Abstract
Bipartite geminiviruses encode a small protein, AC2, that functions as a transactivator of viral transcription and a suppressor of RNA silencing. A relationship between these two functions had not been investigated before. We characterized both of these functions for AC2 from Mungbean yellow mosaic virus-Vigna (MYMV). When transiently expressed in plant protoplasts, MYMV AC2 strongly transactivated the viral promoter; AC2 was detected in the nucleus, and a split nuclear localization signal (NLS) was mapped. In a model Nicotiana benthamiana plant, in which silencing can be triggered biolistically, AC2 reduced local silencing and prevented its systemic spread. Mutations in the AC2 NLS or Zn finger or deletion of its activator domain abolished both these effects, suggesting that suppression of silencing by AC2 requires transactivation of host suppressor(s). In line with this, in Arabidopsis protoplasts, MYMV AC2 or its homologue from African cassava mosaic geminivirus coactivated >30 components of the plant transcriptome, as detected with Affymetrix ATH1 GeneChips. Several corresponding promoters cloned from Arabidopsis were strongly induced by both AC2 proteins. These results suggest that silencing suppression and transcription activation by AC2 are functionally connected and that some of the AC2-inducible host genes discovered here may code for components of an endogenous network that controls silencing.
RNA silencing, also referred to as RNA interference and posttranscriptional gene silencing, is an evolutionarily conserved mechanism that protects cells against invasive nucleic acids, such as viruses, transposons, and transgenes (19). RNA silencing is triggered by double-stranded RNA (dsRNA), effects sequence-specific degradation of cognate viral or endogenous RNA, and, at least in plants, causes de novo methylation of homologous DNA (33). In plants, silencing is increasingly viewed as an adaptive immune system targeting pathogenic RNA and DNA (28, 52). To counteract this defense system, viruses have evolved suppressor proteins (4, 6, 37) that interfere with different steps of the RNA silencing pathway (11), thus allowing efficient viral replication in a single cell and systemic spread of the infection. For example, the coat protein of Turnip crinkle virus blocks generation of small interfering RNAs (siRNAs) (38), derived from dsRNA processing by the RNase III-like enzyme Dicer at an early initiation step of silencing. p19 of tombusviruses binds siRNAs (27, 51), thereby inhibiting a downstream step involving cleavage of cognate RNA by an siRNA-guided, RNA-induced silencing complex. Movement protein P25 of Potato virus X prevents systemic spread of silencing through the vascular system (54). Potyvirus protein HC-Pro might interfere with both the initiation and spread of silencing, although the mechanism of HC-Pro action is still a matter of debate (reference 32 and references therein). Interestingly, HC-Pro and other viral suppressors not only are able to suppress RNA silencing but also can interfere with a related micro-RNA (miRNA) pathway (8, 11, 24, 31) that plays a pivotal role in plant and animal development (3, 7). In plants, the miRNA pathway is similar to RNA silencing in that miRNA precursors are also cleaved by the Dicer-like enzyme DCL1, but the latter is localized in the nucleus (34). It is intriguing that Cucumber mosaic virus, a cytoplasmic RNA virus, codes for a nuclear protein (2b) that suppresses RNA silencing (8, 30) and also interferes with RNA-directed DNA methylation (14).
Thus, viruses seem to exploit various mechanisms of silencing suppression by deploying their specialized proteins to different compartments of the cell. In this paper, we propose a new mechanism of silencing suppression in which a viral, nucleus-targeted protein acts indirectly by activating transcription of host silencing suppressor gene(s).
We are studying the bipartite geminivirus Mungbean yellow mosaic virus-Vigna (MYMV) (23) with the goal of generating resistance to the virus by using an RNAi-based strategy (35). Geminiviruses are small, circular, single-stranded DNA viruses that replicate in the nucleus of an infected cell via double-stranded intermediates that also serve as templates for bidirectional transcription (15, 18). Bipartite geminiviruses of the genus Begomovirus express the small protein AC2 (also called AL2 or TrAP), which transactivates transcription of late viral genes (44, 16). Consistent with its function as a transcriptional activator, three conserved domains have been recognized in this protein: a basic domain with a nuclear localization signal (NLS) at the N terminus, a central DNA-binding domain with a nonclassical Zn-finger motif, and an acidic activator domain at the C terminus (21). Studies on AC2 of African cassava mosaic virus (ACMV) and the homologous C2 of Tomato yellow leaf curl virus-China (TYLCCNV), a related monopartite begomovirus, have implicated these proteins in suppression of RNA silencing (10, 17, 50, 53). Notably, TYLCCN C2 requires functional NLS and Zn-finger domains to suppress silencing (10, 50). In this work, we found that AC2 from MYMV serves as a transactivator of viral promoter and as a suppressor of RNA silencing. These two functions could not be separated by mutations in the three conserved domains including the activator domain, suggesting that suppression of silencing by AC2 might involve activation of transcription of an endogenous silencing suppressor gene(s). By RNA profiling with Affymetrix GeneChips (ATH1) and transient expression assays with Arabidopsis protoplasts, we identified several candidate suppressor genes, whose promoters were dramatically induced in response to AC2 from MYMV and its homologue from ACMV.
MATERIALS AND METHODS
Plasmid construction.
MYMV bidirectional promoter chloramphenicol acetyltransferase (CAT) constructs were generated by replacing the cauliflower mosaic virus 35S promoter and leader sequences between the AflIII and NcoI sites of pKSXAHA (36) with a PCR-amplified, 263-bp segment of MYMV DNA A (accession no. AJ132575), spanning from the AC1 ATG start codon (at position 2609) to the AV2 ATG (at position 141), yielding pAC1AV2CAT (Fig. 1). The AC2 coding region from MYMV (positions 1623 to 1216) was introduced by PCR between the XhoI and SphI sites of pKSXAHA in place of CAT, yielding p35SAC2 (Fig. 1). The following PCR primers were used: 5′TTGTGCTCGAGaaagaatgcggaattctacaccctc (XhoI and AC2 start codon underlined; viral nucleotides in lowercase) and 5′ATTTAGCATGCtcactaaaagtcgataatatcatcccag (SphI and AC2 stop codon underlined). To create a deletion of the AC2 activator domain (AD−), the former primer was used along with 5′gcagtgcAtGcttAaacccgtggttgaacattatc (with SphI and a new stop codon underlined). Point mutations NLS1−, NLS2−, and ZF− were introduced by PCR ligation, using the following pairs of primers containing the respective mutations: 5′caaggttgccGCAGCCGCagcaattcgacgctctcgaattgat and 5′cgaattgctGCGGCTGCggcaaccttgtgttgcgcct, 5′gcgagcaattGCaGCctctGCaattgatttaagctgtgggtgtag and 5′taaatcaattGCagagGCtGCaattgctcgcttcttggcaaccttgtg, and 5′cgaattgatttaagcGCtgggGCtagttattacatccatatcaactgc and 5′ggatgtaataactaGCcccaGCgcttaaatcaattcgagagcgtc.
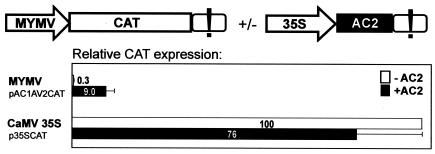
FIG. 1.
MYMV AC2 is a transactivator of viral transcription. Transient expression in N. plumbaginifolia protoplasts of a reporter gene (CAT) driven by the MYMV DNA A rightward (AV2) promoter (scheme of the construct pAC1AV2CAT on top left) and the control CaMV 35S promoter (p35SCAT), in the presence or absence of the AC2-expressing construct (top right), was measured 20 to 24 h posttransfection. Relative expression values indicated are the means for six independent experiments (standard error did not exceed 25% of the mean values; indicated by error bars). The exclamation mark represents CaMV 35S terminator sequences.
The 35S promoter-driven expression cassettes for MYMV proteins AC1 (positions 2611 to 1523; GenBank accession number AJ132575), AC3 (1475 to 1071), AC4 (2460 to 2161), and BC1 (2117 to 1221; GenBank accession number AJ132574) and for ACMV-KE AC2 (1771 to 1364; GenBank accession number NC_001467) were also constructed by replacing CAT between XhoI and SphI of KSXAHA with the respective coding sequences.
To create GFP::ChS::AC2 protein fusions, the wild-type and mutant versions of AC2 were introduced between MluI and XbaI of pEGFPChS: the PCR primer 5′GAGAAACGCGTcggaattctacaccctcaag (MluI underlined, followed by AC2 from the second codon) was used together with either 5′GATTTTCTAGAGCATGCtcactaaaagtcgataatatcatcc (XbaI and AC2 stop codon underlined) or 5′GATTTTCTAGAGCATGCttAaaccccgtggttgaac (XbaI and AD− stop codon underlined).
Particle bombardment of plant seedlings.
Nicotiana benthamiana plants were raised from seeds of the mGFP5ER transgenic line 16c (39); kindly provided by D. Baulcombe) either on agar-solidified Murashige and Skoog medium or in soil at 26°C with 16-h day and 8-h night. Three to four weeks postgermination, seedlings were bombarded using a biolistic particle delivery system (PDS-1000/He; Bio-Rad) with 1-μm gold particles coated with plasmid DNA. For one plate/pot with four to eight seedlings, 2 μg of trigger plasmid p35SmGFP5ER (26) alone or in combination with 2 μg of suppressor plasmid (p35AC2 or its derivatives) was loaded on 750 μg of gold particles and delivered at 1,100 lb/in2, following the manufacturer's recommendations. After bombardment, plants were maintained in a nonstop-light chamber at 26°C. Images of silencing under UV light (100-W longwave mercury spot lamp; OmniLab AG) were taken with a digital camera.
Arabidopsis protoplast preparation.
An Arabidopsis thaliana La-er cell suspension (kindly provided by T. Boller, Institute of Botany, Basel, Switzerland) was maintained in AT medium (4.43 g of Murashige and Skoog basal salts/liter with minimal organics [Sigma], 3% sucrose, 5.4 μM naphthalene acetic acid, 0.23 μM 6-furfurylaminopurine [pH 5.6]) at 25°C and 130 rpm with a 16-h day. Protoplasts were prepared from 50 ml of suspension 1 week after subculturing (1:10) as follows. Cells were harvested by centrifugation (Jouan B4; IG Instrumenten, Zürich, Switzerland) for 2 min at 800 rpm, washed with 0.5 M mannitol (pH 5.8), and transferred to 70 ml of enzyme solution (1% cellulase Onozuka R-10, 0.25% macerozym R-10, 0.5 M mannitol, 10 mM CaCl2 [pH to 5.6]). Following incubation for 16 to 18 h at 26 to 28°C in the dark, protoplasts were filtered through a 100-μm-pore-size sieve, diluted with equal volume of W5 (150 mM NaCl, 5 mM KCl, 125 mM CaCl2, 6 mM glucose [pH 5.6]), and pelleted for 5 min at 1,000 rpm. Cells were resuspended in 10 ml of 0.6 M sucrose-1% morpholineethanesulfonic acid (pH 5.6) and carefully overlaid with 1 ml of W5. Following centrifugation for 10 min at 800 rpm, cells were harvested from the interphase and washed with 10 ml of W5 twice by inverting the tube and spinning for 3 min at 800 rpm; during the second washing, cells were incubated in W5 for 10 to 30 min. Protoplasts were resuspended in 5 ml of MMM (0.5 M mannitol, 0.1% morpholineethanesulfonic acid, 15 mM MgCl2 [pH 5.6]), counted, and adjusted to a density of 2 × 106/ml.
RNA profiling with Arabidopsis protoplasts.
Three-hundred-microliter aliquots (6 × 105 protoplasts), in six replicates for each construct, were mixed with 20 μg of plasmid and incubated for 5 min at room temperature (RT). Three hundred microliters of 40% polyethylene glycol (PEG) 4000 was added, and the contents were mixed and incubated for 20 min at RT and transferred into 4 ml of CMA medium (see the supplemental material). Following incubation for 8 h at 28°C in the dark, protoplasts were diluted with 10 ml of W5 and pelleted for 10 min at 1,000 rpm. The pellets of two replicates were combined (∼80 μl), 800 μl of TRIZOL (GibcoBRL) was added, and the mixture was incubated for 5 min at RT. Protein was extracted with 160 μl of chloroform by vortex mixing and incubation for 3 min at RT followed by centrifugation for 10 min at 13,000 rpm and 4°C. The aqueous phase (∼600 μl) was taken, and total RNA was precipitated by addition of 500 μl of isopropanol for 45 min at RT and pelleted for 15 min at 13,000 rpm and 4°C. The pellet was washed with 75% ethanol, dried in a SpeedVac, and dissolved in 100 μl of sterile bidistilled water. Further purification was performed with an RNeasy Plant minikit (QIAGEN), following the manufacturer's recommendations. The total yield of RNA from the six replicates ranged from 35 to 100 μg.
For each construct, as well as a control mock transfection, two 10-μg total RNA samples derived from two independent batches of Arabidopsis protoplasts were processed for microarray analyses according to the protocol recommended by Affymetrix. Ten micrograms of fragmented cRNA was hybridized to an ATH1 GeneChip (Affymetrix) using standard procedures (45°C, 16 h). Washing and staining were performed in a Fluidics Station 400, using the protocol EukGE-WS2v4, and scanning was carried out with an Affymetrix GeneChip scanner. Analysis of the chips was performed using MicroArraySuite 5 and GeneSpring 5.0 (Silicon Genetics). Changes in gene expression were determined, requiring that a gene was called “present” in one or more conditions and had a Wilcoxon change P value of <0.003 for “increase” or “decrease” in all replicate comparisons. Significance of the changes was assessed in the following ways: the genes passing the expression and severalfold-change filters were subjected to a one-way analysis of variance (P < 0.05) with a Benjamini and Hochberg multiple testing correction. The origin of the differences indicated by the analysis of variance were probed with a Tukey posthoc analysis.
Transient expression in plant protoplasts.
A. thaliana protoplasts were prepared and transfected as described above. Transient expression in Nicotiana plumbaginifolia leaf protoplasts was carried out as described previously (9). A 300-μl protoplast aliquot (6 × 105) was mixed with up to 30 μl of plasmid DNA mixture containing 10 μg of CAT plasmid, 10 μg of viral protein expression plasmid (e.g., AC2 or its derivatives), and 2 μg of β-glucuronidase (GUS) plasmid as an internal control of transfection efficiency. Three hundred microliters of 40% PEG 4000 (A. thaliana) or PEG 6000 (N. plumbaginifolia) was added, and the mixture was incubated for 10 to 30 min at RT and transferred to 4 ml of CMA (A. thaliana) or K3 (N. plumbaginifolia) medium (see the supplemental material). Following incubation for 19 to 24 h at 28°C in the dark, protoplasts were diluted with 10 ml of W5 and pelleted for 10 min at 1,000 rpm. The pellet was diluted with water to a final volume of 90 μl, and 10 μl of 10× GUS buffer (0.5 M NaH2PO4, 0.1 M EDTA, 1% Triton X-100, 1% C15H28NNaO3 [pH 7.0]) was added. The cells were broken by three cycles of freezing in liquid nitrogen and thawing at 37°C. Total protein extract was cleared by centrifugation for 10 min at 15,300 rpm, 4°C. CAT protein accumulation was determined in 30 μl of extract by using a CAT enzyme-linked immunosorbent assay kit (Roche), following the manufacturer's recommendations; GUS activity was determined by a fluorimetric GUS assay (36). Relative GUS activities were taken for normalization of CAT expression levels. For each construct, the values given are the means from at least three independent batches of protoplasts. Deviations from the mean values did not exceed 30%.
Visualization of GFP in plant protoplasts.
N. plumbaginifolia protoplasts were prepared and transfected with 20 μg of plasmid DNA of pEGFPChS or its derivatives as described in the previous section. Twelve hours posttransfection, 500-μl aliquots were mixed with 500 μl of fixation solution (6% paraformaldehyde in phosphate-buffered saline [pH 7.4], 10 mM EGTA) and incubated for 30 min at RT. One hundred fifty- to two hundred-microliter aliquots were applied onto polylysine-coated slides, centrifuged (Cytospin3; Shandon) for 3 min at 1,000 rpm, and air dried for 30 min. Two drops of DAPI-DABCO (Vectashield Hard+Set mounting medium with 1.5 μg of 4′,6′-diamidino-2-phenylindole [DAPI]/ml; Vector Laboratories) was added. Slides were covered with thin glass coverslips and kept for 10 h at 4°C. Fluorescence microscopy was performed with a Nikon Eclipse E800 microscope equipped with Plan Apochromat objectives (Nikon, Tokyo, Japan). Filter set XF100 with excitation at 475 ± 40 nm and emission at 520 ± 30 nm (Omega Optical, Brattleboro, Vt.) was used for visualization of green fluorescent protein (GFP). Protoplasts were visualized by using ×60 oil immersion lens. Images were acquired and processed with an ORCA-100 progressive-scan interline charge-coupled-device camera (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab 3 software (Improvision, Coventry, United Kingdom).
Cloning of AC2-inducible gene promoters from Arabidopsis.
Genomic DNA from the Columbia (Col-0) ecotype of Arabidopsis was used for PCR amplification of AC2-inducible gene sequences. Primer design was based on the complete genome of Col-0 (The Arabidopsis Information Resource database at www.arabidopsis.org). The promoter regions of about 800 to 1,100 bp upstream of the first ATG start codon of each gene (supplemental Table S4) were introduced between AflIII and NcoI of pKSXAHA (36), thus directly fusing the CAT coding sequence to the first ATG.
RESULTS
AC2 transactivates viral promoter.
To study the effect of AC2 on viral transcription, we subcloned an intergenic region of MYMV DNA A, flanked by the ATG start codons of the AC1 and AV2 genes, and fused the coding sequence of a CAT reporter gene to the AV2 ATG (Fig. 1). By analogy with other begomoviruses, this segment of the viral genome should contain a bidirectional promoter and the rightward (AV2) transcription driven by this promoter should be inducible by AC2 (16, 44). Protoplasts from N. plumbaginifolia leaves were transfected with the resulting plasmid by a PEG-mediated transformation method, and after overnight incubation, accumulation of CAT protein was measured in a total protein extract by CAT enzyme-linked immunosorbent assay. A plasmid constitutively expressing a GUS reporter gene was cotransfected to serve as an internal control to monitor transfection efficiency and to normalize CAT expression levels.
The MYMV promoter segment drove very weak expression, which constitutes only 0.3% of CAT expression driven by the strong constitutive CaMV 35S promoter (5), used here as a control. When the MYMV promoter construct was coexpressed with the MYMV AC2 gene driven by the 35S promoter from a separate plasmid (Fig. 1), CAT expression was strongly activated (30-fold; Fig. 1). This dramatic activation elevated expression up to 12% of that driven by the 35S promoter. Note that expression driven by the 35S promoter itself was slightly reduced in the presence of AC2 (Fig. 1).
Similar results were obtained with a second reporter gene (firefly luciferase) and with protoplasts derived from cell suspensions of another dicot plant (Orychophragmus violaceus). However, in a monocot plant protoplast system derived from rice (Oryza sativa) cell suspensions, the MYMV promoters were inactive and could not be activated by AC2 (data not shown).
Precise mapping of the viral transcripts from MYMV-infected Vigna mungo plants by circularization-reverse transcriptase PCR revealed the major rightward transcription start site at positions A137 and A141 (4 nucleotides (nt) upstream of and at the first nucleotide of the AV2 ATG start codon, respectively) at an optimal distance from a consensus TATA box (unpublished data). This finding is consistent with the rightward promoter activity observed here in plant protoplasts. Notably, the reporter CAT mRNA expressed from the MYMV promoter construct should possess at maximum a 4-nt-long 5′-untranslated region (ACGG) of viral origin, the only MYMV sequence on the transcript. Therefore, we consider posttranscriptional regulation by MYMV AC2 highly unlikely.
Taken together, these results establish that, similar to other bipartite geminiviruses (16, 44), MYMV AC2 protein is a strong transactivator of viral transcription.
The three conserved domains of MYMV AC2 referred to above were mutated individually, and the mutants were tested for their ability to transactivate the MYMV promoter in plant protoplasts. Mutation of the basic domain (KKR26AAA [NLS1−] and RRSR31AASA [NLS2−]) or the Zn-finger motif in the DNA-binding domain (C37A and C39A [ZF−]) or deletion of the entire (31-amino-acid) acidic domain (del105-135 [AD−]) all nearly abolished AC2-mediated transactivation, retaining at most 9% of the wild-type activity (Table 1). This suggests that these three AC2 domains are directly or indirectly required to activate viral transcription.
TABLE 1.
Effects of AC2 mutations in N. plumbaginifolia protoplasts
| AC2 | % Transactivation efficiency | No. of DAPI-stained cells | No. of cells with GFP fluorescencea | % Transfection efficiency/GFP accumulation | No. of cells with nuclear GFP | % Cells with nuclear GFP |
|---|---|---|---|---|---|---|
| WT | 100 | 204 | 64 | 31 | 54 | 84 |
| AD− | 4 | 192 | 70 | 36 | 62 | 89 |
| ZF− | 5 | 169 | 56 | 33 | 43 | 77 |
| NLS1− | 4 | 143 | 39 | 27 | 3b | 8 |
| NLS2− | 9 | 263 | 102 | 39 | 32b | 31 |
In nucleus, cytoplasm, or both.
GFP is also (variably) present in cytoplasm.
AC2 is a nuclear protein.
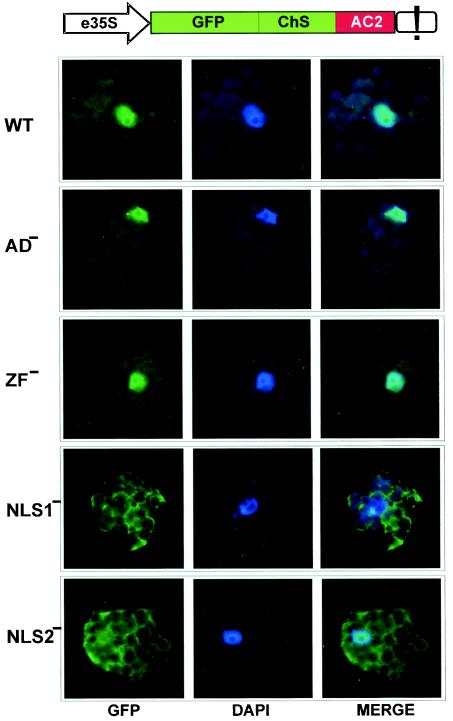
We fused a GFP to the N terminus of AC2 and expressed the resulting fusion under the control of the 35S promoter transiently in N. plumbaginifolia protoplasts. The fusion protein was enlarged by a portion of chalcone synthase (Fig. 2) in order to exceed the exclusion limit of the nuclear pore (∼60 kDa) and thus avoid passive diffusion to the nucleus. As visualized with a confocal fluorescence microscope, bright green fluorescence of GFP-AC2 accumulated predominantly in the nucleus, excluding the nucleolus (Fig. 2, WT). The predominant nuclear localization of AC2 was not affected by the Zn-finger mutation (ZF−) and the C-terminal deletion (AD−), whereas mutation in the basic domain (NLS1−) resulted in predominantly cytoplasmic accumulation of AC2 (Fig. 2 and Table 1). The latter suggests that the KKR motif (mutated in NLS1−) is an essential part of the AC2 NLS. However, in the C2 protein of TYLCCNV, the corresponding KKT26DIT mutation did not abolish nuclear localization, in contrast to mutation RRRR31DVGG in the neighboring motif (10). We therefore also mutated the corresponding motif in the MYMV AC2 (RRSR31AASA [NLS2-]). Although nuclear localization was strongly impaired, it was not abolished by the latter mutation, and in 31% of the GFP-expressing cells the fusion protein was variably distributed between the nucleus and the cytoplasm (Table 1 and Fig. 2, NLS2−). This result suggests that both MYMV AC2 and TYLCCNV C2 possess a split NLS, with the upstream motif playing the more important role in MYMV and the downstream one playing the more important role in TYLCCNV. Notably, accumulation of green fluorescence was similar with both wild-type and mutant AC2 constructs (Fig. 2 and Table 1), indicating that the mutations did not significantly affect protein expression and turnover.
FIG. 2.
MYMV AC2 is a nuclear protein with a split NLS. Subcellular localization of GFP-fused wild type AC2 protein (WT) and its mutant variants (AD−, ZF−, NLS1−, and NLS2−) following transient expression driven by the 35S promoter with double enhancer (e35S; ChS, portion of chalcone synthase gene; see the text) in N. plumbaginifolia protoplasts was detected in individual plant cells (stained with DAPI) by using confocal fluorescence microscopy. Filtered fluorescence images of GFP-AC2 (left) and DAPI-stained nuclear DNA (center) for each cell were merged (right): if GFP-AC2 fusion (green) is localized to the nucleus (dark blue), the latter appears light blue in the merge image (WT, AD−, ZF−, and, less pronouncedly, NLS2−).
These results show that MYMV AC2 is localized to the nucleus, consistent with its function as a transcriptional activator. However, it cannot be excluded that during virus infection this small protein may shuttle between the nucleus and the cytoplasm, as has been reported for the corresponding AL2 protein from Tomato golden mosaic virus (55).
AC2 is a suppressor of silencing.
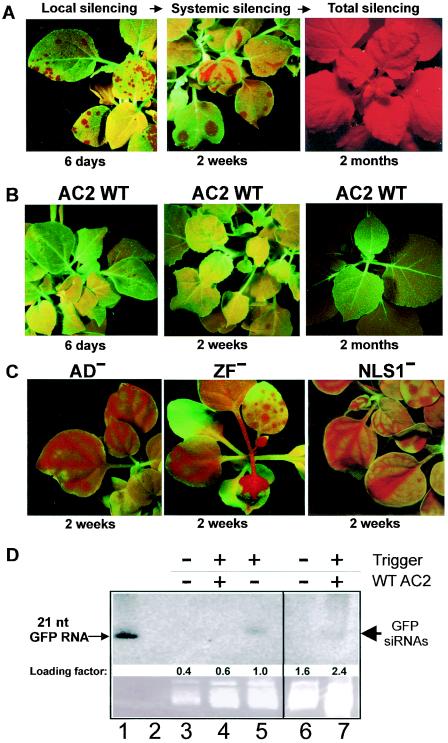
To investigate whether MYMV AC2, like other related geminivurus proteins, is able to suppress RNA silencing, a model system based on the N. benthamiana GFP-transgenic line 16c developed in the laboratory of D. Baulcombe (39) was used. In this system, RNA silencing of the GFP transgene can be triggered by biolistic particle delivery of a GFP-expressing plasmid as well as long or short dsRNA cognate to GFP (26). In our experiments, plant seedlings were bombarded with the GFP plasmid. Around 4 to 6 days postbombardment, local GFP silencing was observed under long-wave UV light as multiple red spots of background chlorophyll fluorescence in the targeted cell areas of otherwise green-fluorescing leaves (Fig. 3A). At around 14 days postbombardment, systemic GFP silencing was observed, which appeared as red veins on nontargeted newly emerging leaves (Fig. 3A). Eventually, most of the plant tissues (data not shown) or the whole plant became red under UV, the manifestation of total GFP silencing (Fig. 3A). These results confirm previous observations by Klahre et al. (26).
FIG. 3.
MYMV AC2 is a suppressor of RNA silencing. (A) GFP silencing in N. benthamiana line 16c seedlings was triggered by biolistic delivery of a GFP plasmid and visualized under UV light after 6 days (local silencing, left image), 2 weeks (systemic silencing, middle image) and 2 months (total silencing, right image). (B) Codelivery of the wild-type MYMV AC2 reduced local silencing (left) and totally suppressed systemic silencing (middle and right images). (C) MYMV AC2 mutants AD−, ZF−, and NLS1− did not exhibit any antisilencing effect. (D) RNA blot hybridization performed as described by Klahre et al. (26). Accumulation of GFP siRNAs (thick arrow) in the presence of MYMV AC2 (lanes 4 and 7; note a fourfold-higher loading of total RNA in lane 7) was lower than in the absence of AC2 (lane 5). No GFP siRNAs could be visualized in “green” tissue in the absence of the silencing trigger (lanes 3 and 6). Besides the siRNAs, the antisense GFP riboprobe hybridized specifically to a synthetic 21-nt sense GFP RNA (lane 1) but not to a synthetic 21-nt antisense GFP RNA (lane 2), both described by Klahre et al. (26). A similar pattern of GFP siRNAs in the plant tissue samples was also detected with a sense GFP probe (data not shown).
To test the effect of AC2 on silencing, line 16c seedlings were bombarded with gold particles carrying both the GFP- and AC2-expressing plasmids in a 1:1 ratio to ensure equivalent delivery and expression levels (due to identical 35S promoters). In the presence of AC2, systemic spread of GFP silencing was totally abolished: in several independent cobombardment experiments, we never observed appearance of red veins on newly emerging leaves of more than 100 plants. At the same time, local silencing was not totally suppressed, although the size of red spots was reduced (Fig. 3B). Moreover, at between 6 and 14 days, those spots did not grow further and merge, as in the absence of AC2, but rather faded out and eventually disappeared (Fig. 3B), suggesting that AC2 might cause a reversal of local silencing.
Previous work on AC2 from ACMV indicated that 21- to 25-nt siRNAs—characteristic of RNA silencing—are reduced but not eliminated in the presence of AC2 (17). We isolated a fraction of small RNAs from leaf tissue samples collected 6 days postbombardment with the silencing trigger alone or with a combination of the trigger and MYMV AC2-expressing plasmids. RNA blot hybridization revealed that the GFP siRNAs could be visualized in the absence but were hardly visible in the presence of MYMV AC2 (Fig. 3D). Thus, the siRNA accumulation roughly correlated with the amount of “red” tissue, reflecting the extent of local silencing in the collected samples. It should be noted that in our particle bombardment assay, precise quantitative analysis of silencing suppression at both the GFP siRNA and the target GFP mRNA levels is not possible because of uneven delivery of the particles over leaf surfaces of young plant seedlings and difficulty in separating small spots of red tissue (hardly visible under UV light in the presence of the suppressor; see Fig. 3B, WT AC2), in which silencing was triggered, from the bulk of “green” tissue (ca. 80 to 95% of the whole sampled tissues), which had not received the silencing trigger at all and therefore contained intact GFP mRNA and no GFP siRNAs.
Other MYMV proteins tested in this system, including AC1, AC3, AC4, and BC1, were not capable of suppressing silencing (data not shown). The inability of AC3 to suppress silencing also rules out the possibility that its C-terminally truncated version, which could potentially be expressed by leaky scanning from the AC2 plasmid, was responsible for the suppression. Furthermore, the observed suppression cannot be explained by an interaction between the 35S promoters of the trigger and suppressor plasmids, because the same expression cassette was used for wild-type and mutant (see below) AC2 as well as other viral proteins.
Mutants NLS1−, ZF−, and AD− were tested for their ability to suppress GFP silencing in the test system. None of the mutants was capable of suppressing silencing (Fig. 3C), indicating that all the three functional domains of AC2 are directly or indirectly required for suppression. The mutant NLS2− exhibited a weak antisilencing activity, causing a delay of several days in the development of systemic silencing (data not shown), which correlates with the residual nuclear localization (Fig. 2) and transcription activation (Table 1) abilities of this mutant.
AC2 up-regulates components of the plant transcriptome.
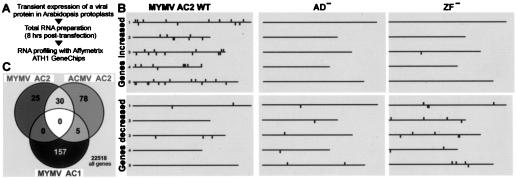
The requirement of all functional domains of AC2 for both activation of viral promoters and suppression of silencing suggested that AC2 might suppress silencing by regulating transcription of host gene(s). To discover such gene(s), we performed RNA profiling with Arabidopsis protoplasts transfected with an AC2-expressing plasmid. Eight hours posttransfection, protoplasts expressing MYMV AC2 or its mutant variants were harvested and total RNA was isolated. Changes in RNA profiles with respect to the control mock-transfected samples were measured on Affymetrix ATH1 GeneChips covering nearly all Arabidopsis protein-coding genes (approximately 24,000). By applying the most stringent criteria with cross-comparison of two biological replicates from two independent batches of protoplasts, 55 genes were found to be up-regulated >2-fold by wild-type AC2, whereas only one (different) gene was induced by the AC2 mutant ZF− and none was induced by AD− (Fig. 4). The highest levels of induction were observed for putative FAD-like oxidase (At5g11540; 72-fold), “expressed protein” (At1g02813; 57-fold), and hypothetical 3′-5′ exonuclease (At3g12460; 43-fold). In contrast, only five genes were down-regulated by AC2 and none of these by more than 1.7-fold. ZF− and AD− down-regulated 21 and 13 genes, respectively, including only one gene in common for all three lists of down-regulated genes (data not shown).
FIG. 4.
Response of the Arabidopsis transcriptome to individual geminivirus proteins. (A) MYMV proteins AC2 (or its mutants AD− and ZF−) and AC1 or ACMV AC2 was expressed individually in Arabidopsis protoplasts, and RNA profiling was performed by using Affymetrix ATH1 GeneChips. (B) The host genes with increased (top panels) or decreased (bottom panels) RNA levels are shown with respect of their physical location on the five chromosomes. (C) Venn diagram showing overlap between the lists of the genes increased >2-fold in response to the respective viral proteins.
AC2 from ACMV, which is also known to activate viral promoters (16) and to be a silencing suppressor (17, 53), was used to test whether our observations are also valid for other begomoviruses. In the RNA profiling experiment, 113 Arabidopsis genes were up-regulated >2-fold by ACMV AC2. Strikingly, 30 of the 55 genes induced >2-fold by MYMV AC2 were also induced >2-fold by ACMV AC2 (Fig. 4), and only 2 of them were not induced at all.
By applying less-stringent criteria, i.e., without cross-comparing the two biological replicates, we could find additional genes that were up-regulated by both of the AC2 proteins (these additional genes appeared to show bigger difference in basal expression levels between the two batches of protoplasts). In this case, a total of 162 genes were induced >2-fold by MYMV AC2, and of these, 139 were also induced by ACMV AC2 but not by the inactive AC2 mutants. A complete list of genes activated by the AC2 proteins, which includes in particular six cold-regulated genes and two members of the Scarecrow-like transcription factor family (see Discussion), can be found in supplemental Table S1.
Notably, AC2-inducible genes were distributed over all five Arabidopsis chromosomes and did not form any obvious cluster (Fig. 4B).
Begomovirus AC1 is also a nuclear protein. It is required for rolling-circle replication of viral DNA and interference with cell cycle regulatory proteins of the retinoblastoma-related/E2F pathway (15). AC1 also acts as a transcriptional repressor of its own promoter (12, 16, 45). When expressed from the 35S promoter plasmid in plant protoplasts, MYMV AC1 repressed its own leftward promoter (unpublished data). Changes in the Arabidopsis transcriptome in response to transient expression of AC1 were absolutely distinct from those caused by AC2. Of 162 genes up-regulated >2-fold by MYMV AC1, none was induced by MYMV AC2 (Fig. 4C). Furthermore, there was no overlap between 97 genes down-regulated by AC1 and those down- or up-regulated by AC2 (data not shown). This shows that the observed changes in the transcriptome are not simply due to the stress caused by introducing a heterologous protein into the nucleus but indeed reflect very specific respective actions of AC2 and AC1. Moreover, genes normally associated with general response to pathogens (e.g., those coding for pathogen-related proteins, heat shock proteins, and WRKY transcriptional factors) were not significantly up-regulated in our protoplast system transiently expressing the geminivirus proteins.
Host promoters induced by AC2.
To validate the GeneChips results and identify candidate silencing suppressor gene(s), we chose six of the genes whose basal transcription level was below that required for reliable detection in control experiments (see supplemental Table S1) and highly elevated in cells expressing both MYMV and ACMV wild-type AC2. The putative promoter regions (about 800 to 1,100 bp preceding the predicted ATG start codons) were introduced upstream of the CAT reporter gene, and their relative activity and responsiveness to AC2 were examined with both Arabidopsis and N. plumbaginifolia protoplasts (Table 2).
TABLE 2.
Transcript levels and promoter activities of selected AC2-inducible genes
| Gene, promoter construct | Basal transcript level on control chips | Fold activation on AC2 chips | AC2 source | Relative CAT expression (%) in N. plumba protoplasts | Relative CAT expression (%) in A. thaliana protoplasts |
|---|---|---|---|---|---|
| CaMV 35S promoter | 100 | 100 | |||
| MYMV | 76 | 99 | |||
| ACMV | 70 | 95 | |||
| At3g12460, hypothetical 3′-5′ exonuclease WEL-1 | 44 (17 to 76) | 0.9 | 0.6 | ||
| 43× | MYMV | 47 | 127 | ||
| 20× | ACMV | 4.3 | 7.1 | ||
| At1g02813, expressed protein | 47 (27 to 62) | 1.1 | 0.4 | ||
| 57× | MYMV | 24 | 154 | ||
| 13× | ACMV | 5.8 | 28 | ||
| At1g13610, hypothetical protein | 6 (3 to 9) | 0.2 | <0.01 | ||
| 56× | MYMV | 65 | 173 | ||
| 27× | ACMV | 25 | 36 | ||
| At5g11540, putative FAD-like oxidase | 11 (4 to 20) | 18 | 4.3 | ||
| 72× | MYMV | 69 | 249 | ||
| 6× | ACMV | 79 | 109 | ||
| At4g39675, expressed protein, similar to HIV-1 p17 | 37 (4 to 116) | 21 | 21 | ||
| 28× | MYMV | 83 | 363 | ||
| 39× | ACMV | 91 | 358 | ||
| At5g15960, cold- and ABA-inducible protein KIN1 | 52 (32 to 84) | 14 | 14 | ||
| 23× | MYMV | 81 | 406 | ||
| 6× | ACMV | 102 | 175 |
Promoters of the hypothetical 3′-5′ exonuclease (At3g12460), expressed protein (At1g02813), and “hypothetical protein” (At1g13610) genes showed similar patterns of expression and responsiveness to AC2 in both systems, which correlates strikingly with the chip data, i.e., very low basal promoter activity and strong induction by both AC2 proteins (Table 2). Interestingly, the correlation was more pronounced in Arabidopsis protoplasts. Also in line with the chip data, MYMV AC2 induced expression much more strongly than ACMV AC2. Notably, in Arabidopsis protoplasts, MYMV AC2-mediated expression from these plant promoters exceeded that driven by the strong 35S promoter.
For other selected genes, coding for putative FAD-like oxidase (At5g11540), expressed protein similar to human immunodeficiency virus type 1 p17 (At4g39675), and cold- and ABA-inducible protein KIN1 (At5g15960), promoter profiles deviated from the above-described pattern. Basal promoter activities were higher, and the degree of induction was less substantial, especially in Nicotiana protoplasts (Table 2). Furthermore, for these genes the degrees of transactivation by the two AC2 proteins were comparable.
To confirm that MYMV AC2 mutated in the three conserved domains loses its ability to transactivate not only the viral promoter (Table 1) but also the host promoters, we coexpressed in Arabidopsis protoplasts each of the four mutant versions of AC2 (AD−, ZF−, NLS1−, and NLS2−) together with the hypothetical 3′-5′ exonuclease promoter- or the “hypothetical protein” promoter-CAT constructs. In contrast to wild-type AC2 (Table 2), all the four mutants failed to transactivate either of the two promoters, exhibiting less than 1% of the wild-type activity.
Taken together, the promoters of all six AC2-inducible plant genes selected on the basis of our GeneChip analysis were strongly responsive to viral transactivator AC2. At least three of these genes, whose promoters exhibited very low (or no) basal activity in the absence of AC2, match our strict criteria for candidate host silencing suppressor genes that are expected to be induced by AC2 at the transcriptional level.
DISCUSSION
Viral suppression of silencing is usually exerted as a secondary function of ordinary viral proteins, e.g., coat protein, movement protein, and protease, which themselves have little in common. This suggests that protein domains responsible for the suppressor activity are, most likely, different from those involved in the primary functions of the viral proteins, although RNA binding has been proposed as a common theme (42).
In begomoviruses, silencing suppression is also a secondary function of the viral transcription activator AC2, but, unexpectedly, in this work we could not separate the two functions of AC2 as viral transcription factor and silencing suppressor by mutagenesis, albeit with a limited number of mutations. This suggests that the suppressor activity is causally coupled to the transcription factor activity.
Silencing suppression through transactivation of other viral genes by AC2 can be excluded, because AC2 acts as a suppressor in a model system in the absence of virus infection, and other MYMV proteins showed no suppressor activity when tested individually.
MYMV AC2 possesses three domains typical of transcription activators: a bipartite NLS, a nonclassical Zn finger, and an acidic activator domain (21; this work). All three of these features are conserved in other begomoviruses. Here we report that these domains in combination are required for both promoter activation and silencing suppression. The fact that the cytoplasmic AC2 variant with mutated NLS (NLS1−) failed to suppress silencing argues for an indirect effect of AC2, because GFP silencing in our model system is most likely a cytoplasm-based mechanism of RNA destruction. Furthermore, the nuclear AC2 requires intact Zn-finger and activator domains to suppress silencing, suggesting that transcription activation of a host suppressor(s) might be involved. Indeed, the loss of suppressor activity of the ZF− and AD− mutants correlates with their failure to induce host genes.
In line with our results, in the case of TYLCCNV C2, a functional NLS and a Zn finger are both required for suppression of RNA silencing (10, 50). However, C2-mediated activation of viral transcription has not been reported for this or other monopartite begomoviruses. Since C2 from the latter genus also possesses a conserved acidic domain at the C terminus, a similar mechanism of silencing suppression via transcription activation of host genes can be proposed.
It has been demonstrated that the AC2 homologue from Tomato golden mosaic virus (TGMV AL2) transactivates late viral genes at the level of transcription (44). However, the molecular mechanism of AC2-mediated transactivation is largely unknown. Attempts to identify any conserved AC2-responsive cis element have met with little success (40, 47). Moreover, sequence-nonspecific and weak binding to double-stranded DNA in vitro (21, 43) suggests that AC2 may rather engage (through its Zn finger) in interaction with one or more cellular factors, which would in turn target it to different promoters.
Recently it has been shown that TGMV AL2 interacts with two cellular proteins, serine/threonine kinase (SNF1) (20) and adenosine kinase (ADK) (55). The first interaction, which is mediated by the AL2 Zn-finger domain, inactivates SNF1 kinase, the key regulator of cell metabolism implicated in the innate antiviral defense, and thereby leads to enhanced susceptibility to infection with DNA and RNA viruses (20, 46). The second interaction inactivates ADK, which may serve as an early activator of SNF1, thus suggesting a dual counter-defensive strategy evolved by geminiviruses to cope with the innate antiviral response (55). It has also been speculated that inactivation of ADK by AL2 may indirectly suppress silencing by interfering with a general methylation pathway that requires ADK (55). Because these two interactions and their effects most likely occur in the cytoplasm and do not require the AL2 transcription activator domain (20, 46, 55), they cannot account for transcription activation of the viral and host genes observed by us. Moreover, suppression of RNA silencing correlates with the nuclear localization of the suppressor proteins TLCCNV C2 (10) and MYMV AC2 (this work) and, in the case of MYMV AC2, requires the transcription activator domain (this work). However, formally we cannot exclude that possible interference with the innate antiviral response by MYMV AC2 may partly contribute to the strong antisilencing effect observed in our experimental system.
Activator domains are believed to enhance transcription by recruiting components of the basal transcriptional machinery (49). In fact, the TGMV AL2 activator domain was able to functionally replace the corresponding domain of the transcriptional activator GAL4 in yeast and human cells (21). Interestingly, attempts to produce transgenic plants constitutively expressing full-length AC2/AL2 proteins have failed, possibly because of toxicity of those proteins, while plants expressing a truncated form of TGMV AL2 lacking the activator domain could be recovered (46). The dramatic changes in the plant transcriptome in response to wild-type AC2 from both MYMV and ACMV described here might help explain those earlier observations. In particular, constitutive up-regulation of endogenous silencing suppressor(s) by AC2 might interfere with normal plant development.
As hypothesized above, host genes involved in silencing suppression might become activated by AC2. Alternatively, to achieve silencing suppression, AC2 may repress genes that are positively involved in the silencing process. Our RNA profiling approach with Arabidopsis protoplasts demonstrates that upon transient expression of wild-type AC2, no dramatic reductions in RNA levels could be detected. This makes transcriptional repression of genes involved in the silencing pathway unlikely, although some of such genes might be missing from the ATH1 chip. On the other hand, RNA profiling revealed a set of genes whose transcripts are elevated considerably in the presence of AC2 from two different begomoviruses, raising the possibility that some of these code for silencing suppressors. An increase in the RNA level could result from either RNA stabilization or activation of transcription. For several selected AC2-inducible genes, we found that the promoter regions cloned from the Arabidopsis genome were highly active only in the presence of AC2 (Table 2), suggesting that the increase in the corresponding RNA levels observed on the chips was due to transcriptional activation.
The promoter sequences tested here include 5′-untranslated regions (UTRs), which often possess important elements regulating transcription (reference 22 and references therein). Such a region may therefore contain (part of) an AC2-responsive element, and formally we cannot exclude that the latter is an RNA-based element. However, we do not favor a scenario in which these 5′-UTR sequences contain any RNA instability determinants that would normally occur either in 3′-UTR or in coding sequence of unstable RNAs.
While the annotations available for most of the AC2-inducible genes give little clue as to whether and how they could exert suppressor functions, at least two such genes, At5g15960 and At3g12460, can be viewed as realistic candidates.
At5g15960 codes for the cold- and abscisic acid-inducible protein KIN1. Interestingly, five additional known or putative cold-regulated genes were also up-regulated by AC2 (supplemental Table S2). It has been reported that low temperatures inhibit RNA silencing (48). One could imagine the existence of a general mechanism limiting silencing at low temperatures and that this mechanism is exploited by AC2 in order to suppress virus silencing.
At3g12460 codes for a hypothetical protein of 242 amino acids with homology to a 3′-5′ exonuclease domain of the Werner syndrome protein, implicated in premature aging in humans (41). Interestingly, in Caenorhabditis elegans and Arabidopsis, genes for Werner-like exonuclease proteins MUT-7 and WEX, respectively, have been identified as positive regulators of RNA silencing (13, 25). Our PSI-BLAST analysis showed that the conserved “DEDDy” signature of Werner-like exonucleases (56) is only partially preserved in the hypothetical WEX-like protein identified by us (hereafter called WEL-1 for Werner exonuclease-like 1). Thus, WEL-1 might exert a dominant-negative effect by interfering with an as yet unknown function of WEX in RNA silencing (13). Alternatively, it might be responsible for degradation of RNA intermediates of the silencing pathway, such as siRNAs. Our unpublished observations indicate that transient expression of a WEL-1 transcription unit is sufficient to suppress RNA silencing in the model N. benthamiana line 16c system. Experiments are currently under way to investigate a possible function of WEL-1 in Arabidopsis in relation to siRNA- and miRNA-generating pathways.
In the Arabidopsis genome, WEL-1 is located within a cluster of seven genes (At3g12470 to At3g12410) coding for highly homologous proteins differing in size due to short deletions and short or long insertions or duplications. Their coding sequences are separated by about 700- to 1,000-bp-long noncoding regions of little similarity, one of which includes the WEL-1 promoter analyzed here. Another gene from this cluster (At3g12440) is also induced by either of the AC2 proteins, albeit rather weakly (2.4- and 2.7-fold), whereas At3g12470 and At3g12420 were not induced. The remaining three genes are not present on the ATH1 chip. It is tempting to speculate that members of the WEL cluster may have similar activities that are induced in response to individual factors.
Why would a host have evolved functions to suppress its own defense system? Besides protecting plant cells from viruses, RNA silencing may also regulate endogenous gene expression, as has been reported for the related miRNA pathway (3, 7). Given the “infectious” nature of RNA silencing, which can amplify and spread systemically throughout the whole plant, a means to down-regulate and/or restrict this process would be desirable. Endogenous silencing suppressors could be involved in this negative regulation either by switching on certain silenced genes according to a developmental program and in response to environmental cues or by preventing the spread of silencing from a single cell or certain tissue where it has initiated. Endogenous silencing suppressors, like their viral counterparts, may act at different steps of RNA silencing and related mechanisms. In fact, viral suppressors might exploit an endogenous pathway by activating its individual components or a combination thereof to block silencing in concert.
Existence of the endogenous pathway of silencing suppression also has been suggested by identification of the calmodulin-related protein rgs-CaM in N. benthamiana, which interacts with the viral suppressor HC-Pro in the yeast two-hybrid system and, like HC-Pro itself, suppresses GFP silencing in N. benthamiana (1). In this case, suppression by HC-Pro might be mediated by activation of rgs-CaM and subsequent amplification of an endogenous pathway that negatively regulates silencing (1).
If, upon silencing suppression by AC2, reactions common to siRNA/miRNA production or action are affected, then some of the genes detected on the chips might have been elevated due to stabilization of their RNAs caused by reduced levels or inactivity of siRNA/miRNAs targeting them. The best candidates for this type of gene are two members of the Scarecrow-like family of plant-specific transcriptional factors, SCL6-II (At2g45160) and SCL8 (At5g52510), whose transcripts were elevated in the presence of AC2 (supplemental Table S3). SCL6-II has been predicted to be a target of miR-171, to which it has perfect complementarity, like its two isoforms, SCL6-III and SCL6-IV, identified as the targets for degradation by RNA-induced silencing complex-mediated cleavage (29). Interestingly, the cleavage can be suppressed by the viral suppressor HC-Pro, leading to elevated levels of SCL6-III and SCL6-IV mRNAs (24). However, we cannot conclude that AC2 is also capable of suppressing the miRNA pathway, because SCL6-III was only slightly elevated in the presence of AC2 and SCL6-IV, and several other predicted miRNA targets were not elevated at all. Yet some of the plant miRNAs might silence target genes by repressing translation of target mRNA without changing RNA accumulation, as documented for AP2-like transcription factors (2). Furthermore, in our transient expression system, potential targets of miRNA/siRNA degradation pathways may become significantly elevated only at later time points (not analyzed here), after AC2-induced suppressor gene(s) have exerted their suppression effect.
Supplementary Material
Acknowledgments
We thank Helen Rothnie for critical reading of the manuscript, Livia Stavolone and Vitaly Boyko for GFP imaging, Dave Kirk and Afzal Dogar for help with plasmid construction, Ueli Klahre for teaching particle bombardment, Herbert Angliker for transcript labeling and chip hybridization, and Matthias Mueller and Monika Fasler for excellent generation of protoplasts.
P.V.S. acknowledges the Council of Scientific and Industrial Research, Government of India, for a fellowship. This work was supported by the Indo-Swiss Collaboration in Biotechnology and European Union framework V grant “VIS.”
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anandalakshmi, R., R. Marathe, X. Ge, J. M. Herr, Jr., C. Mau, A. Mallory, G. Pruss, L. Bowman, and V. B. Vance. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142-144. [DOI] [PubMed] [Google Scholar]
- 2.Aukerman, M. J., and H. Sakai. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel, D. P. 2004. MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe, D. 2002. Viral suppression of systemic silencing. Trends Microbiol. 10:306-308. [DOI] [PubMed] [Google Scholar]
- 5.Benfey, P. N., L. Ren, and N. H. Chua. 1990. Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 9:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18 :1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Tapia, M., A. Himmelbach, and T. Hohn. 1993. Molecular dissection of the cauliflower mosaic virus translation transactivator. EMBO J. 12:3305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, X., R. van Wezel, J. Stanley, and Y. Hong. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77:7026-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunoyer, P., C. H. Lecellier, E. A. Parizotto, C. Himber, and O. Voinnet. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235-1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Eagle, P. A., B. M. Orozco, and L. Hanley-Bowdoin. 1994. A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glazov, E., K. Phillips, G. J. Budziszewski, F. Meins, Jr., and J. Z. Levin. 2003. A gene encoding an RNase D exonuclease-like protein is required for post-transcriptional silencing in Arabidopsis. Plant J. 35:342-349. [DOI] [PubMed] [Google Scholar]
- 14.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez, C. 2000. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 19:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haley, A., X. Zhan, K. Richardson, K. Head, and B. Morris. 1992. Regulation of the activities of African cassava mosaic virus promoters by the AC1, AC2, and AC3 gene products. Virology 188:905-909. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 19.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 20.Hao, L., H. Wang, G. Sunter, and D. M. Bisaro. 2003. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartitz, M. D., G. Sunter, and D. M. Bisaro. 1999. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1-14. [DOI] [PubMed] [Google Scholar]
- 22.He, X., J. Fütterer, and T. Hohn. 2002. Contribution of downstream promoter elements to transcriptional regulation of the rice tungro bacilliform virus promoter. Nucleic Acids Res. 30:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karthikeyan, A. S., R. Vanitharani, V. Balaji, S. Anuradha, P. Thillaichidambaram, P. V. Shivaprasad, C. Parameswari, V. Balamani, M. Saminathan, and K. Veluthambi. 2004. Analysis of an isolate of Mungbean yellow mosaic virus (MYMV) with a highly variable DNA B component. Arch. Virol. 149:1643-1652. [DOI] [PubMed] [Google Scholar]
- 24.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 25.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 26.Klahre, U., P. Crete, S. A. Leuenberger, V. A. Iglesias, and F. Meins, Jr. 2002. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA 99:11981-11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecellier, C. H., and O. Voinnet. 2004. RNA silencing: no mercy for viruses? Immunol. Rev. 198:285-303. [DOI] [PubMed] [Google Scholar]
- 29.Llave, C., Z. Xie, K. D. Kasschau, and J. C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053-2056. [DOI] [PubMed] [Google Scholar]
- 30.Lucy, A. P., H. S. Guo, W. X. Li, and S. W. Ding. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance, and L. H. Bowman. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99:15228-15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallory, A. C., S. Mlotshwa, L. H. Bowman, and V. B. Vance. 2003. The capacity of transgenic tobacco to send a systemic RNA silencing signal depends on the nature of the inducing transgene locus. Plant J. 35:82-92. [DOI] [PubMed] [Google Scholar]
- 33.Matzke, M., W. Aufsatz, T. Kanno, L. Daxinger, I. Papp, M. F. Mette, and A. J. Matzke. 2004. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta 1677:129-141. [DOI] [PubMed] [Google Scholar]
- 34.Papp, I., M. F. Mette, W. Aufsatz, L. Daxinger, S. E. Schauer, A. Ray, J. van der Winden, M. Matzke, and A. J. Matzke. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pooggin, M., P. V. Shivaprasad, K. Veluthambi, and T. Hohn. 2003. RNAi targeting of DNA virus in plants. Nat. Biotechnol. 21:131-132. [DOI] [PubMed] [Google Scholar]
- 36.Pooggin, M. M., T. Hohn, and J. Fütterer. 2000. Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem. 275:17288-17296. [DOI] [PubMed] [Google Scholar]
- 37.Pruss, G., X. Ge, X. M. Shi, J. C. Carrington, and V. B. Vance. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-Medrano, R., R. G. Guevara-Gonzalez, G. R. Arguello-Astorga, Z. Monsalve-Fonnegra, L. R. Herrera-Estrella, and R. J. Rivera-Bustamante. 1999. Identification of a sequence element involved in AC2-mediated transactivation of the pepper huasteco virus coat protein gene. Virology 253:162-169. [DOI] [PubMed] [Google Scholar]
- 41.Shen, J. C., and L. A. Loeb. 2000. The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 16:213-220. [DOI] [PubMed] [Google Scholar]
- 42.Silhavy, D., and J. Burgyan. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76-83. [DOI] [PubMed] [Google Scholar]
- 43.Sung, Y. K., and R. H. Coutts. 1996. Potato yellow mosaic geminivirus AC2 protein is a sequence non-specific DNA binding protein. FEBS Lett. 383:51-54. [DOI] [PubMed] [Google Scholar]
- 44.Sunter, G., and D. M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunter, G., M. D. Hartitz, and D. M. Bisaro. 1993. Tomato golden mosaic virus leftward gene expression: autoregulation of geminivirus replication protein. Virology 195:275-280. [DOI] [PubMed] [Google Scholar]
- 46.Sunter, G., J. L. Sunter, and D. M. Bisaro. 2001. Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285:59-70. [DOI] [PubMed] [Google Scholar]
- 47.Sunter, G., and D. M. Bisaro. 2003. Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305:452-462. [DOI] [PubMed] [Google Scholar]
- 48.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyan. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Triezenberg, S. J. 1995. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5:190-196. [DOI] [PubMed] [Google Scholar]
- 50.van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 51.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. Tanaka Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 52.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 53.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H., L. Hao, C. Y. Shung, G. Sunter, and D. M. Bisaro. 2003. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.