Significance
An increasing number of proteins have been shown to undergo liquid–liquid phase separation in response to changes in their environment, resulting in formation of a dense protein-rich phase (coacervate), and plays an important role in several systems regulating the growth and development of cells and tissues. Determining the effects of phase separation on protein structure and dynamics is critical for understanding how it modulates protein function. However, structural studies have been limited by the intrinsic disorder and decreased mobility of coacervated proteins. We report direct observation of protein structure and dynamics during the phase transition of an elastomeric protein. Despite large changes in dynamics, coacervation has little effect on protein structure, such that intrinsic disorder is retained.
Keywords: phase separation, elastin, NMR, protein structure, dynamics
Abstract
Despite its growing importance in biology and in biomaterials development, liquid–liquid phase separation of proteins remains poorly understood. In particular, the molecular mechanisms underlying simple coacervation of proteins, such as the extracellular matrix protein elastin, have not been reported. Coacervation of the elastin monomer, tropoelastin, in response to heat and salt is a critical step in the assembly of elastic fibers in vivo, preceding chemical cross-linking. Elastin-like polypeptides (ELPs) derived from the tropoelastin sequence have been shown to undergo a similar phase separation, allowing formation of biomaterials that closely mimic the material properties of native elastin. We have used NMR spectroscopy to obtain site-specific structure and dynamics of a self-assembling elastin-like polypeptide along its entire self-assembly pathway, from monomer through coacervation and into a cross-linked elastic material. Our data reveal that elastin-like hydrophobic domains are composed of transient β-turns in a highly dynamic and disordered chain, and that this disorder is retained both after phase separation and in elastic materials. Cross-linking domains are also highly disordered in monomeric and coacervated ELP3 and form stable helices only after chemical cross-linking. Detailed structural analysis combined with dynamic measurements from NMR relaxation and diffusion data provides direct evidence for an entropy-driven mechanism of simple coacervation of a protein in which transient and nonspecific intermolecular hydrophobic contacts are formed by disordered chains, whereas bulk water and salt are excluded.
The liquid–liquid phase separation (LLPS) of molecules has long been exploited to concentrate and encapsulate molecules for drug delivery and food preparation (1, 2). In biology, there is increasing awareness that many proteins exhibit this type of phase behavior, allowing transient microenvironments to be quickly assembled and disassembled in response to changing solution conditions, or the availability of binding partners (3, 4). Protein phase separation occurs intracellularly to generate various membraneless organelles, such as ribonucleoprotein (RNP) bodies involved in nucleic acid processing, transport, and storage (5). A similar phenomenon is observed in the extracellular matrix as a critical step in the synthesis of elastic fibers, which provide extensibility, recoil, and resilience to tissues (6, 7). In the latter system, LLPS of monomeric elastin results in hydrated protein-rich coacervate droplets that are deposited and cross-linked to form an elastic matrix (7, 8). In addition to their fundamental importance to biology, the dynamic reversibility of LLPS makes phase-separated states of proteins an attractive platform for development of responsive biomaterials with broad application, for instance as scaffolds for tissue engineering or as carriers in drug delivery systems (9–11).
Despite the keen interest in proteins that undergo stimulus-responsive phase separations, the highly disordered nature of proteins known to undergo LLPS, as well as unfavorable dynamics caused by increased viscosity in the phase-separated state, has impeded high-resolution studies. Thus, the molecular structure and dynamics of the phase-separated states, as well as the mechanisms through which they form, remain poorly understood aside from a small number of standout examples. In general, LLPS requires that transient protein–protein interactions become favored over interactions with the bulk solvent and takes two general forms: complex and simple coacervation. Although both have been observed for intrinsically disordered proteins (IDPs), the sequence requirements and physical basis of each type of LLPS are distinct (4, 12).
Several recent studies have shown that a combination of electrostatic, cation-π, and hydrophobic interactions can drive complex coacervation of intrinsically disordered RNP-associated proteins, highlighting the enthalpic nature of this type of LLPS (13–15). Conversely, simple coacervation is considered an entropically driven process that depends entirely on hydrophobic interactions between protein chains (4, 16), although few molecular details of this process have been reported. Thus, a detailed understanding of protein structure and dynamics during LLPS is needed to better define the principles governing protein phase separation and to further develop applications of this type of self-assembly in biomaterial design.
The elastin monomer, tropoelastin, undergoes a well-characterized lower critical solution temperature (LCST) phase transition that is promoted by the presence of salt and heat (8, 17, 18). Tropoelastin is an IDP consisting of an alternating arrangement of hydrophobic domains (HDs) and cross-linking domains (CLDs) (7, 19). The HDs are required for coacervation and elasticity and are disordered domains composed of pseudorepetitive sequence motifs enriched in glycine, proline, and hydrophobic amino acids (20–26). In contrast, the CLDs provide structural integrity to polymeric elastin through covalent cross-linking of lysine residues (27, 28). Current knowledge of elastin structure comes primarily from low-resolution spectroscopic and solid-state NMR studies (29–35), or from modeling and theoretical data (20, 22, 24, 36–38), with few molecular details of the elastin LLPS having been reported.
We, and others, have shown that elastin-like polypeptides (ELPs) derived from the native sequence of elastin act as suitable proxies to study phase separation and elasticity (18, 28, 31, 39). In the present study, we highlight the design and characterization of an elastin mimetic that undergoes phase separation and can be cross-linked to create resilient biomaterials whose assembly can be monitored by NMR to obtain site-specific resolution of structure and dynamics of both the phase-separated and cross-linked materials. Here, we demonstrate that, whereas the phase-separated state of an ELP is dynamically heterogeneous, local structural elements present within the hydrophobic domains remain largely unchanged from monomeric protein through coacervation to cross-linked elastomeric material. These results not only provide mechanistic insight into the LLPS behavior of elastin with an unprecedented level of detail, but also deepen our fundamental understanding of simple coacervation.
Results
Design of an ELP for Site-Specific Monitoring of Structure and Dynamics.
Atomic resolution studies of tropoelastin have primarily been inhibited by its intrinsic disorder combined with the degenerate yet repetitive nature of its amino acid sequence. These properties not only preclude the use of crystallography and electron microscopy, but also lead to substantial overlap in NMR spectra, inhibiting resonance assignment. To circumvent these challenges, we designed a simplified elastin mimetic that facilitated NMR studies while retaining the fundamental biochemical properties of tropoelastin: the alternating domain architecture, intrinsic disorder, the ability to undergo liquid–liquid phase separation, and the capacity to form elastic biomaterials when cross-linked at lysine residues.
This recombinantly expressed ELP contains an alternating arrangement of CLDs and HDs; schematically represented as CLD−(HD−CLD)n and represented in the text as ELPn, with n being the number of repeating units (here n = 1–3; SI Appendix, Fig. S1). These CLDs and HDs contain repetitive motifs that are representative of those observed in all amniote tropoelastin sequences analyzed to date, including in humans (19). The CLD sequence is AAAAAKAAKYGA (numbered 1–12 for NMR resonance assignment), whereas each 35-residue HD sequence was composed of GVPGV (numbered G1 through V5) repeated seven times.
ELP3 Recapitulates the Secondary Structure and Material Properties of Tropoelastin.
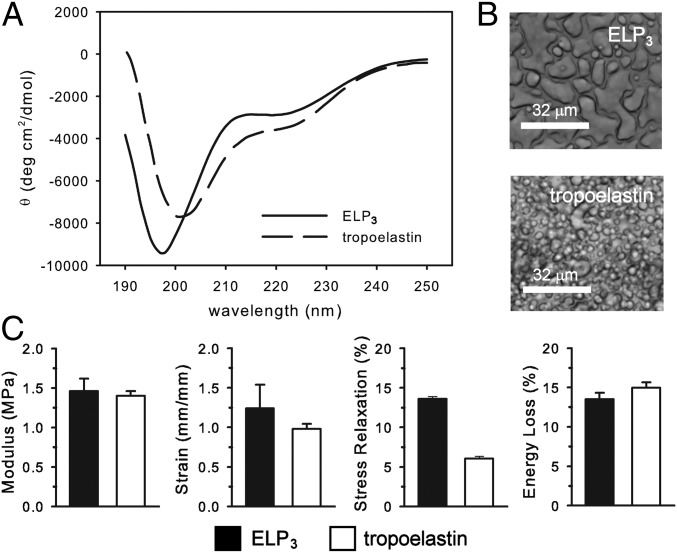
Far UV circular dichroism (CD) spectroscopy of monomeric ELP3 in aqueous solution gave results consistent with previous studies of tropoelastin and elastin-like polypeptides (28, 30, 31) (Fig. 1A). The characteristic weak minimum ellipticity at 222 nm and strong minimum near 200 nm are consistent with the presence of a small amount of α-helical structure in a protein that is otherwise intrinsically disordered.
Fig. 1.
The intrinsically disordered ELP3 recapitulates the phase behavior and elasticity of tropoelastin materials. (A) CD spectra of ELP3 and tropoelastin monomers in 50 mM sodium phosphate, pH 7 at 20 °C, contain features that are indicative of highly disordered proteins. (B) Bright-field microscopy of temperature-induced phase separation of ELP3 and tropoelastin in 1.5 M NaCl solution after 1 h of incubation at 50 °C. (White scale bars, 32 μm.) (C) Mechanical properties of polymeric ELP3 and tropoelastin coacervates cross-linked at lysine residues by genipin. Error bars correspond to the SEM.
ELP3 coacervated and formed a protein-rich liquid phase under conditions of heat and salt similar to those established for other ELPs and for tropoelastin (31, 32) (Fig. 1B). Cross-linking coacervated ELP3 resulted in elastic materials with tensile properties remarkably similar to materials formed from tropoelastin (40, 41): a low elastic modulus, high strain to break, and low energy loss (Fig. 1C). The stress relaxation of ELP3 was greater than that of tropoelastin, but remains in the normal range for elastins and ELPs (41, 42). Taken together, these data demonstrate that ELP3 recapitulates the fundamental macroscopic properties of the monomeric, coacervated, and cross-linked forms of tropoelastin.
NMR Resonance Assignment of Monomeric and Phase-Separated ELP3.
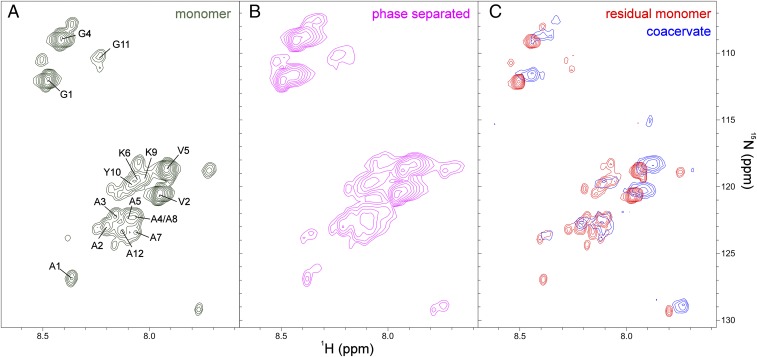
Recombinant ELP3 was uniformly labeled with 15N and 13C to facilitate characterization by NMR spectroscopy. The 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of the monomer showed surprisingly well-resolved peaks with relatively narrow line widths (Fig. 2A). After inducing phase separation, the 1H-15N HSQC spectrum contained two overlapping sets of resonances, one with 1HN and 15N chemical shifts similar to those of the monomer and a second set of broader peaks with a small global downfield shift of the 15N and 1HN resonances (Fig. 2B). To allow independent resolution of these two overlapping sets of resonances, we used HSQC pulse sequences designed to selectively remove either resonances with fast R2 relaxation rates or resonances belonging to proteins with fast translational diffusion rates (Fig. 2C). The spectra clearly demonstrate the presence of two distinct species, one that diffuses quickly with slow R2 relaxation, and a second species with enhanced R2 relaxation but significantly slower diffusion. These two ELP3 populations were identified as residual monomers within the protein-poor bulk phase, and coacervated ELP3 in the protein-rich phase, respectively, through analysis of NMR relaxation and pulsed field gradient (PFG) diffusion rates (detailed below).
Fig. 2.
The unique dynamics of ELP3 monomers and coacervate facilitate NMR spectroscopy of the phase transition. (A) The 1H-15N HSQC NMR spectrum of 4.7 mM ELP3 monomer in 50 mM sodium phosphate, 300 mM NaCl, pH 7 at 37 °C. Labeled peaks, corresponding to the amino acids in the repeating sequence motifs of the HDs and CLDs, were assigned as described in Materials and Methods and SI Appendix, Fig. S1. To remove temperature effects on chemical shift and protein dynamics, phase separation was induced at 37 °C by increasing the concentrations of both ELP and NaCl. (B) HSQC spectrum of a 7.8-mM ELP3 sample undergoing liquid–liquid phase separation in a buffer consisting of 50 mM sodium phosphate, 600 mM NaCl, pH 7 at 37 °C. (C) HSQC spectra of the same phase-separated sample shown in B, acquired using an R2 relaxation rate filter to remove signals with R2 > 5 s−1 (red) or a PFG diffusion filter (removing objects with a diffusion rate faster than 10−7 cm2⋅s−1) (blue) to distinguish residual monomeric ELP3 (red) from coacervate (blue).
Complete backbone resonance assignment of monomeric ELP3 was achieved using standard three-dimensional triple resonance NMR experiments, with additional experiments to aid in assignment of proline-rich segments. Representative NMR spectra, showing partial assignment of CLD resonances, are provided in SI Appendix, Fig. S2, and resonance assignments are listed in SI Appendix, Table S1. Due to the repetitive nature of the sequence and the high degree of disorder, most GVPGV repeats gave rise to an equivalent set of NMR signals, with only those residues adjacent to a CLD showing inequivalence. Similarly, only the N- and C-terminal segments of the first and last CLDs gave rise to distinct resonances, suggesting that the CLDs are also largely equivalent. An additional set of weak resonances was identified in NMR spectra of monomeric ELP3, arising from GVPGV repeats containing cis-proline (SI Appendix, Fig. S3).
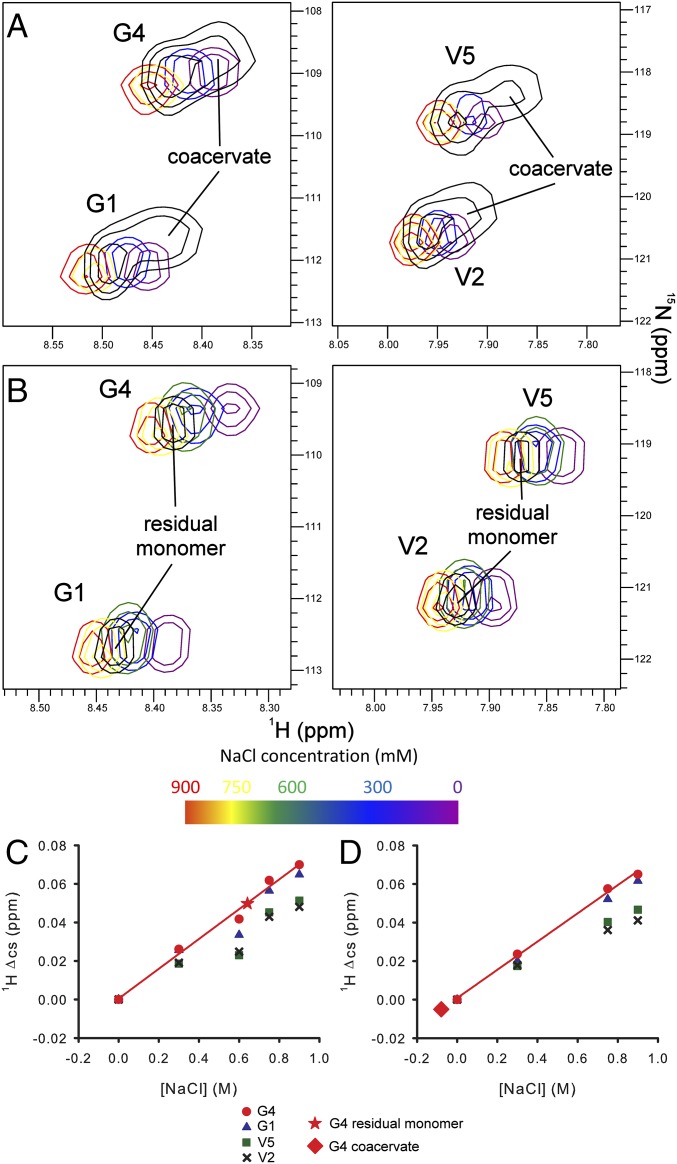
The same methodology was used to obtain near complete assignment of backbone resonances for both the residual monomer and the coacervated species present after LLPS (SI Appendix, Table S2). The increased rates of R2 relaxation exhibited by the slow-diffusing species, combined with a global increase in NMR line widths, led to some ambiguity in sequential assignment of the polyalanine CLD sequences for coacervated ELP3. The global shift in 1HN and 15N chemical shifts observed for the coacervate relative to monomeric ELP3 likely arise from a decreased ionic strength in the coacervate droplets, due to partial exclusion of NaCl (Fig. 3), rather than representing a global structural change.
Fig. 3.
The 1HN chemical shift data reflect altered solution conditions within ELP3 coacervates. The HD peaks from 1H-15N HSQC spectra of ELP3 in 50 mM sodium phosphate, pH 7 with NaCl ranging from 0 to 900 mM are shown in each panel, highlighting the correlation between 1HN chemical shifts and the concentration of NaCl. (A) HSQCs acquired at 37 °C with an ELP3 in 50 mM sodium phosphate, pH 7 and 600 mM NaCl undergoing a phase transition are overlaid in black. (B) HSQCs acquired at 20 °C with residual monomer (coacervate was removed by centrifugation) postcoacervation overlaid in black. Changes in ELP3 monomer 1HN chemical shifts are plotted as a function of NaCl concentration at (C) 37 °C or (D) 20 °C, showing a clear linear dependence of chemical shift on salt concentration. Most HDs also exhibited a small protein concentration-dependent change in chemical shift, as shown by the lower than expected Δcs observed in the 0.6 M NaCl sample with 100 mg/mL ELP3. Linear least squares regression of Δcs (red lines) for the site with the smallest protein concentration dependence (G4) was used to estimate the NaCl concentration experienced by residual monomer (red star) or coacervate (red diamond) after LLPS.
ELP3 Is Highly Disordered in both the Monomeric and Phase-Separated State.
Due to the fast interconversion of conformations in IDPs, the observed NMR chemical shifts represent a weighted average within a structural ensemble (43, 44). Although the presence of a dynamic ensemble precludes determination of a single high-resolution structure, several methods have been developed for prediction of site-specific disorder and secondary structure propensity within a polypeptide sequence, using chemical shift data (45, 46).
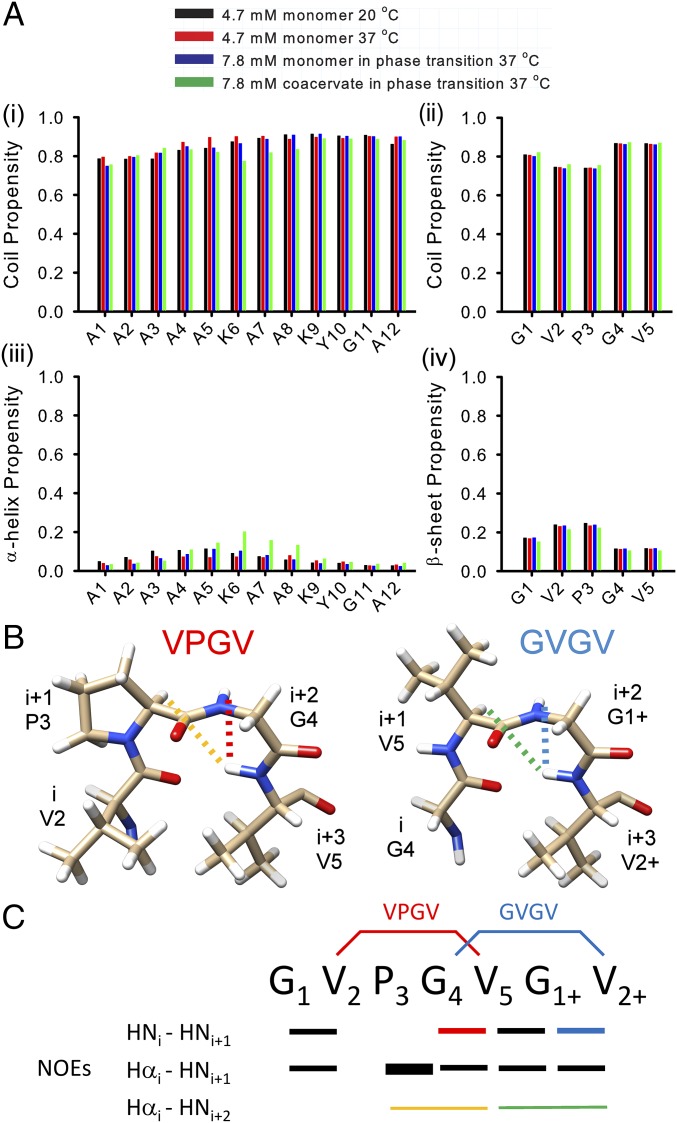
The sequence-specific secondary structure and disorder propensities of monomeric and coacervated ELP3 were determined using the δ2D chemical shift scoring web-server (45). Aside from the salt effect described above, the backbone and side chain chemical shifts were essentially identical in the monomer and coacervate, strongly suggesting that no significant structural changes occurred during LLPS of ELP3. This finding is supported by the δ2D analysis shown in Fig. 4, which predicts a high degree of disorder in both the monomer and coacervate across the entire sequence. The CLDs displayed very low α-helical propensity, as expected based on our CD data, whereas the HD domains had partial β-strand character.
Fig. 4.
The intrinsic disorder of monomeric ELP3 is retained after coacervation. (A) Secondary structure propensities from δ2D analysis of NMR chemical shifts. The random coil propensities for each position in the CLDs (i) and HDs (ii) are shown, as are the α-helix propensities for each position in the CLD sequence (iii) and the β-sheet propensities for HD repeat sequences (iv). (B) Schematic representation of the two β-turns identified in the ELP3 NMR data. The colored dashed lines represent the characteristic connectivities between 1H atoms that were used as identifiers of each turn in NOESY spectra of ELP3. (C) Summary of HN-Hα NOEs observed in the HD sequence of the ELP monomer at 20 °C. The bar thickness in each case represents the relative intensity of the NOE cross-peak, with the colored bars corresponding to the dashed lines in A.
To complement the chemical shift analysis of secondary structure propensity, through-space interactions between 1H nuclei were measured using 2D 1H-1H nuclear Overhauser effect spectroscopy (NOESY) experiments for both the monomer and coacervated ELP3 (Fig. 4 and SI Appendix, Fig. S4). As for the 1H-15N HSQC spectrum, the 1H-1H NOESY of the coacervate displayed broader line widths than the monomer spectra, but otherwise no significant differences were observed, supporting retention of monomer structural properties in the coacervate. Analysis of NOEs arising from short-range interactions between protons in the protein backbone support the formation of two distinct type-II β-turns in the HDs of monomeric and coacervated ELP3, centered on the VPGV and GVGV sequences (Fig. 4 B and C). Importantly, all of the β-turn NOEs were much weaker than expected, relative to reference NOEs, suggesting that these structural elements are only populated 20–40% of the time. Because each site only gives rise to a single, narrow NMR resonance, the rate of exchange between turns and extended conformations is rapid: on the microsecond–nanosecond timescale. Note that, whereas multiple turns could be populated simultaneously, their transient nature and low structural propensity discounts the formation of the β-spiral structure proposed in earlier models of elastin HDs (38). No NOEs characteristic of α-helices were observed for the CLD, supporting the low structure propensity predicted by δ2D.
To probe for intermolecular contacts, 13C and 12C edited 1H-1H NOESY spectra were recorded for monomeric and coacervated samples containing a 1:1 mixture of unlabeled and 13C labeled ELP3 (Fig. 5). Monomeric ELP3 exhibited only intraresidue NOEs in addition to the short-range backbone NOEs identified above. After LLPS, several strong intermolecular NOEs were observed between valine 1Hγ and side chain 1H resonances of alanine, valine, and proline (Fig. 5C). Additional NOEs are also observed between valine 1Hγ and 1Hα of glycine, valine, and proline. These data are consistent with the presence of multiple nonspecific intermolecular hydrophobic contacts within the coacervate. The observed interactions between valines and alanines suggest that, whereas the HDs are required for LLPS of ELPs, CLD sequence composition may influence coacervation, supporting previous studies of CLD mutants (28, 31).
Fig. 5.
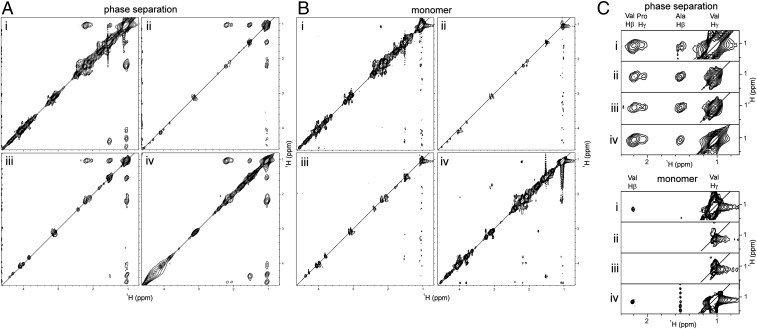
Intermolecular hydrophobic contacts are formed in the phase-separated state of ELP3. The aliphatic regions of 13C double half filtered 1H-1H NOESY spectra, acquired at 37 °C, are shown for phase-separated (A) and monomeric (B) ELP3. For each sample, spectra show NOEs between 1H-13C groups [double 13C selected (i)]; between 1H-12C and 1H-13C groups [13C selected and filtered (ii); 13C filtered and selected (iii)]; or between 1H-12C groups [double 13C filtered (iv)]. Expansions showing NOEs between Val Hγ and Val Hβ, Pro Hγ, or Ala Hβ are shown in C. For the monomeric sample, only intramolecular contacts (i and iv) within Val side chains are observed in C, and some additional weak intraresidue NOEs are observed (B, iv). In the coacervate, intense NOEs are observed in all spectra, indicating the presence of intermolecular Val–Val, Val–Pro, and Val–Ala contacts. The slight increase in coacervate Val–Val NOEs observed in i and iv is due to the additional presence of intraresidue NOEs in these spectra, as seen for the monomeric ELP3.
The Structural Propensities of the ELP3 HD Are Retained in Cross-Linked Materials.
Magic-angle spinning (MAS) solid-state NMR was used to obtain 13C chemical shift data for both coacervated and cross-linked ELP3. Two 13C-13C 2D correlation experiments were used, incorporating either an 1H-13C insensitive nuclei enhanced by polarization transfer (INEPT) transfer for initial 13C excitation, or 1H-13C cross-polarization (CP). The former experiment selects for highly mobile sites within a protein, whereas the latter will preferentially give rise to spectra of relatively immobile segments (47).
The bulk of HD 13C signals are observed after INEPT transfer (SI Appendix, Fig. S5), suggesting that these domains remain highly dynamic in the cross-linked material. The chemical shifts in each state are nearly identical, indicating that the structural propensities of HD in the free monomer are retained even after cross-linking. Highly mobile CLDs observed in the INEPT spectrum of coacervated ELP3 have chemical shifts consistent with a partially helical structure, matching the solution NMR data. In the CP spectrum, a new population of less mobile CLD alanines is observed, with chemical shifts indicative of β-sheets, suggesting a mechanism through which CLD might assemble before cross-linking. As we have reported for another ELP, all CLDs convert to relatively rigid α-helices after cross-linking (31).
Coacervated ELP3 Exhibits Reduced Global Diffusion, but Retains Local Disorder.
Diffusion and R2 relaxation-edited HSQCs showed that LLPS of ELP3 resulted in two distinct populations with very different dynamics. Detailed analysis of these species was carried out to determine the global and local effects of coacervation on the dynamics of the individual protein chains. 15N R1 and R2 relaxation rates and 1H-15N heteronuclear NOEs were obtained for the backbone amide groups of monomeric ELP3, residual monomer post-LLPS, and the coacervate species at 37 °C (SI Appendix, Fig. S6). A general increase in relaxation rates and heteronuclear NOE intensity was observed for all sites in the coacervated species across the sequence, with no discernible local effects, consistent with either a global decrease in fast motions throughout the protein chain, or a reduction in the rate of rotation of the entire protein.
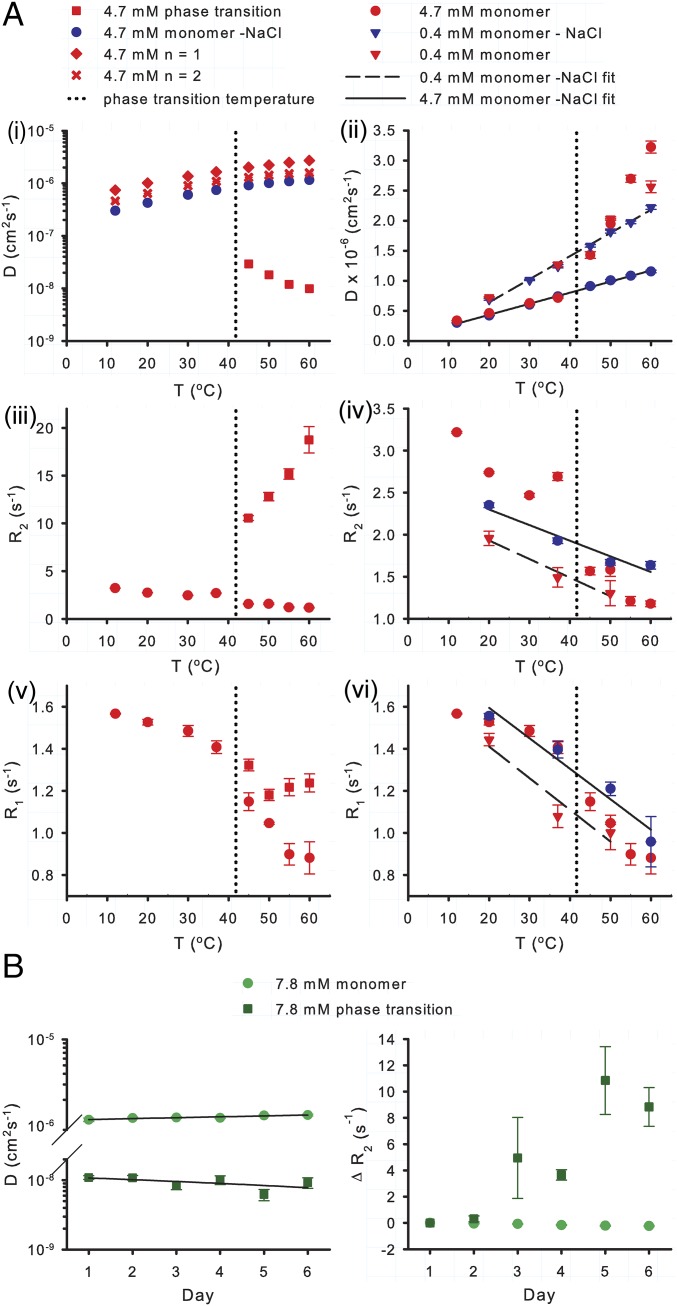
To narrow down the source of increased NMR relaxation rates for coacervated ELP3, R1 and R2 were measured as a function of temperature and both protein and NaCl concentrations. These data were correlated with PFG diffusion measurements made under the same conditions (Fig. 6). The translational diffusion rates of monomeric ELPs of different sizes (ELP1–3, SI Appendix, Fig. S1) and ELP3 coacervate are shown in Fig. 6 A, i. Upon coacervation, the diffusion rate of ELP3 decreased by two orders of magnitude, whereas monomers exhibited increased rates of diffusion with increased temperature and decreased molecular weight. R1 and R2 relaxation (Fig. 6 A, iii and v) follow similar trends, indicating that altered chain dynamics are driven by global rather than local effects. Remarkably, in the coacervated protein, relaxation rates continue to increase at temperatures beyond the phase transition, and diffusion continues to slow. Although this observation runs counter to what is expected for molecules with greater kinetic energy, it is consistent with an entropically driven strengthening of the hydrophobic effect, leading to increased protein–protein interactions even after LLPS has occurred.
Fig. 6.
NMR relaxation and PFG diffusion measurements reveal temperature- and time-sensitive dynamics of phase-separated ELP3. (A) Temperature dependence of ELP3 PFG diffusion rates (i and ii) and 15N R2 or R1 relaxation rates (iii–vi). Odd-numbered panels describe the extreme differences in both relaxation properties and translational diffusion between monomeric ELP3 and ELP3 within a coacervate. Even-numbered panels display the relatively subtle differences in monomer ELP3 dynamics caused by variations in protein or salt concentration. Least squared linear regression fits (solid or dashed lines) of the monomer at high and low protein concentrations in the absence of NaCl show the effect of viscosity on these measurements (ii, iv, and vi). The phase transition temperature for a 4.7-mM sample of ELP3 is indicated in each plot by a vertical dashed line. Error bars indicate relaxation rate fitting error or SEM for diffusion measurements. (B) Time-dependent changes in the diffusion and R2 relaxation rates (mean of G1, V2, G4, and G5 data) of the monomeric and coacervate species in an 8.7-mM sample of ELP3 held at 37 °C (phase transition temperature is 30 °C) for 6 d. Error bars indicate the SEM.
ELP and elastin coacervate droplets have been shown to increase in stability if they are held above the coacervation temperature for an extended period (18, 48, 49). This maturation process has been hypothesized to occur through increased ordering of ELPs at the surface of the droplet, resulting in a less reversible LLPS. In the present work, coacervated ELP3 exhibited an increase in R2 and a decreased rate of diffusion over 6 d following coacervation, suggesting a time-dependent decrease in chain mobility that may be related to maturation (Fig. 6B).
The global reduction in ELP3 dynamics upon LLPS, combined with the lack of structural change, suggests that protein chains within the coacervate experience an environment similar to a highly concentrated monomer solution. Thus, by comparing the diffusion rate of ELP3 within the coacervate with that of high concentrations of monomer, we were able to estimate the protein concentration inside the coacervate phase (SI Appendix, Fig. S7 and Table S3). This estimation gave values of ∼500 mg/mL ELP3 in a protein-rich phase containing ∼62.5% water by mass. This is insufficient water for complete solvation of all ELP3 monomers, highlighting the exclusion of nonessential water and the increase in protein–protein interactions in the coacervate.
A similar increase in HD interactions is observed for monomeric ELP3 chains at elevated temperature and NaCl concentration. When protein concentrations are kept below the threshold for LLPS, the monomer exhibits increased rates of diffusion and reduced R1 and R2 (Fig. 6 A, ii, iv, and vi). These data are consistent with increased compaction of the molecule and suggest that the same forces driving coalescence lead to an intramolecular collapse of the HDs when intermolecular interactions are not present.
Discussion
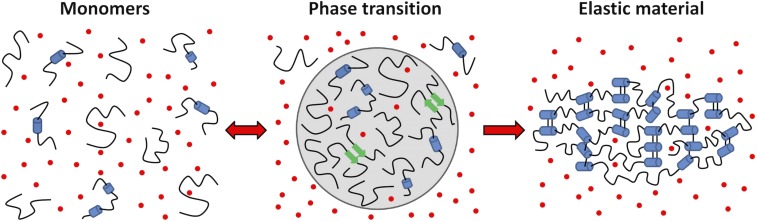
Through a combination of solution and solid-state NMR data, we have tracked the structure and dynamics of an ELP through LLPS and subsequent cross-linking into an elastic material. The resulting data have allowed us to develop a detailed model for ELP self-assembly that builds on early biophysical studies (20–22) and that can serve as a general template for understanding simple coacervation of proteins (Fig. 7). Consistent with this model, recent molecular simulation studies indicate that HDs are disordered and hydrated (25), and that their self-assembly entails formation of intermolecular nonpolar contacts while preserving local secondary structure (50). A frustrated hydrophobic collapse of the HDs is promoted by the increased entropic penalty of solvating these chains under conditions of high salt and temperature. At high protein concentrations, this will lead to intermolecular assembly and formation of a separate phase.
Fig. 7.
Model for ELP self-assembly. Our findings provide support for a comprehensive model for ELP self-assembly in which elevated temperature and salt increase the entropic cost of solvating hydrophobic chains, driving phase separation. Because the protein sequence precludes adoption of a stable folded state, the resulting phase-separated state is maintained through transient and nonspecific HD interactions between monomers. Whereas the coacervate remains hydrated, bulk water is excluded and there is a reduction in Na+ and Cl− ions (red dots), slightly increasing their concentration in the bulk solution. The coacervated ELP can be chemically cross-linked to form an elastic material. HDs remain dynamic and highly disordered throughout the assembly process, whereas CLD motion slows with each step. Partially helical in the monomers (blue cylinders), CLDs exhibit an increased propensity to form β-sheets (green arrows) in the coacervate and become stable α-helices in cross-linked materials.
The inability of Pro/Gly-rich HD sequences to form stable secondary structures (24) results in retention of highly disordered HDs in the coacervated state. This lack of stable structure requires the inclusion of sufficient water to satisfy backbone hydrogen-bonding requirements and potentially to solvate hydrophobic chains in between transient protein–protein interactions mediated by the HDs. All unnecessary water is excluded, as dictated by the entropic cost of its inclusion in the protein-rich phase. The increased ionic strength of the protein-poor solvent phase, due to partial exclusion of salt from the coacervate droplets, also favors coacervation, leading to a reduction in residual monomer. The CLDs remain primarily disordered after coacervation, forming transient helices and small amounts of β-sheet, bringing CLDs together before cross-link formation. After chemical cross-linking, a solid phase is formed in which the HDs retain their essential properties of disorder and high entropy. All structural elements in the HDs remain transient and closely resemble those present in the monomeric ELP, whereas the CLDs adopt a stable helical conformation, as we have previously reported (31).
Whereas the CLD behavior in this model is likely unique to elastin and ELPs, the data presented for the HDs are likely to be more broadly representative of proteins and polymers that undergo LLPS through simple coacervation. Thus, our ability to monitor this process with site-specific resolution not only informs our understanding of elastin assembly, but also allows us to predict how sequence or chemical structure might influence LLPS across many different systems. This approach provides a means to probe important biological questions and also to develop biopolymers with fine-tuned properties of structure and assembly.
Materials and Methods
Protein Expression and Purification.
Expression and purification of recombinant tropoelastin was carried out using previously reported methods (32). All ELPs were expressed in BL21(DE3) Escherichia coli from a pet32-b plasmid (Invitrogen) containing the ELPs fused to an N-terminal thioredoxin, which was removed by cyanogen bromide digestion in 70% formic acid. Each ELP was purified by ion exchange FPLC followed by reverse phase HPLC as described previously for other elastin-like polypeptides (31). When isotopic enrichment of polypeptides was needed for NMR experiments, proteins were expressed from E. coli grown in M9 minimal media supplemented with 15N-NH4Cl and 13C-glucose as the sole sources of nitrogen and carbon.
CD Spectroscopy.
CD experiments were performed using a 1.0-mm cuvette in a Jasco J-810 spectrometer at 20 °C. Protein samples were dissolved in 50 mM sodium phosphate, pH 7.0, to a final concentration of 20 μM or 4 μM for ELP3 and tropoelastin, respectively. Three scans from 260 nm to 190 nm, recorded with 4 s of signal averaging at each point, were performed to obtain the mean spectra shown.
Light Microscopy.
Phase separation of 100 µM tropoelastin and 100 µM ELP3 was monitored in real time using a Zeiss Axiovert 200 inverted epifluorescence microscope with a temperature-controlled Attofluor cell chamber (Molecular Probes). NaCl was added to a final concentration of 1.5 M. The sample chamber was held constant at 50 °C. Samples were imaged for 90 min using differential interference contrast mode, with images saved after 60 min of incubation.
Tensile Mechanical Measurements.
Construction of genipin cross-linked materials from ELP3 and tropoelastin, as well as their mechanical testing and associated data analysis, was carried out as previously described (31, 32, 51). The standard protocol for polymeric material cross-linking was modified to adjust for the phase behavior of ELP3 and maximize the homogeneity of the material thickness. All protein solutions, reagents, and centrifuges were prewarmed to 37 °C. A total of 5 mg of ELP3 was dissolved in 325 μL sodium borate buffer (20 mM, pH 8.0). A total of 125 μL of 5 M NaCl was added to the protein sample in 10-mm2 glass-bottom wells. After 5 min of incubation at 37 °C, 50 μL of genipin (100 mM in ethanol) was added. Samples were immediately spun in a swinging-bucket centrifuge at 1,100 × g for 10 min and then incubated overnight at 37 °C to allow for complete cross-linking.
Solution-State NMR Spectroscopy.
All experiments were carried out on 20-µL samples using a triple-resonance 1-mm TXI MicroProbe with Z gradient and temperature control in a Bruker Avance III NMR spectrometer operating at an 1H frequency of 600 MHz. Samples were buffered with 50 mM phosphate at pH 7.0. Protein and NaCl concentrations were varied to control the desired phase transition temperature. All phase separation was induced in the spectrometer by raising the temperature at the probe. No further mechanical manipulation of the sample outside of the spectrometer was performed. Bruker TopSpin was used for data acquisition. Standard 2D and 3D Bruker pulse sequences with water suppression were used for most experiments. All spectra were processed with the NMRPipe software package (52), then visualized and analyzed with the CcpNmr analysis program (53).
Chemical Shift Assignment and Secondary Structure Prediction.
Chemical shift assignment of monomeric ELP3 was carried out on a sample containing 4.7 mM ELP3 and 300 mM NaCl at 20 °C and 37 °C. Chemical shift assignment of the phase-separated state was performed on 7.8 mM ELP3 with 600 mM NaCl at 37 °C. Both the monomer and coacervate 1H, 13C, and 15N chemical shifts were assigned using standard 2D and 3D NMR methods. The 1H-15N HSQC, HNCA, HN(CO)CA, HNCO, and HN(CA)CO experiments (pulse sequences as provided by Bruker Biospin) were used for sequential backbone assignments, along with an HCAN experiment (54), which was required to obtain the backbone 15N chemical shifts of proline residues and to facilitate assignment of proline-rich segments. A standard 3D HCCH-TOCSY experiment was used to achieve full assignment of side chain chemical shifts for the monomeric protein at 20 °C. The peak volumes observed in the 1H-15N HSQC spectrum of the ELP3 monomer correlated well with the sequence abundance of each assigned backbone amide resonance (SI Appendix, Fig. S3), helping to validate this approach to chemical shift assignment of a polymer-like protein.
The secondary structure propensity of the major repeating units of the cross-linking and hydrophobic domains in both the monomer and coacervate was predicted using the δ2D structure determination web server for intrinsically disordered proteins (45), with the chemical shifts for 1HN, 15N, 13Cα, 13CO, and 1Hα as input parameters. In mixed samples containing both monomer and coacervate, assignment of chemical shifts from each state was facilitated by 1H-15N HSQC experiments incorporating either a PFG diffusion or T2 relaxation-based filter to eliminate signals from monomer or coacervate, respectively. Backbone amide chemical shifts exhibited a linear change with temperature, allowing 1H-15N HSQCs taken during temperature series experiments to be assigned by simple translation of the chemical shift assignments at 20 °C and 37 °C.
1H-1H NOESYs.
Homonuclear 1H NOESY experiments on ELP3 monomer and coacervate samples were recorded using a phase-sensitive experiment with WATERGATE water suppression (55). Experiments with mixing times of 50, 100, and 150 ms were recorded and used to determine the linear region of the NOE buildup curve. Experiments with 100-ms mixing times were then used for integration of peak volumes and determination of NOE intensities. The abundant number of internal proline ring NOEs, which have known 1H-1H distances, were used to establish the relationship between NOE intensity and distance in each sample. NOE intensities of the monomer at 20 °C and 37 °C were grouped as strong (<2.4 Å), medium (≥2.4, ≤3.5 Å), or weak (>3.5 Å, <5 Å). Distance measurements were not performed on the phase-separated sample because chemical shift overlap did not allow for reliable peak assignment and integration analysis.
Intermolecular NOEs were obtained at 37 °C using an 1H-1H NOESY experiment with a 13C double half filter (56), and a 100-ms mixing time. Samples for these experiments contained a 1:1 mixture of unlabeled and 13C-labeled ELP3, giving final conditions of 1.6 mM ELP3 in 50 mM sodium phosphate and 300 mM NaCl at pH 7 (monomer sample) or 7.8 mM ELP3 in 50 mM sodium phosphate and 600 mM NaCl at pH 7 (phase-separated sample).
Pulsed Field Gradient Diffusion.
PFG diffusion data were recorded using a stimulated echo experiment with bipolar gradient pulses and WATERGATE solvent suppression. The 32 1H spectra were acquired at varying gradient strengths between 2% and 98% of the maximum value, while keeping time delays constant throughout each experiment. To obtain the diffusion coefficient D, peak intensity change as a function of gradient strength (g) was fit to a standard equation for bipolar gradients (57):
where I is the observed intensity, I(0) is the unattenuated intensity, γ is the gyromagnetic ratio of 1H, Δ is the diffusion time (100 ms), δ is the total gradient time, and τ is the time between the bipolar gradients (0.2147 ms).
The large difference in the diffusion coefficients of the monomeric and coacervate species allowed for good separation of their observed signal attenuation by running two different experiments with δ-values set to either 6 ms or 15 ms, respectively. When fitting the signal attenuation of the monomer, the remaining signal from the slower diffusing coacervate was omitted. Conversely, when fitting the signal attenuation of the coacervate, the sharp decrease in signal at the beginning of the decay curve due to loss of signal from the faster diffusing monomer was disregarded.
Dynamics Measurements.
The 15N R1 and R2 relaxation times were determined from pseudo-3D experiments using standard inversion recovery or spin-echo experiments, respectively (58, 59). The 13 1H-15N HSQC spectra were acquired in an incremental manner using delay times ranging from 50 to 4,000 μs for R1 experiments, whereas 10 spectra were acquired in an interleaved fashion using delay times of 17–544 μs for R2 experiments. The 1H-15N NOEs were determined using the pulse sequence hsqcnoef3gpsi with interleaved acquisition of spectra recorded with and without proton saturation. Least squares fitting of signal attenuation to obtain R1 and R2 relaxation times, as well as NOE calculations from comparison of saturated and equilibrium spectra, was accomplished using default parameters in CcpNmr. The reported relaxation rates are the inverse of the relaxation times.
Solid-State MAS NMR.
Solid-state experiments were performed using a triple-resonance 4.0-mm MAS probe with temperature control in a Bruker Ascend NMR spectrometer operating at an 1H frequency of 700 MHz. Spectra were externally referenced to the downfield 13C resonance of adamantane at 38.56 ppm relative to tetramethylsilane (60).
All 2D 13C-13C correlation spectra were acquired with 256 points in the indirect dimension. The π/2 pulse widths on all channels ranged from 2 to 4 μs. Cross-polarization was realized using 13C radio frequency field strengths from 40 to 60 kHz with contact times of 1–2 ms. All experiments on the phase-separated and cross-linked samples were performed at an MAS spinning rate of 14 kHz at ∼40 °C.
To identify regions of low mobility within ELP3, 2D 13C-13C correlation spectra were obtained using 1H-13C cross-polarization followed by proton-driven spin diffusion (PDSD) (61, 62) with a mixing time of 50 ms. To identify regions of high mobility, 2D 13C-13C correlation spectra were recorded using INEPT transfer from 1H to 13C followed by 13C-13C recoupling using a total through bond correlation spectroscopy (TOBSY) scheme (63)with a mixing time of 10 ms. The 1H decoupling fields of 100 kHz were applied during all acquisition periods using two-pulse phase-modulated (TPPM) decoupling (64).
Supplementary Material
Acknowledgments
The authors thank Régis Pomès for useful discussions. Research was funded by a grant in aid from the Heart and Stroke Foundation of Ontario (to S.S.), a Canadian Institutes of Health Research operating grant (to S.S. and F.W.K.), and operating funds from the Canadian Consortium on Neurodegeneration in Ageing (to S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701877114/-/DCSupplemental.
References
- 1.Johnson NR, Wang Y. Coacervate delivery systems for proteins and small molecule drugs. Expert Opin Drug Deliv. 2014;11:1829–1832. doi: 10.1517/17425247.2014.941355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shahidi F, Han XQ. Encapsulation of food ingredients. Crit Rev Food Sci Nutr. 1993;33:501–547. doi: 10.1080/10408399309527645. [DOI] [PubMed] [Google Scholar]
- 3.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 4.Uversky VN. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv Colloid Interface Sci. 2017;239:97–114. doi: 10.1016/j.cis.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kielty CM. Elastic fibres in health and disease. Expert Rev Mol Med. 2006;8:1–23. doi: 10.1017/S146239940600007X. [DOI] [PubMed] [Google Scholar]
- 7.Mithieux SM, Weiss AS. Elastin. Adv Protein Chem. 2005;70:437–461. doi: 10.1016/S0065-3233(05)70013-9. [DOI] [PubMed] [Google Scholar]
- 8.Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Adv Colloid Interface Sci. 2011;167:94–103. doi: 10.1016/j.cis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 9.MacEwan SR, Chilkoti A. Elastin-like polypeptides: Biomedical applications of tunable biopolymers. Biopolymers. 2010;94:60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 10.Gagner JE, Kim W, Chaikof EL. Designing protein-based biomaterials for medical applications. Acta Biomater. 2014;10:1542–1557. doi: 10.1016/j.actbio.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeo GC, et al. Fabricated elastin. Adv Healthc Mater. 2015;4:2530–2556. doi: 10.1002/adhm.201400781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergeron-Sandoval LP, Safaee N, Michnick SW. Mechanisms and consequences of macromolecular phase separation. Cell. 2016;165:1067–1079. doi: 10.1016/j.cell.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Pak CW, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bungenberg de Jong HG, Kruyt HR. Coacervation (partial miscibility in colloid systems) Proc K Ned Akad Wet. 1929;32:849–856. [Google Scholar]
- 17.Vrhovski B, Jensen S, Weiss AS. Coacervation characteristics of recombinant human tropoelastin. Eur J Biochem. 1997;250:92–98. doi: 10.1111/j.1432-1033.1997.00092.x. [DOI] [PubMed] [Google Scholar]
- 18.Bellingham CM, et al. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers. 2003;70:445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 19.Chung MIS, et al. Sequences and domain structures of mammalian, avian, amphibian and teleost tropoelastins: Clues to the evolutionary history of elastins. Matrix Biol. 2006;25:492–504. doi: 10.1016/j.matbio.2006.08.258. [DOI] [PubMed] [Google Scholar]
- 20.Urry DW. Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J Protein Chem. 1988;7:1–34. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]
- 21.Luan CH, Harris RD, Prasad KU, Urry DW. Differential scanning calorimetry studies of the inverse temperature transition of the polypentapeptide of elastin and its analogues. Biopolymers. 1990;29:1699–1706. doi: 10.1002/bip.360291403. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Alonso DO, Daggett V. The molecular basis for the inverse temperature transition of elastin. J Mol Biol. 2001;305:581–592. doi: 10.1006/jmbi.2000.4306. [DOI] [PubMed] [Google Scholar]
- 23.Toonkool P, Jensen SA, Maxwell AL, Weiss AS. Hydrophobic domains of human tropoelastin interact in a context-dependent manner. J Biol Chem. 2001;276:44575–44580. doi: 10.1074/jbc.M107920200. [DOI] [PubMed] [Google Scholar]
- 24.Rauscher S, Baud S, Miao M, Keeley FW, Pomès R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure. 2006;14:1667–1676. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Rauscher S, Pomes R. Structural disorder and protein elasticity. In: Fuxreiter F, Tompa P, editors. Structural Disorder in Protein Complexes. Landes Bioscience; Austin, TX: 2011. pp. 159–183. [Google Scholar]
- 26.Muiznieks LD, Weiss AS, Keeley FW. Structural disorder and dynamics of elastin. Biochem Cell Biol. 2010;88:239–250. doi: 10.1139/o09-161. [DOI] [PubMed] [Google Scholar]
- 27.Akagawa M, Suyama K. Mechanism of formation of elastin crosslinks. Connect Tissue Res. 2000;41:131–141. doi: 10.3109/03008200009067665. [DOI] [PubMed] [Google Scholar]
- 28.Miao M, Cirulis JT, Lee S, Keeley FW. Structural determinants of cross-linking and hydrophobic domains for self-assembly of elastin-like polypeptides. Biochemistry. 2005;44:14367–14375. doi: 10.1021/bi0510173. [DOI] [PubMed] [Google Scholar]
- 29.Yao XL, Hong M. Structure distribution in an elastin-mimetic peptide (VPGVG)3 investigated by solid-state NMR. J Am Chem Soc. 2004;126:4199–4210. doi: 10.1021/ja036686n. [DOI] [PubMed] [Google Scholar]
- 30.Tamburro AM, Bochicchio B, Pepe A. Dissection of human tropoelastin: Exon-by-exon chemical synthesis and related conformational studies. Biochemistry. 2003;42:13347–13362. doi: 10.1021/bi034837t. [DOI] [PubMed] [Google Scholar]
- 31.Reichheld SE, et al. Conformational transitions of the cross-linking domains of elastin during self-assembly. J Biol Chem. 2014;289:10057–10068. doi: 10.1074/jbc.M113.533893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao M, et al. Sequence and structure determinants for the self-aggregation of recombinant polypeptides modeled after human elastin. J Biol Chem. 2003;278:48553–48562. doi: 10.1074/jbc.M308465200. [DOI] [PubMed] [Google Scholar]
- 33.Ohgo K, Niemczura WP, Kumashiro KK. Probing the natural-abundance 13C populations of insoluble elastin using 13C-1H heteronuclear correlation (HETCOR) NMR spectroscopy. Macromolecules. 2009;42:7024–7030. [Google Scholar]
- 34.Ohgo K, et al. Resolving nitrogen-15 and proton chemical shifts for mobile segments of elastin with two-dimensional NMR spectroscopy. J Biol Chem. 2012;287:18201–18209. doi: 10.1074/jbc.M111.285163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun C, Mitchell O, Huang J, Boutis GS. NMR studies of localized water and protein backbone dynamics in mechanically strained elastin. J Phys Chem B. 2011;115:13935–13942. doi: 10.1021/jp207607r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauscher S, Pomès R. Molecular simulations of protein disorder. Biochem Cell Biol. 2010;88:269–290. doi: 10.1139/o09-169. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, et al. On the inverse temperature transition and development of an entropic elastomeric force of the elastin mimetic peptide [LGGVG](3, 7) J Chem Phys. 2012;136:085101. doi: 10.1063/1.3685454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urry DW, Venkatachalam C, Long M, Prasad K. Dynamic B-spirals and a librational entropy mechanism of elasticity. In: Srinivasan R, Sarma R, editors. Conformation in Biology. Adenine Press; New York: 1983. pp. 11–27. [Google Scholar]
- 39.Keeley FW, Bellingham CM, Woodhouse KA. Elastin as a self-organizing biomaterial: use of recombinantly expressed human elastin polypeptides as a model for investigations of structure and self-assembly of elastin. Philos Trans R Soc Lond B Biol Sci. 2002;357:185–189. doi: 10.1098/rstb.2001.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vieth S, Bellingham CM, Keeley FW, Hodge SM, Rousseau D. Microstructural and tensile properties of elastin-based polypeptides crosslinked with genipin and pyrroloquinoline quinone. Biopolymers. 2007;85:199–206. doi: 10.1002/bip.20619. [DOI] [PubMed] [Google Scholar]
- 41.Miao M, et al. Sequence and domain arrangements influence mechanical properties of elastin-like polymeric elastomers. Biopolymers. 2013;99:392–407. doi: 10.1002/bip.22192. [DOI] [PubMed] [Google Scholar]
- 42.Miao M, et al. Single nucleotide polymorphisms and domain/splice variants modulate assembly and elastomeric properties of human elastin. Implications for tissue specificity and durability of elastic tissue. Biopolymers. February 19, 2017 doi: 10.1002/bip.23007. [DOI] [PubMed] [Google Scholar]
- 43.Konrat R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J Magn Reson. 2014;241:74–85. doi: 10.1016/j.jmr.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen MR, Ruigrok RWH, Blackledge M. Describing intrinsically disordered proteins at atomic resolution by NMR. Curr Opin Struct Biol. 2013;23:426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 46.Krzeminski M, Marsh JA, Neale C, Choy WY, Forman-Kay JD. Characterization of disordered proteins with ENSEMBLE. Bioinformatics. 2013;29:398–399. doi: 10.1093/bioinformatics/bts701. [DOI] [PubMed] [Google Scholar]
- 47.Etzkorn M, et al. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2007;46:459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- 48.Sato F, et al. Distinct steps of cross-linking, self-association, and maturation of tropoelastin are necessary for elastic fiber formation. J Mol Biol. 2007;369:841–851. doi: 10.1016/j.jmb.2007.03.060. [DOI] [PubMed] [Google Scholar]
- 49.Tu Y, Wise SG, Weiss AS. Stages in tropoelastin coalescence during synthetic elastin hydrogel formation. Micron. 2010;41:268–272. doi: 10.1016/j.micron.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Rauscher S, Pomes R. The liquid structure of elastin. bioRxiv. December 21, 2016 doi: 10.1101/095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muiznieks LD, Reichheld SE, Sitarz EE, Miao M, Keeley FW. Proline-poor hydrophobic domains modulate the assembly and material properties of polymeric elastin. Biopolymers. 2015;103:563–573. doi: 10.1002/bip.22663. [DOI] [PubMed] [Google Scholar]
- 52.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 54.Kanelis V, et al. Sequential assignment of proline-rich regions in proteins: Application to modular binding domain complexes. J Biomol NMR. 2000;16:253–259. doi: 10.1023/a:1008355012528. [DOI] [PubMed] [Google Scholar]
- 55.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 56.Wider G, Weber C, Traber R, Widmer H, Wuthrich K. Use of a double-half filter in two-dimensional 1H nuclear magnetic resonance studies of receptor-bound cyclosporin. J Am Chem Soc. 1990;112:9015–9016. [Google Scholar]
- 57.Stejskal EO, Tanner JE. Spin diffusion measurements: Spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42:288–292. [Google Scholar]
- 58.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 59.Farrow NA, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 60.Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J Magn Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 61.Suter D, Ernst RR. Spin diffusion in resolved solid-state NMR spectra. Phys Rev B Condens Matter. 1985;32:5608–5627. doi: 10.1103/physrevb.32.5608. [DOI] [PubMed] [Google Scholar]
- 62.Szeverenyi NM, Sullivan MJ, Maciel GE. Observation of spin exchange by two-dimensional Fourier transform 13C cross polarization-magic-angle spinning. J Magn Reson. 1982;47:462–475. [Google Scholar]
- 63.Hardy EH, Verel R, Meier BH. Fast MAS total through-bond correlation spectroscopy. J Magn Reson. 2001;148:459–464. doi: 10.1006/jmre.2000.2258. [DOI] [PubMed] [Google Scholar]
- 64.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.