–527 but not to SL-Chk11–502 (SI Appendix, Figs. S4A and 6 B, 1 and 4). Moreover, the Ubr1-recognized region of SL-Chk11–527 may be as small as 10 or fewer C-terminal residues. Specifically, neither SL-Chk11–510 nor SL-Chk11–517 could bind to Ubr1, in contrast to wild-type SL-Chk11–527 (Fig. 6B).
Degron-Specific Ubiquitylation of Chk1 in a Defined in Vitro System.
We also developed a ubiquitylation system that comprised the following purified S. cerevisiae proteins: His6-Ub; the Uba1 E1 enzyme; the Rad6 E2 enzyme; the His6–Ubc4 E2 enzyme; the C-terminally flag-tagged Ufd4f E3; the N-terminally flag-tagged fUbr1 E3; the C-terminally myc–flag-tagged full-length SL-; and also, alternatively, SL-, lacking the last 25 residues of Chk1 (Fig. 7A). This completely defined in vitro system could reproduce the recognition of Chk1 by Ubr1 that was inferred from truncation-degradation, two-hybrid, and split-Ub assays (Fig. 6 A and B and SI Appendix, Fig. S6). SL- was polyubiquitylated if the system contained ATP, Ub, Uba1, and at least Ubr1 and Rad6. Ufd4 and Ubc4 had no effect in this system, and Ufd4–Ubc4 by themselves, in the absence of Ubr1, did not support polyubiquitylation of SL- (Fig. 7 B and C). Crucially, the complete system containing SL-, which lacked the 25 C-terminal residues of SL-, could polyubiquitylate the latter but not the former protein (Fig. 7C, lane 12).
In agreement with degron-mapping results (Figs. 6 A and B and 7 and SI Appendix, Fig. S6), alignments of Chk1 amino acid sequences from several yeasts (S. cerevisiae, Candida glabrata, Lachancea fermentati, and Kluyveromyces lactis) revealed a particularly strong conservation of the last ∼25 residues, in comparison, e.g., with the next ∼30 residues upstream of that region (Fig. 6D). The few divergences over the last 25 residues of Chk1 involved solely changes to similar residues (Fig. 6D). In sum, the minimal degron of SL-Chk11–527 comprises its ∼10 C-terminal residues (Fig. 5B), but the complete degron is larger, as suggested, in particular, by a weak but detectable Ubr1 dependence of the degradation of SL-Chk11–502, which lacked the last 25 residues of Chk1 (SI Appendix, Fig. S8B).
Chk1-Hsp90 Interactions in Wild-Type but Not in naa10Δ Cells.
We found that split-Ub assays (62, 63) could also detect in vivo interactions between full-length Chk1 and Hsc82, the main Hsp90 chaperone (Fig. 6 C, 1 and 2 and SI Appendix, Fig. S4 B, 3). This detection required the absence of obstruction of the C terminus of the Hsc82 moiety. Specifically, a split-Ub configuration in which the C terminus of Hsc82 was linked to the Cub-activator moiety yielded a barely detectable signal (SI Appendix, Fig. S4 B, 3 and 4), in agreement with the importance of the C terminus of Hsc82, including its dimerization and its interactions with cochaperones (8).
Remarkably, the same split-Ub assays that yielded a robust Chk1–Hsc82 interaction signal in wild-type cells produced virtually no interaction signal in naa10Δ cells (Fig. 6C and SI Appendix, Fig. S4C). This striking difference could not be caused by a failure of the split-Ub assay itself, because the interactions between Ubr1 and Chk1 were readily detected by split-Ub in either wild-type or naa10Δ cells (SI Appendix, Fig. S4 B, 1 and 2).
The near-abrogation of the in vivo binding of Chk1 to its protector Hsc82 in naa10Δ cells (in contrast to wild-type cells) illuminated still another aspect of the impairment of the Hsp90 system in the absence of NatA. The mechanism of this effect (Fig. 6C and SI Appendix, Fig. S4C) remains to be understood. It may involve not only Hsc82 and Hsp82, because both they and their ∼15 cochaperones are either identified or predicted substrates of the NatA Nt-acetylase that is absent in naa10Δ cells.
Discussion
This study revealed a major impairment of the Hsp90 chaperone in naa10Δ S. cerevisiae, pinpointed causes of this perturbation, showed that the Arg/N-end rule pathway is strongly up-regulated in the absence of the NatA (Naa10) Nt-acetylase, identified several substrates of the Arg/N-end rule pathway, and suggested specific functions of Nt-acetylation in the physiological regulation of the Hsp90 system. In the absence of the NatA Nt-acetylase, roughly 2,000 normally Nt-acetylated yeast proteins lack this modification (45). In particular, the entire Hsp90 system (Hsc82, Hsp82, and its ∼15 cochaperones) would not be Nt-acetylated in naa10Δ cells (SI Appendix, Fig. S5).
One manifestation of Hsp90 impairment in naa10Δ cells was a much faster degradation of Chk1 (a mitotic checkpoint kinase and Hsp90 client) by the Arg/N-end rule pathway, which was found to recognize a specific C terminus-proximal degron of Chk1 (Figs. 1B, 2, 6, and 7). The protection of Chk1 by Hsp90 could be overridden not only by the absence of the NatA Nt-acetylase but also by overexpression of the Arg/N-end rule pathway in wild-type cells (Fig. 5 A and B). This result indicated that the rate of degradation of Chk1 is determined by its dynamic partitioning between a protected Hsp90-bound state and a free state vulnerable to the Arg/N-end rule pathway. Therefore it was possible that the Arg/N-end rule pathway might be naturally up-regulated in naa10Δ cells, because that upregulation would account, at least in part, for the observed destabilization of Chk1 in these cells. The Arg/N-end rule pathway was indeed found to be much more active in naa10Δ cells than in wild-type cells (Figs. 3 B and C and 5 C–E).
Yet another result that revealed a distinct cause of Hsp90 impairment in the absence of NatA was the degradation of Hsc82, the main Hsp90 chaperone, by the Arg/N-end rule pathway in naa10Δ cells and the corresponding (approximately twofold) decrease in the steady-state level of Hsc82+Hsp82 in the absence NatA, relative to wild-type cells (Fig. 3 D and E and SI Appendix, Fig. S7 C and D). The degradation of the Hsp90 chaperone (Hsc82 and presumably Hsp82 as well) by the Arg/N-end rule pathway in naa10Δ cells accounts, at least in part and independently of other causes, for the decreased protection of Hsp90 clients under these conditions.
These and related results strongly suggested three causes of the Hsp90 impairment in naa10Δ cells that are not mutually exclusive. A fourth cause, also stated below, is hypothetical but verifiable and would be physiologically important if correct.
-
i)
A strongly increased activity of the Arg/N-end rule pathway in the absence of NatA and, consequently, an increased ability of this pathway to compete with Hsp90 for its clients such as, for example, Chk1. This finding (Figs. 3 B and C, and 5 C–E) provides a glimpse of still unexplored circuits that regulate components of the Arg/N-end rule pathway and its proteolytic activity.
-
ii)
An intrinsic weakening of Hsp90 in naa10Δ cells, because nearly all its components (Hsc82, Hsp82, and their ∼15 cochaperones) are NatA substrates (SI Appendix, Fig. S5). The resulting impairment of protein interactions that normally involve the Nt-Ac groups of Nt-acetylated proteins (see the Introduction) is likely to underlie the observed degradation of Hsp90 (Hsc82) by the Arg/N-end rule pathway in naa10Δ cells (Fig. 3 D and E and SI Appendix, Figs. S4 D and E and S5 C and D).
-
iii)
In addition to a decreased steady-state level of the Hsc82 (Hsp90) protein in naa10Δ cells (see item ii above), one would also expect the impairment of the Hsp90 system as a whole in the absence of NatA. The functioning of Hsp90 involves its transient binding to cochaperones, with some of them carrying bound clients (12). Given the role of Nt-Ac groups in modulating (usually enhancing) protein interactions within cognate protein complexes (49), decreased affinities among most components of the Hsp90 system, which are not Nt-acetylated in naa10Δ cells, would be expected to perturb both the delivery of clients to Hsp90 by cochaperones and other aspects of this circuit, based on transient protein interactions.
Our detection, via split-Ub assays in wild-type cells, of the in vivo interaction between Hsp90 (Hsc82) and its client Chk1 and the remarkable near-ablation of this interaction in naa10Δ cells (Fig. 6C and SI Appendix, Fig. S4B, 3 and 4) strongly support this view of the Hsp90 system in cells lacking NatA.
-
iv)
The absence of a normally present Nt-Ac group in a subunit of a protein complex can decrease the subunit’s affinity for the rest of the complex from ∼10-fold to ∼1,000-fold (48, 49). Thus, the numerous and normally Nt-acetylated S. cerevisiae proteins that lack this modification in naa10Δ cells would form otherwise cognate in vivo complexes that would be prone to (reversible) dissociation. Away from their natural protein ligands, at least some dissociated free subunits would be likely to act as conformationally disordered Hsp90 clients, thereby competing with normal Hsp90 clients and, consequently, impairing protein homeostasis for that reason alone. Note that some of these free proteins, dissociated from complexes that would be less stable in naa10Δ cells, may even not require assistance by Hsp90 cochaperones for binding to Hsp90. (The fraction of in vivo Hsp90 clients that can interact with Hsp90 directly, without their initial binding to cochaperones, is not known.) This fourth mechanism of Hsp90 impairment in naa10Δ cells remains to be addressed experimentally.
One feature of naa10Δ S. cerevisiae is the remarkably broad range of this mutant’s abnormal phenotypes. They include cell-cycle and mating defects as well as temperature sensitivity and perturbations of protein synthesis and degradation (27, 46, 64). Our finding of specific connections between the Hsp90-mediated protein homeostasis and the Nt-acetylation of proteins by NatA has revealed a major reason for the multiplicity of naa10Δ effects, given the wide range of Hsp90 functions and their impairment in the absence of NatA. Strong evolutionary conservation of the Hsp90 system, Nt-acetylases, and N-end rule pathways indicates that the results of this study are also relevant to multicellular eukaryotes, including mammals.
Given the advances described above, an important question is whether a perturbation of the Hsp90 system that is similar to the impairment observed with naa10Δ S. cerevisiae can also occur in nonmutational settings. Decreased NatA activity can be caused, for example, by a stress-mediated down-regulation of Ac-CoA, the co-substrate of Nt-acetylases. Indeed, physiologically relevant changes in the levels of Ac-CoA have been shown to affect both the efficacy of Nt-acetylation and the sensitivity of mammalian cells to apoptotic signals (65). It would be illuminating, therefore, to determine a role that the Hsp90 system plays in specific connections between Ac-CoA, Nt-acetylation, and apoptosis, particularly because the Arg/N-end rule pathway has already been shown to be a regulator of apoptosis (22, 29).
Changes in the rate of Nt-acetylation of cellular proteins by NatA can also be caused by altered levels of the NatA Nt-acetylase and/or changes in its enzymatic activity. The expression of NatA in the plant Arabidopsis thaliana and, correspondingly, the in vivo Nt-acetylation of NatA substrates have been found to be controlled by the phytohormone abscisic acid (66). Remarkably, a natural decrease of NatA during drought was shown to be required for plants’ tolerance to drought (66). Analogous regulated changes in the levels of NatA and/or Ac-CoA, as well as connections between these changes and either the Hsp90 system or the Arg/N-end rule pathway, remain to be explored in other organisms, including mammals.
Materials and Methods
For further information, see SI Appendix, SI Materials and Methods.
Yeast Strains, Media, and Genetic Techniques.
The S. cerevisiae strains used in this work are described in SI Appendix, Table S1. Standard techniques were used for strain construction and transformation. S. cerevisiae media included yeast extract/peptone/dextrose (YPD) and Synthetic Complete (SC) medium (Sigma-Aldrich).
Construction of Plasmids.
The plasmids used in this study are described in SI Appendix, Table S2. Construction details are described in SI Appendix, SI Materials and Methods. All final constructs were verified by DNA sequencing.
PRT-Based CHX-Chases, Tc-Chases, and 35S-Pulse-Chase Assays.
The PRT method is described in Fig. 2 A and B. Details of PRT-based CHX-chases and Tc-chases are described in SI Appendix, SI Materials and Methods. Protein bands visualized by immunoblotting were quantified using an Odyssey-9120 Imaging System (LI-COR).
Two-Hybrid Assays.
Yeast-based two-hybrid binding assays (61) were carried out largely as described previously (21). “AD” and “DBD” refer to activation domain and DNA-binding domain, respectively. In both two-hybrid and split-Ub assays (described below), the expression of HIS3 (the ultimate readout of both assays), in otherwise His− cells, was a function of affinity between test proteins.
Split-Ub Assays.
Yeast-based split-Ub binding assays (62, 63) were carried out largely as described previously (21).
In Vitro Ubiquitylation Assay.
Protein components of the assay were expressed in Escherichia coli or S. cerevisiae. The assay was carried as described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Melnykov, I. Printsev, and T. T. M. Vu (California Institute of Technology) for their comments on the manuscript and the present and former members of the A.V. laboratory for their assistance and advice. This work was supported by NIH Grants GM031530 and DK039520 (to A.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705898114/-/DCSupplemental.
References
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and homeostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Horwich AL. Molecular chaperones in cellular protein folding: The birth of a field. Cell. 2014;157:285–288. doi: 10.1016/j.cell.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 3.Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Powers ET, Balch WE. Diversity in the origins of proteostasis networks––a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karagöz GE, Rüdiger SGD. Hsp90 interaction with clients. Trends Biochem Sci. 2015;40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Buchner J. Structure, function and regulation of the hsp90 machinery. Biomed J. 2013;36:106–117. doi: 10.4103/2319-4170.113230. [DOI] [PubMed] [Google Scholar]
- 8.Leach MD, Klipp E, Cowen LE, Brown AJ. Fungal Hsp90: A biological transistor that tunes cellular outputs to thermal inputs. Nat Rev Microbiol. 2012;10:693–704. doi: 10.1038/nrmicro2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D. An interaction network predicted from public data as a discovery tool: Application to the Hsp90 molecular chaperone machine. PLoS One. 2011;6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prodromou C. The ‘active life’ of Hsp90 complexes. Biochim Biophys Acta. 2012;1823:614–623. doi: 10.1016/j.bbamcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer MP, Le Breton L. Hsp90: Breaking the symmetry. Mol Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 12.Verba KA, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352:1542–1547. doi: 10.1126/science.aaf5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiler-Samerotte KA, Zhu YO, Goulet BE, Hall DW, Siegal ML. Selection transforms the landscape of genetic variation interacting with Hsp90. PLoS Biol. 2016;14:e2000465. doi: 10.1371/journal.pbio.2000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 15.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014;24:603–611. doi: 10.1016/j.tcb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 19.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 20.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SJ, Wu X, Wadas B, Oh J-H, Varshavsky A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science. 2017;355:366. doi: 10.1126/science.aal3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eldeeb M, Fahlman R. The-N-end rule: The beginning determines the end. Protein Pept Lett. 2016;23:343–348. doi: 10.2174/0929866523666160108115809. [DOI] [PubMed] [Google Scholar]
- 23.Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: From recognition by N-recognins, to destruction by AAA+proteases. Biochim Biophys Acta. 2012;1823:83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Piatkov KI, Vu TTMH, Hwang CS, Varshavsky A. Formyl-methionine as a degradation signal at the N-termini of bacterial proteins. Microb Cell. 2015;2:376–393. doi: 10.15698/mic2015.10.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cha-Molstad H, et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 2015;17:917–929. doi: 10.1038/ncb3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HK, et al. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 2014;156:158–169. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SE, et al. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347:1249–1252. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA. 2012;109:E1839–E1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brower CS, Piatkov KI, Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell. 2013;50:161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi WS, et al. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol. 2010;17:1175–1181. doi: 10.1038/nsmb.1907. [DOI] [PubMed] [Google Scholar]
- 33.Kim MK, Oh SJ, Lee BG, Song HK. Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc Natl Acad Sci USA. 2016;113:12438–12443. doi: 10.1073/pnas.1612620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wadas B, Piatkov KI, Brower CS, Varshavsky A. Analyzing N-terminal arginylation through the use of peptide arrays and degradation assays. J Biol Chem. 2016;291:20976–20992. doi: 10.1074/jbc.M116.747956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu R-G, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 36.Brower CS, Varshavsky A. Ablation of arginylation in the mouse N-end rule pathway: Loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One. 2009;4:e7757. doi: 10.1371/journal.pone.0007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 38.Hwang CS, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 40.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prasad R, Kawaguchi S, Ng DT. Biosynthetic mode can determine the mechanism of protein quality control. Biochem Biophys Res Commun. 2012;425:689–695. doi: 10.1016/j.bbrc.2012.07.080. [DOI] [PubMed] [Google Scholar]
- 43.Nillegoda NB, et al. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aksnes H, Drazic A, Marie M, Arnesen T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci. 2016;41:746–760. doi: 10.1016/j.tibs.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Dörfel MJ, Lyon GJ. The biological functions of Naa10 - From amino-terminal acetylation to human disease. Gene. 2015;567:103–131. doi: 10.1016/j.gene.2015.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becker T, Weber K, Johnsson N. Protein-protein recognition via short amphiphilic helices; a mutational analysis of the binding site of annexin II for p11. EMBO J. 1990;9:4207–4213. doi: 10.1002/j.1460-2075.1990.tb07868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monda JK, et al. Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure. 2013;21:42–53. doi: 10.1016/j.str.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Kulkarni K, Hanrahan SJ, Thompson AJ, Barford D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010;29:3733–3744. doi: 10.1038/emboj.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menssen R, et al. Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J Biol Chem. 2012;287:25602–25614. doi: 10.1074/jbc.M112.363762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung G-C, Brown CR, Wolfe AB, Liu J, Chiang H-L. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 53.Kötter P, Weigand JE, Meyer B, Entian K-D, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tapia-Alveal C, Calonge TM, O’Connell MJ. Regulation of Chk1. Cell Div. 2009;4:8. doi: 10.1186/1747-1028-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandal AK, et al. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J Cell Biol. 2007;176:319–328. doi: 10.1083/jcb.200604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 57.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McShane E, et al. Kinetic analysis of protein stability reveals age-dependent degradation. Cell. 2016;167:803–815. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 59.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang CS, Varshavsky A. Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc Natl Acad Sci USA. 2008;105:19188–19193. doi: 10.1073/pnas.0808891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vidal M, Fields S. The yeast two-hybrid assay: Still finding connections after 25 years. Nat Methods. 2014;11:1203–1206. doi: 10.1038/nmeth.3182. [DOI] [PubMed] [Google Scholar]
- 62.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dünkler A, Müller J, Johnsson N. Detecting protein-protein interactions with the split-ubiquitin sensor. Methods Mol Biol. 2012;786:115–130. doi: 10.1007/978-1-61779-292-2_7. [DOI] [PubMed] [Google Scholar]
- 64.Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun. 2014;5:4383. doi: 10.1038/ncomms5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi CH, et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell. 2011;146:607–620. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Linster E, et al. Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat Commun. 2015;6:7640. doi: 10.1038/ncomms8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.